Abstract
The Estrogen Related Receptor alpha (ERRα) is an orphan member of the nuclear receptor superfamily of transcription factors whose activity is regulated by the expression level and/or activity of its obligate coregulators, peroxisome proliferator activated receptor gamma coactivator-1 alpha and beta (PGC-1α or PGC-1β). Under normal physiological conditions, and in responding to different environmental stimuli, the ERRα/PGC-1 complex is involved in regulating metabolic homeostasis under conditions of high energy demand in brown adipocytes, proliferating T-cells and in muscle. Interestingly, increased expression/activity of the ERRα/PGC-1 axis has also been shown to correlate with unfavorable clinical outcome in both breast and ovarian tumors. The observation that ERRα activity is manifest in all breast tumor sub-types with particularly high activity being evident in ERα-negative, HER2 positive and triple negative breast cancers has raised significant interest in targeting this receptor for the treatment of those breast cancers where therapeutic options are limited.
Background
The nuclear receptor (NR) superfamily is comprised of a group of structurally-related transcription factors that regulate diverse physiological functions. Whereas the transcriptional activity of many NRs is regulated by small molecule lipophilic ligands i.e. estrogens, glucocorticoids, or androgens, other members of this family have been classified as “orphans” for which a physiologically relevant ligand has not yet been identified. In general, however, upon binding their cognate ligands these receptors undergo a conformational change that increases their DNA binding activity. The DNA bound receptor then nucleates the assembly of a large complex of transcriptional coregulators which results ultimately in an increase or a decrease in target gene transcription.
Dysregulation of NR function and/or the pathways under their control has been causally linked to processes of pathological importance in the reproductive, immune, and cardiovascular systems as well as in a significant number of different cancers. Given this degree of involvement in a diverse range of processes, it is not surprising that 10–15% of the currently approved drugs in the United States target this family of transcription factors (1). One particularly useful target is the estrogen receptor (ER), the intracellular mediator of the actions of the female sex hormone 17β-estradiol, heightened activity of which has been associated with both the initiation and progression of breast cancer. Indeed, therapeutic targeting of this receptor, and pathways that regulate the production of 17β-estradiol and other estrogens, has proven to be extremely useful in both the treatment and prevention of ER-positive breast cancers.
Driven by the success of targeting ER in breast cancer and capitalizing on what has been learned about NR-function in general there continues to be a high level of interest in targeting other NRs in cancer. It was of significance, therefore, that an association between elevated expression of the orphan NR Estrogen-Related Receptor alpha (ERRα) and a poor clinical outcome in both breast and ovarian tumors was observed in several independent studies (2–4). ERRα shares significant sequence homology and structural similarity to ER (5). It was initially considered, therefore, that ERRα might exhibit similar activities as ER and that it would play a role in breast cancer. However, a comprehensive evaluation of the impact of ERRα activation on ERα-dependent transcriptional regulation in MCF-7 breast cancer cells revealed surprisingly few genes that were coregulated by these receptors (6). This was in agreement with other studies, using ChIP-on-chip to evaluate ERα and ERRα binding sites in MCF7 cells, which concluded that only ~18% of ERα target genes also contain an ERRα binding site(s) (7). Interestingly, Gene Ontology analysis of the genes implicated in these studies revealed that while ERα is primarily involved in the regulation of genes involved in tissue development and cell proliferation, ERRα controls the expression of genes involved in the regulation of cellular energy metabolism such as those encoding enzymes in the tricarboxylic acid cycle and oxidative phosphorylation (8). It now appears, therefore, that despite the sequence homology, ERα and ERRα regulate distinct biological processes and should be considered functionally distinct transcription factors.
A causal role for ERRα in breast cancer pathogenesis was first demonstrated in studies which showed that shRNA-mediated knockdown of ERRα expression dramatically inhibited the growth of ERα-negative MDA-MB-231 cells when propagated as xenografts in mice (6). Similar results were observed in other in vitro and in vivo models of breast cancer where ERRα inhibition was accomplished using small molecule antagonists (9–11). Together these definitive findings established a fundamental role for ERRα in tumor growth and confirmed the importance of ER-independent activities of this receptor in breast tumor biology. These findings also provided the rationale for the exploitation of ERRα as a therapeutic target in breast cancer. To further assess the clinical significance of ERRα, we developed a genomic signature designed to evaluate the activity of this receptor and used it to profile more than eight hundred breast tumors. This analysis revealed a shorter disease-free survival in patients with tumors exhibiting elevated ERRα activity (11). Importantly, this signature also revealed that the ERRα activity signature is elevated in tumors that are either HER2 positive, ER-negative or in those that contain P53 mutations. Interestingly, the ability of ERRα antagonists to inhibit proliferation in cellular models of breast cancer correlates with the intrinsic transcriptional activity of this receptor manifest in these cells. Based on these findings it has been proposed that ERRα antagonists may have utility as treatments for breast tumors in which this receptor exhibits elevated transcriptional activity. Not surprisingly, there is considerable interest of late in developing ERRα antagonists that are suitable for clinical use. Furthermore, the information that is emerging on the pathways that are upstream and downstream of this receptor have highlighted other targets the inhibition of which may afford useful therapeutic synergism with ERRα antagonists.
Coregulators function as protein ligands of ERRα
Most NRs contain a complex ligand binding pocket which upon binding small molecule hormones or lipophilic compounds undergo a conformational change that allows them to interact with transcriptional coregulators and regulate target gene transcription. However, despite having a defined “ligand binding domain (LBD)”, crystallographic analyses of ERRα have indicated that the ligand binding pocket of ERRα is filled by bulky amino acid site chains and it has been concluded that the receptor can only accommodate extremely small ligands (~3–4 carbons) (12, 13). In addition, by comparing the structure of ERRα with that of other agonist occupied NRs, it is apparent that the LBD of this receptor is able to adopt an “active” conformation and is capable of binding coactivator proteins in the absence of a bound ligand. Consequently, it has been suggested that the transcriptional activity of ERRα may not be regulated by ligand binding per se but by the expression level and/or activity of its obligate coregulators (coactivators and corepressors) (12, 14–17). Although many coactivators have been shown to interact with and regulate the activity of ERRα, two of its most robust coactivators are the peroxisome proliferative activated receptor gamma coactivator-1 alpha and beta (PGC-1α or PGC-1β) (18–20). Under normal physiological conditions the ERRα/PGC-1 complex is involved in regulating metabolic homeostasis in tissues such as brown adipocytes and muscle in which energy demand is high. PGC-1α expression and/or activity is acutely upregulated in response to cold, fasting, exercise, and hypoxia resulting in the induction of gene expression programs involved in thermogenesis, gluconeogenesis, fatty acid oxidation, mitochondrial biogenesis, and angiogenesis (14, 20–25). Most of these responses have been shown to be mediated by ERRα. PGC-1β expression on the other hand does not seem to be regulated by these acute stresses but rather it is induced by high fat diets and in response to immune challenges. In this manner, it upregulates the expression of genes required for fatty acid synthesis and host immunity (26–28). It remains to be determined if other coregulators can couple with ERRα to regulate transcription. Regardless, however, it is clear that independent of whether or not a small molecule physiological regulator of ERRα exists that this receptor has evolved to respond to “protein ligands” (coregulators) as a means to regulate its transcriptional activity.
Signaling pathways which impinge upon and regulate ERRα/PGC-1 activity in breast cancer
It is likely that some of the stimuli that regulate the ERRα and PGC-1α/β complexes in normal physiology are also involved in regulating ERRα activity in cancer. These pathways are summarized in Figure 1. For instance, the expression of PGC-1α is induced by hypoxia in skeletal muscle resulting in HIF-1-independent but ERRα-dependent expression of VEGF and increased angiogenesis (29). The same mechanism may also be operative in hypoxic regions of tumors. Supporting this hypothesis, we and others have demonstrated that activation of ERRα/PGC-1 induces the expression of VEGF in breast cancer cells in vitro and in a xenograft model of breast cancer (30, 31). Conversely, inhibiting ERRα activity using diethylstilbesterol, a non-selective ERRα antagonist, has been shown to reduce angiogenesis in breast cancer xenografts (32). When taken together these studies highlight a potential role for ERRα in tumor angiogenesis. Interestingly, ERRα and HIF-1 have been shown to physically interact and in doing so to coregulate the expression of several endogenous HIF-1 target genes. The impact of this crosstalk on the regulation of processes of pathological importance in tumors remains to be determined.
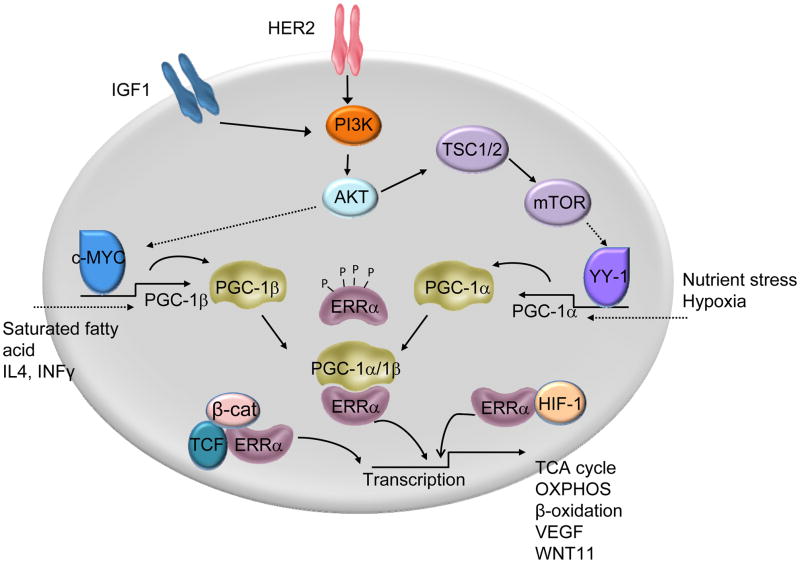
Figure 1. The ERRα/PGC-1 complex is a downstream target of multiple signaling pathways in cancer.
Several signaling pathways relevant to cancer pathogenesis have been shown to converge upon and regulate the expression and activity of PGC-1α and β, two key ERRα coactivators. It has been demonstrated recently that activation of HER2 and IGF-1R signaling pathways increase the expression of PGC-1β through induction of c-MYC. Similarly, activation of the mTOR/YY-1 pathway secondary to PI3K-Akt activation can induce the expression of PGC-1α. In addition, hypoxia and nutrient stress are also known inducers of PGC-1α whereas saturated fatty acids and cytokines have been shown to induce PGC-1β expression under physiological/pathological conditions. The resulting ERRα/PGC-1α/1β complex induces the expression of genes involved in the TCA cycle, OXPHOS and other processes involved in energy metabolism. ERRα has also been shown to interact with the β-catenin/TCF complex and with HIF-1 and reciprocally modulate each others’ transcriptional activities to effect cell migration and angiogenesis. ERRα activity has also been shown to be modulated by phosphorylation downstream of the HER2 signaling.
The expression of PGC-1α, PGC-1β, ERRα and those of their target genes involved in TCA cycle, oxidative phosphorylation and glycolysis, were shown to be upregulated in breast cancer cells that have metastasized to the brain (33). Although the signaling events that leads to the upregulation of the ERRα/PGC-1 axis in these cells have not been defined it has been demonstrated that these changes in gene expression enable the adaptive metabolic events and changes in redox balance that allow the metastasized tumor cells to survive in the nutrient deprived (low glucose) environment of the brain. This result suggests that the ability of ERRα to regulate metabolic function likely contributes to its pathogenic actions in cancer.
The activity of the ERRα/PGC-1 complex is also positively regulated by oncogenic pathways that are highly relevant in breast cancer. HER2 activation, for example, has been shown to initiate a signaling cascade that leads ultimately to the phosphorylation of several serine residues located at the N-terminus of ERRα and these modifications have been shown to increase receptor transcriptional activity (34–36). Furthermore, HER2 activation also increases the expression of PGC-1β. Not surprisingly it has been shown that (a) PGC-1β expression is elevated in HER2 expressing breast cancer cells and (b) knockdown of HER2 reduces PGC-1β expression in HER2 amplified breast cancer models (37). Recently, we demonstrated that PGC-1β is a direct downstream target of c-MYC and that activation of PI3K/AKT, by either heregulin or IGF-1, resulted in increased expression of c-MYC and a subsequent increase in the expression of PGC-1β (11). Importantly, knockdown of PGC-1β reduced the growth of those breast cancer cells in which it was expressed (11, 37). The induction of PGC-1β by c-MYC is particularly significant given that this oncogene is frequently over expressed in a wide variety of tumors. More specifically, it has been shown that 15–45% of breast cancers exhibit overexpression/amplification of MYC and that this overexpression is associated with a poor clinical outcome (38, 39). This likely relates in part to MYC-dependent induction of PGC-1β expression and the subsequent increased expression of those ERRα target genes involved in oxidative phosphorylation and TCA cycle. Indeed, we have proposed that ERRα is a primary integrator of the MYC-induced anapleurotic processes required for biomass synthesis in cancer cells (Figure 2). Whereas most of the pyruvate produced during glycolysis is converted to lactate, a significant amount is diverted into the TCA cycle to form citrate. In most normal cells the TCA cycle operates as a closed system. However in rapidly proliferating cells, including cancer cells, citrate is shuttled out of the mitochondria where it is converted to acetyl CoA and utilized for fatty acid biosynthesis (40, 41). Unless replenished, citrate removal interrupts flux through the TCA cycle resulting in decreased synthesis of the biosynthetic intermediates required for protein and nucleotide synthesis. This situation is mitigated in most cancer cells by increased glutaminolysis; uptake of glutamine, and its subsequent oxidative deamination to form alpha-ketoglutarate. Alpha-ketoglutarate then enters the TCA cycle and replenishes citrate and the other constituent organic acids (42–44). ERRα/PGC-1 regulates the transcription of all of the enzymes that constitute the TCA cycle and supports the biochemical reactions required for biomass synthesis. Given that most MYC transformed cells are dependent on glutaminolysis for growth and survival it is tempting to speculate that this protein may coordinate the refueling of biosynthetic intermediates in part through up regulation of the ERRα/PGC-1 axis. By extension, it is expected that targeting the ERRα/PGC-1 axis would have benefit in treating tumors with MYC amplification/overexpression. Significantly, we have determined that a gene signature enriched for TCA cycle and OXPHOS genes predicts poor survival in multiple cohorts of breast cancer patients. In addition, a potential role for ERRα in glycolysis has also been implicated by studies using ChIP-Chip analyses (45). It was further proposed that ERRα/PGC-1 is an essential component of the processes that enable cells to upregulate glycolysis under conditions where mitochondrial OXPHOS is unable to meet bioenergetics demands.
Figure 2. ERRα/PGC-1 regulates key metabolic processes in cancer.
Activation of ERRα/PGC-1 complex induces the expression of all the enzymes involved in the TCA cycle (highlighted in red), many genes in OXPHOS (not shown), and other processes that regulate energy metabolism i.e. glutaminolysis and glycolysis. Although the TCA cycle and OXPHOS are coupled to production of ATP, a role for the TCA cycle in biomass production in cancer cells has been demonstrated recently. Therefore, in addition to energy production the activity of the ERRα/PGC-1 complex may also be required for the replenishment of the TCA intermediates that are used in biomass production in rapidly proliferating cancer cells. A potential role for ERRα in glycolysis was also revealed in a ChIP-chip analysis performed in in liver cells where all the genes encoding enzymes involved in glycolysis were found to have ERRα binding sites. It was proposed that ERRα is required for the switch from OXPHOS to glycolysis that occurs under conditions where OXPHOS is unable to meet the bioenergetics demands of the cell.
Whereas PGC-1α is likely also to impact tumor cell metabolism through its actions on ERRα, the expression of this coactivator is not directly regulated by MYC. However, its expression has been shown to be upregulated by PI3K and mTOR dependent signaling activities. Specifically, mTOR, through its actions on YY1, induces PGC-1α expression in skeletal muscle cells (46). Whether this mechanism operates in the same way in cancer cells remains to be determined.
Recently, using a gene signature derived from ERRα/PGC-1α/β regulated genes we demonstrated that ERRα/PGC-1α/1β activity is elevated in HER2 positive tumors and those tumors in which elevated MYC activity is apparent (11). The importance of this observation was highlighted by studies which found that (a) the latency of MMTV-Neu (HER2) breast tumors was significantly increased when propagated in an ERRα−/− background and (b) overexpression of PGC-1α increases tumor growth in HER2/Neu-driven xenograft models of breast cancer (31, 47). These results confirm the importance of this signaling axis in breast cancer pathogenesis and suggest that targeting the ERRα/PGC-1 axis should provide additional therapeutic options in these types of cancers.
Similar to other nuclear receptors, ERRα was also found to have significant crosstalk with the WNT signaling pathway. Specifically, it was demonstrated that ERRα physically interacts with β-catenin and TCF/LEF (T cell-specific transcription factor/lymphoid enhancer-binding factor) in breast cancer cells(48). In this manner (a) ERRα influences the activity of TCF-LEF on several endogenous target genes and (b) ERRα transcriptional activity is enhanced by activation of β-catenin (β-cat). Significantly, WNT11, one of the target genes co-regulated by ERRα and β-cat was shown to be involved in breast, prostate, and colon cancer cell migration and that this activity can be inhibited by knockdown of ERRα, β-cat, or WNT11, activities that are completely reversed by adding exogenous recombinant WNT11. Similar crosstalk between ERRα and WNT signaling has also been demonstrated to be involved in osteoblastogenesis (49).
Finally, post-translational modifications of ERRα by phosphorylation (PKCs), sumoylation, and acetylation, at the N-terminus and within the DNA binding domain have been shown to regulate the transcriptional activity of this receptor (34, 36, 50–52), although the biological relevance of these modifications and their regulation has not been established.
Clinical-Translational Advances
Given the ascribed functions of ERRα and PGC-1 in energy metabolism, it is not surprising that the initial efforts to develop modulators of this receptor were focused on the identification of molecules with agonist activity for use in the treatment of metabolic diseases such as diabetes and obesity. However, compounds with significant ERRα agonist activity have yet to be identified (13). Interestingly, several antagonists/inverse agonists have been described that have been shown to inhibit ERRα activity both in vitro and in vivo. Whereas none of these compounds have or are likely to advance to the clinic they have been extremely useful tools to probe ERRα biology. The first selective ERRα antagonist described in the literature, XCT790, ((2E)-3-(4-{[2,4-bis(trifluoromethyl)benzyl]oxy})-3-methoxyphenyl)-2-cyano-N-[5-(trifluoromethyl)-1,3,4-thiadiazol-2-yl]acrylamide) binds ERRα and blocks its interaction with coactivator proteins, down regulates ERRα target gene expression, inhibits breast cancer cell proliferation in vitro, and delays the growth of MCF7 derived xenografts(53–55). In addition to functioning as a classical “antagonist” it has also been observed that XCT790 binding results in a dramatic 26S dependent downregulation of ERRα expression (56). A second ERRα selective antagonist, compound A (N-[(2Z)-3-(4,5-dihydro-1,3-thiazol-2-yl)-1,3-thiazolidin-2-ylidene]-5Hdibenzo[a,d][7]annulen-5-amine) was shown to inhibit the growth of ER+ and ER− breast cancer cells both in vitro and in xenograft models (9, 57, 58). Finally, developed for the treatment of metabolic disease, a diaryl ether based thiazolidenedione (compound 29 in (59)) has also been described and shown to function as a selective and potent ERRα antagonist. It exhibits favorable pharmacokinetic properties and has shown efficacy in animal models of obesity and insulin resistance, although its activity in breast cancer has not yet been tested.
Despite the promising animal studies of some of these ERRα antagonists, no clinical testing of the above-mentioned compounds have been reported. However, driven by recent data which highlights a causal role for ERRα in cancer pathogenesis and the realization that this receptor is druggable, there is of late considerable interest in its pharmaceutical exploitation. Identifying the patients who are most likely to benefit from this type of intervention is now a primary focus of ongoing research in this area.
Acknowledgments
Grant Support: National Institutes of Health (R01DK074652)
Footnotes
Disclosure of Potential Conflicts of interest: no potential conflicts of interest were disclosed
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Fujimoto J, Alam SM, Jahan I, Sato E, Sakaguchi H, Tamaya T. Clinical implication of estrogen-related receptor (ERR) expression in ovarian cancers. J Steroid Biochem Mol Biol. 2007;104:301–4. doi: 10.1016/j.jsbmb.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki T, Miki Y, Moriya T, Shimada N, Ishida T, Hirakawa H, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64:4670–6. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- 4.Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–8. [PubMed] [Google Scholar]
- 5.Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–4. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 6.Stein RA, Chang CY, Kazmin DA, Way J, Schroeder T, Wergin M, et al. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 2008;68:8805–12. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deblois G, Hall JA, Perry MC, Laganiere J, Ghahremani M, Park M, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–57. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 8.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–96. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 9.Chisamore MJ, Wilkinson HA, Flores O, Chen JD. Estrogen-related receptor-{alpha} antagonist inhibits both estrogen receptor-positive and estrogen receptor-negative breast tumor growth in mouse xenografts. Mol Cancer Ther. 2009;8:672–81. doi: 10.1158/1535-7163.MCT-08-1028. [DOI] [PubMed] [Google Scholar]
- 10.Bianco S, Lanvin O, Tribollet V, Macari C, North S, Vanacker JM. Modulating estrogen receptor-related receptor-alpha activity inhibits cell proliferation. J Biol Chem. 2009;284:23286–92. doi: 10.1074/jbc.M109.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang CY, Kazmin D, Jasper JS, Kunder R, Zuercher WJ, McDonnell DP. The metabolic regulator ERRalpha, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell. 2011;20:500–10. doi: 10.1016/j.ccr.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kallen J, Schlaeppi JM, Bitsch F, Filipuzzi I, Schilb A, Riou V, et al. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J Biol Chem. 2004;279:49330–7. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- 13.Hyatt SM, Lockamy EL, Stein RA, McDonnell DP, Miller AB, Orband-Miller LA, et al. On the intractability of estrogen-related receptor alpha as a target for activation by small molecules. J Med Chem. 2007;50:6722–4. doi: 10.1021/jm7012387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–35. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–8. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 16.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends in Endocrinology & Metabolism. 2008;19:269–76. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, Takahashi N, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A. 2003;100:12378–83. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J Biol Chem. 2002;277:13918–25. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta ), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–8. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 20.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 21.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–91. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinsomboon J, Ruas J, Gupta RK, Thom R, Shoag J, Rowe GC, et al. The transcriptional coactivator PGC-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proc Natl Acad Sci U S A. 2009;106:21401–6. doi: 10.1073/pnas.0909131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lustig Y, Ruas JL, Estall JL, Lo JC, Devarakonda S, Laznik D, et al. Separation of the gluconeogenic and mitochondrial functions of PGC-1{alpha} through S6 kinase. Genes Dev. 2011;25:1232–44. doi: 10.1101/gad.2054711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–8. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 25.St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, et al. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 26.Sonoda J, Laganiere J, Mehl IR, Barish GD, Chong LW, Li X, et al. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Dev. 2007;21:1909–20. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J, Yang R, Tarr PT, Wu P-H, Handschin C, Li S, et al. Hyperlipidemic Effects of Dietary Saturated Fats Mediated through PGC-1β Coactivation of SREBP. Cell. 2005;120:261–73. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 28.Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metabolism. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–12. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 30.Stein RA, Gaillard S, McDonnell DP. Estrogen-related receptor alpha induces the expression of vascular endothelial growth factor in breast cancer cells. J Steroid Biochem Mol Biol. 2009;114:106–12. doi: 10.1016/j.jsbmb.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klimcakova E, Chenard V, McGuirk S, Germain D, Avizonis D, Muller WJ, et al. PGC-1alpha promotes the growth of ErbB2/Neu-induced mammary tumors by regulating nutrient supply. Cancer Res. 2012;72:1538–46. doi: 10.1158/0008-5472.CAN-11-2967. [DOI] [PubMed] [Google Scholar]
- 32.Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci U S A. 2008;105:7821–6. doi: 10.1073/pnas.0711677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–86. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 34.Tremblay AM, Wilson BJ, Yang XJ, Giguere V. Phosphorylation-dependent sumoylation regulates estrogen-related receptor-alpha and -gamma transcriptional activity through a synergy control motif. Mol Endocrinol. 2008;22:570–84. doi: 10.1210/me.2007-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ariazi EA, Kraus RJ, Farrell ML, Jordan VC, Mertz JE. Estrogen-related receptor alpha1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol Cancer Res. 2007;5:71–85. doi: 10.1158/1541-7786.MCR-06-0227. [DOI] [PubMed] [Google Scholar]
- 36.Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17:5400–9. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eichner LJ, Perry M-C, Dufour CR, Bertos N, Park M, St-Pierre J, et al. miR-378() Mediates Metabolic Shift in Breast Cancer Cells via the PGC-1β/ERRγ Transcriptional Pathway. Cell Metabolism. 2010;12:352–61. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes Cancer. 2010;1:629–40. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. The Journal of Experimental Medicine. 2012 doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–22. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 41.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–21. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 42.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, et al. The homeobox protein Prox1 is a negative modulator of ERR{alpha}/PGC-1{alpha} bioenergetic functions. Genes Dev. 2010;24:537–42. doi: 10.1101/gad.1871610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–40. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 47.Deblois G, Chahrour G, Perry MC, Sylvain-Drolet G, Muller WJ, Giguere V. Transcriptional control of the ERBB2 amplicon by ERRalpha and PGC-1beta promotes mammary gland tumorigenesis. Cancer Res. 2010;70:10277–87. doi: 10.1158/0008-5472.CAN-10-2840. [DOI] [PubMed] [Google Scholar]
- 48.Dwyer MA, Joseph JD, Wade HE, Eaton ML, Kunder RS, Kazmin D, et al. WNT11 expression is induced by estrogen-related receptor alpha and beta-catenin and acts in an autocrine manner to increase cancer cell migration. Cancer Res. 2010;70:9298–308. doi: 10.1158/0008-5472.CAN-10-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Auld KL, Berasi SP, Liu Y, Cain M, Zhang Y, Huard C, et al. Estrogen-related receptor alpha regulates osteoblast differentiation via Wnt/beta-catenin signaling. Journal of Molecular Endocrinology. 2012;48:177–91. doi: 10.1530/JME-11-0140. [DOI] [PubMed] [Google Scholar]
- 50.Lu N, Wang W, Liu J, Wong CW. Protein kinase C epsilon affects mitochondrial function through estrogen-related receptor alpha. Cell Signal. 2011;23:1473–8. doi: 10.1016/j.cellsig.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha. Mol Endocrinol. 24:1349–58. doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barry JB, Giguère V. Epidermal Growth Factor–Induced Signaling in Breast Cancer Cells Results in Selective Target Gene Activation by Orphan Nuclear Receptor Estrogen-Related Receptor α. Cancer Res. 2005;65:6120–9. doi: 10.1158/0008-5472.CAN-05-0922. [DOI] [PubMed] [Google Scholar]
- 53.Busch BB, Stevens WC, Jr, Martin R, Ordentlich P, Zhou S, Sapp DW, et al. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha. J Med Chem. 2004;47:5593–6. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- 54.Willy PJ, Murray IR, Qian J, Busch BB, Stevens WC, Jr, Martin R, et al. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc Natl Acad Sci U S A. 2004;101:8912–7. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Wang Y, Wong C. Oestrogen-related receptor alpha inverse agonist XCT-790 arrests A549 lung cancer cell population growth by inducing mitochondrial reactive oxygen species production. Cell Proliferation. 2010;43:103–13. doi: 10.1111/j.1365-2184.2009.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lanvin O, Bianco S, Kersual N, Chalbos D, Vanacker JM. Potentiation of ICI182,780 (Fulvestrant)-induced estrogen receptor-alpha degradation by the estrogen receptor-related receptor-alpha inverse agonist XCT790. J Biol Chem. 2007;282:28328–34. doi: 10.1074/jbc.M704295200. [DOI] [PubMed] [Google Scholar]
- 57.Chisamore MJ, Cunningham ME, Flores O, Wilkinson HA, Chen JD. Characterization of a Novel Small Molecule Subtype Specific Estrogen-Related Receptor α Antagonist in MCF-7 Breast Cancer Cells. PLoS ONE. 2009;4:e5624. doi: 10.1371/journal.pone.0005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chisamore MJ, Mosley RT, Cai S-J, Birzin ET, O’Donnell G, Zuck P, et al. Identification of small molecule estrogen-related receptor α–specific antagonists and homology modeling to predict the molecular determinants as the basis for selectivity over ERRβ and ERRγ. Drug Development Research. 2008;69:203–18. [Google Scholar]
- 59.Patch RJ, Searle LL, Kim AJ, De D, Zhu X, Askari HB, et al. Identification of Diaryl Ether-Based Ligands for Estrogen-Related Receptor α as Potential Antidiabetic Agents. J Med Chem. 2011;54:788–808. doi: 10.1021/jm101063h. [DOI] [PubMed] [Google Scholar]