Abstract
Loss of epithelial organization is a hallmark of carcinomas, but whether polarity regulates tumor growth and metastasis is poorly understood. To address this issue we depleted the Par3 polarity gene by RNAi in combination with oncogenic Notch or Ras61L expression in the murine mammary gland. Par3 silencing dramatically reduced tumor latency in both models, and produced invasive and metastatic tumors that retained epithelial marker expression. Par3 depletion was associated with induction of MMP9, destruction of the extracellular matrix, and invasion, all mediated by atypical PKC-dependant JAK/Stat3 activation. Importantly, Par3 expression is significantly reduced in human breast cancers, which correlates with active aPKC and Stat3. These data identify Par3 as a regulator of signaling pathways relevant to invasive breast cancer.
INTRODUCTION
Most solid tumors arise from epithelial cells that have acquired changes in proliferative and organizational capacity. Epithelial cells form characteristic intercellular adhesions and possess apical/basal polarity, which is lost in some invasive and metastatic cancers in a process related to the epithelial-mesenchymal transitions (EMT) that occur during development (Thiery et al., 2009). However, in many cases epithelial features are retained. How epithelial tissues establish their organization in a normal state and how this organization is disrupted during cancer progression are still not well understood. In particular, it is largely unknown if the cell polarity machinery is perturbed during tumorigenesis, and if such disruptions promote metastasis.
Many of the polarity protein complexes localize to distinct domains within the plasma membrane. The Par genes (Par1, 3, 4, 5, 6 and aPKC) encode an evolutionarily conserved group of polarity proteins that play key roles in many aspects of cell polarization (Goldstein and Macara, 2007). To date, only Par4, a protein kinase also known as LKB1, has been identified as a tumor suppressor (Jansen et al., 2009), and it remains uncertain if tumorigenesis in patients with mutant LKB1 is caused by loss of its polarity function.
We have focused on Par3, a multi-domain scaffolding protein required for the spatial organization of several important signaling proteins (Goldstein and Macara, 2007). Par3 is essential for the delivery of aPKC to the apical surface (Harris and Peifer, 2005; McCaffrey and Macara, 2009), through binding of Par3 to the adapter protein Par6, which forms a constitutive complex with aPKC. Furthermore, aPKC can interact directly with Par3, which is essential for apical aPKC localization and epithelial organization (Horikoshi et al., 2009; McCaffrey and Macara, 2009). Loss of aPKC from the apical cortex causes spindle pole orientation defects and epithelial misorganization (Hao et al. 2010). Both the level of aPKC expression and mislocalization correlate with increased invasion and metastasis in breast cancer (Kojima et al., 2008). However, whether loss of Par3 has a role in regulating aPKC during tumorigenesis is unknown.
Some proteins have oncogenic activity when over-expressed. The Notch receptor, an important transcriptional regulator of stem cell fate, is activated by proteolytic cleavage to release an intracellular domain (NICD), which is found at elevated levels in up to 50% of human breast cancers (Pece et al., 2004); and mammary-specific expression of NICD in mice induces breast tumors, though with no metastasis (Hu et al., 2006). Additionally, enhanced growth factor receptor signaling promotes breast cancer. A central effector of growth factor receptor signaling is the Ras oncogene, which, although rarely mutated in breast cancer, is frequently hyperactivated (Clark and Der, 1995). Elevated expression of Neu/ErbB2 or Met receptors are observed in 20–30% and 15–20% of breast cancers, respectively, and can inappropriately stimulate Ras-mediated signaling pathways (Reese and Slamon, 1997; Ponzo and Park, 2010).
Progression of in situ breast carcinomas to metastatic disease requires additional steps, and it is now established that inflammation is necessary for this process (Grivennikov and Karin, 2008). Stat3 has a central role in regulating inflammation in breast cancer through a cytokine loop involving IL-6 (Grivennikov and Karin, 2008; Schafer and Brugge, 2007). Stat3 is Tyr-phosphorylated by Src or JAK kinases, which induces translocation to the nucleus. Stat3 can be hyper-activated in breast cancers, which promotes invasion and metastasis, although Stat3 activation alone is insufficient to induce tumorigenesis (Barbieri et al., 2010b). Therefore, many of the processes that drive tumorigenesis and metastasis are separable, but how they relate to tissue organization is not well understood.
The goal of this study was to determine the role of the apicobasal cell polarity machinery in tumorigenesis, with a focus on the Par3 polarity protein. Using a mouse mammary transplant model coupled with lentiviral transduction, we silenced Par3 expression in the context of two different oncogenes and determined whether loss of Par3 drives tumor growth and/or metastasis. The expression of Par3 was also examined in human breast cancers.
RESULTS
Loss of Par3 cooperates with Notch intracellular domain (NICD) to promote tumorigenesis
We used lentiviral RNAi to deplete Par3 from primary mammary epithelial cells (MECs) and transplanted them orthotopically into the inguinal (#4) mammary fat pads of syngeneic mice. Previously, we reported that Par3-depleted mammary progenitor cells form disorganized ductal outgrowths that resemble early ductal carcinoma in situ (McCaffrey and Macara, 2009). However, over a period of 24 – 37 weeks post-transplantation, Par3 depletion did not lead to tumor formation (Figure 1A), suggesting that Par3 is not a classical tumor suppressor. Next, we asked whether loss of Par3 might enhance tumorigenesis in the context of an oncogene. We initially used NICD, which is upregulated in ~ 50% of human breast cancers (Pece et al., 2004) and drives tumor formation in mice after a latency of ~9 months (Hu et al., 2006). Primary MECs isolated from C3H mice were transduced with lentivirus that expresses active, myc-tagged NICD plus short hairpin RNAs to either Luciferase (control) or murine Par3, using Par3 shRNA that we had validated previously (McCaffrey and Macara, 2009). We refer to the transduced MECs as NICD/shLuc and NICD/shPar3, respectively. For each animal, 10,000 NICD/shLuc or NICD/shPar3 MECs were injected into contra-lateral inguinal (#4) fat pads of the same mouse. Immunoblots of tissue lysates showed that myc-NICD was expressed in the tumors and that Par3 silencing was efficient (Figure 1B).
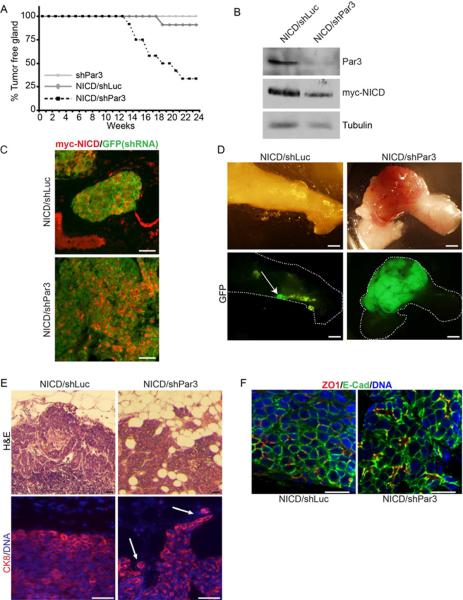
Figure 1.
Loss of Par3 cooperates with NICD to promote mammary tumor formation (A) Kaplan-Meier (KM) curve of tumor-free status in mice transplanted with shPar3 (n=17), NICD/shLuc (n=11), or NICD/shPar3 (n=12) MECs. (B) Tumors arising from orthotopically transplanted myc-NICD/shLuc or myc-NICD/shPar3 MECs were immunoblotted for Par3, myc-NICD and tubulin. (C) Immunofluorescence staining of tumor sections for myc-NICD (red) and GFP (green), which marks cells expressing shRNA. (D) Tumors arising from NICD/shLuc or NICD/shPar3 transduced MECs. GFP is co-expressed with the shRNA and is used as a marker for transduction. Arrow indicates small non-palpable NICD/shLuc tumor, which were found in 7/11 fat pads. (E) Upper panels: Hematoxylin and Eosin (H&E) stained sections of the edges of NICD/shLuc and NICD/shPar3 tumors. Lower panels: Tissue sections of the tumor edge stained with cytokeratin 8 (CK8, red) and Hoechst 33258 (DNA, blue). Arrows show invading cells. (F) Immunofluorescence staining of tumor sections for E-cadherin, ZO1, and Hoeschst 33342 for DNA. Scale bars= 50 μm (C), 2 mm (D), 100 μm (E), 20 μm (F). See also Figure S1.
Strikingly, loss of Par3 caused a dramatic reduction in tumor latency for NICD-transduced MECs, with 50% of NICD/shPar3 animals developing tumors by 18 weeks (Figure 1A). We confirmed that tumors were derived from cells expressing both NICD and shPar3 by imaging the GFP marker for the RNAi lentivirus, and by staining for myc-NICD (Figure 1C, D).
We transplanted NICD/shLuc and NICD/shPar3 MECs into opposite sides of the same mouse, and palpable NICD/shLuc tumors were rarely formed when mice were sacrificed due to the NICD/shPar3 tumor burden. However, in some cases, small NICD/shLuc tumors were found by microscopic examination of the mammary fat pads (Figure 1D). Consistently, all tumors were GFP-positive, and tumors derived from NICD/shPar3 MECs were much larger than those from the NICD/shLuc MECs (Figure 1D). Moreover, while NICD/shLuc tumors possessed well-defined boundaries, the loss of Par3 induced a more invasive phenotype, with cells protruding into the surrounding fat pad (Figure 1E). Both types of tumors retained epithelial characteristics, including expression of E-cadherin at intercellular junctions and the tight junction marker ZO1 at apicolateral boundaries surrounding microlumens (Figures 1E, F). We further examined cellular organization by staining tumor sections for K8 and cytokeratin 14 (K14). In normal murine mammary ducts, K14 is expressed in myoepithelial cells whereas K8 is restricted to luminal cells (Figure S1). Loss of Par3 increased tumor cell heterogeneity in our NICD model (Figures 1E, F, S1). NICD/shLuc tumors were homogeneous and predominantly K8+K14moderate, but the NICD/shPar3 tumors displayed a substantially greater degree of cellular diversity.
Loss of Par3 cooperates with oncogenic H-Ras to promote tumorigenesis
To determine if the promotion of tumor growth by loss of Par3 is specific to NICD or is of more general importance, we asked if Par3 depletion cooperates with a different oncogene, H-Ras61L. Knockdown of Par3 in conjunction with oncogenic GFP-tagged Ras61L significantly reduced tumor latency compared to GFP-Ras61L alone (Figure 2A). Palpable Ras61L/shPar3 tumors had an average latency of 114 d ± 68 compared to Ras61L/shLuc, which had a latency of >230 d. By 37 weeks, 92% of Ras61L/shLuc transplant mice remained tumor free compared to 54% Ras61L/shPar3 transplant mice (Figure 2A). We confirmed comparable Ras expression levels and efficient Par3 knockdown by immunoblotting tumor lysates (Figure 2B).
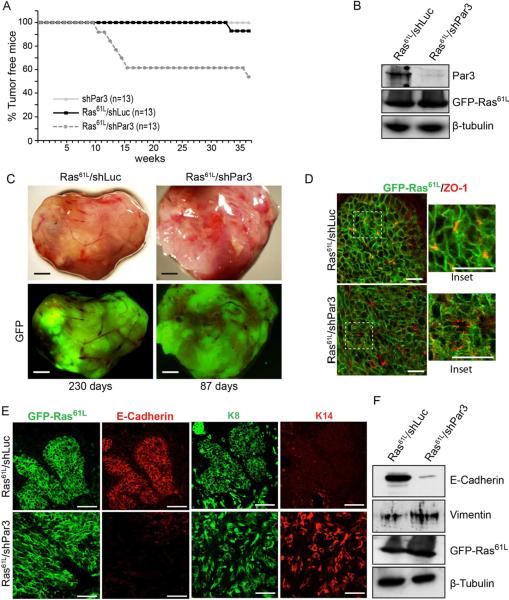
Figure 2.
Loss of Par3 cooperates with Ras61L to promote mammary tumor formation (A) KM curve for mice transplanted with MECs expressing GFP/shPar3 (n=13), Ras61L/shLuc (n=13), or Ras61L/shPar3 (n=13). (B) Imunoblot of primary Ras61L /shLuc and Ras61L /shPar3 tumor lysates. (C) Micrographs of tumors from transplanted Ras61L/shLuc or Ras61L/shPar3 MECs. GFP indicates tumor cells are transduced with lentivirus. (D) Immunofluorescence staining of tumor sections for GFP-Ras61L (green) and ZO1 (red). (E) Immunofluorescence staining of tumor sections for GFP-Ras61L (green) and E-cadherin (red), or cytokeratin 8 (green) and cytokeratin 14 (red). (F) Western blot of primary Ras61L /shLuc and Ras61L /shPar3 tumor cell lysates. Scale bars = 500 μm (C), 50 μm (D) and (E). See also Figures S2.
While both Ras61L/shLuc and Ras61L/shPar3 tumors expressed GFP and were able to grow to comparable sizes (Figures 2C), Ras61L/shPar3 tumors grew more rapidly and were consistently more aggressive than Ras61L/shLuc tumors; they invaded through the peritoneum, with the bulk of the tumors growing inside the body cavity, and were not detected during palpation (Figure S2A). Additionally, ~30% of the Ras61L/shPar3 tumors invaded through the skin (not shown). Consistent with these differences in invasiveness, Ras61L/shLuc tumors were more organized and retained regions that possessed a lobular organization with distinct boundaries, while Ras61L/shPar3 tumors exhibited no discernable organization and cells appeared more spindle-shaped (Figures 2D, E). In contrast, NICD/shPar3 tumors were restricted to the fat pad, with occasional invasion into the peritoneum.
Ras tumors depleted of Par3 retained expression of ZO1, which remained localized at sites of cell-cell contacts marking the boundaries of minilumens (Figure 2D). Unexpectedly, although Ras61L is concentrated at intercellular junctions in the Ras61L/shLuc tumors, loss of Par3 results in the partial redistribution of the oncoprotein into the cytoplasm (Figure 2D, insets), which might have consequences for downstream signaling. The tumors also retained expression of the luminal epithelial marker cytokeratin 8 (K8) (Figure 2E), demonstrating that, as with the NICD model, the Ras61L tumor cells are disorganized but retain epithelial characteristics in the absence of Par3. The Ras61L/shLuc tumors also expressed E-cadherin (Figure 2E). In contrast, however, E-cadherin (Figure 2E) and β-catenin (Figure S2B) were almost undetectable in Ras61L/shPar3 tumors. The absence of staining reflects down-regulation of expression rather than mislocalization (Figures 2F, S2C). Although Ras61L/shPar3 tumor cells were more spindle-shaped, there was no increase in vimentin, a mesenchymal marker (Figure 2F). Thus, loss of Par3, specifically in the context of the Ras oncogene, represses E-cadherin expression, though not the loss of other luminal epithelial markers.
Interestingly, Ras61L/shLuc tumor cells were primarily K8+K14−, whereas the Ras61L/shPar3 tumors were more heterogeneous, and included K8+K14− and K8+K14+ dual-positive cells (Figure 2E). K8+K14+ dual positive cells may be undifferentiated progenitors (Raouf et al., 2008; Shackleton et al., 2006; Villadsen et al., 2007). Together, these data are consistent with our previous identification of a role for Par3 in driving progenitor cell differentiation in the mammary gland (McCaffrey and Macara, 2009). They also indicate that loss of Par3 causes tissue misorganization rather than a simple loss of apical/basal polarity.
Par3 acts as an invasion and metastasis suppressor
To determine if loss of Par3 promotes metastasis we examined the lungs of mice after orthotopic injection of NICD/shLuc or NICD/shPar3 into mammary fat pads. None of the NICD/shLuc mice had lung metastases (n=14), consistent with published data on NICD transgenic mice (Hu et al., 2006). However, >80% of NICD/shPar3 mice (n=17) displayed extensive colonization, with an average of 32 colonies visible per lung (Figures 3A, B). These values are significantly different (p = 0.0001). Importantly, lung metastases from both NICD/shPar3 and Ras61L/shPar3 tumors were comprised of similar epithelial cell types as the primary tumors (Figure S3A, B). While Ras61L alone was sufficient to induce metastasis, loss of Par3 increased the number and size of the colonies (Figure S3C).
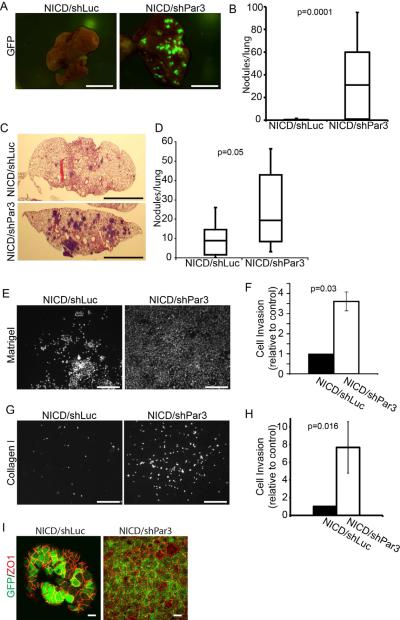
Figure 3.
Suppression of Par3 increases tumor invasion and metastasis
(A) Whole mount GFP fluorescent images of lung metastases from tumor-bearing mice following orthotopic mammary gland transplants of MECs transduced with NICD/shLuc (n=14) and NICD/shPar3 (n=17; p=0.0001). (B) Box plots showing the number of metastatic nodules in lungs from (A). (C) H&E-stained sections of lungs following tail vein injections of 3×105 MECs transduced with NICD/shLuc or NICD/shPar3. (D) Box plots showing the number of metastatic nodules in lungs (n=10) following systemic injections of transduced cells from (C). (E) Hoechst stained nuclei of NICD/shLuc or NICD/shPar3 MECs that invaded through the Matrigel pad and 8μm filter inserts after 72 h. (F) Quantification of (E) and results are average of 3 independent experiments. Error bars are 1 sem. (G) Same as (E), except invasion through collagen I gels. (H) Quantification of (G); results are means of 3 independent experiments. Error bars = sem. (I) Immunofluorescence staining of NICD/shLuc or NICD/shPar3 MECs that migrated through the Matrigel for GFP (green) and ZO1 (red). Scale bars = 1cm (A, C), 100 μm (E, G), 10 μm (I). See also Figures S3.
As a further test of metastatic potential, we injected equal numbers of NICD/shLuc or NICD/shPar3 mammary cells systemically via the tail veins (n=10), and after 3 weeks the lungs were sectioned and examined for metastases. In all cases, colonization of the lungs was detected (Figure 3C), but metastases produced by NICD/shPar3 cells were significantly larger and more numerous compared to NICD/shLuc cells (Figure 3D). These results are consistent with increased efficiency of invasion, dissemination and colonization, and support the hypothesis that Par3 normally can suppress metastatic progression.
To further examine the potential for Par3 to suppress tumor invasion, we asked if MECs transduced with or without Par3 shRNA and an oncogene would show increased migration in vitro, using 3-dimensional Matrigel or collagen I invasion assays (Figures 3E–H, S3D–E). Previously, loss of Par3 has been reported to inhibit cell migration in 2-dimensional scratch wound assays (Schmoranzer et al., 2009). The non-transduced cells showed no detectable invasion through Matrigel pads over the time course of the experiment, and silencing of Par3 alone did not enable invasion (not shown). Expression of either the NICD or Ras61L oncogenes caused a significant number of cells to penetrate through the Matrigel. Strikingly, however, the silencing of Par3 in the context of either oncogene caused a substantial increase in invasion efficiency. Loss of Par3 caused a 3.5-fold (NICD) and 3.9-fold increase (Ras61L) in cells that invaded through the 3D matrix, compared to controls (Figures 3E–F, S3D–E). Invasion of NICD cells through collagen I was also stimulated more than 7-fold by loss of Par3 (Figure 3G–H).
Although most metastatic carcinomas retain epithelial characteristics, it has been proposed that tumor cells might undergo a transient EMT during dissemination then revert to an epithelial phenotype when they colonize an ectopic site (Guarino et al., 2007). We examined the expression of EMT markers in our cultures and found a modest increase in ZEB1 expression, but no overall changes in gene expression that would indicate a complete EMT (Figure S3F). Interestingly, the NICD/shPar3 cells that had migrated through the matrix to the filter retained expression of the epithelial marker ZO1 (Figure 3I). We conclude that in the context of an activated oncogene, loss of Par3 expression increases invasive behavior, and these cells retain the ability to express epithelial characteristics.
Loss of Par3 induces MMP expression and cell detachment in transformed mammary cells
Migration through 3D matrices often requires expression of matrix metalloproteinases (MMPs), which degrade the ECM (Rorth, 2009). To test whether loss of Par3, in the context of an oncogene, might alter MMP expression, or expression of other adhesion-related genes, we performed QRT-PCR array analysis of adhesion-related genes on primary NICD/shLuc and NICD/shPar3 MECs in vitro, in the absence of selection. In the context of NICD/shPar3, MMP9 showed the most robust increase in expression over NICD /shLuc, and MMP9 induction was second highest in the Ras model, of all genes analyzed (Tables S1–S4). Changes in MMP9 expression were confirmed by RT-PCR using different primers (Figure 4A). The expression of 3 other genes was upregulated and 10 genes were reduced in both models (Figure S4A). Expression of other genes differed between the two models, and the expression of some other MMPs was reduced (MMPs 1a, 12, 14 for NICD; and 2, 15 for Ras). NICD/shPar3 cells also showed significant decreases in the protease inhibitors, TIMP1 and TIMP2. Therefore, in both models, metalloproteinase expression was altered by Par3 depletion, with MMP9 being the most consistently up-regulated gene.
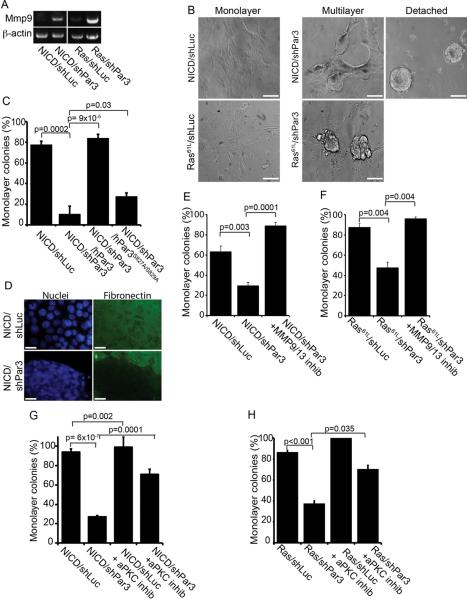
Figure 4.
Loss of Par3 induces MMP expression and cell detachment in transformed mammary epithelial cells through activation of aPKC
(A) RT-PCR on total RNA from tumor tissues expressing either NICD or Ras61L +/− shRNA against Par3, using primers for MMP9 and β-actin (control). (B) Primary MECs stably expressing NICD/shLuc, NICD/shPar3, Ras61L/shLuc or Ras61L/shPar3 were plated on fibronectin for 72 h, and imaged by DIC. Representative images of the various colony phenotypes are shown. Scale bars = 50 μm. (C) Quantification of cell detachment by MECs expressing NICD/shLuc, NICD/shPar3, and NICD/shPar3 with RNAi-resistant full-length human Par3, or mutant Par3S827A/S829A that does not bind aPKC. (D) IF staining of fibronectin under colonies of NICD/shLuc and NICD/shPar3 MECs. Scale bars = 10 μm. (E) Quantification of cell detachment for NICD/shLuc and NICD/shPar3 MECs +/− 400 pM of MMP Inhibitor I. (F) Quantification of cell detachment for Ras61L/shLuc and Ras61L/shPar3 MECs grown +/− 400 pM MMP9/13 inhibitor I. (G) Quantification of cell detachment for NICD/shLuc and NICD/shPar3 MECs +/− 40μg/ml of aPKC pseudosubstrate inhibitor. (H) Detachment of Ras61L/shLuc and Ras61L/shPar3 MECs grown +/− 40μg/ml aPKC inhibitor. Results are means of at least 3 independent cultures. Error bars = +/− 1sd. See also Figure S4 and Tables S1–S4.
Consistent with induction of MMPs, there was a dramatic change in colony morphology when Par3 expression was suppressed. Most NICD/shLuc cells grew as monolayers on fibronectin-coated dishes (~60–80%; Figure 4B, C). In contrast, only ~10–30% of NICD/shPar3 cells formed monolayers, with the rest detaching as spheroid colonies after several days culture (Figure 4B). Importantly, all cultures adhered normally during the first 24 – 48 h demonstrating that maintenance rather than initial adhesive ability of the NICD/shPar3 cells is defective. Moreover, after trypsinization, detached cells were able to re-adhere to new plates, and again the cells began to detach after 24 – 48 h. To determine if loss of Par3 causes defective attachment to specific types of ECM we also plated cells on collagen I. Whereas ~67% of NICD/shLuc cells grew as monolayer colonies on collagen I, less than 1% of cells lacking Par3 remained as monolayer colonies on collagen I (Figure S4B).
Ras61L-transduced MECs also grew as monolayers, and silencing of Par3 increased multiple layering, while detachment occurred as single cells, rather than as multicellular spheroids (Figures 4B and S4C, D). The inability of Ras61L/shPar3 to form spheroids may be due to reduced E-cadherin expression as noted above, which would prevent cells from maintaining intercellular adhesions.
Consistent with the idea that ECM degradation is responsible for cell detachment, staining for fibronectin showed that while the ECM was intact beneath cells that express NICD alone, it was absent from patches where clusters of detached NICD/shPar3 cells had formed (Figure 4D). Finally, we asked if MMP activity is required for cell detachment. A MMP inhibitor almost completely blocked detachment induced by Par3 depletion in the context of either NICD or Ras (Figure 4E, F). To confirm the involvement of MMP9 in the invasive behavior of the NICD/shPar3 cells, we used shRNA-lentivirus that target the murine MMP9 and transduced them together with the NICD and shPar3 viruses into primary MECs. The two most effective shRNAs (shMMP9-1 and -3) significantly reduced invasion through Matrigel (Figures S4E, F). The MMP inhibitor also efficiently blocked invasion (Figure S4G). Together, these data identify a mechanism whereby loss of Par3 induces MMP9, which triggers degradation of ECM with consequent cell detachment and increased invasive migration.
Cell detachment is mediated through inappropriate activation of aPKC
To confirm that the adhesion defects were caused by loss of Par3, we first performed rescue experiments using shRNA-resistant human Par3, which efficiently restored the ability of cells (85%) to remain attached to the ECM (Figure 4C). Notably, however, a mutant (Par3S827A/S829A) that is unable to directly bind and be phosphorylated by aPKC could not rescue the adhesion defects, with only 28% of colonies growing as monolayers (Figure 4C). Next, to determine more directly whether aPKC activity is required for adhesion, we plated cells +/− an aPKC inhibitor. This myristoylated pseudosubstrate peptide penetrates cell membranes and specifically inhibits aPKC isoforms. Inhibition of aPKC completely restored cell-ECM adhesion to NICD/shPar3 and reduced Ras61L/shPar3 multilayering (Figure 4G, H).
Our previous work showed that Par3 is required for normal localization of aPKC to the apical surface of luminal epithelial cells (McCaffrey and Macara, 2009). To determine if aPKC is also mislocalized in NICD/shPar3 cells, we stained for aPKC and ZO1. In the NICD/shLuc control, ZO1 and aPKC both formed a tight border around the cells (Figure S4H). Thus, expression of the NICD oncogene alone is not sufficient to disrupt tight junctions and aPKC localization. In contrast, aPKC was completely lost from the apical junctions of Par3-depleted NICD MECs. Cortical ZO1 persisted in Par3-depleted cells, but was more punctate (Figure S4H). This result is consistent with our previous studies showing that loss of Par3 negatively affects tight junction formation (Chen and Macara, 2005).
Since inhibiting aPKC can reverse cell detachment, we asked whether aPKC activity was altered in NICD/shPar3 cells. Active atypical PKC is phosphorylated on T403/T410, and immunoblots of cell lysates for total and p-aPKC revealed a substantial increase in p-aPKCT410 levels in NICD/shPar3 cells (Figure 5A, left panels). Thus, in these oncogene-transformed mammary cells, loss of Par3 induces aPKC activation. Notably, however, in normal epithelial cells, although loss of Par3 causes mislocalization of aPKC it does not alter T410 phosphorylation (Hao et al. 2010).
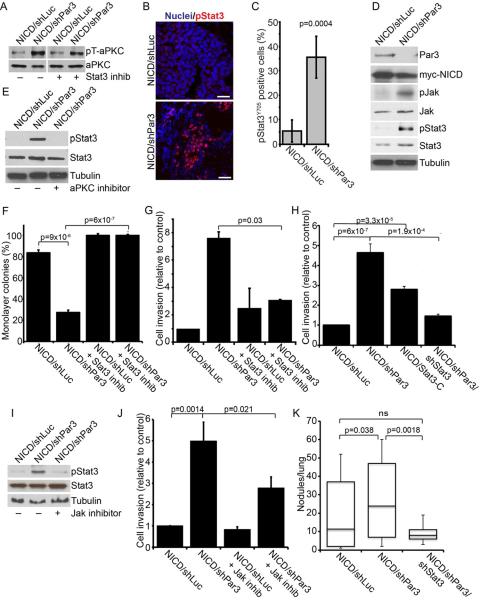
Figure 5.
Loss of Par3 activates Stat3 signaling through aPKC
(A) Immunoblot of NICD/shLuc and NICD/shPar3 MEC lysates showing active phospho-aPKCT410/403 (pT-aPKC) and total aPKC protein levels +/− 50nM Stat3 inhibitor, Cucurbitacin I. (B) Immunofluorescence staining of NICD/shLuc or NICD/shPar3 tumor sections for pStat3Y705 (red) and nuclei (blue). Scale bars = 50 μm. (C) Quantification of pStat3Y705 positive cells in tumor sections (n=8). (D) Immunoblots of lysates from NICD/shLuc or NICD/shPar3 primary MECs. Phospho-antibodies were used to detect pJAKY1007/8 and pStat3Y705. (E) Immunoblots of NICD/shLuc or NICD/shPar3 MEC lysates +/− 40μg/ml aPKC inhibitor. (F) Quantification of cell detachment for NICD/shLuc and NICD/shPar3 MECs grown +/− 50 nM Cucurbitacin I. Results from 2 independent experiments. (G) Quantification of invasion of NICD/shLuc and NICD/shPar3 MECs through Matrigel, +/− 50 nM Cucurbitacin I. (H) Quantification of invasion of NICD/shLuc, NICD/shPar3, constitutively active Stat3-C, and NICD/shPar3/shStat3 MECs through Matrigel. (I) Immunoblots of NICD/shLuc or NICD/shPar3 MEC lysates +/− 10 nM JAK inhibitor, Pyridone 6. (J) Quantification of invasion of NICD/shLuc and NICD/shPar3 MECs through Matrigel, +/− 10 nM Jak inhibitor, Pyridone 6. (K) Quantification of lung nodules arising from NICD/shLuc, NICD/shPar3, and NICD/shPar3/shStat3 transformed MECs injected systemically. Five sections were examined from each lung of 5 mice for each treatment. Results are means of at least 3 independent cultures, unless otherwise noted. Error bars = +/− 1sd. See also Figure S5.
Loss of Par3 activates Stat3 signaling through aPKC
To determine the mechanism by which Par3 controls adhesion and invasion, we examined potential downstream signaling pathways. Stat3 activation drives MMP expression in multiple cancer cells, and promotes invasive behavior (Song et al., 2008; Xie et al., 2004). Active Stat3 is also frequently present at the invasive edge of tumors (Bromberg and Wang, 2009). Therefore, we stained sections from the NICD tumors for phospho-Stat3 (pStat3Y705). Although few cells were pStat3Y705-positive in the NICD/shLuc tumors, silencing Par3 increased the number of pStat3Y705-positive cells throughout the tumor mass (Figure 5B, C).
In many tumors, Stat3 activation can be induced indirectly through cytokine secretion by infiltrating hematopoietic cells (Yu et al., 2009). To test whether Stat3 activation is intrinsic to NICD/shPar3 epithelial cells, we examined transduced, unselected MECs. Only ~5% of NICD/shLuc cells expressed detectable (but weak) levels of pStat3Y705. Significantly, however, pStat3Y705 was present in 18% of NICD/shPar3 MECs, with a higher intensity of staining compared to NICD/shLuc cells (Figure S5A, B). Silencing Par3 in Ras61L MECs also caused a substantial increase in pStat3Y705 as judged by immunofluorescence (Figure S5C, D). We confirmed that loss of Par3 caused a marked increase in pStat3Y705 levels as determined by immunoblot (Figure 5D). Furthermore, conditioned medium from NICD/shPar3 cultures failed to induce Stat3 activation when added to NICD/shLuc cultures (not shown). We conclude that Stat3 activation is cell autonomous and does not depend on paracrine cytokine signaling from immune cells.
To determine whether aPKC acts upstream or downstream of Stat3 activation, we inhibited Stat3 and examined aPKC activation by blotting for p-aPKCT403/410. Atypical PKC activity was independent of Stat3 activity (Figure 5A). In contrast, treatment of mammary NICD/shPar3 cells with the aPKCps inhibitor diminished pStat3Y705 to control levels (Figure 5E), demonstrating that aPKC acts upstream of Stat3 induction.
As Stat3 can induce MMP expression in some cancer cells, we next tested the effect of a selective Stat3 inhibitor, Cucurbitacin-I (JSI-124), on mammary cell detachment. Cells were treated with 50nM Cucurbitacin, 24 h after being plated on ECM, and were examined 48 h later. Cucurbitacin completely reversed the adhesion defect in NICD/shLuc, NICD/shPar3, Ras61L/shLuc and in ~80% of Ras61L/shPar3 colonies (Figures 5F and S5E). Moreover, treatment of NICD/shLuc and NICD/shPar3 MECs with Cucurbitacin significantly reduced the invasive potential of the MECs through Matrigel (Figure 5G). In addition, we asked if reducing Stat3 expression could block the effects of Par3 depletion in the invasion of MECs through Matrigel. As shown in Figures 5H and S5F, knockdown of Stat3 significantly reduced the invasiveness of the oncogene-transduced cells that lack Par3. These data suggest that activation of Stat3 is required for the cell detachment and invasive phenotypes caused by loss of Par3.
As a direct test of this hypothesis, we expressed a constitutively active mutant, Stat3-C (Bromberg et al., 1999), in MECs together with NICD or Ras61L. This mutant phenocopied the loss of Par3, by reducing cell attachment and increasing invasion (Figures 5H, 5SF). Furthermore, we confirmed that in murine MECs active Stat3-C also induces MMP9 (Figure S5G).
Jak phosphorylation was also substantially increased by loss of Par3 (Figure 5D). To test whether Jak is upstream of Stat3 activation in Par3-depleted cells we used a specific Jak inhibitor, Pyridone 6 (Thompson et al., 2002). This inhibitor blocked shPar3-dependent Stat3 activation and significantly reduced invasion of the cells through Matrigel (Figures 5I, J).
Importantly, all of the effects on signaling caused by loss of Par3 in the context of oncogenic activation could be fully reversed by expression of a GFP fusion of human Par3, (Figure S5H). GFP-hPar3 did not induce re-expression of the endogenous Par3 but inhibited the phosphorylation of aPKC and Stat3, and blocked the induction of MMP9. Therefore, these signaling responses depend specifically on the loss of Par3 and are not off-target effects of the shRNA.
Finally, to determine whether the induction of Stat3-mediated invasive behavior is significant for shPar3-dependent metastasis in vivo, we measured lung colonization after tail vein injections of NICD-transformed cells with shRNAs against Par3 alone or Par3 and Stat3. As described above (Figure 3) loss of Par3 caused a significant increase in lung metastasis, which was completely suppressed by co-silencing Stat3 (Figure 5K). These data identify an unanticipated pathway in which loss of Par3 results in the aPKC-dependent activation of JAK/STAT signaling, which induces MMP9 expression and consequent destruction of the ECM, increased invasion and lung metastasis by oncogene-activated mammary cells.
Par3 expression is frequently lost in human breast cancers
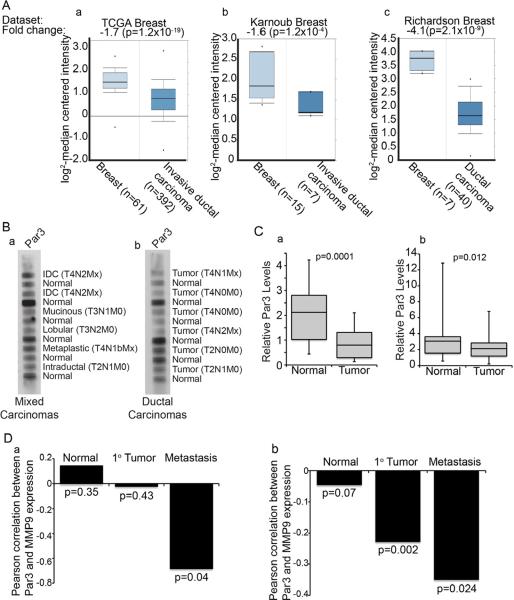
To address the relevance of Par3 loss to human breast cancer, we explored the expression of the PARD3 transcript in tumors from selected cohorts of patients. Significant reductions in PARD3 gene expression were apparent in invasive ductal and lobular carcinomas compared to normal breast tissue (Figure 6A). Notably, PARD3 expression was also reduced in other epithelial cancers (Supplementary Figure S6A). We next analyzed a human tumor lysate array by immunoblot and found a significant reduction in PAR3 protein for 50% of the tumors as compared to matched samples of normal breast tissue from the same patients (Figure 6B, left panel). A second, independent matched group of 52 patient samples also showed significant reduction in PAR3 expression (Figure 6B, right panel). The same membranes were also probed for RanGTPase as a loading control and the PAR3/RAN ratios were measured, to provide a corrected level of PAR3 (Figure 6C). To validate the specificity of the Par3 antibody, we probed breast cancer cell line lysates under similar buffer conditions to those used in generating the commercial membranes. The antibody specifically detected PAR3, with low background and no non-specific bands, indicating that the signal detected on the arrays is PAR3-specific (Figure S6B). In addition, we immunoblotted a limited number of freshly isolated normal and breast tumor samples including 3 invasive carcinomas (IDC) and one ductal carcinoma in situ (DCIS), which confirmed a decrease in PAR3 protein expression in the invasive breast cancer samples compared to normal tissue (Figure S6C).
Figure 6.
PAR3 expression is reduced in human breast cancer, correlating with elevated MMP9 expression
(A) Microarray data showing relative levels of PARD3 gene expression for invasive ductal carcinomas versus matched normal breast tissue from TCGA (a), Karnoub (b) and Richardson (c) datasets (see Supplementary Methods for information on datasets). (B) Human tumor protein array membranes of mixed carcinomas (a) and ductal carcinomas (b) with matched normal adjacent tissue were immunoblotted for PAR3. IDC=Intraductal carcinoma. TNM classification values are given for each tumor sample. (C) Box and whisker plots showing quantification of band intensities for PAR3 that were normalized to RAN expression levels in 52 matched human normal and breast tumor lysates from mixed carcinomas (a) and ductal carcinomas (b). (D) Spearman's coefficients of correlation between PAR3 and MMP9 expression in normal breast, primary breast tumor and metastatic human breast cancers from two independent data sets (a; accession number: GSE1477; b, accession number: GSE7390). See also Figure S6.
To determine if PARD3 expression is associated with any change in survival probability we compared Kaplan-Meier plots for high and low expression of PARD3, using a validated Jetset probe (Figure S6D). For a set of 2,324 breast cancer patients, low PARD3 correlated with a modest but statistically significant reduction in survival probability (p = 9.8 × 10−5) (Gyorffy et al., 2010). Our mouse models had revealed that loss of Par3 triggers the induction of MMP9 and invasion. Therefore, we compared the expression of PARD3 and MMP9 in cohorts of normal human breast, primary breast cancers and metastases. Consistent with a role for loss of PAR3 in regulating metastasis through MMP9, a significant anti-correlation exists between PARD3 and MMP9 expression in metastases (Figure 6D). One dataset also showed a significant anti-correlation in primary tumors, although to a lesser degree than in metastases (Figure 6Db).
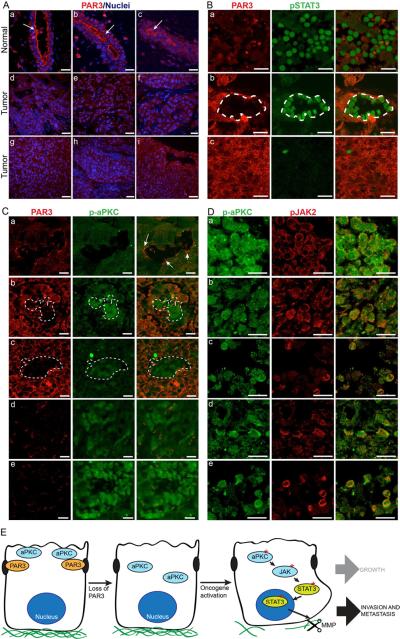
Par3 protein is localized to tight junctions at the apical/lateral boundary in murine mammary epithelia (McCaffrey and Macara, 2009), and immunofluorescence of tissue arrays revealed a similar distribution for human mammary ducts (Figure 7A a – c). Strikingly, however, PAR3 localization was completely lost in many of the human breast cancer samples (Figure 7A d– i). From a total of 76 tumor samples examined, 63 lacked strong cortex-associated PAR3 staining. Importantly, co-staining of tissue sections of 166 human invasive ductal carcinomas showed that p-aPKC and pSTAT3 were frequently prominent in regions with weak Par3 staining, whereas regions with more intense Par3 staining were negative for pSTAT3Y705 and p-aPKC (Figure7B, C). Furthermore, many regions were dual-positive for p-aPKC and pJAK2 (Figure 7D), consistent with a role for PAR3 in regulating aPKC, JAK, and STAT3 activity in human breast cancers.
Figure 7.
PAR3 protein expression is reduced and aPKC/STAT3 signaling is activated in human breast cancers
(A) Tissue sections of human normal and breast cancers stained for PAR3 (red) and nuclei (blue). Arrows show PAR3 enriched at the apical membrane. Images represent normal (a – c), infiltrating ductal carcinoma (d – f) and metastatic carcinoma (g – i). Scale bars = 100 μm. Arrows show PAR3 enriched at the apical membrane. (B) Tissue sections of human invasive breast cancers stained for PAR3 (red) and pSTAT3Y705 (green), representative of 166 stained tumor samples. Tumors with weak (top; 64% of tumors), mixed (middle; 28% of tumors), and intense (bottom, 8% of tumors) PAR3 staining. (C) Tissue sections of human invasive breast cancers stained for PAR3 (red) and active phospho-aPKCT560 (green). Arrows show PAR3 enriched at apical membrane and weak p-aPKC staining. Images show representative weak (a, 69% of tumors) mixed (b – c, 25% of tumors), and intense (d – e; 6% of tumors) p-aPKC staining. (D) Tissue sections of human invasive breast cancers co-stained for p-aPKCT560 (green) and pJAK2Y1007/8 (red). Representative images are shown for 5 tumor samples (a–e). (E) Model for cooperative effects of loss of Par3 in NICD or Ras61L tumors. Loss of Par3 in the context of either oncogene results the mis-localization and inappropriate activation of aPKC, which drives the induction and activation of Jak/Stat3 signaling, thereby causing increased MMP expression and ECM degradation. Scale bars = 30 μm.
DISCUSSION
The apical/basal polarity of epithelial cells is controlled in part by the Par proteins together with a group of epithelial-specific proteins first identified in Drosophila (Crumbs, Scribble, Lgl, Dlg). Loss of such proteins, or their misregulation, might therefore be expected to play a pivotal role in carcinogenesis, an idea that has been discussed in numerous reviews (Dow and Humbert, 2007; Feigin and Muthuswamy, 2009; Januschke and Gonzalez, 2008; St Johnston and Ahringer, 2010). Yet there is surprisingly little published data to support this view. In Drosophila Scrb, Lgl, and Dlg can behave as tumor suppressors (Bilder et al., 2000; Brumby and Richardson, 2003), and can cooperate with oncogenes to drive metastasis (Pagliarini and Xu, 2003; Wu et al., 2010). Deletion of Scrb in prostate epithelium predisposed mice to neoplasia (Pearson et al., 2011). Silencing of Scrb in murine mammary cells stimulated tumor growth driven by c-myc but had no effect on tumor latency and did not induce metastasis (Zhan et al., 2008). Nor does silencing of Scrb disrupt apical/basal polarity in mammalian epithelial cells (Dow and Humbert, 2007; Qin et al., 2005). Additionally, increased aPKC activity or Par6 levels have been linked to TGFβ signaling, and to breast cancer invasiveness (Viloria-Petit et al., 2009; Zhan et al., 2008). However, to date the only bona fide tumor suppressor among the polarity proteins in humans is PAR4/LKB1, (Jansen et al., 2009) and we are not aware of evidence that other Par proteins function as invasion suppressors, or that disruption of a polarity gene can induce tumorigenesis through loss of epithelial polarity.
To examine this issue, we used lentiviruses to deplete Par3 and express activated Notch or Ras in primary murine mammary cells, which were implanted orthotopically into normal immuno-competent mice. An advantage of this approach is that multiple changes in gene expression can be manipulated simultaneously, for instance to couple RNAi with ectopic expression. Additionally, the same cells can be used in vitro, without prior selection, to elucidate the molecular mechanisms underlying their phenotypes. This cancer model revealed several unexpected effects of silencing Par3. Most remarkably, despite the fact that the Ras and NICD oncogenes function through distinct mechanisms, the phenotypes induced by loss of Par3 are very similar. Loss of Par3 potently reduced tumor latency in both contexts and increased lung colonization, consistent with in vitro increases in invasiveness and detachment from the ECM. These effects were independent of any consistent pattern of EMT. For example, both the NICD and Ras tumors and metastases retained K8 and ZO1 expression and did not express the classical mesenchymal markers vimentin or N-cadherin, consistent with a classification as luminal-type tumors. Moreover, loss of Par3 caused the activation of Stat3 and induction of MMP9 expression in both tumor models. Indeed, the only consistent difference was that Ras/shPar3 tumors had lost expression of E-cadherin, whereas cadherin expression was retained in the context of NICD.
What are the underlying mechanisms through which loss of Par3 triggers rapid tumor growth and invasion? We propose the following model (Figure 7E). First, Par3 spatially restricts aPKC at the apical membrane (McCaffrey and Macara, 2009), and apical localization requires the binding and phosphorylation of Par3 by aPKC. Notably, over-expression or mislocalization of aPKC is commonly found in invasive human breast tumors (Kojima et al., 2008; Regala et al., 2005). Loss of Par3 triggers both the mislocalization and – in the context of at least some oncogenes – the activation of aPKC, which, unexpectedly, triggers JAK-dependent activation of Stat3. Stat3 in turn induces MMP expression, resulting in degradation of the ECM and permitting escape from the primary tumor. Our results generally agree with previous data that aPKCs were necessary for STAT3 activity and MMP1/13 expression in cytokine-stimulated chondrocytes (Litherland et al., 2010). However, Litherland et al. found a dependence on ERK phosphorylation, whereas we saw no changes in ERK activation in Par3-depleted cells (data not shown). Instead, loss of Par3 in an epithelial tumor context induces Stat3 activation through JAK by a cytokine-independent mechanism.
Stat3 is normally expressed only at low levels in the developing mammary gland, and is upregulated during involution to regulate cell death. Nonetheless, active STAT3 is often found at the invasive edges of tumors (Bromberg and Wang, 2009) and is known to promote metastasis in breast cancer (Barbieri et al., 2010a). Brugge and colleagues found that NICD is sufficient to activate STAT3 in MCF10A cells (Mazzone et al., 2010). However, these cells cannot form normal tight junctions and do not exhibit cortical polarity (Fogg et al., 2005). Most likely, therefore, aPKC is not restricted to the apical surface, and NICD expression is sufficient to induce Stat3 activation.
The coupling between polarity proteins, oncogenes, and Stat3 is paralleled to a remarkable degree by Drosophila, in which clones of epithelial cells lacking the Scribble polarity protein become highly metastatic in the context of oncogenic Ras (Pagliarini and Xu, 2003; Wu et al., 2010). Loss of Scribble alone triggers a JNK-dependent apoptotic response through the cell competition pathway, by which neighboring wild type cells eliminate the mutant cells; however, the expression of RasV12 switches this response from apoptosis to uncontrolled proliferation, via a compensatory mechanism dependent on JAK/STAT activation. Tantalizingly, loss of a polarity protein in the mammary gland also stimulates apoptosis, just as in Drosophila (McCaffrey and Macara, 2009). An important question for the future is whether this response is mediated by cell competition, and whether an aberrant compensatory mechanism drives tumor growth and dissemination when polarity proteins are lost from oncogene-transformed cells.
The striking effects of Par3 deficiency in the context of activated oncogenes suggest that polarity proteins might be suppressors of tumorigenesis in human carcinomas. PAR3 protein levels are significantly reduced in 50% of breast cancer samples compared with matched normal tissue, and a large majority of breast cancers lack normal PAR3 localization. Importantly, loss of Par3 was tightly correlated with increased p-aPKC and pSTAT3 across multiple breast tumor samples. Our data demonstrate that in addition to promoting metastasis through Stat3/MMPs, reduction of Par3 also reduces tumor latency, indicating that Par3 may suppress several steps in tumorigenesis. Microarray data support a broad decrease in PARD3 gene expression across multiple epithelial cancers, including invasive ductal carcinoma of the breast. Deletions in the PARD3 locus have also been identified. For human esophageal small cell carcinoma, the PARD3 gene was homozygously deleted in 15%, and expression was reduced in 90% of cell lines tested, compared to normal esophageal epithelial cells (Zen et al., 2009). Additionally, small deletions have been identified within the PARD3 locus in a variety of cancer types (Rothenberg et al., 2010). As Par3 functions in a polarity signaling network, mutations in other components of this network might also contribute to metastasis by human carcinomas. Together, our data establish that the Par3 polarity protein is an important suppressor of tumorigenesis and metastasis, and that it may play a significant role in human breast cancer progression.
EXPERIMENTAL PROCEDURES
Culture Conditions, Antibodies, Immunostaining, and Inhibitors
Orthotopic mammary transplants
All animal procedures were performed in accordance with protocols approved by the Animal Use Committees at the University of Virginia and McGill University, Montreal. Freshly isolated mammary cells were transduced with lentivirus expressing myc-NICD, Ras61L, and control shRNA (shLuc; against luciferase) or shRNA specific to murine Par3 (shPar3) (Zhang and Macara, 2006). Cells were transduced at an MOI=5, for oncogenes, and MOI=10 for shRNA. For in vivo tumorigenesis, 1×104 transduced MECs, for NICD experiments or 1×105 MECs for Ras61L, were injected into cleared fat pads, as described previously (McCaffrey and Macara, 2009). For assessing tumor incidence and growth, cells expressing NICD/shLuc were injected in the contralateral side to NICD/shPar3 of each mouse. Mice were examined biweekly for palpable tumors. To assess metastasis, cells were transplanted into paired inguinal (#4) fat pads. Once palpable tumors were found, they were measured with calipers weekly. Mice were sacrificed when the calculated tumor volume reached 1cm3.
Invasion assays
2.5×104 primary mammary epithelial cells (MECs)/well were transduced with NICD/shLuc, or NICD/shPar3, Ras61L/shLuc or Ras61L/shPar3 lentivirus and 2.5×104 Comma-D1 cells/well transduced with Ras61L/shLuc or Ras61L/shPar3 were plated in 8 μm pore Transwell inserts (Corning) in a 24-well plate format on top of 100 μl of 50% growth factor reduced Matrigel or 100 μl of Collagen I gels (rat tail collagen I, Gibco), prepared as described (Estecha et al., 2009). Culture medium was changed twice a day for 3 d. After 72 h, cells that had migrated through the Matrigel to the filter were stained with Hoechst 33342 and counted.
Tumor Array
Commercially available membranes (ST2-6X-1 and ST2-6X-2) and SomaPlex Breast Cancer Tissue Lysate Protein Microarray (PMA2-001-L) from Protein Biotechnologies were probed according to the manufacturer's protocol (See Extended Experimental Procedures). Breast tissue was provided by the University of Virginia Biorepository and Tissue Research Facility. All human samples were de-identified and are exempt from informed consent.
Statistical analyses
A Wilcoxon Signed-Rank test was used to determine the significance (p-value) of tumor-free status in mice for the in vivo tumorigenesis. P-values were determined using unpaired, 2-tailed Student's t-test for all assays except the tumor arrays, which used paired 2-tailed Student's t-tests.
Supplementary Material
Highlights
Par3 protein expression is significantly reduced in human breast cancers.
Loss of Par3 cooperates with oncogenes to trigger rapid tumor growth and metastasis.
Loss of Par3 enhances oncogene-dependent invasion in the absence of overt EMT.
Atypical PKC-dependent activation of Stat3 is essential for increased invasiveness.
SIGNIFICANCE.
Although loss of cell polarity is often considered a hallmark of invasive cancers there is little experimental evidence for any role of the polarity machinery in tumor suppression. Here, we demonstrate that Par3 polarity protein expression is frequently lost in human breast cancers. In the context of different oncogenes, loss of Par3 increases primary tumor growth and metastatic colonization of the lungs through the production of MMP9 downstream of Jak/Stat3 signaling, which is responsible for the invasive behavior of the tumors. We find that expression of Par3 is anti-correlated with phospho-aPKC, phospho-JAK, phospho-Stat3 and MMP9 expression in human breast cancers, establishing Par3 as a potent tumor growth and invasion suppressor.
ACKNOWLEDGEMENTS
We thank Didier Trono (Lausanne, Switzerland) for lentivectors, Shinya Yamanaka (Kyoto Univ.) for Stat3-C, Connie Cepko (Harvard Univ.) for myc-NICD, Jim Fawcett (Dalhousie Univ., Canada) for Par3 antibody, Deborah Lannigan (Univ. of Virginia) for breast tissue samples, and acknowledge grant support from the NIH GM070902 and CA132898 (to IGM), F32CA139950 (to JM), the Terry Fox Research Institute, Project #1009 (to LMM), CIHR 200602MFE-159430-14-900 (to LMM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barbieri I, Pensa S, Pannellini T, Quaglino E, Maritano D, Demaria M, Voster A, Turkson J, Cavallo F, Watson CJ, et al. Constitutively active Stat3 enhances neu-mediated migration and metastasis in mammary tumors via upregulation of Cten. Cancer Res. 2010a;70:2558–2567. doi: 10.1158/0008-5472.CAN-09-2840. [DOI] [PubMed] [Google Scholar]
- Barbieri I, Quaglino E, Maritano D, Pannellini T, Riera L, Cavallo F, Forni G, Musiani P, Chiarle R, Poli V. Stat3 is required for anchorage-independent growth and metastasis but not for mammary tumor development downstream of the ErbB-2 oncogene. Mol Carcinog. 2010b;49:114–120. doi: 10.1002/mc.20605. [DOI] [PubMed] [Google Scholar]
- Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. Scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. Embo J. 2003;22:5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Der CJ. Aberrant function of the Ras signal transduction pathway in human breast cancer. Breast Cancer Res Treat. 1995;35:133–144. doi: 10.1007/BF00694753. [DOI] [PubMed] [Google Scholar]
- Dow LE, Humbert PO. Polarity regulators and the control of epithelial architecture, cell migration, and tumorigenesis. Int Rev Cytol. 2007;262:253–302. doi: 10.1016/S0074-7696(07)62006-3. [DOI] [PubMed] [Google Scholar]
- Estecha A, Sanchez-Martin L, Puig-Kroger A, Bartolome RA, Teixido J, Samaniego R, Sanchez-Mateos P. Moesin orchestrates cortical polarity of melanoma tumour cells to initiate 3D invasion. J Cell Sci. 2009;122:3492–3501. doi: 10.1242/jcs.053157. [DOI] [PubMed] [Google Scholar]
- Feigin ME, Muthuswamy SK. Polarity proteins regulate mammalian cell-cell junctions and cancer pathogenesis. Curr Opin Cell Biol. 2009;21:694–700. doi: 10.1016/j.ceb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg VC, Liu CJ, Margolis B. Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. J Cell Sci. 2005;118:2859–2869. doi: 10.1242/jcs.02412. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13:7–9. doi: 10.1016/j.ccr.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–823. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi Y, Suzuki A, Yamanaka T, Sasaki K, Mizuno K, Sawada H, Yonemura S, Ohno S. Interaction between PAR-3 and the aPKCPAR-6 complex is indispensable for apical domain development of epithelial cells. J Cell Sci. 2009;122:1595–1606. doi: 10.1242/jcs.043174. [DOI] [PubMed] [Google Scholar]
- Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Ten Klooster JP, Offerhaus GJ, Clevers H. LKB1 and AMPK family signaling: the intimate link between cell polarity and energy metabolism. Physiol Rev. 2009;89:777–798. doi: 10.1152/physrev.00026.2008. [DOI] [PubMed] [Google Scholar]
- Januschke J, Gonzalez C. Drosophila asymmetric division, polarity and cancer. Oncogene. 2008;27:6994–7002. doi: 10.1038/onc.2008.349. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Akimoto K, Nagashima Y, Ishiguro H, Shirai S, Chishima T, Ichikawa Y, Ishikawa T, Sasaki T, Kubota Y, et al. The overexpression and altered localization of the atypical protein kinase C lambda/iota in breast cancer correlates with the pathologic type of these tumors. Hum Pathol. 2008;39:824–831. doi: 10.1016/j.humpath.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Litherland GJ, Elias MS, Hui W, Macdonald CD, Catterall JB, Barter MJ, Farren MJ, Jefferson M, Rowan AD. Protein kinase C isoforms zeta and iota mediate collagenase expression and cartilage destruction via STAT3- and ERK-dependent c-fos induction. J Biol Chem. 2010;285:22414–22425. doi: 10.1074/jbc.M110.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, Pandya D, Lu Y, Mills GB, Aster JC, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci U S A. 2010;107:5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 2009;23:1450–1460. doi: 10.1101/gad.1795909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- Pearson HB, Perez-Mancera PA, Dow LE, Ryan A, Tennstedt P, Bogani D, Elsum I, Greenfield A, Tuveson DA, Simon R, et al. SCRIB expression is deregulated in human prostate cancer, and its deficiency in mice promotes prostate neoplasia. J Clin Invest. 2011;121:4257–4267. doi: 10.1172/JCI58509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzo MG, Park M. The Met receptor tyrosine kinase and basal breast cancer. Cell Cycle. 2010;9:1043–1050. doi: 10.4161/cc.9.6.11033. [DOI] [PubMed] [Google Scholar]
- Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J Cell Biol. 2005;171:1061–1071. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Reese DM, Slamon DJ. HER-2/neu signal transduction in human breast and ovarian cancer. Stem Cells. 1997;15:1–8. doi: 10.1002/stem.150001. [DOI] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, Fields AP. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- Rorth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- Rothenberg SM, Mohapatra G, Rivera MN, Winokur D, Greninger P, Nitta M, Sadow PM, Sooriyakumar G, Brannigan BW, Ulman MJ, et al. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 2010;70:2158–2164. doi: 10.1158/0008-5472.CAN-09-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Song Y, Qian L, Song S, Chen L, Zhang Y, Yuan G, Zhang H, Xia Q, Hu M, Yu M, et al. Fra-1 and Stat3 synergistically regulate activation of human MMP-9 gene. Mol Immunol. 2008;45:137–143. doi: 10.1016/j.molimm.2007.04.031. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Cubbon RM, Cummings RT, Wicker LS, Frankshun R, Cunningham BR, Cameron PM, Meinke PT, Liverton N, Weng Y, et al. Photochemical preparation of a pyridone containing tetracycle: a Jak protein kinase inhibitor. Bioorg Med Chem Lett. 2002;12:1219–1223. doi: 10.1016/s0960-894x(02)00106-3. [DOI] [PubMed] [Google Scholar]
- Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D, Roncari L, Narimatsu M, Bose R, Moffat J, et al. A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci U S A. 2009;106:14028–14033. doi: 10.1073/pnas.0906796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, Huang S. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zen K, Yasui K, Gen Y, Dohi O, Wakabayashi N, Mitsufuji S, Itoh Y, Zen Y, Nakanuma Y, Taniwaki M, et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 2009;28:2910–2918. doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.