Abstract
Introduction
Amplification of FGFR1 has been reported in squamous cell lung carcinoma and may be a molecular target for therapy. Little is known, however, about the clinical and demographic correlates of FGFR1 amplification.
Methods
The study is an institutional review board-approved retrospective analysis of 226 patients with squamous cell lung cancer seen at the Massachusetts General Hospital (MGH) from 2005–2011. Clinical and demographic characteristics were obtained on all patients, as well as treatment details including surgery, radiation, and chemotherapy, and overall survival. FISH was performed for FGFR1 on formalin fixed paraffin-embedded tumor tissue. Clinical genotyping results were also reviewed where available.
Results
37 of 226 (16%) patients with squamous cell lung cancer were positive for amplification using a definition of amplification of a gene to copy number control ratio >/= 2.2. FGFR1 amplification status was not associated with age, sex, stage, histologic subtype within squamous cell, smoking history or pack-years of smoking. We found no significant difference in overall survival by FGFR1 amplification status as a whole; in the advanced stage subset, our findings are inconclusive due to the small sample size.
Conclusions
FGFR1 amplification was found in 16% of a clinical cohort of squamous cell lung cancer patients. The lack of any specific clinicodemographic features that correlates with FGFR1 amplification suggests that all squamous cell patients should be tested for this genomic change.
Keywords: squamous cell lung cancer, FGFR1, amplification
Introduction
Lung cancer is the leading cause of cancer-related death in the United States, with over 220,000 new cases and over 157,000 deaths annually [1]. Approximately 85% of newly diagnosed lung cancers are non-small cell lung cancer (NSCLC), and of these, approximately 30% are squamous cell carcinoma. Squamous cell carcinoma and adenocarcinoma of the lung are increasingly recognized as harboring different molecular profiles [2–3] and requiring different treatment strategies [4–9]. In adenocarcinoma, effective molecularly-targeted therapies such as erlotinib and crizotinib have dramatically improved the clinical course for patients with sensitizing EGFR mutations and ALK translocations [10–15]. Further progress in NSCLC treatment will require the identification and successful targeting of molecular alterations in all subtypes of lung cancer, including squamous cell.
One potential molecular target in squamous cell lung cancer is FGFR1. Amplification at 8p12 was observed in multiple studies of squamous cell lung cancer [16–18], and FGFR1 has been identified as a potential candidate gene in this region. FGFR1 is a member of the FGFR family of receptor tyrosine kinases; activation leads to downstream signaling via the PI3K/AKT and RAS/MAPK pathways which are central to growth, survival migration and angiogenesis in many cancers. Dysregulation of FGFR family signaling has been described in multiple cancers, with amplification, translocation, and point mutations being described in a broad range of tumor types, including breast, prostate, myeloma, sarcoma, bladder, and endometrial cancers, among others [19–23].
In lung cancer, FGFR1 amplification is found in approximately 20% of squamous cell cancers, but rarely in adenocarcinoma [17]. Inhibition of FGFR1 in amplified cell lines and in mouse models with FGFR1 amplified engrafted tumors showed growth inhibition and induced apoptosis [17]. Multiple FGFR inhibitors are in development; many of these are multitargeted tyrosine kinase inhibitors with activity against other targets in addition to FGFR1.
Other than the association with squamous histology, little is known about clinical or demographic correlates of FGFR1 amplification. In this study we describe the rate of FGFR1 amplification, co-localization with other potential oncogenic changes, and clinical and demographic correlates, in our cohort of squamous cell lung cancer patients.
Materials and Methods
Patient Population
The study is an institutional review board-approved retrospective analysis of 226 patients with squamous cell lung cancer seen at the Massachusetts General Hospital (MGH) from 2005–2011. For all patients, medical records were reviewed to obtain clinical and demographic characteristics, including age, sex, stage, histology, smoking history, treatment details including surgery, radiation, and chemotherapy, and overall survival. In addition, clinical genotyping results were reviewed. Since 2009, molecular testing of tumors for genomic changes has been integrated into the MGH oncology clinic as a part of standard clinical care [2]. We employ the SNaPshot platform, a validated, CLIA-certified, multiplex tumor genotyping assay that utilizes formalin-fixed paraffin-embedded tissue to identify commonly mutated loci in many key oncogenes [2, 24–25]. FISH has been routinely performed for ALK in adenocarcinoma cases since 2009 for FGFR1 and PDGFRA in squamous cell carcinoma since 2011.
In this cohort we reviewed all patients with squamous cell lung cancer seen during the study period and included those who have been prospectively genotyped as part of their clinical care as well as those with available tissue blocks for retrospective analysis of FGFR1 amplification by FISH.
FISH for FGFR1 and PDGFR
Hematoxylin and eosin (H&E) staining was performed on 5-µm sections from formalin-fixed paraffin-embedded (FFPE) tumor tissue and areas for hybridization marked with a diamond-tipped pen. FFPE slides were de-paraffinized, treated with protease, and co-denatured with FISH probes using a Hybrite slide processor (Abbott Molecular), washed, counterstained, cover-slipped and analyzed using an Olympus BX61 fluorescence microscope equipped with orange, green, and DAPI filters. Images were captured and analyzed using Cytovision software (Genetix Inc., San Jose, CA). Positive cases were defined as tumors harboring a gene:copy number control ratio >/= 2.2 [26]. The bacterial artificial chromosome (BAC) clone CTD-2288L6 (chromosome 8p FGFR1 locus labeled orange; Invitrogen, Carlsbad, CA) was paired with a centromere 8 copy number control (CEP8 SpectrumAqua, Abbott-Vysis 06J54-018). BAC clone RP11-58C6 (chromosome 4q12 PDGFRA locus labeled orange; Invitrogen, Carlsbad, CA) was paired with a centromere 4 copy number control (CEP4 SpectrumGreen, Abbott-Vysis 06J37-014).
Statistical Considerations
Demographic and clinical information were compared across tumor genotypes using Pearson chi-squared tests (for categorical variables) and Kruskal-Wallis tests (for continuous variables), where appropriate. Standard methods for time-to-event data such as Kaplan-Meier methods and log rank test were used to compare survival outcomes by genotype. All statistical testing was done using SAS software version 9.3 (SAS Institute, Cary, NC).
Results
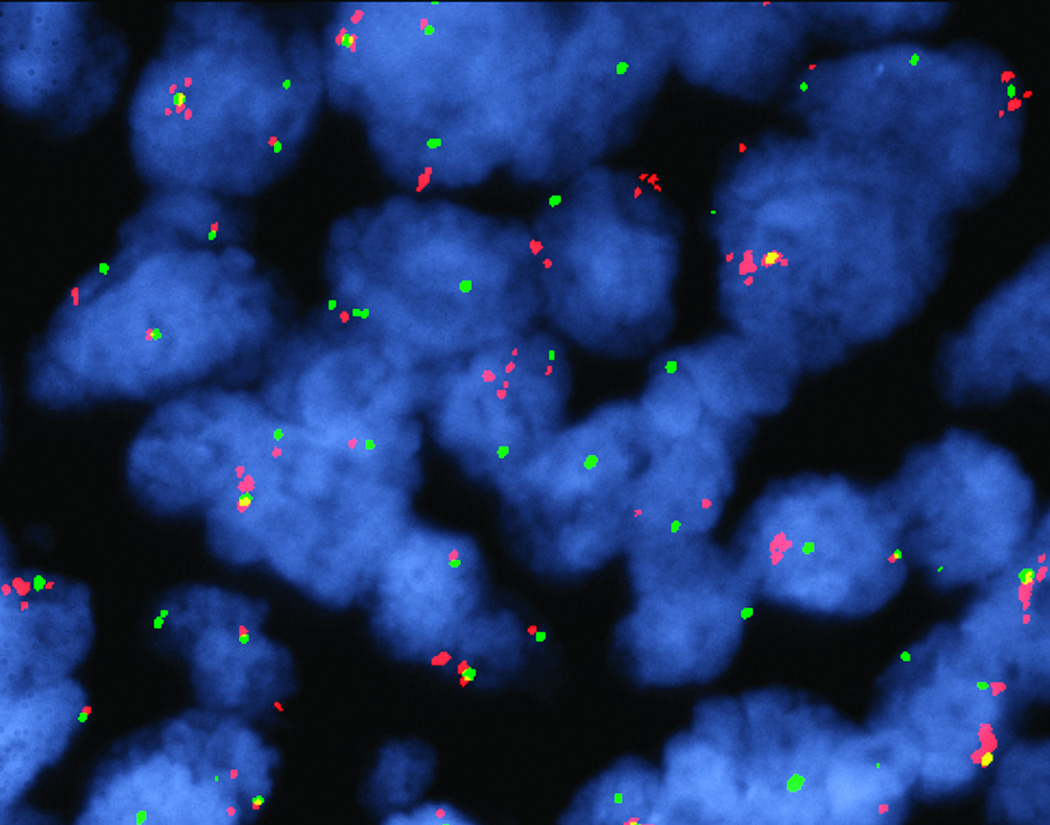
A total of 226 patients with squamous cell lung cancer were analyzed for FGFR1 amplification and 37 (16%) were positive for amplification using a definition of amplification of a gene to copy number control ratio >/= 2.2. Figure 1 shows a representative image of a tumor with FGFR1 amplification. FGFR1 amplification was a focal event, involving only a subset of tumor cells, in the majority (> 2/3) of the positive cases, in accordance with prior reports [17]. Table 1 depicts basic demographic and clinical features of the cohort. FGFR1 amplification status was not associated with age, sex, stage, histologic subtype within squamous cell, smoking history or pack-years of smoking.
Figure 1. FGFR amplification.
FISH analysis of FGFR1 demonstrates FGFR1 gene amplification in this tumor. Amplified FGFR1 appears as a cluster of isolated red signals; note that this is present in some and not all tumor cells.
Table 1.
Patient Characteristics
| Total | FGFR1 amp | FGFR not amp | ||
|---|---|---|---|---|
| N = 226 | N = 37 | N = 189 | p | |
| Age (median, range) | 69 (38–91) | 67 (39–87) | 69 (38–91) | 0.53 |
|
Gender Male Female |
128 (57%) 98 (43%) |
25 (68%) 12 (32%) |
103 (55%) 86 (45%) |
0.14 |
|
Histology Squamous Squamous with basaloid Squamous with small cell Squamous with clear cell Squamous with focal adeno component |
200 (89%) 17 (8%) 3 (1%) 3 (1%) 3 (1%) |
33 (89%) 3 (8%) 0 1 (3%) 0 |
167 (89%) 14 (7%) 3 (2%) 2 (1%) 3 (1%) |
0.77 |
|
Stage 1A 1B 2A 2B 3A 3B 4 |
57 (25%) 54 (24%) 19 (8%) 25 (11%) 34 (15%) 10 (4%) 27 (12%) |
10 (27%) 6 (16%) 6 (16%) 5 (14%) 6 (16%) 2 (5%) 2 (5%) |
47 (25%) 48 (25%) 13 (7%) 20 (11%) 28 (15%) 8 (4%) 25 (13%) |
0.39 |
|
Smoking status Neversmoker Former smoker Current smoker |
9 (4%) 172 (76%) 45 (20%) |
2 (5%) 29 (78%) 6 (16%) |
7 (4%) 143 (76%) 39 (20%) |
0.76 |
| Packyears (median, range) | 50 (0–180) | 53 (0–162) | 50 (0–180) | 0.79 |
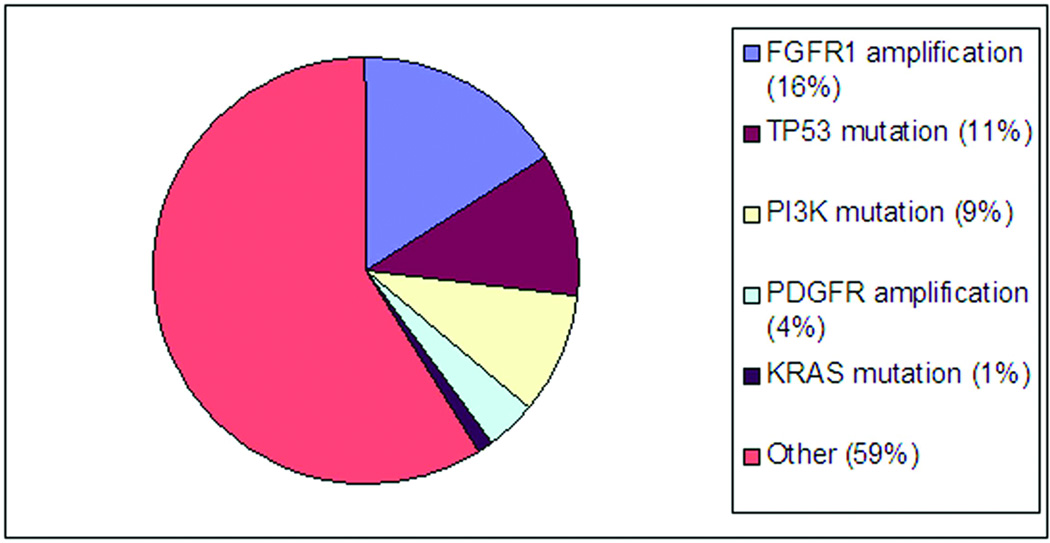
Of the 37 patients who were FGFR1 amplified, 18 also had clinical genotyping performed with SNaPshot. Three of 18 cases had p53 mutations and one had a PIK3CA mutation. It should be noted that the SNaPshot platform underestimates alterations in p53 as it tests for hotspot mutations only. Overall, 130 squamous cell lung cancer patients from the cohort had clinical genotyping performed via SNaPshot. Just as in the FGFR1-amplified group, the most common abnormality seen in the overall group was p53 mutation (n=14), followed by PIK3CA mutation (n=12). Two patients had KRAS mutations; both of these cases were squamous histology (p63+, TTF1−), with focal adenocarcinoma component on histology. PDGFRA amplification was also tested and found in 7 of 160 cases (4%), including 2 patients who had both FGFR1 and PDGFRA amplification. Figure 2 depicts the distribution of genotype findings in squamous cell carcinoma.
Figure 2. Molecular alterations in squamous cell.
*observed frequency of p53 in our cohort is likely an underestimate of true p53 alterations because of the limitations of the SNaPshot assay, which focuses on hotspot mutations
There were no statistically significant differences in overall survival by FGFR1 status in the population as a whole (log rank p = 0.36, see Table 2 and Figure 3a). This held true for those with Stage I and II squamous cell lung cancer, the vast majority of whom were treated with primary surgical resection (Figure 3b). Although patients with advanced stage (Stage 3–4) squamous cell lung cancer appeared to do better if FGFR1 amplified, the sample size is small and has a moderate rate of censoring, making definite conclusions from this observation challenging. We attempted to account for treatment heterogeneity among the group by including only those with advanced disease treated with primary platinum-doublet based chemoradiation or chemotherapy, and observed a similar trend, though again conclusions are limited due to lack of power.
Table 2.
Overall Survival by FGFR1 amplification
| FGFR1 status | Events/ Total |
Median OS (years) (95% CI) |
Log rank p |
|---|---|---|---|
| Entire cohort | |||
|
amplified not amplified |
8/37 45/189 |
5.9 (2.7 – NR) 4.6 (3.7 – 6.0) |
0.36 |
| Stage I–II | |||
|
amplified not amplified |
7/27 24/128 |
4.9 (2.7-NR) 5.6 (3.8-NR) |
0.66 |
| Stage III–IV | |||
|
amplified not amplified |
1/10 21/61 |
NR 2.5 (1.3–4.6) |
0.07 |
| Platinum chemoRT/chemo | |||
|
amplified not amplified |
0/4 11/25 |
NR 1.0 (0.7 – 4.3) |
0.09 |
NR = not reached
Figure 3.
a. Overall survival by FGFR1 amplification status
b. Overall Survival by FGFR1 amplification status in Stage 1–2
Discussion
We have reported the findings from a North American single-center series of squamous cell lung cancer patients undergoing molecular genotyping and identified a 16% rate of FGFR1 amplification. Contrary to other molecular changes in lung cancer such as EGFR mutations or ALK translocations, we did not find a distinct pattern of clinical characteristics that could help identify those likely to harbor FGFR1 amplification. There was no correlation with age or sex. In terms of smoking history, Weiss et al had previously reported that they had found FGFR1 amplification only in smokers, and not in never smokers [17]. While the majority of patients with squamous lung cancer are former or current smokers, in our cohort of patients there was a small proportion that were never smokers, and FGFR1 amplification was found among this group as well as those with a smoking history.
Similar to Weiss et al, we found concurrent p53 mutations among our patients who had FGFR1 amplification. The observed frequency of p53 in our cohort is likely an underestimate of true p53 alterations because of the limitations of the SNaPshot assay, which focuses on hotspot mutations only while a large proportion of p53 alterations are known to occur at other loci. We also found co-occurrence of FGFR1 amplification with other tumor genetic alterations, including PIK3CA mutation and PDGFRA amplification. As would be expected in squamous cell lung cancer, we found a relatively frequent occurrence of p53 and PI3K mutations in our cohort. KRAS mutation was rare and only found in cases which were squamous cell carcinoma but exhibited glandular differentiation in a small fraction (<10% of tumor cells).
We found no significant difference in overall survival by FGFR1 amplification status. This is contrary to Weiss et al, who reported a trend toward inferior survival among those with FGFR1 amplification. Of note, the lack of an OS difference remains when we stratify our population into those with early stage squamous cell lung cancer treated with surgical resection alone; here, without the confounding variables of other therapies we are likely to have the best assessment of prognostic impact. We find no evidence that FGFR1 amplification is a poor prognostic marker in patients with squamous cell lung cancer.
Our finding that FGFR1 amplified patients with advanced stage lung cancer treated with platinum doublet based chemoradiation or chemotherapy appeared to have a trend toward improved survival should be interpreted cautiously. The sample size in this series is too small to draw definitive conclusions, and larger studies or combinations of multiple institutions’ experiences will be helpful to clarify whether this is a true association.
It should be noted that FGFR1 amplification has been reported to be associated with worse clinical outcomes in other cancers. However, it remains unclear whether these studies truly reflect an association with poor prognosis or relative endocrine resistance. Amplification of 8p12 has been reported in approximately 10–20% of breast cancer [27–28]. Multiple studies have reported that FGFR1 amplification in breast cancer is associated with worse metastasis-free survival [28–29]. However, FGFR1 amplification was also found to be associated with resistance to endocrine therapy [29]. In one of the studies, all the patients whose tumors were investigated had received adjuvant tamoxifen; in the other, no clinical treatment data is reported, but given that the study comprised of a majority of ER+ localized breast cancer cases, adjuvant endocrine therapy seems likely to have been given in a significant proportion of patients. Therefore, it remains unclear whether the worse outcomes noted were due to the truly prognostic effects of FGFR1 amplification itself, or due to differences in responsiveness to adjuvant endocrine therapy. Interestingly, FGFR1 amplification may be associated with anti-androgen resistance as well. In prostate cancer, overexpression of FGFR1 has been associated with increased risk of developing castrate-resistant prostate cancer when FGFR1 expression is elevated in hormone-naïve tumors, higher levels in castrate-resistant versus hormone-naïve tumors, as well as shorter time to death in castrate resistant tumors [30].
Defining therapeutic targets for subgroups of squamous cell lung cancer patients is critical to advancing treatment of this disease. Currently, FGFR1 amplification defines the largest such subgroup, and efforts to target this population are already ongoing with some genotype-specific trials already in progress. Whether FGFR1 amplification will be a predictive marker for FGFR-targeted therapies remains to be proven. A Phase 1b trial of BGJ398, an oral pan FGFR kinase inhibitor, is underway in patients with advanced solid tumors that harbor specific FGFR abnormalities (FGFR1 or FGFR2 amplification or FGFR3 mutation). Multiple other FGFR inhibitors are in clinical development (see Table 3); many of these are multi-targeted, frequently hitting other receptors, including PDGFR and VEGFR, in addition to FGFR, and most current ongoing clinical trials are not genotype-specific [22–23]. Some kinase inhibitors such as ponatinib appear to have activity when tested against a variety of cell lines with either FGFR amplification or mutation [31]. There are also attempts to try to more selectively target FGFR while minimizing VEGFR effects [32].
Table 3.
Selected FGFR inhibitors in clinical development
| Drug | Company | Main Targets | Clinical Dev |
|---|---|---|---|
| Small molecule TKIs | |||
| E3810 | Ethical Oncology Science | FGFR, VEGFR | I |
| BGJ398 | Novartis | FGFR | I |
| TSU-68 | Taiho Pharmaceutical | FGFR, PDGFR, VEGFR | I/II |
| AZD4547 | Astrazeneca | FGFR | I/II |
| AP24534 (Ponatinib) | Ariad | FGFR, BCR-ABL | II (in CML) |
| E7080 | Eisai | FGFR, PDGFR, VEGFR | II/III |
| BMS-582,664 (Brivanib) | Bristol Myers Squibb | FGFR, VEGFR | II/III |
| TKI258 (Dovitinib) | Novartis | FGFR, PDGFR, VEGFR | II/III |
| BIBF1120 | Boehringer Ingelheim | FGFR, PDGFR, VEGFR | III |
| LY2874455 | Lilly | FGFR | n/a |
| FGFR antibodies and FGF ligand traps | |||
| R3Mab | Genentech | FGFR3-specific Ab | II |
| FP-1039 | Five Prime Therapeutics | FGF ligand trap (multiple FGFs) | I/II |
One noteworthy point is that while we identified FGFR1 amplification in 16% of our squamous cell lung cancer cases, the amplification level was generally not very high, and amplification was focal. The heterogeneity of amplification within a tumor sample, where not all tumor cells exhibit FGFR1 amplification, may suggest that targeting FGFR1 alone may not yield adequate tumor regression or control given this focality. The results of clinical trials testing FGFR1 inhibitors in amplified tumors with correlation to level and extent of amplification will be critical to further understand this issue.
Our data confirm a robust rate of FGFR1 amplification in squamous cell lung cancer patients. Further clinical investigations to target this molecularly defined subgroup of squamous cell patients will be of great interest.
Acknowledgments
Supported by: SPORE Career Development Award
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.ACS Facts and Figures [Google Scholar]
- 2.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small cell lung cancers into routine clinical practice. Ann Oncol. 2011;22(12):2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heist RS, Sequist LV, Engelman JA. Genetic Changes in Squamous Cell Lung Cancer: A Review. JTO. 2012;7(5):924–933. doi: 10.1097/JTO.0b013e31824cc334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandler A, Gray R, Perry MC, et al. Paclitaxel-carbolatin alone or with bevacizumab for non-small cell lung cancer. NEJM. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 5.Hainsworth JD, Fang L, Huang JE, et al. BRIDGE : An open label Phase II trial evaluating the safety of bevacizumab + carboplatin/paclitaxel as first-line treatment for patients with advanced, previously untreated, squamous non-small cell lung cancer. JTO. 2011;6(1):109–114. doi: 10.1097/JTO.0b013e3181f94ad4. [DOI] [PubMed] [Google Scholar]
- 6.Scagliotti G, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naïve patients with advanced stage non-small cell lung cancer. JCO. 2008;26:3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 7.Hanna N, Sheperd FA, Fossella FV, et al. Randomized Phase III trial of pemetrexed versus docetaxel in patients with non-small cell lung cancer previously treated with chemotherapy. JCO. 2004;22:1589–1597. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 8.Cieuleanu T, Brodowicz T, Zielinski C, et al. Maintenance pemetrexed plus best supportive care versus placebo plus best supportive care for non-small cell lung cancer : a randomised, double-blind, phase 3 study. Lancet. 2009;374:1432–1440. doi: 10.1016/S0140-6736(09)61497-5. [DOI] [PubMed] [Google Scholar]
- 9.Scagliotti G, Hanna N, Fossella F, et al. The differential efficacy of pemetrexed according to NSCLC histology : a review of two phase III studies. Oncologist. 2009;14:253–263. doi: 10.1634/theoncologist.2008-0232. [DOI] [PubMed] [Google Scholar]
- 10.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small cell lung cancer to gefitinib. NEJM. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 11.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2005;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. PNAS. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. NEJM. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 14.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small cell lung cancer. Nature. 2007;448:561–567. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 15.Kwak EL, Bang YJ, Camidge R, et al. Anaplastic lymphoma kinase inhibition in non-small cell lung cancer. NEJM. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage survival oncogene in lung and esophageal squamous cell carcinomas. Nature Genetics. 2009;41(11):1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Science Translational Medicine. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nature Reviews Neuroscience. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- 20.Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res. 2010;8:1439–1452. doi: 10.1158/1541-7786.MCR-10-0168. [DOI] [PubMed] [Google Scholar]
- 21.Acevedo VD, Ittman M, Spencer DM. Paths of FGFR-driven tumorigenesis. Cell Cycle. 2009;8:580–588. doi: 10.4161/cc.8.4.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner N, Grose R. Fibroblast growth factor signaling: from development to cancer. Nature Reviews. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 23.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochemo J. 2011;437:199–212. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 24.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumors: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;(2):146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pao W, Kris MG, Iafrate AJ, et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009;15(17):5317–5322. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 26.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 27.Courjal F, Cuny M, Simony-Lafontaine J, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypical groups. Cancer Res. 1997;57:4360–4367. [PubMed] [Google Scholar]
- 28.Letessier A, Sircoulomb F, Ginestier C, et al. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer. 2006;6:245. doi: 10.1186/1471-2407-6-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70(5):2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong K, Ahamad I, Kalna G, et al. Upregulated FGFR1 expression is associated with the transition of hormone-naïve to castrate-resistant prostate cancer. B J Cancer. 2011;105:1362–1369. doi: 10.1038/bjc.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gozgit JM, Wong MJ, Moran L, et al. Ponatinib (AP24523), a multi-targeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Therapeutics. 2012 doi: 10.1158/1535-7163.MCT-11-0450. epub. [DOI] [PubMed] [Google Scholar]
- 32.Zhao G, Li W, Chen D, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10:2200–2210. doi: 10.1158/1535-7163.MCT-11-0306. [DOI] [PubMed] [Google Scholar]