Abstract
There has been a recent resurgence of interest in New World monkeys within the biomedical research community, driven both by the sequencing of the common marmoset (Callithrix jacchus) genome as well as a growing demand for alternatives to Old World primates. New World monkeys offer attractive advantages over Old World species including cheaper and simpler husbandry while still maintaining a greater evolutionary proximity to humans than other animal models. Although numerous commonalities across primate species exist, there are also important genetic and reproductive differences that can and should play a critical role in selecting appropriate animal models. Common marmosets in particular have significantly reduced diversity at the major histocompatibility complex loci and are born as hematopoietic chimeras. New World primates can make ideal translational models for research, but scientists must necessarily incorporate complete understandings of their genetic and phenotypic differences from humans and other model organisms.
Keywords: Platyrrhines, Callitrichidae, Callithrix jacchus, Saimiri sp, Saguinus oedipus, Animal Models
Revisiting the New World primate model
Animal models are an important tool for biomedical research, but establishing animal models of human disease is inundated with challenges. Rodents, the most extensively used animal models, present difficulties both in accurately representing the disease under study as well as predicting the response to treatment in humans. This is particularly true for neuropsychiatric disorders [1,2] and infectious diseases [3,4]. Primate models, because of their greater genetic, behavioral, and physiological similarities to humans ameliorate some of these issues. Traditionally, Old World monkeys have been the primary non-human primate model, both for historical reasons of access as well as evolutionary proximity. The cost of maintaining non-human primate colonies is high, however, and available resources are strained under constant pressure by both the growing demands of scientists and increasingly restricted facilities and support. One potential solution to these shortages is increasing the availability of New World species. The most commonly used New World monkey species today, the common marmoset (Callithrix jacchus) and the squirrel monkey (Saimiri sp.), are smaller and easier to manage than their Old World brethren and have shorter generational times. We believe that with the recently released marmoset genome and pending release of the squirrel monkey genome, these and other New World monkeys will become more important and more commonly used in biomedical research.
Historically non-human primates, including the New World monkeys, have been used across a varied and complex field of research, encompassing infectious disease, neuroscience, drug development, reproductive biology, and behavioral sciences (Figure 1). Relative to Old World primates, the reduced body mass of New World monkeys brings particular advantages. In pharmacological research, smaller body sizes decrease the costs of compound synthesis. Their size also reduces feeding costs and lessens the square-footage required to house them, which lowers caging costs. A small body size also allows more easily for social group housing and makes the implementation of enrichment programs more manageable for researchers and facilities managers [5]. Generally, New World monkeys also have high reproductive efficiency and reach sexual maturity as early as 12 to 13 months [6], although squirrel monkeys do not sexually mature until 30 months [7]. Marmosets also have shorter lifespans, averaging about six years with a maximum of 15 years in captivity [6]. This helps enable the study of aging and age-related illness in these primate models and improves the feasibility of longitudinal studies [8]. In addition, New World monkeys do not naturally harbor herpes B virus (Herpesvirus simiae), alleviating some biosafety concerns relative to Old World species [9].
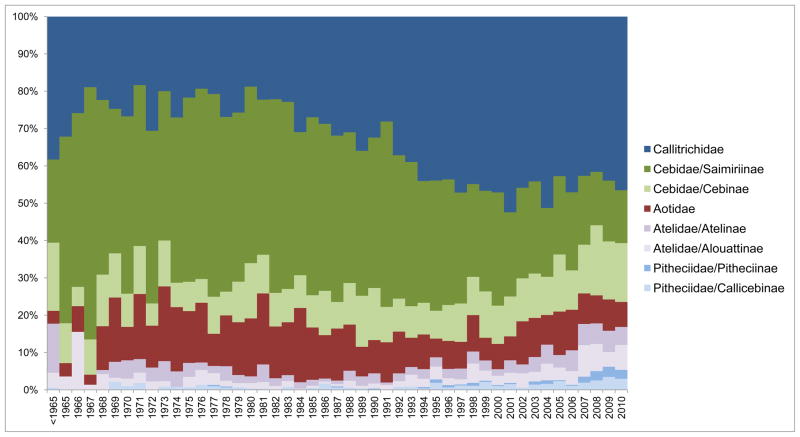
Figure 1.
Distribution timeline of New World monkey publications in the last fifty years. The percentage of total publications featuring New World monkeys is shown by year grouped by Family or Family/Subfamily following the taxonomy of Rylands and Mittermeier [62]: Callitrichidae, including marmosets and tamarins; Cebidae/Caimiriinae, including squirrel monkeys; Cebidae/Cebinae, including capuchin monkeys; Aotidae, including owl monkeys; Atelidae/Atelinae, including spider monkeys; Atelidae/Alouattinae, including howler monkeys; Pitheciidae/Pitheciinae, including sakis; and Pitheciidae/Callicebinae, including titi monkeys.
Although the suitability of New World monkeys as models will necessarily be dependent on the specifics of the study, there are many situations where the evolutionary proximity to humans holds advantages over rodents and the husbandry benefits offer advantages over Old World non-human primates. New World primates (platyrrhines) are found in the Americas and diverged from Old World primates (catarrhines) approximately 43 million years ago and saw widespread radiation 20–25 million years ago [10]. This has resulted in distinct differences across species, many of which are relevant to their use in biomedical research. One major barrier to the adoption New World monkeys as modern animal models was the lack of a reference genome, but the genome of the common marmoset (Callithrix jacchus) has been recently completed (Marmoset Genome Sequencing Consortium, Baylor College of Medicine – Human Genome Sequencing Center, personal communication), and the genome of the Bolivian squirrel monkey (Saimiri boliviensis boliviensis) is currently assembled (Vertebrate Genome Biology Group, Broad Institute, unpublished). Selecting capable animal models for translational studies must be done with an appreciation for the genetic and physiological differences that separate New and Old World primates and that separate various New World primate species from one another. This is especially true when considering the unique physiology of the callitrichids.
Hematopoietic chimerism in Callitrichidae
Callitrichids, tamarins, and marmosets are notable for their natural dizygotic twinning. Embryologists in the first half of the twentieth century observed that in early development, blastocysts of these twins fuse, giving rise to a single shared placenta [11,12]. This placental anastomosis results in the exchange of genetically distinct hematopoietic precursor cells during embryonic development [13,14]. This hematopoietic chimerism can cause challenges for genetics work in callitrichids. The most common and straightforward methods for gathering genomic DNA from blood result in effectively mixed samples, complicating attempts at genotyping and necessitating collection from other tissue sources. Even then, cryptic blood or lymphocytic infiltrates may complicate particularly sensitive molecular studies [15]. But although this chimerism may complicate genetic studies, it also allows for unique opportunities.
Historically, naturally occurring bone marrow-chimeric species have proven useful models for examining allotolerance, the regulation of reactivity toward the host environment [16–18]. In humans, placental anastomosis is extremely rare [19] and chimerism is seldom demonstrated [20–23], but callitrichids provide a natural primate model for studying transplantation biology. A study of skin grafts in tamarins (Saguinus spp.) demonstrated that intraspecies allografts were tolerated for extended periods of time (mean survival ranging between 38 and 70 days), and interspecies xenografts could also be well-tolerated (mean survival times between 14 and 23 days) [24]. As graft rejection is largely mediated by a major histocompatibility complex (MHC) class I response [25], this immunotolerance in callitrichids may reflect important genetic constraints. Indeed, it has been suggested that the bone marrow chimerism may be responsible for selective pressures on identity at the MHC class I loci in twins [26] and in turn suggests an important role for callitrichids in understanding MHC gene evolution.
MHC diversity and evolution in Callitrichidae
Today the forces that both generate and maintain the high MHC allelic diversity are of considerable debate [27,28]. Despite very common MHC class I polymorphisms across vertebrates, some callitrichids present limited MHC diversity. For example, Saguinus oedipus is restricted to 11 allelic products at only three MHC class I loci, [29]. Restriction fragment length polymorphism analysis has recently showed limited diversity of common marmoset MHC class I alleles [30]. In addition, the expressed MHC class I molecules of marmosets have been found to be 94% similar at the nucleotide level to those of Saguinus oedipus [31]. In comparison, over 5,518 distinct alleles at six class I loci have been identified in humans [32]. The limited MHC class I polymorphism of Callitrichidae species may contribute to their high susceptibility to a number of pathogens, leading researchers to explore their potential use as models of human disease. Up to 60% of captive S. oedipus are reported to spontaneously develop ulcerative colitis, of which 15% develop adenocarcinoma of the colon [33]. Additionally, S. oedipus develops fatal lymphoproliferative syndromes after infection with Epstein-Barr virus and Herpesvirus saimiri, as well as being highly susceptible to retrovirus-induced sarcomas and measle virus challenges [34,35]. This unusual immunogenetic profile, in addition to providing increased understanding of human disease, can also be studied to clarify both the mechanisms underlying the maintenance of bone marrow-chimerism and the evolution of restricted MHC class I gene products.
There are at least five potential explanations for the origin and maintenance of the unusually restricted MHC class I polymorphism found in callitrichids. A recent founder effect was first considered, but findings that demonstrate relatively polymorphic MHC class II loci have made this explanation unlikely [36]. Second, the geographic isolation of callitrichid species was thought to have contributed to low MHC class I diversity; however, other primates with restricted habitats such as the owl monkey (Aotus spp.) maintain polymorphic MHC class I loci, thus this explanation alone is insufficient [37]. Third, it may be possible that pressures in the natural habitat are driving down MHC class I loci diversity through event- or pathogen-driven selection. It is impossible to exclude event-driven selection without supporting evidence to the contrary; however, selective pressure for MHC class I identity between virally infected target cells and thymus-educated MHC-restricted cytotoxic T lymphocytes in these primates might play a role in the evolution of low levels of MHC class I polymorphism [29]. A fourth possibility, that the limited number of wild-caught animals in biomedical facilities may have created a founder effect in captivity, has been explored as an explanation for low MHC class I loci polymorphism. Although it is impossible to rule out this phenomenon completely, it has been suggested that the MHC class I profile of the over forty thousand S. oedipus exported from Columbia mirror that of S. oedipus genotyped directly from wild-caught primates [38]. The high frequency of twinning and bone marrow-chimerism in callitrichids may provide a fifth potential explanation [26]. Relatively non-polymorphic MHC class I genes might be used as a mechanism by which MHC class I alleles can still be served with a minimum stimulation of alloreactivity [31]. If true, this finding would have implications on current research into the mediating role of MHC in reproductive success and spontaneous abortions. A definitive explanation for the restricted MHC class I polymorphism in callitrichidshas not yet been reached.
Expressed S. oedipus MHC class I genes are more closely related to the human non-classical HLA-G gene than they are to the genes of the classical HLA-A, -B, or -C loci, suggesting that these New World monkey MHC class I genes may have evolved from ancestral human non-classical homologues [39]. Because the species show normal CD8+ cytotoxic T lymphocyte responses, it suggests that their HLA-G-related MHC class I genes constitute the classical MHC class I genes of this primate [39]. If accepted, this finding would indicate that classical and non-classical MHC genes do not constitute mutually exclusive groups over evolutionary time.
Squirrel monkey provenance and genetics
Squirrel monkeys (Saimiri spp.) are another commonly used New World primate in biomedical research. They share many of the same advantages to researchers common to other New World monkeys, including small body mass, preference for grouped housing, and a relatively short time required to reach sexual maturity (2.5 to 3 years of age) [7]. Additionally, they have been reported to be more tolerant to some environmental stressors commonly associated with the modern research facility, particularly noise, than marmosets [40]. First formally elaborated upon as a research animal in 1968 [41], these monkeys have historically served as models for studying parasitic disease [42] as well as a host of other clinical and behavioral science research [43,44]. This utility and common usage was recognized in the decision to sequence the squirrel monkey genome and to introduce post-genomic tools to squirrel monkey research.
Unlike the callitrichids where genetic confounds arising from hematopoietic chimerism are found in individual animals, the main difficulty with squirrel monkey studies is cryptic, but significant, differences between populations. Squirrel monkeys in biomedical research tend to be an amalgam of at least three populations, Saimiri sciureus sciureus, Saimiri boliviensis boliviensis, and Saimiri boliviensis peruviensis, each of which is genetically distinct [45,46] and shows phenotypic variability [47]. Historically, the biomedical community using these resources has been ignorant of or ambivalent about these differences leading to hybridization within research colonies and calls for greater a priori characterization [48–50]. A number of PCR-based assays were developed for distinguishing among the populations [45,48,49,51] but generally these have only been implemented by savvy researchers already focused on squirrel monkey genetics and not among the more general community of behavioral and infectious disease researchers for whom the squirrel monkey is a valuable model.
More than two decades ago, Abee [50] reviewed some of the species differences in phenotype including those in physiology [52], behavior [53], and development [54]. Since that time additional differences have emerged, notably in susceptibility to the malarial vector Plasmodium falciparum [55], and these trait differences have been elaborated upon [47]. Given the likelihood that these phenotypic differences are driven in part, and likely large part given the similarities in husbandry between the squirrel monkey populations in captive colonies, by genetic differences, it is important that these be considered more explicitly in study design. These studies have, to date, been fairly limited and rarely extend into biomedical research. Even the most basic understanding of relationships and differences within the Saimiri genera has been elusive, and although progress has been made towards it [56], many questions remain [57].
Even when the phenotype under study is not divergent between the populations, cryptic substructure can affect genetic studies. Differences in composition between “control” and “experimental” groups can lead to spurious genetic associations in much the same way as unaccounted for ethnic differences can impede human studies. Further, many of the squirrel monkey species can hybridize both in captivity and the wild [48]. Because of this, understanding the provenance and genetic heritage of these animals prior to use in study is imperative. Unfortunately it is all too common to see species or subspecies of squirrel monkeys unidentified in the literature or worse, misidentified.
Concluding remarks
Although there are many advantages to using New World primates, the genetic differences that separate them from Old World non-human primates need to be carefully considered when selecting the appropriate animal models across a host of research areas. For instance, certain research on infectious diseases, notably HIV, can only be studied in specific Old World monkey species. The publication of the Callithrix jacchus genome, combined with increasing demands from scientists, will mark a resurgence of New World monkeys in biomedical research that will only be furthered by the increasingly restrictive use of Old World primates [58] and the development of transgenic New World monkeys [59–61], perhaps aided by the species’ natural tolerance for chimerism. This resurgence must be met by improved characterization and coordination of New World primate polymorphism data, providing researchers with a more definitive guide on whether the use of alternative primate species is both appropriate and translational for their research program. Another feature that should be considered is the unusually restricted MHC class I profile in callitrichids, which makes New World primates a likely candidate model for shedding light on the controversy surrounding the potential role of MHC-mediated behavior in both the maintenance of high MHC polymorphism and reproductive success.
New World monkey models have yet wide-spread enough to achieve the same level of collaboration and cross-fertilization that Old World primate research enjoys (Figure 2). However, the continued and renewed interest in these species necessitates a better and more complete understanding of their genetic structure and its implications for research. The technologies now available allow us to address these concerns, producing genetically-characterized, more translationally relevant New World monkey models of human disease.
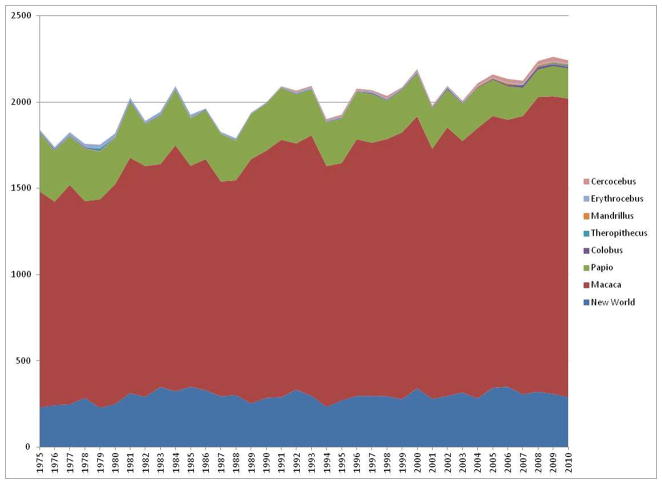
Figure 2.
New World monkey publications as a percentage of all non-human primate publications. PubMed was used as the source resource capture in order to standardize the publication calculation, which includes primarily literature relating to biomedical research. We anticipate a future surge in the use of New World primates as models for disease research in the coming years.
Text Box 1. A brief history of MHC.
Research leading to the discovery of MHC genes dates to the early 20th century, when Little and Tyzzer discovered that tumors could be successfully transplanted among some strains of mice, but not in others [63]. Research on tissue rejection continued and, eleven years later, it was discovered that tissue transplants were not rejected if the donor and the recipient were identical twins [64]. This led many to believe that histocompatibility might be genetically controlled, and it was later suggested that immune responses may be the cause of tissue transplant rejection [65]. This was subsequently confirmed by the demonstration of an immune response attacking foreign tissue grafts in rabbits [66]. George Snell published more definitive evidence showing a genetic role in tissue graft compatibility beginning in the late 1940s. Snell’s laboratory bred congenic strains of mice which differed at a single genetic region that controlled the rejection of foreign tissue. Snell coined these genes “histocompatibility (H) genes” [67], from which the contemporary name “major histocompatibility complex” is derived, modified due to later discoveries of the many genes present that are all found on the same chromosome. A better understanding of the immunological functions of the MHC gene products began about twenty years following Snell’s naming of the H genes when Zinkernagel and Doherty demonstrated that T-cell responses were not only restricted to specific antigens, but also to specific MHC molecules [68]. Later, the understanding of the role of MHC genes in immune regulation was extended to antibody-mediated responses [69–71]. It was at this time that the codominant expression of MHC class I and class II gene products in an individual was first thought to confer a survival advantage to heterozygotes by increasing the variety of pathogens an immune system could interact with.
Acknowledgments
We appreciate helpful conversations with Carolyn Sweeney, Lisa Ogawa, and Greg Miller. This work was supported by NIH grants AA019688 (to EJV) and OD011103.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Markou A, et al. Removing obstacles in neuroscience drug discovery: the future path for animal models. Neuropsychopharmacology. 2009;34:74–89. doi: 10.1038/npp.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 4.Roep BO, et al. The problems and promises of research into human immunology and autoimmune disease. Nat Med. 2012;18:48–53. doi: 10.1038/nm.2626. [DOI] [PubMed] [Google Scholar]
- 5.Mansfield K. Marmoset models commonly used in biomedical research. Comp Med. 2003;53:383–392. [PubMed] [Google Scholar]
- 6.Tardif SD, et al. Reproduction in captive common marmosets (Callithrix jacchus) Comp Med. 2003;53:364–368. [PubMed] [Google Scholar]
- 7.Baldwin JG. The Behavior of Squirrel Monkeys (Saimiri) in Natural Environments. In: Rosenblum LA, Coe CL, editors. Handbook of Squirrel Monkey Research. Plenum Press; 1985. pp. 35–53. [Google Scholar]
- 8.Tardif SD, et al. The marmoset as a model of aging and age-related diseases. ILAR J. 2011;52:54–65. doi: 10.1093/ilar.52.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy K, Tosolini FA. A review of primate herpes viruses. Proc R Soc Med. 1975;68:145–150. [PMC free article] [PubMed] [Google Scholar]
- 10.Perelman P, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill JP. Croonian Lecture: The Developmental History of the Primates. Philosophical Transactions of the Royal Society of London. Series B, Containing Papers of a Biological Character. 1932;221:45–178. [Google Scholar]
- 12.Wislocki GB. Observations on twinning in marmosets. American Journal of Anatomy. 1939;64:445–483. [Google Scholar]
- 13.Benirschke K, et al. Marrow Chimerism in Marmosets. Science. 1962;138:513–515. doi: 10.1126/science.138.3539.513. [DOI] [PubMed] [Google Scholar]
- 14.Benirschke K, Brownhill LE. Further observations on marrow chimerism in marmosets. Cytogenetics. 1962;1:245–257. doi: 10.1159/000129734. [DOI] [PubMed] [Google Scholar]
- 15.Sweeney CG, et al. Quantitative molecular assessment of chimerism across tissues in marmosets and tamarins. BMC Genomics. 2012;13:98. doi: 10.1186/1471-2164-13-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bevan MJ. In a radiation chimaera, host H-2 antigens determine immune responsiveness of donor cytotoxic cells. Nature. 1977;269:417–418. doi: 10.1038/269417a0. [DOI] [PubMed] [Google Scholar]
- 17.Ildstad ST, et al. Characterization of mixed allogeneic chimeras. Immunocompetence, in vitro reactivity, and genetic specificity of tolerance. J Exp Med. 1985;162:231–244. doi: 10.1084/jem.162.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summers PM, Shelton JN. Long-term acceptance of full thickness body skin grafts between Bos taurus-Bos indicus chimeric twins. Aust J Exp Biol Med Sci. 1985;63 (Pt 3):329–332. doi: 10.1038/icb.1985.38. [DOI] [PubMed] [Google Scholar]
- 19.Benirschke K. Accurate Recording of Twin Placentation: A Plea to the Obstetrician. Obstetrics & Gynecology. 1961;18:334–347. [Google Scholar]
- 20.Booth PB, et al. Blood chimerism in a pair of twins. Br Med J. 1957;1:1456–1458. doi: 10.1136/bmj.1.5033.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunsford I, et al. A human blood-group chimera. Br Med J. 1953;2:81. doi: 10.1136/bmj.2.4827.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholas JW, et al. Human blood chimeras a study of surviving twins. Br Med J. 1957;1:1458–1460. doi: 10.1136/bmj.1.5033.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueno S, et al. Human chimerism in one of a pair of twins. Acta Genet Stat Med. 1959;9:47–53. [PubMed] [Google Scholar]
- 24.Gengozian N, Porter RP. Transplantation immunology in the marmoset. In: Goldsmith EI, Moor-Jankowski J, editors. Medical Primatology. Karger; 1971. p. 165. [Google Scholar]
- 25.Gorer PA, et al. Studies on the Genetic and Antigenic Basis of Tumour Transplantation. Linkage between a Histocompatibility Gene and ‘Fused’ in Mice. Proceedings of the Royal Society of London. Series B - Biological Sciences. 1948;135:499–505. [Google Scholar]
- 26.Watkins DI, et al. Genetically distinct cell populations in naturally occurring bone marrow-chimeric primates express similar MHC class I gene products. J Immunol. 1990;144:3726–3735. [PubMed] [Google Scholar]
- 27.Garamszegi LZ, Nunn CL. Parasite-mediated evolution of the functional part of the MHC in primates. J Evol Biol. 2011;24:184–195. doi: 10.1111/j.1420-9101.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- 28.Sutton JT, et al. Disentangling the roles of natural selection and genetic drift in shaping variation at MHC immunity genes. Mol Ecol. 2011;20:4408–4420. doi: 10.1111/j.1365-294X.2011.05292.x. [DOI] [PubMed] [Google Scholar]
- 29.Watkins DI, et al. A primate species with limited major histocompatibility complex class I polymorphism. Proc Natl Acad Sci U S A. 1988;85:7714–7718. doi: 10.1073/pnas.85.20.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang H, et al. Non-human primate model of Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2009;5:e1000606. doi: 10.1371/journal.ppat.1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watkins DI, et al. Unusually limited nucleotide sequence variation of the expressed major histocompatibility complex class I genes of a New World primate species (Saguinus oedipus) Immunogenetics. 1991;33:79–89. doi: 10.1007/BF00210819. [DOI] [PubMed] [Google Scholar]
- 32.Robinson J, et al. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalifoux LV, Bronson RT. Colonic adenocarcinoma associated with chronic colitis in cotton top marmosets, Saguinus oedipus. Gastroenterology. 1981;80:942–946. [PubMed] [Google Scholar]
- 34.Miller G, et al. Lymphoma in cotton-top marmosets after inoculation with Epstein-Barr virus: tumor incidence, histologic spectrum antibody responses, demonstration of viral DNA, and characterization of viruses. J Exp Med. 1977;145:948–967. doi: 10.1084/jem.145.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfe LG, Deinhardt F. Overview of viral oncology studies in Saguinus and Callithrix species. Primates Med. 1978;10:96–118. [PubMed] [Google Scholar]
- 36.Alvarez M, et al. Description of a new kind of MHC DNA sequence in Saguinus oedipus (cotton-top tamarin) Eur J Immunogenet. 1998;25:287–292. doi: 10.1046/j.1365-2370.1998.00099.x. [DOI] [PubMed] [Google Scholar]
- 37.Cadavid LF, et al. Evolutionary instability of the major histocompatibility complex class I loci in New World primates. Proc Natl Acad Sci U S A. 1997;94:14536–14541. doi: 10.1073/pnas.94.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyllensten U, et al. The cotton-top tamarin revisited: Mhc class I polymorphism of wild tamarins, and polymorphism and allelic diversity of the class II DQA1, DQB1, and DRB loci. Immunogenetics. 1994;40:167–176. doi: 10.1007/BF00167076. [DOI] [PubMed] [Google Scholar]
- 39.Watkins DI, et al. Evolution of the MHC class I genes of a New World primate from ancestral homologues of human non-classical genes. Nature. 1990;346:60–63. doi: 10.1038/346060a0. [DOI] [PubMed] [Google Scholar]
- 40.Tardif S, et al. Preparing New World monkeys for laboratory research. ILAR J. 2006;47:307–315. doi: 10.1093/ilar.47.4.307. [DOI] [PubMed] [Google Scholar]
- 41.Lang CM. The Laboratory Care and Clinical Management of Saimiri (Squirrel Monkeys) In: Rosenblum LA, Cooper RW, editors. The Squirrel Monkey. Academic Press; 1968. [Google Scholar]
- 42.Galland GG. Role of the squirrel monkey in parasitic disease research. ILAR J. 2000;41:37–43. doi: 10.1093/ilar.41.1.37. [DOI] [PubMed] [Google Scholar]
- 43.Brady AG. Research techniques for the squirrel monkey (Saimiri sp.) ILAR J. 2000;41:10–18. doi: 10.1093/ilar.41.1.10. [DOI] [PubMed] [Google Scholar]
- 44.Williams L, Glasgow M. Squirrel monkey behavior in research. ILAR J. 2000;41:26–36. doi: 10.1093/ilar.41.1.26. [DOI] [PubMed] [Google Scholar]
- 45.Vandeberg JL, et al. Genetic relationships among three squirrel monkey types: Implications for taxonomy, biomedical research, and captive breeding. American Journal of Primatology. 1990;22:101–111. doi: 10.1002/ajp.1350220204. [DOI] [PubMed] [Google Scholar]
- 46.Boinski S, Cropp SJ. Disparate Data Sets Resolve Squirrel Monkey (Saimiri) Taxonomy: Implications for Behavioral Ecology and Biomedical Usage. International Journal of Primatology. 1999;20:237–256. [Google Scholar]
- 47.Zimbler-DeLorenzo HS, Stone AI. Integration of field and captive studies for understanding the behavioral ecology of the squirrel monkey (Saimiri sp.) American Journal of Primatology. 2011;73:607–622. doi: 10.1002/ajp.20946. [DOI] [PubMed] [Google Scholar]
- 48.Lavergne A, et al. Genetic analysis of the Saimiri breeding colony of the Pasteur Institute (French Guiana): development of a molecular typing method using a combination of nuclear and mitochondrial DNA markers. Journal of Medical Primatology. 2003;32:330–340. doi: 10.1046/j.1600-0684.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- 49.Schreiber A, et al. Captive breeding of squirrel monkeys, Saimiri sciureus and Saimiri boliviensis: The problem of hybrid groups. Zoo Biology. 1998;17:95–109. [Google Scholar]
- 50.Abee CR. The Squirrel Monkey in Biomedical Research. ILAR News. 1989;31:11–20. [Google Scholar]
- 51.Osterholz M, et al. A PCR-based marker to simply identify Saimiri sciureus and S. boliviensis boliviensis. American Journal of Primatology. 2008;70:1177–1180. doi: 10.1002/ajp.20606. [DOI] [PubMed] [Google Scholar]
- 52.Coe CL, et al. The Endocrine System of the Squirrel Monkey. In: Rosenblum LA, Coe CL, editors. Handbook of Squirrel Monkey Research. Plenum Press; 1985. pp. 35–53. [Google Scholar]
- 53.Mendoza SP, et al. Social organization and social behavior in two subspecies of squirrel monkeys (Saimiri sciureus) Folia Primatol (Basel) 1978;30:126–144. doi: 10.1159/000155859. [DOI] [PubMed] [Google Scholar]
- 54.Ausman LM, et al. Nutrition and Metabolism of the Squirrel Monkey. In: Rosenblum LA, Coe CL, editors. Handbook of Squirrel Monkey Research. Plenum Press; 1985. pp. 35–53. [Google Scholar]
- 55.Whiteley HE, et al. Pathologic Changes Associated with Fatal Plasmodium falciparum Infection in the Bolivian Squirrel Monkey. The American Journal of Tropical Medicine and Hygiene. 1987;37:1–8. doi: 10.4269/ajtmh.1987.37.1. [DOI] [PubMed] [Google Scholar]
- 56.Hershkovitz P. Taxonomy of squirrel monkeys genus Saimiri (cebidae, platyrrhini): A preliminary report with description of a hitherto unnamed form. American Journal of Primatology. 1984;6:257–312. doi: 10.1002/ajp.1350060402. [DOI] [PubMed] [Google Scholar]
- 57.Chiou KL, et al. Pleistocene diversification of living squirrel monkeys (Saimiri spp.) inferred from complete mitochondrial genome sequences. Mol Phylogenet Evol. 2011;59:736–745. doi: 10.1016/j.ympev.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 58.Ringach DL, Jentsch JD. We must face the threats. J Neurosci. 2009;29:11417–11418. doi: 10.1523/JNEUROSCI.3738-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan AW, Yang SH. Generation of transgenic monkeys with human inherited genetic disease. Methods. 2009;49:78–84. doi: 10.1016/j.ymeth.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasaki E, et al. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 61.Shiozawa S, et al. Gene targeting and subsequent site-specific transgenesis at the beta-actin (ACTB) locus in common marmoset embryonic stem cells. Stem Cells Dev. 2011;20:1587–1599. doi: 10.1089/scd.2010.0351. [DOI] [PubMed] [Google Scholar]
- 62.Rylands AB, Mittermeier RA. The Diversity of the New World Primates (Platyrrhini): An Annotated Taxonomy. In: Garber PA, et al., editors. South American Primates: Comparative Perspectives in the Study of Behavior, Ecology, and Conservation. Springer; 2009. [Google Scholar]
- 63.Little CC, Tyzzer EE. Further experimental studies on the inheritance of susceptibility to a Transplantable tumor, Carcinoma (J. W. A.) of the Japanese waltzing Mouse. J Med Res. 1916;33:393–453. [PMC free article] [PubMed] [Google Scholar]
- 64.Bover KH. Homoisotransplantation van Epidermis bei eineigen Zwilligen. Beitrage Zur Klinischen Chirugica. 1927;141:393–453. [Google Scholar]
- 65.Haldane JBS. The Part Played by Recurrent Mutation in Evolution. The American Naturalist. 1933;67:5–19. [Google Scholar]
- 66.Medawar PB. The behaviour and fate of skin autografts and skin homografts in rabbits: A report to the War Wounds Committee of the Medical Research Council. J Anat. 1944;78:176–199. [PMC free article] [PubMed] [Google Scholar]
- 67.Snell GD. Methods for the study of histocompatibility genes. J Genet. 1948;49:87–108. doi: 10.1007/BF02986826. [DOI] [PubMed] [Google Scholar]
- 68.Zinkernagel RM, Doherty PC. H-2 compatability requirement for T-cell-mediated lysis of target cells infected with lymphocytic choriomeningitis virus. Different cytotoxic T-cell specificities are associated with structures coded for in H-2K or H-2D. J Exp Med. 1975;141:1427–1436. doi: 10.1084/jem.141.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benacerraf B, McDevitt HO. Histocompatibility-linked immune response genes. Science. 1972;175:273–279. doi: 10.1126/science.175.4019.273. [DOI] [PubMed] [Google Scholar]
- 70.Levine BB, et al. Studies on Artificial Antigens. Iii. The Genetic Control of the Immune Response to Hapten-Poly-L-Lysine Conjugates in Guinea Pigs. J Exp Med. 1963;118:953–957. doi: 10.1084/jem.118.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McDevitt HO, Tyan ML. Genetic control of the antibody response in inbred mice. Transfer of response by spleen cells and linkage to the major histocompatibility (H-2) locus. J Exp Med. 1968;128:1–11. [PMC free article] [PubMed] [Google Scholar]