Abstract
Kaposi sarcoma (KS), the most common cancer in HIV-positive individuals, is caused by endothelial transformation mediated by the KS herpes virus (KSHV)-encoded G-protein coupled receptor (vGPCR). Infection of blood vascular endothelial cells (BECs) by KSHV reactivates an otherwise silenced embryonic program of lymphatic differentiation. Thus, KS tumors express numerous lymphatic endothelial cell (LEC)-signature genes. A key unanswered question is how lymphatic reprogramming by the virus promotes tumorigenesis leading to KS formation. In this study, we present evidence that this process creates an environment needed to license the oncogenic activity of vGPCR. We found that the G-protein regulator RGS4 is an inhibitor of vGPCR that is expressed in BECs, but not in LECs. RGS4 was downregulated by the master regulator of LEC differentiation PROX1, which is upregulated by KSHV and directs KSHV-induced lymphatic reprogramming. Moreover, we found that KSHV upregulates the nuclear receptor LRH1, which physically interacts with PROX1 and synergizes with it to mediate repression of RGS4 expression. Mechanistic investigations revealed that RGS4 reduced vGPCR-enhanced cell proliferation, migration, VEGF expression and Akt activation and to suppress tumor formation induced by vGPCR. Our findings resolve long-standing questions about the pathological impact of KSHV-induced reprogramming of host cell identity, and they offer biological and mechanistic insights supporting the hypothesis that a lymphatic microenvironment is more favorable for KS tumorigenesis.
Keywords: Kaposi sarcoma, lymphatic reprogramming, RGS4, viral GPCR, endothelial cells
INTRODUCTION
Kaposi’s sarcoma (KS) is a multifocal, highly proliferative soft-tissue cancer that is most prevalent in HIV-infected patients. The causative agent of KS has been identified as Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus-8 (HHV8). The characteristic spindle cells in KS tumors result from KSHV-induced transformation of vascular endothelial cells. Notably, KSHV G-protein coupled receptor (vGPCR) was found to be both necessary and sufficient to induce endothelial cell transformation(1, 2). Although vGPCR is constitutively active, it can be further stimulated by cellular ligands such as IL-8 and Gro-α(3). vGPCR in turn triggers several intracellular signaling cascades to promote cell survival and sarcomagenesis, and additionally, induces the secretion of angiogenic growth factors, such as VEGF, IL-8 and Gro-α, playing essential roles in host cell transformation(1–4).
The regulator of G-protein signaling (RGS) family proteins function as GTPase-activating proteins (GAP) for Gα subunits and have been found to attenuate the signaling of GPCRs through rapid deactivation of Gα subunits(5, 6). The RGS proteins regulate a wide range of cellular functions, including cell migration, proliferation and survival, and suppress epithelial and endothelial cell tubulogenesis by inhibiting G-protein and VEGF-mediated activation of MAPK(7).
KS tumor cells have been shown to express lymphatic endothelial cell (LEC)-signature molecules, such as vascular endothelial growth factor receptor 3 (VEGFR3), LYVE-1 and PROX1, and are thus hypothesized to be derived from lymphatic lineage cells(8). However, recent studies by us and other groups have shown that KSHV infection induces lymphatic reprogramming of blood vascular endothelial cells (BECs), which is characterized by the upregulation of lymphatic-signature genes and simultaneous downregulation of BEC-associated molecules(9–11). This pathological lymphatic reprogramming of host cells was found to be mediated by KSHV-induced upregulation of PROX1 (9), a master regulator of lymphatic differentiation (12), suggesting that the oncogenic virus is capable of reactivating the otherwise silenced embryonic lymphatic differentiation program. Although this interesting discovery of the virus-induced endothelial cell fate re-specification has raised the possibility that LECs, as opposed to BECs, may be more favorable for KSHV-mediated endothelial transformation, experimental evidence supporting such a notion are not available to date. In the current study, we investigated how PROX1-mediated lymphatic differentiation contributes to KSHV-mediated endothelial transformation, supporting the possibility that the lymphatic compartment provides more favorable microenvironment for KSHV pathogenesis.
MATERIALS AND METHODS
Cells and Animals
Normal C57BL/6, RGS4-GFP BAC (B6. Cg-Tg(Rgs4-EGFP)4Lvt/J) and immunodeficient NOD-SCID IL2Rγnullmice (NSG; NOD. Cg- Prkdcscid Il2rgtm1Wjl /SzJ) were purchased from the Jackson Laboratory. Athymic nude mice (Crl:NU-Foxn1<nu>/Foxn1<+>) were purchased from Charles River Laboratories. All mouse experiments have been pre-approved by the University of Southern California Institutional Animal Care and Use Committee (IACUC). Primary blood and lymphatic endothelial cells were isolated from deidentified human foreskins and cultured in endothelial basal medium (EBM, Lonza) supplemented with 10% fetal bovine serum (FBS) and other supplements as previously described (13, 14). Isolation and culture of human endothelial cells were pre-approved by the University of Southern California Institutional Review Board (IRB). Primary human BECs and LECs were transfected by electroporation (Nucleofector II, Amaxa Biosystems) and other cell lines were transfected using Lipofectamine 2000 (Invitrogen). SV40 large T-antigen immortalized murine endothelial cells (SVECs) and its vGPCR-expressing derivative cell line (SVEC-vGPCR) were generously provided by Dr. Silvia Montaner (University of Maryland) and cultured as previously described (15). We have authenticated SVECs and SVEC-vGPCR to be a mouse endothelial cell line based on their mouse endothelial cell-specific gene expression pattern determined by quantitative real-time RT-PCR (qRT-PCR), semi-quantitative RT-PCR, western blot and immunofluorescent analyses (Supplemental Figure 4 data not shown). We authenticated these cell lines once every six months and the last test was performed in July, 2012. SVEC-vGPCR cells were transfected with either a human RGS4-expressing vector (Cat. No. RGS040TN00, Missouri S&T cDNA Resource Center) or pcDNA3. 1 (Invitrogen), along with a hygromycin-resisant vector (pIRESHyg2, Clontech) at a molar ratio of 10:1. Transfected cells were selected with hygromycin to obtain RGS4-expressing SVEC-vGPCR cells(SVEC-vGPCR/RGS4) and corresponding control cells (SVEC-vGPCR/CTR). The PROX1-expressing adenovirus was previously described (13).
Cell proliferation assay, scratch assay and chromatin immunoprecipitation (ChIP)
Proliferation and scratch assays were performed as previously described (16). Cells were seeded and various time points were analyzed (24,48and 72 hours) using WST-1 assay (TaKaRa MK400). For scratch assay, cells were grown in a 6-cm dish until they reached 90–95% confluency, where the cell monolayer was then scratched using a 1ml pipette tip. The scratched monolayer was pre-treated with mitomycin C (10 µg/mL) prior to activation with Gro-α (50mg/ml) or not in serum-free media for 24 hours. The scratched area was photographed at 0, 2, 4, 8, 12, and 24 hours and measured using NIH ImageJ software. ChIP assay was performed as previously described (13) using a rabbit anti-PROX1 antibody (generated by the authors) or normal rabbit IgG (Sigma) against LEC cell lysates (in vitro ChIP) or mouse organ lysates (in vivo ChIP). Primers used for in vitro ChIP were as follows: human RGS4 (#1, TGACATTGGTGGAGACATTGA/GTGAACGAGCAGAGAAAATCC; #2, TGACATTGGTGGAGACATTGA/ TGACGCATCAGCAATGTTAAGTG), human FER (CACCCTCGAATAATGACGCATA/ AACCCAAACGGGTCTGCTCT) and human HS3ST2 (CCCTGGTAGGTGGTCTTTGA/ GCACTTCAGAAAAGCCTTGG). Primers used for in vivo ChIP were against mouse RGS4 (AACGCCAAAGCTGGACTAGA/ ACACGGAGGGATGTGGATAG) and mouse ROSA26 (AAGGGAGCTGCAGTGGAGTA/ CCGAAAATCTGTGGGAAGTC). To prepare the organ lysates, brain and intestine isolated from a normal C57BL/6 mouse were minced using a homogenizer, filtered through 70-µm strainers and centrifuged. The pellets were resuspended in DMEM with 10% calf-serum, incubated with 1% Formaldehyde at 37°C for 10 minutes and subjected to the standard ChIP protocol.
Western blot, luciferase and Immunofluorescence (IF) assays
Western blot assays were performed as previous described (13), with a modified cell lysis buffer (RIPA; 20mM Tris-Cl, 1% Nonidet P-40, 0. 1% sodium dodecyl sulfate, 1% sodium deoxycholate, 150mM sodium chloride, and 1mM ethylene glycol tetraacetic acid), supplemented with phenylmethylsulfonyl fluoride, phosphatase inhibitors and proteasome inhibitor MG-132 (20 µM, Sigma). Antibodies used are PROX1 (ReliaTech GmbH, Germany), RGS4 (Santa Cruz), total AKT and phospho-AKT(S473) (Cell Signaling) and total and phospho-ERK1/2 (Cell Signaling). The human RGS4 promoter luciferase constructs(17) were kindly provided by Dr. Vishwajit L. Nimgaonkar (University of Pittsburgh). Luciferase reporter assays were carried out at 48-hours post transfection and luciferase activity was measured in triplicate using Bright-Glo reagent (Promega), followed by normalization to total protein amount used. Immunofluorescent staining was performed as previously described (13) and antibody sources are CD31 (Upstate Biotech), RGS4 (Santa Cruz) and LANA (Advanced Biotechnologies Inc. , Columbia, MD).
Isolation of infectious KSHV
BCBL-1 cells were cultured to a density of 10 million cells/ml and subsequently activated with TPA (20 ng/ml) and sodium butyrate (NaB, 3 mM). At 48-hours post TPA/NaB virion induction, culture media was replaced with normal media and cells were incubated for an additional 3 days. Culture media was then collected and filtered through 0. 45-mm filter and centrifuged for 30 minutes at 4°C at 4,000 rpm to remove cell debris. Supernatant was then further centrifuged for 5 hours at 4°C at 10,000 rpm to pellet the virus. Virus-containing pellet was resuspended in endothelial cell media. Infectivity was measured by immunohistochemistry for LANA after a 5 day infection. GFP-labeled KSHV was prepared from iSLK cells carrying GFP-KSHV after stimulation with Doxycyclin (1µg/mL) and sodium butyrate (0. 1 mM) for 4~5 days, where the virus was then harvested from the culture media as previously described (18).
Electro-mobility shift assay (EMSA
EMSA was performed as previously described (13). Double-stranded oligonucleotides with the following DNA sequences spanning the 0. 3-kb RGS4 promoter were annealed and extended with the Klenow fragment (New England Biolabs) in the presence of 32P dCTP: (P1, TTTTCAGAAGGATTTTCTCTGCTCGTTCACTTAACATTGC,TGACGCATCAGCAATGTTAAGTGAACGAGCAGAGAAAATC; P2, ATTTTTTCCCATATCCCTACTTTTCAGAAGGATTTTCTCT, GTGAACGAGCAGAGAAAATCCTTCTGAAAAGTAGGGATAT; P3,TGATGCGTCAGTCTTTTCTTCCTCATCTCTTTCAGGGGCT, CTGCCTCTCCAGCCCCTGAAAGAGATGAGGAAGAAAAGAC; P4,GGAGAGGCAGAGGGAGACAGAGGAGCTGGTACTGCAGAGC, TCAGACGACCGCTCTGCAGTACCAGCTCCTCTGTCTCCCT). Fully extended labeled probes were purified by polyacrylamide gel electrophoresis, followed by dialysis. Full-length PROX1 recombinant protein produced in bacteria was incubated with the labeled probes in the presence of excessive poly-dIdC and subjected to non-denaturing polyacrylamide gel electrophoresis.
Statistical Analysis
Results are expressed as average ± standard deviation for each experiment. All analyses were performed in quadruplicates and each experiment was repeated more than two times. Student t-test was used to determine whether the differences between the experimental and control groups for both in vitro and in vivo studies were statistically significant. All reported P values were two-sided at a significance level of less than 0. 05. The analyses were performed using Microsoft Excel (Microsoft Office).
RESULTS
KSHV selectively represses the expression of RGS4
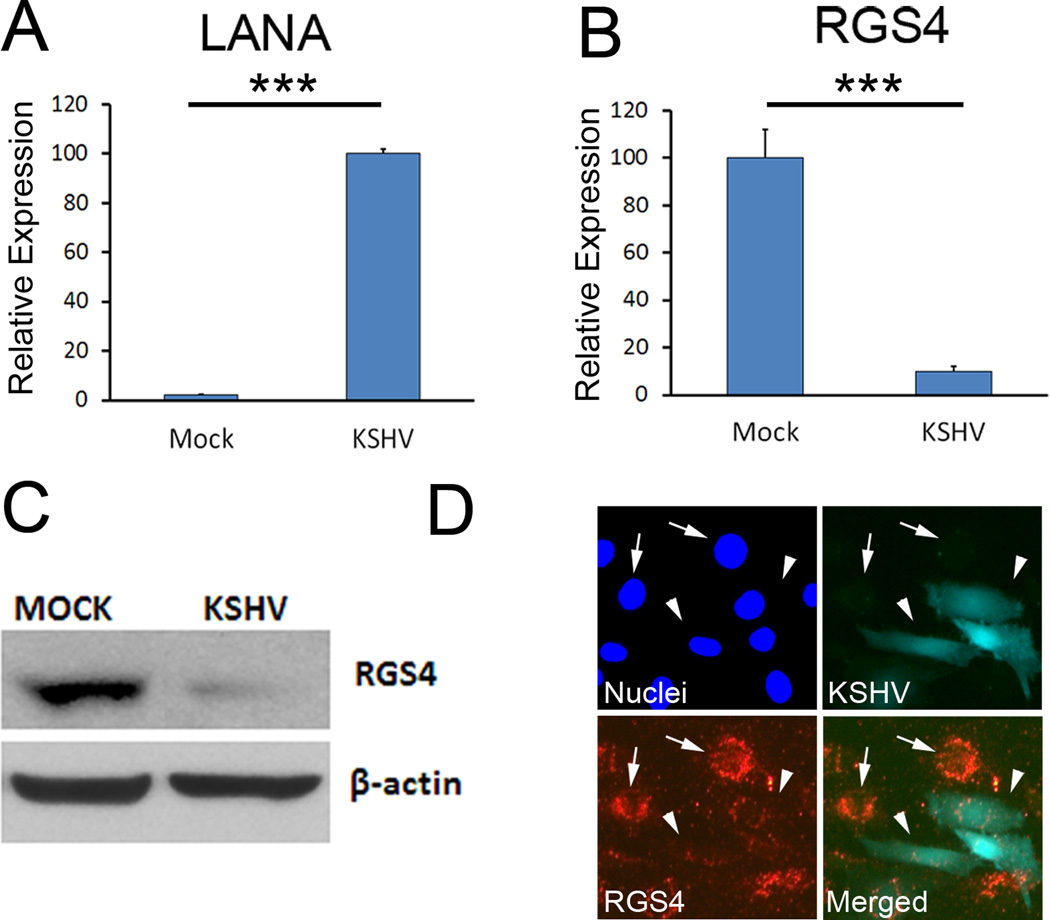
To better understand the pathological benefits of KSHV-induced lymphatic reprogramming, we performed a genome-wide comparative survey to search for endothelial-lineage genes that are commonly regulated by KSHV and PROX1, using the two independent sets of microarray data (9, 14). This comprehensive survey identified RGS4 to be of particular interest for its profound role in regulating the activity of cellular GPCRs. RGS4 was significantly downregulated by KSHV, whereas other RGS proteins tested were not controlled by KSHV (Supplemental Table 1). We further confirmed this KSHV-mediated downregulation of RGS4 mRNA and protein by quantitative real-time RT-PCR (qRT-PCR) and western blot analyses, respectively (Fig. 1A–C). In addition, we asked whether RGS4 is downregulated solely in KSHV-infected cells, or in uninfected neighboring cells as well, via a paracrine effect. Immunofluorescent analyses performed on BECs infected with GFP-labeled KSHV(18)revealed that RGS4 downregulation was only in KSHV-infected cells that were GFP-positive, but not in neighboring cells (Fig. 1D). In addition, we evaluated RGS4 expression in KS tumor lesions from HIV-positive patients and found that RGS4 was not expressed in KS cells (Supplemental Figure 1). Together, these data demonstrate that KSHV downregulates RGS4 at the transcriptional level and this repression occurs only in KSHV-infected cells, suggesting that viral infection is required for RGS4 regulation.
Figure 1. RGS4 is downregulated in BECs during KSHV infection.
Primary human dermal BECs were infected with KSHV for two days and the expression of LANA (A) and RGS4 (B) was determined by qRT-PCR. Results are shown as average relative expression ± standard deviation. ***, p < 0. 001. (C) Western blot analyses showing downregulation of RGS4 protein in KSHV-infected BECs. (D) Immunofluorescent staining against RGS4 in BECs that were infected with GFP-labeled KSHV revealed that RGS4 was downregulated in only GFP-positive, KSHV-infected BECs (arrowhead), but not in uninfected neighboring cells (arrow). All experiments were repeated three times and comparable outcome was obtained.
RGS4 is predominantly expressed in BECs, but not LECs
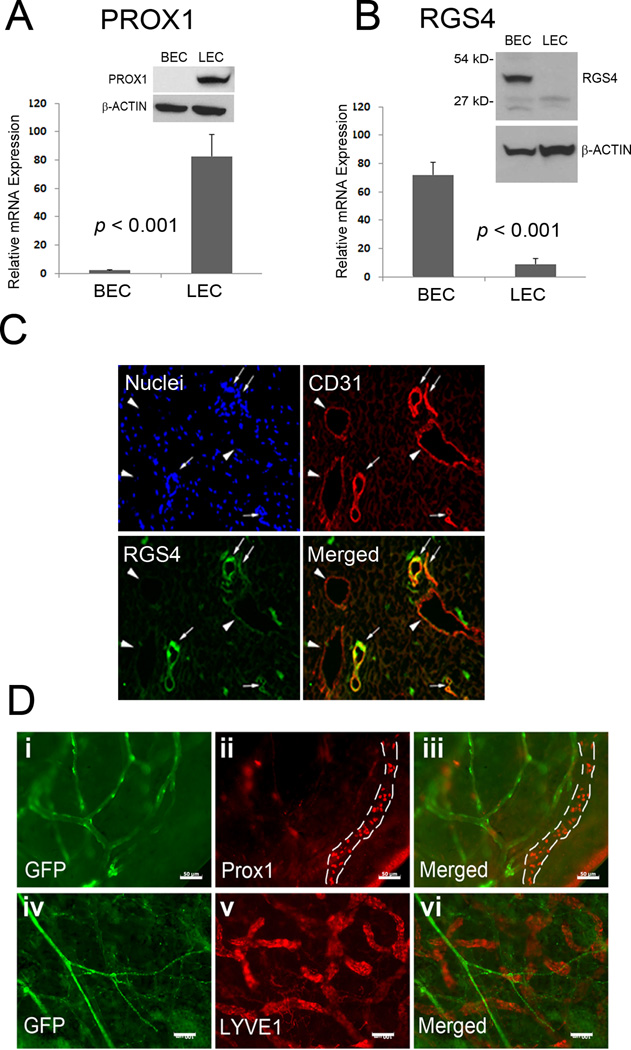
We next studied the expression pattern of RGS4 in human primary BECs and LECs isolated from the same donor. A series of qRT-PCR and western blot analyses revealed that while PROX1 was selectively expressed in LECs, and not BECs, RGS4 was predominantly expressed in BECs, and not LECs (Fig. 2 A,B). To validate these in vitro findings, human foreskin skin sections were stained for RGS4 along with CD31, an endothelial marker that is highly expressed in BECs, but only weakly in LECs (19). Consistent with our in vitro data, RGS4 was predominantly expressed in the CD31-high blood vessels, but not in the CD31-low lymphatic vessels (Fig. 2C). In addition, we utilizeda transgenic reporter mouse that harbors RGS4-GFP(20) to visualize GFP expression directed by exogenous RGS4 promoter. Indeed, whole-mount staining of the ears of RGS4-GFP adult mice clearly marks blood vascular networks with GFP, which do not overlap with Prox1- and LYVE1- positive lymphatic vessels (Fig. 2D). Moreover, GFP-positive vasculature in RGS4-GFP mice was morphologically distinct from GFP-positive lymphatic vessels that were visualized using lymphatic-specific PROX1-GFP transgenic mice that we have recently reported(21) (Supplemental Figure 2). Taken together, our in vitro and transgenic animal data demonstrate that RGS4 is predominantly expressed in blood vessels, and not lymphatic vessels.
Figure 2. RGS4 is predominantly expressed in BECs, compared to LECs.
Expression of PROX1 (A) and RGS4 (B) in human primary dermal BECs and LECs was determined by qRT-PCR and western blot analyses. Data are shown as average relative expression ± standard deviation. qRT-PCR and western blot analyses were repeated at least two times. (C) Immunofluorescent analyses with DAPI (Nuclei), anti-CD31, and anti-RGS4 antibodies on neonatal human foreskin section show that RGS4 is mainly expressed in CD31-high blood vessels (arrows), but not in CD31-low lymphatic vessels (arrowheads). Scale bar, 20 µm. (D) Whole mount staining of the ears of RSS4-GFP adult mice for two lymphatic markers Prox1 (i–iii) and LYVE-1 (iv–vi). (i, iv) GFP expression from transgenic RGS4-GFP mice; (ii, v) antibody staining for Prox1 and LYVE1, respectively; (iii, vi) merged images. A Prox1-positive lymphatic vessel was marked with dotted lines in panel (ii, iii). Scale bars: 50 µm (i–iii) & 100 µm (iv–vi). Immunofluorescent analyses were performed more than twice with consistent results.
PROX1 downregulates RGS4 expression by binding to the RGS4 promoter
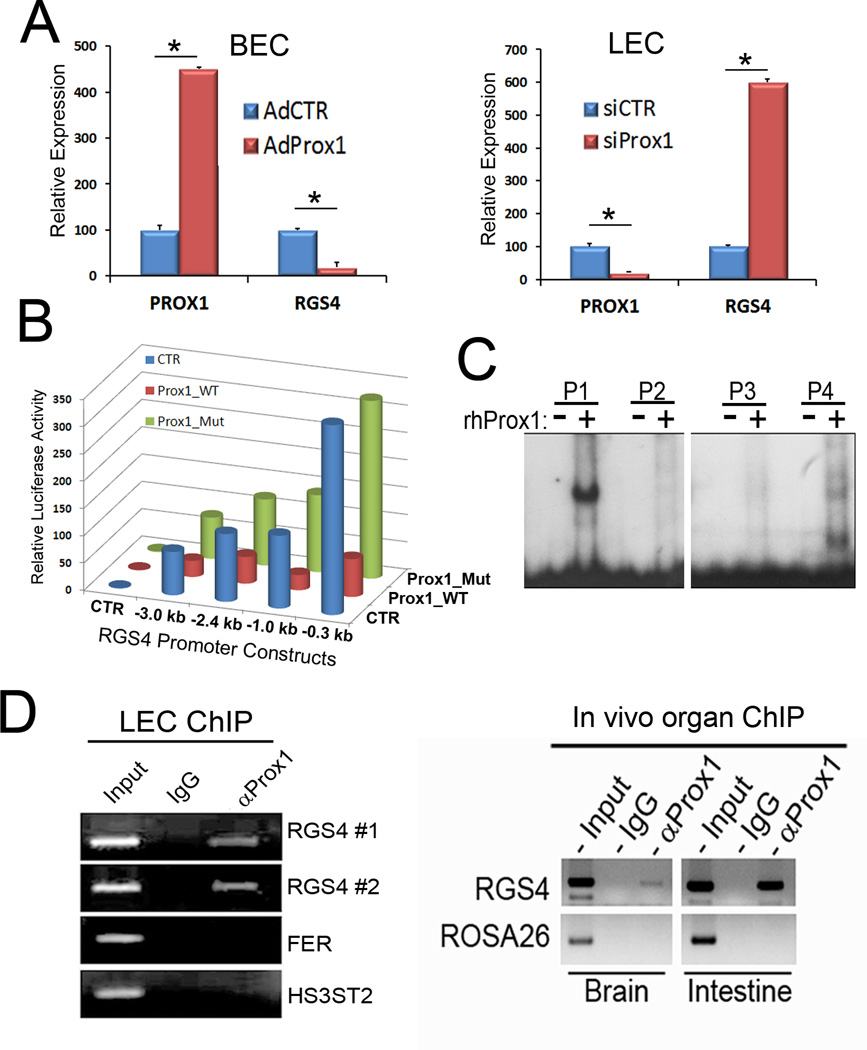
Because PROX1 is known to regulate a number of endothelial lineage genes (22–24), we asked whether downregulation of RGS4 by KSHV is mediated by PROX1, which is upregulated by KSHV-infection in BECs (9–11). Our qRT-PCR studies demonstrated that adenoviral overexpression of PROX1 in BECs strongly repressed RGS4 expression (Fig. 3A). Conversely, siRNA-mediated knockdown of PROX1 in LECs resulted in a significant upregulation of RGS4 (Fig. 3A). We next studied the responsiveness of the RGS4 promoter to PROX1-mediated repression by using luciferase reporter constructs containing various lengths of the RGS4 promoter (17). All RGS4 promoter-constructs tested were responsive to PROX1-mediated repression, and, in particular, the proximal 0. 3-kb region of the RGS4 promoter was sufficient to deliver PROX1-mediated repression (Fig. 3B). Importantly, the repression was detected only with wild-type PROX1, but not a DNA-binding defective mutant of PROX1 (25), indicating that physical interaction of PROX1 protein with the RGS4 promoter is required for transcriptional repression of RGS4. We further investigated the molecular interaction between recombinant PROX1 protein and the RGS4 minimal promoter by electro-mobility shift assay (EMSA) and found that PROX1 protein indeed binds to the RGS4 promoter with high affinity (Fig. 3C). To corroborate this molecular interaction, we carried out PROX1-chromatin immunoprecipitation (ChIP) against the RGS4 promoter region using cultured human primary LECs (in vitro) and mouse whole organ lysates prepared from the brain and intestine (Fig. 3D). Both in vitro and in vivo ChIP assays confirmed that PROX1 protein physically associates with the RGS4 promoter region in cultured human LECs and in mouse brain and intestine. Taken together, these data demonstrate that PROX1 binds to the promoter of RGS4 and directly represses its transcription.
Figure 3. PROX1 downregulates the expression of RGS4 by binding to the RGS4 promoter.
(A)Left Panel: Relative expression of PROX1 and RGS4 was determined by qRT-PCR in BECs that were transduced with a control or PROX1-expressing adenovirus for 48 hours. Right Panel: Relative expression of PROX1 and RGS4 was determined by qRT-PCR in LECs that were transfected with fire-fly luciferase siRNA as a control (siCTR) or PROX1 siRNA (siPROX1), for 48 hours. Experiment was performed three times and representative results are shown. (B) Luciferase assays performed in HEK293 cells showing PROX1-mediated repression of RGS4 promoter constructs. Prox1_WT, wild-type PROX1; Prox1_Mut, DNA-binding defective mutant; CTR, control empty plasmid. Luciferase assay was repeated three times with comparable outcomes. (C) EMSA showing binding of recombinant PROX1 protein to isotope-labeled DNA probes (P1~P4) spanning the RGS4 proximal promoter. See Materials and Methods for the sequences of the probes. (D) Left Panel: PROX1 ChIP performed against the RGS4 promoter in cultured LECs (LEC ChIP). Two sets of primers (RGS4 #1 and RGS4 #2) were used to detect endogenous RGS4 promoter. As negative controls, a normal IgG antibody and a set of primers for unrelated FER and HS3ST2 genes were used. Right Panel: PROX1 ChIP performed against the RGS4 promoter in brain and intestinal cells (In vivo organ ChIP). A normal IgG and a set of primers for ROSA26 locus were used as negative controls. The outcome of qRT-PCR A,B) and luciferase (C) assays are displayed as relative expression or activity ± standard deviation. *, p < 0. 05. ChIP experiments were repeated twice with comparable outcomes.
KSHV downregulates RGS4 through cooperative action of PROX1 and LRH1
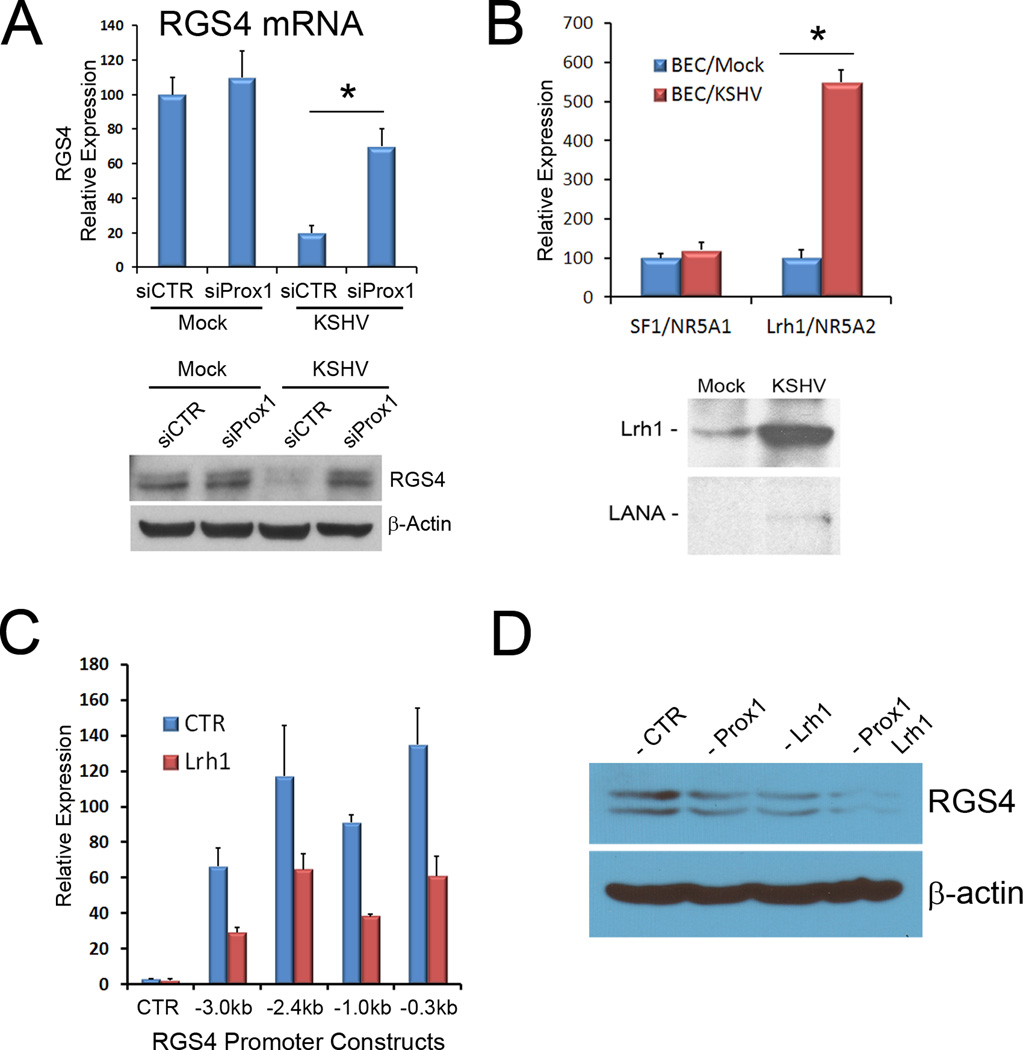
To investigate whether PROX1 is responsible for KSHV-mediated RGS4 downregulation in BECs, we transfected PROX1 siRNA into BECs prior to KSHV infection and determined the expression of RGS4. Indeed, inhibition of KSHV-induced PROX1 upregulation clearly abrogated KSHV-mediated RGS4 repression (Fig. 4A), indicating that RGS4-downregulation in KSHV-infected BECs requires PROX1 expression. Moreover, PROX1 was previously reported to interact with various nuclear receptors such as COUP-TFII/NR2F2, LRH1/NR5A2, and HNF4A/NR2A1, to function as a co-regulator (14, 26–28). This prompted us to ask whether any of the known PROX1-interacting nuclear receptors are regulated by KSHV and whether they are involved in KSHV-mediated RGS4 downregulation by collaborating with PROX1. Interestingly, we found from our previous KSHV-microarray study (9) that, among the 24 different nuclear receptors tested, only LRH1/NR5A2 was significantly upregulated upon KSHV-infection of BECs (Supplemental Table 2). We confirmed this KSHV-mediated upregulation of LRH1 by qRT-PCR and western blot analyses (Fig. 4B). Moreover, LRH1 was able to repress all RGS4 promoter-constructs examined (Fig. 4C). Finally, when PROX1 and LRH1 were co-transfected into BECs, RGS4 was synergistically downregulated (Fig. 4D). Taken together, our study demonstrates that the nuclear receptor LRH1, in addition to its interacting coregulator PROX1, is upregulated by KSHV and that these two proteins collaboratively repress the expression of RGS4 in BECs.
Figure 4. KSHV-upregulated PROX1 and LRH1 act cooperatively to repress RGS4 expression.
(A)PROX1 is required for KSHV to repress RGS4 expression. PROX1 expression was inhibited by transfecting siRNA into BECs 18-hour prior to KSHV infection. After 48 hours of KSHV infection, RGS4 expression was determined by qRT-PCR or western blot analyses. (B) KSHV infection upregulated LRH1 mRNA and protein in BECs based on qRT-PCR and western blot analyses, respectively. Expression of viral LANA protein was detected to confirm KSHV infection. (C) Repression of RGS4 promoter constructs by LRH1. A set of RGS4 promoter-reporter plasmids was transfected with either a LRH1-expressing or a control vector into HEK293 cells and luciferase activity was determined after 48 hours. (D) Concerted repression of RGS4 expression by PROX1 and LRH1 in BECs. A vector expressing either PROX1 or LRH1 was transfected alone or in combination into BECs for 48 hours and RGS4 protein level was detected by western blot. The outcome of qRT-PCR A, B) and luciferase (C) assays are displayed as relative expression ± standard deviation. *, p < 0. 05. All experiments were performed more than two times with consistent outcomes.
RGS4 attenuates vGPCR-mediated activation of Akt and its downstream effects
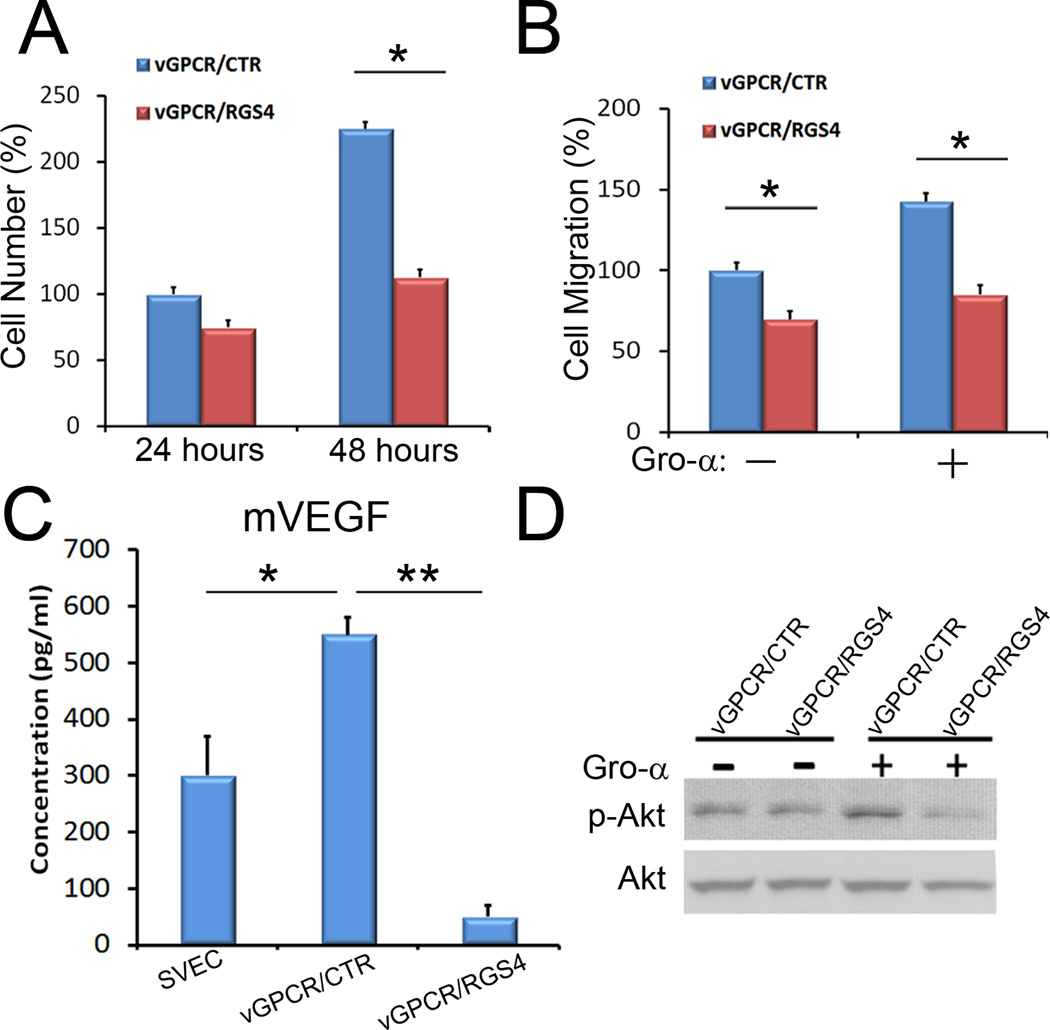
Because RGS4 inhibits activation signaling mediated by the Gαi and Gαq classes of Gα subunits(5, 6), we asked whether RGS4 could act as a negative regulator of vGPCR. To address this question, we stably transfected an RGS4-expressing vector or a control vector into a vGPCR-expressing mouse endothelial cell line(SVEC/vGPCR)(15) and established multi-clonal populations of SVEC/vGPCR/RGS4 and SVEC/vGPCR/CTR cells, respectively. We then evaluated the effect of RGS4 expression in the vGPCR-expressing cells and found that RGS4 significantly reduced cell proliferation of vGPCR-expressing cells (Fig. 5A): While population doubling time of SVEC/vGPCR/CTR cells was approximately 20~24 hours, that of SVEC/vGPCR/RGS4 cells was about 72 hours. Moreover, RGS4 inhibited cell migration of vGPCR-expressing cells in the presence or absence of Gro-α, a potent ligand for vGPCR (Fig. 5B). Previously, the expression and secretion of VEGF was shown to be increased by a stable expression of vGPCR (29). We found here that vGPCR-induced secretion of VEGF was significantly diminished upon expression of RGS4 (Fig. 5C). Lastly, we observed that RGS4 expression strongly reduced Gro-α-induced phosphorylation of Akt (Fig. 5D). Together, these data demonstrate that RGS4 can serve as a potent inhibitor of vGPCR-induced activation of Akt signaling and the accompanying cellular effects.
Figure 5. RGS4 inhibits the proliferation, migration, VEGF secretion, and Akt activation of vGPCR-expressing cells.
(A) Expression of RGS4 resulted in a strong inhibition of proliferation of the vGPCR-expressing SVECs. vGPCR/CTR and vGPCR/RGS4 are pooled populations of SVEC/vGPCR cells that were stably transfected with a control and an RGS4-expressing vector, respectively. (B) Expression of RGS4 reduced cell migration of vGPCR-expressing cells in the presence or absence of Gro-α. (C) RGS4 inhibited the secretion of vascular endothelial growth factor (VEGF) by the vGPCR-expressing cells based on ELISA. (D) Stable expression of RGS4 inhibited Gro-α-induced phosphorylation of Akt (S473) in the vGPCR-expressing cells. The outcome of proliferation and migration assays are displayed as a percent average of cell number/migration ± standard deviation. *, p <0. 05; **, p <0. 01. Each experiment was performed twice with consistent results.
RGS4 antagonizes tumor formation by vGPCR
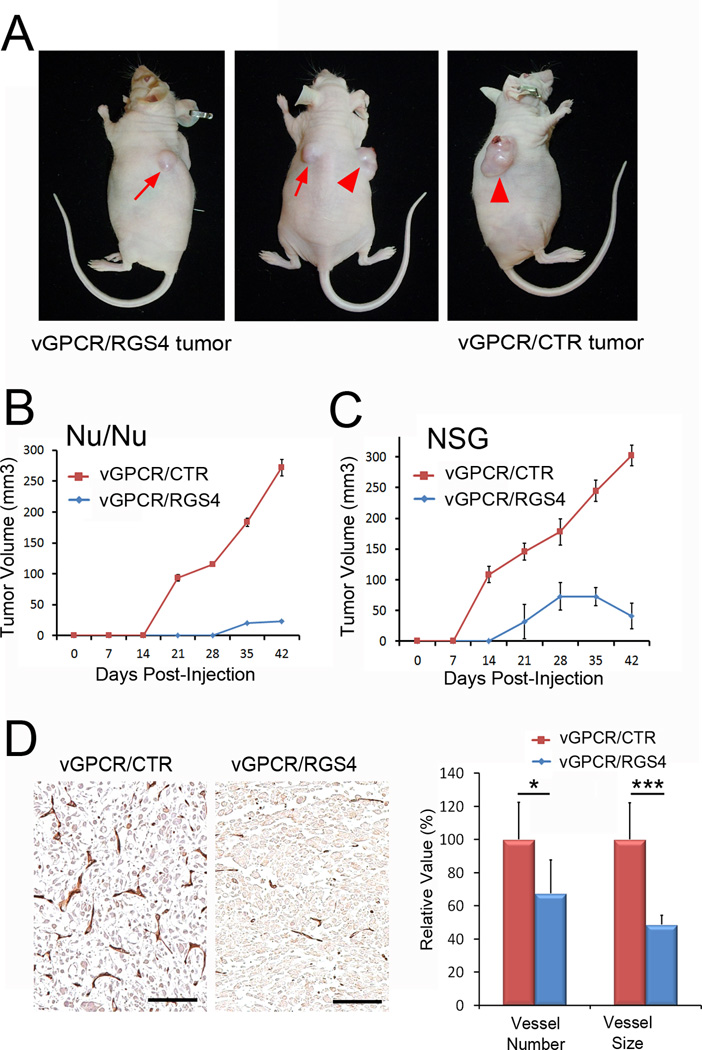
To further corroborate our in vitro data showing the RGS4-mediated inhibition of vGPCR activity, we next investigated whether RGS4 could inhibit vGPCR-induced tumor formation using two different immunodeficient mouse models, athymic nude mice (Nu/Nu) and NOD-SCID IL2Rγnullmice (NSG). As reported in a previous study(15), control parental SVECs did not form any detectable tumors in athymic nude mice (data not shown). We thus grafted vGPCR/CTR and vGPCR/RGS4 cells on the right and left flank areas, respectively, of the same mouse (Fig. 6A). In athymic nude mice, while control vGPCR-expressing SVEC cells (vGPCR/CTR) formed palpable tumors in two weeks, RGS4- and vGPCR-expressing SVEC cells(vGPCR/RGS4)formed significantly smaller tumors only after the 4th week (Fig. 6B). Similarly, in NOD-SCID IL2Rγnullmice, control vGPCR-expressing SVEC cells (vGPCR/CTR)began to form tumors after day 7, whereas RGS4- and vGPCR-expressing SVEC cells(vGPCR/RGS4) started to form visible tumors only after day 14, and these tumors were much smaller than control tumors in the same mouse (Fig. 6C). Subsequent immunohistochemistry analyses of these tumors revealed that RGS4 strongly repressed tumor-associated angiogenesis induced by vGPCR (Fig. 6D). Notably, both number and size of CD31-positive vessels were significantly reduced in RGS4-expressing tumors as compared to control tumors. Moreover, we performed vascular analyses in the tumors of comparable size (~50 mm3): vGPCR/CTR tumors were harvested from NSG mice at day 10 and their vascularity was compared to that of vGPCR/RGS4 tumors collected from NSG mice at day 42. Vascular analyses revealed that while vessel size was comparable, the number of vessels was much less in vGPCR/RGS4 tumors (Supplemental Figure 3). Taken together, these in vivo studies demonstrate that RGS4 can antagonize vGPCR-mediated tumor formation by suppressing tumor-associated angiogenesis.
Figure 6. RGS4 inhibits vGPCR-induced tumor formation.
(A) Tumor formation by SVECs expressing vGPCR alone (vGPCR/CTR) or vGPCR and RGS4 (vGPCR/RGS4) was evaluated in athymic nude mice. Arrow indicates vGPCR/RGS4 tumor and arrowhead indicates vGPCR/CTR tumor in the same mouse at 6 weeks post subcutaneous inoculation. (B,C) Growth curves of vGPCR/CTR and vGPCR/RGS4 tumors in athymic nude (Nu/Nu) (B) or NOD-SCID IL2Rγnullmice (NSG) (C). Equal numbers of vGPCR/CTR and vGPCR/RGS4 cells were subcutaneously injected at the right and left flank area, respectively, five female mice per group (n=5). Experiments were performed twice with similar results and tumor volume is shown as average tumor volume ± standard deviation of one representative experiment. (D) Reduced tumor-associated angiogenesis by RGS4-expressing tumors. Sections prepared from paraffin-embedded tumors harvested from athymic nude mice at Day 42 were subjected to immunohistochemistry analysis for CD31. Number and size of CD31-positive vessels were analyzed using the NIH ImageJ program and are displayed as relative value ± standard deviation. Scale bars, 100 µm; *, p <0. 05; ***, p <0. 001. Animal studies were performed three times with consistent results.
DISCUSSION
Host cells are generally equipped with various defense mechanisms against viral infection and propagation. On the other hand, many pathogenic viruses have acquired a list of counteracting mechanisms that can nullify or constrain the host defense mechanisms. Numerous studies have demonstrated that KSHV follows this model of virus-host interaction. It is believed that KSHV vGPCR plays a necessary and sufficient role in KSHV-mediated endothelial cell transformation by activating important signal cascades such as AKT, ERK1/2 and p38, and additionally, by stimulating production of pro-angiogenic factors including VEGF, angiopoietin-2, and angiopoietin-like (Angptl)-4, (1–6, 30–32). Although vGPCR is constitutively active, its activity can be further stimulated or inhibited by agonists or inverse agonists of CXCR2. While the CXC chemokines CXCL1/Gro-α and CXCL8/IL-8 function as agonists to activate vGPCR-mediated downstream signaling, other CXC chemokines such as CXCL10/IP-10 and CXCL12/SDF-1α can act as inverse agonists to inhibit downstream signaling by vGPCR(33). In addition, various desensitization mechanisms for cellular GPCR can be considered as another layer of control point in restricting the activity of vGPCR. This GPCR desensitization is sequentially carried out by two molecular players: the GPCR kinases (GRKs), which phosphorylate intracellular serine and threonine residues of activated GPCRs, and the arrestins, which uncouple phosphorylated GPCRs from heterotrimetic G protein complexes (34). The sequential action of GRKs and the arrestins result in rapid attenuation of GPCR-mediated signaling, followed by receptor internalization. The carboxy terminal tail of vGPCR was found to be the target site for GRK/arrestin-mediated receptor desensitization (35, 36). Consistently, PMA-induced activation of protein kinase C (PKC) and the expression of GRK4 were shown to inhibit the KSHV vGPCR-mediated signaling (2, 37).
In this study, we have identified a novel cellular desensitization mechanism for KSHV vGPCR. RGS4 is known to function as a GTPase-activating protein (GAP) for the Gα subunits and to suppress the signaling of cellular GPCRs by promoting hydrolysis of Gα-GTP to Gα-GDP(5, 6). We expect that RSG4 would exercise the same mechanism to suppress the activity of KSHV GPCR: Rather than directly targeting vGPCR, it would act on various Gα subunits that are associated with vGPCR and promote hydrolysis of the Gα-GTP to inhibit the vGPCR signaling cascades.
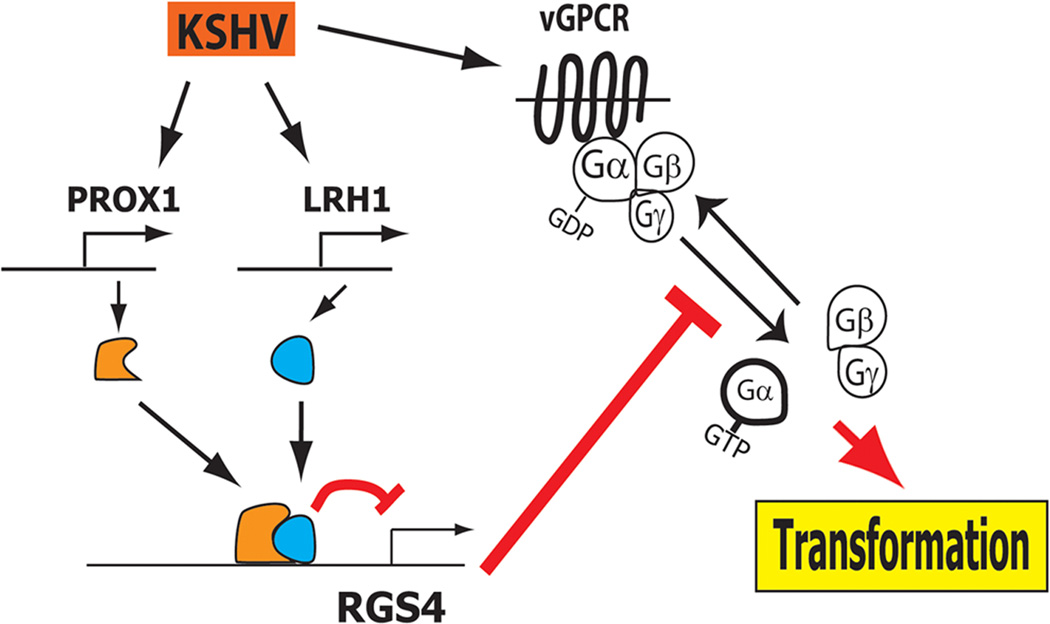
RGS4 was found to be expressed in vascular endothelial cells and to inhibit cell proliferation, migration, and invasion(7). Importantly, we discovered that RGS4 is predominantly expressed in BECs, but not in LECs, and that KSHV infection of BECs significantly inhibits RGS4 expression. Moreover, this repression of RGS4 by KSHV was mediated by the lymphatic-specific regulator PROX1 and its interacting nuclear receptor LRH1. Our study revealed that PROX1 directly binds to the promoter of RGS4 and represses the transcription of RGS4 through collaboration with LRH1. It is worth noting that our microarray study found that KSHV does not alter the expression of GRK5 or arrestins (9) and also that RGS4 is selectively regulated by KSHV. Taken together, our previous and current studies put forward an interesting hypothesis that KSHV-mediated upregulation of PROX1 results in suppression of RGS4 expression, which may otherwise antagonize the activity of vGPCR and thus inhibit KSHV-induced endothelial transformation (Fig. 7).
Figure 7. Working hypothesis of PROX1/LRH1-mediated inhibition of RGS4 expression to protect vGPCR activity for KS tumorigenesis.
Acting as aGTPase-activating protein (GAP)of cellular GPCRs, RGS4 can also antagonize KSHV viral GPCR activity. In order to ensure the maximum activity of vGPCR, KSHV-mediated upregulation of a nuclear receptor LRH1 and its interacting coregulator PROX1 leads to cooperative suppression of the expression of RGS4, a newly identified inhibitor of vGPCR.
PROX1 was isolated as an interacting protein of LRH1 through yeast two hybrid screens(26, 28). Interestingly, while PROX1 has gained its alias as the master regulator of lymphatic development(38), LRH1 has been known as a master regulator of cholesterol homeostasis (39). It is important to note that LRH1 has not been previously associated with KSHV pathology or KS tumorigenesis, and additionally, KSHV selectively upregulates these two key regulators for cell differentiation and metabolism. The tumor-promoting role of LRH1 has been reported in various cancers. In particular, a genome-wide association study of pancreatic cancer has identified polymorphisms in the LRH1 gene as a common susceptibility locus for pancreatic cancer (40). Supporting this finding, LRH1 siRNA inhibited pancreatic cancer cell proliferation, partly due to the downregulation of its transcriptional targets that control cell growth, proliferation and differentiation (41). In addition, LRH1 was shown to contribute to intestinal tumor formation through effects on cell cycle and inflammation(42) and to promote breast cancer cell motility and invasion (43). Interestingly, a recent study showed that inactivation of RGS4 strongly promotes breast cancer cell migration and invasion (44). Therefore, the suppression of RGS4 and consequent promotion of GPCR activities are largely consistent with the tumor-promoting role of LRH1. It will be interesting to further investigate how LRH1 contributes to KS tumor development, other than its interaction with PROX1.
Notably, our immunostaining and transgenic mouse studies revealed that RGS4 expression varies significantly in different vascular compartments, as well as in different tissues. This may be attributable to the fact that smooth muscle cells (SMCs) express RGS4(45)and different vascular compartments have different densities of SMCs. In the context of the physiological role of RSG4 in endothelial cells, another interesting question remains: Why is RGS4 down-regulated in LECs, compared to BECs. Although many explanations are possible, our favorite is that lymphatic vessels may need a different set of GPCR-modifiers to carry out its functions, which are distinct from blood vessels. There are more than 20 RGS family members, which can function as regulators for one or multiple GPCR proteins (5, 6, 46). It would be reasonable to speculate that BECs and LECs are equipped with a different array of GPCR regulators. For example, Wick et al. reported that RGS-3 is predominantly expressed in LECs, compared to BECs (47). Notably, according to our microarray study, RGS-3 was not regulated by KSHV (Supplemental Table 1). We asked another interesting question: whether ectopic expression of RGS4 can induce BEC-phenotypes and found that overexpression of RGS4 in KS tumor cells and primary LECs did not induce expression of BEC-signature genes (data not shown).
A recent study has utilized SVECs as murine LECs, based on their findings that SVECs express some lymphatic-signature genes such as LYVE-1, VEGFR-3, and podoplanin (48). In the same study, however, the authors found that SVECs express BEC-associated genes such as CD105, vinculin, neuropilin-1 and STAT6, and noted that SVECs have been variably used as lymphatic, high endothelial venule or blood microvascular endothelium(48). Most importantly, SVECs were found to not express PROX1 by the authors(48). Consistent with this finding, our western blot and immunofluorescence analyses revealed the absence of Prox1 expression in SVECs and derivative cells (Supplemental Figure 4). Together, these findings suggest that SVECs may have lost their original cell identity due to SV40-mediated immortalization and/or prolonged in vitro cell culturing, acquiring both blood and lymphatic cell phenotypes.
In summary, our study demonstrates that RGS4 downregulation is one significant beneficial aspect of lymphatic reprogramming during KS tumor development. Given the enormous body of evidence pointing to vGPCR as the viral oncogene driving KS tumorigenesis, it is quite exciting to understand how the cancer-causing virus could find its way to ensure and maximize the oncogenic potential of vGPCR by manipulating host gene regulatory networks. Moreover, RGS4-dependent desensitization mechanism against vGPCR may present a novel therapeutic target and further investigation should be warranted.
Supplementary Material
ACKNOWLEDGEMENT
This study was supported by NIH/NIDDK (5K08DK078589, CJK), American Cancer Society (RGS-08-194-01-MBC, YKH), NIH/NICHD (HD059762, YKH), NIH (CA31363, CA082057, CA115284 and DE019085, JJ) and Fletcher Jones Foundation (JJ).
Footnotes
Present Address: Inho Choi, Hoseo University, Asan, Korea
The authors declare no conflict of interest.
REFERENCES
- 1.Sodhi A, Montaner S, Gutkind JS. Does dysregulated expression of a deregulated viral GPCR trigger Kaposi's sarcomagenesis? FASEB J. 2004;18:422–427. doi: 10.1096/fj.03-1035hyp. [DOI] [PubMed] [Google Scholar]
- 2.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, et al. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 3.Couty JP, Gershengorn MC. Insights into the viral G protein-coupled receptor encoded by human herpesvirus type 8 (HHV-8) Biol Cell. 2004;96:349–354. doi: 10.1016/j.biolcel.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Martin D, Galisteo R, Molinolo AA, Wetzker R, Hirsch E, Gutkind JS. PI3Kgamma mediates kaposi's sarcoma-associated herpesvirus vGPCR-induced sarcomagenesis. Cancer Cell. 2011;19:805–813. doi: 10.1016/j.ccr.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 2009;78:1289–1297. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther. 2007;116:473–495. doi: 10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albig AR, Schiemann WP. Identification and characterization of regulator of G protein signaling 4 (RGS4) as a novel inhibitor of tubulogenesis: RGS4 inhibits mitogen-activated protein kinases and vascular endothelial growth factor signaling. Mol Biol Cell. 2005;16:609–625. doi: 10.1091/mbc.E04-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilar B, Hong YK. Kaposi Sarcoma A Model of Oncogenesis: Research Signpost. 2010. The origin of Kaposi sarcoma tumor cells; pp. 123–138. [Google Scholar]
- 9.Hong YK, Foreman K, Shin JW, Hirakawa S, Curry CL, Sage DR, et al. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma-associated herpesvirus. Nat Genet. 2004;36:683–685. doi: 10.1038/ng1383. [DOI] [PubMed] [Google Scholar]
- 10.Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–693. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 11.Carroll PA, Brazeau E, Lagunoff M. Kaposi's sarcoma-associated herpesvirus infection of blood endothelial cells induces lymphatic differentiation. Virology. 2004;328:7–18. doi: 10.1016/j.virol.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- 13.Kang J, Yoo J, Lee S, Tang W, Aguilar B, Ramu S, et al. An exquisite cross-control mechanism among endothelial cell fate regulators directs the plasticity and heterogeneity of lymphatic endothelial cells. Blood. 2010 doi: 10.1182/blood-2009-11-252270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, Kang J, Yoo J, Ganesan SK, Cook SC, Aguilar B, et al. Prox1 physically and functionally interacts with COUP-TFII to specify lymphatic endothelial cell fate. Blood. 2009;113:1856–1859. doi: 10.1182/blood-2008-03-145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montaner S, Sodhi A, Ramsdell AK, Martin D, Hu J, Sawai ET, et al. The Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor as a therapeutic target for the treatment of Kaposi's sarcoma. Cancer Res. 2006;66:168–174. doi: 10.1158/0008-5472.CAN-05-1026. [DOI] [PubMed] [Google Scholar]
- 16.Choi I, Lee S, Kyoung Chung H, Suk Lee Y, Eui Kim K, Choi D, et al. 9-cis retinoic Acid promotes lymphangiogenesis and enhances lymphatic vessel regeneration: therapeutic implications of 9-cis retinoic Acid for secondary lymphedema. Circulation. 2012;125:872–882. doi: 10.1161/CIRCULATIONAHA.111.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdari KV, Bamne M, Joel W, Talkowski ME, Mirnics K, Levitt P, et al. Linkage Disequilibrium Patterns and Functional Analysis of RGS4 Polymorphisms in Relation to Schizophrenia. Schizophr Bull. 2007 doi: 10.1093/schbul/sbm042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, et al. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus Bacterial artificial chromosome clone. J Virol. 2012 doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baluk P, Tammela T, Ator E, Lyubynska N, Achen MG, Hicklin DJ, et al. Pathogenesis of persistent lymphatic vessel hyperplasia in chronic airway inflammation. J Clin Invest. 2005;115:247–257. doi: 10.1172/JCI22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebert PJ, Campbell DB, Levitt P. Bacterial artificial chromosome transgenic analysis of dynamic expression patterns of regulator of G-protein signaling 4 during development I. Cerebral cortex. Neuroscience. 2006;142:1145–1161. doi: 10.1016/j.neuroscience.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi I, Chung HK, Ramu S, Lee HN, Kim KE, Lee S, et al. Visualization of lymphatic vessels by Prox1-promoter directed GFP reporter in a bacterial artificial chromosome-based transgenic mouse. Blood. 2011;117:362–365. doi: 10.1182/blood-2010-07-298562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrova TV, Makinen T, Makela TP, Saarela J, Virtanen I, Ferrell RE, et al. Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. EMBO J. 2002;21:4593–4599. doi: 10.1093/emboj/cdf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong YK, Harvey N, Noh YH, Schacht V, Hirakawa S, Detmar M, et al. Prox1 is a master control gene in the program specifying lymphatic endothelial cell fate. Dev Dyn. 2002;225:351–357. doi: 10.1002/dvdy.10163. [DOI] [PubMed] [Google Scholar]
- 24.Hirakawa S, Hong YK, Harvey N, Schacht V, Matsuda K, Libermann T, et al. Identification of vascular lineage-specific genes by transcriptional profiling of isolated blood vascular and lymphatic endothelial cells. Am J Pathol. 2003;162:575–586. doi: 10.1016/S0002-9440(10)63851-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin JW, Min M, Larrieu-Lahargue F, Canron X, Kunstfeld R, Nguyen L, et al. Prox1 promotes lineage-specific expression of fibroblast growth factor (FGF) receptor-3 in lymphatic endothelium: a role for FGF signaling in lymphangiogenesis. Mol Biol Cell. 2006;17:576–584. doi: 10.1091/mbc.E05-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffensen KR, Holter E, Bavner A, Nilsson M, Pelto-Huikko M, Tomarev S, et al. Functional conservation of interactions between a homeodomain cofactor and a mammalian FTZ-F1 homologue. EMBO Rep. 2004;5:613–619. doi: 10.1038/sj.embor.7400147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin J, Gao DM, Jiang QF, Zhou Q, Kong YY, Wang Y, et al. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-alpha-hydroxylase gene. Mol Endocrinol. 2004;18:2424–2439. doi: 10.1210/me.2004-0009. [DOI] [PubMed] [Google Scholar]
- 28.Liu YW, Gao W, Teh HL, Tan JH, Chan WK. Prox1 is a novel coregulator of Ff1b and is involved in the embryonic development of the zebra fish interrenal primordium. Mol Cell Biol. 2003;23:7243–7255. doi: 10.1128/MCB.23.20.7243-7255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bais C, Van Geelen A, Eroles P, Mutlu A, Chiozzini C, Dias S, et al. Kaposi's sarcoma associated herpesvirus G protein-coupled receptor immortalizes human endothelial cells by activation of the VEGF receptor-2/ KDR. Cancer Cell. 2003;3:131–143. doi: 10.1016/s1535-6108(03)00024-2. [DOI] [PubMed] [Google Scholar]
- 30.Mutlu AD, Cavallin LE, Vincent L, Chiozzini C, Eroles P, Duran EM, et al. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV: a cell and animal model of virally induced Kaposi's sarcoma. Cancer Cell. 2007;11:245–258. doi: 10.1016/j.ccr.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vart RJ, Nikitenko LL, Lagos D, Trotter MW, Cannon M, Bourboulia D, et al. Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer Res. 2007;67:4042–4051. doi: 10.1158/0008-5472.CAN-06-3321. [DOI] [PubMed] [Google Scholar]
- 32.Ma T, Jham BC, Hu J, Friedman ER, Basile JR, Molinolo A, et al. Viral G protein-coupled receptor up-regulates Angiopoietin-like 4 promoting angiogenesis and vascular permeability in Kaposi's sarcoma. Proc Natl Acad Sci U S A. 2010;107:14363–14368. doi: 10.1073/pnas.1001065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenkilde MM, Kledal TN, Brauner-Osborne H, Schwartz TW. Agonists and inverse agonists for the herpesvirus 8-encoded constitutively active seven-transmembrane oncogene product, ORF-74. Journal of Biological Chemistry. 1999;274:956–961. doi: 10.1074/jbc.274.2.956. [DOI] [PubMed] [Google Scholar]
- 34.Sherrill JD, Miller WE. Desensitization of herpesvirus-encoded G protein-coupled receptors. Life Sci. 2008;82:125–134. doi: 10.1016/j.lfs.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Sandford G, Fei G, Nicholas J. Galpha protein selectivity determinant specified by a viral chemokine receptor-conserved region in the C tail of the human herpesvirus 8 g protein-coupled receptor. J Virol. 2004;78:2460–2471. doi: 10.1128/JVI.78.5.2460-2471.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verzijl D, Pardo L, van Dijk M, Gruijthuijsen YK, Jongejan A, Timmerman H, et al. Helix 8 of the viral chemokine receptor ORF74 directs chemokine binding. J Biol Chem. 2006;281:35327–35335. doi: 10.1074/jbc.M606877200. [DOI] [PubMed] [Google Scholar]
- 37.Geras-Raaka E, Arvanitakis L, Bais C, Cesarman E, Mesri EA, Gershengorn MC. Inhibition of constitutive signaling of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor by protein kinases in mammalian cells in culture. J Exp Med. 1998;187:801–806. doi: 10.1084/jem.187.5.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Choi I, Hong YK. Heterogeneity and plasticity of lymphatic endothelial cells. Semin Thromb Hemost. 2010;36:352–361. doi: 10.1055/s-0030-1253457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YK, Moore DD. Liver receptor homolog-1, an emerging metabolic modulator. Front Biosci. 2008;13:5950–5958. doi: 10.2741/3128. [DOI] [PubMed] [Google Scholar]
- 40.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1-1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benod C, Vinogradova MV, Jouravel N, Kim GE, Fletterick RJ, Sablin EP. Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc Natl Acad Sci U S A. 2011;108:16927–16931. doi: 10.1073/pnas.1112047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoonjans K, Dubuquoy L, Mebis J, Fayard E, Wendling O, Haby C, et al. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc Natl Acad Sci U S A. 2005;102:2058–2062. doi: 10.1073/pnas.0409756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chand AL, Herridge KA, Thompson EW, Clyne CD. The orphan nuclear receptor LRH-1 promotes breast cancer motility and invasion. Endocr Relat Cancer. 2010;17:965–975. doi: 10.1677/ERC-10-0179. [DOI] [PubMed] [Google Scholar]
- 44.Xie Y, Wolff DW, Wei T, Wang B, Deng C, Kirui JK, et al. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69:5743–5751. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grillet N, Pattyn A, Contet C, Kieffer BL, Goridis C, Brunet JF. Generation and characterization of Rgs4 mutant mice. Mol Cell Biol. 2005;25:4221–4228. doi: 10.1128/MCB.25.10.4221-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribas C, Penela P, Murga C, Salcedo A, Garcia-Hoz C, Jurado-Pueyo M, et al. The G protein-coupled receptor kinase (GRK) interactome: Role of GRKs in GPCR regulation and signaling. Biochim Biophys Acta. 2006 doi: 10.1016/j.bbamem.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 47.Wick N, Saharinen P, Saharinen J, Gurnhofer E, Steiner CW, Raab I, et al. Transcriptomal comparison of human dermal lymphatic endothelial cells ex vivo and in vitro. Physiol Genomics. 2007;28:179–192. doi: 10.1152/physiolgenomics.00037.2006. [DOI] [PubMed] [Google Scholar]
- 48.Ledgerwood LG, Lal G, Zhang N, Garin A, Esses SJ, Ginhoux F, et al. The sphingosine 1-phosphate receptor 1 causes tissue retention by inhibiting the entry of peripheral tissue T lymphocytes into afferent lymphatics. Nat Immunol. 2008;9:42–53. doi: 10.1038/ni1534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.