SUMMARY
Dynamic actin cytoskeletal reorganization is integral to cell motility. Profilins are well-characterized regulators of actin polymerization; however, functional differences among co-expressed profilin isoforms are not well defined. Here, we demonstrate that profilin-1 and profilin-2 differentially regulate membrane protrusion, motility, and invasion; these processes are promoted by profilin-1 and suppressed by profilin-2. Compared to profilin-1, profilin-2 preferentially drives actin polymerization by the Ena/VASP protein, EVL. Profilin-2 and EVL suppress protrusive activity and cell motility by an actomyosin contractility-dependent mechanism. Importantly, EVL or profilin-2 downregulation enhances invasion in vitro and in vivo. In human breast cancer, lower EVL expression correlates with high invasiveness and poor patient outcome. We propose that profilin-2/EVL-mediated actin polymerization enhances actin bundling and suppresses breast cancer cell invasion.
INTRODUCTION
Cell motility requires precisely orchestrated regulation of multiple cellular processes that involve dynamic actin cytoskeletal reorganization. Reorganization of the actin cytoskeletal is controlled by actin-binding proteins that regulate nucleation, branching, elongation, bundling, severing and capping of actin filaments (DesMarais et al., 2005; Insall and Machesky, 2009; Pollard and Borisy, 2003). In particular, profilins are key actin polymerization regulators that promote the conversion of ADP-actin to ATP-actin and interact with poly-L-proline domains (PPP[A/P]PPLP; abbreviated as “PLP”) found in a variety of actin nucleation promotion factors, actin nucleators, and actin filament barbed end elongation factors. These include WASp/WAVE/SCAR, formins and Ena/VASP proteins (Mena, VASP and EVL) (Ferron et al., 2007; Gertler et al., 1996; Jockusch et al., 2007; Lambrechts et al., 2000; Mahoney et al., 1997; Reinhard et al., 1995). Orchestrated regulation of these actin polymerization factors leads to distinct changes in the actin cytoskeleton architecture. These cytoskeletal changes regulate cellular processes that impact cell motility, and can also promote or suppress invasive migration. Four profilin isoforms have been identified; while profilin-1 is ubiquitously expressed, other isoforms show more selective expression in specific tissues. It is currently unclear whether profilin-2/3/4 isoforms have evolved to carry out distinct functional activities with respect to actin polymerization or to protect against loss of one isoform by genetic or epigenetic alterations.
Previously, in a siRNA high-throughput cell migration screen, we found that suppression of PFN1, the ubiquitously expressed profilin isoform, inhibited cell migration in MCF10A mammary epithelial cells whereas PFN2 downregulation enhanced migration in these cells (Simpson et al., 2008). Although profilin-2 has been considered a neuronal-specific isoform (Honore et al., 1993; Witke et al., 1998), it is expressed in many other tissues, including breast epithelium (EST Profile Viewer at www.ncbi.nlm.nih.gov/unigene/ESTProfileViewer;cgap.nci.nih.gov/sage). Despite structural similarities in their PLP binding sites (Kursula et al., 2008; Lambrechts et al., 1997; Witke, 2004), profilin-1 and -2 show variation in surface charge distribution at these sites (Nodelman et al., 1999). Differences in ligand binding preferences have been reported using in vitro binding assays or mass spectrometry analysis of profilin-1 and profilin-2 binding proteins (Lambrechts et al., 2000; Miki et al., 1998; Nodelman et al., 1999; Veniere et al., 2009; Witke et al., 1998); however the extent to which these binding differences affect actin-based cellular processes has not been explored mechanistically.
In this report, we demonstrate that altering profilin-1 or -2 levels has dramatically different effects on actin cytoskeletal organization, affecting cell migration and invasion. Profilin-2 controls protrusive activity and migratory behavior of normal and tumor cells by promoting EVL-mediated polymerization of long actin filaments that assemble into contractile bundles. In addition, downregulation of profilin-2 or EVL markedly enhances invasion in vitro and in vivo, and the expression profiles of these actin regulators in human tumors is significantly correlated with tumor grade and invasiveness.
RESULTS
Profilin-1 and Profilin-2 Have Differential Effects on Cell Motility and Invasion
To investigate profilin-1 and profilin-2 contributions to cell motility, we depleted each isoform and analyzed changes in the motile behavior of MCF10A cells. SMARTpool siRNAs selectively downregulated profilin-1 and -2 levels, and knockdown (KD) of one isoform did not affect the levels of the other (Figure 1A–C). PFN2 KD increased migration speed and scattering, whereas PFN1 KD decreased migration speed and promoted cell clustering (Figures 1A–D and Movie S1). We confirmed the siRNA specificity using two shRNAs, targeting each isoform; these shRNAs displayed the same specificity and efficacy in knockdown, and induced the same migratory phenotypes (Figure S1A). Using these shRNAs, we evaluated the effects of profilin-1 and -2 depletion in 3D cultures. MCF-10A cells form cyst-like acinar structures reminiscent of mammary gland alveoli (Muthuswamy et al., 2001; Petersen et al., 1992). PFN2 KD induced the formation of dysmorphic 3D structures not observed in control cultures (Figures 1E); and about 5% of PFN2 KD structures displayed an invasive phenotype exhibiting cell dissemination into the matrix (Figure 1E). This invasive behavior is noteworthy since overcoming structural restraints within acini is highly atypical; most genes that have been implicated in tumor progression fail to induce invasion in MCF-10A cells, requiring additional genetic or epigenetic alterations to promote this phenotype (Debnath and Brugge, 2005; Witt et al., 2006). PFN2 KD also significantly induced MCF10A cell invasion through Matrigel™ in a Boyden chamber assay (Figure 1F).
FIGURE 1. Profilin-1 and -2 Differentially Regulate Cell Migration and Invasion.
(A) Montages of DIC from Movie S1. Time stamps are hrs:min:sec. Scale bar is 50μm. (B) Migration tracks of the cells indicated by red and yellow arrows in A. (C) Western blot analysis of profilin-1 and -2. (D) Quantitation of migration speed; values are averages of mean speed from at least 30 cells ± SEM from three experiments. (E) MCF10A 3D cultures. Left panels are phase-contrast images (arrows indicate cell invasion) and right panels are 3D reconstruction of confocal z- series. Scale bar is 50μm. (F) Boyden chamber invasion assays; values are averages of mean number of invading cells (normalized over control) from three independent experiments ± SEM. (G) Quantitation of profilin-1 (a) and 2 (b) intracellular concentrations. Values are means from three independent experiments ± Std Dev. (H) Wound-healing assay of SUM159 control, PFN1 KD and PFN2 KD cells; plot shows wound areas (normalized over control) from a representative experiment (of three experiments); and right panels show cells at the edge of the wound highlighted in green. Scale bar is 50μm. See also Figure S1 and Movie S1.
To examine the contrasting functions of profilin-1 and 2 in cancer cell migration, we screened a large number of breast tumor cell lines for expression of the two profilin isoforms; we chose SUM159 cells for analysis because they express levels of profilin-1 and profilin-2 comparable to MCF-10A cells (Figure 1G and S1B). Similarly to MCF10A cells (Simpson et al., 2008), PFN2 KD enhanced SUM159 cell migration in a wound-healing assay and increased cell scattering at the wound edge, whereas PFN1 KD suppressed wound closure (Figure 1H and S1C).
To determine the contribution of each profilin isoform to the total pool of profilin, we quantified profilin-1 and profilin-2 cellular concentrations in MCF10A and SUM159 cells. Profilin-1 concentration, approximately 13 μM was about 15-fold higher than that of profilin-2, 0.8 μM(Figure 1G and S1B); this suggests that cell migration is sensitive to changes in profilin-2 levels specifically since altering profilin-2 alone would not significantly alter the combined concentration of both isoforms.
To examine whether alterations in profilin-1 and 2 levels affect invasive behavior of SUM159 cells, we embedded them in Matrigel™, in which they form 3D clusters with cells protruding into the surrounding matrix. PFN2 KD enhanced migratory and invasive behavior of SUM159 cells in 3D cultures, exhibiting enhanced protrusive activity and cell dissemination into the matrix, whereas overexpression of PFN2 (HA-profilin-2) resulted in the opposite phenotypes (Figure 2A and S2A–B); PFN1 KD diminished protrusive activity and suppressed migration and invasion into the matrix, whereas PFN1 overexpression enhanced them (Figure 2A and S2A–B). Consistent with these results, PFN2 KD increased invasion in Boyden chambers, whereas PFN1 KD significantly decreased it (Figure 2B).
FIGURE 2. PFN2 Knockdown Enhances Invasion of SUM159 Breast Tumor Cells.
(A) 3D reconstruction of confocal z-series of 3D cultures. Scale bar is 40μm. (B) Boyden chamber invasion assays; values are averages of mean number of invading cells (normalized over control) from three independent experiments ± SEM. (C) Macroscopic view of representative tumors from mammary fat pads injections. (D) (Top row) H and E staining of sections from control, PFN1 KD and PFN2 KD SUM159 tumors. White arrow indicates local invasion. (Bottom row) Left panel shows box plots of the quantitation of invasive foci in tumors from three experiments; middle panel shows an invasive focus at the sentinel lymph node in PFN2 KD tumor (inset is a magnification of the region indicated by a white arrow); and right panel shows a tumor edge with dissociated invasive cells in a PFN2 KD tumor. Scale bar is 50μm. See also Figure S2.
To investigate whether profilin-1 and 2 influence invasion of breast cancer cells in vivo, we injected control, PFN1 KD or PFN2 KD SUM159 cells orthotopically into the mammary fat pad of NOD/SCID mice. While there were no discernable differences in tumor growth between PFN1 KD, PDN2 KD and control tumors (Figure S2C), PFN2 KD significantly increased the number of extra-tumoral invasive foci, particularly, in the surrounding stroma and muscle tissue and occasionally in the sentinel lymph node (Figure 2C–D and S2D). These studies indicate that profilin-2 downregulation enhances stromal infiltration in SUM159-derived tumors.
Profilin-2-Mediated Actin Polymerization Promotes Actin Bundling and Suppresses Protrusive Activity
Given its low cellular concentration, the significant effects of profilin-2 downregulation on migration and invasion suggest that its contribution to actin cytoskeletal reorganization is distinct from that of profilin-1. To characterize the contribution of each profilin to actin cytoskeletal remodeling, we examined PFN1 KD and PFN2 KD SUM159 cells microinjected with labeled actin by time-lapse microscopy. PFN1 KD increased in F-actin bundles; PFN2 KD, however, decreased bundling, especially at regions of the leading edge undergoing increased protrusive activity (Figure 3A and Movie S2).
FIGURE 3. Profilin-2 Promotes Actin Bundling in SUM159 cells.
(A) Montage from time-lapse movies of cells microinjected with Alexa Fluor® 568-conjugated actin (Movie S2); colored lines indicate the positions of the leading edge in the corresponding cells. Indicated time is in minutes. Scale bar is 10μm. (B) Electron micrographs of cortical F-actin cytoskeletons. Arrow indicates a small protrusion (green shade) next to an actin bundles (red shade). Insets are higher resolution images. Scale bar is 1μm. (C) Quantitation of cells with prominent stress fibers (~200 cells were analyzed per group) and (D) Representative images; arrow indicate small protrusion and scale bar is 10 μm. See also Figure S3 and Movie S2.
To compare the ultrastructure of the actin cytoskeleton in control and KD cells, we examined metal cast cytoskeletons generated by rapid freezing and rotary-shadowing. Consistent with the light microscopy data, PFN1 KD cytoskeletons showed marked increase in actin filament bundles (Figure 3B). To quantify the change in actin bundling, we assessed the percentage of cells with prominent stress fibers by light microscopy; PFN1 KD enhanced stress fiber formation in SUM159 cells, whereas PFN2 KD diminished it (Figure 3C–D and S3A). Importantly, PFN2 KD in PFN1 KD cells (double knockdown) resulted in dramatic reduction in actin polymerization and in collapse of the actin cytoskeleton (Figure S3B); these data suggest that profilin-1 and -2 are the two major profilin isoforms regulating actin polymerization in SUM159 cells and that profilin-2 promotes actin bundling. Indeed, similarly to PFN1 KD, overexpression of PFN2 enhanced stress fiber formation, a phenotype that was reversed by the concomitant KD of PFN2; moreover, overexpression of a siRNA-resistant PFN2 mutant prevented the reversal of the PFN2 overexpression phenotype by PFN2 KD, further validating the specificity of profilin-2 effects on actin bundling (Figure S3C–D).
Since these alterations in the actin cytoskeleton correlated with changes in protrusive activity, we examined protrusion and retraction dynamics after altering profilin-1 and -2 levels. PFN2 KD in SUM159 cells enhanced protrusion and retraction as compared to control cells, while PFN1 KD suppressed these activities (Figure 4A and Movie S3). Changes in protrusive activity were quantified in kymographs generated from highly resolved time-lapse image series (3600 frames at a rate of 1 frame/sec). PFN2 KD increased speed and frequency of protrusion and retraction by 1.6-2-fold, and decreased protrusion persistence by 32% as compared to control (p<0.05) (Figures 4B–E, S4A–D and Movie S3); additionally, PFN2 KD decreased the idle time between protrusion/retraction events, as compared to control (Figure S4C). In contrast, PFN1 KD diminished protrusion speed, but did not affect persistence or frequency (Figures 4B–D and S4A–D). To observe the direct effects of increased intracellular concentrations of profilin-1 or 2, we microinjected the respective purified proteins into SUM159 cells (Figure 4F and Movie S4). Profilin-1 injection enhanced protrusion/retraction speed and frequency, whereas profilin-2 injection suppressed these activities (Figures 4G–J, S4D–F and Movie S4); overexpression of PFN2a also suppressed protrusive activity significantly (Figure S4G–H). Together these results suggest that profilin-2 suppresses protrusive activity in SUM159 cells, consistent with its suppressive effects on migration and invasion of these cells.
FIGURE 4. Profilin-2 Suppresses Protrusive Activity in SUM159 Cells.
(A) Montages of DIC images selected from a segment of Movie S3 at one-hour intervals. Scale bar is 20μm. (B) Kymography analysis: (left) DIC images from Movie S3 at time 0; and (right) minimum projections (showing regions of protrusive activity) of entire time series (acquired at a rate of one frame/sec). Lines indicate the position at which kymographs were registered. Scale bar is 20μm. (C) kymographs from the corresponding movies in (B). Vertical scale bar is 20μm. Horizontal scale bar is two minutes. (D) Average retraction and protrusion speeds; and (E) frequency. Values are averages of means from at least 30 cells (pooled from 3 different experiments) ± SEM. (F) Montages of DIC images selected from a segment of Movie S4 at one- hour intervals. Yellow dextran marks the injected cells. Scale bar is 20μm. (G) Kymography analysis: (left) DIC images from Movie S4 at time 0; and (right) minimum projections of entire time series (acquired at a rate of one frame/sec). Labeled dextran (cyan) was co-injected to identify microinjected cells. Lines indicate the position at which kymographs were registered. Scale bar is 20μm. (H) kymographs from the corresponding movies in (E). Vertical scale bar is 10μm. Horizontal scale bar is one minute. (I) Average retraction and protrusion speed; and (J) frequency. Values are averages of means from at least 30 cells (pooled from three different experiments) ± SEM. See also Figure S4 and Movie S3 and S4.
The Suppressive Effects of Profilin-2 Are Dependent on Myosin Contractility
To investigate the involvement of myosin motor activity in profilin-2- induced actin bundling, we examined myosin light chain (MLC) phosphorylation. In PFN1 KD cells, phospho-MLC decorated F-actin bundles at the leading edge (Figure 5A); PFN2 KD cells displayed reduced total phospho-MLC, consistent with sparser cortical actin bundles (Figure 5A–B and S5A). Conversely, overexpression of PFN2a increased phospho-MLC levels (Figure S5A). Moreover, we examined the effects of overexpression of PFN2b; PFN2b is a PFN2 splice isoform that does not bind to G-actin and has low affinity to poly-L- proline (Di Nardo et al., 2000), and was used as negative control lacking the capacity to promote actin polymerization. Overexpression of PFN2b did not increase phospho-MLC levels, suggesting that profilin-2-mediated polymerization is required for the generation of the contractile actin bundles (Figure S5A).
FIGURE 5. Profilin-2 Suppressive Effects Are Dependent on Acto-myosin Contractility in SUM159 cells.
(A) Phospho-MLC staining (phospho-Ser19). Insets are magnifications of the areas in the white boxes; white lines trace the cell edge. Scale bar is 10μm. (B) Phospho-MLC WB analysis. (C) Speed (a) and frequency (b) of retraction/protrusion in control and PFN2a overexpressing SUM159 cells with or without ROCK inhibition (Movie S5). Values are averages of means from at least 30 cells (pooled from 3 different experiments) ± SEM. (D) 3D reconstruction of confocal z-series of SUM159 3D cultures. Scale bar is 40μm. See also Figure S5 and Movie S5.
To examine the connection between the generation of contractile actin bundles and the regulation of protrusive activity by profilin-2, we assessed the effect of altering myosin activity on profilin-2’s suppressive effects. For that purpose, we used Y27632, a pharmacological inhibitor of ROCK-mediated MLC phosphorylation and myosin motor activity. ROCK inhibition reversed the suppressive effects of profilin-2 on protrusive activity, thus increasing the frequency and speed of protrusion and retraction (Figure 5C and Movie S5); this increase was significantly greater than baseline, suggesting that profilin-2 has a positive effect on protrusion in the absence of myosin activity. In a wound-healing assay, ROCK inhibition reversed the suppression of cell migration by PFN1 KD (Figure S5B); not surprisingly, this ‘rescue’ was only partial since the KD cells are depleted of profilin-1, and the endogenous levels of profilin-2 are significantly lower than 1. In agreement with these data, ROCK inhibition increased protrusion and invasion into the surrounding matrix in 3D cultures, mimicking the effects of PFN2 KD, and partially ‘rescued’ protrusive activity in PFN1 KD cell clusters (Figure 5D and S5C). Moreover, blebbistatin (an inhibitor of myosin ATPase activity) also increased protrusion and invasion into the matrix and partially reversed PFN1 KD phenotype (Figure S5C). Collectively, these results suggest that suppression of protrusive activity, migration, and invasion of SUM159 cells by profilin-2 is dependent on the generation of contractile actomyosin bundles.
Profilin-2-Induced Contractile Actin Bundles Are Generated by EVL, Which Suppresses Protrusive Activity, Migration and Invasion
The structural differences in actin filaments generated by profilin-1 and 2 (Figure 6A and S6A) suggest that they interact with distinct actin polymerization regulators. Using mass-spectrometry, we identified high-confidence interactions between profilin-1/2 and Ena/VASP proteins; in particular, EVL was found to preferentially bind profilin-2 as compared to 1 (personal communication, L. L. G. and Mathew Sowa), consistent with previous studies (Ferron et al., 2007; Kursula et al., 2008; Lambrechts et al., 2000; Nodelman et al., 1999; Veniere et al., 2009).
FIGURE 6. EVL Exhibits Preferential Binding to Profilin-2 and Suppresses Protrusive Activity by Generating Actin Bundles in a Profilin-2 Dependent Manner.
(A) High magnification micrographs of the cell edge actin of control, PFN1 KD and PFN2 KD SUM159 cells. Scale bar is 500 nm. (B) (left) Analysis of EVL binding to profilin-1 and 2 in SUM159 cells: HA immuno- precipitation, followed by profilin-1 and 2 western blot (WB); HA-GFP was used as negative control, and HA WB shows the expression levels of HA-GFP and HA-EVL; (right) quantitation of the relative levels of profilin-1 and 2 bound to HA-EVL in SUM159 cells. These data are representative of three independent experiments. Values are averages ± SEM (C) Sedimentation equilibrium analytical ultracentrifugation used to determine the solution molecular weight of monomeric EVL in the presence of profilin-1 or 2a. The sedimentation profile of Cy3-mEVL1-235aa (5μM) alone or combined with either profilin-1 or 2a (40μM) was determined by monitoring the absorbance at 527/550 nm. Global fitting of three equilibrium traces (at three different centrifugation speeds: 10, 14, 20K rpm) for each condition was performed (see Methods for more detail). An extinction coefficient of 79,982 M-1cm-1 (Cy3, 527nm) was used to determine the protein concentration as a function of the radial position. A monomer-dimer model was used to determine the molecular weight for a single ideal species (top panels). (D) (left) Image sequence of filaments polymerizing in vitro in the presence of 2 μM actin (10% Alexa488), plus 2 μM profilin-2a. Barbed end growth of actin filaments was visualized using TIRF microscopy. Top row, 0 nM EVL; bottom row, plus 200 nM EVL. Yellow arrowhead tracks the growth of a single actin filament barbed end. Scale bar is 5 μm. (right) Average barbed-end polymerization rates (subunits/seconds) for single actin filaments in the presence of profilin-1 or 2, plus or minus EVL. In the presence of 1uM actin (10% Alexa488) alone and 100 nM EVL, barbed ends elongated at a rate of 31.1 ± 2.9 sub/sec (unpublished data). Values are averages of polymerization rates from at least 30 filaments pooled from 2–3 slides ± SEM. (E) Control or PFN2 KD SUM159 cells expressing GFP-EVL; (a) still images from Movie S6, scale bar is 50μm; speed (b) and frequency (c) of retraction/protrusion in control or PFN2 KD cells with or without GFP-EVL expression as calculated from corresponding kymographs. Values are averages of means from at least 30 cells (pooled from 3 different experiments) ± SEM. (F) (Right) F-actin staining in control and PFN2 KD cells with or without EVL overexpression. Scale bar is 50μm. (Left) % of EVL overexpressing (GFP positive) cells with prominent stress fibers (G) 3D reconstruction of confocal z-series of 3D cultures of control and PFN2 KD cells with or without EVL overexpression. Scale bar is 50μm. (H) Q-PCR showing decreased EVL expression after long-term selection in control cells and cells expressing the two shRNAs targeting EVL. (I) F-actin staining and quantitation of reduced bundling. Scale bar is 20μm. (J) Representative wound-healing assay. Values are averages ± Std Dev. (K) Kymograph analyses (images are from Movie S6; scale bar is 20μm); values are averages of means from at least 30 cells (pooled from 3 different experiments) ± SEM. See also Figure S6 and Movie S6.
To characterize the binding preferences of profilin-2 to the three ENA/VASP family members, we examined the relative binding capacity of Mena, EVL and VASP to profilin-2 as compared to profilin-1 (Figure 6B, S6B–C). Both EVL and VASP preferentially immunoprecipitated with profilin-2 (7-fold and 1.8-fold, respectively); Mena showed no preferential binding to either profilin isoform (data not shown). Direct measurement of profilin binding capacity with purified monomeric EVL and VASP using sedimentation equilibrium confirmed that both proteins show preferential binding to profilin-2 relative to 1 (Figure 6C and S6D). In addition, both monomeric EVL and VASP had higher affinity and binding capacity for profilin-2 compared to 1 (Figure S6D). Similarly, 10-fold more profilin- 2 than profilin-1 immunoprecipitated with HA-EVL (Figure 6B) and, vice versa, 7-fold more EVL were immunoprecipitated with profilin 2 compared to profilin-1 (Figure S6B). Consistent with the mass-spectrometry analysis, these results indicate that VASP and EVL interact preferentially with profilin-2 in vivo and in vitro.
Although VASP has been previously shown to enhance barbed end filament elongation in the presence of profilin-1 and cytoplasmic actin (Hansen and Mullins, 2010), the profilin isoform specificity for Ena/VASP proteins remains poorly understood. To compare the relative ability of EVL and VASP to enhance actin barbed end polymerization in the presence of profilin-1 and 2, we visualized the assembly of single actin filaments in vitro using TIRF microscopy. EVL-dependent barbed end polymerization was significantly faster in the presence of profilin-2, compared to profilin-1 (Figure 6D). Furthermore, although EVL could bind to profilin-1 in our sedimentation equilibrium experiments, elongation of actin filaments in the presence of profilin-1 was inefficient (Figure 6D). VASP exhibited a similar preference for profilin-2 binding in vitro; however, we observed only a marginal difference between the rate of VASP-mediated barbed end filament elongation in the presence of profilin-1 versus profilin-2 (Figure S6E). Based on observations reported by Breitsprecher et al. (2011), the rate of Ena/VASP-dependent barbed end elongation is directly related to the affinity for monomeric actin. Since profilin enhances actin monomer binding to Ena/VASP proteins (Ferron et al., 2007), the differences in filament elongation rates in the presence of the two profilin isoforms can be attributed to the different affinities and binding capacity of EVL and VASP. Based on these differences between profilin-1 and 2, it is feasible that profilin-2 could compete effectively with profilin-1 for binding to EVL despite the 15-fold higher concentrations of profilin-1, since only the former would appreciably participate in generating EVL-mediated actin structures. This is supported by the immunoprecipitation of EVL with profilin-1 and profilin-2 from cell lysates.
To investigate the effect of EVL activity on membrane protrusion and the requirement of profilin-2 for this activity, we examined membrane dynamics in control and PFN2 KD SUM159 cells overexpressing GFP-EVL (Figure 6E, and Movie S6). Overexpression of GFP-EVL significantly inhibited protrusion and retraction in control but not in PFN2 KD cells (Figure 6E). In addition, GFP-EVL increased the abundance of stress fibers in control cells in a profilin-2 dependent manner (Figure 6F and S6F); this EVL-induced increase in stress fibers correlated with suppression of protrusion and invasion in 3D cultures (Figure 6G).
In addition, EVL KD suppressed stress fiber formation in SUM159 cells (Figures 6H–I and S6G). This decrease in stress fibers correlated with significantly weakened matrix adhesion, especially in cells strongly depleted of EVL (data not shown); moderate EVL depletion, which did not weaken adhesion to the same extent, increased wound closure rate (Figure 6J) and protrusion and retraction frequency and speed in SUM159 cells, similarly to PFN2 KD (Figure 6K and Movie S6). In addition, EVL KD increased migration speed of MCF10A cells, which also exhibited compromised adhesion at high KD levels (Figure S6H–J, and data not shown). These results suggest that EVL-induced actin polymerization is dependent on profilin-2, and that this polymerization mode promotes actin bundling leading to diminished protrusive activity.
VASP overexpression, on the other hand, increased stress fiber generation only weakly (Figure S6K–L). In addition, VASP KD did not have a significant effect on protrusive activity (Figure S6M–N). These data are consistent with VASP having only a marginal preference for binding to profilin-2 relative to profilin-1.
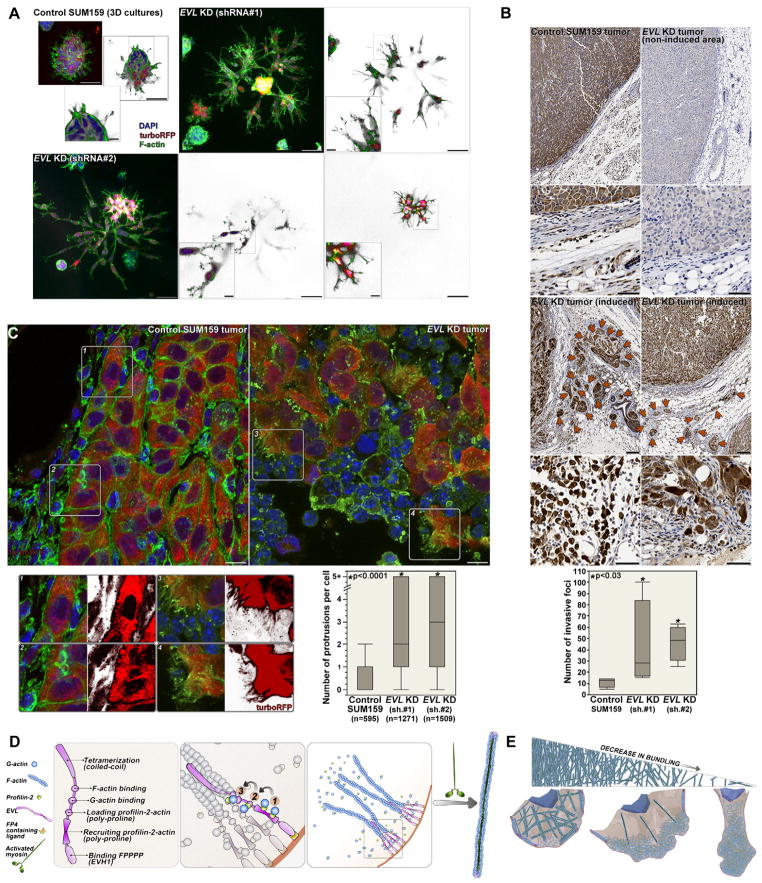
To investigate the effects of EVL KD on invasion, we established a inducible shRNA system in SUM159 cells, which made it feasible to achieve high knockdown levels - 70–80% depletion after 48 hours of induction (Figure S7A); this approach allowed us to circumvent any effect the KD might have on tumor initiation and growth due to altered adhesion properties. In 3D cultures, induction of EVL KD increased invasion, similarly to PFN2 KD (Figure 7A). To examine invasion in vivo, we induced EVL KD in SUM159 tumors derived from orthotopic fat pad injections. EVL KD did not affect tumor volume over the course of two- week induction, after which the tumors were harvested (Figure S7B). Induction of either of two different shRNAs targeting EVL increased in the number of extra-tumoral invasive foci significantly (Figure 7B). Moreover, staining for turboRFP, which is expressed as a cytoplasmic marker upon induction, allowed us to examine subcellular structures in tumor cells. Interestingly, EVL KD increased the number of protrusions per cell as compared to control (Figure 7C). Together, these studies indicate that EVL downregulation is associated with increased protrusion and invasion in vitro and in vivo.
FIGURE 7.
(A) Confocal microscopy (maximum projection images) of 3D cultures of SUM159 cells on day 8 and after 4 days of induction with doxycycline of control and EVL KD cells using two different inducible shRNA targeting EVL (scale bar is 50 μm); large insets are mid-sections of the confocal z-series and small insets are magnification of the region in the box (scale bar is 10 μm). (B) TurboRFP staining of sections from control, non-induced area of EVL KD SUM159 tumors and of induced EVL KD tumors; arrows indicate extratumoral invasive foci; box-and-whisker plot shows quantitation of invasive foci in the corresponding tumors. Scale bar is 50μm. (C) Confocal microscopy (maximum projection images) of control and EVL KD tumors. Scale bar is 10 μm. Insets are magnified areas within the designated boxes; red channels are shown separately to visualize tumor cell morphology; box-and-whisker plot shows quantitation of the number of protrusions per cell. (D) Model representing the generation of actin bundles by profilin-2/EVL-mediated linear actin polymerization and activated myosin: right panel illustrates the process of actin polymerization mediated by profilin-2, which is summarized in three major steps (middle panel): recruitment (1) and loading (2) of profilin-2:actin by interaction with the profilin-2 PLP binding site; followed by addition of one actin monomer (G-actin) to the barbed end (3). Left panel shows the domain structure of EVL. (E) Model representing the correlation between protrusive activity and the level of actin bundling. See also Figure S7 and Movie S7 and S8.
To examine the effects of altering profilin-2/EVL-mediated actin polymerization on protrusive activity and cell migration in other cell lines, we examined the expression profile of PFN2 and EVL in an array of cancer cell lines using published datasets (Neve et al., 2006). We chose MCF7 cells because they express relatively high levels of PFN2 and EVL (Neve et al., 2006). Consistent with the results from SUM159 cells, PFN2 KD or EVL KD enhanced MCF7 cell migration in wound-healing assays and increased protrusive activity as measured by kymography (Figures S7C–F and Movie S7). In addition, PFN2 KD or EVL KD in colorectal adenocarcinoma Caco-2 cells also increased protrusive activity (Figure S7G and Movie S8).
Our findings support a model in which three elements are important for EVL-mediated suppression of protrusive activity (Figure 7D–E): (1) profilin-2 to specifically deliver polymerization-competent actin monomers to EVL, (2) EVL to assemble unbranched actin filaments and (3) myosin contractility to generate actin bundles.
PFN2 and EVL Are Differentially Expressed in Human Breast Cancer
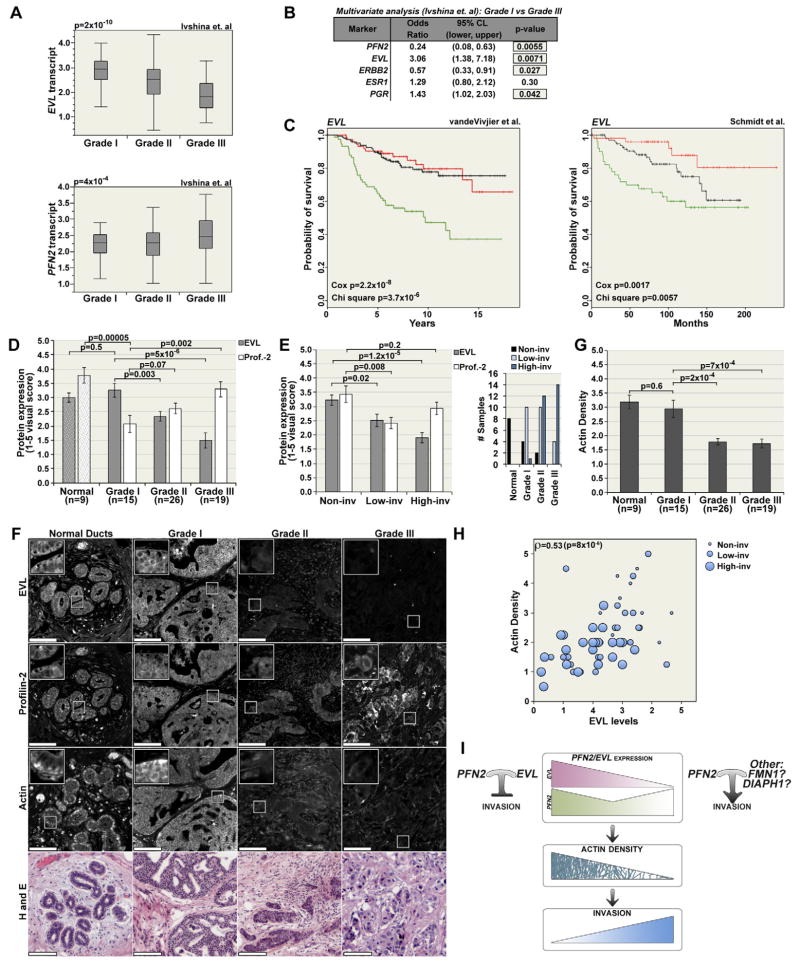
To examine whether the expression of PFN2 and EVL is linked to clinical aspects of breast tumors, we examined the relationship between these markers and tumor grade. In five datasets (Desmedt et al., 2007; Ivshina et al., 2006; Loi et al., 2007; Lu et al., 2008; Minn et al., 2005), EVL transcript levels were significantly lower in grade II and III tumors as compared to grade I, whereas PFN2 expression was higher in high-grade tumors (Figure 8A, S8A and data not shown). In addition, multiple logistic regression analyses of two large datasets (Ivshina et. al, 2006 and Lu et. al, 2008) determined that EVL is a significant predictor of tumor grade, independent of other known markers, namely ESR1, PGR and ERBB2; PFN2, on the other hand did not show a consistent pattern in both studies (Figure 8B and S8B).
FIGURE 8. PFN2 and EVL Are Differentially Expressed in Human Breast Tumors.
(A) Box-and-whisker plots showing relative levels of EVL and PFN2 transcript in grade I, II and III; p values are from ANOVA analysis. (B) Logistic regression analysis of the relationship between transcript level and tumor grade. (C) Kaplan-Meier curves representing the probability of survival of breast cancer patients based on relative levels of EVL expression (green: tumors in the lowest quartile; red: tumors in the highest quartile; and black: the interquartile range). Chi square p values evaluate whether there are significant differences between any of the three groups. Cox p values evaluate the association of expression with survival by treating EVL levels as a continuous variable. (D) Quantitation of EVL and profilin-2 protein expression in normal breast tissue, and in grade I, II and III tumors (n is the number of patients per group; and triplicate sections from each patient were analyzed). Values are averages from visual scores (scale: 1–5) ± SEM. (E) Representation of protein expression versus tumor invasion (invasion was assessed in a blinded fashion: Non-inv is non-invasive group and low-inv and high inv are low and high invasion groups, respectively. Right panel shows the distribution of each group in terms of tumor grade. (F) Representative images of normal and tumor breast tissue. Scale bar is 100μm. Insets are magnifications of the boxed areas. (G) Analysis of actin density in normal and tumor breast tissue (actin density was assessed in a blinded fashion, as described in D). Values are averages from visual scores (scale: 1–5) ± SEM. (H) Correlation of EVL levels with actin density (Spearman’s ρ =0.53; p<8×10-6); size of the circles represents the invasive activity in the corresponding tumors. (I) Schematic representation of the correlation between EVL/profilin-2 expression, actin density and invasive behavior, and the potential effect of profilin-2 activity on invasion with or without EVL. See also Figure S8.
To investigate the prognostic significance of EVL and PFN2 expression, we examined the association of EVL and PFN2 expression with the probability of survival in two large breast cancer patient cohorts with long term follow up (Schmidt et al. 2008; van de Vijver et al., 2002). Patients who had tumors with low EVL expression exhibited a significantly lower probability of survival (Figure 8C). Moreover, when treated as a continuous variable, EVL expression proved to be a significant prognostic marker in both cohorts. On the other hand, patients whose tumors expressed either high or low PFN2 expression had a significantly lower probability of survival (Figure S8C). Importantly, in both studies, the high-PFN2 group of patients with poor outcome was significantly enriched in tumors with low EVL expression (Schmidt: 1.4-fold enriched, p=0.0318; Vandevijver: 1.5-fold enriched, p=0.00134).
To examine profilin-2 and EVL protein expression in tumors, we used an array of normal and tumor tissue samples donated by 65 breast cancer patients (spotted in triplicates). Consistent with mRNA levels, profilin-2 protein expression was higher in Grade III tumors compared to other tumor grades (Figure 8D). EVL protein levels, on the other hand, were significantly lower in grade II and III tumors as compared to normal breast tissue (mammary ducts) and grade I tumors (Figure 8D); these results were validated using a second tissue array of samples from 48 patients (spotted in duplicates) (Figure S8D).
Moreover, we grouped tumors based on the extent of tumor infiltration into stroma using a visual score consisting of three categories: “non-inv”, including normal breast tissue and DCIS tumors with no infiltrating tumor cells, “low-inv” with minimal tumor infiltration, and “high-inv”, with extensive infiltration into the stroma as small clusters and single cells (Figure 8E). Profilin-2 levels were lower in the low-inv group compared to the non-inv group; however, as predicted based on the high percentage of grade III tumors in the high-inv group, the levels of profilin-2 were higher in this group compared to the low-inv group (Figure 8E). EVL expression, on the other hand, was strongly anti-correlated with invasion in all categories (Figure 8E); these results were confirmed in the second tissue array (Figure S8D). Consistent with the multivariate analyses, these results suggest that EVL is a potential biomarker for invasion, in addition to tumor grade.
In addition, we stained for actin (β and γ-1) in the same tumors in which we assessed profilin-2 and EVL levels. Grade II and III tumors exhibited significantly lower staining intensity as compared to normal ducts and grade I tumors (Figure 8F–G). Moreover, EVL expression significantly correlated with actin staining intensity regardless of tumor grade (Figure 8H). Importantly, tumors from different grades express equivalent levels of actin (ACTB and ACTG1) and, in tissue culture cells, EVL KD did not alter actin expression (Figure S8E); therefore, the observed differences in actin staining intensity are likely due to differences in actin density possibly caused by changes in the structure of the actin cytoskeleton, such as decreased actin bundling.
Together, these results suggest that EVL is an independent biomarker for tumor grade, and could serve as a potential predictor of prognosis in breast cancer.
DISCUSSION
Our results demonstrate that profilin-2 regulates actin-based cellular processes in a distinct manner compared to the ubiquitous and well-characterized profilin-1 isoform. Profilin-2 preferentially promotes the activity of the Ena/VASP protein EVL, generating unbranched filaments that, when bundled by myosin- dependent contractility, suppress protrusive activity. Down regulation of either profilin-2 or EVL enhances cell migration and invasion in vitro and in vivo. In human tumors, relatively low levels of EVL correlate with low actin density and high invasive activity. Critically, EVL expression is an independent biomarker for tumor grade, and predictive of poor patient outcome.
Profilin Isoforms Regulate the Architecture of the Actin Cytoskeleton
Modulation of the actin cytoskeleton architecture by altering the relative levels of profilin-1 and -2 is due, in part, to a shift in the dominant mode of actin polymerization. Our data suggest that decreasing the relative levels of profilin-1 and increasing those of profilin-2 result in a shift toward a less branched and more linear actin polymerization. Profilin-2 displays higher binding affinity for EVL (and to a lesser extent VASP) than profilin-1, making it more efficient in promoting EVL-mediated linear polymerization; this is in agreement with previous reports showing that EVL and VASP preferentially interact with profilin-2 as compared to profilin-1 (Kursula et al., 2008; Lambrechts et al., 2000; Nodelman et al., 1999; Veniere et al., 2009). Therefore, when profilin-2 is more abundant, EVL activity becomes more dominant in driving actin polymerization; the generation of long unbranched actin filaments by EVL, when coupled with myosin contractility, could suppress protrusive activity.
Moreover, previous reports suggest that profilin-1 binds with higher affinity than profilin-2 to WAVE-2, which promotes branched polymerization by Arp2/3 activation (Miki et al., 1998); this difference in binding affinity renders profilin-1 more critical for Arp2/3-mediated polymerization. Therefore, down regulation of profilin-1 could significantly suppress filament branching and make profilin-2/EVL-dependent polymerization the more dominant mode of polymerization. In addition, dominant EVL activity at the leading edge could directly reduce branched polymerization by suppressing Arp2/3 activity through anti-capping and anti-branching (Bear and Gertler, 2009). Conversely, down regulation of profilin-2 expression could increase filament branching by the Arp2/3 complex and decrease actin bundling, leading to highly dynamic protrusions.
In previous studies using the MDA-MB-231 breast tumor cell line, PFN1 KD was reported to decrease speed of protrusion but increase persistence and enhance directionality of cell migration (Bae et. al 2009; Zou et. al 2007). However, PFN1 KD in MDA-MB-231 cells used in our studies decreased the number of protrusions per cell and suppressed cell migration and invasion (data not shown). In these cells, the profilin-1 concentration (30 uM) was more than 100-fold higher than that of profilin-2 (0.27 uM), consistent with the correlation between highly dynamic cell migration and high profilin-1/low profilin-2 levels. Interestingly, the phenotypic changes to protrusive activity described in Bae et al are similar to those induced by PFN1 KD in SUM159 cells, which express higher levels of profilin-2 than MDA-MB-231 cells. We speculate that the discrepancy between our data and the published studies could be due to differences in the expression of profilin-2 between the MDA-MB-231 variants employed in these studies, or other experimental variables.
Actomyosin Contractility is Important for the Suppression of Protrusive Activity by Profilin-2/EVL-mediated Actin Polymerization
Our data revealed that the suppression of protrusive activity by profilin-2/EVL-mediated actin polymerization requires the aggregation of actin filaments into contractile bundles in a myosin-dependent mechanism. Inhibition of myosin contractility is sufficient to reverse the suppressive effects of profilin-2 and EVL. Interestingly, in the absence of myosin activation, PFN2 overexpression enhances protrusive activity instead of suppressing it. In addition, down regulation of either PFN2 or EVL dramatically decreases actin bundling and increases protrusive activity; this suggests that the polymerization events driven by profilin-2 and EVL are involved in the regulation of protrusion and cell migration by contractile activity.
Importantly, VASP do not exhibit the same capacity to generate profilin-2 mediated actin bundles capable of suppressing protrusive activity; this suggests that the generation of such actin bundles, which might represent a distinct subpopulation of stress fibers, could be unique to EVL. The specificity of EVL involvement in this type of actin cytoskeletal remodeling could be due to distinct set of binding partners that affect EVL function spatially and temporally; currently, we are analyzing the components of the EVL complex by mass spectrometry.
In addition to our data, the suppressive effects of cortical actomyosin bundling on protrusive activity and cell migration have been previously reported in endothelial cells (Fischer et al., 2009). However, other studies showed that some cancer cells are dependent on contractility for migration (Sanz-Moreno et al., 2011). We hypothesize that the effects of contractility vary depending on many factors, including the spatial organization of contractile actin filaments, the nature and strength of matrix adhesion and the cortical actin organization and linkage to the membrane (Friedl and Wolf, 2010; Nakamura et al., 2011; Sheetz et al., 2006). In our model, EVL could play a role in the regulation of several of these factors, thus affecting the outcome of increased contractility.
Profilin-2/EVL-mediated Regulation of Actin Assembly Influences Invasion in Vitro and in Vivo
Our data show that elevated levels of profilin-2 or EVL suppress invasion in 3D matrices in a manner dependent on myosin contractility; and down regulation of profilin-2 or EVL or inhibition of contractility increases 3D invasion significantly. Moreover, in mammary fat-pad xenograft tumors, down regulation of PFN2 or EVL increases the number of extra-tumoral invasive foci.
The increase in invasion associated with EVL KD, correlates with increased protrusions and decreased contractility. Nonetheless, the mechanism by which EVL/profilin-2-mediated polymerization regulates invasion also involves alteration of other cellular processes, such as cell:cell and cell:matrix adhesion. In fact, we did observe that down regulation of PFN2 or EVL decreases both types of adhesion in normal and cancer cells. The characteristic changes in cellular processes that contribute to promoting invasion vary based on genetic and epigenetic alterations within tumor cells, and on alterations in the tumor microenvironment; therefore, the mode of invasion could vary within the same tumor and over the course of tumor progression. In fact, some types of tumor cells are able to switch between different modes of single-cell invasion, such as: amoeboid, characterized by round morphology, high contractility, weak matrix-adhesion, decreased protrusive activity and increased membrane blebbing; and mesenchymal, characterized by elongated morphology, low contractility, strong matrix-adhesion and increased protrusive activity (Sanz-Moreno et al., 2008; Friedl and Wolf, 2010).
In the model we present here, increased invasion is associated with increased protrusion, decreased contractility and also decreased adhesion. Therefore, the invasive phenotype that is induced by downregulation of EVL does not fit the characteristics of either mesenchymal or amoeboid modes of invasion. This suggests that invading cancer cells may display phenotypes along a continuum between the mesenchymal and amoeboid states, in which multiple cellular processes are continuously altered.
PFN2 and EVL Distinctive Expression Profiles Are Predictive of Invasiveness and Poor Prognosis in Human Breast Cancer
EVL expression is significantly lower in highly invasive human breast tumors, in particular high-grade tumors. Moreover, we discovered a strong correlation between EVL expression and actin density in human tumors; highly invasive tumors are characterized by low EVL expression and low actin density. More importantly, lower levels of EVL correlate with poor prognosis and higher mortality in patients. Our analyses demonstrate that EVL is a significant independent biomarker of invasiveness and tumor grade, and could be predictive of prognosis.
Downregulation of EVL has also been implicated in progression of other epithelial tumors. In two large-scale studies investigating genetic and epigenetic alterations in colon cancer, DNA methylation of EVL was frequently observed and correlated with poor prognosis (Grady et al., 2008; Yi et al., 2011). Consistent with these studies, we found that PFN2 and EVL KD enhance protrusive activity in colorectal carcinoma Caco-2 cells. This suggests that downregulation of EVL might be a common feature of more aggressive tumors in multiple types of cancer. Paradoxically, a previous study has shown that EVL mRNA levels are correlated positively with clinical stage (Hu et al., 2008); however the number of tumors analyzed therein was very low (i.e. 3 stage III tumors) and neither invasiveness nor tumor grade were assessed in the context of EVL expression.
Low PFN2 expression has been reported to be associated with poor prognosis in a study of 88 patients with oral squamous cell carcinomas (ma et. al, 2011). In breast adenocarcinomas, we found that both high and low levels of PFN2 expression correlate with poor prognosis in two major clinical studies including data from 495 patients followed over the course of 15 years. This dichotomous correlation of profilin-2 expression with outcome may reflect differences in the phenotypic effects of profilin-2 interactions with distinct binding partners. For example, in tumors that express EVL, profilin-2 could suppress invasive activity, whereas in tumors with low EVL expression, profilin-2 could promote invasive behavior through interactions with other actin polymerization regulators, such as formins (Figure 8I). This may explain why profilin-2 alone does not serve as a significant biomarker of outcome, whereas EVL, which may have more specialized activities that suppress invasion, serves as a better biomarker. More generally, this highlights how differences in expression of a given protein in tumors can lead to distinct outcomes depending on the expression of collaborating proteins. Thus, the assessment of certain biomarkers of clinical outcome may require analysis of interacting proteins that together regulate a biological process.
EXPERIMENTAL PROCEDURES
Quantitation of Intracellular Concentration of Profilin-1 and Profilin-2
Total protein concentration was measured in MCF10A, SUM159 and MDA231 cells using purified profilin-1 and 2 as standards (see supplemental methods).
Kymography Analysis
Kymographs were generated in Nikon Elements along the axis of protrusion/retraction, perpendicular to the cell membrane. Minimum intensity projections were used to determine the areas of high membrane dynamics. Average velocity and frequency of retractions and protrusions, as well as the percent of time the membrane spent retracting, protruding, or resting, were calculated. Persistence (the average duration of protrusion) was also calculated for all conditions, but it was discussed in Results only when significant.
Analytical Ultracentrifugation
We determined the solution molecular weight of monomeric hVASP and EVL in the presence of human profilin-1 and mouse profilin-2a using sedimentation equilibrium analytical ultracentrifugation. Samples containing 5μM Cy3-hVASP1-240aa or Cy3-mEVL1-235aa were combined with 20- 40μM human profilin-1 and/or mouse profilin-2a. A buffer composition of 10mM HEPES pH 7.5, 50mM KCl, and 1mM TCEP was used for all experiments. Proteins were centrifuged until they reached equilibrium at three different speeds (e.g. 10, 14, and 20K rpm) in a Beckman Coulter XL-I ultracentrifuge. The sedimentation profile of Cy3-VASP and Cy3-EVL was determined by monitoring the absorbance at 550 nm (Cy3 fluorophore) every two hours. Global fitting of three equilibrium traces for each condition was performed using NIH Sedphit and Sedphat software. An extinction coefficient of 150,000 M-1cm-1 (Cy3, 550 nm) was used to determine the protein concentration as a function of the radial position. Using a monomer-dimer model, we determined the molecular weight for a single ideal species. Proteins were purified and characterized as previously described (Hansen and Mullins, 2010). See supplemental methods for determination of equilibrium dissociation constants.
Single Actin Filament TIRF Assays
Biotin pegylated TIRF-M imaging chambers used for the visualization of single actin filament polymerization kinetics were generated as previously described by Hansen and Mullins, (2010) and Bieling et al., (2010). See supplemental methods for details.
SUM159 Tumor Model
106 SUM159 cells re-suspended in 30 μl MatrigelTM were injected into the fat pad of 6 to 8 week-old female NOD/SCID Balb/C mice. Three independent experiments were performed; the first and the second experiment consisted of injections of 5 mice per group in each, and the thirds experiment consisted of injections of 10 mice per group. For invasion analysis tumors were collected at 8 to 10 weeks. Invasive foci were defined as clusters of tumor cells outside the margin of the tumor. Invasion was quantified in 9 control, 10 PFN1 KD and 10 PFN2 KD tumors, which account for all tumors of similar size and incubation time. (See supplemental information for the experimental procedure using to generate and induce the EVL KD tumors). All experiments were performed according to the guidelines of the IACUC committee of Harvard Medical School.
Analysis Human Tumor Array
Tumor grade is obtained from the patient pathology report associated with each sample. Protein expression was assessed based on a visual scale ranging from 1 (lowest) to 5 (highest), in triplicate (array#1; 65 patients) or duplicate (array#2; 48 patients) sections from each patient; each analysis was performed in a blinded fashion by at least two different individuals. Human samples used in our studies are exempt from informed consent.
Supplementary Material
SIGNIFICANCE.
The actin cytoskeletal architecture impacts many cellular processes associated with cancer invasion. We show that changing the relative levels of two profilin paralogs alters actin cytoskeletal architecture and influences normal and cancer cell behavior. Our studies revealed functional divergence between, profilin-1, which promotes membrane protrusion, motility, and invasion, and profilin-2, which suppresses these processes. These contrasting phenotypic effects involve distinct actin cytoskeletal remodeling by the two profilins. Profilin-2 exerts these previously unrecognized suppressive effects through selective interaction with the actin polymerization regulator, EVL, which promotes profilin-2-mediated actin cytoskeletal remodeling and suppresses migratory and invasive behaviors. Importantly, analyses of breast tumors revealed that lower EVL expression corresponds to high invasiveness and poor prognosis, making EVL a potential biomarker for patient outcome.
HIGHLIGHTS.
Profilin-2 preferentially drives EVL-mediated actin cytoskeletal remodeling.
Profilin-2 and EVL suppress protrusion, cell motility and invasion.
Profilin-2 and EVL suppressive effects are dependent on actomyosin contractility.
Low EVL expression predicts poor patient outcome in breast cancer.
Acknowledgments
We thank: James Bui and Wa Xian for technical assistance; Rachel Davidowitz, Taru Muranen and Scott Valastyan for their blinded analysis of the human tumor tissue arrays; the Nikon Imaging Center, particularly Jennifer Waters and Wendy Salmon; and the Rodent Histopathology Core Facility, particularly Roderick Bronson. This work was funded by: the Breast Cancer Research Foundation, the NIGMS Cell Migration Consortium, and a gift from the Lee Jeans Foundation through the Entertainment Industry Foundation (JSB); NIH grant P01 HL059561 (JHH); NIH grant ROI #GM61010, UCSF/UC Berkeley Nanomedicine Development Center, and the National Science Foundation (RDM and SDH); and NIH #GM58801 (FBG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bae YH, Ding Z, Zou L, Wells A, Gertler F, Roy P. Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J Cell Physiol. 2009;219:354–364. doi: 10.1002/jcp.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ghosh M, Eddy R, Condeelis J. Cofilin takes the lead. J Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d’Assignies MS, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- Di Nardo A, Gareus R, Kwiatkowski D, Witke W. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J Cell Sci. 2000;113(Pt 21):3795–3803. doi: 10.1242/jcs.113.21.3795. [DOI] [PubMed] [Google Scholar]
- Falet H, Hoffmeister KM, Neujahr R, Italiano JE, Jr, Stossel TP, Southwick FS, Hartwig JH. Importance of free actin filament barbed ends for Arp2/3 complex function in platelets and fibroblasts. Proc Natl Acad Sci U S A. 2002;99:16782–16787. doi: 10.1073/pnas.222652499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. The EMBO journal. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19:260–265. doi: 10.1016/j.cub.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM, et al. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- Honore B, Madsen P, Andersen AH, Leffers H. Cloning and expression of a novel human profilin variant, profilin II. FEBS Lett. 1993;330:151–155. doi: 10.1016/0014-5793(93)80262-s. [DOI] [PubMed] [Google Scholar]
- Hu LD, Zou HF, Zhan SX, Cao KM. EVL (Ena/VASP-like) expression is up-regulated in human breast cancer and its relative expression level is correlated with clinical stages. Oncol Rep. 2008;19:1015– 1020. [PubMed] [Google Scholar]
- Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- Kursula P, Kursula I, Massimi M, Song YH, Downer J, Stanley WA, Witke W, Wilmanns M. High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: the formin homology 1 domain of mDia1 and the proline-rich domain of VASP. J Mol Biol. 2008;375:270–290. doi: 10.1016/j.jmb.2007.10.050. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, Gertler FB. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J Biol Chem. 2000;275:36143–36151. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Janmey PA, Almo SC. Structure of the profilin- poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodelman IM, Bowman GD, Lindberg U, Schutt CE. X-ray structure determination of human profilin II: A comparative structural analysis of human profilins. J Mol Biol. 1999;294:1271–1285. doi: 10.1006/jmbi.1999.3318. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kolbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gaggioli C, Yeo M, Albrengues J, Wallberg F, Viros A, Hooper S, Mitter R, Féral CC, Cook M, Larkin J, Marais R, Meneguzzi G, Sahai E, Marshall CJ. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell. 2011;20:229–245. doi: 10.1016/j.ccr.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Sable JE, Dobereiner HG. Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:417–434. doi: 10.1146/annurev.biophys.35.040405.102017. [DOI] [PubMed] [Google Scholar]
- Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, Brugge JS. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nature cell biology. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Veniere S, Ampe C, Vandekerckhove J, Lambrechts A. The interaction of proline-rich ligands with profilin probed with an enzyme- linked immunosorbent assay. J Biomol Screen. 2009;14:350–359. doi: 10.1177/1087057109332594. [DOI] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt AE, Hines LM, Collins NL, Hu Y, Gunawardane RN, Moreira D, Raphael J, Jepson D, Koundinya M, Rolfs A, et al. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. J Proteome Res. 2006;5:599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JM, Dhir M, Van Neste L, Downing SR, Jeschke J, Glockner S, Calmon M, Hooker C, Funes JM, Boshoff C, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011;17:1535–1545. doi: 10.1158/1078-0432.CCR-10-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Jaramillo M, Whaley D, Wells A, Panchapakesa V, Das T, Roy P. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br J Cancer. 2007;97:1361–1371. doi: 10.1038/sj.bjc.6604038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.