Abstract
The Comparative Toxicogenomics Database (CTD) contains manually curated literature that describes chemical–gene interactions, chemical–disease relationships and gene–disease relationships. Finding articles containing this information is the first and an important step to assist manual curation efficiency. However, the complex nature of named entities and their relationships make it challenging to choose relevant articles. In this article, we introduce a machine learning framework for prioritizing CTD-relevant articles based on our prior system for the protein–protein interaction article classification task in BioCreative III. To address new challenges in the CTD task, we explore a new entity identification method for genes, chemicals and diseases. In addition, latent topics are analyzed and used as a feature type to overcome the small size of the training set. Applied to the BioCreative 2012 Triage dataset, our method achieved 0.8030 mean average precision (MAP) in the official runs, resulting in the top MAP system among participants. Integrated with PubTator, a Web interface for annotating biomedical literature, the proposed system also received a positive review from the CTD curation team.
Background
The Comparative Toxicogenomics Database (CTD) is a publicly available resource that manually curates a triad of chemical–gene, chemical–disease and gene–disease relationships from biomedical literature (1). Although previous tasks in the BioCreative competition were focused on gene/protein name tagging and protein–protein interactions (PPIs) (2,3), this new task addresses the problem of finding articles that include the triad of three entities: gene, chemical and disease that have important relationships (4). One can expect that effective approaches to this task will be beneficial for manual curation in CTD. Compared with previous BioCreative tasks, the CTD Triage task has the following differences: (i) target chemicals are explicitly given for training and test sets; (ii) entities to be identified are chemical, gene and disease names and (iii) the available training set is quite limited.In the BioCreative PPI article classification tasks (ACTs), protein names of interest were not given as parameters of the search. However, the CTD dataset consists of multiple groups categorized by their target chemicals, that is, a set of documents includes entity–entity relationship information relevant to a specific chemical name. Ideally, one can extract an entity–entity relationship directly from text and use this information for deciding whether an article is of interest, but this is impossible for a system without the relation extraction capability.
The second problem is that chemical and disease mentions should be identified along with gene mentions. Named entity recognition (NER) has been a main research topic for a long time in the biomedical text-mining community. The common strategy for NER is either to apply certain rules based on dictionaries and natural language processing techniques (5–7) or to apply machine learning approaches such as support vector machines (SVMs) and conditional random fields (8–10). However, most NER systems are class specific, i.e. they are designed to find only objects of one particular class or set of classes (11). This is natural because chemical, gene and disease names have specialized terminologies and complex naming conventions. In particular, gene names are difficult to detect because of synonyms, homonyms, abbreviations and ambiguities (12,13). Moreover, there are no specific rules of how to name a gene that are actually followed in practice (14). Chemicals have systematic naming conventions, but finding chemical names from text is still not easy because there are various ways to express chemicals (15,16). For example, they can be mentioned as IUPAC names, brand names, generic names or even molecular formulas. However, disease names in literature are more standardized (17) compared with gene and chemical names. Hence, using terminological resources such as Medical Subject Headings (MeSH) and Unified Medical Language System (UMLS) Metathesaurus help boost the identification performance (17,18). But, a major drawback of identifying disease names from text is that they often use general English terms.
The last problem in the Triage task is that the size of the training set is relatively small. For the Triage task, the numbers of positive and negative examples are only 1031 and 694, respectively. This is much smaller than the 20 000 training documents available for PPI ACTs. The small dataset is especially critical for data-driven systems utilizing machine learning methods.
Here, we assume the Triage task is an extension of the BioCreative III ACT, where PPI information is the only concern for prioritizing PubMed documents. Because both tasks are data driven and their goals are to find interaction information among specific entities, we basically follow the same framework (19,20) developed for ACT. However, new issues in the Triage task are addressed by changing feature types and entity recognition approaches. We first assume that target chemicals can be mined through machine learning procedures if we seed correct features from PubMed, for example, MeSH and substance fields in PubMed citations. This is based on the fact that major topics are likely to appear in those fields. Second, a Semantic Model is introduced to identify multiple entities simultaneously. The Semantic Model obtains semantic relationships from PubMed and the UMLS semantic categories and other sources (21). Assuming the evidence describing entity–entity relationships can be found from multiple sentences, this new approach provides a simple way to determine relevant sentences. Third, latent topics are analyzed using Latent Dirichlet Allocation (LDA) (22) and used as a new feature type. The small number of training examples is not trivial for machine learning and, in particular, is harder to handle in a sparse data type such as text documents. The LDA method provides a semantic view of what is latent or hidden in text and enriches features for better separation between positive and negative examples.
In the official runs, our updated method achieved 0.857, 0.824 and 0.728 average precision scores for ‘cyclophosphamide’, ‘phenacetin’ and ‘urethane’ test sets, respectively, which allowed our system to be a top performer (23). This prioritization scheme was also integrated with a Web interface, PubTator (24,25), for potentially assisting curators and received a positive review from the CTD curation team (23).
Materials and methods
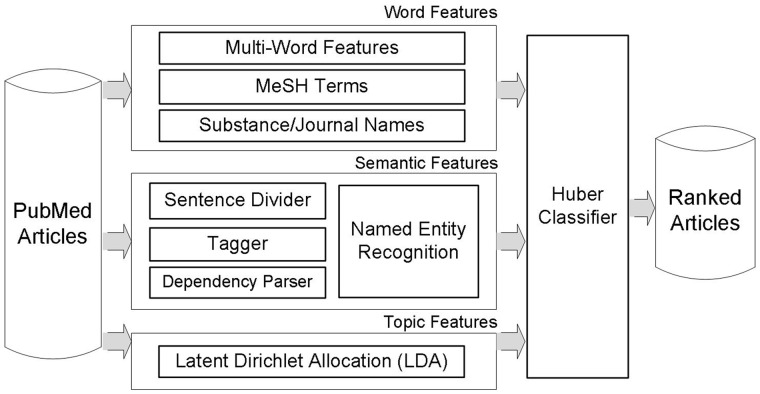
Figure 1 depicts the overview of our article prioritization method. For input articles, features are extracted in three different ways. One is word features including multiwords, MeSH terms and substance/journal names. The second is syntactic features based on dependency relationships between words. The third is topic features obtained from LDA. After feature extraction procedures, a large margin classifier with Huber loss function (26) is utilized for learning and prioritizing articles. The following subsections describe these feature types.
Figure 1.
Our article prioritization method for the BioCreative 2012 Triage task. For input articles, features are generated in three different ways: word features including multiwords, MeSH terms and substance/journal names; semantic features utilizing dependency relations and a Semantic Model; topic features are extracted by LDA topic modeling.
Word features from PubMed
Multiwords are known as n-grams, where n consecutive words are considered as features. Here, we use unigrams and bigrams from titles and abstracts. MeSH is a controlled vocabulary for indexing and searching biomedical literature. These terms are included as features because MeSH terms are used to indicate the topics of an article. In detail, MeSH terms are also handled as unigrams and bigrams. In the Triage task, target chemicals are designated for a set of articles, and journals are treated differently in the CTD rule-based system (http://www.biocreative.org/tasks/bc-workshop-2012/Triage). Therefore, substances and journal names are extracted from PubMed and used as word features.
Semantic features for identifying entity relationships
This feature identifies interactions or relationships among entities by syntactically analyzing sentences. By using a dependency parser (27), a head word and a dependent word are determined as a two-word combination. Because our goal is to find relationships between two entities, any words indicating relations are likely placed in the head position, whereas their corresponding entities will be placed in the dependent position. Thus, we only consider dependent words as candidate entities. For example, verbs and conjunctions are removed from this process.
For the NER method, we use a vector space approach to modeling semantics (28) and compute our vectors as described in (29) except we ignore the actual mutual information and just include a component of 1 if the dependency relation occurs at all for a word, else the component is set to 0. We constructed our vector space from all single tokens (a token must have an alphabetic character) throughout the titles and abstracts of the records in the whole of the PubMed database based on a snapshot of the database taken in January 2012. We included only tokens that occurred in the data sufficient to accumulate 10 or more dependency relations. There were just over 750 000 token types that satisfied this condition and are represented in the space. We then took all the single tokens and all head words from multitoken strings in the categories ‘chemical’, ‘disease’ and ‘gene’ from an updated version of the SemCat database (21) and placed all the other SemCat categories similarly processed into a category ‘other’. We considered only the tokens in these categories that also occurred in our semantic vector space and applied SVM learning to the four resulting disjoint semantic classes in a one-against-all strategy to learn how to classify into the different classes.
The Semantic Model is an efficient and general way to identify words indicating an entity type. Unlike other NER approaches, this model decides a target class solely based on a single word. However, evaluating only single tokens may increase false positives. To overcome this pitfall, we assume that a relevant document mentions entity–entity relationships multiple times at the sentence level. Hence, if two different entity types are found in a sentence, we assume this sentence includes an entity–entity relationship. By counting the number of entity–entity relationship sentences, c, discretized numbers are obtained as follows: 1 for c < 2, 2 for c = 2, 3 for c = 3, 4 for c = 4 and 5 for c > 4. These numbers are then used as nominal features.
Topic features
Along with semantic features, topic features are newly added to address the Triage problem. LDA is a generative probabilistic model in which documents are represented as random mixtures over latent topics, and each topic is characterized by a distribution over words (22). There is some evidence that LDA topics can provide features with better generalization properties when there is little training data (30). We pooled the whole CTD (http://ctdbase.org) and the Triage training set. In our application of LDA, we used the model as put forward in (22) and calculated the model using Markov Chain Monte Carlo simulation as described in (31). For LDA topic modeling, we took the parameters based on the setting used in (31) as follows:
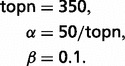 |
Here, ‘topn’ is the number of topics, α is the Dirichlet prior on topic distributions, and β is the Dirichlet prior on word distributions. The small value of β is chosen so that these topics are well filled. This choice of β and number of topics seemed to produce topics of the right size to make useful features for the classification problem we are dealing with. A larger choice of β tended to produce many sparse topics and a few that contained most of the terminology.
Huber classifiers
The Huber classifier (32) is a variant of SVM. This method determines feature weights that minimize the modified Huber loss function (26), which is a function that replaces the hinge loss function commonly used in SVM learning.
Let T denote the size of the training set, the binary feature vector of the ith pair in the training set be denoted by Xi, yi = 1 if the pair is annotated as positive and yi = −1 otherwise, w denote a vector of feature weights, of the same length as Xi, θ denote a threshold parameter and λ denote a regularization parameter. Then the cost function is given by
where the function h is the modified Huber loss function. The values of the parameters, w and θ minimizing C are determined using a gradient descent algorithm. The regularization parameter λ is computed from the training set as follows:
where  is the average Euclidean norm of the feature vectors in the training set. The parameter
is the average Euclidean norm of the feature vectors in the training set. The parameter 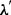 was tuned to maximize average precisions for the CTD Triage training set, and it was set to 0.0001 for official runs.
was tuned to maximize average precisions for the CTD Triage training set, and it was set to 0.0001 for official runs.
Entity annotation and user interface
As a requirement for the Triage task, chemical, gene and disease actors should be annotated for result submission. Although entity annotation can be combined with an article prioritization method, our approach does not use fully annotated names for genes, chemicals and diseases. As mentioned earlier, the proposed method makes its decision based on the features of single words obtained from dependency parsing. As a result, we currently cannot obtain gene/chemical/disease actors directly from the proposed system. However, our experimental setup makes individual processes independent. Thus, each module can be replaced with other similar approaches as desired. This applies to our feature selection, machine learning classifiers and even entity/actor annotations.
Because official runs should be submitted with actor information as well as prioritized articles, we used PubTator (24) for annotating entities and providing a Web interface for the Triage task. PubTator is a Web-based tool that is developed for creating, saving and exporting annotations. PubTator was customized to have a tailored output for combining the results of article ranking and bioconcept annotation. The CTD curation team also rated this Web interface outstanding (23).
Results and discussion
Dataset
The CTD Triage set is categorized by 11 target chemicals, which contain ‘2-acetylaminofluorene’, ‘amsacrine’, ‘aniline’, ‘aspartame’, ‘doxorubicin’, ‘indomethacin’, ‘quercetin’ and ‘raloxifene’ for training and ‘cyclophosphamide’, ‘phenacetin’ and ‘urethane’ for testing. Even though the total number of documents is 1725 (1031 positives and 694 negatives), each subset has a different ratio in the number of positive and negative examples. In this setup, it is not easy to tune a data-driven system for addressing both balanced and unbalanced datasets. Thus, we optimize our system to achieve the best performance on averaged ranking scores, i.e. for each run, the proposed system is trained by using seven target chemicals in turn and the eighth is used for testing. The parameters are tuned to obtain the best MAP (Mean Average Precision) as an average for the eight runs. Mean Average Precision (MAP) is the mean of average precision scores. For a given ranking, the average precision is the average of all precisions computed at ranks containing relevant documents. Higher MAP scores indicate a system places more relevant documents in top ranks. Table 1 shows the target chemicals and the number of positive and negative examples in the CTD Triage set. Note that the three test chemicals shown in the table were not known during the system development period.
Table 1.
Dataset
| Dataset | Chemical names | Positives | Negatives | Total |
|---|---|---|---|---|
| Training | 2-Acetylaminofluorene | 81 | 97 | 178 |
| Amsacrine | 37 | 32 | 69 | |
| Aniline | 100 | 126 | 226 | |
| Aspartame | 46 | 110 | 156 | |
| Doxorubicin | 138 | 61 | 199 | |
| Indomethacin | 76 | 9 | 85 | |
| Quercetin | 392 | 150 | 542 | |
| Raloxifene | 161 | 109 | 270 | |
| Test | Cyclophosphamide | 107 | 47 | 154 |
| Phenacetin | 65 | 21 | 86 | |
| Urethane | 106 | 98 | 204 |
The training and test sets include eight and three target chemicals, respectively. Because the ratio of positive and negative examples varies with target chemicals, our system is tuned to achieve high MAP score on the training chemicals.
Utilizing semantic and topic features
The proposed method in the Triage task includes new feature types: semantic and topic features. The semantic feature utilizes a new NER scheme termed a Semantic Model, and the topic feature uses LDA for obtaining latent topics.
The Semantic Model classifies single words to ‘gene’, ‘chemical’, ‘disease’ or ‘other’. Table 2 presents the number of strings in each class and the NER performance on the four different classes. From a 10-fold cross-validation, the Semantic Model produces 0.914, 0.868, 0.706 and 0.912 MAP scores for ‘gene’, ‘chemical’, ‘disease’ and ‘other’, respectively. This does not mean the Semantic Model can produce a good performance in general; however, it shows that the Semantic Model has a reasonably good discriminative power on this four-class dataset. Although this procedure is efficient for identifying multiple entities in text, it may produce incorrect predictions even with our assumption that a positive document has multiple evidences at the sentence level. For this reason, it is important to include the other features that we consider to obtain good triage performance.
Table 2.
Semantic classes and the classification performance for the semantic model
| Class name | Gene | Chemical | Disease | Other |
|---|---|---|---|---|
| Number of strings | 70 832 | 49 800 | 7589 | 113 815 |
| Mean average precision | 0.914 | 0.868 | 0.706 | 0.912 |
The second row contains the number of unique strings in the four different classes. The last row shows the MAP scores from a 10-fold cross-validation to learn how to distinguish each class from the union of the other three.
Tables 3 and 4 show the average precision changes when semantic and topic features are added to word features. ‘BASE’ means word features without substance and journal names from PubMed. ‘IXN’ and ‘TOPIC’ mean semantic and topic features, respectively. All feature combinations in the tables use the ‘BASE’ feature type, but add ‘IXN’ and ‘TOPIC’ alternatively. The difference between Tables 3 and 4 is whether the full CTD set is used to augment training. All PubMed IDs were downloaded from the CTD database and used as positives. Due to some duplicates, PubMed IDs appeared in both training and testing are removed from the training set. From the averaged ranking performance, it is difficult to say which feature type contributes more. Table 3 shows more performance improvement when semantic features are used. In Table 4, adding topic feature provides better performance improvement. However, these two feature types are important because the ranking performance reaches top scores only when both features are used.
Table 3.
Average precision changes with Triage (training) + Triage (testing)
| Chemical names | BASE | IXN | TOPIC | IXN + TOPIC |
|---|---|---|---|---|
| 2-Acetylaminofluorene | 0.6702 | 0.6742 | 0.6969 | 0.6956 |
| Amsacrine | 0.6980 | 0.6956 | 0.6773 | 0.6848 |
| Aniline | 0.7765 | 0.7891 | 0.7887 | 0.8006 |
| Aspartame | 0.4845 | 0.5096 | 0.4687 | 0.4859 |
| Doxorubicin | 0.8610 | 0.8627 | 0.8690 | 0.8689 |
| Indomethacin | 0.9758 | 0.9766 | 0.9748 | 0.9751 |
| Quercetin | 0.9315 | 0.9313 | 0.9310 | 0.9313 |
| Raloxifene | 0.8060 | 0.8107 | 0.8152 | 0.8191 |
| Average performance | 0.7754 | 0.7812 | 0.7777 | 0.7827 |
The Triage dataset is used for training and testing in a leave-one (chemical)-out approach. ‘BASE’ means word features without substance/journal names. ‘IXN’ and ‘TOPIC’ mean semantic and topic features, respectively. ‘BASE’ features are used for all the experiments.
Table 4.
Average precision changes with CTD (training) + Triage (testing)
| Chemical names | BASE | IXN | TOPIC | IXN + TOPIC |
|---|---|---|---|---|
| 2-Acetylaminofluorene | 0.6776 | 0.6776 | 0.6814 | 0.7096 |
| Amsacrine | 0.7202 | 0.7308 | 0.7468 | 0.7577 |
| Aniline | 0.7625 | 0.7542 | 0.7477 | 0.7677 |
| Aspartame | 0.4902 | 0.4958 | 0.5269 | 0.5388 |
| Doxorubicin | 0.8767 | 0.8828 | 0.8871 | 0.8937 |
| Indomethacin | 0.9608 | 0.9610 | 0.9621 | 0.9604 |
| Quercetin | 0.9186 | 0.9190 | 0.9162 | 0.9189 |
| Raloxifene | 0.7820 | 0.7803 | 0.7737 | 0.7661 |
| Average performance | 0.7736 | 0.7752 | 0.7802 | 0.7891 |
Again a leave-one-out train and test procedure is followed. The full dataset was downloaded from the CTD database and used to augment the training. Any duplicates appearing in both training and testing sets were removed from the training set. ‘BASE’ uses word features without substance/journal names. ‘IXN’ and ‘TOPIC’ mean semantic and topic features, respectively. ‘BASE’ features are used for all the experiments.
Table 5 shows overall performance changes for different dataset, feature and classifier combinations. The last column is the configuration we used for the official run. Compared with Bayes classifiers (first column), the proposed method improves average precisions up to 5% on average. Note that test examples were always excluded from the training set in both ‘Triage’ and ‘CTD’ experiments. ‘All Proposed Features’ in Table 5 includes the substance/journal name features, and this accounts for the improvements seen over Table 4 results.
Table 5.
Overall performance (average precision) changes for different dataset, feature and classifier combinations
| Training set | Triage |
CTD |
||
|---|---|---|---|---|
| Feature | Multiword features |
All proposed features |
||
| Classifier | Bayes | Huber | Huber | Huber |
| 2-Acetylaminofluorene | 0.7151 | 0.6812 | 0.7055 | 0.6932 |
| Amsacrine | 0.5880 | 0.6676 | 0.6850 | 0.7411 |
| Aniline | 0.7589 | 0.7646 | 0.8000 | 0.7708 |
| Aspartame | 0.3755 | 0.4520 | 0.4890 | 0.5902 |
| Doxorubicin | 0.8434 | 0.8718 | 0.8689 | 0.8895 |
| Indomethacin | 0.9599 | 0.9699 | 0.9761 | 0.9626 |
| Quercetin | 0.9068 | 0.9176 | 0.9321 | 0.9227 |
| Raloxifene | 0.7913 | 0.7940 | 0.8175 | 0.7759 |
| Average performance | 0.7424 | 0.7648 | 0.7843 | 0.7933 |
‘Triage’ means the Triage training set is used for training. ‘CTD’ means the full CTD set is used to augment the positive set and negatives are from the Triage set. Again a leave-one-out train and test scenario are used. ‘Bayes’ and ‘Huber’ indicate Bayes and Huber classifiers, respectively.
Official performance on the Triage test set
For the official run, we trained the proposed system by enriching positive examples from the CTD database. Even though the prediction in this setup favors the positive label more, it improves ranking performance. Table 6 presents the performance on the official Triage test data. Our method obtained 0.857, 0.824 and 0.728 MAP scores for ‘cyclophosphamide’, ‘phenacetin’ and ‘urethane’, respectively. Because our system produces only a ranking result, the gene, chemical and disease name detection was performed by PubTator. For entity recognition, PubTator also produced a good result by obtaining 0.426, 0.647 and 0.456 hit rates for gene, chemical and disease names, respectively.
Table 6.
Official performance on the Triage test set
| Chemical names | AP | Hit rate |
||
|---|---|---|---|---|
| Gene | Chemical | Disease | ||
| Cyclophosphamide | 0.857 | 0.339 | 0.593 | 0.646 |
| Phenacetin | 0.824 | 0.627 | 0.667 | 0.333 |
| Urethane | 0.728 | 0.311 | 0.681 | 0.389 |
| Average performance | 0.803 | 0.426 | 0.647 | 0.456 |
AP, average precision. ‘Hit Rate’ is the fraction of extracted terms that are matched with manually curated entities (precision).
Table 7 shows the MAP scores for top-ranking teams (23). Team 130 basically uses co-occurrences between entities, which concept is similar to our semantic features. Team 133 applies a simple strategy utilizing a number of entities and a number of sentences in a document. From these results, it is clear that relation extraction is not necessary to achieve high MAP scores. The effectiveness of using co-occurrence between entities, however, needs to be explored more because not all teams using co-occurrence obtained high MAP scores in BioCreative 2012. Even though the top three teams achieved the best score on different target chemicals, our method produced the best overall score on test set. The average performances of over all participants were 0.7617, 0.8171 and 0.6649 for ‘cyclophosphamide’, ‘phenacetin’ and ‘urethane’, respectively.
Table 7.
Average precision comparison among top MAP scoring teams
| Chemical names | Teams |
||
|---|---|---|---|
| Our team | Team 130 | Team 133 | |
| Cyclophosphamide | 0.8570 | 0.7740 | 0.7220 |
| Phenacetin | 0.8240 | 0.8020 | 0.8750 |
| Urethane | 0.7280 | 0.7600 | 0.6660 |
| Mean average precision | 0.8030 | 0.7787 | 0.7543 |
Team 130 uses co-occurrences between entities and their network centralities for document ranking. Team 133 uses document scores obtained from entity frequencies and the number of sentences for ranking. The average performance over all participants was 0.7617, 0.8171 and 0.6649 for ‘cyclophosphamide’, ‘phenacetin’ and ‘urethane’, respectively.
Conclusions
Here, we present our updated system framework for the CTD Triage task. The Triage task is a newly introduced topic, where documents should be prioritized in terms of chemical–gene interactions, chemical–disease relationships and gene–disease relationships. This task is especially challenging because of multiple entities and the small number of training examples. To tackle these issues, a semantic model is used to obtain semantic features and LDA is used to produce latent topics. Applied to the Triage test set, our official run ranked the first in MAP score. A customized interface using PubTator also received a positive review by achieving the second ranking performance on NER.
Even though the current setup provides good performance on article prioritization and entity recognition, there are still some difficulties to be overcome. Our Semantic Model does not produce fully annotated predictions for gene, chemical and disease names. As in BioCreative III, we also found that accurate NER is a critical component for this Triage task. Therefore, an integrated solution for finding relevant articles and identifying full entity names is an important subject for future research. For topic features, the number of topics is manually chosen considering the size of the dataset. However, it would be desirable to have a systematic way to automatically assign the number of topics.
Funding
Funding for open access charge: The Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest. None declared.
References
- 1.Davis AP, King BL, Mockus S, et al. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 2011;39:D1067–D1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krallinger M, Morgan A, Smith L, et al. Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge. Genome Biol. 2008;9(Suppl. 2):S1. doi: 10.1186/gb-2008-9-s2-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arighi CN, Lu Z, Krallinger M, et al. Overview of the BioCreative III Workshop. BMC Bioinformatics. 2011;12(Suppl. 8):S1. doi: 10.1186/1471-2105-12-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiegers TC, Davis AP, Mattingly CJ. Collaborative biocuration-text mining development task for document prioritization for curation. Database. 2012;2012 doi: 10.1093/database/bas037. doi:10.1093/database/bas037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuason O, Chen L, Liu H, et al. Biological nomenclatures: a source of lexical knowledge and ambiguity. Pac. Symp. Biocomput. 2004:238–249. doi: 10.1142/9789812704856_0023. [DOI] [PubMed] [Google Scholar]
- 6.Ananiadou S, Sullivan D, Black W, et al. Named entity recognition for bacterial Type IV secretion systems. PLoS One. 2011;6:e14780. doi: 10.1371/journal.pone.0014780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen QL, Tikk D, Leser U. Simple tricks for improving pattern-based information extraction from the biomedical literature. J. Biomed. Semantics. 2010;1:9. doi: 10.1186/2041-1480-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitsumori T, Fation S, Murata M, et al. Gene/protein name recognition based on support vector machine using dictionary as features. BMC Bioinformatics. 2005;6(Suppl. 1):S8. doi: 10.1186/1471-2105-6-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Lin H, Li Y. Exploiting the contextual cues for bio-entity name recognition in biomedical literature. J. Biomed. Inform. 2008;41:580–587. doi: 10.1016/j.jbi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac. Symp. Biocomput. 2008:652–663. [PubMed] [Google Scholar]
- 11.Leser U, Hakenberg J. What makes a gene name? Named entity recognition in the biomedical literature. Brief. Bioinform. 2005;6:357–369. doi: 10.1093/bib/6.4.357. [DOI] [PubMed] [Google Scholar]
- 12.Alako BT, Veldhoven A, van Baal S, et al. CoPub Mapper: mining MEDLINE based on search term co-publication. BMC Bioinformatics. 2005;6:51. doi: 10.1186/1471-2105-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frisch M, Klocke B, Haltmeier M, et al. LitInspector: literature and signal transduction pathway mining in PubMed abstracts. Nucleic Acids Res. 2009;37:W135–W140. doi: 10.1093/nar/gkp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschman L, Morgan AA, Yeh AS. Rutabaga by any other name: extracting biological names. J. Biomed. Inform. 2002;35:247–259. doi: 10.1016/s1532-0464(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 15.Rocktaschel T, Weidlich M, Leser U. ChemSpot: a hybrid system for chemical named entity recognition. Bioinformatics. 2012;28:1633–1640. doi: 10.1093/bioinformatics/bts183. [DOI] [PubMed] [Google Scholar]
- 16.Klinger R, Kolarik C, Fluck J, et al. Detection of IUPAC and IUPAC-like chemical names. Bioinformatics. 2008;24:i268–i276. doi: 10.1093/bioinformatics/btn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimeno A, Jimenez-Ruiz E, Lee V, et al. Assessment of disease named entity recognition on a corpus of annotated sentences. BMC Bioinformatics. 2008;9(Suppl. 3):S3. doi: 10.1186/1471-2105-9-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury MFM, Lavelli A. Proceedings of the 2010 Workshop on Biomedical Natural Language Processing. Uppsala, Sweden: Association for Computational Linguistics; 2010. Disease mention recognition with specific features; pp. 83–90. [Google Scholar]
- 19.Kim S, Wilbur WJ. Classifying protein-protein interaction articles using word and syntactic features. BMC Bioinformatics. 2011;12(Suppl. 8):S9. doi: 10.1186/1471-2105-12-S8-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Kwon D, Shin SY, et al. PIE the search: searching PubMed literature for protein interaction information. Bioinformatics. 2012;28:597–598. doi: 10.1093/bioinformatics/btr702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanabe L, Thom LH, Matten W, et al. SemCat: semantically categorized entities for genomics. AMIA Annu. Symp. Proc. 2006:754–758. [PMC free article] [PubMed] [Google Scholar]
- 22.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J. Mach. Learn. Res. 2003;3:993–1022. [Google Scholar]
- 23.Wiegers TC, Davis AP, Mattingly CJ. 2012 BioCreative Workshop. Washington, DC: 2012. Collaborative biocuration-text mining development task for document prioritization for curation; pp. 2–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei C-H, Kao H-Y, Lu Z. 2012 BioCreative Workshop. Washington, DC: 2012. PubTator: A PubMed-like interactive curation system for document triage and literature curation; pp. 145–150. [Google Scholar]
- 25.Wei C-H, Harris BR, Li D, et al. Accelerating literature curation with text mining tools: a case study of using PubTator to curate genes in PubMed abstracts. Database. 2012;2012 doi: 10.1093/database/bas041. doi:10.1093/database/bas041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T. Proceedings of the Twenty-First International Conference on Machine Learning. Canada: ACM, Banff, Alberta; 2004. Solving large scale linear prediction problems using stochastic gradient descent algorithms; pp. 919–926. [Google Scholar]
- 27.Curran JR, Clark S, Bos J. Proceedings of the 45th Annual Meeting of the ACL on Interactive Poster and Demonstration Sessions. Prague, Czech Republic: Association for Computational Linguistics; 2007. Linguistically motivated large-scale NLP with C&C and boxer; pp. 33–36. [Google Scholar]
- 28.Turney PD, Pantel P. From frequency to meaning: vector space models of semantics. J. Artif. Intell. Res. 2010;37:141–188. [Google Scholar]
- 29.Pantel P, Lin D. Proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Alberta, Canada: ACM, Edmonton; 2002. Discovering word senses from text; pp. 613–619. [Google Scholar]
- 30.Halpern Y, Horng S, Nathanson LA, et al. NIPS 2011 Workshop on from Statistical Genetics to Predictive Models in Personalized Medicine. Spain: Sierra Nevada; 2011. Patient surveillance algorithms for the emergency department. [Google Scholar]
- 31.Griffiths TL, Steyvers M. Finding scientific topics. Proc. Natl Acad. Sci. USA. 2004;101:5228–5235. doi: 10.1073/pnas.0307752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith LH, Wilbur WJ. Finding related sentence pairs in MEDLINE. Inf. Retr. 2010;13:601–617. doi: 10.1007/s10791-010-9126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]