Abstract
UDP-glucuronosyltransferase (UGT) 2A1 is a respiratory and aerodigestive tract-expressing phase II detoxifying enzyme that metabolizes various xenobiotics including polycyclic aromatic hydrocarbons (PAHs). In the present study, a novel exon 3 deletion splice variant was identified for UGT2A1 (UGT2A1Δexon3). As determined by reverse transcription-polymerase chain reaction (PCR), UGT2A1Δexon3 was shown to be expressed in various tissues including lung, trachea, larynx, tonsil, and colon. The ratio of UGT2A1Δexon3/wild-type UGT2A1 expression was highest in colon (0.79 ± 0.08) and lung (0.42 ± 0.12) as determined by real-time PCR; an antibody specific to UGT2A1 showed splice variant protein (UGT2A1_i2) to wild-type protein (UGT2A1_i1) ratios in the range of 0.5 to 0.9 in these tissues. Using ultra-pressure liquid chromatography, we found that homogenates prepared from UGT2A1_i2-overexpressing human embryonic kidney 293 cells exhibited no glucuronidation activity against PAHs, including benzo[a]pyrene-7,8-dihydrodiol (B[a]P-7,8-diol). An inducible in vitro system was created to determine the effect of UGT2A1_i2 expression on UGT2A1_i1 activity. Increasing UGT2A1_i2 levels resulted in a significant (p < 0.01) decrease in the UGT2A1_i1 Vmax against 1-hydroxy (OH)-pyrene, 3-OH-benzo[a]pyrene, and B[a]P-7,8-diol; no significant changes in KM were observed for any of the three substrates. Coimmunoprecipitation experiments suggested the formation of UGT2A1_i1 and UGT2A1_i2 hetero-oligomers and UGT2A1_i1 homo-oligomers; coexpression of UGT2A1_i1 or UGT2A1_i2 with other UGT1A or UGT2B enzymes caused no change in UGT1A or UGT2B glucuronidation activity. These data suggest that a novel UGT2A1 splice variant regulates UGT2A1-mediated glucuronidation activity via UGT2A1-specific protein-protein interactions, and expression of this variant could play an important role in the detoxification of carcinogens within target tissues for tobacco carcinogenesis.
Introduction
The UDP-glucuronosyltransferase (UGT) family of enzymes catalyzes the metabolism of various xenobiotics, including endogenous substrates such as steroid hormones and exogenous substrates such as various drugs and carcinogens (Wells et al., 2004; Nagar and Remmel, 2006). UGT enzymes are classified into families based on sequence and structural homology (Mackenzie et al., 2005). Enzymes in the UGT1A and UGT2B families are the most well known and studied UGTs, with nine functional UGT1A isoforms and seven functional UGT2B isoforms characterized previously (Mackenzie et al., 2005). In comparison with the UGT1A and UGT2B enzyme families, the UGT2A family has been relatively understudied. Recent evidence suggests that one member of the UGT2A subfamily, UGT2A1, may be an important enzyme for tobacco carcinogen metabolism in respiratory and aerodigestive tract tissues (Bushey et al., 2011).
UGT2A1 was initially cloned from the olfactory epithelium, with enzyme expression also reported in fetal lung and brain (Jedlitschky et al., 1999). A later study reported UGT2A1 expression in lung and trachea by quantitative real-time polymerase chain reaction (PCR) (Nishimura and Naito, 2006). More recent quantitative PCR studies have shown UGT2A1 expression is highest in lung followed by trachea, tonsil, larynx, and colon (Bushey et al., 2011). UGT2A1 enzymatic activity was initially reported against substrates such as androgens, estrogens, drugs, and phenol odorants (Jedlitschky et al., 1999; Itäaho et al., 2008; Sten et al., 2009). Recent studies have shown significant UGT2A1 enzyme activity against polycyclic aromatic hydrocarbons (PAHs) implicated in lung and aerodigestive tract carcinogenesis (Bushey et al., 2011). Although other UGTs such as UGT1A6, UGT1A7, and UGT1A10 have been reported to exhibit some activity against certain PAHs, UGT2A1 is the only UGT well expressed in both the respiratory and aerodigestive tracts that exhibits significant glucuronidation activity against a broad range of PAHs and their metabolites (Jin et al., 1993; Zheng et al., 2002; Dellinger et al., 2006; Nishimura and Naito, 2006; Bushey et al., 2011). Based on the expression and activity profile of UGT2A1, this understudied enzyme potentially has a crucial role in the local detoxification of activated PAHs in tobacco target tissues.
Through the process of alternative splicing, single genes can produce multiple mRNA and protein isoforms with varying function. Alternative splicing was initially proposed to affect half of all genes, but other reports suggest that alternative splicing may occur in more than 90% of all genes (Lander et al., 2001; Wang et al., 2008). Other reports suggest that alternative splicing creates UGT1A diversity at the 3′ end of the transcript, through alternative splicing of a novel exon 5b (Girard et al., 2007; Lévesque et al., 2007). The mRNAs resulting from alternative splicing of exon 5b have been shown to have widespread tissue expression, and functional assays have suggested that the proteins resulting from exon 5b splice variants are inactive (Girard et al., 2007). Proteins resulting from UGT1A exon 5b splice variants have also been shown to interact with UGT1A wild-type proteins and negatively modulate wild-type UGT1A glucuronidation activity through a protein-protein interaction (Girard et al., 2007; Lévesque et al., 2007; Bellemare et al., 2010). Whereas UGT2B enzymes are comprised of six independent exons, alternative splicing has also been described for UGT2B4 and UGT2B7, resulting in various nonfunctional alternatively spliced UGT2B4 (Lévesque et al., 2010) or UGT2B7 (Ménard et al., 2011) variants. These splice variants have also been found to be well expressed in various tissues, and the resulting splice variant proteins of UGT2B4 and UGT2B7 have been shown to negatively regulate wild-type UGT2B4 and UGT2B7 glucuronidation activity (Lévesque et al., 2010; Ménard et al., 2011).
Similar to the UGT1A locus, diversity in the UGT2A family is created through exon sharing. UGT2A1 and UGT2A2 transcripts are comprised of a unique first exon joined to common exons 2 to 6 (Mackenzie et al., 2005; Sneitz et al., 2009). Unique first exons in UGT2A1 and UGT2A2 transcripts create variable aglycone-binding domains, altering the substrate specificity for each UGT isoform (Nagar and Remmel, 2006). In the present study, a novel exon 3 deletion splice variant is described for UGT2A1. This variant was hypothesized to negatively regulate UGT2A1 activity, as has been reported for other UGT splice variants. Described are studies examining the expression and function of this variant (UGT2A1Δexon3) and its potential importance in altering overall UGT2A1 glucuronidation activity.
Materials and Methods
Materials.
UDP-glucuronic acid (UDPGA), alamethicin, β-glucuronidase, 4-methylumbelliferone, 1-hydroxy (OH)-pyrene, and 1-naphthol were purchased from Sigma-Aldrich (St. Louis, MO). 1-OH-benzo[a]pyrene (B[a]P), 3-OH-B[a]P, 7-OH-B[a]P, 8-OH-B[a]P, 5-methyl-chrysene-1,2-diol, dibenzo[a,l]pyrene-11,12-diol, and B[a]P-7,8-diol were synthesized in the Organic Synthesis Core Facility at the Penn State College of Medicine (Hershey, PA). High-pressure liquid chromatography-grade ammonium acetate, acetonitrile, a BCA Protein Assay kit, and agarose were purchased from Thermo Fisher Scientific (Waltham, MA). Real-time PCR probes and gene expression assays were acquired from Applied Biosystems (Foster City, CA). Complete Control Inducible Mammalian Expression System kits, ponasterone A (PonA), and Pfu polymerase were obtained from Agilent Technologies (Santa Clara, CA). Dulbecco's modified Eagle's medium, Dulbecco's phosphate-buffered saline (minus calcium-chloride and magnesium-chloride), fetal bovine serum, penicillin-streptomycin, geneticin (G418), and blasticidin were purchased from Invitrogen (Carlsbad, CA). The pcDNA 3.1/V5-His-TOPO and pcDNA6.2/V5/GW/D-TOPO mammalian expression vectors, Superscript II reverse transcription (RT) kit, and hygromycin B were obtained from Invitrogen. The RNeasy kit, QIAquick gel extraction kit, Plasmid Mini kit, and Plasmid Maxi kit were purchased from QIAGEN (Valencia, CA). All oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA).
Determination of UGT2A1Δexon3 Tissue Expression.
The UGT2A1 exon 3 deletion splice variant was initially discovered after RT-PCR using pooled lung RNA from three individuals. The primers used to amplify full-length UGT2A1Δexon3 from lung cDNA were 5′-CATCAAATCTTCTGCATCAAGCCAC-3′ (sense; UGT2A1_S1) and 5′-TGACAGGAAGAGGGTATAGTCAGC-3′ (antisense; UGT2A1_AS1), corresponding to nucleotides −28 to −4 and +1834 to +1811, respectively, relative to the UGT2A1 translation start site. For all PCRs, RNA was acquired and RT-PCR was completed as described previously (Bushey et al., 2011). In brief, 2 μg of RNA from each tissue was used with oligo(dT)s for RT, with cDNA corresponding to 100 ng of RNA used in the subsequent PCRs. Unless otherwise noted, all PCRs were performed with Pfu polymerase, with an initial denaturing temperature of 94°C for 2 min, 40 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 2 min, followed by a final cycle of 10 min at 72°C.
After the initial discovery of UGT2A1Δexon3 from pooled lung cDNA, additional tissues were screened for UGT2A1Δexon3 mRNA expression by RT-PCR and real-time PCR using pooled RNAs from at least three individuals. Primers specific to exon 1, 5′-GACATGGCTGGAAAATAGACC-3′ (sense), and exon 5, 5′-CCATAGGGACTCCGTGGTAAAT-3′ (antisense), of UGT2A1, corresponding to nucleotides +276 to +296 and +1165 to +1144, respectively, relative to the translation start site were used to screen for UGT2A1Δexon3 expression. RT was completed by using RNAs from HEK293 cell lines overexpressing wild-type UGT2A1 or UGT2A1Δexon3 (see Generation of a UGT2A1_i2 Overexpressing Cell Line and Determination of UGT2A1_i2 Protein Expression for methods), and cDNAs after RT were used as positive controls for PCR amplification, and water was used as a negative control. PCR products were gel-purified by using a QIAquick gel extraction kit (QIAGEN) and sequenced by dideoxy sequencing at the Penn State University Nucleic Acid Facility (State College, PA) and then compared with the UGT2A1 sequence in GenBank (NM_006798.3). To verify UGT2A1Δexon3 expression in the tissues analyzed, PCRs were run multiple times with all positive and negative controls. Genomic DNA from five individuals was PCR-amplified to determine whether prevalent splice site polymorphisms exist in introns 2 or 3 of UGT2A1. A sense primer specific for the 3′ end of UGT2A1 exon 2 and an antisense primer specific for UGT2A1 exon 3 were used to amplify intron 2, whereas a sense primer specific for exon 3 and an antisense primer specific for the 5′ end of UGT2A1 exon 4 were used to amplify intron 3. After PCR amplification the PCR products were gel-extracted and sequenced.
A real-time PCR assay was developed to quantitatively assess the relative levels of wild-type UGT2A1 and UGT2A1Δexon3 transcripts in tissues that were determined previously to have UGT2A1 expression. Separate assays were designed to specifically amplify wild-type UGT2A1 or UGT2A1Δexon3 (Fig. 1A). A sense primer specific to exon 1 (5′-CTACATGTTTGAAACTCTTTGGAAATC-3′) and a 5′-labeled VIC probe (Applied Biosystems) specific to exon 2 (5′-TCCGAACATATTGGGATT-3′), corresponding to nucleotides +660 to +686 and +767 to +784, respectively, relative to the UGT2A1 translation start site, were used to detect both wild-type UGT2A1 and UGT2A1Δexon3. An antisense primer specific to UGT2A1 exon 3 (5′-TTACCTGAGCTCTGGAT-AAATTCTTC-3′), corresponding to nucleotides +896 to +871 relative to the UGT2A1 translation start site, was used to specifically amplify the wild-type UGT2A1 transcript, and an antisense primer specific to the UGT2A1Δexon3 exon 2 and 4 junction (5′-TTTCCTTTGTATCTCCATAAAACCTTAG-3′), corresponding to nucleotides +887 to +860 relative to the UGT2A1Δexon3 translation start site, was used to specifically amplify the UGT2AΔexon3 transcript. Reactions were completed by using the standard Applied Biosystems thermal cycling parameters, with human large ribosomal protein (RPLPO; Hs99999902_m1) used as a housekeeping gene. cDNAs, corresponding to 20 ng of RNA, were used for each real-time assay, and reactions were performed in triplicate. Real-time PCR experiments were carried out at the Penn State College of Medicine Functional Genomics Core Facility (Hershey, PA) with an ABI 7900 HT thermal cycler (Applied Biosystems), and data were analyzed by using SDS 2.2 software (Applied Biosystems). Assay specificity for wild-type UGT2A1 or UGT2A1Δexon3 transcripts was confirmed through agarose gel electrophoresis and dideoxy sequencing of real-time PCR products. Real-time PCR data were corrected to account for the amplification efficiency of each real-time PCR assay as described previously (Jones et al., 2012). The relative tissue expression of UGT2A1Δexon3 transcript was calculated by using the ΔΔCt method relative to the amount of wild-type UGT2A1 transcript for each tissue specimen examined.
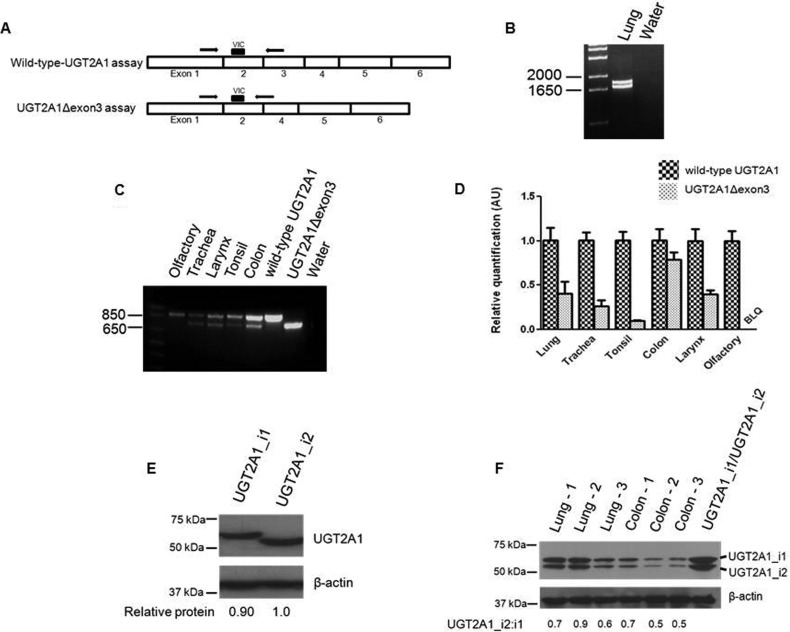
Fig. 1.
Determination of UGT2A1Δexon3 expression. A, schematic of quantitative real-time PCR assay developed to specifically detect either wild-type UGT2A1 or UGT2A1Δexon3. B, full-length UGT2A1 was PCR-amplified after RT of pooled lung RNA. A UGT2A1 mRNA variant lacking exon 3 (UGT2A1Δexon3), in addition to wild-type UGT2A1 mRNA, was discovered after gel extraction and dideoxy sequencing of the PCR products. C, a sense primer specific to exon 1 and an antisense primer specific to exon 5 of UGT2A1 were used in RT-PCR to determine tissue-specific expression of UGT2A1Δexon3 in tissues that were determined previously to express wild-type UGT2A1 (Bushey et al., 2011). For both RT-PCR experiments (B and C), the cDNA equivalent of 100 ng of RNA was used. RNAs from HEK293 cell lines overexpressing wild-type UGT2A1 or UGT2A1Δexon3 were used as a positive control, and water in place of cDNA was used as a negative control. D, quantitative real-time PCR was completed to determine UGT2A1Δexon3 expression levels relative to wild-type UGT2A1 expression in human tissues. cDNAs, corresponding to 20 ng of RNAs from tissues exhibiting UGT2A1Δexon3 expression (B and C), were used in conjunction with the custom-designed real-time PCR assay described in A. Relative UGT2A1Δexon3 expression in each tissue was determined by comparing UGT2A1Δexon3 mRNA levels to wild-type UGT2A1 mRNA levels, set to 1.0 as a reference. Results, expressed as the mean ± S.D. of triplicates, were normalized to RPLPO RNA expression for each tissue. BLQ, below limit of quantification. E, representative Western blots showing UGT2A1_i2 expression in an overexpressing HEK293 cell line. Fifty micrograms of total protein homogenate from cell lines overexpressing UGT2A1_i1 or UGT2A1_i2 was loaded to each lane for a Western blot, and expression was determined by using an anti-UGT2A1 antibody. β-Actin was used as a loading control. The relative ratio of UGT2A1_i1 or UGT2A1_i2 protein to β-actin was used to determine relative protein expression in each overexpressing cell line. F, lung and colon tissue homogenates (250 μg) were screened for UGT2A1_i1 and UGT2A1_i2 expression; 60 μg of protein (30 μg of each UGT2A1 isoform) from HEK293 cell lines overexpressing UGT2A1_i1 or UGT2A1_i2 was mixed in a 1:1 ratio and used as a positive control. β-Actin was used as a loading control.
Generation of a UGT2A1_i2 Overexpressing Cell Line and Determination of UGT2A1_i2 Protein Expression.
A HEK293 cell line overexpressing wild-type UGT2A1 (UGT2A1_i1) was established previously (Bushey et al., 2011). A HEK293 cell line overexpressing the isoform corresponding to UGT2A1exonΔ3 (UGT2A1_i2) was created, and cell homogenates were made by using a similar protocol (described in Supplemental Methods). An antibody designed to recognize UGT2A1 protein was created previously by using a peptide encoded by exon 1, a region common to both UGT2A1_i1 and UGT2A1_i2 (Bushey et al., 2011). Levels of UGT2A1_i1 and UGT2A1_i2 protein were determined by Western blot analysis using the anti-UGT2A1 antibody at a 1:500 dilution as recommended by the manufacturer (Open Biosystems, Huntsville, AL) and 50 μg of UGT2A1-overexpressing cell homogenate protein. The monoclonal β-actin antibody (Sigma-Aldrich) was used as a loading control, and the Western blot was done in triplicate. The intensity of the UGT2A1 signal was measured with the ImageJ program (National Institutes of Health, Bethesda, MD). Because a UGT2A1 standard is not commercially available, the relative protein expression of UGT2A1_i1 and UGT2A1_i2 in homogenate from each cell line was calculated relative to the β-actin loading control. UGT2A1 protein expression was also analyzed in normal human lung and colon homogenates prepared from tissues from independent subjects (obtained from the Banner Sun Health Research Institute, Sun City, AZ) by a similar Western blot procedure, using 250 μg of tissue homogenate prepared by using an Omni TH rotor-stator homogenizer (Omni International, Kennesaw, GA) in 50 mM Tris, pH 7.5, 1.15% KCl, and 1 mM disodium EDTA. For the determination of UGT2A1 expression in colon and lung tissue homogenates, protein from cell lines overexpressing UGT2A1_i1 and UGT2A1_i2 was mixed in a 1:1 ratio and loaded (60 μg of total protein) as a positive control. The relative amount of UGT2A1_i2 to UGT2A1_i1 in each lung or colon specimen was determined relative to β-actin as described above.
Glucuronidation Assays.
Glucuronidation assays using homogenate from HEK293 cells overexpressing UGT2A1_i1 and UGT2A1_i2 were completed essentially as described previously (Fang et al., 2002; Wiener et al., 2004a). In brief, after an initial incubation of 100 μg of protein homogenate with alamethicin (50 μg/mg protein) for 15 min on ice, glucuronidation reactions were performed in a final reaction volume of 25 μl at 37°C with 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 4 mM UDPGA, and between 6 and 800 μM substrate. Reactions were terminated by the addition of 25 μl of cold acetonitrile on ice. Reaction mixtures were centrifuged for 10 min at 16,100g before the collection of supernatant. Glucuronide formation was determined by using an Acquity ultra-pressure liquid chromatography (UPLC) System (Waters, Milford, MA) as described previously (Fang et al., 2002; Wiener et al., 2004b; Dellinger et al., 2007; Balliet et al., 2009; Olson et al., 2009; Bushey et al., 2011). The flow rate was maintained at 0.5 ml/min, and a reverse-phase Acquity UPLC BEH C18 1.7-μm 2.1 × 100-mm column (Waters) was used to separate free substrate and the conjugated glucuronide. A gradient of solution A (5 mM NH4OAc, pH 5.0 and 10% acetonitrile) and solution B (100% acetonitrile) was used to elute the glucuronide and substrate from the column. The initial solvent gradients and UV absorbance wavelengths used to detect glucuronidation of various substrates were described previously (Bushey et al., 2011). Reactions with nontransfected HEK293 cell homogenate, no substrate added to the reaction mixture, or only substrate and no homogenate added to the reaction mixture were used as negative controls. Homogenate from a HEK293 cell line overexpressing UGT2A1_i1 was used as a positive control.
Creation of a UGT2A1_i2 Inducible System.
Wild-type UGT2A1 was cloned into the V5-tagged, blasticidin resistance gene-containing pcDNA6.2/V5/GW/D-TOPO vector by using a sense primer 5′-CACCATGTTAAACAACCTTCTGC-3′ (UGT2A1_S2) and antisense primer 5′-TTCTCTTTTTTTCTTCTTTCCTATCTTACC-3′ (UGT2A1_AS2) corresponding to nucleotides +1 to +19 and +1581 to +1552, respectively, relative to the UGT2A1 translation start site, amplifying the entire coding region of wild-type UGT2A1 minus the stop codon (underlined nucleotides in sense primer indicate a CACC anchor). An ecdysone-analog-inducible mammalian expression system was used to regulate UGT2A1_i2 levels, with UGT2A1Δexon3 cloned into the FLAG-tagged, hygromycin resistance gene-containing pEGSH vector by using a similar sense primer to that described above but containing a XhoI restriction site (underlined) on the 5′ end (5′-GCACTCGAGATGTTAAACAACCTTCTGC-3′; UGT2A1_S3), and a similar antisense primer to that described above with a XbaI restriction site (underlined) on the 5′ end (5′-GATTCTAGACGTTCTCTTTTTTTCTTCTTTCCTATCTTACC-3′; UGT2A1_AS3). After plasmid preparations for each clone, vector sequences were verified by direct dideoxy sequencing. Eight micrograms each of the pEGSH_UGT2A1Δexon3 vector and the G418 resistance gene-containing pERV regulatory vector, comprising the inducible system, as well as the pcDNA6.2/V5/GW/D-TOPO_wtUGT2A1 vector, were stably transfected simultaneously into HEK293 cells by using a standard Lipofectamine protocol. Selection of HEK293 cells overexpressing the three vectors was completed by using a combination of 400 μg/ml G418, 9 μg/ml blasticidin, and 75 μg/ml hygromycin B in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Multiple clones were analyzed for inducible gene expression, and a stable clone overexpressing all three vectors simultaneously was chosen based on the efficiency of UGT2A1_i2 induction.
After the creation of the stable UGT2A1_i1_V5/UGT2A1_i2_FLAG/pERV-overexpressing cell line, UGT2A1_i2 expression was induced with varying levels of the ecdysone-analog PonA. HEK293 cells at 50% confluence were treated with 2, 6, or 10 μM PonA in ethanol for 12 h. Vehicle (0.01% ethanol) was added to these cells as a negative control. Cells were harvested, and homogenates were made as described previously (Dellinger et al., 2006; Sun et al., 2006). Protein homogenates from the various treatment groups were screened for UGT2A1_i1_V5 and UGT2A1_i2_FLAG expression by Western blot analysis using 50 μg of total protein per sample, with the analysis of UGT2A1_i2_FLAG induction performed in triplicate. UGT2A1_i1_V5 expression was determined by using a monoclonal mouse V5-HRP antibody (Invitrogen) at a 1:5000 dilution, and UGT2A1_i2_FLAG was determined by using a monoclonal mouse anti-Flag antibody (Sigma-Aldrich) at a 1:1000 dilution. UGT2A1_i1_V5 and UGT2A1_i2_FLAG expression levels were also confirmed by using the anti-UGT2A1 antibody described above, with 100 μg of protein homogenate from both the control and 10-μM PonA treatment groups used with the anti-UGT2A1 antibody at a 1:500 dilution. In all cases, the monoclonal β-actin antibody was used as a loading control.
After verification of UGT2A1_i1 and UGT2A1_i2 protein levels, homogenate was prepared and used for activity assays as described previously (Dellinger et al., 2007; Sun et al., 2007; Balliet et al., 2009, 2010; Olson et al., 2009; Bushey et al., 2011). Cell lysates were homogenized for 10 s on ice by using a Bio-Vortexer (Biospec Products, Bartlesville, OK). Activity was determined against three PAHs that were previously shown to be substrates of UGT2A1 (Bushey et al., 2011). Activity assays were completed in triplicate for each substrate examined, using the control cell line and cell lines treated with 2, 6, or 10 μM PonA. For each substrate, the glucuronidation rate was determined at eight concentrations that encompassed the KM of the substrate. For glucuronidation rate determinations, cell homogenate protein levels and incubation times for each substrate were determined experimentally to ensure that substrate utilization was less than 10% and maximized the levels of detection while in a linear range of glucuronide formation. Cell lines overexpressing either UGT2A1_i1_V5 or UGT2A1_i2_FLAG alone were also created by using an identical protocol for nontagged UGT2A1_i1 and UGT2A1_i2-overexpressing cell lines, with cell homogenates prepared as described above.
UGT2A1_i1 and UGT2A1_i2 Coimmunoprecipitation Assays.
HEK293 cells overexpressing UGT2A1_i1_V5/UGT2A1_i2_FLAG were treated with 10 μM PonA for 12 h before washing with phosphate-buffered saline and homogenate preparation as described previously (Bellemare et al., 2010). A Dynabead Protein G Immunoprecipitation Kit (Invitrogen) using the standard protocol was used to determine potential protein interactions. Three micrograms of a mouse monoclonal anti-FLAG antibody or 1.5 μg of mouse monoclonal V5 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) were incubated with Dynabeads for 15 min with rotation at room temperature. Protein lysates at a concentration of 2.5 μg/μl (1250 μg of total protein) from vehicle- or PonA-treated UGT2A1_i1_V5/UGT2A1_i2_FLAG-inducible cells were then incubated with the same Dynabeads for 30 min with rotation at room temperature. After three washes in phosphate-buffered saline at room temperature, immunoprecipitated proteins were eluted with the provided elution buffer (Invitrogen) under denaturing conditions, heated at 90°C for 10 min in loading buffer, and subjected to Western blot analysis using either a monoclonal mouse V5-HRP antibody at a 1:5000 dilution or a mouse FLAG-HRP antibody (Cell Signaling Technology, Danvers, MA) at a 1:1000 dilution. All coimmunoprecipitation (coIP) experiments were repeated four times. UGT2A1_i1 homo-oligomerization was investigated by using a similar protocol (described under Supplemental Methods).
Coexpression of UGT2A1_i1 and UGT2A1_i2 with Other UGT Isoforms.
UGT2A1Δexon3 was cloned into the pcDNA6.2/V5/GW/D-TOPO vector by using UGT2A1_S2 and UGT2A1_AS2 primers as described above. The newly created pcDNA6.2/V5/GW/D-TOPO_UGT2A1Δexon3 vector was transfected by using a standard Lipofectamine protocol into previously established stable HEK293 cell lines overexpressing UGT1A7, UGT1A10, or UGT2B17 (Ren et al., 2000; Dellinger et al., 2006; Sun et al., 2006; Chen et al., 2008). The pcDNA6.2/V5/GW/D-TOPO_wtUGT2A1 vector described above was also transfected into UGT1A10- and UGT2B17-overexpressing cell lines by using a standard Lipofectamine protocol. Selection of HEK293 cells coexpressing UGT2A1_i1 or UGT2A1_i2 and UGT1A7, UGT1A10, or UGT2B17 was completed by using blasticidin (9 μg/ml) and G418 (400 μg/ml).
After RNA extraction and RT of RNA from the UGT-coexpressing cell lines, real-time PCR was performed to determine the relative levels of UGT1A7, UGT1A10, or UGT2B17 expression versus wild-type UGT2A1 or UGT2A1Δexon3 expression in each coexpressed cell line. Applied Biosystems gene expression assays for UGT1A7 (Hs02517015_s1), UGT1A10 (Hs02516990_s1), UGT2B17 (Hs00854486_sH), and UGT2A1 (Hs00792016_m1) were used to determine relative transcript levels. The UGT2A1 Applied Biosystems gene expression assay is specific for UGT2A1 exon 1, enabling the assay to detect transcripts from both the wild-type UGT2A1 and exon 3-deleted UGT2A1 splice variant. Reactions were completed by using the standard Applied Biosystems protocol, with RPLPO used as a housekeeping gene. cDNA corresponding to 20 ng of RNA was used for each real-time reaction, and reactions were performed in triplicate by using standard Applied Biosystems thermal cycling parameters. Real-time-PCR data were corrected to account for the amplification efficiency of each real-time PCR assay as described previously (Jones et al., 2012). The relative level of UGT2A1 transcript in each cell line was calculated by using the ΔΔCt method, relative to the amount of UGT1A7, UGT1A10, or UGT2B17 in each coexpressed cell line. An anti-UGT1A antibody (BD Biosciences, San Jose, CA) at 1:3000 dilution and the V5-HRP antibody described above at 1:5000 dilution were used in Western blot analyses to verify that coexpression had no impact on UGT1A7, UGT1A10, or UGT2A1_V5 expression.
Homogenate was created from each of the HEK293 coexpressed overexpressing cell lines, and glucuronidation assays were completed as described above. Glucuronidation assays were performed by using homogenates from HEK293 overexpressing either UGT1A7/UGT2A1_i2, UGT1A10/UGT2A1_i2, or UGT2B17/UGT2A1_i2, with 3-OH-B[a]P and/or 1-naphthol as substrates, because PAHs were shown previously to be glucuronidated by these UGTs (Zheng et al., 2002; Turgeon et al., 2003; Dellinger et al., 2006). Because UGT2A1 exhibits activity against PAHs (Bushey et al., 2011), homogenates from HEK293 cells overexpressing UGT1A10/UGT2A1_i1 and UGT2B17/UGT2A1_i1 were used in glucuronidation assays with the tobacco-specific nitrosamine (TSNA) 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), which is not a substrate for UGT2A1 but is metabolized by both UGT1A10 and UGT2B17 (Lazarus et al., 2005; Balliet et al., 2010; Bushey et al., 2011). Because previous coexpression studies have reported activity changes to be substrate-dependent (Fujiwara et al., 2007; Finel and Kurkela, 2008), additional activity reactions were performed by using other non-UGT2A1 substrates, including the flavonoid chrysin for UGT1A7/UGT2A1_i2 (Webb et al., 2005), the heterocyclic amine (HCA) metabolite N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine (N-OH PhIP) for UGT1A10/UGT2A1_i1 and UGT1A10/UGT2A1_i2 (Dellinger et al., 2007), and ibuprofen for UGT2B17/UGT2A1_i1 and UGT2B17/UGT2A1_i2 (Turgeon et al., 2003). For chrysin, N-OH PhIP, and ibuprofen glucuronidation rate determinations, substrate concentrations were chosen that approximated the reported KM from previous experiments.
Data Analysis and Statistics.
Three independent experiments were performed for kinetic analyses, and Prism 5 software (GraphPad Software, Inc., San Diego, CA) was used to calculate kinetic values. Kinetic constants Vmax and KM for all substrates were calculated by using the Michaelis-Menten equation for the rate of product formation versus substrate concentration, with data transformed into linear Eadie-Hofstee plots. An analysis of variance, followed by a post test for linear trend, was used to compare the KM and Vmax of glucuronide formation for the various UGT2A1_i1/UGT2A1_i2 PonA treatment groups. Student's t test was used to compare mRNA expression levels and enzyme kinetics of homogenate activities after coexpression of UGT2A1 with UGT1A7, UGT1A10, or UGT2B17.
Results
Relative UGT2A1Δexon3 Expression in Multiple Tissues.
Since its original discovery in olfactory epithelium (Jedlitschky et al., 1999), UGT2A1 has been shown to be expressed in multiple respiratory, digestive, and aerodigestive tract tissues (Bushey et al., 2011). In the present study, RT-PCR amplification of UGT2A1 from pooled lung RNA yielded two distinct products: wild-type UGT2A1 and a novel variant of UGT2A1, which upon direct sequencing was shown to be a splice variant lacking exon 3 (UGT2A1Δexon3; Fig. 1B). The deletion of this exon creates a transcript that is 132 nucleotides shorter, with the open reading frame of the gene remaining intact. Independent PCR amplification and sequencing of genomic DNA from five individual lung specimens containing both the wild-type and variant isoforms showed that UGT2A1 had no polymorphisms between exons 2 and 4, inclusive of the exons 2 and 4 splice sites; the lack of prevalent polymorphisms in these regions was confirmed by analyzing this sequence in HapMap (International HapMap Consortium, 2003). After the discovery of this splice variant in lung tissue, additional aerodigestive tract tissues known to express wild-type UGT2A1 were screened for UGT2A1Δexon3 expression. As shown in Fig. 1C, UGT2A1Δexon3 was also expressed in the trachea, larynx, tonsil, and colon; no UGT2A1Δexon3 expression was observed in pooled olfactory RNA. Using a custom real-time PCR assay (Fig. 1A), the relative expression levels of UGT2A1Δexon3 were determined relative to wild-type UGT2A1 in each tissue, with wild-type UGT2A1 expression in each tissue set to 1.0 as a reference. The relative UGT2A1Δexon3 expression was demonstrated to be the highest in colon (0.79 ± 0.08), followed by lung (0.42 ± 0.12) > larynx (0.39 ± 0.05) > trachea (0.27 ± 0.07) > tonsil (0.10 ± 0.02) (Fig. 1D). In agreement with a previous study analyzing wild-type UGT2A1 expression (Bushey et al., 2011), no UGT2A1Δexon3 expression was detected after multiple RT-PCR attempts or real-time PCR in tissues of the prostate, liver, pancreas, kidney, esophagus, whole brain, olfactory, or breast (results not shown).
UGT2A1_i2 Protein Expression in a HEK293-Overexpressing Cell Line and Tissue Homogenates.
An antibody against UGT2A1 was previously created and used to assess wild-type UGT2A1 (UGT2A1_i1′) protein levels in HEK293-overexpressing cell lines (Bushey et al., 2011). Likewise, a stable HEK293 cell line transfected with UGT2A1Δexon3 demonstrated UGT2A1 splice variant protein (UGT2A1_i2′) overexpression using the same antibody (Fig. 1E), with the mean level of UGT2A1_i1 protein expression calculated to be 0.92 ± 0.07 versus that observed for UGT2A1_i2 (set as 1.0 as a reference) in the two UGT2A1-overexpressing cell lines. The UGT2A1_i2 protein lacks 44 amino acids that are derived from exon 3, creating a protein that is 483 amino acids in length compared with the 527 amino acids that comprise UGT2A1_i1. Expression of both UGT2A1_i1 and UGT2A1_i2 was shown in protein homogenates made from normal lung tissues from three donors and normal colon tissues from three donors (Fig. 1F). For the lung and colon specimens analyzed, UGT2A1_i1 was expressed at marginally higher levels than UGT2A1_i2, with the ratios of UGT2A1_i2/UGT2A1_i1 ranging from 0.6 to 0.9 for lung and 0.5 to 0.7 for colon.
Enzymatic Activity of UGT2A1_i2.
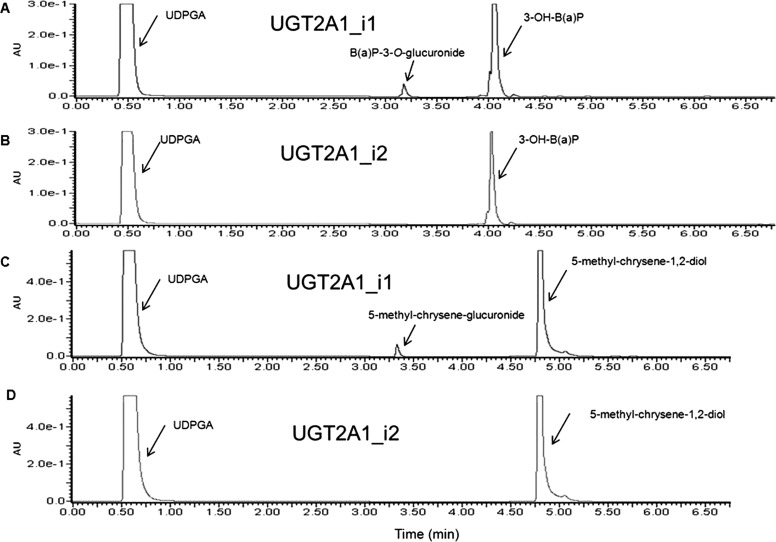
Because UGT2A1Δexon3 RNA and UGT2A1_i2 protein were shown to be expressed in lung and a variety of other human tissues including tissues of the aerodigestive tract, and UGT2A1_i1 was previously shown to be active against PAHs involved in tobacco carcinogenesis (Bushey et al., 2011), UGT2A1_i2 activity against PAHs and other tobacco carcinogens was investigated. Homogenate from UGT2A1_i2-overexpressing HEK293 cells, which was shown to express UGT2A1_i2 protein at slightly higher relative levels than the stable HEK293 cell line overexpressing UGT2A1_i1 (Fig. 1E), was used in glucuronidation activity assays to determine the enzymatic activity of UGT2A1_i2. Although cell homogenates overexpressing UGT2A1_i1 demonstrated glucuronidation activity against both 3-OH-B[a]P (Fig. 2A) and 5-methyl-chrysene-1,2-diol (Fig. 2C), no glucuronide was detected when UGT2A1_i2 protein was used in the assay (Fig. 2, B and D). No detectable glucuronidation activity was observed for UGT2A1_i2-overexpressing cell homogenates against all other PAHs examined including 1-OH-pyrene, 1-naphthol, 1-OH-B[a]P, 7-OH-B[a]P, 8-OH-B[a]P, B[a]P-7,8-diol, and dibenzo[a,l]pyrene-11,12-diol, using up to 400 μg of cellular homogenate and 750 μM substrate in a 18-h incubation (results not shown). The UGT2A1_i2 variant also lacked activity against 4-methylumbelliferone, a known UGT2A1 substrate and a common substrate of most UGT isoforms (Uchaipichat et al., 2004). UGT2A1_i1 was shown previously to have no detectable glucuronidation activity against TSNAs and HCAs (Bushey et al., 2011); UGT2A1_i2 also had no detectable glucuronidation activity against these substrates (results not shown).
Fig. 2.
UGT2A1_i2 exhibits no detectable glucuronidation activity against various PAHs. UGT2A1_i2 activity was determined against PAHs that were determined previously to be substrates of UGT2A1_i1. Shown are representative UPLC chromatograms of UGT2A1_i1 activity against 3-OH-B[a]P (A), UGT2A1_i2 activity against 3-OH-B[a]P (B), UGT2A1_i1 activity against 5-methyl-chrysene-1,2-diol (C), and UGT2A1_i2 activity against 5-methyl-chrysene-1,2-diol (D).
UGT2A1_i2 Modulates UGT2A1_i1 Activity.
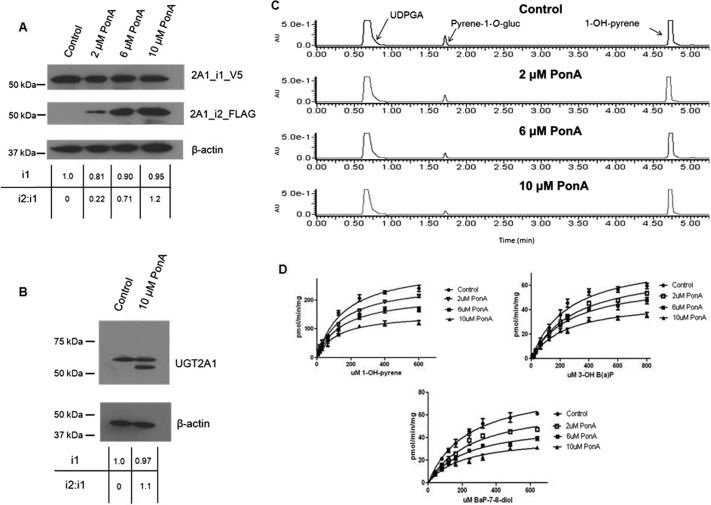
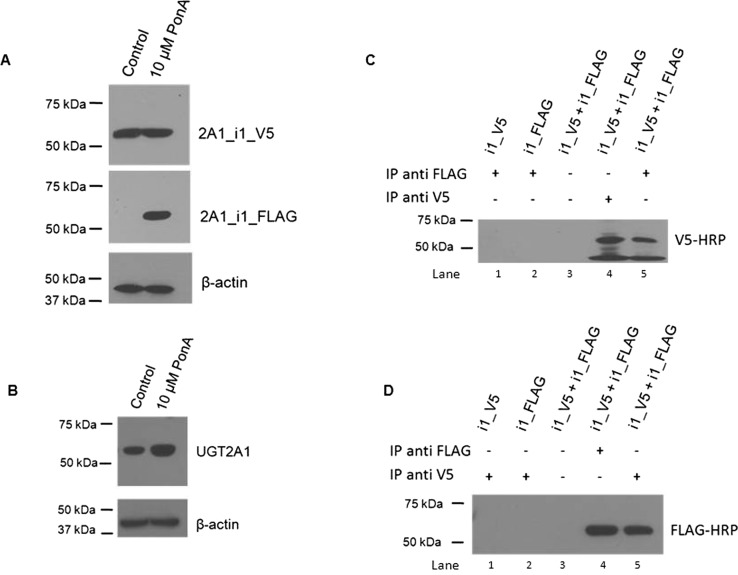
To examine the potential effects of increasing levels of UGT2A1_i2 expression on UGT2A1_i1 activity, a coexpression system was generated to allow for stable UGT2A1_1 protein levels and UGT2A1_2 expression levels regulated by the ecdysone analog PonA. To more easily differentiate between the UGT2A1 isomers in coIP experiments, UGT2A1_i1 was V5-tagged, and UGT2A1_i2 was FLAG-tagged, both at the C terminus of the protein. As shown by Western blot analysis (Fig. 3A), no detectable expression of UGT2A1_i2_FLAG was observed in the vehicle control group. After the addition of increasing dosages of PonA there was a corresponding increase in UGT2A1_i2_FLAG levels, whereas UGT2A1_i1_V5 levels remained relatively constant. The UGT2A1_i2 induction and corresponding protein quantification was completed in triplicate: 2 μM PonA treatment induced the mean UGT2A1_i2 levels to 0.28 ± 0.05 relative to UGT2A1_i1 (set as 1.0 as a reference in all cases), 6 μM PonA treatment induced the mean UGT2A1_i2 levels to 0.76 ± 0.06 relative to UGT2A1_i1, and 10 μM PonA treatment induced the mean UGT2A1_i2 levels to 1.18 ± 0.09 relative to UGT2A1_i1. Relative UGT2A1_i1_V5 protein levels were calculated for each treatment group and used for normalization of kinetic data, with UGT2A1_i1 expression in the control group set as the reference at 1.0. UGT2A1_i1 expression was relatively consistent in all treatment groups (Fig. 3A). Mean UGT2A1_i1_V5 levels were determined to be 0.89 ± 0.08 in the 2-μM PonA treatment group, 0.95 ± 0.06 in the 6-μM PonA treatment group, and 0.94 ± 0.04 in the 10-μM PonA treatment group. UGT2A1_i1_V5 and UGT2A1_i2_FLAG expression levels were confirmed by Western blot using the anti-UGT2A1 antibody (Fig. 3B). Using this antibody, no UGT2A1_i2 was detected in the control group, and the ratio of UGT2A1_i2/UGT2A1_i1 in the 10-μM treatment group (1.1) was approximately equal to that observed using the anti-FLAG and anti-V5 antibodies (1.18 ± 0.09) described above. In addition, the relative expression of UGT2A1_i1 in the 10-μM PonA treatment group using the anti-UGT2A1 antibody (0.97) was similar to that observed using the anti-FLAG and anti-V5 antibodies (0.94 ± 0.04) described above.
Fig. 3.
Effect of increasing UGT2A1_i2 expression on UGT2A1_i1 glucuronidation activity. A, representative Western blots showing stable expression of UGT2A1_i1_V5 and induction of UGT2A1_i2_FLAG expression in HEK293 cells. UGT2A1_i2_FLAG expression was induced through the addition of increasing doses of PonA for 12 h; 50 μg of total protein homogenate from each PonA treatment group was loaded per lane for each Western blot. B, Western blots with 100 μg of protein from the control and 10-μM PonA treatment groups was performed with an anti-UGT2A1 antibody to confirm UGT2A1 expression levels detected by the anti-V5 and anti-FLAG antibodies. For A and B the relative expression of UGT2A1_i1, and the relative UGT2A1_i2/UGT2A1_i1 ratio, was determined for each treatment group. Induction of UGT2A1_i2 expression and Western blots were performed in triplicate. In all cases, β-actin was used as a loading control. C, representative chromatograms showing the effect of increasing UGT2A1_i2 expression on UGT2A1_i1 activity against 1-OH-pyrene. D, Michaelis-Menten kinetic curves for 1-OH-pyrene, 3-OH-B[a]P, and B[a]P-7,8-diol glucuronidation by UGT2A1_i1, showing the effect of increasing expression levels of UGT2A1_i2 on UGT2A1_i1 enzyme activity.
Activity assays and enzyme kinetics were completed to determine the potential impact of UGT2A1_i2 expression on UGT2A1_i1 enzyme activity. The effects of UGT2A1_i2 expression on UGT2A1_i1 activity were assessed in glucuronidation assays using the PAH substrates 1-OH-pyrene, 3-OH-B[a]P, and B[a]P-7,8-diol. These substrates represent PAHs of varying complexity and were shown to be substrates for UGT2A1_i1 in previous studies (Bushey et al., 2011). Representative chromatograms using cell homogenates from the control group and the three PonA treatment groups against 1-OH-pyrene (Fig. 3C) and Michaelis-Menten kinetics curves for three PAHs examined (Fig. 3D) show that increases in UGT2A1_i2 expression caused a decrease in the rate of UGT2A1_i1 glucuronide formation. Upon kinetic analysis, significant (p < 0.01) trends were observed between increasing levels of UGT2A1_i2 expression and decreasing PAH-glucuronide formation as determined by Vmax and Vmax/KM for each of the three substrates analyzed (Table 1). Homogenate from cells with the highest UGT2A1_i2 expression, which exhibited a UGT2A1_i2/UGT2A1_i1 ratio of approximately 1.2, had a ∼50% reduction in glucuronide formation for all three substrates examined. No significant changes in KM values were observed when UGT2A1_i2 expression was induced, regardless of UGT2A1_i2 expression levels. PonA treatment had no effect on the glucuronidation activity of homogenates expressing UGT2A1_i1 against 1-OH-pyrene (results not shown). The C-terminal V5 tag caused no significant changes in UGT2A1 enzyme activity (data not shown), with the enzyme kinetics (Vmax and KM) for the control UGT2A1_i1_V5-overexpressing cell homogenates in these experiments similar to those reported previously for untagged UGT2A1 against PAHs (Bushey et al., 2011).
TABLE 1.
Kinetic analysis of the effect of UGT2A1_i2 coexpression on UGT2A1_i1 activity against PAH substrates
Data are expressed as milligram of total protein homogenate, corrected for relative UGT2A1_i1 expression. KM and Vmax represent the mean of three independent experiments.
| 1-OH-pyrene |
3-OH B[a]P |
B[a]P-7,8-diol |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| KM | Vmax | Vmax/KM | KM | Vmax | Vmax/KM | KM | Vmax | Vmax/KM | |
| μM | pmol/min/mg | μl/min/mg | μM | pmol/min/mg | μl/min/mg | μM | pmol/min/mg | μl/min/mg | |
| Vehicle control | 159 ± 16 | 319 ± 12* | 2.0 ± 0.08* | 256 ± 32 | 82 ± 4.3* | 0.32 ± 0.04* | 230 ± 22 | 86 ± 3.6* | 0.37 ± 0.04* |
| 2 μM PonA | 140 ± 16 | 261 ± 11 | 1.9 ± 0.09 | 276 ± 20 | 74 ± 2.2 | 0.27 ± 0.02 | 253 ± 37 | 70 ± 4.6 | 0.28 ± 0.05 |
| 6 μM PonA | 145 ± 17 | 219 ± 8.8 | 1.5 ± 0.1 | 261 ± 24 | 64 ± 2.4 | 0.24 ± 0.005 | 224 ± 19 | 53 ± 2.0 | 0.24 ± 0.02 |
| 10 μM PonA | 138 ± 12 | 153 ± 5.6 | 1.1 ± 0.08 | 238 ± 24 | 47 ± 1.9 | 0.20 ± 0.01 | 205 ± 32 | 41 ± 2.6 | 0.20 ± 0.01 |
p(trend) < 0.01.
UGT2A1_i1 and UGT2A1_i2 Hetero-Oligomerization.
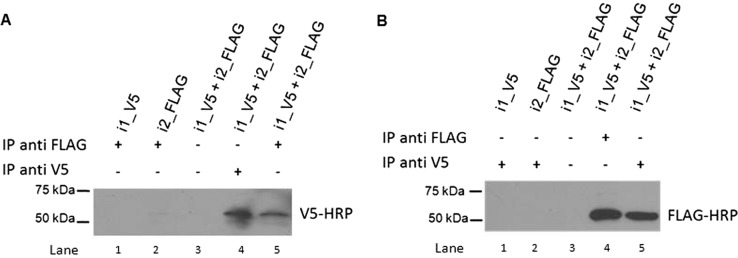
To test whether UGT2A1_i2 potentially modulates UGT2A1_i1 glucuronidation activity by a direct protein-protein interaction, protein lysate from cells treated with 10 μM PonA was used in coIP experiments. Using an anti-FLAG antibody to immunoprecipitate UGT2A1_i2_FLAG and an anti-V5 antibody to detect UGT2A1_i1_V5 by Western blot analysis, a band corresponding to UGT2A1_i1_V5 (∼57 kDa) was observed (Fig. 4A, lane 5). As expected, a UGT2A1_i1_V5 band was observed in Western blot analysis when an anti-V5 antibody was used to pull down UGT2A1_i1_V5 (Fig. 4A, lane 4); no bands were observed when no antibody was added to the immunoprecipitation lysate (Fig. 4A, lane 3) or when lysate was used from HEK293 cell lines overexpressing either UGT2A1_i1_V5 or UGT2A1_i2_FLAG alone (Fig. 4A, lanes 1 and 2). Likewise, a band corresponding to UGT2A1_i2_FLAG (∼52 kDa) was detected when an anti-V5 antibody was used to immunoprecipitate UGT2A1_i1_V5 and an anti-Flag antibody was used to detect UGT2A1_i2_FLAG by Western blot (Fig. 4B, lane 5). Again, as expected, a UGT2A1_i2_FLAG band was observed by Western blot analysis when an anti-FLAG antibody was used to pull down UGT2A1_i2_FLAG (Fig. 4B, lane 4); no bands were observed when no antibody was added to the immunoprecipitation lysate (Fig. 4B, lane 3) or when lysate was used from HEK293 cell lines overexpressing either UGT2A1_i1_V5 or UGT2A1_i2_FLAG alone (Fig. 4B, lanes 1 and 2).
Fig. 4.
Hetero-oligomerization between UGT2A1_i1 and UGT2A1_i2 demonstrated by coIP. A, the UGT2A1_i1_V5/UGT2A1_i2_FLAG complex was immunoprecipitated with an anti-FLAG antibody, then visualized with a HRP-labeled V5 antibody (lane 5). Homogenates from cells overexpressing UGT2A1_i1_V5 (lane 1) or UGT2A1_i2_FLAG (lane 2) alone were used as negative controls. Dynabeads without antibody conjugation were used as an additional negative control (lane 3). The UGT2A1_i1_V5/UGT2A1_i2_FLAG complex was immunoprecipitated with an anti-V5 antibody, and then visualized with an anti-V5-HRP antibody as a positive control (lane 4). B, the UGT2A1_i1_V5/UGT2A1_i2_FLAG complex was immunoprecipitated with an anti-V5 antibody, then visualized with a HRP-labeled FLAG antibody (lane 5). Homogenates from cells overexpressing UGT2A1_i1_V5 (lane 1) or UGT2A1_i2_FLAG (lane 2) alone were used as negative controls. Dynabeads without antibody conjugation were used as an additional negative control (lane 3). The UGT2A1_i1_V5/UGT2A1_i2_FLAG complex was immunoprecipitated with an anti-FLAG antibody, and then visualized with an anti-FLAG-HRP antibody as a positive control (lane 4). All coIP experiments were repeated four to six times to verify the protein-protein interactions between UGT2A1_i1 and UGT2A1_i2.
UGT2A1_i1 Homo-Oligomerization.
Because UGT2A1_i1 and UGT2A1_i2 were shown to form hetero-oligomers, experiments were conducted to determine whether UGT2A1_i1 homo-oligomerization also occurs. As described under Materials and Methods, a stable HEK293 cell line was created to coexpress UGT2A1_i1_V5 and UGT2A1_i1_FLAG. After treatment with 10 μM PonA, the levels of UGT2A1_i1_FLAG were induced to approximately the same levels as UGT2A1_i1_V5, whereas no UGT2A1_i1_FLAG expression was observed in the vehicle control group (Fig. 5A). UGT2A1_i1_V5 and UGT2A1_i1_FLAG expression levels were confirmed to be relatively equal after Western blot analysis using the anti-UGT2A1 antibody (Fig. 5B). Using an anti-FLAG antibody to immunoprecipitate UGT2A1_i1_FLAG and an anti-V5-HRP antibody to detect UGT2A1_i1_V5 by Western blot analysis, a band corresponding to UGT2A1_i1_V5 was detected (Fig. 5C, lane 5). Background bands at an approximate size of 45 kDa were observed in all cases when homo-oligomerization was investigated by using an anti-FLAG antibody to immunoprecipitate the complex and an anti-V5-HRP antibody for Western blot; the cause of these background signals is unknown. Likewise, a band corresponding to UGT2A1_i1_FLAG was detected when an anti-V5 antibody was used to immunoprecipitate UGT2A1_i1_V5 and an anti-Flag-HRP antibody was used to detect UGT2A1_i1_FLAG by Western blot (Fig. 5D, lane 5). Positive and negative controls, identical to those described in detail for UGT2A1_i1 and UGT2A1_i2 hetero-oligomerization, were used.
Fig. 5.
Homo-oligomerization of UGT2A1_i1 demonstrated by coIP. A, representative Western blots showing stable expression of UGT2A1_i1_V5 and induction of UGT2A1_i1_FLAG expression in an inducible coexpressed cell line. UGT2A1_i1_FLAG expression was induced through the addition of 10 μM PonA for 12 h. Fifty micrograms of control and PonA-treated protein were used for each Western blot, with the ratio of UGT2A1_i1 to UGT2A1_i1 expression determined to be approximately 1:1. B, Western blots using 100 μg of protein from the control and 10-μM PonA treatment groups were performed by using an anti-UGT2A1 antibody to confirm UGT2A1 expression levels detected by the anti-V5 and anti-FLAG antibodies in A. C, the UGT2A1_i1_V5/UGT2A1_i1_FLAG complex was immunoprecipitated with an anti-FLAG antibody, then visualized with a HRP-labeled V5 antibody (lane 5). Homogenates from cells overexpressing UGT2A1_i1_V5 (lane 1) or UGT2A1_i1_FLAG (lane 2) alone were used as negative controls. Dynabeads without antibody conjugation were used as an additional negative control (lane 3). The UGT2A1_i1_V5/UGT2A1_i1_FLAG complex was immunoprecipitated with an anti-V5 antibody, and then visualized with an anti-V5-HRP antibody as a positive control (lane 4). D, the UGT2A1_i1_V5/UGT2A1_i1_FLAG complex was immunoprecipitated with an anti-V5 antibody, then visualized with a HRP-labeled FLAG antibody (lane 5). Homogenates from cells overexpressing UGT2A1_i1_V5 (lane 1) or UGT2A1_i1_FLAG (lane 2) alone were used as negative controls. Dynabeads without antibody conjugation were used as an additional negative control (lane 3). The UGT2A1_i1_V5/UGT2A1_i1_FLAG complex was immunoprecipitated with an anti-FLAG antibody, and then visualized with an anti-FLAG-HRP antibody as a positive control (lane 4). All coIP experiments were repeated four to six times to verify UGT2A1_i1 homo-oligomerization.
Effect of UGT2A1_i1 or UGT2A1_i2 Coexpression on the Glucuronidation Activities of Other UGT Isoforms.
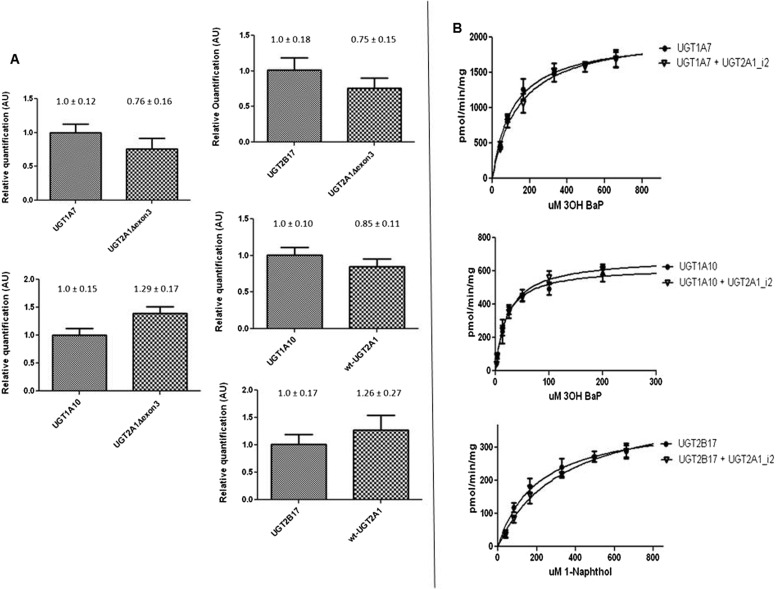
To examine whether UGT2A1 isoforms potentially modulate the activity of other UGTs, studies were performed coexpressing either UGT2A1_i1_V5 or UGT2A1_i2_V5 with UGT1A7, UGT1A10, or UGT2B17. These UGTs were chosen because they are expressed in the respiratory and/or aerodigestive tract and are active against tobacco carcinogens (Zheng et al., 2002; Dellinger et al., 2006; Gallagher et al., 2007). As determined by real-time PCR, the relative levels of transcript in each UGT-coexpressing cell line were not significantly different for the UGT2A1 isoform compared with its UGT1A or UGT2B counterpart (Fig. 6A). Western blots demonstrated that UGT1A7 and UGT1A10 were expressed at similar levels when overexpressed alone or coexpressed with UGT2A1, and similar levels of UGT2A1_i1_V5 and UGT2A1_i2_V5 protein were observed in each coexpressed overexpressing cell line (data not shown).
Fig. 6.
Glucuronidation activity of UGT1A7, UGT1A10, or UGT2B17 against 3-OH-B[a]P and 1-naphthol after coexpression with UGT2A1_i1 or UGT2A1_i2. A, real-time PCR was used to determine the approximate mRNA levels of each UGT in stable coexpressed HEK293-overexpressing cell lines. Applied Biosystems gene expression assays were used to quantitatively detect levels of UGT2A1, UGT1A7, UGT1A10, or UGT2B17 transcript. Relative UGT2A1Δexon3 levels were determined by comparing mRNA levels of UGT2A1Δexon3 transcript with UGT1A7, UGT1A10, and UGT2B17 mRNA by using the ΔΔCt method. Similar experiments were completed to determine wild-type UGT2A1 mRNA levels relative to UGT1A10 or UGT2B17 levels in additional coexpressed cell lines. Data are expressed as the mean ± S.D. of quadruplicate experiments and were normalized to RPLPO protein levels in each cell line and corrected for differences in assay efficiencies. B, representative Michaelis-Menten kinetics curves summarizing activity data by using homogenates from HEK293 cell lines overexpressing UGT1A7 or UGT1A7 + UGT2A1_i2 against 3-OH-B[a]P, UGT1A10 UGT1A10 + UGT2A1_i2 against 3-OH-B[a]P, and UGT2B17 or UGT2B17 + UGT2A1_i2 against 1-naphthol.
Glucuronidation assays were performed by using protein from cells overexpressing UGT1A or UGT2B enzymes alone or after coexpression with UGT2A1 isoforms. Representative Michaelis-Menten curves for UGT1A7- and UGT1A7/UGT2A1_i2-overexpressing cell homogenates show similar kinetics against 3-OH-B[a]P (Fig. 6B). A similar pattern was observed for UGT1A10- and UGT1A10/UGT2A1_i2-overexpressing cell homogenates against 3-OH-B[a]P and for UGT2B17- and UGT2B17/UGT2A1_i2-overexpressing cell homogenates against 1-naphthol (Fig. 6B). Glucuronidation assays also showed no significant changes in glucuronidation activity for UGT1A10- and UGT1A10/UGT2A1_i1-overexpressing cell homogenates and UGT2B17- and UGT2B17/UGT2A1_i1-overexpressing cell homogenates against the non-UGT2A1 substrate NNAL (results not shown). No significant differences in KM or Vmax were observed for any substrate analyzed after UGT1A or UGT2B coexpression with either UGT2A1_i2 or UGT2A1_i1. No significant differences in glucuronidation rates of additional substrates were observed by using homogenates from cells coexpressing UGT1A7, UGT1A10, or UGT2B17 and UGT2A1; this included glucuronidation of the flavonoid chrysin by the UGT1A7-coexpressing cell line, glucuronidation of the HCA metabolite N-OH PhIP by the UGT1A10-coexpressing cell lines, and glucuronidation of ibuprofen by the UGT2B17-coexpressing cell lines (data not shown). In addition, UGT2A1_i1 activity against testosterone and the PAH 5-methylchrysene-1,2-diol was unchanged after coexpression with UGT1A10 or UGT2B17, respectively (data not shown).
Discussion
The present study is the first to identify and characterize a novel splice variant of UGT2A1, demonstrating its expression in multiple tissues at both the level of mRNA and protein and its functional importance as an inhibitor of UGT2A1 activity. This variant consists of a deletion of exon 3 that results in an inactive UGT2A1 isoform. UGT2A1 exon 3 is a conserved region throughout all UGT family members (Nagar and Remmel, 2006) and is also highly conserved with the corresponding region of the mouse ortholog UGT2a1 after sequence alignment. The exon 3-encoded protein region of all UGTs is hypothesized to form a portion of the UDPGA cosubstrate binding pocket necessary for UGT function, and similar to the activity results for the splice variant UGT2A1 isoform in the present study, amino acid point mutations in this region have been shown previously to ablate UGT enzyme activity (Miley et al., 2007; Bushey et al., 2011; Laakkonen and Finel, 2010).
In addition to possessing no detectable glucuronidation activity, UGT2A1_i2 was shown to act as a negative regulator of UGT2A1_i1 activity. This negative regulatory effect is similar to that observed for other UGT splice variants, including inactive UGT1A splice variants with an alternate exon 5b at the 3′ end of the UGT1A transcript (Girard et al., 2007; Lévesque et al., 2007). This negative inhibition seems to be caused by direct binding of the inactive UGT2A1_i2 variant with UGT2A1_i1, a mechanism similar to that observed for other UGTs including the UGT1A exon 5b variant (Bellemare et al., 2010). Furthermore, a Gln331Stop polymorphism in UGT1A1 leads to a truncated and inactive protein, and this protein isoform has been reported to bind to and inhibit wild-type UGT1A1 activity (Koiwai et al., 1996). Ghosh et al. (2001) showed that an inactive mutant form of UGT1A1, caused by a single amino acid change at codon 127, could bind to and inhibit wild-type UGT1A1 activity in a dominant-negative manner. In addition, other work has identified inactive UGT2B4 splice variant isoforms that negatively regulate wild-type UGT2B4 activity (Lévesque et al., 2010), and inactive UGT2B7 splice variant isoforms negatively modulate wild-type UGT2B7 activity (Ménard et al., 2011).
In the present study, a PonA-inducible coexpression system was designed to control UGT2A1_i2 expression so that its effects on UGT2A1_i1 activity could be examined. A linear correlation was observed between the ratio of UGT2A1_i2/UGT2A1_i1 expression and overall glucuronidation activity of homogenates from coexpressed cells. At the highest level of UGT2A1_i2 induction, with the expression of UGT2A1_i2 approximately equal to that of UGT2A1_i1, there was an approximately 50% reduction in the rate of glucuronide formation for three PAH substrates examined. As the UGT2A1_i2/UGT2A1_i1 ratio decreased, proportional increases in UGT2A1_i1 activity were observed against all three PAH substrates. These data suggest that UGT2A1_i1 interacts with UGT2A1_i2 in a 1:1 stoichiometry, probably as a dimer as proposed for other UGTs (Finel and Kurkela, 2008; Bellemare et al., 2010). The in vitro inducible UGT2A1_i2/UGT2A1_i1 ratios observed in this study approximated the ratios observed physiologically in several human tissues including lung and colon. These UGT2A1_i2/UGT2A1_i1 ratios corresponded with 10 to 50% decreases in the Vmax for UGT2A1_i1-mediated glucuronidation activities against PAH substrates; no significant change in UGT2A1_i1 KM caused by UGT2A1_i2 coexpression was observed. These data suggest that the inhibitory effect of UGT2A1_i2 expression is mediated by the removal of UGT2A1_i1 from the active protein pool, rather than an effect on UGT2A1_i1 enzyme-substrate affinity.
UGTs have been shown to reside in the endoplasmic reticulum to form both homo- and hetero-oligomers (Fujiwara et al., 2007; Operaña and Tukey, 2007; Finel and Kurkela, 2008). In vitro UGT coexpression and oligomerization studies have shown that UGT interactions are complex, with kinetic changes dependent on the specific UGT isoforms interacting and the substrates undergoing glucuronidation (Fujiwara et al., 2007; Operaña and Tukey, 2007; Finel and Kurkela, 2008). The coIP experiments presented in this study suggested hetero-oligomerization occurs between the UGT2A1_i1 and UGT2A1_i2 isoforms, and UGT2A1 homo-oligomerization was also investigated. In vitro coIP experiments showed for the first time that UGT2A1_i1 homo-oligomerization does, in fact, occur. UGT2A1 coexpression studies were completed with two members of the UGT1A subfamily, UGT1A7 and UGT1A10, and UGT2B17 to determine any UGT2A1-mediated changes in UGT1A or UGT2B glucuronidation activity. Like UGT2A1, UGT1A7, UGT1A10, and UGT2B17 are expressed in certain tissues of the respiratory and aerodigestive tract and exhibit glucuronidation activity against multiple tobacco carcinogens (Zheng et al., 2002; Dellinger et al., 2006, 2007; Gallagher et al., 2007). UGT2A1 was determined to have no effect on the glucuronidation activity of UGT1A7, UGT1A10, or UGT2B17 against tobacco carcinogens, including PAHs, TSNAs, and HCAs, suggesting that UGT2A1 does not affect the ability of these UGTs to detoxify tobacco carcinogens in vivo and UGT2A1_i2 regulation of UGT activity is UGT2A1-specific.
The entire UGT2A family has been understudied, with limited information reported on the expression and activity of these enzymes. UGT2A3 has been shown to be well expressed in the colon, small intestine, and liver, with activity reported against bile acids (Court et al., 2008). UGT2A2 has been reported to be expressed in the nasal mucosa, with broad substrate selectivity reported against various estrogen and phenylphenol metabolites (Sneitz et al., 2009). Neither of these enzymes has been carefully investigated for aerodigestive tract expression or metabolism against carcinogens, so this will be investigated in future studies. UGT2A1 and UGT2A2 both have been shown to exhibit glucuronidation activity against similar substrates, with UGT2A1 generally having higher glucuronidation rates (Sneitz et al., 2009). Based on exon sharing of exon 1 of UGT2A1 or UGT2A2 to common exons 2 to 6, the same exon 3 deletion splice variant for UGT2A2 is likely to occur. Future studies are planned to determine whether the UGT2A2Δexon3 variant exists and, if so, the expression and functional implications of this UGT2A2 variant will be examined.
The novel UGT2A1_i2 regulatory mechanism described here may allow for tighter control of UGT2A1 glucuronidation activity. The expression of both UGT2A1_i1 and UGT2A1_i2 in multiple human tissues suggests widespread physiological importance, particularly as it relates to tobacco-related cancer susceptibility. We hypothesize that the balance between active UGT2A1_i1 and inactive UGT2A1_i2 could influence carcinogen metabolism in local tissues susceptible to tobacco-induced carcinogenesis, with individuals expressing higher amounts of UGT2A1_i2 potentially exhibiting higher susceptibility to tobacco-related cancers of the respiratory and aerodigestive tracts. The potential influence of a UGT2A1 splicing variant on cancer risk could also complicate the findings of genetic epidemiologic studies (including genome-wide association studies) of UGT2A1, which exhibits at least two prevalent (>4%) polymorphisms that alter UGT2A1 enzymatic activity (Bushey et al., 2011). Further studies examining the involvement of UGT2A1 genetic and splicing variants on cancer susceptibility are currently underway.
Supplementary Material
Acknowledgments
We thank the staffs at the Functional Genomics Core Facility at the Penn State University College of Medicine for real-time PCR services, the Nucleic Acid Facility at Penn State University for DNA sequencing services, and the Organic Synthesis Core Facility at the Penn State University College of Medicine for providing tobacco carcinogens for this study.
This work was supported by the National Institutes of Health National Institute of Dental and Craniofacial Research [Grant R01-DE13158] (to P.L.); the National Institutes of Health, National Cancer Institute [Grant R01-CA164366] (to P.L.); and the Pennsylvania Department of Health's Health Research Formula Funding Program [Grant 4100038714] (to P.L.).
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
 The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- UGT
- UDP-glucuronosyltransferase
- UDPGA
- UDP-glucuronic acid
- PAH
- polycyclic aromatic hydrocarbon
- PonA
- ponasterone A
- RT
- reverse transcription
- PCR
- polymerase chain reaction
- B[a]P
- benzo[a]pyrene
- B[a]P-7,8-diol
- benzo[a]pyrene-7,8-dihydrodiol
- coIP
- coimmunoprecipitation
- UPLC
- ultra-pressure liquid chromatography
- HEK
- human embryonic kidney
- HRP
- horseradish peroxidase
- HCA
- heterocyclic amine
- N-OH PhIP
- N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine
- TSNA
- tobacco-specific nitrosamine
- NNAL
- 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- AU
- arbitrary units.
Authorship Contributions
Participated in research design: Bushey and Lazarus.
Conducted experiments: Bushey.
Contributed new reagents or analytic tools: Bushey and Lazarus.
Performed data analysis: Bushey and Lazarus.
Wrote or contributed to the writing of the manuscript: Bushey and Lazarus.
References
- Balliet RM, Chen G, Dellinger RW, Lazarus P. (2010) UDP-glucuronosyltransferase 1A10: activity against the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol, and a potential role for a novel UGT1A10 promoter deletion polymorphism in cancer susceptibility. Drug Metab Dispos 38:484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balliet RM, Chen G, Gallagher CJ, Dellinger RW, Sun D, Lazarus P. (2009) Characterization of UGTs active against SAHA and association between SAHA glucuronidation activity phenotype with UGT genotype. Cancer Res 69:2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemare J, Rouleau M, Harvey M, Guillemette C. (2010) Modulation of the human glucuronosyltransferase UGT1A pathway by splice isoform polypeptides is mediated through protein-protein interactions. J Biol Chem 285:3600–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey RT, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. (2011) Characterization of UDP-glucuronosyltransferase 2A1 (UGT2A1) variants and their potential role in tobacco carcinogenesis. Pharmacogenet Genomics 21:55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Dellinger RW, Gallagher CJ, Sun D, Lazarus P. (2008) Identification of a prevalent functional missense polymorphism in the UGT2B10 gene and its association with UGT2B10 inactivation against tobacco-specific nitrosamines. Pharmacogenet Genomics 18:181–191 [DOI] [PubMed] [Google Scholar]
- Court MH, Hazarika S, Krishnaswamy S, Finel M, Williams JA. (2008) Novel polymorphic human UDP-glucuronosyltransferase 2A3: cloning, functional characterization of enzyme variants, comparative tissue expression, and gene induction. Mol Pharmacol 74:744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. (2007) Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis 28:2412–2418 [DOI] [PubMed] [Google Scholar]
- Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. (2006) Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos 34:943–949 [DOI] [PubMed] [Google Scholar]
- Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, Lazarus P. (2002) Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res 62:1978–1986 [PubMed] [Google Scholar]
- Finel M, Kurkela M. (2008) The UDP-glucuronosyltransferases as oligomeric enzymes. Curr Drug Metab 9:70–76 [DOI] [PubMed] [Google Scholar]
- Fujiwara R, Nakajima M, Yamanaka H, Katoh M, Yokoi T. (2007) Interactions between human UGT1A1, UGT1A4, and UGT1A6 affect their enzymatic activities. Drug Metab Dispos 35:1781–1787 [DOI] [PubMed] [Google Scholar]
- Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, Chase GA, Richie J, Lazarus P. (2007) The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev 16:823–828 [DOI] [PubMed] [Google Scholar]
- Ghosh SS, Sappal BS, Kalpana GV, Lee SW, Chowdhury JR, Chowdhury NR. (2001) Homodimerization of human bilirubin-uridine-diphosphoglucuronate glucuronosyltransferase-1 (UGT1A1) and its functional implications. J Biol Chem 276:42108–42115 [DOI] [PubMed] [Google Scholar]
- Girard H, Lévesque E, Bellemare J, Journault K, Caillier B, Guillemette C. (2007) Genetic diversity at the UGT1 locus is amplified by a novel 3′ alternative splicing mechanism leading to nine additional UGT1A proteins that act as regulators of glucuronidation activity. Pharmacogenet Genomics 17:1077–1089 [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 [DOI] [PubMed] [Google Scholar]
- Itäaho K, Mackenzie PI, Ikushiro S, Miners JO, Finel M. (2008) The configuration of the 17-hydroxy group variably influences the glucuronidation of β-estradiol and epiestradiol by human UDP-glucuronosyltransferases. Drug Metab Dispos 36:2307–2315 [DOI] [PubMed] [Google Scholar]
- Jedlitschky G, Cassidy AJ, Sales M, Pratt N, Burchell B. (1999) Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochem J 340:837–843 [PMC free article] [PubMed] [Google Scholar]
- Jin CJ, Miners JO, Burchell B, Mackenzie PI. (1993) The glucuronidation of hydroxylated metabolites of benzo[a]pyrene and 2-acetylaminofluorene by cDNA-expressed human UDP-glucuronosyltransferases. Carcinogenesis 14:2637–2639 [DOI] [PubMed] [Google Scholar]
- Jones NR, Sun D, Freeman WM, Lazarus P. (2012) Quantification of hepatic UGT1A splice variant expression and correlation of UGT1A1 variant expression with glucuronidation activity. J Pharmacol Exp Ther 342:720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai O, Aono S, Adachi Y, Kamisako T, Yasui Y, Nishizawa M, Sato H. (1996) Crigler-Najjar syndrome type II is inherited both as a dominant and as a recessive trait. Hum Mol Genet 5:645–647 [DOI] [PubMed] [Google Scholar]
- Laakkonen L, Finel M. (2010) A molecular model of the human UDP-glucuronosyltransferase 1A1, its membrane orientation, and the interactions between different parts of the enzyme. Mol Pharmacol 77:931–939 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. (2005) Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics 15:769–778 [DOI] [PubMed] [Google Scholar]
- Lévesque E, Girard H, Journault K, Lépine J, Guillemette C. (2007) Regulation of the UGT1A1 bilirubin-conjugating pathway: role of a new splicing event at the UGT1A locus. Hepatology 45:128–138 [DOI] [PubMed] [Google Scholar]
- Lévesque E, Ménard V, Laverdière I, Bellemare J, Barbier O, Girard H, Guillemette C. (2010) Extensive splicing of transcripts encoding the bile acid-conjugating enzyme UGT2B4 modulates glucuronidation. Pharmacogenet Genomics 20:195–210 [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. (2005) Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 15:677–685 [DOI] [PubMed] [Google Scholar]
- Ménard V, Eap O, Roberge J, Harvey M, Lévesque E, Guillemette C. (2011) Transcriptional diversity at the UGT2B7 locus is dictated by extensive pre-mRNA splicing mechanisms that give rise to multiple mRNA splice variants. Pharmacogenet Genomics 21:631–641 [DOI] [PubMed] [Google Scholar]
- Miley MJ, Zielinska AK, Keenan JE, Bratton SM, Radominska-Pandya A, Redinbo MR. (2007) Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J Mol Biol 369:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar S, Remmel RP. (2006) Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene 25:1659–1672 [DOI] [PubMed] [Google Scholar]
- Nishimura M, Naito S. (2006) Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab Pharmacokinet 21:357–374 [DOI] [PubMed] [Google Scholar]
- Olson KC, Dellinger RW, Zhong Q, Sun D, Amin S, Spratt TE, Lazarus P. (2009) Functional characterization of low-prevalence missense polymorphisms in the UDP-glucuronosyltransferase 1A9 gene. Drug Metab Dispos 37:1999–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Operaña TN, Tukey RH. (2007) Oligomerization of the UDP-glucuronosyltransferase 1A proteins: homo- and heterodimerization analysis by fluorescence resonance energy transfer and co-immunoprecipitation. J Biol Chem 282:4821–4829 [DOI] [PubMed] [Google Scholar]
- Ren Q, Murphy SE, Zheng Z, Lazarus P. (2000) O-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos 28:1352–1360 [PubMed] [Google Scholar]
- Sneitz N, Court MH, Zhang X, Laajanen K, Yee KK, Dalton P, Ding X, Finel M. (2009) Human UDP-glucuronosyltransferase UGT2A2: cDNA construction, expression, and functional characterization in comparison with UGT2A1 and UGT2A3. Pharmacogenet Genomics 19:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sten T, Bichlmaier I, Kuuranne T, Leinonen A, Yli-Kauhaluoma J, Finel M. (2009) UDP-glucuronosyltransferases (UGTs) 2B7 and UGT2B17 display converse specificity in testosterone and epitestosterone glucuronidation, whereas UGT2A1 conjugates both androgens similarly. Drug Metab Dispos 37:417–423 [DOI] [PubMed] [Google Scholar]
- Sun D, Chen G, Dellinger RW, Duncan K, Fang JL, Lazarus P. (2006) Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res 8:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Sharma AK, Dellinger RW, Blevins-Primeau AS, Balliet RM, Chen G, Boyiri T, Amin S, Lazarus P. (2007) Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos 35:2006–2014 [DOI] [PubMed] [Google Scholar]
- Turgeon D, Carrier JS, Chouinard S, Bélanger A. (2003) Glucuronidation activity of the UGT2B17 enzyme toward xenobiotics. Drug Metab Dispos 31:670–676 [DOI] [PubMed] [Google Scholar]
- Uchaipichat V, Mackenzie PI, Guo XH, Gardner-Stephen D, Galetin A, Houston JB, Miners JO. (2004) Human udp-glucuronosyltransferases: isoform selectivity and kinetics of 4-methylumbelliferone and 1-naphthol glucuronidation, effects of organic solvents, and inhibition by diclofenac and probenecid. Drug Metab Dispos 32:413–423 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb LJ, Miles KK, Auyeung DJ, Kessler FK, Ritter JK. (2005) Analysis of substrate specificities and tissue expression of rat UDP-glucuronosyltransferases UGT1A7 and UGT1A8. Drug Metab Dispos 33:77–82 [DOI] [PubMed] [Google Scholar]
- Wells PG, Mackenzie PI, Chowdhury JR, Guillemette C, Gregory PA, Ishii Y, Hansen AJ, Kessler FK, Kim PM, Chowdhury NR, et al. (2004) Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab Dispos 32:281–290 [DOI] [PubMed] [Google Scholar]
- Wiener D, Doerge DR, Fang JL, Upadhyaya P, Lazarus P. (2004a) Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab Dispos 32:72–79 [DOI] [PubMed] [Google Scholar]
- Wiener D, Fang JL, Dossett N, Lazarus P. (2004b) Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res 64:1190–1196 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Fang JL, Lazarus P. (2002) Glucuronidation: an important mechanism for detoxification of benzo[a]pyrene metabolites in aerodigestive tract tissues. Drug Metab Dispos 30:397–403 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.