SUMMARY
It remains unclear how cancer cells coordinate glycolysis and biosynthesis to support rapidly growing tumors. We found that glycolytic enzyme phosphoglycerate mutase 1 (PGAM1), commonly upregulated in human cancers due to loss of TP53, contributes to biosynthesis regulation in part by controlling intracellular levels of its substrate 3-phosphoglycerate (3-PG) and product 2-phosphoglycerate (2-PG). 3-PG binds to and inhibits 6-phosphogluconate dehydrogenase in the oxidative pentose phosphate pathway (PPP), while 2-PG activates 3-phosphoglycerate dehydrogenase to provide feedback control of 3-PG levels. Inhibition of PGAM1 by shRNA or a small molecule inhibitor PGMI-004A results in increased 3-PG and decreased 2-PG levels in cancer cells, leading to significantly decreased glycolysis, PPP flux and biosynthesis, as well as attenuated cell proliferation and tumor growth.
INTRODUCTION
The Warburg effect in cancer cells consists of an increase in aerobic glycolysis and enhanced lactate production, which generates more ATPs more quickly than in normal cells that overwhelmingly rely on oxidative phosphorylation (Kroemer and Pouyssegur, 2008). In addition, tumor tissue traps more glucose than normal tissue does, as cancer cells use elevated amounts of glucose as a carbon source for anabolic biosynthesis of macromolecules. These include nucleotides, amino acids and fatty acids, to produce RNA/DNA, proteins and lipids, respectively, which are necessary for cell proliferation and to fulfill the request of the rapidly growing tumors (Kroemer and Pouyssegur, 2008). Interestingly, leukemia cells are also highly glycolytic (Elstrom et al., 2004; Gottschalk et al., 2004), despite the fact that such cells reside within the bloodstream at higher oxygen tensions than cells in most normal tissues.
During glycolysis, glycolytic intermediates including glucose-6-phosphate (G6P) can be diverted into the pentose phosphate pathway (PPP), which contributes to macromolecular biosynthesis by producing reducing potential in the form of reduced nicotinamide adenine dinucleotide phosphate (NADPH) and/or ribose-5-phosphate (R5P), the building blocks for nucleotide synthesis. NADPH is the most crucial metabolite produced by the PPP, because NADPH not only fuels macromolecular biosynthesis such as lipogenesis, but also functions as a crucial antioxidant, quenching the reactive oxygen species (ROS) produced during rapid proliferation of cancer cells. Glycolysis and glutaminolysis supply the carbon input required for the TCA cycle to function as a biosynthetic ‘hub’ and permit the production of other macromolecules including amino acids and fatty acids (Cairns et al., 2011). Thus, cancer cells appear to coordinate glycolysis and anabolism to provide an overall metabolic advantage to cancer cell proliferation and disease development. However, the detailed mechanisms underlying this coordination remain largely unknown.
Phosphoglycerate mutase 1 (PGAM1) catalyzes the conversion of 3-phosphoglycerate (3-PG) to 2-phosphoglycerate (2-PG) during glycolysis. PGAM1 regulates a unique step in glycolysis, and most of the glycolytic intermediates that are used as precursors for anabolic biosynthesis are upstream of this step. In many cancers, including hepatocellular carcinoma and colorectal cancer, PGAM1 activity is increased compared to that in the normal tissues (Liu et al., 2008; Ren et al., 2010). PGAM1 gene expression is believed to be upregulated due to loss of TP53 in cancer cells, as TP53 negatively regulates PGAM1 gene expression (Corcoran et al., 2006; Tennant et al., 2009; Tennant et al., 2010). Here we study the role of PGAM1 in coordination of glycolysis and anabolic biosynthesis, as well as the mechanism by which PGAM1 promotes cancer cell proliferation and tumor growth.
RESULTS
PGAM1 controls intracellular 3-PG and 2-PG levels, and is important for glycolysis and anabolic biosynthesis in cancer cells, as well as cancer cell proliferation and tumor growth
To better understand how cancer cells coordinate glycolysis and anabolic biosynthesis, we examined the effects of targeted downregulation of the glycolytic enzyme PGAM1. Stable knockdown of PGAM1 in lung cancer H1299, breast cancer MDA-MB231, acute myeloid leukemia Molm14 and head and neck cancer 212LN cells resulted in decreased PGAM1 activity (Figure S1). We next performed Global Metabolic Profiling (Metabolon) using cell lysate samples of parental H1299 cells and cells with stable knockdown of PGAM1. The results indicate that PGAM1 knockdown results in altered intracellular concentrations of 118 biochemicals (61 upregulated and 57 downregulated) with p<0.05 using Welch’s Two Sample t-tests. Among these biochemicals, we observed that the PGAM1 substrate 3-PG levels are increased in PGAM1 knockdown compared to control cells (Table S1–S2). In consonance with this observation, we found that attenuation of PGAM1 by shRNA in diverse cancer cells leads to not only increased 3-PG (Figure 1A) but also decreased 2-PG (Figure 1B) levels compared to corresponding control cells harboring an empty vector (detailed data are shown in Table S3). The intracellular levels of 3-PG and 2-PG determined using different methods are comparable (Figure S1B–S1C). In addition, stable overexpression of PGAM1 in 3T3 cells results in increased 2-PG and decreased 3-PG levels, compared to control parental 3T3 cells (Figure S1D). These results suggest a crucial role for PGAM1 controlling the metabolite levels of its substrate 3-PG and product 2-PG in cancer cells.
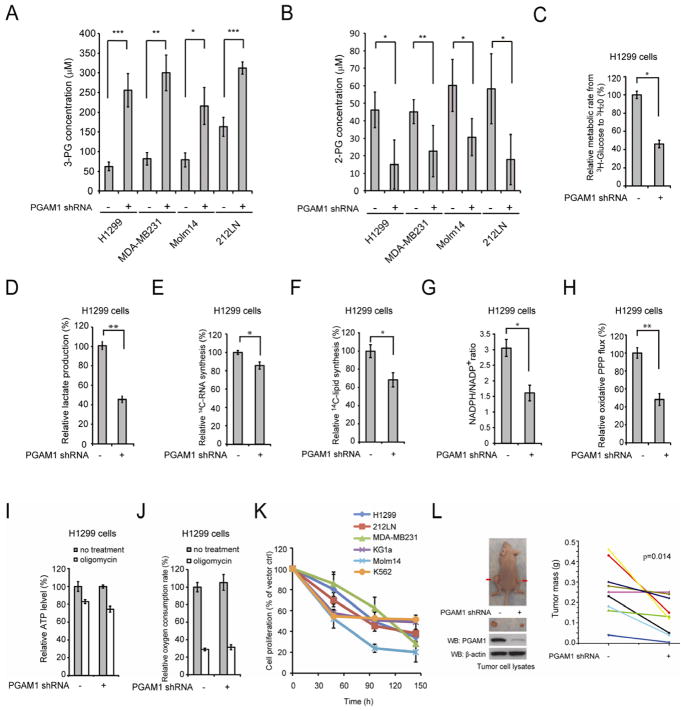
Figure 1. PGAM1 controls intracellular 3-PG and 2-PG levels in cancer cells and is important for glycolysis and anabolic biosynthesis, as well as cell proliferation and tumor growth.
(A–B) Intracellular concentrations of 3-PG and 2-PG were determined in diverse PGAM1 knockdown cancer cells and compared to control cells. Detailed concentrations are listed in Table S3.
(C–J) H1299 cells with stable knockdown of PGAM1 and control cells harboring an empty vector were tested for glycolytic rate (C), lactate production (D), RNA biosynthesis (E), lipogenesis (F), NADPH/NADP+ ratio (G), and oxidative PPP flux (H). The intracellular ATP levels (I) and oxygen consumption rate (J) in the presence or absence of 100nM oligomycin (ATP synthase inhibitor) were also tested.
(K) Cell proliferation rates were determined by cell counting in diverse human cancer (H1299, 212LN and MDA-MB231) and leukemia (KG1a, Molm14 and K562) cells with stable knockdown of PGAM1, which were normalized to the corresponding control cells harboring an empty vector.
(L) Stable knockdown of PGAM1 by shRNA attenuates tumor growth potential of H1299 cells in xenograft nude mice. Left: Dissected tumors (indicated by red arrows) in a representative nude mouse and expression of PGAM1 in tumor lysates are shown. Right: PGAM1 knockdown cells show significantly reduced tumor formation in xenograft nude mice compared to cells harboring empty vector control (p values were determined by a two-tailed paired Student’s t test).
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.01<p<0.01; ***: p<0.001).
See also Figure S1 and Table S1–S3.
We next examined the role of PGAM1 in cancer cell metabolism. We found that, compared to vector control cells, stable knockdown of PGAM1 results in decreased glycolytic rate (Figure 1C) and lactate production (Figure 1D), as well as reduced glucose-dependent biosynthesis of RNA and lipids, accompanied by reduced NADPH/NADP+ ratio (Figure 1E–1G, respectively). Since the PPP produces NADPH and R5P to contribute to macromolecular biosynthesis, we next examined whether PGAM1 contributes to PPP flux. Indeed, we found that oxidative PPP flux is reduced in PGAM1 knockdown compared to control vector cells (Figure 1H). Interestingly, attenuation of PGAM1 in cancer cells does not affect glucose uptake rate (Figure S1E), or intracellular ATP levels (Figure 1I) or O2 consumption rate (Figure 1J) in either the presence or absence of ATP synthase inhibitor oligomycin. These results suggest that downregulation of PGAM1 attenuates glycolysis, PPP and biosynthesis, but does not significantly affect glucose uptake or intracellular ATP levels.
In addition, we found that stable knockdown of PGAM1 results in decreased cell proliferation in diverse human cancer and leukemia cells (Figure 1K). Moreover, we performed a xenograft experiment in which nude mice were subcutaneously injected with control H1299 cells harboring an empty vector on the left flank and PGAM1 knockdown H1299 cells on the right flank (Figure 1L; left). The mice were monitored for tumor growth over a 6-week time period. The masses of tumors derived from PGAM1 knockdown H1299 cells were significantly reduced compared to those of tumors formed by vector control cells (Figure 1L; right).
PGAM1 knockdown results in elevated levels of 3-PG, which binds to and inhibits 6PGD by competing with its substrate 6-phosphogluconate (6-PG)
We next explored the molecular mechanism by which PGAM1 regulates the PPP. Our data suggest that the abnormally high levels of 3-PG in PGAM1 knockdown cells may be accounted for inhibition of oxidative PPP flux (Figure 1). To test this hypothesis, we examined the effect of 3-PG on glucose-6-phosphate dehydrogenase (G6PD), the first and most important enzyme of the oxidative PPP, which produces NADPH, and 6-phosphogluconate dehydrogenase (6PGD), an enzyme that also produces NADPH while converting 6-phosphogluconate into ribulose 5-phosphate in the presence of NADP+. We performed in vitro 6PGD and G6PD assays in the presence of increasing concentrations of 3-PG. Physiological concentrations of 3-PG in human cells are reported to be approximately 50–80μM (Feig et al., 1971; Minakami et al., 1964; Mulquiney and Kuchel, 1999). As shown in Table S3, we determined that, in H1299, MDA-MB231 and Molm14 cells, the 3-PG levels are approximately 60–80μM in control vector cells and 200–300μM in PGAM1 knockdown cells, while the 3-PG concentrations are approximately 160μM and 310μM in 212LN control and PGAM1 knockdown cells, respectively. Thus, we next examined the effects of increasing concentrations of 3-PG on G6PD and 6PGD enzyme activities according to the aforementioned physiological 3-PG levels in tumor cells.
We found that treatment with 3-PG concentrations analogous to those in PGAM1 knockdown H1299 cells (~250μM) results in decreased enzyme activity of 6PGD (Figure 2A) in H1299 cell lysates or recombinant 6PGD (r6PGD) (Figure 2B), whereas the physiological 3-PG concentrations determined in control H1299 cells (~60μM) do not significantly affect 6PGD enzyme activity in both experiments. In control experiments, treatment with increasing concentrations of 3-PG did not significantly affect G6PD activity in H1299 cell lysates or rG6PD activity (Figure S2A). In addition, 2-PG did not affect 6PGD enzyme activity in H1299 cell lysates or r6PGD activity (Figure S2B). These results suggest that abnormally high levels of 3-PG, as in PGAM1 knockdown cells, may selectively and directly inhibit 6PGD but not G6PD.
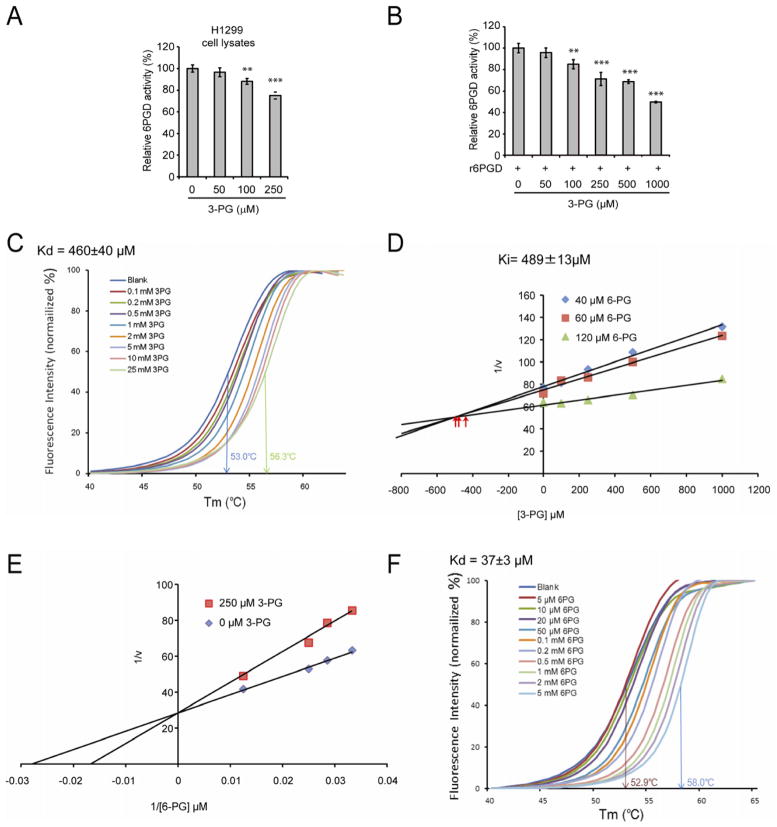
Figure 2. Attenuation of PGAM1 results in increased intracellular levels of 3-PG, which binds to and inhibits 6PGD by competing with its substrate 6-PG.
(A–B) Enzyme activity of 6PGD in H1299 cell lysates (A) or recombinant 6PGD (r6PGD) (B) was determined in the presence of increasing concentrations of 3-PG. Relative 6PGD activity was normalized to the control samples without 3-PG treatment. 3-PG levels in control H1299 cells with empty vector and PGAM1 knockdown are 62.5±10.8μM and 256±41.9μM, respectively. The error bars represent mean values +/− SD from three replicates of each sample (**: 0.01<p<0.01; ***: p<0.001).
(C) Thermal shift melting curves of 6PGD and 3PG. Thermal shift assay was performed to examine the protein (6PGD) and “ligand” (3PG) interaction. Change of melting temperature (Tm) in a dose-dependent manner at concentrations from 100 μM to 25 mM demonstrates that 3-PG directly binds to the protein. Kd for 6PGD-3-PG interaction was determined to be 460±40 μM.
(D) The Dixon plot shows that 3-PG inhibits 6PGD and the dissociation constant (Ki) was determined.
(E) The Lineweaver-Burk plot shows that 3-PG functions as a competitive inhibitor of 6PGD.
(F) Thermal shift melting curves of 6PGD and 6PG. Thermal shift assay was performed to examine the protein (6PGD) and ligand (6PG) interaction. Change of melting temperature (Tm) in a dose-dependent manner at concentrations from 5 μM to 5 mM demonstrates that 6-PG directly binds to the protein. Kd for 6PGD-6PG interaction was determined to be 37±3μM.
See also Figure S2.
To examine whether 3-PG binds to and inhibits 6PGD, we performed a thermal melt shift assay to examine the interaction of protein (6PGD) and “ligand” (3-PG). Incubation of increasing concentrations of 3-PG raises 6PGD melting temperature (Tm) in a dose-dependent manner, suggesting that 3-PG directly binds to the protein (Figure 2C). The Kd value for protein-“ligand” interaction was calculated to be 460±40 μM. Moreover, we performed kinetics studies on the inhibition of 6PGD by 3-PG. As shown in Figure 2D, the Dixon plot indicates that 3-PG binds and inhibits 6PGD. The inhibition constant (Ki) was determined to be 489±13μM, in agreement with the Kd determined.
We next determined the intracellular concentration of 6-PG in H1299, MDA-MB231 and 212LN cells to be 34.9±2.1μM, 37.6±0.7μM and 24.9±0.4μM, respectively. We performed additional enzyme kinetics assays to test whether 3-PG at a concentration analogous to that in PGAM1 knockdown H1299 cells (~250μM) functions as a competitive or non-competitive inhibitor of 6PGD in the presence of physiological concentrations of 6-PG (~35μM). As shown in Figure 2E, the Lineweaver-Burk plot demonstrates that 3-PG functions as a competitive inhibitor of 6PGD. Since the Kd value for protein (6PGD)-ligand (6-PG) interaction was calculated to be 37±3 μM in a thermal melt shift assay (Figure 2F), these data together suggest that at physiological concentrations, 3-PG (~60–80μM) cannot effectively compete with 6-PG (~35μM) to inhibit 6PGD in cancer cells; however, upon attenuation of PGAM1, elevated cellular 3-PG levels (~250–300μM) result in reduced 6PGD enzyme activity.
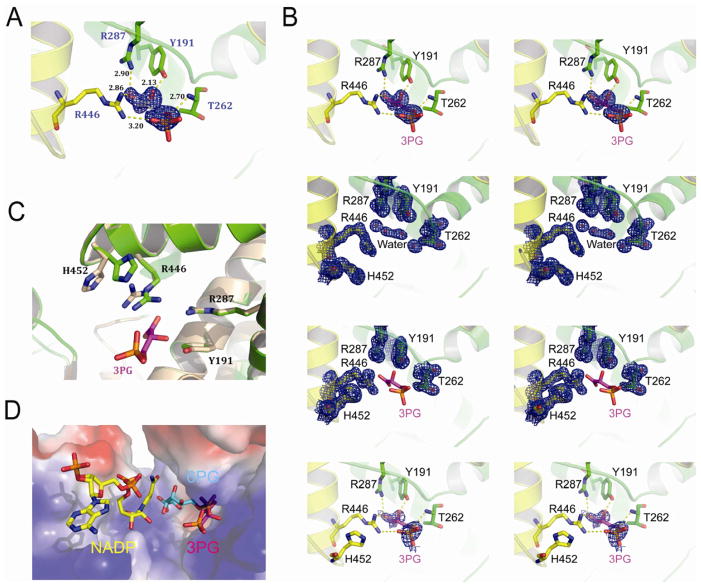
To further understand the structural properties of 3-PG mediated inhibition of 6PGD, we crystallized the apo-form of 6PGD (1.39 Å), which was also soaked with 3-PG to obtain the 3-PG-bound form of 6PGD (1.53 Å) (Table S4). The Fo-Fc density analysis revealed that the electron density of 3-PG was located in the active site of the 3-PG-bound 6PGD structure (Figure 3A) but not in the apo-6PGD structure (Figure 3B). 3-PG interacts with several residues (Y191, T262, R287, R446) in the active site of 6PGD that are important for substrate binding and enzymatic activity of 6PGD (Li et al., 2006) (Figure 3A). Different conformations were observed for Arg 446 and His 452 in the 3-PG-bound 6PGD structure compared to the apo-form 6PGD structure (Figure 3C). An alignment of three different 6PGD structures with bound NADP, 6-PG and 3-PG shows an overlap of 3-PG and 6-PG in the active site (Figure 3D). Together, these results demonstrate that 3-PG directly binds to 6PGD and inhibits 6PGD enzyme activity by competing with the cognate substrate 6-PG, representing a molecular mechanism to explain how PGAM1, as a glycolytic enzyme, contributes to the regulation of the oxidative PPP and consequently anabolic biosynthesis.
Figure 3. Co-crystallization analysis of 3-PG mediated inhibition of 6PGD.
(A) Stereo view of the Fo-Fc electron density map contoured at 3.0 σ around 3-PG. The Fo-Fc density map is shown as blue mesh. Residues of 6PGD interact with 3-PG are shown in stick.
(B) Upper top: Stereo view of the unbiased Fo-Fc electron density map contoured at 3.0 σ around 3PG. The Fo-Fc density map is shown as blue mesh. Residues interact with 3-PG are shown in stick. Lower top: Stereo view of the 2Fo-Fc electron density map of 6PGD apo-form contoured at 1.2 σ at 3-PG binding pocket in the same orientation as in Figure 2A. The 2Fo-Fc density map is shown as blue mesh. Upper bottom: Stereo view of the 2Fo-Fc electron density map of 6PGD-3-PG complex contoured at 1.2 σ at 3-PG binding pocket in the same orientation as in Figure 3A and 3C. The 2Fo-Fc density map is shown as blue mesh. Lower bottom: Stereo view of simulated-annealing omit map contoured at 0.8 σ around 3-PG. The omit density map is shown as blue mesh.
(C) Structure comparison of the 6PGD apo-form (wheat) and the 6PGD-3-PG complex (green). Arg 446 and His 452 in the 6PGD-3-PG complex structure show different conformation.
(D) Surface electrostatic potential of the substrate-binding pocket of 6PGD. The bound 3-PG (pink) competes with 6-PG (blue) but not NADP (yellow) in the active site. The model was built by aligning structures of 6PGD-NADP (PDB code: 2JKV), 6PGD-6-PG (PDB code: 3FWN) and 6PGD-3PG.
See also Table S4.
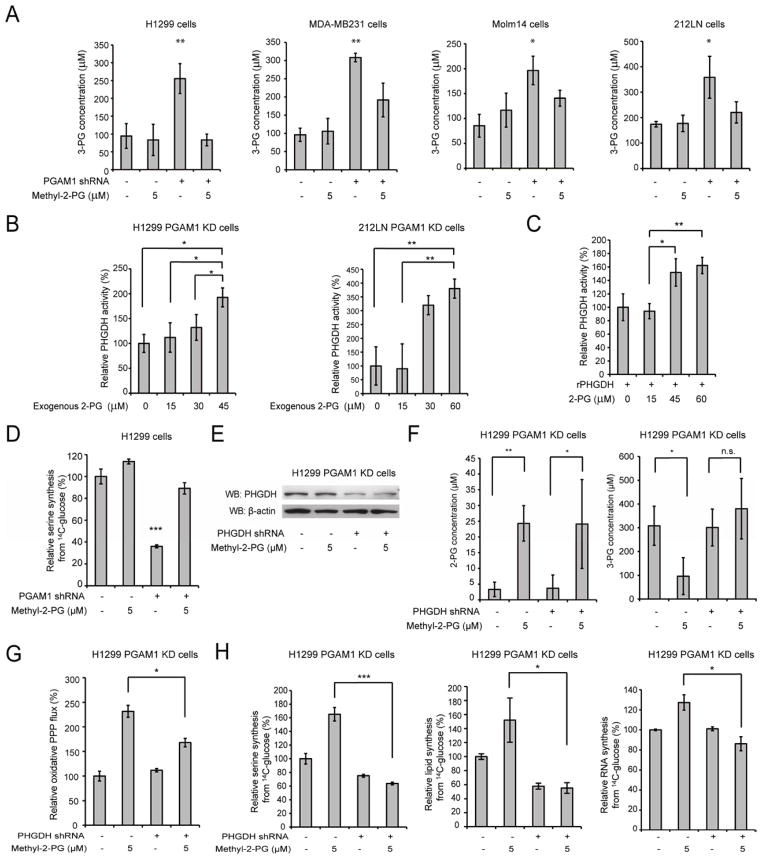
Rescue of reduced 2-PG levels in PGAM1 knockdown cells results in decreased 3-PG levels by activating 3-phosphoglycerate dehydrogenase (PHGDH)
In order to examine the effect of decreased 2-PG levels on cancer cell metabolism, we treated the aforementioned PGAM1 knockdown cancer cells with a cell permeable agent, methyl-2-PG, which converts to 2-PG in cells. We verified that in diverse PGAM1 knockdown cancer cells, treatment with methyl-2-PG results in increased 2-PG cellular levels comparable to those in the corresponding control vector cells (Figure 4A). We also observed that methyl-2-PG treatment rescues the reduced lactate production (Figure 4B) but has no significant effect on intracellular ATP levels (Figure S3A) in H1299 cells with stable knockdown of PGAM1 compared to control vector cells. This result suggests that rescuing cellular 2-PG levels reverses the inhibitory effect of PGAM1 knockdown on glycolysis and allows downstream glycolytic reactions to resume and ultimately produce lactate. However, such rescued glycolytic activity does not affect ATP levels, which is consistent with our previous observation (Figure 1I–1J).
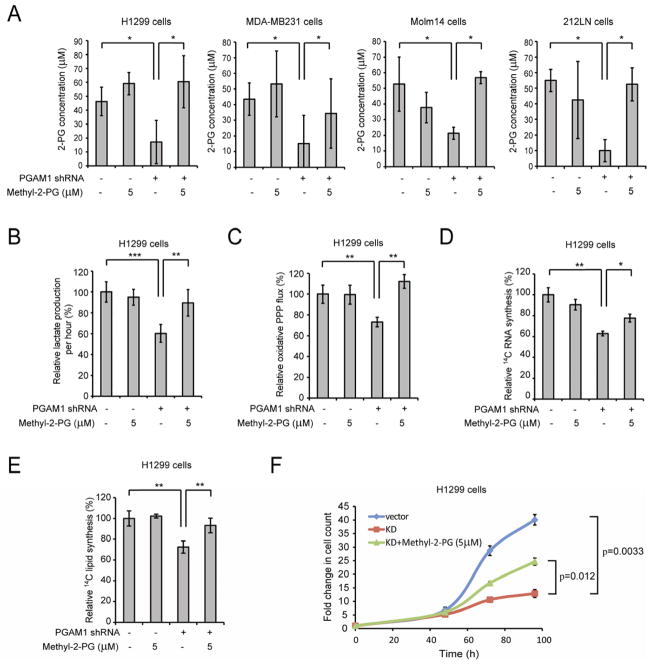
Figure 4. Rescue of reduced 2-PG levels in PGAM1 knockdown cells reverses the phenotypes due to attenuation of PGAM1.
(A) 2-PG levels in diverse cancer cells with stable knockdown of PGAM1 were determined in the presence and absence of cell permeable methyl-2-PG.
(B–F) H1299 cells with stable knockdown of PGAM1 were tested for lactate production (B), oxidative PPP flux (C) and biosynthesis of RNA (D) and lipids (E), as well as cell proliferation
(F) in the presence and absence of methyl-2-PG.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.01<p<0.01; ***: p<0.001).
See also Figure S3.
Surprisingly, we also found that methyl-2-PG treatment rescues the decreased oxidative PPP flux and biosynthesis of RNA and lipids, as well as partially restores the reduced cell proliferation in H1299 PGAM1 knockdown cancer cells compared to the corresponding control vector cells (Figure 4C–Figure 4F). Similar results were obtained using MDA-MB231 vector and PGAM1 knockdown cells (Figure S3B–S3E). These data suggest that the increased 2-PG levels in PGAM1 knockdown cells provide a feedback mechanism to rescue the abrogated PPP and anabolic biosynthesis upstream of PGAM1.
We tested this hypothesis by examining the effect of rescued 2-PG levels on 3-PG concentrations in PGAM1 knockdown cells. We found that treatment with methyl-2-PG results in decreased 3-PG concentrations in diverse PGAM1 knockdown cells to levels that are comparable to the 3-PG concentrations in the corresponding control vector cells (Figure 5A). These results further suggest that PGAM1 controls 2-PG levels in cancer cells, which contributes to PGAM1-dependent coordination of glycolysis and anabolic biosynthesis by adjusting 3-PG levels.
Figure 5. Rescue of reduced 2-PG levels due to PGAM1 attenuation results in decreased 3-PG levels by activating PHGDH.
(A) 3-PG levels in diverse cancer cells with stable knockdown of PGAM1 were determined in the presence and absence of methyl-2-PG.
(B–C) Enzyme activity of PHGDH in PGAM1 knockdown H1299 (B; left) or 212LN (B; right) cell lysates and recombinant PHGDH (rPHGDH) (C) were determined in the presence of increasing concentrations of 2-PG. Relative enzyme activity was normalized to the control samples without 2-PG treatment. 2-PG levels in control H1299 cells with empty vector and PGAM1 knockdown cells are 46.2±10.2μM and 15.0±14.1μM, respectively, while 2-PG levels in 212LN cells with empty vector and stable knockdown of PGAM1 are 58.3±20.1μM and 17.8±14.4μM, respectively.
(D) Serine biosynthesis rate of H1299 cells with stable knockdown of PGAM1 was determined by measuring 14C incorporation into serine from 14C-glucose in the presence and absence of methyl-2-PG. Relative serine biosynthesis was normalized to control cells harboring an empty vector without methyl-2-PG treatment.
(E) Western blot result shows shRNA-mediated knockdown of PHGDH in H1299 cells with stable knockdown of PGAM1 in the presence or absence of methyl-2-PG treatment.
(F) 2-PG (left) and 3-PG (right) levels in PGAM1 knockdown cells upon PHGDH knockdown were determined in the presence and absence of methyl-2-PG.
(G–H) PGAM1 stable knockdown cells treated with or without shRNA targeting PHGDH were tested for PPP flux (G) as well as biosynthesis of serine, lipids and RNA (H; left, middle and right, respectively) in the presence and absence of methyl-2-PG.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.01<p<0.01; ***: p<0.001; n.s.: not significant).
We next determined the molecular mechanism underlying 2-PG dependent feedback regulation of intracellular 3-PG levels. Besides conversion to 2-PG catalyzed by PGAM1 in glycolysis, 3-PG also serves as a precursor for serine synthesis and can be converted to 3-phosphohydroxypyruvate (pPYR) by PHGDH. Since PGAM1 activity is attenuated in PGAM1 knockdown cells, it is possible that the rescued cellular 2-PG levels by methyl-2-PG treatment decreases 3-PG levels by activating PHGDH. We tested this hypothesis by examining the effect of 2-PG on PHGDH activity and we used PGAM1 knockdown cells to exclude the endogenous PGAM1 effect on 3-PG and 2-PG in the PHGDH enzyme activity reactions. Indeed, we found that treatment with 2-PG concentrations equivalent to those determined in control H1299 cells (~45μM) or methyl-2-PG treated PGAM1 knockdown cells (~60μM) results in higher PHGDH enzyme activity in H1299 PGAM1 knockdown cell lysates (Figure 5B; left). Similar results were obtained by treating 212LN PGAM1 knockdown cell lysates with increasing concentrations of 2-PG (Figure 5B; right). Moreover, treatment with increasing concentrations of 2-PG results in increased enzyme activity of recombinant PHGDH (rPHGDH) (Figure 5C). In contrast, 2-PG concentrations that correspond to those determined in PGAM1 knockdown cells (~15μM) did not significantly affect PHGDH activity. Together, these studies reveal a feedback mechanism by which cellular 2-PG levels contribute to control of 3-PG levels in cells through regulation of PHGDH.
In addition, we found that stable knockdown of PGAM1 results in significantly decreased serine biosynthesis, while treatment with methyl-2-PG rescues the phenotype (Figure 5D). Moreover, shRNA-mediated knockdown of PHGDH (Figure 5E) does not affect rescued 2-PG levels in PGAM1 knockdown cells upon treatment with methyl-2-PG, while PHGDH knockdown abolishes the methyl-2-PG dependent decrease of the elevated 3-PG levels in H1299 PGAM1 knockdown cells (Figure 5F, left and right, respectively). These data support our hypothesis that PGAM1 controls 2-PG levels to regulate PHGDH, which consequently regulates 3-PG levels by diverting 3-PG in serine biosynthesis. Furthermore, knockdown of PHGDH in PGAM1 stable knockdown cells reverses the methyl-2-PG treatment dependent rescue of oxidative PPP flux as well as biosynthesis of serine, lipids and RNA (Figure 5G–5H, respectively). These data together suggest that, besides being a glycolytic metabolite, 2-PG may also signal through PHGDH to provide regulation of PPP flux and anabolic biosynthesis, at least in part by regulating 3-PG levels.
PGAM1 enzyme activity strikes a balance between 3-PG and 2-PG levels, which coordinates glycolysis and biosynthesis to promote cancer cell proliferation
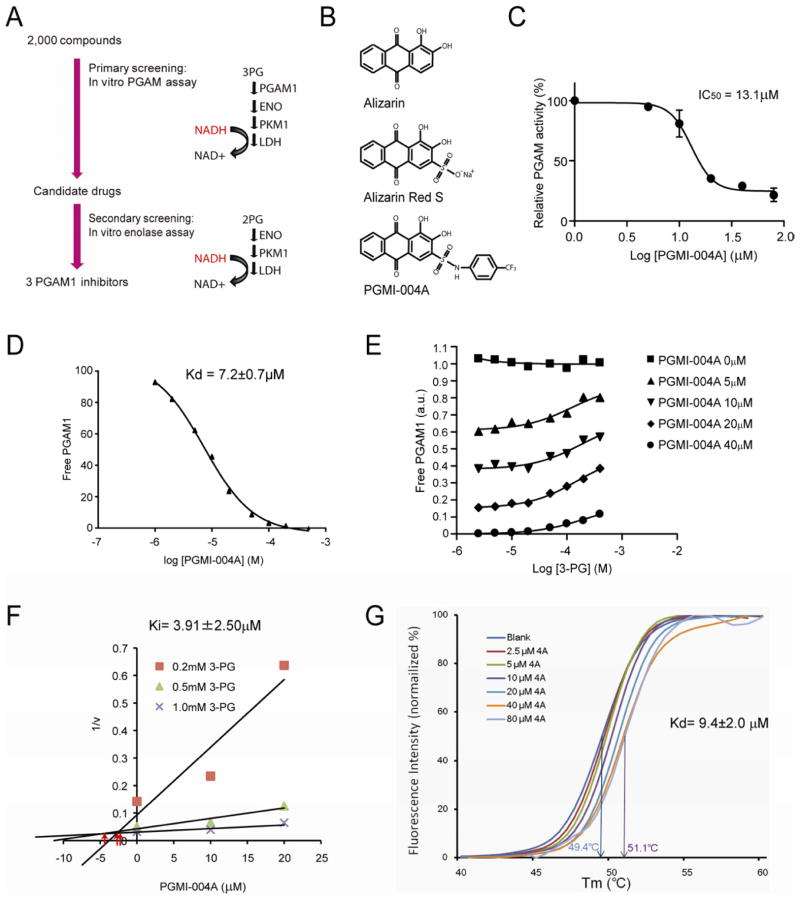
In order to study the role of PGAM1 enzyme activity in cancer metabolism and tumor development, we screened and developed a small molecule inhibitor of PGAM1. Currently the only reported PGAM1 inhibitor is MJE3, which specifically inhibits PGAM1 activity exclusively in intact cells, probably by targeting the active site of PGAM1 with certain modifications in vivo (Evans et al., 2007; Evans et al., 2005). We designed a screening strategy using coupled PGAM1 and enolase assays and identified three lead small molecule compounds, including alizarin, as PGAM1 inhibitors from a library of FDA approved 2,000 small molecule compounds (Figure 6A; Figure S4A–S4B). We focused on 1,2-dihydroxyanthraquinone a.k.a. alizarin (C14H8O4) (Figure 6B; top), which is an organic compound that is historically important as a prominent dye, originally derived from the roots of plants of the Madder genus. Treatment with alizarin results in decreased proliferation of human leukemia KG1a cells in a dose-dependent manner (Figure S4C–S4D).
Figure 6. Identification and characterization of small molecule PGAM1 inhibitor, PGMI-004A.
(A) Schematic representation of the primary and secondary screening strategies to identify lead compounds as PGAM1 inhibitors.
(B) Structure of alizarin and its derivatives alizarin Red S and PGAM inhibitor (PGMI)-004A.
(C) PGMI-004A inhibits PGAM1 with an IC50 of 13.1 μM, which was determined by incubating purified human PGAM1 proteins with increasing concentrations of PGMI-004A. The error bars represent mean values +/− SD from three replicates of each sample.
(D) Kd value was determined as 7.2±0.7 μM by incubating purified human PGAM1 proteins with increasing concentrations of PGMI-004A. The fluorescence intensity (Ex: 280nm, Em: 350nm) from Tryptophan was measured (Schauerte and Gafni, 1989).
(E) Competitive binding assay of PGMI-004A with recombinant PGAM1 protein in the presence of increasing concentrations of PGAM1 substrate 3-PG. Increased free PGAM1 was determined by an increase in fluorescence intensity.
(F) Dixon plot analysis of PGAM1 enzyme assay in the presence of different concentrations of PGMI-004A and 3-PG. The reaction velocity (v) was determined by the rate of the decrease in fluorescence (ex: 340nm, em: 460nm) by NADH oxidation. Ki was determined to be 3.91±2.50μM.
(G) Thermal shift melting curves of PGAM1 and PGMI-004A. Thermal shift assay was performed to examine the protein (PGAM1) and “ligand” (inhibitor PGMI-004A) interaction. Change of melting temperature (Tm) in a dose-dependent manner at concentrations from 2.5μM to 80μM demonstrates that PGMI-004A directly binds to the protein. Kd for PGAM1-PGMI-004A interaction was determined to be 9.4±2.0μM.
See also Figure S4.
We next identified alizarin Red S (Figure 6B; middle) as a more potent PGAM1 inhibitor from a group of alizarin derivatives (Figure S4E–S4F). We designed a group of alizarin Red S derivatives by adding hydrophobic groups through a sulfonamide bond (Figure S5A). Among these compounds, we focused on PGAM1 inhibitor 004A (PGMI-004A)(Figure 6B; bottom), which, although less potent than Red S in vitro, demonstrates enhanced potency to inhibit PGAM1 in leukemia KG1a cells compared to its parental compounds (Figure S5B–S5C). This may be due to the fact that PGMI-004A is more hydrophobic than alizarin and alizarin Red S, which confers better cell permeability.
PGMI-004A inhibits PGAM1 with an IC50 of approximately 13.1μM (Figure 6C) and the Kd value of the PGMI-004A-PGAM1 interaction was determined to be 7.2±0.7μM from fluorescence-based binding assay (Figure 6D). In a competitive binding assay where PGMI-004A was incubated with recombinant PGAM1 proteins in the presence of different concentrations of PGAM1 substrate 3-PG, we found that increasing concentrations of 3-PG caused an increase in the fluorescence intensity from PGMI-004A-unbound form of PGAM1 in the presence of different concentrations of PGMI-004A, but not in the absence of PGMI-004A (Figure 6E). This suggests that PGMI-004A may allosterically modulate the enzyme activity of PGAM1. The Ki value was determined to be 3.91±2.50μM using Dixon plot analysis (Figure 6F). In addition, we performed a thermal melt shift assay to examine the interaction of protein (PGAM1) and ligand (PGMI-004A). Incubation of increasing concentrations of PGMI-004A raises PGAM1 melting temperature (Tm) in a dose-dependent manner, suggesting that PGMI-004A directly binds to the protein (Figure 6G). The Kd value for protein-ligand interaction was calculated to be 9.4±2.0 μM. Together, these results suggest that PGMI-004A directly binds to PGAM1 and inhibits its enzyme activity.
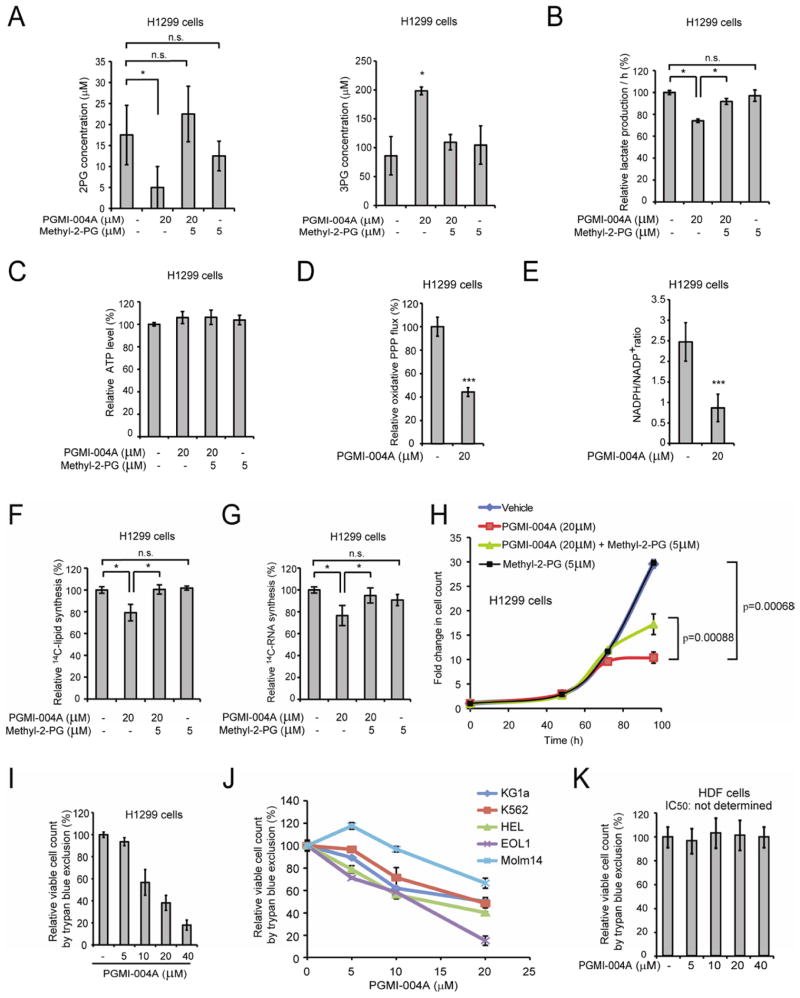
We found that inhibition of PGAM1 activity by PGMI-004A treatment results in decreased 2-PG and increased 3-PG levels in H1299 cells, which could be rescued by treatment with methyl-2-PG (Figure 7A). Moreover, treatment with PGMI-004A results in significantly reduced lactate production that was rescued by methyl-2-PG treatment (Figure 7B), but has no significant effect on intracellular ATP levels (Figure 7C). In consonance with these observations, the rescued lactate production due to methyl-2-PG treatment was abolished when enolase was knocked down or inhibited by specific inhibitor NaF in PGMI-004A treated cells (Figure S5D). These results also suggest that rescued 2-PG derived from methyl-2-PG is metabolized by cells to restore the decreased glycolysis due to PGAM1 inhibition in cancer cells. We also found that PGMI-004A treatment results in decreased oxidative PPP flux (Figure 7D) and NADPH/NADP+ ratio (Figure 7E), as well as reduced biosynthesis of lipids and RNA (Figure 7F–7G, respectively) and cell proliferation (Figure 7H) in H1299 cells. These phenotypes are similar to those observed in PGAM1 knockdown cells, which could be significantly rescued by treatment with methyl-2-PG, suggesting that PGMI-004A targets PGAM1 to inhibit cancer cell metabolism and proliferation.
Figure 7. Inhibition of PGAM1 by PGMI-004A reveals that PGAM1 enzyme activity is important for regulation of 3-PG and 2-PG levels and coordination of glycolysis and biosynthesis to promote cancer cell proliferation.
(A) 2-PG (left) and 3-PG (right) levels in H1299 cells treated with or without PGMI-004A were determined in the presence and absence of methyl-2-PG.
(B–C) Lactate production (B) and intracellular ATP levels (C) in H1299 cells treated with or without PGMI-004A were determined in the presence and absence of methyl-2-PG.
(D–E) H1299 cells treated with or without PGMI-004A were tested for oxidative PPP flux (D) and NADPH/NADP+ ratio (E).
(F–H) H1299 cells treated with or without PGMI-004A were tested for biosynthesis of lipids (F) and RNA (G), as well as cell proliferation (H) in the presence and absence of methyl-2-PG.
(I–K) Cell viability of H1299 cells (I), diverse human leukemia cells (J) and control human dermal fibroblasts (HDF) cells (K) in the presence of increasing concentrations of PGMI-004A. Cell viability was determined by trypan blue exclusion.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; ***: p<0.001; n.s.: not significant).
See also Figure S5.
In addition, we observed that PGMI-004A treatment results in decreased cell proliferation of diverse human cancer and leukemia cells (Figure 7I–7J; S5E–S5H), but not control human dermal fibroblasts (HDF), human foreskin fibroblasts (HFF), human HaCaT keratinocyte cells and human melanocyte PIG1 cells (Figure 7K and S5I), suggesting minimal non-specific toxicity of PGMI-004A in normal, proliferating human cells.
Targeting PGAM1 by PGMI-004A treatment inhibits cancer cell proliferation and tumor growth, and alters 3-PG and 2-PG levels in primary leukemia cells from human patients, leading to attenuated leukemia cell proliferation
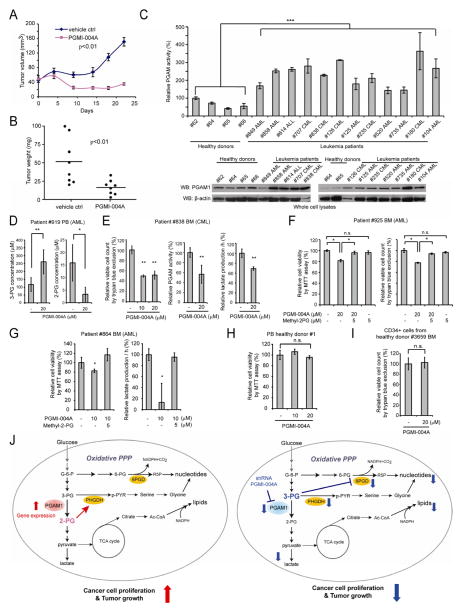
We next performed an in vivo drug treatment experiment. Initial toxicity studies by chronic injection of PGMI-004A to nude mice for 4 weeks revealed that 100mg/kg/day administered intraperitoneally is a well-tolerated dose. In addition, continuous treatment with PGMI-004A (100mg/kg/day) for 7 days did not result in significant alteration in body weight, complete blood counts (CBC) or hematopoietic properties of nude mice (Table S5). Histopathological analyses revealed that no notable differences between the vehicle-treated and PGMI-004A-treated groups were evident (Figure S6A–S6D). We performed xenograft experiment by injecting H1299 cells to nude mice as described (Hitosugi et al., 2009). Six days post-injection, mice were divided into two groups (n=8/group) and treated with either PGMI-004A (100mg/kg/day) or vehicle for 21 days. We found that PGMI-004A treatment results in significantly decreased tumor growth and tumor size in treated mice compared with mice receiving vehicle control (Figure 8A–8B, respectively; Figure S6E–S6F). Moreover, treatment with PGMI-004A effectively inhibits PGAM1 enzyme activity in tumors in vivo in resected tumors from xenograft nude mice (Figure S6G). These data together suggest that targeting PGAM1 by PGMI-004A inhibits PGAM1 in vivo, and that this inhibition causes specific toxicity to tumor cells.
Figure 8. PGMI-004A treatment results in increased 3-PG and decreased 2-PG levels, and reduced cell proliferation of primary leukemia cells from human patients, as well as attenuated tumor growth in xenograft nude mice in vivo.
(A–B) Tumor growth (A) and tumor size (B) in xenograft nude mice injected with H1299 cells were compared between the group of mice treated with PGMI-004A and the control group treated with vehicle control. p values were determined by a two-tailed Student’s t test.
(C) PGAM1 protein expression (lower) and enzyme activity (upper) levels were examined using primary leukemia cells from diverse human patients with AML, CML and B-ALL and compared to control peripheral blood cells from healthy donors.
(D) Effect of PGMI-004A treatment on 3-PG (left) and 2-PG (right) levels in human primary leukemia cells isolated from peripheral blood samples from a representative AML patient.
(E) Effect of PGMI-004A treatment on cell viability (left), PGAM1 activity (middle) and lactate production (right) in human primary leukemia cells from a representative CML patient.
(F–G) Effect of methyl-2-PG treatment on decreased cell viability (F; G left) and lactate production (G right) in PGMI-004A-treated human primary leukemia cells from AML patients.
(H–I) PGMI-004A shows no toxicity in treatment (120h) of peripheral blood cells (H) and CD34+ cells isolated from bone marrow samples (I) from representative healthy human donors.
(J) Proposed model: role of PGAM1 in cancer cell metabolism.
Left: PGAM1 activity is upregulated in cancer cells to promote glycolysis and keep the intracellular 3-PG levels low, which in turn permits high levels of the PPP and biosynthesis to fulfill the request of rapidly growing tumors. PGAM1 also maintains the physiological levels of 2-PG to sustain PHGDH activity, which diverts 3-PG from glycolysis to serine synthesis and contributes to maintaining relatively low levels of 3-PG in cancer cells. These effects in concert provide a metabolic advantage to cancer cell proliferation and tumor growth.
Right: When PGAM1 is inhibited, 3-PG levels are elevated, which in turn inhibit 6PGD and consequently the oxidative PPP and anabolic biosynthesis. At the same time, 2-PG is decreased to levels below the physiological concentrations, leading to decreased PHGDH activity, which facilitates 3-PG accumulation. Such metabolic changes result in attenuated cell proliferation and tumor development.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.01<p<0.01; ***: p<0.001; n.s.: not significant).
We found that PGAM1 protein expression and enzyme activity levels are commonly upregulated in primary leukemia cells from diverse AML, CML and B-ALL patients (n=12), compared to control peripheral blood cells from healthy donors (n=4) (Figure 8C). We next found that, consistent with our observations in cancer cell lines, inhibiting PGAM1 by PGMI-004A treatment results in increased 3-PG and decreased 2-PG levels in primary leukemia cells from a representative AML patient (Figure 8D). PGMI-004A treatment also results in decreased cell viability and reduced PGAM1 activity and lactate production in the samples from 7 (1 CML and 6 AML) out of 8 leukemia patients. Figure 8E and S6H–S6I show results using samples from CML and AML patients as representatives, respectively. Moreover, methyl-2-PG treatment rescues the decreased cell viability (Figure 8F; 8G left) and lactate production (Figure 8G right) in primary leukemia cells from representative AML patients. In addition, PGMI-004A treatment did not affect cell viability of mononucleocytes in peripheral blood samples from two healthy human donors (Figures 8H and S6J) and CD34+ cells isolated from bone marrow samples from four healthy donors (Figures 8I and S6K), suggesting promising anti-cancer potential of PGMI-004A with minimal toxicity to human blood cells. These results of translational studies together suggest that PGAM1 is a promising therapeutic target in treatment of human malignancies.
DISCUSSION
Our findings suggest that upregulation of PGAM1 by increased gene expression in cancer cells provides a metabolic advantage to cancer cell proliferation and tumor growth; PGAM1 coordinates glycolysis and anabolic biosynthesis, at least in part by controlling intracellular levels of its substrate 3-PG and product 2-PG (Figure 8J). Our results revealed a molecular mechanism by which 3-PG inhibits 6PGD by directly binding to the active site of 6PGD and competing with its substrate 6-PG. Attenuation of PGAM1 results in abnormal accumulation of 3-PG, which in turn inhibits 6PGD and consequently the oxidative PPP and anabolic biosynthesis. Moreover, our findings suggest that PGAM1 controls the intracellular levels of its product 3-PG not only directly through substrate consumption but also indirectly by controlling levels of its product 2-PG. Physiological concentrations of 2-PG promote the enzyme activity of PHGDH, which converts 3-PG to pPYR, reducing the cellular 3-PG levels. Upon attenuation of PGAM1, 2-PG is decreased to levels below the physiological concentrations, leading to decreased PHGDH activity, which facilitates 3-PG accumulation. This represents a regulatory mechanism by which 2-PG activates PHGDH to provide feedback control of 3-PG levels. Thus, we suggest that PGAM1 activity is upregulated in cancer cells to promote glycolysis and keep the intracellular 3-PG levels low, which in turn permits high levels of the PPP and biosynthesis to fulfill the request of rapidly growing tumors. This is consistent with previous report that expression of TP53 suppresses oxidative PPP in cancer cells (Jiang et al., 2011). In addition, PGAM1 may also be responsible for maintaining the physiological levels of 2-PG to sustain PHGDH activity, which diverts 3-PG from glycolysis to serine synthesis and contributes to maintaining relatively low levels of 3-PG in cancer cells.
Inhibition of PGAM1 by shRNA or treatment with a small molecule inhibitor PGMI-004A results in altered glycolysis and anabolic biosynthesis, and reduced cancer cell proliferation and tumor growth. Interestingly, targeting PGAM1 does not significantly affect intracellular ATP levels. Decreased ATP production due to attenuated glycolysis in PGAM1 knockdown cells may be compensated by alternative mechanisms other than mitochondrial oxidative phosphorylation, or perhaps the ATP consumption in PGAM1 knockdown cells is decreased accordingly. Methyl-2-PG treatment rescues most of the aforementioned phenotypes. Rescued 2-PG levels in cells with attenuated PGAM1 reversed decreased lactate production by rescuing the glycolytic process downstream of PGAM1, as well as reduced oxidative PPP flux and biosynthesis of RNA and lipids, at least in part by decreasing elevated 3-PG levels. However, methyl-2-PG treatment only partially rescues the attenuated cell proliferation in PGAM1 knockdown cells or cells treated with PGMI-004A. This result suggests that PGAM1 may contribute to cell proliferation in both 2-PG-dependent and independent manners.
The current understanding of the connection between glycolysis and PPP/biosynthesis is based upon a model in which glycolytic intermediates can be diverted into PPP and biosynthesis pathways as precursors. Our results show that the concentrations of glycolytic metabolites such as 3-PG and 2-PG can directly affect the catalytic activity of enzymes involved in PPP and biosynthesis, which represents an additional link between glycolysis, PPP and biosynthesis. Metabolites have been suggested to function as signaling molecules in the past. Examples include AMP, which is an allosteric activator for AMP-Activated Protein Kinase (AMPK), a kinase that senses intracellular energy levels (ATP/AMP ratio) (Shackelford and Shaw, 2009), and glutamine, which activates leucine uptake, leading to mTOR activation (Nicklin et al., 2009). We found that the cellular levels of 3-PG and 2-PG, two key intermediates in glycolysis, have additional regulatory impact on metabolic enzymes to affect cell metabolism and consequently proliferation, which provides an example to suggest that glycolytic metabolites could also serve as signaling molecules to control cell metabolism and cellular responses. Moreover, our findings also describe a feedback mechanism by which the product levels (2-PG) of a metabolic enzyme (PGAM1) can regulate its substrate levels (3-PG) by affecting an alternative enzyme (PHGDH) that is involved in production of this substrate. Thus, this study showcases the complexity of cellular metabolism, demonstrating that control of the intracellular levels of a particular metabolite may involve diverse enzymes in different metabolic reactions, such that the balance of the intracellular levels of various metabolites may exert regulatory functions on enzymes in different pathways to control cellular metabolism. Such a mechanism can be explored for anti-cancer therapies.
Previous reports describe that targeting PGAM1 by a PGAM1-derived inhibitory peptide or PGAM inhibitor MJE3 attenuates cancer cell proliferation (Engel et al., 2004; Evans et al., 2005). In consonance with these observations, our studies suggest that protein expression and enzyme activity levels of PGAM1 are important for cancer cell proliferation and tumor growth. Our compound PGMI-004A exhibits promising efficacy in treatment of xenograft nude mice in vivo with minimal toxicity, as well as in diverse human cancer cells and primary leukemia cells from human patients in vitro with no obvious off target effect and minimal toxicity to human cells. These translational studies provide “proof of principle” to suggest anti-PGAM1 as a promising therapy in clinical treatment of tumors that heavily rely on the Warburg effect. However, the potential toxicity of PGAM inhibitors in vivo to normal post-mitotic, metabolically active organs such as brain, liver, skeletal muscle and heart, all of which are glycolytic, remains undetermined. This warrants further detailed toxicity and pharmacokinetics studies to improve the proposed anti-PGAM1 therapy in cancer treatment.
EXPERIMENTAL PROCEDURES
Cellular metabolites extraction and measurement
Cellular metabolites were extracted and spectrophotometrically measured as described previously (Kauffman et al., 1969; Minakami et al., 1965) with some modifications. To determine cellular concentration of 2-PG and 3-PG, 100 μL of packed cell pellets were homogenized in 1.5 ml of hypotonic lysis buffer (20 mM HEPES (pH 7.0), 5 mM KCl, 1 mM MgCl2, 5 mM DTT, and protease inhibitor cocktail). The homogenates were centrifuged in a cold room at 4 °C for 10 minutes at maximum speed, and the supernatants were applied to Amicon Ultra tubes with 10KDa cut off filter (Millipore). The flow through containing the metabolites was used for the measurement. NADH, ADP, and MgCl2 were added to the flow through to final concentrations of 0.14 mM, 1 mM, and 50 mM, respectively. Recombinant LDH and PKM1 proteins were added to final concentrations of 5 μg/ml and 10 μg/ml, respectively. Recombinant enolase protein was added to a final concentration of 50 μg/ml to measure cellular 2PG. Once the reaction was initiated by enolase, a decrease in absorbance at 340nm from NADH oxidation was measured by a DU800 spectrophotometer (Beckman). After termination of the enolase reaction, recombinant PGAM1 protein was added to a final concentration of 25 μg/ml and the decrease in absorbance at 340 nm was immediately monitored to measure cellular 3PG. 100 μL of 2-PG and 3-PG (Sigma) diluted with 1.5 ml of hypotonic lysis buffer were used as the standards.
Protein crystallization, data collection and structure determination
For protein crystallization, 6PGD was crystallized by using the hanging drop vapor diffusion method. To soak in 3-PG, 0.2 μL of 5 mM 3-phosphoglycerate acid were added into the mixture and incubated for 2 hours. The crystals were then flash-frozen in liquid nitrogen with the same cryoprotectant solution. Since a high concentration of 3-PG causes the crystal to crack, a final 3-PG concentration of 833 μM was applied and an occupancy of 50% was obtained after refinement. For data collection, the crystal diffraction data of 6PGD apo-form and 6PGD-3-PG complex were collected at the macromolecular crystallography for life science beamline GM/CA-CAT (23-ID-F) and NE-CAT (24-ID-E) respectively at the Advanced Photon Source, Argonne National Laboratory. The data were processed with HKL2000 and the scaled data were used for molecular replacement. For phasing, model building and refinement, the structure of 6PGD apo-form was determined by molecular replacement using Molrep in the CCP4 suite, with the protein portion of the solved structure of 6PGD-6-PG as the search model (pdb code: 3FWN)(Chen et al., 2010). The structure was then refined by using Phenix (Haddadian et al., 2011). Manual rebuilding of the model was carried out using the molecular graphics program COOT based on electron density interpretation. Water molecules were incorporated into the model if they gave rise to peaks exceeding 3σ in Fo-Fc density maps. The structure of 6PGD-3-PG complex was also determined by molecular replacement using Molrep with the solved structure of 6PGD apo-form as the search model, and then refined by using Phenix. Manual rebuilding of the model was carried out using the molecular graphics programs COOT. The final structures of apo-form and 6PGD-3-PG complex were visualized by using PyMol software.
Xenograft studies
Approval of use of mice and designed experiments was given by the Institutional Animal Care and Use Committee of Emory University. Nude mice (Athymic Nude-Foxn1nu, female 6–8-week-old, Harlan) were subcutaneously injected with 10 × 106 H1299 cells harboring empty vector on the left flank, and cells with stable knockdown of endogenous hPGAM1 on the right flank, respectively. The tumors were harvested and weighed at the experimental endpoint, and the masses of tumors (g) derived from cells with and without stable knockdown of endogenous hPGAM1 in both flanks of each mouse were compared. Statistical analyses were performed by comparison in relation to the control group with a two-tailed paired Student’s t test. For drug evaluation of PGMI-004A using xenograft mice, the drug was administered by daily i.p. injection at a dose of 100 mg/kg from 6 days after subcutaneous injection of H1299 cells on right flank of each mouse. Tumor growth was recorded by measurement of two perpendicular diameters of the tumors over a 3-week course using the formula 4π/3 × (width/2)2 × (length/2). The tumors were harvested and weighed at the experimental endpoint. The masses of tumors (g) treated with vehicle control (DMSO:PEG400:PBS at a ratio of 4:3:3) and PGMI-004A were compared and p values were determined by a two-tailed Student’s t test.
Primary tissue samples from human patients with leukemia and healthy donors
Approval of use of human specimens was given by the Institutional Review Board of Emory University School of Medicine. All clinical samples were obtained with informed consent with approval by the Emory University Institutional Review Board. Clinical information for the patients was obtained from the pathological files at Emory University Hospital under the guidelines and with approval from the Institutional Review Board of Emory University School of Medicine and according to the Health Insurance Portability and Accountability Act. Only samples from patients that were not previously treated with chemotherapy or radiation therapy were used. Mononuclear cells (MNCs) were isolated from peripheral blood and bone marrow samples from human leukemia patients or peripheral blood samples from healthy donors using lymphocyte separation medium (Cellgro). Cells were cultured in RPMI 1640 medium supplemented with 10% FBS and penicillin/streptomycin and incubated with increasing concentrations of PGMI-004A for up to 72 or 120 hours.
Supplementary Material
Significance.
The current understanding of the Warburg effect consists of an increase in aerobic glycolysis in cancer cells. The connection between glycolysis and PPP/biosynthesis is based upon a model in which glycolytic intermediates can be diverted into PPP and biosynthesis pathways as precursors. Our findings demonstrate that PGAM1 regulates the concentrations of glycolytic metabolites 3-PG and 2-PG, which function as signaling molecules to directly affect the catalytic activity of enzymes involved in PPP and biosynthesis, representing an additional link between glycolysis, PPP and biosynthesis. PGAM1 inhibitor PGMI-004A exhibits promising efficacy and minimal toxicity in treatment of xenograft nude mice and human primary leukemia cells, providing “proof of principle” for the development of PGAM1 inhibitors as anti-cancer agents.
Highlights.
PGAM1 controls 3-PG and 2-PG levels to coordinate glycolysis and biosynthesis
3-PG binds to and inhibits 6PGD in the oxidative PPP
2-PG potentiates PHGDH to provide feedback control of 3-PG levels
PGAM1 is a promising anti-cancer target
Acknowledgments
We thank Susan Sunay at the Hematology Division Tissue Bank, Winship Cancer Institute of Emory for providing primary tissue samples from leukemia patients, Drs. Sagar Lonial and Lawrence Boise for providing peripheral blood samples from healthy donors, and Dr. Yoke Wah Kow for human HaCaT and PIG1 cell lines. This work was supported in part by NIH grants CA120272 (J.C.), CA140515 (J.C.), GM071440 (C.H.) and the Pharmacological Sciences Training Grant T32 GM008602 (S.E.). J.X. and T.-L.G. are employees of Cell Signaling Technology, Inc. T.H. is a Fellow Scholar of the American Society of Hematology. S. E. is an NIH pre-doctoral fellow and an ARCS Foundation Scholar. G.Z.C., D.M.S., F.R.K., S.K. and J.C. are Georgia Cancer Coalition Distinguished Cancer Scholars. S. K. is a Robbins Scholar. S.K. and J.C. are American Cancer Society Basic Research Scholars. J.C. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Supplemental Information includes six figures, four tables, Supplemental Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Chen YY, Ko TP, Chen WH, Lo LP, Lin CH, Wang AH. Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism. J Struct Biol. 2010;169:25–35. doi: 10.1016/j.jsb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Corcoran CA, Huang Y, Sheikh MS. The regulation of energy generating metabolic pathways by p53. Cancer biology & therapy. 2006;5:1610–1613. doi: 10.4161/cbt.5.12.3617. [DOI] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- Engel M, Mazurek S, Eigenbrodt E, Welter C. Phosphoglycerate mutase-derived polypeptide inhibits glycolytic flux and induces cell growth arrest in tumor cell lines. J Biol Chem. 2004;279:35803–35812. doi: 10.1074/jbc.M402768200. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Morris GM, Wu J, Olson AJ, Sorensen EJ, Cravatt BF. Mechanistic and structural requirements for active site labeling of phosphoglycerate mutase by spiroepoxides. Molecular bioSystems. 2007;3:495–506. doi: 10.1039/b705113a. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol. 2005;23:1303–1307. doi: 10.1038/nbt1149. [DOI] [PubMed] [Google Scholar]
- Feig SA, Shohet SB, Nathan DG. Energy metabolism in human erythrocytes. I. Effects of sodium fluoride. The Journal of clinical investigation. 1971;50:1731–1737. doi: 10.1172/JCI106662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk S, Anderson N, Hainz C, Eckhardt SG, Serkova NJ. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clin Cancer Res. 2004;10:6661–6668. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- Haddadian EJ, Gong H, Jha AK, Yang X, Debartolo J, Hinshaw JR, Rice PA, Sosnick TR, Freed KF. Automated real-space refinement of protein structures using a realistic backbone move set. Biophys J. 2011;101:899–909. doi: 10.1016/j.bpj.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nature cell biology. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman FC, Brown JG, Passonneau JV, Lowry OH. Effects of changes in brain metabolism on levels of pentose phosphate pathway intermediates. J Biol Chem. 1969;244:3647–3653. [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Li L, Dworkowski FS, Cook PF. Importance in catalysis of the 6-phosphate-binding site of 6-phosphogluconate in sheep liver 6-phosphogluconate dehydrogenase. J Biol Chem. 2006;281:25568–25576. doi: 10.1074/jbc.M601154200. [DOI] [PubMed] [Google Scholar]
- Liu L, Wang S, Zhang Q, Ding Y. Identification of potential genes/proteins regulated by Tiam1 in colorectal cancer by microarray analysis and proteome analysis. Cell Biol Int. 2008;32:1215–1222. doi: 10.1016/j.cellbi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Minakami S, Suzuki C, Saito T, Yoshikawa H. Studies on erythrocyte glycolysis. I. Determination of the glycolytic intermediates in human erythrocytes. J Biochem. 1965;58:543–550. doi: 10.1093/oxfordjournals.jbchem.a128240. [DOI] [PubMed] [Google Scholar]
- Minakami S, Takayasu S, Suzuki C, Yoshikawa H. The hydrogen ion concentrations and erythrocyte glycolysis. Biochem Biophys Res Commun. 1964;17:748–751. [Google Scholar]
- Mulquiney PJ, Kuchel PW. Model of 2,3-bisphosphoglycerate metabolism in the human erythrocyte based on detailed enzyme kinetic equations: equations and parameter refinement. Biochem J. 1999;342(Pt 3):581–596. [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao Y, Chen X, Zeng D, Tong A, Chen L, et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Molecular cancer. 2010;9:81. doi: 10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauerte JA, Gafni A. Long-lived tryptophan fluorescence in phosphoglycerate mutase. Biochemistry. 1989;28:3948–3954. doi: 10.1021/bi00435a048. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Boulahbel H, Gottlieb E. Metabolic transformation in cancer. Carcinogenesis. 2009;30:1269–1280. doi: 10.1093/carcin/bgp070. [DOI] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- Willis JE, Sallach HJ. The occurrence of D-3-phosphoglycerate dehydrogenase in animal tissues. Biochim Biophys Acta. 1964;81:39–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.