Abstract
The nonsteroidal antiestrogen tamoxifen was introduced as a treatment for breast cancer 3 decades ago. It has also been approved as a chemopreventive agent and is prescribed to women at high risk for this disease. However, several studies have shown that use of tamoxifen leads to increased risk of endometrial cancer in humans. One potential pathway of tamoxifen toxicity could involve metabolism via hydroxylation to give 4-hydroxytamoxifen (4OHtam), which may be further oxidized to form a quinone methide. CYP2B6 is a highly polymorphic drug-metabolizing enzyme, and it metabolizes a number of clinically important drugs. Earlier studies from our laboratory have shown that tamoxifen is a mechanism-based inactivator of CYP2B6. The aim of the current study was to investigate the possible formation of reactive intermediates through detection of protein covalent binding and glutathione ethyl ester adduct (GSHEE) formation. The incubation of tamoxifen with 2B6 gave rise to an adduct of 4OHtam with glutathione, which was characterized as the 4OHtam quinone methide + GSHEE with an m/z value of 719, and the structure was characterized by liquid chromatography-tandem mass spectrometry. The metabolic activation of tamoxifen in the CYP2B6 reconstituted system also resulted in the formation of an adduct to the P4502B6 apoprotein, which was identified using liquid chromatography mass spectrometry. The site responsible for the inactivation of CYP2B6 was determined by proteolytic digestion and identification of the labeled peptide. This revealed a tryptic peptide 188FHYQDQE194 with the site of adduct formation localized to Gln193 as the site modified by the reactive metabolite formed during tamoxifen metabolism.
Introduction
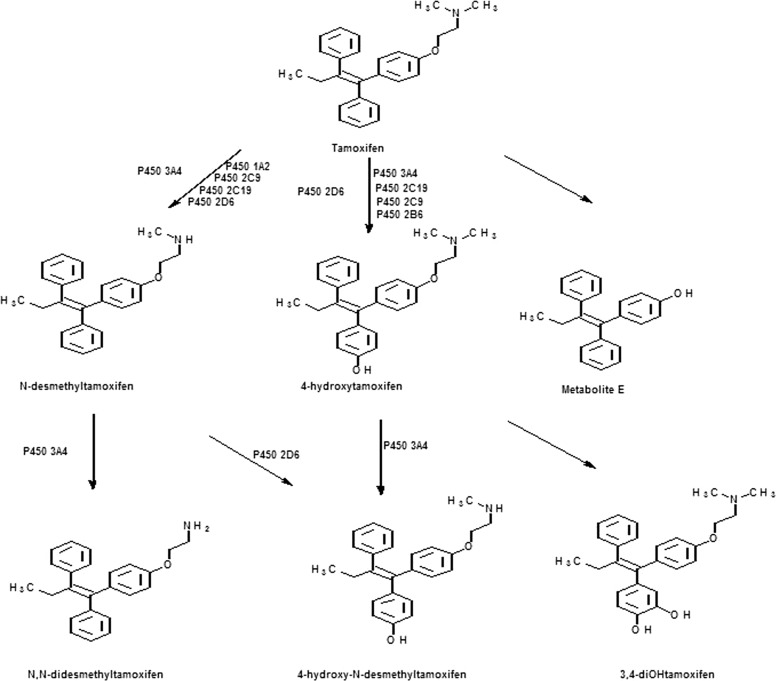
Tamoxifen, a selective estrogen receptor modulator, is widely used drug for the treatment of estrogen-receptor positive breast cancer (Furr and Jordan, 1984). CYP3A4 is the major enzyme responsible for the N-demethylation of tamoxifen, and 4-hydroxylation is primarily mediated by CYP2D6 (Fig. 1), although a number of human P450s can metabolize tamoxifen (Mani et al., 1993; Crewe et al., 2002). Tamoxifen is a prodrug because metabolic activation is required for maximal biological activity (Mani et al., 1993; Crewe et al., 2002) and two of its metabolites, 4OHtam and 4-hydroxy-N-demethyltamoxifen, have markedly higher affinity for estrogen receptor than tamoxifen itself (Jordan, 1982; Johnson et al., 2004). Tamoxifen undergoes extensive metabolism to a variety of primary and secondary metabolites by various cytochrome P450 enzymes (Furr and Jordan, 1984). Bioactivation of tamoxifen has been found to result in the formation of covalent adducts of its metabolites with rat and human microsomal proteins. At least three different reactive intermediates are known to be formed, including a carbocation, an O-quinone, and a quinone methide (Phillips et al., 1994a,b; Dehal and Kupfer, 1996; Moorthy et al., 1996), and studies have indicated it is the covalent binding of the metabolites to DNA or protein that is responsible for toxicity (Dehal and Kupfer, 1996). Treatment with tamoxifen has also been associated with a relatively high incidence of endometrial cancer in women (Andersson et al., 1991; Fisher et al., 1994), and tamoxifen has been shown to be a hepatocarcinogen in rats (Divi et al., 1999).
Fig. 1.
Major pathways for the metabolism of tamoxifen.
Since the approval of tamoxifen by the U.S. Food and Drug Administration as a chemopreventive agent, studies have been done on the formation and identification of its reactive metabolites (Fisher et al., 1998). Research has identified a variety of functional groups that can be bioactivated to give reactive intermediates leading to toxicity. Formation of a quinone methide is known to be one of the major routes of metabolism of tamoxifen that results in covalent binding to microsomal proteins. It has been shown that tamoxifen or 4OHtam can undergo hydroxylation to give 3,4-dihydroxytamoxifen-O-quinone, which is the penultimate metabolite that generates the reactive intermediate that binds to proteins (Dehal and Kupfer, 1996). The quinone methide can also be formed by the initial hydroxylation of tamoxifen to give 4OHtam, which can then undergo two electron oxidation to form the quinone methide (Thompson et al., 1995).
Cytochrome P4502B6 is involved in the metabolic activation and inactivation of a variety of endogenous substrates as well as a large number of clinically important drugs that include bupropion, efavirenz, cyclophosphamide, and tamoxifen (Ekins and Wrighton, 1999; Hesse et al., 2000). Two studies have reported higher levels of expression of CYP2B6 in estrogen receptor-positive breast tumors (Hellmold et al., 1998; Lo et al., 2010), and that could lead to vulnerability to damage through bioactivation of compounds metabolized by this enzyme. Tamoxifen has also been shown to act as a phenobarbital-like inducer in rats causing up-regulation of the CYP2B and 3A enzymes at doses comparable to doses normally administered in humans (White et al., 1993; Nuwaysir et al., 1995; Ekins and Wrighton, 1999). It has been suggested that treatment with tamoxifen over time might change the rates and relative proportions of tamoxifen metabolites formed as a result of induction of CYP2B6 (Crewe et al., 2002). Relatively little is known about the contribution of CYP2B6 to the overall metabolism of tamoxifen. However, it has been shown that 4OHtam formation is inhibited 13% by a monoclonal antibody against CYP2B6, suggesting a minor role of CYP2B6 in 4OHtam formation (Coller et al., 2002). CYP2B6 has been shown to have significant catalytic activity for the metabolism of tamoxifen at high substrate concentrations. Metabolism of 250 μM tamoxifen by CYP2B6 in a reconstituted system exhibited a rate of formation of 4OHtam of 2.6 pmol/mg protein/min (Crewe et al., 2002). We have previously shown that purified CYP2B6 can metabolize tamoxifen to N-desmethyl tamoxifen, 4OHtam, and 4′-OH tamoxifen and that tamoxifen is a mechanism-based inactivator of CYP2B6, and the kinetic parameters for 2B6 inactivation were determined (Sridar et al., 2002). However, the mechanism of inactivation was not investigated. This has prompted us to conduct further studies on the bioactivation of tamoxifen catalyzed by CYP2B6. In the presence of glutathione ethyl ester, 4OHtam quinone methide conjugate was detected when CYP2B6 was incubated with tamoxifen. In addition, the site of covalent modification of CYP2B6 by a reactive intermediate leading to the loss of catalytic activity was identified by digesting the adducted protein with trypsin and analyzing the resulting peptides by LC-MS and Sequest software (Thermo Fisher Scientific, Waltham, MA).
Materials and Methods
Materials.
Tamoxifen, catalase, glutathione ethyl ester (GSHEE), and NADPH were obtained from Sigma-Aldrich (St. Louis, MO). 4OHtam, N-desmethyl-4-OH tamoxifen, and tamoxifen-N-oxide were obtained from Toronto Research Chemicals Inc. (Toronto, ON, Canada). Human liver microsomes were obtained from BD Biosciences (Woburn, MA). All other chemicals were of highest quality and available from commercial sources.
Protein Purification.
CYP2B6 was expressed in Escherichia coli C41 DE3 cells and purified according to published protocols (Hanna et al., 2000). P450 reductase and cytochrome b5 were purified as described previously (Hanna et al., 1998; Lin et al., 2005).
In Vitro Trapping of Reactive Metabolites of Tamoxifen with GSHEE.
The reaction mixture for the trapping and identification of the glutathione ethyl ester adducts contained 0.25 nmol of recombinant purified 2B6, 0.5 nmol of reductase, 0.25 nmol of b5, 100 units of catalase, and 10 mM GSHEE in 50 mM potassium phosphate buffer, pH 7.4, in a final volume of 500 μl. The final concentration of tamoxifen was 60 μM. The reaction was initiated by the addition of 1.2 mM NADPH, with control samples receiving the same volume of water. The reaction samples were incubated at 37°C for 45 min, and the reactions were terminated by the addition of 5 volumes of acetonitrile. The reaction mixtures were centrifuged at 13,200g at room temperature for 10 min. The supernatants were transferred to new tubes and dried under a stream of nitrogen gas. The residue was suspended in 50% acetonitrile in water (75 μl), and 50-μl aliquots were injected onto the LC-MS for further analysis. HPLC separation of the tamoxifen glutathione ethyl ester adducts was carried out using a Phenomenex Luna C18 reverse phase column (4.6 × 100 mm; Phenomenex, Torrance, CA). The flow rate was 300 μl/min. For HPLC-ESI-MS/MS analysis, initial conditions were 95% of 0.1% (v/v) acetic acid in water (solvent A) and 5% of 0.1% acetic acid in acetonitrile (solvent B). The percentage of B was maintained for 5 min followed by a linear gradient to 30% B from 5 to 15 min, to 80% B from 15 to 35 min and, to 90 min from 35 to 40 min. The column was then returned to initial conditions and equilibrated for 10 min before the next injection. ESI-LC-MS/MS was carried out using a ThermoQuest LCQ ion trap mass spectrometer (Thermo Fisher Scientific) interfaced with a Hewlett Packard 1100 series HPLC system (Agilent Technologies, Palo Alto, CA). The sheath gas was set at 80 arbitrary units, and the auxiliary gas was set at 15 arbitrary units. The spray voltage was 5 kV, and the capillary temperature was 180°C. Data were acquired in the positive ion mode using the Xcalibur software (Thermo Fisher Scientific) with two data dependent scans for the two most intense ions.
Trapping of 4OHtam Metabolites by GSHEE.
The reconstitution and incubation conditions for the metabolism of 4OHtam by CYP2B6 were carried out in the presence of GSHEE exactly as described above. The final concentration of 4OHtam used in the incubation reaction was 90 μM. Reactions and analyses of the products were carried out as described above.
Quinone Methide Trapping Using Human Liver Microsomes.
The in vitro formation of hydroxytamoxifen quinone methides and trapping with GSHEE were investigated in human liver microsomes by incubating tamoxifen with human liver microsomal protein obtained from BD Biosciences. Microsomes (5 μg) were diluted to a volume of 500 μl with 0.5 M potassium phosphate buffer, pH 7.4, catalase (100 units), and 90 μM tamoxifen. The incubation and inactivation of the samples were carried out as described above. Extraction and analysis conditions were carried out as described for reactive metabolite trapping.
ESI-LC-MS Analysis of the Apoprotein-Tamoxifen Adducts.
Inactivation of CYP2B6 by tamoxifen was carried out as indicated above. The primary reaction mixture contained 0.2 nmol of 2B6, 0.4 nmol of reductase, and 0.2 nmol of b5. The final concentration of tamoxifen was 60 μM. Incubations were carried out at 37°C for 10 min in the absence of NADPH as a negative control or in the presence of NADPH. The samples were then resolved on an Agilent Zorbax C3 column (3.0 × 150 mm, 3.5 μm; Agilent Technologies) eluted with 40% acetonitrile, 0.1% TFA at a flow rate of 300 μl/min. The proteins were eluted using a linear gradient to 90% B over 40 min before returning the column to equilibration conditions. The samples were analyzed using a ThermoQuest LCQ ion trap mass spectrometer (Thermo Fisher Scientific) interfaced with a Hewlett Packard 1100 series high-performance liquid chromatography system (Hewlett Packard, Palo Alto, CA), the spectra were obtained for the control, and the inactivated samples were then deconvoluted using ThermoQuest Bioworks (Thermo Fisher Scientific) to determine the masses of the proteins. The ESI conditions were as follows: auxiliary gas was set at 90 arbitrary units, sheath gas was set at 30 arbitrary units, the spray voltage was 4.2 kV, and the capillary temperature was 230°C.
Identification of the Site of Modification by Tamoxifen Activation on CYP2B6.
The inactivation of CYP2B6 was performed as described above. The protein mixture contained 1 nmol of CYP2B6, 2 nmol of reductase, and 1 nmol of b5 in a final volume of 500 μl. The final concentration of the tamoxifen used was 100 μM. The samples were incubated with tamoxifen in the presence or absence of NADPH, and the inactivation reaction was carried out by incubating the samples at 37°C for 30 min. The control and tamoxifen-inactivated samples were then denatured by incubation in 8 M urea at 55°C for 30 min followed by dilution with 50 mM ammonium bicarbonate to a final volume of 7 ml (to reduce the urea concentration). The samples were then concentrated using Amicon Ultracentrifuge filter devices (Millipore Corporation, Billerica, MA) at 4°C. The volume of the samples was reduced to 100 μl. The samples were then reduced with 5 mM dithiothreitol at 60°C for 30 min. Digestion was carried out with trypsin using a 1:20 trypsin/protein ratio. The samples were incubated overnight in a shaking water bath at 37°C. The reactions were terminated by the addition of 1 μl of 10% trifluoroacetic acid. The digested samples were centrifuged at 13,000g in an Eppendorf centrifuge (Eppendorf North America, New York, NY) for 10 min, and the supernatants were used for analysis.
High-pressure liquid chromatography separation of the tryptic peptides was performed on a Shimadzu LC-10AD systems (Shimadzu Corporation, Kyoto, Japan). The mobile phase consisted of 0.025% TFA, 0.025% formic acid in water (solvent A) and 0.025% TFA, 0.025% formic acid in acetonitrile (solvent B). The flow rate was 300 μl/min. The proportion of solvent B was increased from 10% B to 35% B over 45 min and then to 90% B in 90 min. It was maintained at 90% B for 10 min followed by re-equilibration for 15 min. The LC-MS system was a Thermo LCQ-Deca XP ion trap mass spectrometer interfaced to a Phenomenex Jupiter C18 column (150 × 2.00 mm, 5 μm). The instrument was operated in positive electrospray ionization mode with the following parameters: activation time was 30 ms, activation Q was 0.3, and the normalized collision energy was set at 35%. The dynamic exclusion width was set at 1.5 ppm. The mass spectrometer was operated at a capillary temperature of 210°C and capillary voltage of 2 V. The analysis was performed in the data-dependent acquisition mode in which a full scan was recorded over the mass range of 150 to 2000 mass units followed by collision-induced dissociation of the six most abundant ions. Data acquisition and analysis were performed using both Xcalibur software (v. 1.2; Thermoquest, Manchester, UK) and Sequest Bioworks (Thermo Fisher Scientific).
The peptides were identified automatically using the Sequest Bioworks computer program, which correlated the experimental tandem mass spectra against the theoretical tandem mass spectra from amino acid sequences obtained from the National Center for Biotechnology Information (NCBI) sequence database. The following criteria were applied for filtering the results. Peptides were identified on the basis of XCorr values, which were set at 2.0, 2.2, and 3.75 for single, double, or triply charged ions, respectively, and a Sp value greater than 400 and 600 for singly and doubly charged ions, respectively. The Sp value, which indicates the preliminary score of the peptide, shows the number of fragment ions that matched the total number of fragment masses for the peptide. XCorr is the cross-correlation score between the actual and the predicted MS/MS spectrum. Peptides that had a probability score less than 1.0 × 10−4 were not considered, and sequence assignments were based on selecting the peptide that displayed a ΔCn greater than 0.1 (ΔCn is the relative difference between the correlation score for the top matching peptide and the second best matching peptide). In addition, manual inspection of the peptide identifications from the MS/MS spectrum was performed to ensure that the major MS/MS fragmentation peaks matched the theoretical peaks.
Results
In Vitro Trapping of Reactive Metabolites of Tamoxifen with GSHEE.
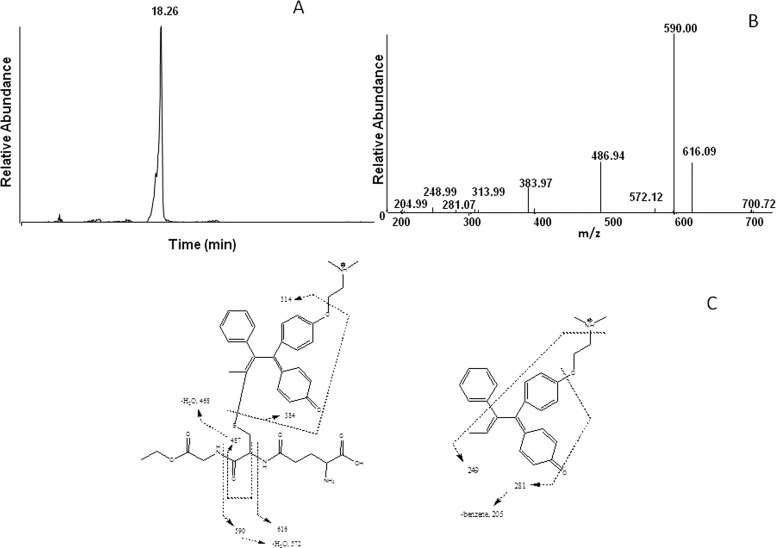
The electrophilic reactive metabolite generated by incubating tamoxifen with CYP2B6 in the presence of NADPH was trapped with GSHEE and analyzed using LC-MS/MS. The GSHEE adduct exhibited a molecular ion at m/z 719 identifying the daughter ion of the reactive intermediate as having m/z 386. The extracted ion chromatogram at m/z 719 with the GSHEE adduct eluting at 18.3 min is shown in the Fig. 2A. This is proposed to be derived from tamoxifen (m/z 372) metabolizing to 4OHtam (m/z 388), which then is oxidized to 4OHtam quinone methide (m/z 386). 4OHtam quinone methide (m/z 386) binds with GSHEE (m/z 335) to form the adduct (m/z 721), which further undergoes oxidation with an m/z 719. Similar oxidized forms of quinone adducts have previously been reported for (glutathione-S-yl)-1,4-benzoquinone when incubated with cytochrome c (Person et al., 2005) and the diclofenac quinone imine GSH conjugate as an oxidation product of hydroxyl diclofenac (Waldon et al., 2010). Figure 2B shows the MS/MS fragmentation for the ion with m/z 719. The MS/MS spectrum of the adduct exhibits prominent ions at m/z 616, 590, and 487, which are characteristic of a loss of 103, 129, and 232 mass units and which correspond to the loss of the glycylethyl ester, pyroglutamate, and a combined loss of pyroglutamate and glycylethyl ester from the GHSEE moiety, respectively. Likewise, the fragment ions at m/z 701 and 572 were formed by the loss of a water molecule from the ions at m/z 719 and 590, respectively. The ion with m/z of 205 is from the loss of benzene from the product ion with m/z 280. The ion with m/z 314 is from the loss of CH2CH2NCH3CH3. The ion with m/z 281 could not be assigned. The fragmentation pattern for the major ions observed in the MS/MS spectrum for the hydroxylated quinone methide adduct is shown in Fig. 2C. The fragmentation pattern in Fig. 2C revealed that the GSHEE bound at the α-position and the hydroxylation of the ring potentially occurred at the 4-position as determined by the incubation of 4-OH tamoxifen with CYP2B6 as described below. Control incubations using tamoxifen in the absence of NADPH did not show formation of this adduct.
Fig. 2.
LC-MS analysis of the GSHEE adduct formed by incubation of tamoxifen with CYP2B6 in the presence of NADPH. CYP2B6 was reconstituted with reductase and incubated with tamoxifen and GHSEE. The separation and analysis of the adduct are as described under Materials and Methods. A, the extracted ion chromatogram for the hydroxytamoxifen quinone methide adduct with m/z 719. B, the MS2 of the peak eluting at 18.3 min with an m/z 719. C, the primary sites of fragmentation of the tamoxifen-GSHEE adduct.
Trapping of the Reactive Intermediate from Metabolism of 4-Hydroxytamoxifen Using GSHEE.
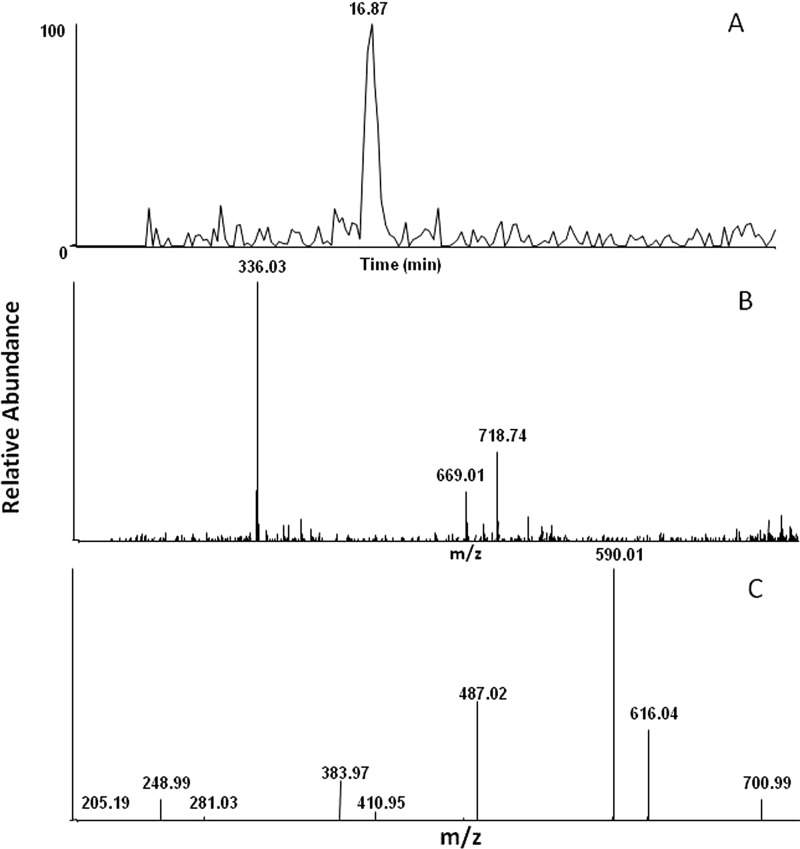
Verification of the quinone methide responsible for forming the GSHEE adduct was obtained when 4OHtam was incubated with CYP2B6 in the presence of NADPH. A GSHEE adduct exhibiting a molecular ion at m/z 719 was observed similar to that seen when tamoxifen was incubated with CYP2B6, but it exhibited a retention time of approximately 2 min faster compared with the samples incubated with tamoxifen, indicating that the adduct could be with a different regioisomer. The ability of CYP2B6 to catalyze 4-hydroxylation and trans-4-OH tamoxifen isomerization has previously been shown by Crewe et al. (2002). The peak eluting at 16.9 min with m/z 719 was not seen in control samples. Figure 3A shows the extracted ion chromatogram for the GSHEE adducts with m/z 719. The peak with m/z 336 is M+H of GSHEE, and the peak with m/z 669 was also observed in the control sample. The parent ion had an m/z 719, and the retention time differed by 2 min as seen with 4OHtam incubation with CYP2B6. This peak could again represent a different regioisomer of the same parent peak. Interindividual variation in the isomerization of 4OHtam by human liver microsomes has been shown by Williams et al. (1994).
Fig. 3.
LC-MS chromatogram of the GSHEE adduct formed when 4OHtam was incubated with P4502B6 in the presence of NADPH. CYP2B6 was reconstituted with reductase and incubated with 4OHtam, GSHEE, and NADPH. Incubation conditions and analysis of the adduct are as described under Materials and Methods. A, the extracted ion chromatogram of the 4OHtam quinone methide with m/z 719. B, the mass spectrum of the peak eluting at 16.9 min with m/z 719. C, the MS2 of the m/z 719 ion.
Figure 3, B and C, shows the MS spectrum of the GSHEE adduct and the MS/MS fragmentation patterns, respectively. The MS/MS fragmentation of the peak with m/z 719 was similar to that seen for the adduct identified when CYP2B6 was incubated with tamoxifen. This indicates that hydroxylation of tamoxifen occurs initially at the 4-position of tamoxifen, and this product is then further oxidized to form the quinone methide. Together, these findings support the formation of hydroxytamoxifen quinone methides by CYP2B6.
Trapping of the Quinone Methide-Reactive Intermediate Formed by Human Liver Microsomes.
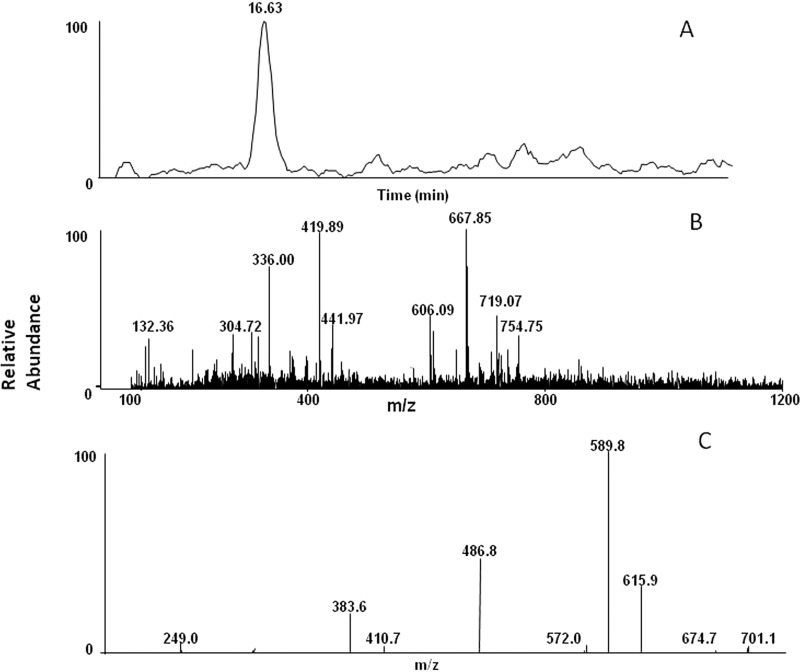
To see whether formation of quinone methide is relevant in clinical samples, human liver microsomes were incubated with tamoxifen. Human liver microsomes obtained from BD Biosciences were incubated with tamoxifen and GSHEE in the presence and absence of NADPH. A GSHEE adduct exhibiting a molecular ion at m/z 719 and eluting at 16.6 min was observed similar to that seen when tamoxifen was incubated with CYP2B6 in a reconstituted system. The peak eluting at 16.6 min with m/z 719 was not seen in control samples. Figure 4A shows the extracted ion chromatogram of the GSHEE adduct with m/z 719, and Fig. 4B shows the MS/MS spectrum of the GSHEE adduct. The MS/MS fragmentation of the peak with m/z 719 was similar to that seen with the adduct identified when CYP2B6 was incubated with tamoxifen (Fig. 4C).
Fig. 4.
LC-MS/MS spectrum of the GSHEE adduct formed by human liver microsomes. Human liver microsomes were incubated with tamoxifen and GSHEE in the presence or absence of NADPH. The incubation conditions and the analysis are as described under Materials and Methods. A, the extracted ion chromatogram for the OHtam quinone methide-GSHEE adduct eluting at 16.6 min. B, the MS/MS spectrum of the peak eluting at 16.6 min with m/z 719. C, the MS2 of the m/z 719 ion.
ESI-LC-MS Analysis of the Tamoxifen Adduct to the CYP2B6 Apoprotein.
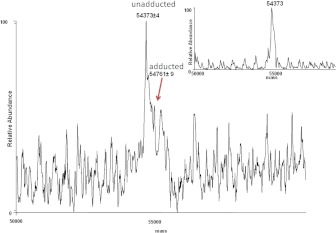
CYP2B6 was incubated with tamoxifen in the presence or absence of NADPH, and the apoprotein was analyzed by LC-MS to determine whether any of the reactive intermediates formed during metabolism formed covalent adducts with the P450 apoprotein. The inactivated sample showed a 60% loss in enzyme activity as measured by the 7-EFC-deethylation assay. Figure 5 shows the deconvoluted spectrum of CYP2B6 incubated with tamoxifen and NADPH. The two peaks correspond to the nonadducted apo-P450 with a mass of 54,373 Da and a small peak of modified apo-P450 2B6 with a mass of 54,761 Da. The inactivation by tamoxifen resulted in the appearance of a new peak with a mass difference of 388 ± 9, which is consistent with the calculated mass for the hydroxytamoxifen. This mass difference could also be attributed to the binding of the 4OHtam quinone methide (mass of 386) to the apoprotein. The instrument is not sensitive to pick up the 2Da-mass difference between the OHtam (388) and OHtam quinone methide (386). The inset in Fig. 5 shows the deconvoluted spectrum of the control sample of CYP2B6 incubated with tamoxifen in the absence of NADPH, with a mass of 54,373 Da. The presence of the trapping agent GSHEE did not affect the ability of tamoxifen to inactivate CYP2B6, and studies on the formation of the tamoxifen adduct with the apoprotein was done in the absence of GSHEE.
Fig. 5.
ESI-LC-MS analysis of CYP2B6 after incubation with tamoxifen. CYP2B6 was incubated with 60 μM tamoxifen in the presence and absence of NADPH. The incubation conditions and analyses are described under Materials and Methods. The figure shows the deconvoluted mass spectrum of the modified (54,761 Da) and the unmodified CYP2B6 apoprotein (54,373 Da). The inset shows the deconvoluted spectrum of the apo2B6 from a control incubation in the presence of tamoxifen and in the absence of NADPH.
Identification of the Amino Acid Residue Modified by Tamoxifen during the Inactivation of CYP2B6.
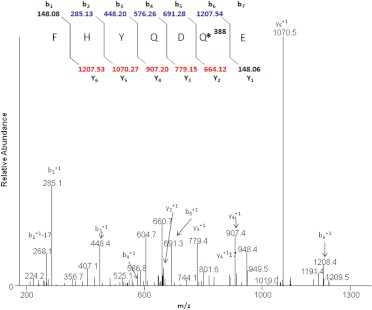
CYP2B6 that had been inactivated by incubation with tamoxifen was digested with trypsin and analyzed by LC-MS. Sequest Bioworks 3.2 software was used to determine the identity of the modified peptide and the amino acid modified by the inactivator. Tryptic peptides obtained from the control and the inactivated samples were compared with each other and with the theoretically expected digests. The Sequest search identified the human 2B6 isozyme, with sequence coverage of ∼61%. Analysis of the tryptic digest revealed a peptide adduct having a mass corresponding to the addition of hydroxytamoxifen and not the mass of the quinone methide of hydroxytamoxifen. A peptide with MH+ at m/z 1355 was identified as the peptide modified by 4OHtam. As shown in Fig. 6, the modified peptide with m/z 1355 corresponded to the sequence 188FHYQDQE194, and on the basis of the MS/MS data, the site modified by adduct formation with 4OHtam was identified to be the Gln193 as demonstrated by the 388 shift of the y2 ion. The MS/MS spectrum of the doubly charged [M + 2H]2+ ion at m/z 678.2 from the peptide modified by the hydroxytamoxifen metabolite is shown in Fig. 6. The identities of the b and y ions for the modified peptide were predicted using ProteinProspector, and the spectrum indicates an increase in the mass of the y2 to y5 ions by 388 Da, whereas the mass of the b fragment ions showed no increase from b1 to b5 and a mass increase of 388 Da for the b6 ion. A mass shift of 388 Da between that expected for b6 and the observed b6 ion indicated that the modified residue is glutamine at position 193. Many of the singly charged fragment ions of both the y and b ions were observed with good intensity in the MS/MS analysis. In the Sequest search, the peptide was identified with a probability score of 2.8 × 10−8, an Sp value of 643, and XCorr of 3.9, which exceeded the criteria specified in under Materials and Methods. Manual interpretation of the MS/MS spectra was done using Xcalibur to verify the peptide identification. These data also support the conclusion that modification of the peptide occurred at Gln193. The mass of the corresponding unmodified parent peptide was seen at MH+ 967 in the control sample (data not shown).
Fig. 6.
LC-MS/MS spectrum of the CYP2B6 tryptic peptide modified by tamoxifen. CYP2B6 was reconstituted with reductase and tamoxifen in the presence or absence of NADPH and digested with trypsin. The samples were analyzed by LC-MS/MS and Sequest software as described under Materials and Methods. The figure shows the profile for the doubly charged ion with m/z 678.2 for the modified peptide 188FHYQDQE194. The inset shows the calculated b and y ions for the modified peptide, and the b and y ions identified in the MS/MS spectrum are shown in blue and red, respectively.
Discussion
Numerous metabolic pathways have been proposed for the formation of reactive intermediates responsible for leading to the toxicity of tamoxifen. One of the proposed pathways includes aromatic hydroxylation of tamoxifen forming 4-hydroxytamoxifen, which can then undergo further oxidation resulting in the formation of quinone methide (Fan et al., 2000). Studies previously have shown that the 4-hydroxytamoxifen quinone methide reacts to form covalent adducts with DNA in vitro (Marques and Beland, 1997). Two studies have shown that P450s 3A4, 2D6, and 2C19 as well as several other forms of P450 have the potential to produce catechol metabolites from both tamoxifen and 4-hydroxytamoxifen (Dehal and Kupfer, 1996; Notley et al., 2002). Earlier studies from our laboratory have shown that tamoxifen is a mechanism-based inactivator of CYP2B6 (Sridar et al., 2002). The loss in the catalytic activity was associated with modification of the protein. The studies here are focused on identifying the reactive intermediate of tamoxifen responsible for mechanism-based inactivation of 2B6.
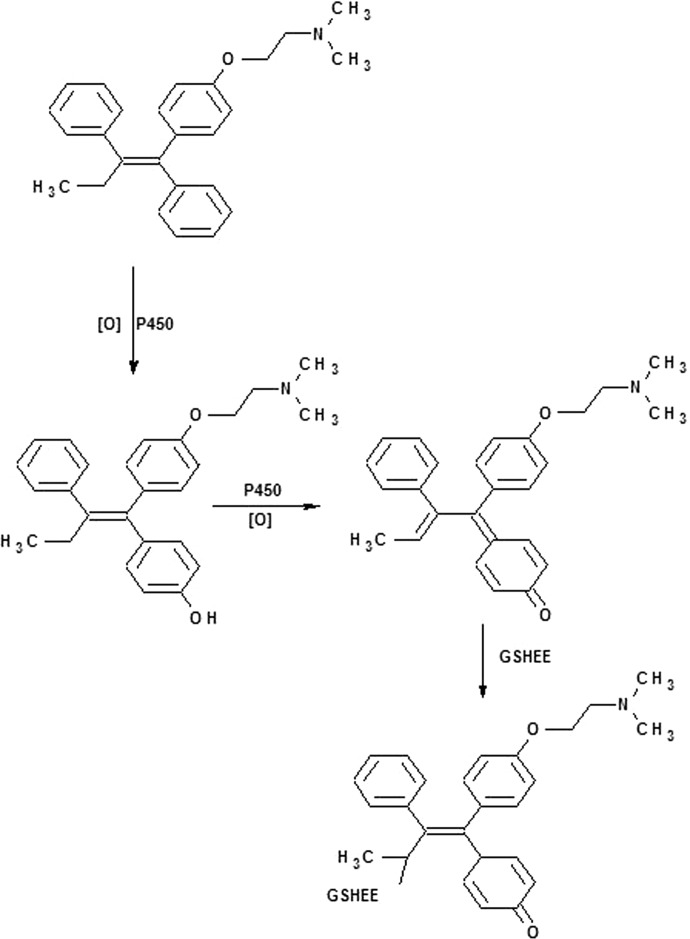
Fan and Bolton (2001) have previously demonstrated the oxidation of metabolite E to a quinone methide, which was then trapped as a GSH adduct when tamoxifen was incubated with rat liver microsomes. The metabolism involved the formation of stable epimers of the trans form of the metabolite E-GSH adduct and formation of three glutathione conjugates of 4-hydroxytamoxifen (Fan and Bolton, 2001). Here, in vitro studies were undertaken to probe the bioactivation of tamoxifen by CYP2B6 and identify the possible reactive species using GSHEE as a trapping agent, which led to the identification of a GSHEE adduct formed by reaction with quinone methides. The electrophilic intermediate detected in our studies was the GSHEE adduct of the OHtam quinone methide having an m/z 719, with GSHEE binding at the aliphatic carbon (Fig. 2). To explore the site of hydroxylation and quinone methide formation, the formation of adducts from the metabolism of 4OHtam by 2B6 was investigated. The mass spectrum with the molecular ion at m/z 719 suggested the formation of a GSHEE adduct, and the MS/MS fragmentation pattern of this adduct indicated that the hydroxylation occurred at the 4-position (Fig. 3). Analysis of the adduct formed when CYP2B6 is incubated with tamoxifen or 4OHtam suggested that the 4OHtam quinone methide trapped by GSHEE underwent a further two-electron oxidation. Both oxidized and reduced forms of quinone adducts previously have been observed when (glutathione-S-yl)-1,4-benzoquinone was incubated with cytochrome c (Person et al., 2005) and when an oxidized quinone imine containing glutathione conjugate was isolated from rat bile following exposure to diclofenac (Waldon et al., 2010), and this may be why the adduct has an m/z of 719 instead of 721.
Fan et al. (2000) have shown that quinone methides formed from 4OHtam are unlikely to contribute to the cytotoxic or genotoxic effects of the parent drugs given that no conjugates were observed in a breast cancer cell line. It has also been established that the reaction between the 4OHtam quinone methide and GSH is a reversible process, with the conjugate decomposing over time to regenerate the quinone methide with a half-life of approximately 4 min (Fan et al., 2000). It is noteworthy that treatment with tamoxifen induces CYP2B and 3A in rats (White et al., 1993). Tamoxifen tends to be easily metabolized to form 4OHtam. Thus, it is presumed that the quinone methide of 4OHtam may either bind to GHSEE or revert back to the 4OHtam quinone methide. The sequence proposed for the reactions for the metabolism of tamoxifen by CYP2B6 in the presence of GSHEE and NADPH is depicted in Scheme 1. Investigation of a panel of 50 human liver samples demonstrated that CYP2B6 plays a role in 4OHtam formation and that the 2B6 genotype has an influence on the rates of formation of 4OHtam (Coller et al., 2002). We also investigated the oxidation of tamoxifen to quinone metabolites in human liver microsomes obtained from BD Biosciences in the presence of GSHEE as the trapping agent. LC-MS analysis revealed the presence of a peak with m/z 719 eluting at 16.6 min, and MS/MS analysis of the peak suggested that the peak was the OHtam quinone methide (Fig. 4).
Scheme 1.
Proposed sequence of reactions for the metabolism of tamoxifen by CYP2B6 in the presence of GSHEE and NADPH.
Many compounds are known to be oxidized by cytochrome P450s to form quinones or epoxides that have the potential to modify cellular macromolecules and bind to the active site of the enzyme that produces them. Quinone methides can also inhibit enzymes irreversibly through covalent modifications of critical residues. The formation of quinone methides in vitro has been linked to cytotoxicity in studies with hepatocytes, liver slices, bronchiolar Clara cells, and keratinocytes from mice or rats (Bolton et al., 1997). In an attempt to identify the potential site for inactivation due to adduct formation with the protein, inactivated protein was digested with trypsin and analyzed by LC-MS/MS. In our studies, we could only identify a peptide adduct resulting from modification by hydroxytamoxifen, and none was identified as a result of modification by the hydroxytamoxifen quinone methide. This is not surprising, because Fan et al., 2000 have shown that 1,8-Michael or 1,6-Michael addition of GSH to the quinone methide is a reversible process and that the adduct has a short half-life. In addition, 4-hydroxytamoxifen has been shown to form adducts with deoxynucleosides (Yao et al., 2001), and Notley et al. (2002) have shown that recombinant P450s 2B6 and 2C19 catalyzed drug-protein adduct formation through 4OHtam and not catechol formation, whereas P450s 2D6 and 3A5 generated catechol metabolite formation and not drug-protein adduct formation. We were able to successfully identify and characterize a P450 adduct using recombinant purified CYP2B6. CYP2B6 apoprotein in control samples eluted at 24 min, and scanning across this peak yielded a protein envelope that was deconvoluted to give a mass of 54,373 Da (Fig. 5, inset). Deconvolution of the protein envelope for the tamoxifen-inactivated CYP2B6 yielded a pattern that included an unadducted CYP2B6, with a mass of 54,373 Da, plus a small peak that deconvoluted to a mass of 54,761 Da and that was not observed in control samples. The mass difference for this additional peak is 388, which are close to the mass of hydroxytamoxifen quinone methide (mass 386), providing evidence for adduct formation with the apoprotein that may be responsible for the inactivation by tamoxifen. Sequest analysis of the tryptic digest of the modified protein revealed that the peptide 188FHYQDQE194 was the tryptic peptide modified by hydroxytamoxifen and that the site of adduct formation was glutamine 193 (Fig. 6). According to the 2B6 crystal structure, residue Gln193 is located in the F helix (Gay et al., 2010). Various structural studies have identified the F helix as a topological element that has a number of conserved residues, although their spatial disposition relative to heme, as well as the size and role, may vary among different P450s (Pikuleva et al., 2001). In P4502C5, the F helix forms the ceiling of the active site (Williams et al., 2000), and in mitochondrial P450s 27A1 and 11A1, the F helix forms a part of the substrate access channel and may play a role in the control of the regioselectivity of hydroxylation (Pikuleva et al., 2001). The amide nitrogen of Gln is a weak nucleophile, and the adduct probably forms because the binding in the substrate pocket orients the Gln residue and the electrophilic center on the reactive metabolite in close proximity to each other. Gln has also been shown to form covalent adducts with 5-hydroxytryptamine and dopamine (Johnson et al., 2010). Although our studies identified only binding of the OHtam to a single amino acid residue, this does not preclude the possibility of OHtam quinone methide binding to other residues in CYP2B6 that are modified, which may have eluded detection by our techniques. Alternatively, formation of 4OHtam quinone methide may not lead to covalent binding of CYP2B6 but could still lead to binding to other proteins or DNA contributing to toxicity or carcinogenesis, respectively.
In conclusion, we have shown that incubation of tamoxifen with recombinantly expressed CYP2B6 results in oxidation to a quinone methide and that this electrophile reacts with the GSHEE to form a corresponding conjugate. In human liver microsomal incubations, tamoxifen was metabolized to form the 4OHtam quinone methide, and thus quinone methide formation may be one of the pathways involved in the mechanism of inactivation of CYP2B6 and may also contribute to the potential carcinogenicity of tamoxifen. The fact that the 4-hydroxylation of tamoxifen has been suggested to be an activating route of metabolism (King, 1995) and similar adducts have been found in rat and human liver microsomes argues for the further investigation of this drug, especially in cases of multiple drug therapies where CYP2B6 is required for clearance.
Acknowledgments
We thank Dr. Judy Bolton for providing us with metabolite E and 3,4-dihydroxytamoxifen. We also thank Hsia-Lien Lin for the purification of reductase.
This work was supported in part by the National Institutes of Health National Cancer Institute [Grant CA16954].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
- 4OHtam
- 4-hydroxytamoxifen
- LC-MS
- liquid chromatography-mass spectrometry
- GSHEE
- glutathione ethyl ester
- HPLC
- high-performance liquid chromatography
- ESI
- electrospray ionization
- MS/MS
- tandem mass spectrometry
- TFA
- trifluoroacetic acid
- P450
- cytochrome P450
- 7-EFC
- 7-ethoxy-4-(trifluoromethyl)coumarin O-deethylation
- OHtam
- hydroxytamoxifen.
Authorship Contributions
Participated in research design: Sridar, D'Agostino, and Hollenberg.
Conducted experiments: Sridar.
Contributed new reagents or analytic tools: Sridar.
Performed data analysis: Sridar, D'Agostino, and Hollenberg.
Wrote or contributed to the writing of the manuscript: Sridar, D'Agostino, and Hollenberg.
References
- Andersson M, Storm HH, Mouridsen HT. (1991) Incidence of new primary cancers after adjuvant tamoxifen therapy and radiotherapy for early breast cancer. J Natl Cancer Inst 83:1013–1017 [DOI] [PubMed] [Google Scholar]
- Bolton JL, Turnipseed SB, Thompson JA. (1997) Influence of quinone methide reactivity on the alkylation of thiol and amino groups in proteins: studies utilizing amino acid and peptide models. Chem Biol Interact 107:185–200 [DOI] [PubMed] [Google Scholar]
- Coller JK, Krebsfaenger N, Klein K, Endrizzi K, Wolbold R, Lang T, Nüssler A, Neuhaus P, Zanger UM, Eichelbaum M, et al. (2002) The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol 54:157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crewe HK, Notley LM, Wunsch RM, Lennard MS, Gillam EM. (2002) Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4′-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos 30:869–874 [DOI] [PubMed] [Google Scholar]
- Dehal SS, Kupfer D. (1996) Evidence that the catechol 3,4-Dihydroxytamoxifen is a proximate intermediate to the reactive species binding covalently to proteins. Cancer Res 56:1283–1290 [PubMed] [Google Scholar]
- Divi RL, Osborne MR, Hewer A, Phillips DH, Poirier MC. (1999) Tamoxifen-DNA adduct formation in rat liver determined by immunoassay and 32P-postlabeling. Cancer Res 59:4829–4833 [PubMed] [Google Scholar]
- Ekins S, Wrighton SA. (1999) The role of CYP2B6 in human xenobiotic metabolism. Drug Metab Rev 31:719–754 [DOI] [PubMed] [Google Scholar]
- Fan PW, Bolton JL. (2001) Bioactivation of tamoxifen to metabolite E quinone methide: reaction with glutathione and DNA. Drug Metab Dispos 29:891–896 [PubMed] [Google Scholar]
- Fan PW, Zhang F, Bolton JL. (2000) 4-Hydroxylated metabolites of the antiestrogens tamoxifen and toremifene are metabolized to unusually stable quinone methides. Chem Res Toxicol 13:45–52 [DOI] [PubMed] [Google Scholar]
- Fisher B, Costantino JP, Redmond CK, Fisher ER, Wickerham DL, Cronin WM. (1994) Endometrial cancer in tamoxifen-treated breast cancer patients: findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst 86:527–537 [DOI] [PubMed] [Google Scholar]
- Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, et al. (1998) Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst 90:1371–1388 [DOI] [PubMed] [Google Scholar]
- Furr BJ, Jordan VC. (1984) The pharmacology and clinical uses of tamoxifen. Pharmacol Ther 25:127–205 [DOI] [PubMed] [Google Scholar]
- Gay SC, Shah MB, Talakad JC, Maekawa K, Roberts AG, Wilderman PR, Sun L, Yang JY, Huelga SC, Hong WX, et al. (2010) Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution. Mol Pharmacol 77:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna IH, Reed JR, Guengerich FP, Hollenberg PF. (2000) Expression of human cytochrome P450 2B6 in Escherichia coli: characterization of catalytic activity and expression levels in human liver. Arch Biochem Biophys 376:206–216 [DOI] [PubMed] [Google Scholar]
- Hanna IH, Teiber JF, Kokones KL, Hollenberg PF. (1998) Role of the alanine at position 363 of cytochrome P450 2B2 in influencing the NADPH- and hydroperoxide-supported activities. Arch Biochem Biophys 350:324–332 [DOI] [PubMed] [Google Scholar]
- Hellmold H, Rylander T, Magnusson M, Reihnér E, Warner M, Gustafsson JA. (1998) Characterization of cytochrome P450 enzymes in human breast tissue from reduction mammaplasties. J Clin Endocrinol Metab 83:886–895 [DOI] [PubMed] [Google Scholar]
- Hesse LM, Venkatakrishnan K, Court MH, von Moltke LL, Duan SX, Shader RI, Greenblatt DJ. (2000) CYP2B6 mediates the in vitro hydroxylation of bupropion: potential drug interactions with other antidepressants. Drug Metab Dispos 28:1176–1183 [PubMed] [Google Scholar]
- Johnson KB, Thompson JM, Watts SW. (2010) Modification of proteins by norepinephrine is important for vascular contraction. Front Physiol 1:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC. (2004) Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat 85:151–159 [DOI] [PubMed] [Google Scholar]
- Jordan VC. (1982) Metabolites of tamoxifen in animals and man: identification, pharmacology, and significance. Breast Cancer Res Treat 2:123–138 [DOI] [PubMed] [Google Scholar]
- King CM. (1995) Tamoxifen and the induction of cancer. Carcinogenesis 16:1449–1454 [DOI] [PubMed] [Google Scholar]
- Lin HL, Kent UM, Hollenberg PF. (2005) The grapefruit juice effect is not limited to cytochrome P450 (P450) 3A4: evidence for bergamottin-dependent inactivation, heme destruction, and covalent binding to protein in P450s 2B6 and 3A5. J Pharmacol Exp Ther 313:154–164 [DOI] [PubMed] [Google Scholar]
- Lo R, Burgoon L, Macpherson L, Ahmed S, Matthews J. (2010) Estrogen receptor-dependent regulation of CYP2B6 in human breast cancer cells. Biochim Biophys Acta 1799:469–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani C, Gelboin HV, Park SS, Pearce R, Parkinson A, Kupfer D. (1993) Metabolism of the antimammary cancer antiestrogenic agent tamoxifen. I. Cytochrome P-450-catalyzed N-demethylation and 4-hydroxylation. Drug Metab Dispos 21:645–656 [PubMed] [Google Scholar]
- Marques MM, Beland FA. (1997) Identification of tamoxifen-DNA adducts formed by 4-hydroxytamoxifen quinone methide. Carcinogenesis 18:1949–1954 [DOI] [PubMed] [Google Scholar]
- Moorthy B, Sriram P, Pathak DN, Bodell WJ, Randerath K. (1996) Tamoxifen metabolic activation: comparison of DNA adducts formed by microsomal and chemical activation of tamoxifen and 4-hydroxytamoxifen with DNA adducts formed in vivo. Cancer Res 56:53–57 [PubMed] [Google Scholar]
- Notley LM, De Wolf CJ, Wunsch RM, Lancaster RG, Gillam EM. (2002) Bioactivation of tamoxifen by recombinant human cytochrome P450enzymes. Chem Res Toxicol 15:614–622 [DOI] [PubMed] [Google Scholar]
- Nuwaysir EF, Dragan YP, Jefcoate CR, Jordan VC, Pitot HC. (1995) Effects of tamoxifen administration on the expression of xenobiotic metabolizing enzymes in rat liver. Cancer Res 55:1780–1786 [PubMed] [Google Scholar]
- Person MD, Mason DE, Liebler DC, Monks TJ, Lau SS. (2005) Alkylation of cytochrome c by (glutathion-S-yl)-1,4-benzoquinone and iodoacetamide demonstrates compound-dependent site specificity. Chem Res Toxicol 18:41–50 [DOI] [PubMed] [Google Scholar]
- Phillips DH, Carmichael PL, Hewer A, Cole KJ, Poon GK. (1994a) alpha-Hydroxytamoxifen, a metabolite of tamoxifen with exceptionally high DNA-binding activity in rat hepatocytes. Cancer Res 54:5518–5522 [PubMed] [Google Scholar]
- Phillips DH, Potter GA, Horton MN, Hewer A, Crofton-Sleigh C, Jarman M, Venitt S. (1994b) Reduced genotoxicity of [d5-ethyl]-tamoxifen implicates alpha-hydroxylation of the ethyl group as a major pathway of tamoxifen activation to a liver carcinogen. Carcinogenesis 15:1487–1492 [DOI] [PubMed] [Google Scholar]
- Pikuleva IA, Puchkaev A, Björkhem I. (2001) Putative helix F contributes to regioselectivity of hydroxylation in mitochondrial cytochrome P450 27A1. Biochemistry 40:7621–7629 [DOI] [PubMed] [Google Scholar]
- Sridar C, Kent UM, Notley LM, Gillam EM, Hollenberg PF. (2002) Effect of tamoxifen on the enzymatic activity of human cytochrome CYP2B6. J Pharmacol Exp Ther 301:945–952 [DOI] [PubMed] [Google Scholar]
- Thompson DC, Perera K, Krol ES, Bolton JL. (1995) o-Methoxy-4-alkylphenols that form quinone methides of intermediate reactivity are the most toxic in rat liver slices. Chem Res Toxicol 8:323–327 [DOI] [PubMed] [Google Scholar]
- Waldon DJ, Teffera Y, Colletti AE, Liu J, Zurcher D, Copeland KW, Zhao Z. (2010) Identification of quinone imine containing glutathione conjugates of diclofenac in rat bile. Chem Res Toxicol 23:1947–1953 [DOI] [PubMed] [Google Scholar]
- White IN, Davies A, Smith LL, Dawson S, De Matteis F. (1993) Induction of CYP2B1 and 3A1, and associated monoxygenase activities by tamoxifen and certain analogues in the livers of female rats and mice. Biochem Pharmacol 45:21–30 [DOI] [PubMed] [Google Scholar]
- Williams ML, Lennard MS, Martin IJ, Tucker GT. (1994) Interindividual variation in the isomerization of 4-hydroxytamoxifen by human liver microsomes: involvement of cytochromes P450. Carcinogenesis 15:2733–2738 [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. (2000) Microsomal cytochrome P450 2C5: comparison to microbial P450s and unique features. J Inorg Biochem 81:183–190 [DOI] [PubMed] [Google Scholar]
- Yao D, Zhang F, Yu L, Yang Y, van Breemen RB, Bolton JL. (2001) Synthesis and reactivity of potential toxic metabolites of tamoxifen analogues: droloxifene and toremifene o-quinones. Chem Res Toxicol 14:1643–1653 [DOI] [PubMed] [Google Scholar]