Abstract
Purpose
Leukemias with MLL gene rearrangement are associated with a poor prognosis. Natural killer (NK) cell therapy is a potential treatment, but leukemia cells may be resistant. Here, we sought to determine the susceptibility of MLL-rearranged leukemia cells to NK cell lysis and to develop a novel immunotherapeutic approach to optimize NK cell therapy, including the use of an antibody against leukemia-associated antigen and the elimination of killer-cell immunoglobulin-like receptor (KIR)–mediated inhibition.
Experimental Design
Three MLL-rearranged leukemia cell lines (RS4;11, SEM, and MV4-11) and primary leukemia blasts were assessed for surface phenotype and susceptibility to NK cell lysis with or without antibodies against CD19 (XmAb5574), CD33 (lintuzumab), or KIR ligands.
Results
All three cell lines were resistant to NK cell lysis, had some inhibitory KIR ligands and protease inhibitor-9, and expressed low levels of NKG2D activating ligands and adhesion molecules. After treatment with XmAb5574 or lintuzumab, MLL-rearranged leukemia cells were efficiently killed by NK cells. The addition of pan–major histocompatibility complex class I antibody, which blocked inhibitory KIR-HLA interaction, further augmented degranulation in all three KIR2DL1, KIR2DL2/3, and KIR3DL1 subsets of NK cells based on the rule of missing-self recognition. A mouse model showed a decreased rate of leukemia progression in vivo as monitored by bioluminescence imaging and longer survival after antibody treatment.
Conclusion
Our data support the use of a triple immunotherapy approach, including an antibody directed against tumor-associated antigen, KIR-mismatched NK cell transplantation, and inhibitory KIR blockade, for the treatment of NK cell–resistant MLL-rearranged leukemias.
Keywords: Antibody-dependent cell-mediated cytotoxicity, natural killer cells, killer-cell immunoglobulin-like receptors, mixed-lineage leukemia, targeted therapy
Introduction
Recurrent translocations that involve chromosome 11 band q23 have been observed in acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), and biphenotypic (mixed lineage) leukemia; thus, the gene has been named MLL (for myeloid/lymphoid, or mixed lineage, leukemia) (1). The MLL gene is a member of the trithorax group and consists of 36 exons encoding a DNA-binding methyltransferase that contains 3,969 amino acids with a molecular weight of 430 kDa (2). The protein methylates histone H3 on lysine residue 4 (H3K4) for epigenetic control of early embryonic development and hematopoiesis (3, 4). Chromosomal translocations during leukemogenesis usually involve an 8.3 kb breakpoint cluster region spanning exons 5–11 of MLL which then join the amino terminal of MLL to the carboxy terminal of one of 70 partner proteins in frame (2, 4). The common translocations include t(4;11) and t(11;19) in ALL and t(9;11) and t(6;11) in AML, resulting in the formation of fusion proteins, including MLL-AF4, MLL-ENL, MLL-AF9, and MLL-AF6, all of which have lost H3K4 methyltransferase activity (3). Instead, the chimeric fusion proteins lead to the aberrant expression of many downstream target genes, including HOX, MEIS1, BCL2, C-MYC, and CDK6 (2, 5).
MLL-rearranged leukemias have unique clinical features and are often associated with a poor prognosis (6). MLL rearrangements are found in approximately 80% of infant leukemias and in 10% of AML in adults (3). A very high proportion of patients with therapy-related acute leukemia after treatment with topoisomerase II inhibitors have MLL abnormalities involving AF4, AF9, and ENL, as well as CBP, that are characteristic of therapy-related AML (2, 7). Patients with MLL-rearranged leukemia have a low probability of survival, in the 30% to 40% range, even with contemporary chemotherapy and hematopoietic stem cell transplantation (6, 8).
Because many MLL-rearranged leukemias express mixed-lineage or biphenotypic markers including B and myeloid antigens, targeted therapy using monoclonal antibodies against these antigens is an attractive alternative treatment. Rituximab is an FDA-approved chimeric antibody against human CD20, an antigen expressed beginning at the pre–B-cell stage. Unfortunately, most MLL-rearranged leukemias are stem-cell–like and CD20-negative (3, 4). Therefore, CD19 is a better target as a pan–B-cell antigen. XmAb5574 is a humanized anti-CD19 antibody with its Fc domain engineered for higher affinity to FcγRIIIa of effector cells and diminished non-specific binding to FcγIIb. In chronic lymphoblastic leukemia, ALL, and mantle cell lymphoma, it may mediate more effective antibody-dependent cell-mediated cytotoxicity (ADCC) than its parental counterpart as well as other therapeutic antibodies such as rituximab, ofatumumab and alemtuzumab (9–11). For pan-myeloid antigens, CD33 is an attractive target. Lintuzumab (also known as SGN-33 and huM195) is an anti-CD33 therapeutic antibody in clinical development (12). It was reported to promote survival in preclinical mouse models of AML (13, 14).
Natural killer (NK) cells are the primary lymphocytes that are involved in ADCC through the activation of high-affinity FcγRIIIa (CD16) on their cell surfaces. Human NK cell transplantation has become feasible recently and has therefore generated much interest in augmenting cancer antibody therapy (15). In a clinical study, NK cell therapy alone was found to be safe and beneficial in AML patients (16); however, NK cell transplantation may not always be effective, because immune escape is possible. Biologically, NK cell functions are regulated by two sets of surface molecules: activating and inhibitory receptors. Killer-cell immunoglobulin-like receptors (KIR) and NKG2D are two of the receptor families that are known to be important as inhibitory and activating receptors, respectively, in human leukemia cell recognition. Thus, leukemia cells may escape from NK cell immunosurveillance by upregulation of the expression of a KIR inhibitory ligand or downregulation of NKG2D activation signals. In this study, we characterized the NK cell ligand expression in MLL-rearranged leukemia and provided evidence that ADCC mediated by XmAb5574 or lintuzumab was useful to overcome its inherent NK cell-resistance. We found that either KIR blockade or selection of a KIR-incompatible donor could further augment ADCC activity in vitro and in vivo.
Materials and Methods
Cell purification
Human peripheral blood mononuclear cells (PBMCs) from healthy donors and primary AML blasts from patients were obtained following informed consent under the approval of the institutional review board of St. Jude Children’s Research Hospital. Untouched NK cells were enriched using an NK Cell Isolation Kit (Miltenyi, Auburn, CA) according to the manufacturer’s instructions. The purity of the NK cells was more than 95% in all experiments. Human leukemia cells RS4;11 and MV4-11 were from American Type Culture Collection (Manassas, VA). SEM cells were from DSMZ (Braunschweig, Germany). All cell lines were cultured as recommended and passaged with no more than three months. Routine authentication was performed by typical morphology observation. The expression of CD19 and CD33 were continuously monitored by flow cytometry throughout the experimental period to ensure the expression levels were consistent with those at the time of cell line acquisition from suppliers.
Phenotypic analysis by flow cytometry
The following antibodies were purchased from commercial suppliers and used for phenotypic analysis: fluorescein isothiocyanate (FITC)-conjugated anti-ULBP1 (170818), anti-PVR (TX21), anti-CD48 (J4.57), anti-NTB-A (292811), anti-Fas (CH-11), anti-ICAM-1 (84H10), anti-CD33 (HIM3-4), anti-CD158ah (11PB6), and anti-NKB1 (DX9); phycoerythrin-conjugated anti-MICA (159227), anti-ULBP2 (165903), anti-nectin-2 (R2.525), anti-TRAIL-R1 (DJR1), anti-TRAIL-R2 (71908), anti-CD19 (SJ25C1), and anti-CD158b (GL183); phycoerythrin-Texas red–conjugated anti-CD3 (UCHT1); allophycocyanin-conjugated anti-CD56 (N901(NKH-1)) and anti-MICB (236511); Alexa Fluor 488-conjugated anti-PI-9 (7D8); and unconjugated anti-ULBP3 (166510). Flow cytometry analyses were performed with LSRII (BD Bioscience, San Jose, CA), and the data were analyzed with FlowJo 8.8.6 (Tree Star, OR).
KIR genotyping and KIR ligand allele typing
Genotyping of KIR was performed using an Olerup SSP KIR genotyping kit (Olerup SSP AB, Stockholm, Sweden) according to the manufacturer’s instructions. Genomic DNA from PBMCs was extracted with a QIAamp DNA blood mini kit (Qiagen Inc., Valencia, CA). The KIRs were PCR–amplified, and the products were electrophoresed in 2% gel. The amplified KIR alleles were determined according to the expected product sizes. KIR ligand typing was performed using a single-nucleotide polymorphism assay following an allelic discrimination assay protocol (Applied Biosystems, Life Technologies, Carlsbad, CA) as previously described (17). The probes used were as follows: HLA-Bw4-associated HLA-B and -A: 6FAM-CCGCTAC TACAACCAG-MGBNFQ; HLA-Bw6: VIC-CGGCTACTACAACCAG-MGBNFQ; HLA-C1: 6FAM-CCGAGTGAGCCTGC-MGBNFQ; and HLA-C2: VIC-CCGAGTGA ACCTGC-MGBNFQ.
Cytotoxicity assay
Target cells, including those from cell lines and primary patient samples, were labeled with DELFIA BATDA reagent (PerkinElmer Life and Analytical Sciences, Waltham, MA) according to the manufacturer's instructions. After labeling, the cells were coated with either anti-CD19 XmAb5574 (Xencor Inc., Monrovia, CA) or anti-CD33 lintuzumab (Seattle Genetics Inc., Bothell, WA) with or without anti-panMHCI (LEAF clone W6/32, Biolegend, San Diego, CA) antibodies for 15 min at 37°C. The cells were washed and then co-cultured with effector cells in a 20:1 ratio (effector to target) for 2 h at 37°C. Cell supernatants were then reacted with DELFIA Europium solution, and the fluorescence was measured using a Wallac Victor 2 Counter Plate Reader (both from PerkinElmer Life and Analytical Sciences). Specific lysis was calculated by a formula: (experimental lysis count minus spontaneous lysis count) divided by (maximum lysis count [by lysis buffer] minus spontaneous lysis count) times 100%.
CD107a degranulation assay
NK cells were co-cultured with antibody-coated target cells at a ratio of 1:2 in the presence of FITC-conjugated anti-CD107a antibody (H4A3, BD Bioscience). Monensin (BD Bioscience) was added to the cells 1 h after co-culture. After 5 h, the cells were harvested and stained with anti-CD3, CD56, and particular KIR antibodies to differentiate the percentage of CD107a+ cells by flow cytometric analysis. Anti-CD19 (10 ng/mL of XmAb5574) or anti-CD33 (1000 ng/mL of lintuzumab) was used. Resting NK cells were used for all experiments, except in Figure 2 in which NK cells were pretreated overnight with 10 U/mL IL-2 (Aldesleukin (Proleukin), Novartis Pharmaceutials, Emeryville, CA).
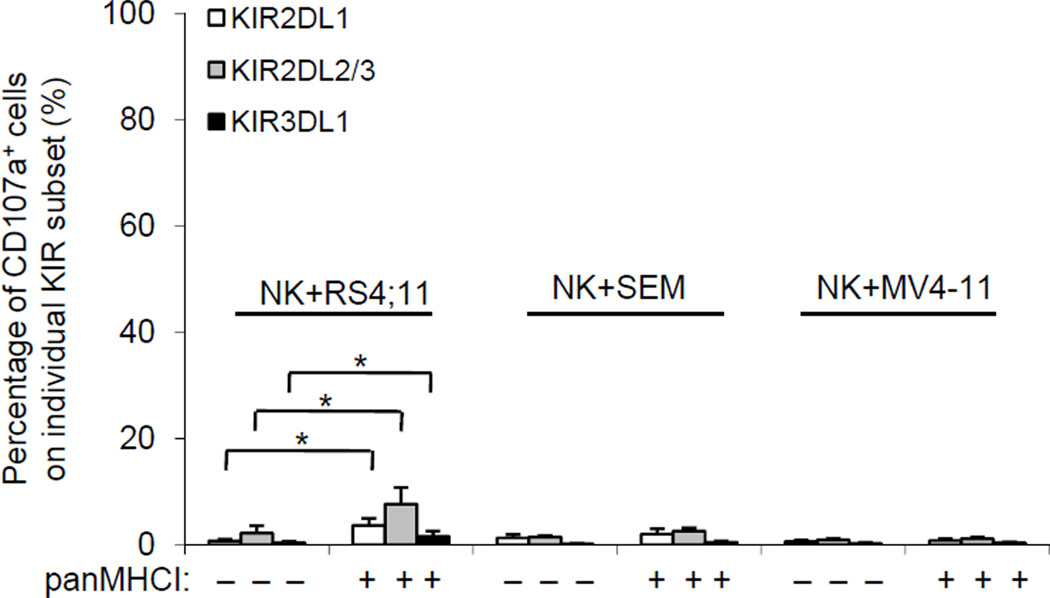
Figure 2.
Degranulation of NK cells with or without inhibition of KIR-HLA interaction. Target MLL-rearranged leukemia cells were left untreated or treated with 10 µg/mL of panMHCI antibody before co-culture with 10 U/mL IL-2 pretreated NK cells at a ratio of 2:1 (targets to effectors) for 5 h. The percentages of CD107a+ KIR2DL1, KIR2DL2/3, and KIR3DL1 subsets of NK cells were analyzed and plotted. Data represent mean ± S.E.M. of three independent experiments. *p<0.05.
In vivo model
Twelve- to 20-week-old nonobese diabetic/severe combined immunodeficient interleukin-2 receptor γnull (NOG) mice (Jackson Laboratory, Bar Harbor, ME) were used in compliance with institutional animal care and use committee regulations. Large-scale human NK cell isolation was performed using the CliniMACS (Miltenyi Biotec, Auburn, CA) device according to the manufacturer’s instructions. First, 5×106 leukemic cells were injected into the mice intravenously. For treatment groups, 5×106 primary NK cells were injected intravenously (one injection only to mimic a prior clinical trial (16)) with or without antibody treatment every 4 days for a total of four doses. Disease progression in the mice was measured and evaluated by the IVIS Xenogen system (Caliper Life Sciences, Hopkinton, MA). Physical disease and survival were also monitored and recorded. Mice were euthanized immediately upon observable signs of disease, including rear limb paralysis, weight loss >20%, or severe lethargy. Gross autopsy confirmed leukemia infiltration in all diseased mice at multiple sites including spleen, liver, kidneys, and bone marrow. The experiments were terminated by euthanasia for surviving mice without any signs of disease by day 60.
Statistical analysis
Comparisons between means in two groups were based on a 2-tailed nonparametric test. For comparisons of means in more than two groups, one-way ANOVA was used. The difference was considered statistically significant if p < 0.05.
Results
NK-resistant phenotype in MLL-rearranged leukemia cell lines
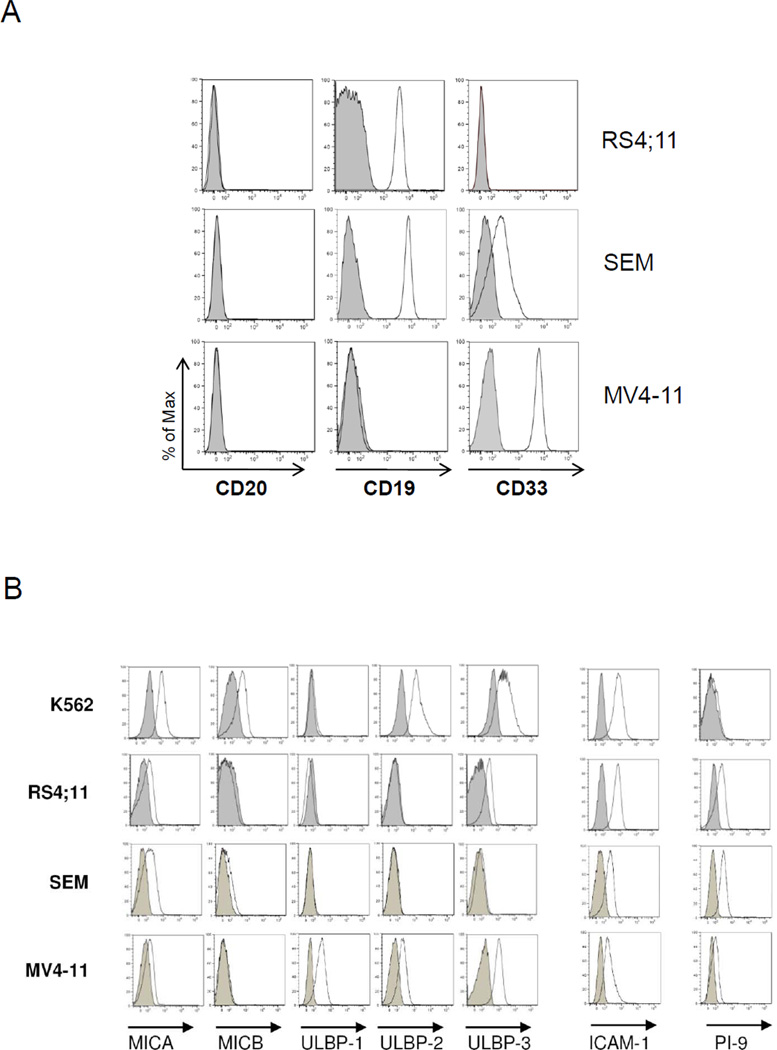
The absence of CD20 in leukemia cells is one of the known mechanisms for rituximab resistance. In this study, we used three MLL-rearranged cell lines (RS4;11, SEM, and MV4-11) that were CD20− (Figure 1A) with variable CD19 and CD33 expression: RS4;11 was CD20− CD19+ CD33−, MV4-11 was CD20− CD19+ CD33+, and SEM was CD20− CD19+ CD33dim (Figure 1A). All cell lines had some inhibitory KIR ligands (Table 1): RS4;11 cells expressed all three KIR ligands (HLA-C2 for KIR2DL1, HLA-C1 for KIR2DL2/3, and HLA-Bw4 for KIR3DL1); SEM cells did not express KIR2DL1 ligand; and MV4-11 cells did not express KIR3DL1 ligand. For NKG2D ligands (MICA/B and ULBP1-3) (Figure 1B), RS4;11 cells only weakly expressed MICA and ULBP3; SEM cells were weakly positive for MICA; and MV4-11 cells were positive for ULBP1-3. The expression levels of the NKG2D ligands on the three MLL cell lines were in general lower than those found on the NK-sensitive target cell, K562. All three cell lines had low expression of the adhesion molecule ICAM-1, which is important for the formation of immune synapse. Serine protease inhibitor PI-9 was positive in the 3 cell lines, rendering them resistance to granzyme-induced apoptosis.
Figure 1.
Characterization of MLL-rearranged cell lines by flow cytometric analyses. Expression of (A) CD20, CD19, and CD33; (B) NKG2D ligands (including MICA, MICB, ULBP-1, ULBP-2, and ULBP-3), adhesion molecule ICAM-1, and intracellular PI-9. Standard NK cell target K562 was used as a positive control for comparison. Data are representative of three independent experiments. Shaded histograms represent the isotype control.
Table 1.
Summary of KIR ligands in the three MLL-rearranged leukemia cell lines
| KIR ligand | RS4;11 | SEM | MV4-11 |
|---|---|---|---|
| Bw4 | + | + | − |
| C1 | + | + | + |
| C2 | + | − | + |
MLL-rearranged leukemia was NK resistant, and neutralizing KIR inhibition alone did not activate NK cells
In light of the multiple NK resistant features observed in the MLL-rearranged leukemia cell lines (presence of inhibitory KIR ligands, lack of NKG2D activation and adhesion molecules, and expression of PI-9), we hypothesized that these leukemia cells would have NK-resistance, of which blockade of the interaction between inhibitory KIR and HLA ligand alone would not be sufficient to activate NK cells. As shown in Figure 2 none of the KIR2DL1, 2DL2/3, and 3DL1 subsets of NK cells degranulated (as measured by an increase in CD107a+ cells) when NK cells were co-cultured with RS4;11, SEM, or MV4-11 cells. Even after blockade with a pan–major histocompatibility complex (MHC) class I (panMHCI) antibody, the proportion of CD107a+ cells in the KIR subsets was only marginally greater in RS4;11 cells (which had the most inhibitory ligands) but remained low for the other 2 cell lines. Thus, neutralizing the inhibitory signals through KIR was insufficient to trigger a substantial NK cell response to lyse any of these three MLL-rearranged cell lines.
Anti-CD19 and anti-CD33 antibodies induced ADCC effectively against MLL-rearranged leukemia
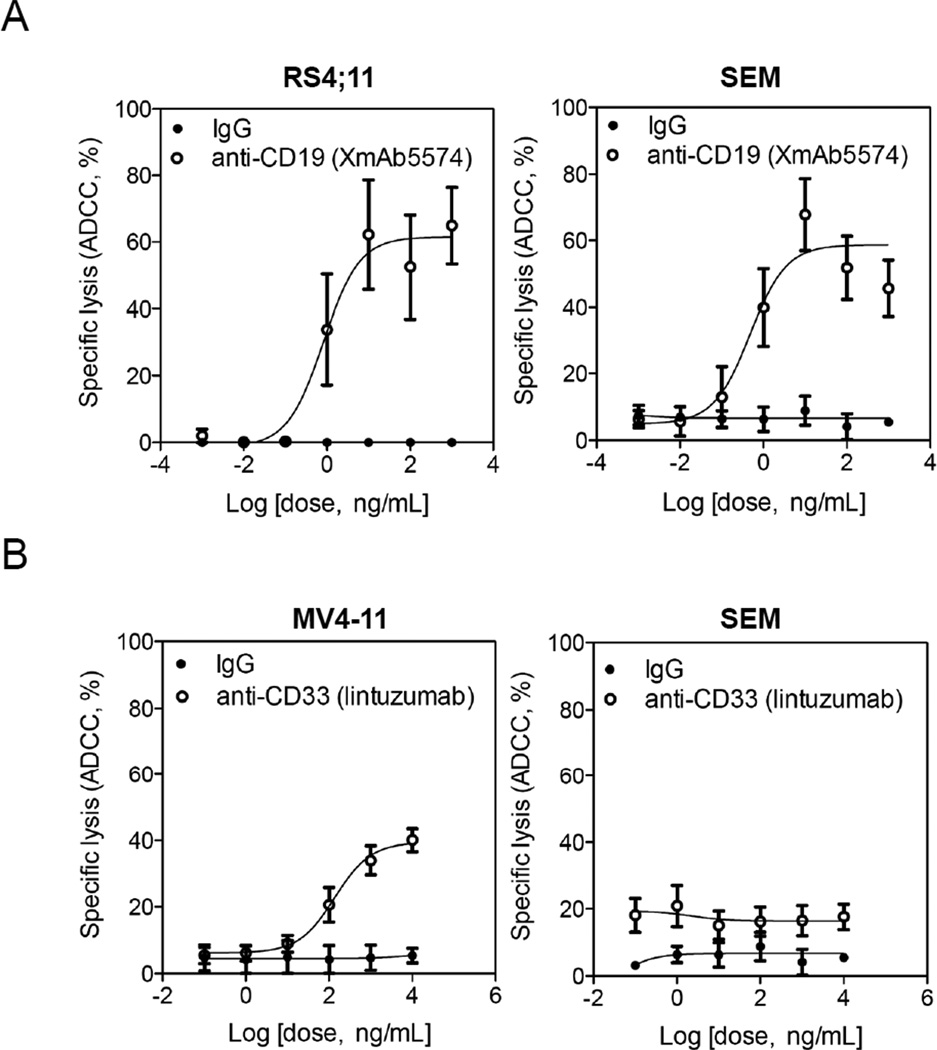
Because FcγIIIa is a potent NK cell activating receptor, we hypothesized that all of the NK-resistant mechanisms in MLL-rearranged leukemia could be overridden by activating FcγIIIa through monoclonal antibodies against leukemia-associated antigens. To test the hypothesis, we used two therapeutic antibodies that can trigger FcγIIIa, including one against CD19 and the other against CD33 antigen. With the anti-CD19 antibody (XmAb5574), there was a dose-dependent cytotoxicity against CD19+ RS4;11 cells with a plateau at ~60% lysis (Figure 3A). A similar dose-dependent cytotoxicity was observed when CD19+ SEM cells were treated with XmAb5574. When the anti-CD33 antibody (lintuzumab) was applied to induce ADCC activity on CD33bright MV4-11, a dose-dependent increase in cytotoxicity was also seen with a plateau at ~40% lysis. As expected, a lesser cytotoxic effect was observed with the CD33dim SEM cells (Figure 3B).
Figure 3.
Dose-dependent increase in ADCC induced by anti-CD19 XmAb5574 or anti-CD33 lintuzumab. (A) MLL-rearranged leukemia cells were treated with XmAb5574 antibody at doses ranging from 10−3 to 103 ng/mL in the presence of NK cells (1:20 targets to effectors). (B) MLL-rearranged leukemia cells were treated with lintuzumab antibody at doses from 10−1 to 104 ng/mL. Isotype IgG antibody was used as a negative control. Data are mean ± S.E.M. from five independent experiments.
KIR blockade augmented ADCC
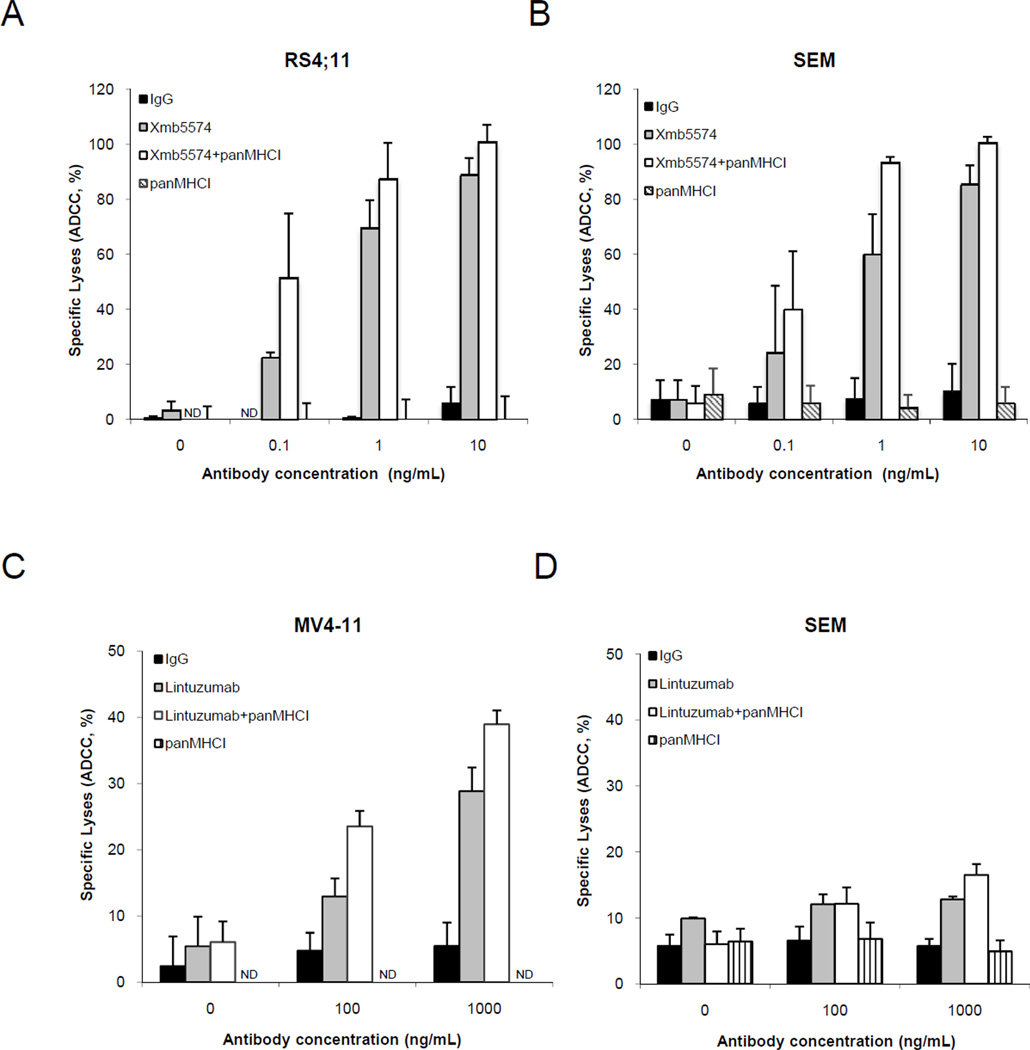
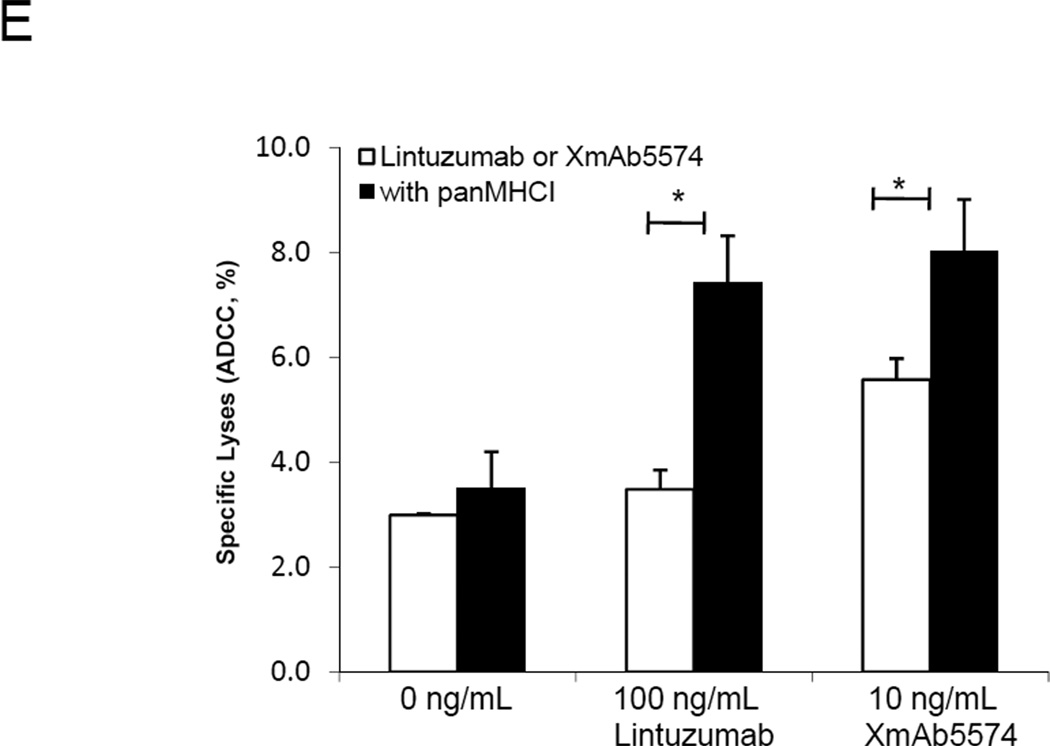
We then sought to determine whether blocking inhibitory KIR-HLA interaction might further increase ADCC activity. NK-mediated cytotoxicity to RS4;11 cells averaged 22.4% with 0.1 ng/mL and 69.9% with 1 ng/mL of XmAb5574 (Figure 4A); the corresponding cell lysis increased with the addition of panMHCI antibody to 51.5% and 99.8% (p<0.01 and p<0.001), respectively. Similar increases in cytotoxicity with panMHCI antibody were observed in XmAb5574-treated SEM cells (Figure 4B) and in lintuzumab-treated MV4-11 cells (Figure 4C). For CD33dim SEM cells, only a modest increase in cytotoxicity was observed when a high dose of lintuzumab (1000 ng/mL) was used together with the panMHCI (Figure 4D). The panMHCI antibody could enhance ADCC even when the effector:target ratio decreased from 20:1 to 5:1 (Supplementary Figure 1). When panMHCI antibody was used alone, there was minimal lysis of MLL-rearranged leukemia cells in all experiments, confirming prior observations that removing KIR inhibition alone was insufficient for NK cell activation (Figure 2). We also examined the effect of XmAb5574 or lintuzumab with or without panMHCI on a primary biphenotypic leukemia sample using saturating dose of the two antibodies (Figure 4E). Phenotyping showed that the surface expression of CD19 in the blasts was lower than that of SEM, but the expression of CD33 was similar. We observed minimal cytotoxicity on the blasts when XmAb5574 or lintuzumab was used alone. However, when panMHCI antibody was added, the cytotoxicity from lintuzumab- or XmAb5574-induced ADCC significantly increased (p<0.05 for both cases).
Figure 4.
ADCC in MLL-rearranged leukemia cells augmented by inhibition of KIR-HLA interactions. (A) RS4;11 cells and (B) SEM cells were pre-coated with XmAb5574 at the indicated doses with or without 10 µg/mL panMHCI antibody before being co-cultured with NK cells (1:20 targets to effectors). (C) Lintuzumab at indicated doses with or without 10 µg/mL panMHCI antibody was used to coat MV4-11 cells or (D) SEM cells before they were co-cultured with NK cells (1:20 targets to effectors). Isotype IgG antibody was used as a negative control. Data represent the mean ± S.E.M. from five independent experiments. (E) XmAb5574 or lintuzumab with panMHCI antibody induced ADCC on fresh primary leukemia blasts. Either 10 ng/mL XmAb5574 or 100 ng/mL lintuzumab was used to induce ADCC activity on primary leukemia blasts with or without 10 µg/mL panMHCI. Data represent the mean ± S.E.M. from three independent NK cell donors and one primary leukemia patient. *p<0.05. ND: not detectable.
Cytotoxicity followed the rule of missing-self recognition
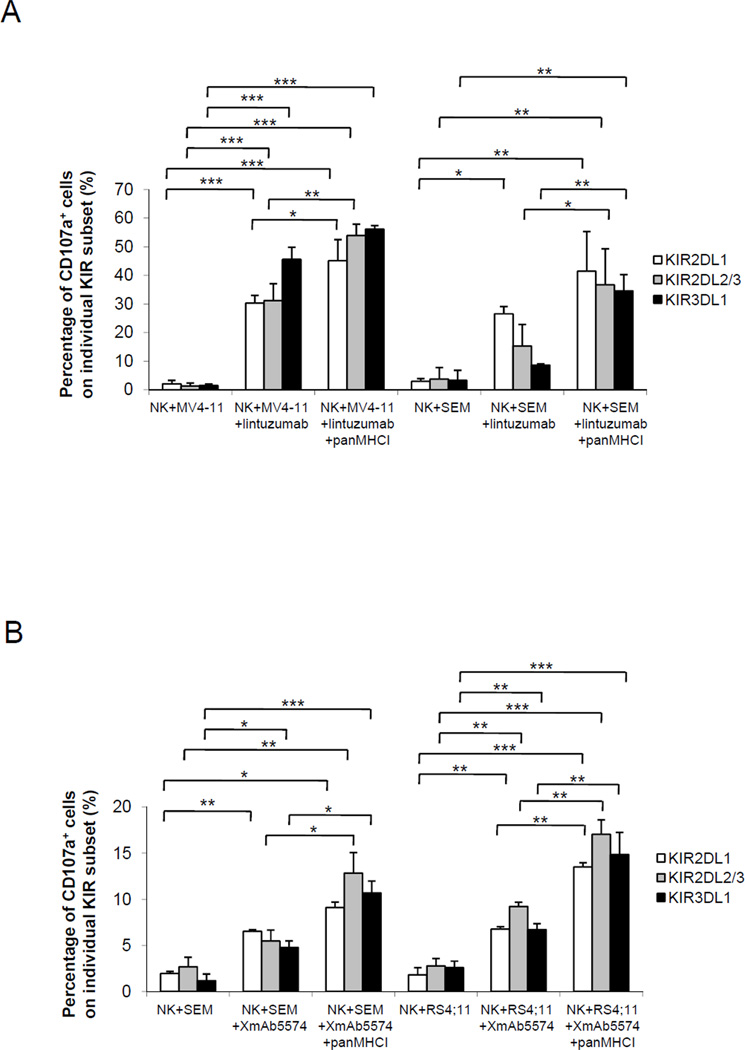
Because a KIR-specific antibody is not yet available clinically, we sought to determine whether the ADCC mediated by lintuzumab or XmAb5574 followed the rule of missing-self recognition and whether a KIR-mismatched NK cell donor might therefore be preferable. In the subsequent experiments, NK cells were obtained from donors positive for all three classes of KIR (KIR2DL1, KIR2DL2/3, and KIR3DL1). The MV4-11 cells had the ligands for KIR2DL1 and KIR2DL2/3 but not for KIR3DL1 (Table 1). When MV4-11 cells were treated with 1000 ng/mL lintuzumab, the KIR3DL1 subset degranulated the most, with minimal change after the addition of panMHCI antibody (Figure 5A). Both KIR2DL1 and KIR2DL2/3 subsets degranulated less, but when panMHCI antibody was included, the degranulation for these groups increased to a level comparable to that of the KIR3DL1 subset (p<0.05 and p<0.01, respectively).
Figure 5.
Lysis followed the rule of missing-self recognition. (A) CD107a degranulation was measured on KIR2DL1, KIR2DL2/3, and KIR3DL3 subsets of NK cells co-cultured with MLL-rearranged leukemia cells pre-coated with 1000 ng/mL lintuzumab or (B) 10 ng/mL XmAb5574 with or without 10 µg/mL panMHCI antibody. The data are mean ± S.E.M. of four independent experiments. *p<0.05 **p<0.01 ***p<0.001.
SEM cells did not have the ligand for KIR2DL1, and degranulation was highest in the KIR2DL1 subset in the presence of lintuzumab (Figure 5A) or XmAb5574 (Figure 5B) without KIR-HLA blockade. With panMHCI antibody, the degranulation in both KIR2DL2/3 and KIR3DL1 subsets increased to a level similar to that of KIR2DL1 (p<0.05 and p<0.01, respectively).
RS4:11 cells had ligands for the 3 groups of inhibitory KIR. When the panMHCI antibody was added to XmAb5574, the percentages of degranulated cells increased significantly in all three KIR subsets (p<0.01 for all cases). Collectively, the data from all 3 cell lines suggested that the use of NK cells from a KIR-mismatched donor or the use of KIR-blocking antibody could synergize with ADCC therapy targeting CD19 or CD33.
Role of FcγIIIa polymorphism and activating KIR
A single-nucleotide polymorphism is present at amino acid position 158 (Val or Phe) of the FcγRIIIa receptor, with 158V reported to have a higher affinity for IgG and associated with increased ADCC efficacy (18). We genotyped our cohort of donor NK cells and found that there was no correlation between 158V and ADCC activity in our experiments (data not shown). We also found no statistical correlation between donor activating KIR-S repertoire and ADCC activity (data not shown).
Blocking KIR ligand increased disease-free survival induced by ADCC in a mouse model
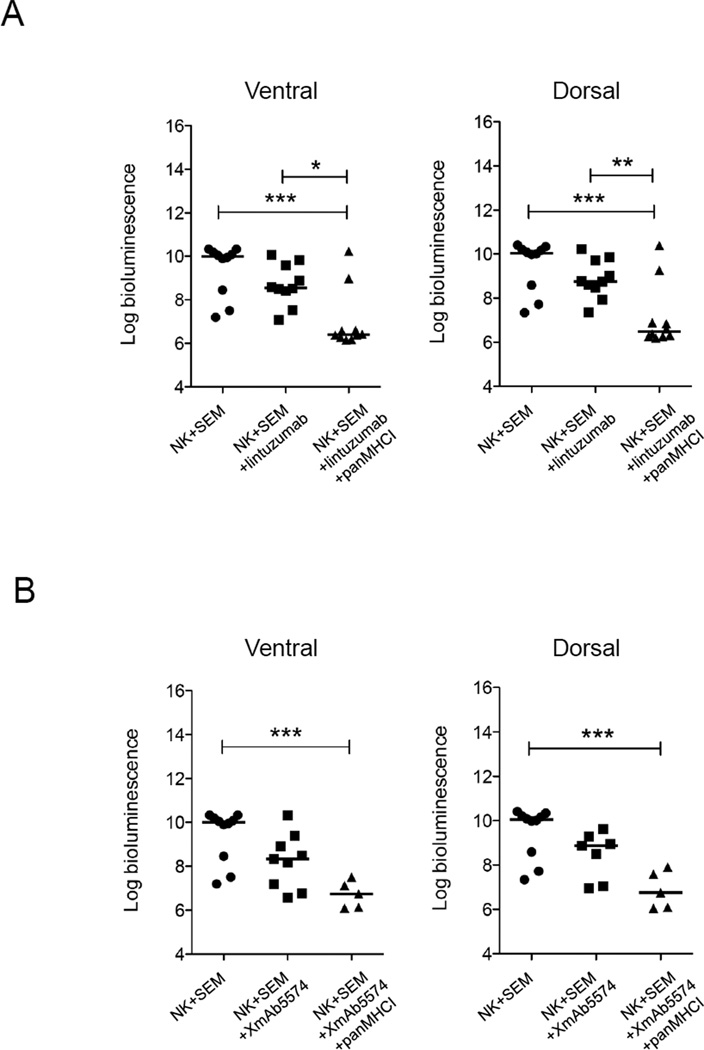
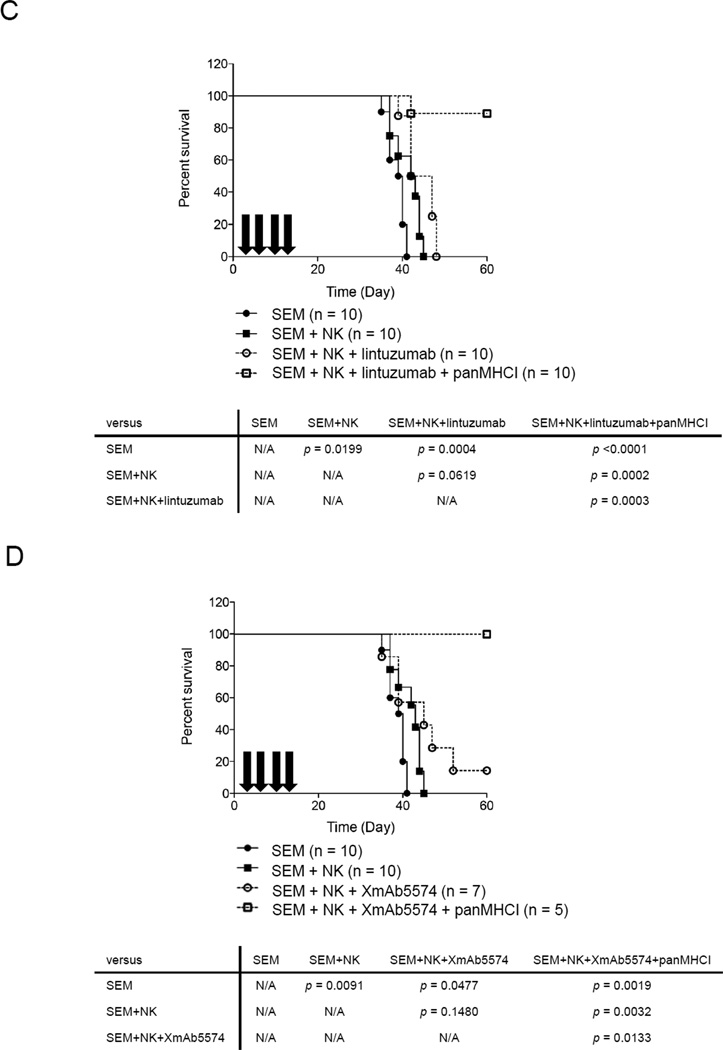
We used a mouse model to determine whether the ADCC induced by XmAb5574 or lintuzumab could benefit host survival in MLL-rearranged leukemia. To accurately estimate the leukemia burden during disease progression, we used the luciferase bioluminescence system. The susceptibility of the luciferase-transduced SEM cell line to NK cell lysis in vitro remained the same as that of untransduced cells (Supplementary Figure 2A and B). We chose SEM as the cell model because it is a typical MLL cell line that expresses the least activating NK cell ligands and expressed both CD19 and CD33 antigens. We injected 5×106 SEM and 5×106 NK cells intravenously on day 0. The next day, we injected intraperitoneally 3 mg/kg lintuzumab or 1 mg/kg XmAb5574. The antibody injection was repeated every 4 days for a total of 4 doses. To determine the optimal time for imaging, we used control mice given injections of SEM alone to establish the linearity of the bioluminescence signals over time (Supplementary Figure 2C). We found that day 15 was the earliest time suitable for cross-sectional analysis and for comparison of bioluminescence intensity among the treatment groups. For the mice treated with lintuzumab, the median bioluminescence signal from the ventral side of the animals was 1.44 logs lower than that of untreated control mice (Figure 6A, left panel). However, a statistically significant decrease (3.59 logs) was observed when panMHCI antibody was injected as well (p<0.001 compared with the NK-alone group). Similar results were obtained on the bioluminescence signals from the dorsal side of the mice (Figure 6A, right panel). For the mice treated with XmAb5574, the median bioluminescence signal from the ventral side of the mice decreased 1.66 logs compared with that from untreated mice. A significant decrease in signals (3.25 logs) was observed in the mice that also received panMHCI antibody (p<0.001, Figure 6B). A similar pattern was observed on the dorsal side of the mice. In terms of survival, the mice treated with NK cells alone (n=10) or NK+lintuzumab (n=10) survived significantly longer than untreated mice (SEM alone) (p=0.02 and p=0.0004, respectively) (Figure 6C). However, all of these mice eventually succumbed to disease. Remarkably, 9 of 10 mice treated with NK cells+lintuzumab+panMHCI antibodies survived long-term; thus their survival was significantly longer than that of the mice given SEM alone (p<0.0001), SEM+NK cells (p<0.0002), or SEM+NK cells+lintuzumab (p=0.0003). Additionally, mice that received NK cells+XmAb5574 also survived significantly longer (p=0.0477, Figure 6D). With the addition of panMHCI, 5 of 5 mice survived long-term (p=0.0019 compared with the SEM-alone group).
Figure 6.
ADCC with or without interruption of KIR-HLA interaction decreased the rate of leukemia progression and improved disease-free survival in an MLL-rearranged leukemia mouse model. The bioluminescence signals (photons/second) from the ventral and dorsal side of mice of different treatment groups 15 days after treatment with (A) NK (n=10), NK+lintuzumab (n=9), or NK+lintuzumab+panMHCI (n=10). (B) Treatment with NK (n=10), NK+XmAb5574 (n=9), or NK+XmAb5574+panMHCI (n=5). In all cases, 0.5 mg/kg panMHCI antibody was used together with lintuzumab and XmAb5574 as indicated. The bioluminescence signals values were log10 transformed. One-way ANOVA was used to compare the statistical significance between groups. *p<0.05 **p<0.01 ***p<0.001. (C) Kaplan-Meier survival curve of SEM-engrafted mice that received no treatment (closed circle), NK cells alone (closed square), NK+lintuzumab (open circle), or NK+lintuzumab+panMHCI (open square). Arrows represent the timing of antibody injections. Mouse survival was followed for 60 days. (D) The Kaplan-Meier survival curve for the mice that received NK cells with or without XmAb5574 and panMHCI are shown. N/A: not applicable.
Discussion
The susceptibility of cancer cells to NK cells depends on a balance of activation and inhibitory signals. Cancer cells may escape the immune surveillance by NK cells through the expression of inhibitory ligands and lower levels of activation ligands. Herein, we found that MLL-rearranged leukemias, which carry a poor prognosis clinically, expressed generally limited amounts of NK activation ligands and had MHCI, which can inhibit NK cells through the three common KIR2DL1, KIR2DL2/3, and KIR3DL1 receptors. NK-resistance, however, could be optimally overcome by simultaneously triggering ADCC and eliminating KIR inhibition.
XmAb5574 is a humanized anti-CD19 antibody with an Fc domain engineered to have higher binding affinity to the FcγIIIa receptor (10). It induces more potent ADCC against B-lymphoma and leukemia cell lines (9, 11). This antibody could be used for salvage or maintenance treatment of leukemias that do not express CD20, the target of rituximab. The efficacy of this antibody depends in part on triggering the FcγIIIa receptor that is expressed primarily on NK cells. Both our in vitro and in vivo data showed potent activity of XmAb5574 against CD19+ MLL-rearranged leukemia in the presence of NK cells. XmAb5574 induced dose-dependent ADCC from 10−2 to 101 ng/mL against the MLL cell lines tested in this study. In our mouse model, treatment with XmAb5574 plus NK cells significantly prolonged survival.
Lintuzumab is a humanized anti-CD33 antibody. It prolonged the survival of mice with acute myeloid leukemia in preclinical studies (14). In a recent phase III multicenter clinical trial, it was found that it was safe but failed to improve patient survival (12). However, the therapeutic effects of lintuzumab could be improved when it was used together with 5-azacytidine (13). Gemtuzumab ozogamicin, another anti-CD33 antibody, has been showed to produce complete molecular remission in a patient with MLL-AF9 AML (19). In this study, lintuzumab triggered dose-response ADCC activity on CD33bright MV4-11 cells from 100 to 104 ng/mL. Significantly prolonged survival was also observed in mice treated with lintuzumab and NK cells. Future studies are warranted to further elucidate the recognition, migration, and cytotoxic interactions between NK cells and MLL-leukemia cells in a time-dependent manner, such as those by using time-lapsed fluorescence microscopy, in the presence of anti-CD19 and anti-CD33 antibodies.
Besides monoclonal antibodies against leukemia-associated antigens (e.g., anti-CD19 and anti-CD33), our findings suggest that the addition of KIR interruption could be a useful strategy to further augment ADCC mediated by NK cells. We used a panMHCI antibody to disrupt the interaction of HLA-A, B, and C with inhibitory KIR2DL1, KIR2DL2/3, and KIR3DL1. A substantial synergistic effect was observed when panMHCI antibody was added to lintuzumab or XmAb5574 in the treatment of MLL-rearranged leukemia cells and fresh primary biphenotypic blasts. The combination resulted in long-term survival in our mouse experiments. Although a specific antibody against KIR, 1-7F9 (20), was shown recently to preferentially augment killing of malignant cells but not normal cells, our findings showed that MLL-rearranged leukemia cells did not have sufficient activation signals. Therefore, blockade against inhibitory KIR alone will be ineffective in breaking their resistance to NK cells. An activating agent, such as XmAb5574 or lintuzumab, is necessary to trigger NK cell lysis of MLL-rearranged leukemia cells.
In the clinical setting, the immune status of the patients must be considered. The administration of XmAb5574 or lintuzumab plus KIR blockade may not be sufficient to elicit a response, as many leukemia patients undergoing chemotherapy have inadequate functional NK cells to mediate ADCC activity. Therefore, allogeneic NK cell transplantation from a healthy donor may be necessary during antibody treatment. In this regard, selection of a KIR-incompatible donor may be crucial. Patients with an inhibitory KIR-ligand mismatch had a lower relapse rate and longer event-free survival than those without (15). Furthermore, we provide direct evidence here that the ADCC of NK cells in the presence of XmAb5574 or lintuzumab followed the rule of missing-self recognition. NK cells with KIR3DL1 degranulated more against MV4-11 cells, which did not have ligands for KIR3DL1. Similar findings were observed for the KIR2DL1 subset against SEM cells that did not have ligands for KIR2DL1. Our findings underscore the desirability of KIR-mismatched NK cell donors, if available, during ADCC-based targeted therapy in future clinical settings.
In conclusion, our study supports a triple immunotherapy approach, including antibody against tumor-associated antigens, KIR-mismatched NK cell transplantation, and inhibitory KIR blockade, for the treatment of MLL-rearranged leukemia. Existing clinical data suggest that all three treatments have minimal side effects.
Supplementary Material
Translational relevance.
MLL-rearranged leukemias have a poor response to standard treatments, including chemotherapy and hematopoietic stem cell transplantation. Unfortunately, these leukemia cells often have an NK-resistant phenotype. We found that this resistance could be overcome by antibody-dependent cell-mediated cytotoxicity (ADCC) against specific antigens expressed by a wide spectrum of clinical MLL-rearranged leukemia, including the pan-B cell marker CD19 and the myeloid marker CD33. Blocking inhibitory KIR signaling further augmented the ADCC activity. This underscores the desirability of KIR-mismatched NK cell donors during ADCC-based targeted therapy. Our findings suggest the use of a triple immunotherapy approach for the treatment of MLL-rearranged leukemias.
Acknowledgments
We thank David Galloway (Department of Scientific Editing, St. Jude Children’s Research Hospital) for scientific editing of the manuscript.
Grant Support
This work was supported in part by research grants from NIH P30 CA-21765, the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Conflict of Interests: WKC, YL, SS, and WL declared no conflict of interest; MKS is an employee of Seattle Genetics, Inc., and JZ was an employee of Xencor, Inc.
References
- 1.Ziemin-van der Poel S, McCabe NR, Gill HJ, Espinosa R, 3rd, Patel Y, Harden A, et al. Identification of a gene-MLL that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci U S A. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harper DP, Aplan PD. Chromosomal rearrangements leading to MLL gene fusions: clinical and biological aspects. Cancer Res. 2008;68:10024–10027. doi: 10.1158/0008-5472.CAN-08-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 4.Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113:6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pui CH, Gaynon PS, Boyett JM, Chessells JM, Baruchel A, Kamps W, et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet. 2002;359:1909–1915. doi: 10.1016/S0140-6736(02)08782-2. [DOI] [PubMed] [Google Scholar]
- 7.Pui CH. Acute leukemias with the t(4;11)(q21;q23) Leuk Lymphoma. 1992;7:173–179. doi: 10.3109/10428199209053620. [DOI] [PubMed] [Google Scholar]
- 8.Woodard P, Barfield R, Hale G, Horwitz E, Leung W, Ribeiro R, et al. Outcome of hematopoietic stem cell transplantation for pediatric patients with therapy-related acute myeloid leukemia or myelodysplastic syndrome. Pediatr Blood Cancer. 2006;47:931–935. doi: 10.1002/pbc.20596. [DOI] [PubMed] [Google Scholar]
- 9.Awan FT, Lapalombella R, Trotta R, Butchar JP, Yu B, Benson DM, Jr, et al. CD19 targeting of chronic lymphocytic leukemia with a novel Fc-domain-engineered monoclonal antibody. Blood. 2008;115:1204–1213. doi: 10.1182/blood-2009-06-229039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton HM, Bernett MJ, Pong E, Peipp M, Karki S, Chu SY, et al. Potent in vitro and in vivo activity of an Fc-engineered anti-CD19 monoclonal antibody against lymphoma and leukemia. Cancer Res. 2008;68:8049–8057. doi: 10.1158/0008-5472.CAN-08-2268. [DOI] [PubMed] [Google Scholar]
- 11.Rafiq S, Cheney C, Mo X, Jarjoura D, Muthusamy N, Byrd JC. XmAb-5574 antibody demonstrates superior antibody dependent cellular cytotoxicity as compared to CD52 and CD20 targeted antibodies in adult acute lymphoblastic leukemia cells. Leukemia. 2012 doi: 10.1038/leu.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman EJ, Brandwein J, Stone R, Kalaycio M, Moore J, O'Connor J, et al. Phase III randomized multicenter study of a humanized anti-CD33 monoclonal antibody, lintuzumab, in combination with chemotherapy, versus chemotherapy alone in patients with refractory or first-relapsed acute myeloid leukemia. J Clin Oncol. 2005;23:4110–4116. doi: 10.1200/JCO.2005.09.133. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland MK, Yu C, Anderson M, Zeng W, van Rooijen N, Sievers EL, et al. 5-azacytidine enhances the anti-leukemic activity of lintuzumab (SGN-33) in preclinical models of acute myeloid leukemia. MAbs. 2010:2. doi: 10.4161/mabs.2.4.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland MK, Yu C, Lewis TS, Miyamoto JB, Morris-Tilden CA, Jonas M, et al. Anti-leukemic activity of lintuzumab (SGN-33) in preclinical models of acute myeloid leukemia. MAbs. 2009;1:481–490. doi: 10.4161/mabs.1.5.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung W. Use of NK cell activity in cure by transplant. Br J Haematol. 2011;155:14–29. doi: 10.1111/j.1365-2141.2011.08823.x. [DOI] [PubMed] [Google Scholar]
- 16.Rubnitz JE, Inaba H, Ribeiro RC, Pounds S, Rooney B, Bell T, et al. NKAML: a pilot study to determine the safety and feasibility of haploidentical natural killer cell transplantation in childhood acute myeloid leukemia. J Clin Oncol. 2010;28:955–959. doi: 10.1200/JCO.2009.24.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bari R, Leung M, Turner VE, Embrey C, Rooney B, Holladay M, et al. Molecular determinant-based typing of KIR alleles and KIR ligands. Clin Immunol. 2011;138:274–281. doi: 10.1016/j.clim.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the FcγRIIIa-158 V/V and V/F polymorphism. Blood. 2007;110:2561–2564. doi: 10.1182/blood-2007-01-070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asano H, Yamamoto G, Hosoi M, Takahashi T, Hangaishi A, Kurokawa M. Complete molecular remission in refractory acute myeloid leukemia with MLL/AF9 treated with gemtuzumab ozogamicin. Leuk Res. 2010;34:e152–e153. doi: 10.1016/j.leukres.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Romagne F, Andre P, Spee P, Zahn S, Anfossi N, Gauthier L, et al. Preclinical characterization of 1-7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells. Blood. 2009;114:2667–2677. doi: 10.1182/blood-2009-02-206532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.