Abstract
There are many potential RNA drug targets in bacterial, viral, and the human transcriptomes. However, there are few small molecules that modulate RNA function. This is due, in part, to a lack of fundamental understanding about RNA-ligand interactions including the types of small molecules that bind to RNA structural elements and the RNA structural elements that bind to small molecules. In an effort to better understand RNA-ligand interactions, we diversified the 2-aminobenzimidazole core (2AB) and probed the resulting library for binding to a library of RNA internal loops. We chose the 2AB core for these studies because it is a privileged scaffold for binding RNA based on previous reports. These studies identified that N-methyl pyrrolidine, imidazole, and propylamine diversity elements at the R1 position increase binding to internal loops; variability at the R2 position is well tolerated. The preferred RNA loop space was also determined for five ligands using a statistical approach and identified trends that lead to selective recognition.
INTRODUCTION
Ribonucleic acid (RNA) is involved in many cellular processes including regulation of gene expression, protein synthesis, and bio-catalysis (1–3). The discovery of small molecule chemical probes or lead therapeutic agents that affect RNA function has wide-ranging implications. Few small molecules modulate RNA function, with the exception of antibacterials that target the bacterial ribosome. It is difficult to identify small molecules that bind a validated RNA target with high affinity and selectivity, as evidenced by the lower hit rate for RNAs than proteins in high throughput screening campaigns. This suggests that sub-optimal chemical space is being screened for binding RNAs. Thus, we have taken a bottom-up approach to develop strategies to target RNA. That is, chemical space is screened for binding to discrete RNA secondary structure (motif) libraries to uncover preferred/privileged chemical space and RNA space simultaneously (4, 5).
It has been previously reported that the benzimidazole scaffold (Figure 1A) is a privileged pharmacophore. NMR spectroscopy-based screening identified a benzimidazole that competes with aminoglycosides for binding the bacterial rRNA A-site. Another benzimidazole binds the Hepatitis C internal ribosomal entry site (IRES) and affects the RNA’s fold. This structural change precludes the IRES from binding the human ribosome and inhibits propagation. A bis-benzimidazole binds the expanded r(CUG) repeats that cause myotonic dystrophy type 1 and improves DM1-associated defects in cellular and animal models (6).
Figure 1.
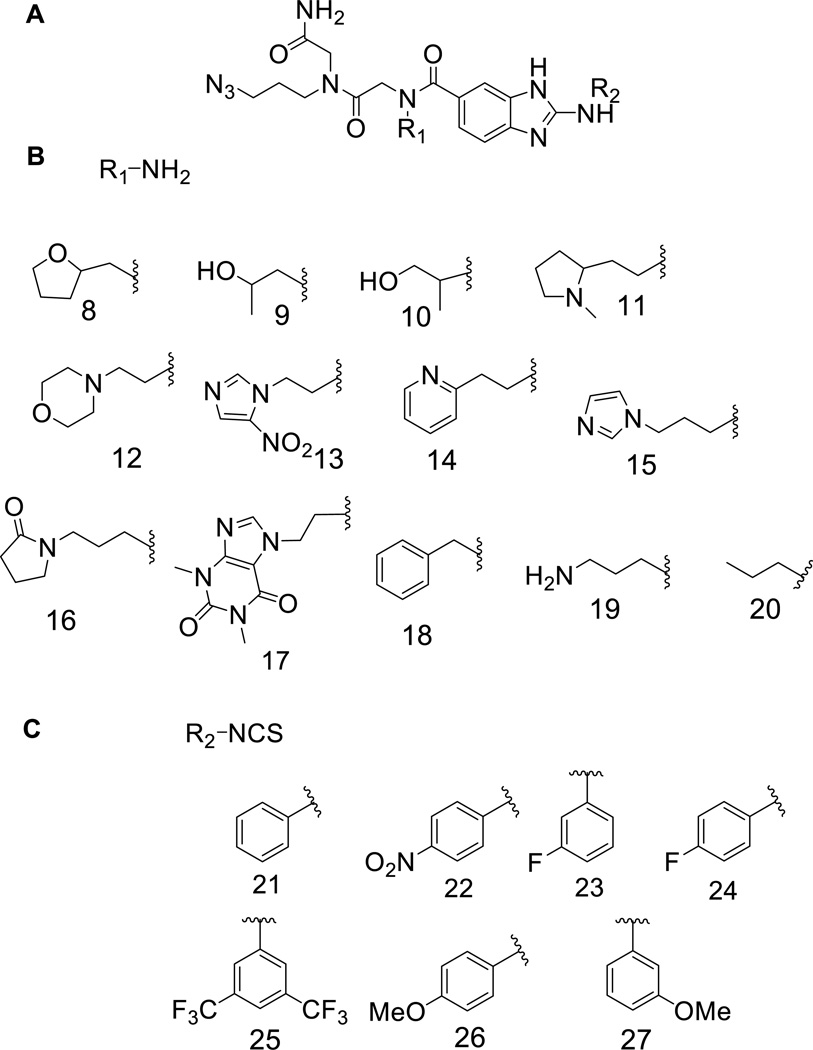
General structure of the 2-aminobenzimidazole library and the submonomers used to construct it. (A) General structure of 2-aminobenzimidazole library members. (B) Submonomers used at the R1 position. (C) Submonomers used at the R2 position. Each compound has an invariant azido group for conjugation onto alkyne-functionalized agarose microarray surfaces.
In order to identify the optimal RNA targets for benzimidazoles, we synthesized a small molecule library using a 2-aminobenzimidazole core (2AB) and probed it for binding to a library of RNA internal loops (1, Figure 2). Diversity elements (Figure 1B and 1C) were chosen based on their potential to stack or hydrogen bond with RNA bases. These studies uncovered diversity elements that impart affinity and specificity for binding RNA internal loops and the RNA loops that are targetable by 2AB’s.
Figure 2.
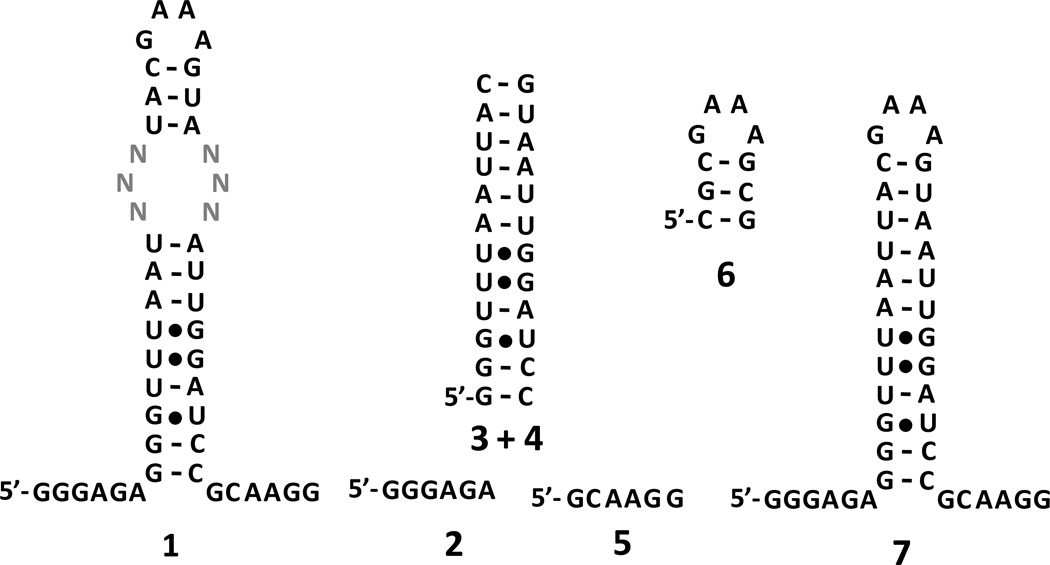
The 3×3 nucleotide internal loop library (1) and competitor oligonucleotides (2–6) that restrict selected interactions to the randomized region. Oligonucleotides 2 and 5 mimic the single stranded regions; 3+4 mimic base paired regions; and 6 mimics the GNRA hairpin. Oligonucleotide 7 is cassette into which the randomized region was inserted.
RESULTS & DISCUSSION
2-Aminobenzimidazole Library Design and Synthesis
The benzimidazole scaffold consists of a pharmacophore with hydrogen bond donors and acceptors combined with an aromatic ring (Figure 1A). We chose two positions to introduce diversity into the 2AB core, R1 (Figure 1B) and R2 (Figure 1C). The benzimidazole library was synthesized according to a modified version of a previously described solid-phase approach (7, 8). The first point of diversity (R1), installed as a peptoid submonomer adjacent to the 2-aminobenzimidazole core, can interact with RNA through stacking and hydrogen bonding (Figure 1B). Most of the submonomers used are the same or variants of ones used in the synthesis of RNA-focused peptoids (9). The second point of diversity, R2, was introduced via reaction of an isothiocyanate and an aromatic diamine, which adds an aromatic ring that could stack with RNA bases (Figure 1C). These elements have a central phenyl group surrounded by substituents that can affect the partial charges of the aromatic ring. The nomenclature used to describe these compounds is R1–AB–R2 where AB indicates the 2-aminobenzimidazole core.
Each library member also contained an azide tag to allow for site-specific conjugation onto an alkyne-functionalized agarose microarray surface via a Huisgen dipolar cycloaddition reaction (HDCR). The HDCR reaction and the agarose microarray surface have been previously utilized (4, 10–13). The azide and alkyne reactive groups that are required for an HDCR are desirable because they are orthogonal to the reaction conditions that are used to construct the 2AB library. Successful synthesis was confirmed by mass spectrometry (ESI-MS). By screening arrays of quality-controlled compounds, confidence can be placed in the binding results, and structure-activity relationships can be constructed to enable the design of second-generation compounds that could have improved potencies and specificities (9). In all, 79 diversified benzimidazoles were synthesized.
Screening of the 2-Aminobenzimidazole via Microarray
We used a previously reported library-on-library screen named Two-Dimensional Combinatorial Screening (2DCS) in order to identify privileged chemical and RNA spaces simultaneously (4, 5). Prior to our selection experiment, the 2AB library was screened to identify the compounds that bind RNA internal loops. The library of small molecules was spatially arrayed onto agarose microarray surfaces to create small molecule microarrays (12, 13). All compounds (79) were arrayed at a single concentration of 4.5 mM, or 1.4 nmoles (300 nL spot volume).
After conjugation, the array was hybridized with [32P]-labeled RNA library 1 (Figure 2). This library has a six nucleotide randomized region displayed in a 3×3 nucleotide internal loop pattern. Thus, 1 has 4,096 unique members. RNA library 1 was used because it displays small, discrete RNA motifs that are likely to be found as components of larger RNAs that have biological functions. The library displays 3×3 nucleotide internal loops, 2×2 nucleotide internal loops, 1×1 nucleotide loops, bulges, and base paired RNAs (4). Since 4,096 members of 1 were screened for binding to 79 small molecules, 323,584 interactions were probed simultaneously in this single experiment. Hybridization was completed in the presence of Torula yeast bulk RNA at three times the amount of total moles of small molecule delivered to the microarray surface. (We also synthesized RNA-focused peptoid and peptide libraries and probed them for binding 1 under the same conditions. No significant binding was observed to any of the compounds. Details can be found in the Supporting Information.)
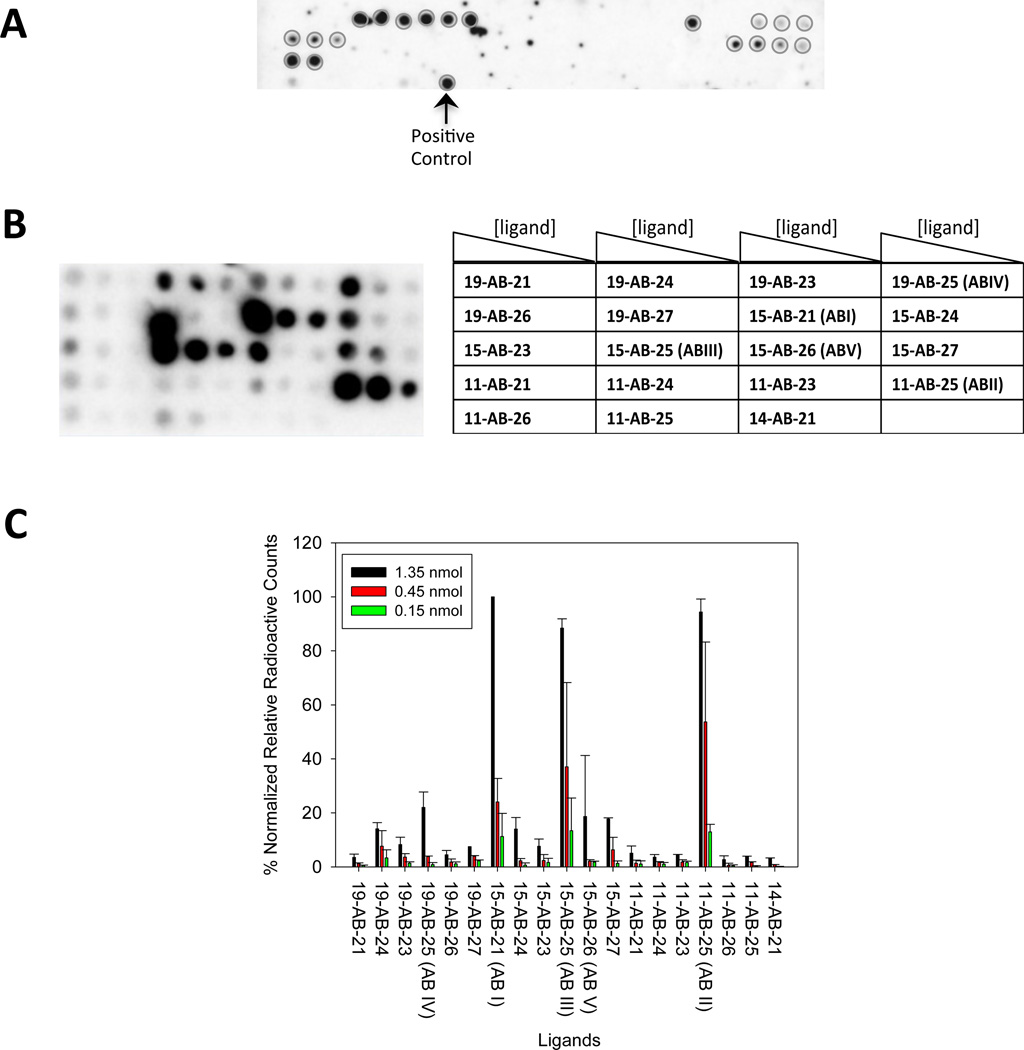
We designated “hits” by comparing the radioactive signal of a compound of interest to the compound that had the highest signal, or 15-AB-21. Hit compounds have >20% of the signal of 15-AB-21. A total of 19 compounds were scored as hits (Figure 3B), or 24% of the 2AB library. Thus, the aminobenzimidazole scaffold alone is not sufficient for binding 1 under these conditions. That is, certain diversity elements impart binding affinity for members of 1.
Figure 3.
Microarray screening of the 2-aminobenzimidazole library for binding 1. (A) Image of the microarray with all 79 ligands after hybridization with 32P-labeled 1 in the presence of Torula yeast bulk RNA (competitor RNA used to increase stringency). Approximately 1.4 nmoles of each compound were delivered to the slide surface. The 19 hit compounds are indicated with circles. (B) A secondary assay for the binding of the 19 hit compounds. The compounds were purified to homogeneity by HPLC and then immobilized onto alkyne-functionalized agarose microarrays as serial dilutions (1.35 nmoles, 0.45 nmoles, and 0.15 nmoles delivered to the surface). The arrays were then hybridized with 1 in the presence of Torula yeast bulk RNA (competitor RNA used to increase stringency). (C) Plot of the data from the experiment shown in B.
In order to identify the diversity elements that are important for binding 1, the hit compounds were analyzed using the Privileged Chemical Space Predictor Program (PCSP) (14). The program uses statistical analysis to determine the features in compounds that impart binding affinity (or some other activity), or privileged chemical space. In this case, PCSP normalized the radioactive signal from our microarray (binding value) and determined which features bias the compounds for binding 1 by comparing the proportion of a submonomer in the hit compounds to the proportion of the submonomer in the entire 2AB library. This pooled population comparison is represented as Z-scores that are converted into two-tailed p-values. A two-tailed p-value represents the probability (confidence level) that the observed proportion of a specific feature could occur if in fact there is no such bias for binding. A confidence level >95% is considered statistically significant.
PCSP determined that submonomers at both the R1 and R2 positions impart binding affinity. For the R1 position, 11 (N-methyl pyrrolidine) and 19 (propylamine) bias the 2AB core for binding members of 1 with two-tailed p-values of 0.0004 and <0.0001, respectively (Figure 1B). For the R2 position, 25 (bis-trifluoromethyl-benzene) positively contributes to binding 1 (two tailed p-value = 0.0366) while 23 (para-nitrobenzene) is deleterious to binding (selected against; two tailed p-value = 0.0050) (Figure 1C).
In order to identify the RNA loops that bind each compound, the 19 2-aminobenzimdazoles were purified by HPLC and tested in a secondary assay for binding to 1 (Figure 3B). Of these 19 compounds, 15-AB-21 (ABI), 11-AB-25 (ABII), 15-AB-25 (ABIII), 19-AB- 25 (ABIV), and 15-AB-26 (ABV) gave the highest signal on the array surface (Figures 3b and 3c). These compounds display 19 (propylamine), 15 (imidazole) or 11 (N-methyl pyrrolidine) side chains at the R1 position (Figure 1B) and 21 (benzene), 25 (bis-trifluoromethyl-benzene) or 26 (para-methoxy-benzene) at the R2 position (Figure 1C).
Selection of the RNA Motifs that Bind Lead Compounds (2DCS)
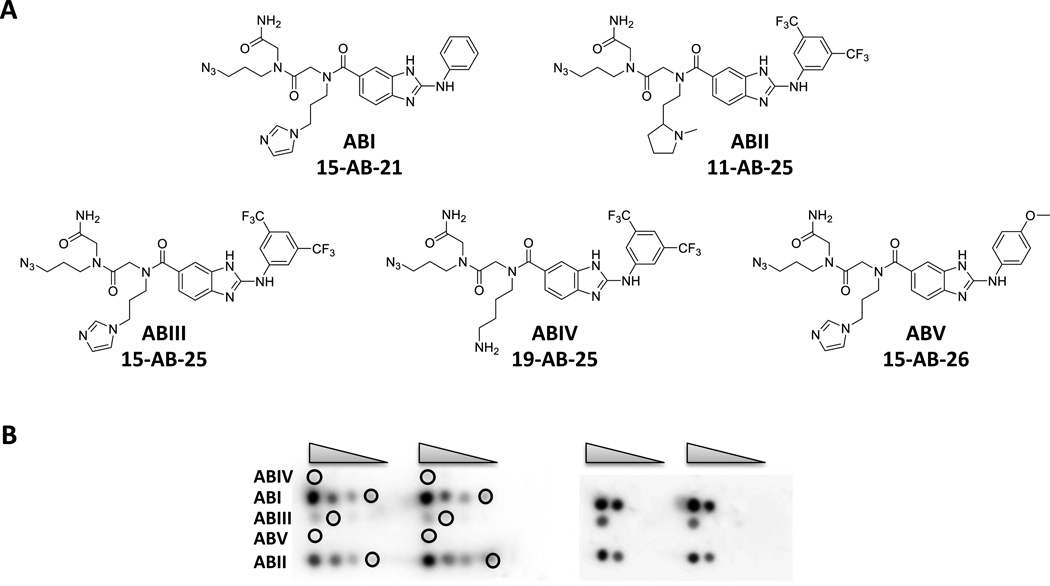
Based on the above results, the top five compounds were used in a 2DCS selection. In these experiments, 32P-labeled 1 and competitor oligonucleotides 2 – 6 (Figure 2) were hybridized with arrays displaying serially diluted ABI - ABV. Oligonucleotides 2 – 6 mimic the regions in 1 that are constant to all library members (Figure 2). This ensures that the selected interactions occur between the ligand and the randomized region. The competitor oligonucleotides are each present in 3-fold excess over the amount of moles of ligand delivered to a microarray surface and in >3000-fold excess over 1 such that the selection conditions are highly stringent. Under these conditions, ABI and ABII had the highest signal (Figure 4B). The lowest loading spots that gave signal over background were excised, amplified, and sequenced (Figure 4B, Tables S-1 & S-2). The lowest loading spot that gave sufficient signal was used because previous studies have shown that RNAs harvested from lower ligand loadings bind with higher affinities (4).
Figure 4.
Selection of the RNAs in 1 that bind ABI–ABV via Two-Dimensional Combinatorial Screening (2DCS). (A) Structures of 2-aminobenzimidazoles that were used in 2DCS selection. (B) Left, image of a microarray that displays all five compounds after hybridization with 32P-labeled 1. Serial dilutions of ABI – ABV were delivered to the slide surface (1.4, 0.7, 0.4, and 0.2 nmoles) and then hybridized with 1 in the presence of competitor oligonucleotides 2 – 6 (each present at 3 times the number of moles of ligand spotted and at >3000 times the concentration of 1). Circles indicate the positions where bound RNA was excised. Right, image of the array after bound RNA was excised.
The mixtures of RNAs that bound each ligand were RT-PCR amplified and transcribed in vitro. The affinities of the ligands for the corresponding mixtures of selected RNAs were determined using an in-solution, fluorescence-based binding assay. The ligands were fluorescently labeled by conjugating them to fluorescein (4). Saturable binding (micromolar range) was only observed for ABI and ABII when up to 1 mM RNA was added. Therefore, further analysis was only completed for ABI and ABII. The sequences selected for all ligands are available in the Supporting Information (Tables S-1 – S-5).
Informatic Analysis and Binding Studies for Members of 1 Selected to Bind ABI and ABII
The RNA motifs that bound ABI and ABII were cloned and sequenced as previously described (15). We obtained 71 sequences for ABI (Table S-1) and 77 sequences for ABII (Table S-2). The sequences were analyzed by using a statistical approach in order to identify features that afford high affinity binding. Analysis was completed by using the RNA Privileged Space Predictor (RNA-PSP) computer program (4, 5, 15–19). In RNA-PSP, the rate of occurrence of a feature in the selected RNAs is compared to the rate of occurrence in the entire library 1. The confidence that the selected feature did not occur randomly is assigned a Z-score and the corresponding two-tailed p-value. Only features that are statistically significant (p ≤ 0.05 or ≥95% confidence) are further considered. It should be noted that this analysis identifies features that contribute positively (positive Z-score) and negatively (negative Z-score) to binding. The features that have the highest confidence for binding ABI and ABII (two tailed p-values <0.0001) are shown in Table 1. Table 1 reports the following information for each trend that has a two tailed p-value <0.0001: the number of selected RNAs that have the trend of interest (present in sequencing data), the number of RNAs in 1 that have the trend of interest, and the corresponding Z-score. Interestingly, ABI has only 14 features compared to 49 features for ABII. Thus, ABI is more selective in the RNA motif space that it recognizes compared to ABII.
Table 1.
Features in selected sequences that impart binding affinity for ABI and ABII. Only features with two tailed p-values <0.0001 are reported.
| Trends for ABII with two tailed p-values <0.0001 | |||||||
|---|---|---|---|---|---|---|---|
| Trend | #of Selected Sequences |
#of Sequences in 1 |
Z-Score | Trend | #of Selected Sequences |
#of Sequences in 1 |
Z-Score |
| 5'ACA 3'CNN |
4/78 (5.1%) |
16/4096 (0.4%) |
6.00 | 5'NAA 3'NGG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'CGN |
4/78 (5.1%) |
16/4096 (0.4%) |
6.00 | 5'NAU 3'NCC |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CNG 3'NGG |
4/78 (5.1%) |
16/4096 (0.4%) |
6.00 | 5'NUC 3'NCC |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'CNN |
8/78 (10.3%) |
64/4096 (1.6%) |
5.84 | 5'NAU 3'CNC |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CNN 3'CGN |
7/78 (8.9%) |
64/4096 (1.6%) |
5.01 | 5'NAC 3'CNG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'NNN 3'CCC |
7/78 (8.9%) |
64/4096 (1.6%) |
5.01 | 5'NCU 3'CNU |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'ACA 3'NGN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NAU 3'CCN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'ACA 3'NNG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NAC 3'CAN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CUC 3'NUN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NCA 3'CGN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'NGU |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NCG 3'CCN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'NGG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NGU 3'GCN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'CNU |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NAN 3'CCC |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'CNG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NCN 3'CGU |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'CNG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NNA 3'CGG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'AUN 3'GCN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NNU 3'CCC |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'CCN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NNG 3'AGG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 |
| 5'CAN 3'CGN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'UNN 3'NCC |
6/78 (7.7%) |
64/4096 (1.6%) |
4.17 |
| 5'UNU 3'NCC |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'CNN 3'NGG |
6/78 (7.7%) |
64/4096 (1.6%) |
4.17 |
| 5'CNC 3'NUU |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NCN 3'CCN |
6/78 (7.7%) |
64/4096 (1.6%) |
4.17 |
| 5'CNGv 3'ANG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NCN 3'CGN |
6/78 (7.7%) |
64/4096 (1.6%) |
4.17 |
| 5'CAN | 3/78 | 16/4096 | 4.49 | 5'NNU | 6/78 | 64/4096 | 4.17 |
| 3'CGN | (3.8%) | (0.4%) | 3'NCC | (7.7%) | (1.6%) | ||
| 5'CNG 3'AGN |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'CAN 3'NGN |
6/78 (7.7%) |
64/4096 (1.6%) |
4.17 |
| 5'UNN 3'CCC |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NCN 3'CNN |
14/78 (17.9%) |
256/4096 (6.3%) |
4.16 |
| 5'CNN 3'AGG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | 5'NNN 3'NCC |
14/78 (17.9%) |
256/4096 (6.3%) |
4.16 |
| 5'CNN 3'CGG |
3/78 (3.8%) |
16/4096 (0.4%) |
4.49 | ||||
| Trends for ABI with two tailed p-values <0.0001 | |||||||
|---|---|---|---|---|---|---|---|
| Trend | #of Selected Sequences |
#of Sequences in 1 |
Z-Score | Trend | #of Selected Sequences |
#of Sequences in 1 |
Z-Score |
| 5'GCU 3'NNC |
4/71 (5.6%) |
16/4096 (0.4%) |
6.33 | 5'GNU 3'NCC |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 |
| 5'ANN 3'CNG |
7/71 (9.9%) |
64/4096 (1.6%) |
5.35 | 5'ANG 3'CNG |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 |
| 5'UAU 3'NAN |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 | 5'UNU 3'UAN |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 |
| 5'AUA 3'GNN |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 | 5'ANN 3'CUG |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 |
| 5'UCN 3'NGG |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 | 5'GNN 3'GGU |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 |
| 5'ACN 3'CNG |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 | 5'NGA 3'CNC |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 |
| 5'GAN 3'ACN |
3/71 (4.2%) |
16/4096 (0.4%) |
4.75 | 5'GNU 3'NNC |
6/71 (8.5%) |
64/4096 (1.6%) |
4.47 |
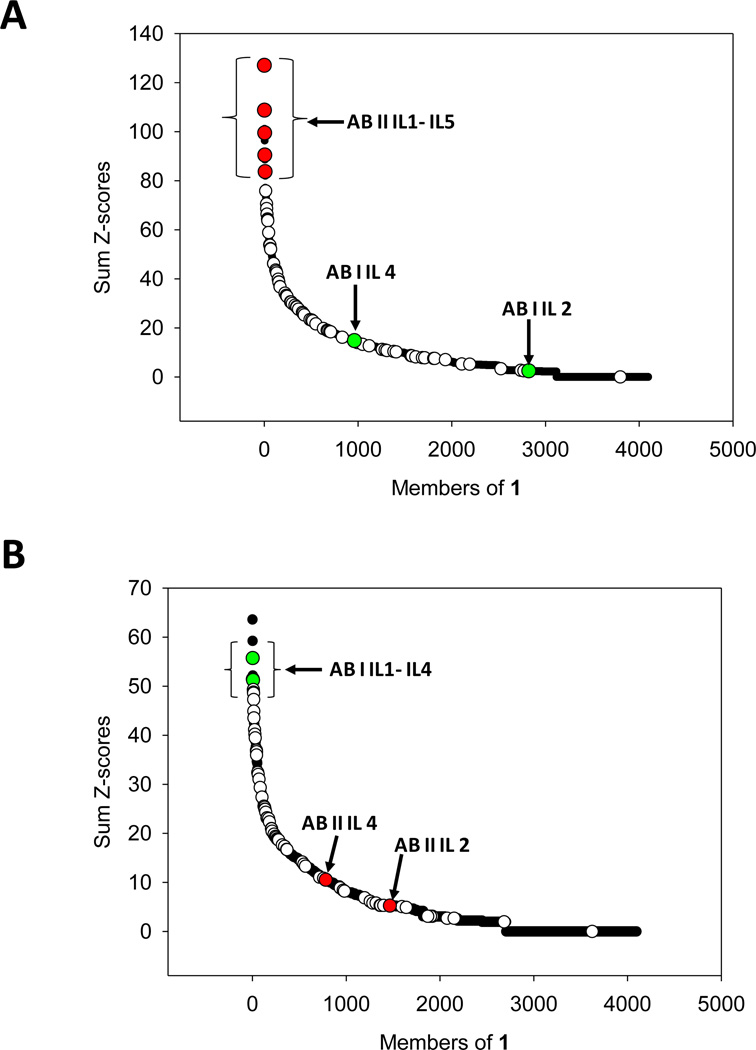
Each RNA can have more than one statistically significant feature. Therefore, the Z-scores for each feature are summed to afford a Sum Z-score (15, 16). Previous studies have shown that RNA motif-ligand interactions that have the highest Sum Z-scores have the highest affinity (15, 16). Moreover, by combining these statistical parameters with experimentally determined binding affinities, the affinity and selectivity of every RNA displayed in 1 can be predicted (15, 16). The selectivity of an RNA can be predicted by comparing its Sum Z-score for one ligand to its Sum Z-score for another. An RNA that has a large Sum Z-score for one ligand and a small Sum Z-score for another ligand is selective. This entire approach is called Structure-Activity Relationships Through Sequencing, or StARTS (16). The Sum Z-scores for all selected sequences are shown graphically in Figure 5A and 5B (open, red, and green circles) and summarized in Tables S-1 and S2. The black circles in the plots are for the RNA loops that are represented in 1 but were not selected.
Figure 5.
Structure-Activity Relationships Through Sequencing (StARTs) approach to predict the affinity and selectivity of RNA-ligand interactions. (A) Plot of the Sum Z-score for each member of 1 and ABI. The RNAs with the largest Sum Z-scores are predicted to be the highest affinity (15, 16). The open circles represent RNAs from sequencing data. The red circles represent the RNAs selected for ABI for which binding affinities were measured (Figure 6). The green circles represent the RNAs selected to bind ABII that have large Sum Z-scores (see panel B). Since these RNA have small Sum Z-scores for ABI, they are predicted to bind weakly to ABI. (B) Plot of the Sum Z-score for each member of 1 and ABII. The open circles represent RNAs from sequencing data. The green circles represent the RNAs selected to bind ABII for which binding affinities were measured (Figure 6). These RNAs have large Sum Z-scores. Red circles represent the RNAs that were selected to bind ABI shown in panel A. Since these RNA have small Sum Z-scores for ABII, they are predicted to bind weakly to ABII.
Binding Affinities of ABI and ABII for Selected RNAs
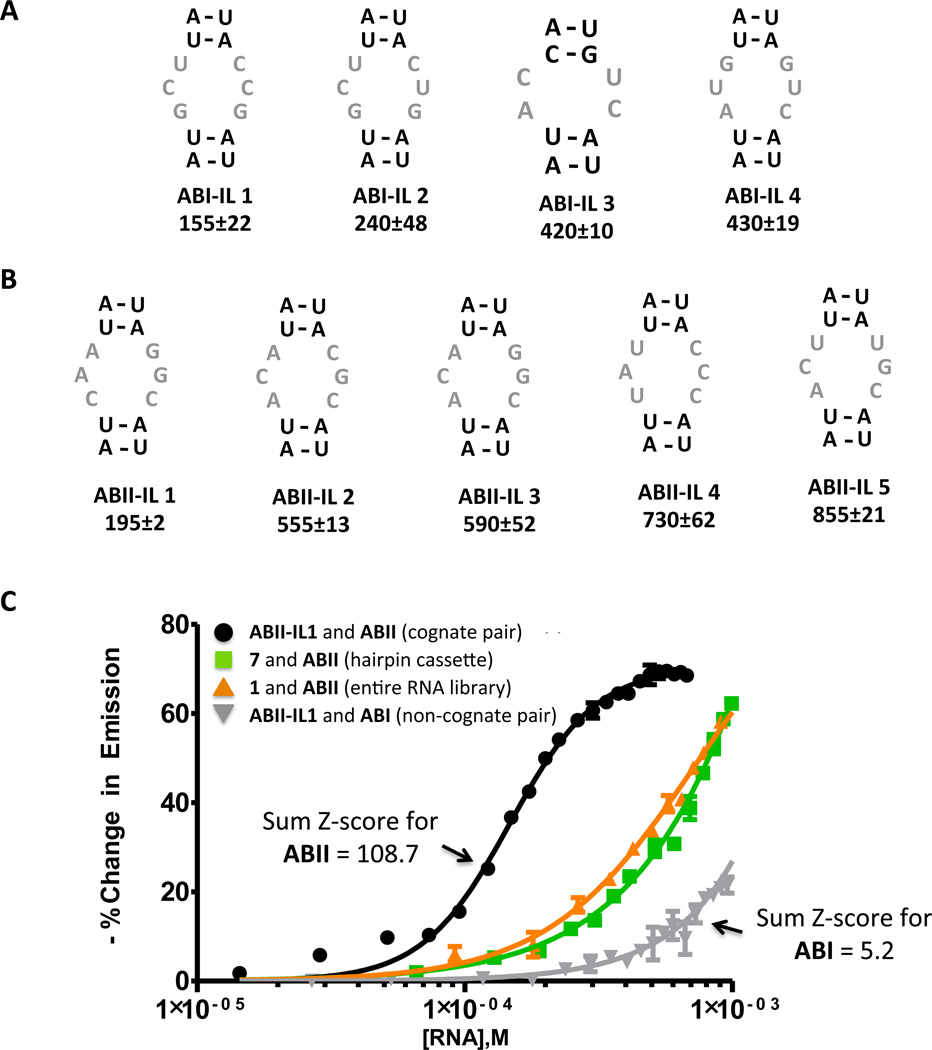
Next, the affinities of RNAs from our sequencing data for the corresponding ligand were measured (Figure 6). Each RNA is given a unique identifier: ABX-IL Y where ABX denotes the ligand for which the RNA was selected and IL Y indicates a unique internal loop (IL) identifier. We chose the RNAs with the highest Sum Z-scores from our statistical analysis (ABI-IL 1 – ABI-IL 4 (green circles) and ABII-IL 1 – ABII-IL 5 (red circles); Figure 5). Based on our previous studies, these RNAs are predicted to have the highest affinities (15, 16). As mentioned above, binding affinities were determined using a fluorescence-based assay with fluorescently labeled derivatives of the compounds, or ABI-Fl and ABII-Fl (4).
Figure 6.
Secondary structures of RNAs selected to bind ABI and ABII that have the highest Sum Z-scores as predicted by RNAstructure (20) and their corresponding dissociation constants. (A) The internal loops selected to bind ABI that have large Sum Z-scores (red circles, Figure 5A). The values below the loop identifier are the Kd’s for the RNA-ligand complex (µM). (B) The internal loops selected to bind ABII that have large Sum Z-scores (green circles, Figure 5B). The values below the loop identifier are the Kd’s for the RNA-ligand complex (µM). (C) Representative binding curves that show selected interactions are specific. Cognate pairs with large Sum Z-scores (ABII and ABII-IL 1) are the highest affinity. Non-cognate pairs with small Sum Z-scores (ABI and ABII-IL 1) interact weakly (no saturation is observed). Importantly, the ligands bind weakly to the entire RNA library, 1, or the hairpin cassette into which it was inserted, 7 (Figure 2). (No saturable binding is observed when up to 3 mM RNA is added; the ligand concentration is 50 nM.) Selectivity is achieved in our 2DCS selection by using competitor oligonucleotides (2–6, Figure 2) that mimic regions constant to all library members in large excess over 1 and the number of moles of ligand delivered to the surface.
The predicted secondary structures of the RNA loops (20) with the highest Sum Z-scores selected in our 2DCS experiment (i.e., in the sequencing data) and their corresponding Kd’s (µM) are shown in Figure 6. The highest affinity interactions were obtained with ABI, with Kd’s ranging from about 150–400 µM (Figure 6A). Interestingly, ABI gave the highest signal on the array in the 2DCS experiment (Figure 5). The ligand with second highest signal on the array (ABII) selected RNAs with a slightly wider range of Kd’s, from about 200–850 µM (Figure 6B). Both ligands bind weakly to 1 (the entire library) and the hairpin cassette in which the randomized region was inserted (7, Figure 1) (Kd’s >> 1 mM) (Figure 6C), supporting the highly stringent nature of our selection and that selected interactions to the randomized regions in 1.
The StARTS method can also predict the selectivity of an RNA (15, 16). RNAs with large Sum Z-scores bind tightly whereas RNAs with small Sum Z-scores bind weakly (15, 16). Thus, an RNA with a large Sum Z-score for ABI and a small Sum Z-score for ABII should bind ABI tightly and ABII weakly (Figure 5). ABI-IL 2 and ABII-IL 2 have Sum Z-scores in the ~25th percentile (small Sum Z-score) for their non-cognate ligand and in the top 1% for the ligand for which it was selected (large Sum Z-score) (Figure 5). Therefore, we determined the affinities of ABI-IL 2 for ABII and ABII-IL 2 for ABI (non-cognate pairs). In each case, no saturable binding was observed up to 1.5 mM RNA concentration. Thus, although the identified RNA-ligand partners have mid-micromolar Kd’s, the RNAs are specific for the ligands they were selected to bind, and selectivity can be predicted via StARTS as has been described previously (15).
Comparison to Previous Studies
Previously, the preferred RNA motifs for aminoglycosides and bis-benzimidazole small molecules were determined via 2DCS (4, 5, 15–17, 19). In general, internal loops bind to aminoglycosides with low nanomolar Kd’s while the more drug-like bis-benzimidazoles bind to RNA internal loops with high nanomolar to low micromolar Kd’s. In the present study, the 2-aminobenzimdazoles bind to single RNA motifs with mid-micromolar Kd’s, however, these affinities are similar to that of an extensively optimized benzimidazole that binds to the HCV IRES (21). Importantly, the RNAs that we selected to bind ABI and ABII are specific despite their modest affinities. The affinities and the selectivities of these RNA-binding modules can be improved using a modular assembly approach. That is, a multivalent ligand can be designed that binds multiple targetable motifs simultaneously, as has been demonstrated with repeating transcripts (22–26).
Knowing the RNA secondary structural elements that bind a small molecule is both important and useful. This information can be used to: (i) identify potential off-targets prior to in vivo testing; and (ii) design more selective compounds using a medicinal chemistry approach.
EXPERIMENTAL
General
N-methyl-2-pyrrolidinone (NMP) was purchased from Fisher. Ultra dry dimethylformamide (DMF) was purchase from Acros while ACS grade DMF was from EMD. Dichloromethane (DCM) was purchased from J.T. Baker. Piperidine and bromoacetic acid were purchased from Sigma-Aldrich. HPLC grade methanol and acetonitrile were purchased from Honeywell. PyBOP was purchased from Nova-Biochem. All chemicals were used without further purification. Water was obtained from a Barnstead NANOpure Diamond Water Purification System operating at 18.2 MΩ-cm.
Instrumentation
Preparative and analytical HPLC separations were performed on a Waters 1525 binary pump system with a Waters 2487 dual wavelength absorbance detector set to 218 and 254 nm. For preparative HPLC runs, a Waters Symmetry Prep©, C18, 7 µm, 19×150 mm column and a flow rate of 10 mL/min were used. For analytical HPLC separations, a Waters Symmetry, C18, 5µm, 4.6×150mm column and a flow rate of 1 mL/min were used. The solvents were water (solvent A) and methanol (solvent B) containing 0.1% trifluoroacetic acid (TFA) (v/v). All HPLC separations were conducted at room temperature.
Mass spectra were recorded on a LCQ Advantage IonTrap LC/MS equipped with a Surveyor HPLC system. NMR spectra were recorded on a Varian NMR operating at 500, 400, or 300 MHz on proton. Solid phase synthesis was performed on an APEX 396 synthesizer unless otherwise noted.
2-Aminobenzimidazole Library Synthesis
The library with the 2-(arylamino)benzimidazole scaffold (Figure 1) was synthesized on the solid-phase as previously described (7, 8). The building blocks utilized are shown in Figure 1B and 1C. Solid-phase synthesis of the benzimidazole library was carried out in a J-Kem heating block installed on a J-Kem shaker using 4 mL glass vials. Initially, an azidopropylamine-modified resin was synthesized in a common reaction flask, and then it was split into individual vials for further modifications.
Vials were charged with the azide-modified resin (15 mg, 10 µmol) and installed in a heating block. Solutions of corresponding amines R1NH2 (0.35 mL, 1M in NMP) were loaded into each vial and the displacement was carried out at 50°C for 20 h (shaking speed = 75 rpm). The liquids were evacuated, and the beads were washed with DMF (2×1 mL) at 28°C for 20 min with shaking at 150 rpm (2×1 mL). The beads were washed ethyl acetate (1.5 mL). Next, a solution of 3,4-diaminobenzoic acid (0.8 g, 5.3 mmol), N-hydroxybenztriazole (0.85 mg, 6.3 mmol) and N,N'-diisopropylcarbodiimide (DIC; 1.9 mL, 12 mmol) in NMP (23 mL) was distributed into each vial (0.35 mL). The reaction was carried out at 37°C for 36 h (shaking speed = 75 rpm). The liquids were evacuated, and the beads were washed with DMF (2×1 mL; 28°C for 20 min with shaking at 150 rpm) and then with ethyl acetate (1.5 mL). Into each vial, NMP (0.35 mL), the corresponding isothiocyanate R2NCS (8 eq) and N,N-diisopropylethylamine (DIPEA, 20 µL) were added. The reaction was carried out at 37°C for 20 h (shaking speed = 75 rpm). The liquids were evacuated, and the beads were washed with DMF (2×1 mL; 28°C for 20 min with shaking at 150 rpm) and then with ethyl acetate (1.5 mL). Then into each vial, DIC (0.5 mL, 0.5M in NMP) was loaded, and the reaction was carried out at 37°C for 20 h (shaking speed = 75 rpm). The liquids were evacuated, the beads were washed with DMF (2×1 mL; 28°C for 20 min with shaking at 150 rpm) and with ethyl acetate (1.5 mL). Finally, the beads were washed with 20% piperidine in DMF (1 mL, 1 h), DMF, and ethyl acetate. The product was cleaved from the resin with a TFA/DCM/water mixture (60:40:2) at 28°C for 5 h (shaking speed = 75 rpm). The liquids were saved, and the beads were washed with methanol (1 mL). The combined liquids were evaporated under stream of air. The residues were dissolved in methanol (1 mL), and analyzed by MS-ESI. The samples were dried, and then dissolved in 1.2 mL of a methanol- water-glycerol (1:1:1, v/v/v) mixture, providing approximately 5.5 mM concentration. Out of 91 reactions, 79 compounds were successfully synthesized.
Re-synthesis of 2-Aminobenzimidazole Hit Compounds
Preparation of the corresponding methyl esters: To a stirred solution of methyl 3,4-diaminobenzoate (0.5 g, 3 mmol) in DCM (20 mL), the corresponding phenyl isothiocyanate (3.6 mmol) was slowly added. The reaction was stirred at room temperature for 36 h (monitored by TLC using ethyl acetate). Then, DIC (1.4 mL) was added, and the reaction was stirred at room temperature for additional 36 h (monitored by TLC using ethyl acetate). Once reactions were complete, the mixtures were diluted with a DCM-hexane mixture (1:3, 20 mL), and precipitates of the clean products were filtered.
Saponification of ester: The corresponding methyl esters (3 mmol) were dissolved in 10% (w/v) potassium hydroxide (KOH) in methanol. The reactions were refluxed for 2 h. The solvent was then evaporated to dryness, and 10 mL of water were added. The product was then acidified with 2 M HCl to pH 2, forming a white precipitate. The product was filtered, washed with ether (2×10 mL) and dried. The residues were dissolved in 60% aq. methanol, and the products were purified by HPLC using a gradient of 5% to 100% B in A over 40 min.
Synthesis of ABI-Fl and ABII-Fl
N-(2-propynyl) 5-fluoresceincarboxamide (Fl) was synthesized according to a previously published method (15). A 100 nmol sample of Fl in methanol was added to a solution containing 300 nmol of the corresponding 2-aminobenzimidazole, 1 µmol CuSO4, 5 µmol freshly prepared solution of ascorbic acid in water, and 60 nmoles tris(benzyltriazolylmethyl) amine (TBTA; dissolved in DMSO). The final volume was brought to 1 mL with methanol. The reaction mixture was added to a microwave reaction vessel, which as placed in an Emrys™ microwave system (Biotage). The reaction was maintained at 110°C for 4 h with stirring. The product was purified by preparative HPLC, and characterized by ESI-MS and analytical HPLC as described above. ABI-Fl: HR-MS calculated mass: 971.3583 (M+H); observed mass: 971.3591; tR = 29 min; 30% yield as determined by absorbance at 496 nm in 1X PBS, pH 7.4 using an extinction coefficient of 45,000 M−1cm−1 (27). ABII-Fl: HR-MS calculated mass: 1110.3691 (M+H); observed mass: 1110.3688; tR = 38 min; 25% yield as determined by absorbance at 496 nm in 1X PBS, pH 7.4 using an extinction coefficient of 45,000 M−1cm−1 (27).
Construction of Small Molecule Microarrays
Alkyne-functionalized microarrays were prepared as previously described (5, 28). Ligands were spotted onto alkyne-functionalized slides in 1X Spotting Solution (10 mM Tris-HCl (pH 8.5), 100 µM ascorbic acid, 100 µM TBTA (dissolved in 4:1 2-butanol/dimethyl sulfoxide), 1 mM CuSO4 and 10% glycerol). The slides were placed in a humidity chamber for 3 h and then washed with NANOpure water. Arrays were allowed to dry on the bench top.
Initial Small Molecule Library Screen
Internally 32P-labeled 1 (0.4 nmol) and Torula yeast bulk RNA (1.15 µmol; 3-fold excess of ligand spotted) were folded separately in 1X Hybridization Buffer (HB1; 20 mM (4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (Hepes), pH 7.5, 150 mM NaCl, and 5 mM KCl) at 60°C for 5 min and allowed to cool slowly to room temperature. MgCl2 was then added to a final concentration of 1 mM. The folded RNAs were mixed together and then carefully pipetted onto a microarray. (Microarrays are pre-equilibrated with HB1 supplemented with 1 mM MgCl2 and 40 µg/mL bovine serum albumin (BSA) (HB2) prior to hybridization). The solution was evenly spread across the slide surface with a piece of parafilm, and the slides were hybridized at room temperature for 45 min. The hybridization solution was removed, and the slides were washed by submerging them in HB2 for 5 min and then in NANOpure water three times for 5 min each. The arrays were exposed to a phosphorimager screen and imaged using a BioRad FX phosphorimager.
Secondary Microarray Screening of Leads Ligands
The slides were hybridized as described above with 80 pmol of internally 32P-labeled 1 and 117 nmol of yeast bulk RNA (3-fold excess of ligand spotted).
RNA Selection via 2DCS
Serial dilutions of ABI – ABV were delivered to the slide surface and immobilized as described above. The ligand loadings were 1.4, 0.7, 0.4, and 0.2 nmoles. The arrays were then hybridized as described above with 20 pmol of internally 32P- labeled 1 and 79 nmol of each competitor oligonucleotide 2 - 6 (3-fold excess of the moles of ligand spotted).
Cloning and Sequencing
The RT-PCR products were cloned into pUC-19 via double digestion with EcoR1 and BamH1 restriction enzymes. Plasmids were sequenced at Functional Biosciences, Inc.
Binding Affinity Measurements
Dissociation constants for the binding of RNAs to ABI-Fl and ABII-Fl were determined using an in solution, fluorescence-based assay. A selected RNA was folded in 20 mM HEPES, 150 mM NaCl, 5 mM KCl and 40 µg/mL BSA at 60°C for 5 min and allowed to slowly cool to room temperature. MgCl2 was then added to a final concentration of 1 mM (affording 1X HB2). The RNA solution was then titrated into a well of a 96-well plate containing 50 nM of the corresponding ligand in 50 µL of HB2. Fluorescence intensity was measured on a Bio-Tek FLX-800 plate reader. The change in fluorescence intensity as a function of RNA concentration was fit to the following equation (5):
where I and I0 are the observed fluorescence intensity in the presence and absence of RNA respectively, ΔE is the difference between the fluorescence intensity in the absence of RNA and in the presence of infinite RNA concentration, [FL]0 and [RNA]0 are the concentrations of ABI-Fl or ABII-Fl and RNA, respectively, and Kt is the dissociation constant. All measurements were performed in duplicate.
Supplementary Material
Footnotes
Supporting Information
Synthetic procedures, small molecule screening, and computational analysis. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Gallego J, Varani G. Targeting RNA with small molecule drugs: therapeutic promise and chemical challenges. Acc. Chem. Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 2.Magnet S, Blanchard JS. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 2005;105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 3.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 4.Childs Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop ligand interactions. ACS Chem. Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 5.Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. Two dimensional combinatorial screening identifies specific aminoglycoside RNA internal loop partners. J. Am. Chem. Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 6.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman DE, Thornton CA, Disney MD. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif ligand database & chemical similarity searching. J. Am. Chem. Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith JM, Gard J, Cummings W, Kanizsai A, Krchnak V. Necklace coded polymer supported combinatorial synthesis of 2 arylaminobenzimidazoles. J. Comb. Chem. 1999;1:368–370. [Google Scholar]
- 8.Carpenter RD, DeBerdt PB, Lam KS, Kurth MJ. Carbodiimide-based benzimidazole library method. J. Comb. Chem. 2006;8:907–914. doi: 10.1021/cc060106b. [DOI] [PubMed] [Google Scholar]
- 9.Labuda LP, Pushechnikov A, Disney MD. Small molecule microarrays of RNA focused peptoids help identify inhibitors of a pathogenic group I intron. ACS Chem. Biol. 2009;4:299–307. doi: 10.1021/cb800313m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vegas AJ, Fuller JH, Koehler AN. Small molecule microarrays as tools in ligand discovery. Chem. Soc. Rev. 2008;37:1385–1394. doi: 10.1039/b703568n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffner JL, Clemons PA, Koehler AN. A pipeline for ligand discovery using small molecule microarrays. Curr. Opin. Chem. Biol. 2007;11:74–82. doi: 10.1016/j.cbpa.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 12.Hergenrother PJ, Depew KM, Schreiber SL. Small molecule microarrays: Covalent attachment and screening of alcohol containing small molecules on glass slides. J. Am. Chem. Soc. 2000;122:7849–7850. [Google Scholar]
- 13.MacBeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein ligand interactions en masse. J. Am. Chem. Soc. 1999;121:7967–7968. [Google Scholar]
- 14.Seedhouse SJ, Labuda LP, Disney MD. The Privileged Chemical Space Predictor (PCSP): a computer program that identifies privileged chemical space from screens of modularly assembled chemical libraries. Bioorg. Med. Chem. Lett. 2010;20:1338–1343. doi: 10.1016/j.bmcl.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velagapudi SP, Seedhouse SJ, French J, Disney MD. Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. J. Am. Chem. Soc. 2011;133:10111–10118. doi: 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velagapudi SP, Seedhouse SJ, Disney MD. Structure activity relationships through sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew. Chem. Int. Ed. Engl. 2010;49:3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aminova O, Paul DJ, Childs Disney JL, Disney MD. Two dimensional combinatorial screening identifies specific 6'acylated kanamycin A and 6' acylated neamine-RNA hairpin interactions. Biochemistry. 2008;47:12670–12679. doi: 10.1021/bi8012615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran T, Disney MD. Two dimensional combinatorial screening of a bacterial rRNA A site like motif library: defining privileged asymmetric internal loops that bind aminoglycosides. Biochemistry. 2010;49:1833–1842. doi: 10.1021/bi901998m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul DJ, Seedhouse SJ, Disney MD. Two dimensional combinatorial screening and the RNA Privileged Space Predictor program efficiently identify aminoglycoside RNA hairpin loop interactions. Nucleic Acids Res. 2009;37:5894–5907. doi: 10.1093/nar/gkp594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth PP, Miyaji A, Jefferson EA, Sannes Lowery KA, Osgood SA, Propp SS, Ranken R, Massire C, Sampath R, Ecker DJ, Swayze EE, Griffey RH. SAR by MS: discovery of a new class of RNA binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J. Med. Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 22.Childs-Disney JL, Hoskins J, Rzuczek S, Thornton C, Disney MD. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem. Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pushechnikov A, Lee MM, Childs Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MM, Childs Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J. Am. Chem. Soc. 2009;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Disney MD, Lee MM, Pushechnikov A, Childs-Disney JL. The role of flexibility in the rational design of modularly assembled ligands targeting the RNAs that cause the myotonic dystrophies. Chembiochem. 2010;11:375–382. doi: 10.1002/cbic.200900716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MM, Pushechnikov A, Disney MD. Rational and modular design of potent ligands targeting the R NA that causes myotonic dystrophy 2. ACS Chem Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durroux T, Peter M, Turcatti G, Chollet A, Balestre MN, Barberis C, Seyer R. Fluorescent pseudo peptide linear vasopressin antagonists: design, synthesis, and applications. J. Med. Chem. 1999;42:1312–1319. doi: 10.1021/jm9804782. [DOI] [PubMed] [Google Scholar]
- 28.Disney MD, Barrett OJ. An aminoglycoside microarray platform for directly monitoring and studying antibiotic resistance. Biochemistry. 2007;46:11223–11230. doi: 10.1021/bi701071h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.