Abstract
The RecQ family of helicases has been shown to play an important role in maintaining genomic stability. In humans, this family has five members and mutations in three of these helicases, BLM, WRN and RECQL4, are associated with disease. Alterations in RECQL4 are associated with three diseases, Rothmund-Thomson syndrome, Baller-Gerold syndrome, and RAPADILINO syndrome. One of the more common mutations found in RECQL4 is the RAPADILINO mutation, c.1390+delT which is a splice-site mutation leading to an in-frame skipping of exon 7 resulting in 44 amino acids being deleted from the protein (p.Ala420-Ala463del). In order to characterize the RAPADILINO RECQL4 mutant protein, it was expressed in bacteria and purified using an established protocol. Strand annealing, helicase, and ATPase assays were conducted to characterize the protein's activities relative to WT RECQL4. Here we show that strand annealing activity in the absence of ATP is unchanged from that of WT RECQL4. However, the RAPADILINO protein variant lacks helicase and ssDNA-stimulated ATPase activity. These observations help explain the underlying molecular etiology of the disease and our findings provide insight into the genotype and phenotype association among RECQL4 syndromes.
Keywords: RecQ helicase, RECQL4, RAPADILINO, Rothmund-Thomson syndrome, ATPase
1.0 Introduction
The RecQ helicase family of proteins is critically important for genome maintenance and in human cells there are five RecQ family homologues: RECQL1, WRN, BLM, RECQL4 and RECQL5 (for recent reviews see [1,2]. Mutations in WRN or BLM genes give rise to Werner (OMIM ID 277700) or Bloom syndromes (OMIM ID 210900), respectively, whereas mutations within RECQL4 give rise to three related but distinct diseases: RAPADILINO (OMIM ID 266280), Rothmund-Thomson (RTS, OMIM ID 268400), and Baller-Gerold (BGS, OMIM ID 218600) syndromes. Not all reported RTS patients have identifiable mutations within the RECQL4 gene, and therefore, there are two classes of RTS patients: Type I, which lack RECQL4 mutations and type II which have confirmed RECQL4 mutations [3]. At this time it is not clear why mutations in the RECQL4 gene gives rise to three different clinical syndromes.
All three RECQL4-associated diseases are rare, autosomal recessive and characterized by growth retardation and bone malformations, especially radial ray defects (for recent reviews see [4,5]). The RAPADILINO syndrome was first identified in 1989, and the name is an acronym for the features observed in patients: RAdial hypoplasia/aplasia, PAtellar hypoplasia/aplasia, cleft or highly arched PAlate, DIarrhea and DIslocated joints, LIttle size and LImb malformation, and slender NOse and NOrmal intelligence [6]. Poikiloderma, a cutaneous rash, is one of the distinguishing features of RTS and BGS but is lacking in RAPADILINO. Another feature found in RTS patients but missing in RAPADILINO patients is alopecia, which includes deficiencies in hair growth including the absence of eyelashes and eyebrows. More recently it has been recognized that RAPADILINO and type II RTS patients share a common predisposition for osteosarcomas and lymphomas [4]. An elevated risk for osteosarcoma is also a characteristic feature seen in WRN syndrome patients [7]. While the three RECQL4 associated syndromes share some common features, their unique spectrum of clinical manifestations highlights the complexity of deciphering the clinical phenotypes associated with patient RECQL4 genotypes.
The RECQL4 gene encompasses 21 exons, and exons 8–14 encode the conserved RecQ helicase domain. The most common RECQL4 mutation in RAPADILINO patients is the c.1390+delT (p.Ala420-Ala463del) [4,8]. This mutation is found in the majority of reported RAPADILINO patients, especially those of Finnish decent, and importantly, it is one of a few homozygous RECQL4 mutations. The mutation destroys a splice acceptor site resulting in skipping of exon 7, which immediately precedes the exons responsible for encoding the helicase domain of RECQL4. Therefore, loss of this exon could alter the protein's function. The region of RECQL4 encoded by exon 7 has been previously reported to be important for nuclear import and retention, because the GFP fusion of RECQL4 protein harboring the major RAPADILINO patient mutation (RAPA) was overwhelmingly mislocalized to the cytoplasm [9]. More recently, our laboratory has shown that this region of RECQL4 is also important for localization to focal laser-induced DNA damage [10]. In the literature, the major RAPA mutation is thought to spare the helicase domain [8], however there is no published information about whether this is true or if this mutation alters the biochemical activities of the RECQL4 protein.
RECQL4 is the least well characterized protein among the mammalian RecQ helicases, whose deficiencies cause human disease. RecQ helicase proteins typically share three structural elements: the conserved helicase domain, a RecQ helicase Conserved domain, (RQC), and a Helicase and RNase D C-terminal domain (HRDC). RECQL4 is unique among the mammalian proteins because it lacks the RQC and HRDC domains. In the other RecQ helicase proteins, these two domains have been shown to be important for DNA binding and for mediating protein:protein interactions (reviewed in [1]and [2]). In the original characterization of RECQL4, it was described as lacking helicase activity [11]. However, more recent work has demonstrated that RECQL4 in fact does possess helicase activity, although with a very limited substrate specificity [12–14]. It was proposed that the robust strand annealing activity of RECQL4 was responsible for masking the helicase activity and only upon addition of excess single stranded DNA was the activity revealed [12]. Upon further characterization, our laboratory has shown that by employing short forked DNA substrates RECQL4's inherent helicase activity can be readily observed without the need for excess single stranded DNA [14]. Thus, like other RecQ helicases, RECQL4 possesses multiple biochemical functions including: DNA binding, DNA strand annealing, the classic 3' to 5' helicase and DNA-stimulated ATPase activities [11–14].
The in vivo biochemical functions and pathways in which RECQL4 plays a role are not yet fully characterized. The protein has been shown to localize to multiple cellular compartments such as the nucleus, nucleolus, cytoplasm and mitochondria [15–19]. The precise localization of RECQL4 is cell type specific as, some cells show more cytoplasmic than nuclear localization [15]. Acetylation by p300 may also contribute to the differential localization patterns of RECQL4 [20]. Additionally, RECQL4 has been shown to localize to sites experiencing DNA damage including foci found following oxidatively generated DNA damage and double strand break DNA damage [10,16,21,22]. There is evidence that RECQL4 modulates base excision DNA repair [21] and that it plays a role in DNA double strand break repair [10,16]. However, its role in DNA repair is much less clear than the roles of the other human disease-causing RecQ helicases (WRN and BLM). RECQL4 has also been found to associate with sites of DNA replication and DNA replication initiation proteins [23–25].
Here we have characterized the biochemical properties of the major RAPA patient mutant protein, RAPA RECQL4. This mutation is a RECQL4 gene mutation found in the homozygous state in patients. The mutation is the result of a splice-site mutation resulting in the in-frame skipping of exon 7. Consequently, the RAPA RECQL4 protein lacks 44 amino acids just prior to the helicase domain. The WT and RAPA RECQL4 recombinant proteins were purified from bacteria and analyzed in vitro for strand annealing, helicase, and ssDNA-stimulated ATPase activities. Our analysis showed that the RAPA patient mutation compromised both the helicase and ATPase activities of the protein while sparing the strand annealing activity. Thus, not only does the RAPA mutation cause mislocalization of the RECQL4 protein, it also severely impairs the protein's catalytic functions. Thorough biochemical characterization of RECQL4 patient mutations may contribute to a better understanding of the genotype:phenotype relationships between RECQL4 mutations and the three RECQL4-associated syndromes RAPADILINO, RTS and BGS. Only through such analysis are we likely to unravel how RECQL4 mutations contribute to the disease phenotypes.
2.0 Results
2.1 Production of RAPA RECQL4
RECQL4 is found on chromosome 8q, and its genomic organization is comprised of 21 exons across 6.5 kb of DNA with a high number of unusually short intervening introns [26]. The intron-exon boundaries of WT and RAPA RECQL4 are shown in Fig. 1A. The N-terminus of the protein shares sequence similarity to Sld2/DRC1, a yeast protein involved in DNA replication [23]. In the middle of the protein, exons 8–14 encode the conserved helicase domain. The RAPA mutation, c.1390+delT, causes deletion of exon 7 from RECQL4's mRNA, but it does not result in the introduction of a premature termination codon, thus the truncated protein coded by this mRNA variant lacks 44 amino acids just upstream from the conserved helicase domain.
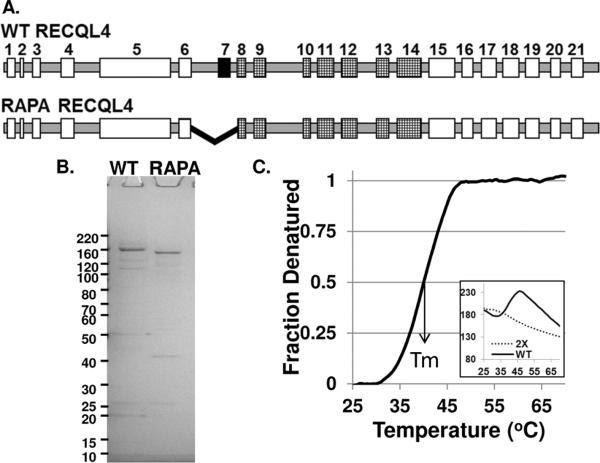
Figure 1. Graphic representation WT and RAPA RECQL4.
A, The intron-exon boundaries of RECQL4 gene. Exon 7 is deleted by the RAPA patient mutation, black filled rectangles. Exons 8–14, hatched, encode the RecQ helicase domain. B, Simply Blue stained SDS-PAGE gel of bacterially expressed and purified WT and RAPA RECQL4 proteins, 500 ng of each full length protein. C, Denaturation curves for 2× Sypro Orange control, dashed line, and WT RECQL4 plus 2× Sypro Orange, solid line. Insert shows original raw fluorescence values versus temperature, which the denaturation curve is extrapolated from.
The WT and RAPA RECQL4 proteins were expressed and purified from bacteria as previously described [11]. Deletion of exon 7 caused the protein's pI to change from 8.5 to 8.8, and therefore the RAPA RECQL4 eluted slightly later on the SP sepharose column than WT RECQL4 (data not shown). This unfortunately resulted in RAPA RECQL4 co-eluting with a major GST-RECQL4 fragment. Thus, only those fractions of RAPA RECQL4 which eluted before the major GST-RECQL4 fragment were pooled and applied to the glutathione resin. Consequently, the overall yield for RAPA RECQL4 was not as high as WT RECQL4, but the final protein concentrations were similar (1 and 2–3 μM for RAPA and WT RECQL4, respectively). A Simply Blue stained gel containing the RECQL4 proteins (500 ng of each full length protein) is shown Fig. 1B. As can be seen, WT RECQL4 shows slightly more impurities than RAPA RECQL4. As expected the RAPA RECQL4 protein has a slightly lower molecular weight than WT RECQL4.
2.2 Protein structural stability
WT and RAPA RECQL4 proteins were subjected to differential scanning fluorimetry to determine their structural stability [27,28]. The assay takes advantage of the elevated fluorescence emissions of Sypro Orange dye, as it interacts with hydrophobic sites on denatured proteins. In these assays, 1–2 μg of protein were assayed with various concentrations of Sypro Orange (0.5–2×). Denaturation curves for WT could be readily obtained whereas the RAPA protein did not respond in this assay (see Fig. 1C). Under these reaction conditions, the Tm for WT RECQL4 was 39.5 ± 0.05 °C. Thus, the 44-amino acid deletion within RAPA RECQL4 does apparently alter the structure of the protein.
2.3 Stand annealing activity is comparable between the WT and RAPA RECQL4 proteins
WT RECQL4 is known to possess robust strand annealing activity [11,12], therefore we compared the relative strand annealing activities of WT and RAPA RECQL4 proteins in Fig. 2. For the strand annealing assay, a 37-mer single stranded DNA substrate was employed. The product of the strand annealing reaction is a forked DNA molecule which is composed of a 22 bp duplex region and two 15-mer polydT splayed arms. The proteins were tested for their ability to anneal the radiolabeled T1 substrate to its corresponding partially complementary oligonucleotide, B1. Fig. 2A and B show the results of the strand annealing when the assay is done in the absence of ATP. The RAPA RECQL4 protein showed wild type levels of strand annealing activity. Approximately 3 nM of RECQL4 was necessary to anneal 50% of the 0.5 nM substrate for both proteins. Given the robust strand annealing activity observed from the RAPA RECQL4 variant, it is unlikely that the protein is grossly misfolded.
Figure 2. RAPA RECQL4 possesses wild type levels of strand annealing.
The strand annealing activities of the RECQL4 proteins were determined using radiolabeled T1 (0.5 nM) and cold B1 (1 nM), as described in the Material and Methods. A, a representative phosphorimage of the gel showing the relative single strand annealing activity of WT (lanes 2–5) and RAPA RECQL4 (lanes 6–9) in the absence of ATP. The position of the single stranded and forked dsDNAs are shown. B, graph of the quantification of three independent experiments of strand annealing in the absence of ATP. C, a representative phosphorimage of a gel showing the relative single strand annealing activity of WT (lanes 11–14) and RAPA RECQL4 (lanes 15–18) in the presence of ATP. D, graph of the quantification of three independent experiments of strand annealing in the presence of ATP. The mean and standard deviation are reported in B and D.
It has been shown that ATP inhibits the strand annealing activity of RECQL4 and of the other RecQ helicases [12,29–32]. Therefore, we wanted to assess if the proteins behaved similarly in the presence and absence of ATP. The strand annealing activity of both proteins was tested in the presence of 5 mM ATP (Fig. 2C and D). As can be seen, at high protein concentrations, both proteins' strand annealing activity was inhibited by the presence of ATP (compare panels B and D, at 5 and 10 nM). Interestingly, the WT RECQL4 activity plateaued between 5–10 nM protein whereas RAPA RECQL4's strand annealing activity continued to rise between 5–10 nM protein, compare 2C lanes 8–9 with 17–18. Since we used a splayed arm DNA substrate, it is possible that the helicase activity from WT RECQL4 started to compete for the substrate. As expected, when a full duplex oligonucleotide was incubated with WT RECQL4, WT RECQL4 no longer showed this inhibition of strand annealing (data not shown). These results suggest that the RAPA mutant protein may have a helicase deficiency.
2.4 RAPA RECQL4 lacks helicase activity
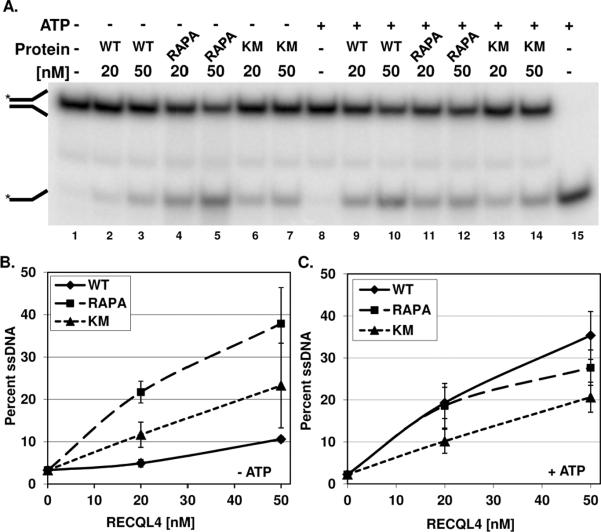
We elected to test the helicase activity of WT and RAPA RECQL4 on the short fork substrate (B1/T1, B1 labeled) reported previously by Rossi et al. since this substrate allowed us to assess helicase activity in the absence of excess ssDNA. Fig. 3 shows the results of the helicase assay. WT RECQL4 protein showed a protein concentration-dependent increase in activity and was able to unwind 36% of the substrate at the highest protein concentration used (Fig. 3, lanes 5–6). In contrast, the RAPA RECQL4 protein liberated no ssDNA (Fig. 3, lanes 7–11). Therefore, even though the RAPA mutation is positioned outside of the conserved helicase domain, the protein lacks helicase activity.
Figure 3. RAPA RECQL4 lacks helicase activity.
The helicase activity of WT and RAPA RECQL4 was determined using the forked duplex B1/T1 (0.5 nM) as described in the Materials and Methods. A, a representative phosphorimage of a gel displaying the helicase activity of WT (lanes 2–6) and RAPA RECQL4 (lanes 7–11) is shown. The Δ symbol denotes the heat denatured DNA substrate. The position of the single stranded and forked dsDNAs are denoted by the images on the left side of the gel. B, a graph of the quantification of three independent helicase gels. The mean and standard deviation are reported.
2.5 Strand separating activity in the presence of excess ssDNA
It has been reported that the presence of excess single stranded DNA can suppress the robust ssDNA annealing activity of RECQL4, and therefore aid in the visualization of the helicase products [12,14]. Additionally, helicase dead mutants of RECQL4, K508R or D605A, have been shown to possess strand separating activity when ssDNA is present, and thus RECQL4 was proposed to have two DNA unwinding domains [12]. The domain responsible for this second strand separating activity was mapped to the N-terminus of the protein [12]. We have shown that in the absence of ssDNA, the K508M RECQL4 mutant displays no ATPase activity and no helicase activity, but is competent to anneal single stranded DNA as efficiently as WT RECQL4 [14]. Therefore, RAPA RECQL4's strand separating activities, in the presence and absence of ATP, were compared to both WT and K508M RECQL4 proteins. The same duplex as in Fig. 3 was used and the reactions were done in the presence of 25-fold excess of cold B1. The left side of Fig. 4 (A, lanes 2–7) shows that all the proteins possess DNA strand separating activities in the absence of ATP and with excess ssDNA present. Curiously, the WT protein shows the least amount of unwinding activity, and the helicase dead protein displays a level of activity between RAPA and WT RECQL4 (Fig. 4B).
Figure 4. Strand separating activity in the presence of excess single stranded DNA.
The strand separating activity of WT and RAPA RECQL4 was determined using the labeled forked duplex B1/T1 (0.5 nM) in the presence of 12.5 nM cold B1 as described in the Materials and Methods. A, a representative phosphorimage of a gel displaying the strand separating activity of WT (lanes 2–3 and 9–10), RAPA RECQL4 (lanes 4–5 and 11–12), and K508M RECQL4 (lanes 6–7 and 13–14) in the absence and presence of ATP is shown. The Δ symbol denotes the heat denatured DNA substrate. The position of the single stranded and forked dsDNAs are denoted by the images on the left side of the gel. B and C, graph of the quantification of three independent gels each in the absence and presence of ATP, respectively. The mean and standard deviation are reported.
With ATP present, WT and RAPA RECQL4 showed a similar level of activity at 20 nM whereas RAPA RECQL4 had slightly less activity at 50 nM protein (Fig. 4A, lanes 9 & 10 compared to lanes 11& 12, and Fig. 4C). The helicase dead K508M mutant showed reduced strand separating activity, but was clearly active in the assay despite the inactivating mutation within the helicase domain (Fig. 4C). The results with our K508M mutant are consistent with those reported for the K508R and D605A mutants discussed above. Thus, the addition of excess single stranded DNA appears to have restored the apparent unwinding activity of RAPA RECQL4. As will be discussed below, this apparent strand separating activity could have been derived from the N-terminal domain of RECQL4 since this region is unchanged in RAPA RECQL4.
2.6 Loss of ssDNA-stimulated ATPase activity from RAPA RECQL4
RecQ helicases traditionally display ssDNA-dependent ATPase activity. Macris et al. previously reported that the minimum length of DNA required for ATPase activation of RECQL4 was 60 nucleotides [11]. Therefore, we measured both WT and RAPA RECQL4 ATPase activity in the absence or presence of 61-mer ssDNA (Table I). This oligonucleotide substrate was previously used by Rossi et al. to demonstrate that the K508M mutation in the conserved ATP binding pocket of RECQL4, the Walker A motif, abolished the ATPase activity of the RECQL4 [14]. In the absence of ssDNA, neither WT nor RAPA RECQL4 displayed ATPase activity, as to be expected, since RECQL4 is a DNA-stimulated ATPase (data not shown). In the presence of ssDNA, WT RECQL4 displayed a linear increase in ATPase activity proportional to the increase in protein concentration (Fig. 5A, lanes 2–4 and 5B). In contrast, the RAPA RECQL4 showed little or no ATPase activity above spontaneous ATP hydrolysis.
Table I.
Oligonucleotide Sequences
| Tl | GTAGTGCATGTACACCACACTCTTTTTTTTTTTTTTT |
| Bl | TTTTTTTTTTTTTTTGAGTGTGGTGTACATGCACTAC |
| 61 mer | GGGTGAACCTGCAGGTGGGCAAAGATGTCCTAGCAAGGCACTGGTAGAATTCGGCAGCGTC |
Figure 5. RAPA RECQL4 lacks ATPase activity.
A, a representative phosphorimage of a TLC plate showing the relative ATPase activity of WT and RAPA RECQL4. For this assay a 61-mer ssDNA (8 pmol) was incubated with either WT or RAPA RECQL4 and 1 μCi [γ-32P] ATP (PerkinElmer Life Sciences) in helicase assay buffer with 50 μM cold ATP for 1 h at 37 °C. The protein concentrations tested were 10, 50 and 100 nM. B, Graphic representation of the quantification of three independent ATPase experiments is shown. The mean and standard deviation are reported.
3.0 DISCUSSION
There has been a great deal of increased interest in RECQL4 due to the recent discovery that RECQL4 has helicase activity and because this protein is associated with three human diseases. Helicases are a family of proteins that are essential in most aspects of DNA metabolism, including DNA packaging, DNA replication, transcription and DNA repair. RECQL4 is one of five human RecQ helicases, whose exact role in the various DNA metabolic processes has yet to be determined. However, recent work supports a role for RECQL4 in DNA replication and multiple DNA repair pathways (reviewed in [33]). Mutations within RECQL4 gives rise to three separate but distinct syndromes RTS, BGS and RAPADILINO syndrome. A common RECQL4 mutation, c.1390+delT, is prevalent in RAPADILINO syndrome and some patients are homozygous for this mutation [8]. The mutation destroys a splice acceptor site, causing elimination of exon 7 from the mRNA. The resulting protein variant, RAPA RECQL4, is lacking 44 amino acids just upstream from the conserved helicase domain.
In this study, we sought to compare the biochemical properties of WT and RAPA RECQL4. Our analysis of the RAPA RECQL4 protein indicates that it has a structural change, which prevents it from functioning as a helicase or ATPase (Figs. 3 and 5). Interestingly, the strand annealing activity of the protein is left intact, with activity levels comparable to that of WT RECQL4 (Fig. 2).
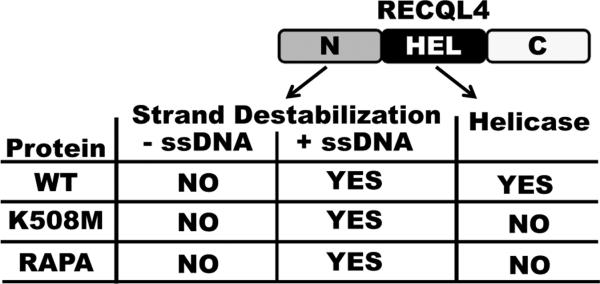
As discussed above, we have shown that on a short forked duplex substrate the RAPA RECQL4 cannot perform unwinding activity (Fig. 3). However, upon addition of excess ssDNA the protein appears to have strand separating activity (Fig. 4). Additionally, we have shown that the RAPA RECQL4 lacks ATPase activity (Fig. 5). Taken together the results support the conclusion that RAPA RECQL4 lacks helicase activity and that another domain is responsible for the apparent strand separating activity in the presence of excess ssDNA. To reconcile the strand separating assay results in the absence and presence of excess ssDNA, we propose that the N-terminal fragment 1–492 of RECQL4 is likely responsible for the observed activity when excess ssDNA is present. This domain has been previously characterized as a second DNA unwinding domain within RECQL4 [12]. Xu and Liu demonstrated that this fragment bound ATP, interacted with both ssDNA and duplex DNA and that it was able to function in the DNA unwinding assay when excess ssDNA was present [12]. Additionally, Xu and Liu reported that RECQL4 proteins harboring mutations in either of the conserved helicase Walker A (K508R) or DEAH (D605A) motifs still showed ATP-dependent strand separating activity in the presence of excess ssDNA [12]. Like these RECQL4 mutants, RAPA RECQL4 does not hydrolyze ATP, but shows apparent strand separating activity when excess ssDNA is present. However, this activity cannot be considered a true helicase because ATP hydrolysis does not occur. Therefore, we suggest it is RECQL4's ATP-independent strand annealing or exchange activity that is responsible for the apparent unwinding activity in the presence of excess ssDNA. As shown in Fig. 6, without excess ssDNA, the domain responsible for the strand destabilization is inactive. Whereas when excess ssDNA is present, the strand annealing activity works and would collaborate with the helicase domain since both give rise to the same endpoint, liberated ssDNA. In contrast, for both the K508M and RAPA proteins, without ssDNA, the proteins appear to be devoid of activity because neither the helicase nor strand destabilization domain are functional. The collective conclusion is that ATP-dependent helicase activity of RECQL4 stems from its conserved helicase domain. Secondarily, there is another unwinding activity from N-terminal domain that is distinctly ATP-independent and excess ssDNA-dependent. It is possible that the N-terminal domain of RECQL4 initiates destabilization of the substrate when no excess ssDNA is present, leading to incomplete unwinding and subsequent re-annealing of the substrate. As long as the oligos re-anneal the proteins would appear to have no apparent helicase activity. In contrast, when the substrate is destabilized in the presence of excess ssDNA, this allows for hybridization of the cold strand and then strand switching can occur. All of which occurs in an ATP-independent manner. This is why the RAPA RECQL4 and the helicase dead RECQL4 proteins appear to have helicase activity in the presence of excess ssDNA (Fig. 4).
Figure 6. Comparison of RECQL4 activities in absence or presence of ssDNA.
An image of RECQL4 protein is shown. The N-terminal and helicase domains are depicted as being responsible for the strand destabilization or helicase activity, respectively. The chart below image shows which activities are functional in the RECQL4 proteins, WT, K508M and RAPA, minus or plus ssDNA.
Strand annealing is common among all human RecQ helicases, and it has been suggested that it is intrinsic to the RecQ helicase domain, because it is inhibited by ATP and non-hydrolysable ATP analogues ([2] and references sited therein). However, there is accumulating evidence that several of the RecQ helicases have domains outside of the helicase region that are capable of performing strand annealing. As discussed above, the N-terminal fragment of RECQL4 (amino acids 1–492) displays strand annealing activity [12]. A 79 amino acid region between the RQC and HRDC domains in WRN was reported to have strand annealing activity as well [32]. In a recent publication, the N-terminal domain of BLM was shown to possess strand annealing and strand exchange activity [34]. Additionally, the unique C-terminal domain of RECQL5 has been reported to have strand annealing activity [29]. Thus, four out of the five human RecQ proteins possess strand annealing domains outside of their respective helicase domains. Given that human RecQ helicases only share sequence similarity within the conserved helicase domain, it is somewhat surprising to find that the proteins share a functional attribute, such as strand annealing, without any apparent sequence similarity. Nonetheless, more work is required to determine the exact biological roles that strand annealing plays in vivo.
Altogether, patients that are homozygous for the common RAPADILINO mutation will suffer from the mislocalization of RECQL4 to the cytoplasm [9], from a failure of RAPA RECQL4 to respond to DNA damage [10], and from RAPA RECQL4's lack of helicase and ATPase activity (this paper). Type II RTS and RAPADILINO patients share short stature, radial ray defects, feeding, vomiting diarrhea and a predisposition for cancer, especially osteosarcoma [4,5]. Almost half of all the patients with RAPADILINO syndrome have developed cancer, either osteosarcomas or lymphomas [4]. Given our finding that the RAPA RECQL4 lacks helicase and ATPase activity, this lends support to the proposal that lack of helicase activity contributes to the cancer predisposition seen in both RTS and RAPADILINO patients.
Our finding may also have implications for the overlapping and discordant phenotypes observed between RTS and RAPADILINO patients. Poikiloderma and alopecia are symptoms found in RTS patients but lacking in RAPADILINO patients [4,5]. This is intriguing and thus further analysis of other patient mutations derived from both RTS and RAPADILINO patients may help elucidate the biochemical differences in RECQL4 proteins that give rise to the disorders.
4.0 MATERIALS AND METHODS
4.1 Cloning of the RAPADILINO mutation into bacterial and mammalian expression vectors
The major RAPA mutation was constructed by deleting 132 bp, encoded by exon 7, from the RECQL4 gene. A vector containing the human RECQL4 gene lacking exon 7 (pLP-exon7 del-eGFP RECQL4) was obtained from Dr. Sharon Plon (Baylor, TX) and was previously described in Burks et al. [9]. A bacterial expression construct for the human RECQL4 gene, pGEX6p1-WT RECQL4, with an N-terminal glutathione S-transferase and C-terminal 9-histidine tag was obtained from Patrick Sung (Yale, CT). Both vectors, pLP-exon7 del-eGFP RECQL4 and pGEX6p1-RECQL4-His, were digested with HindIII and KpnI. The HindIIIKpnI fragment containing the RAPA deletion was cut from pLP-exon7 del-eGFP-RECQL4 vector, gel purified then ligated into the pGEX6p1-WT RECQL4-His bacterial expression vector, thereby creating the bacterial expression vector pGEX6p1-RAPA RECQL4-His.
4.1 Protein expression and purification
WT, K508M and RAPA RECQL4 were expressed and purified from E. coli Rosetta2 (DE3) (Novagen, Gibbstown, NJ) as described previously [14]. The purity and concentrations of the proteins were determined by Simply Blue staining of polyacrylamide gels. The RECQL4 proteins were aliquoted, and stored at −80°C until use.
4.3 Protein structural stability assay
Protein stabilities were determined using differential scanning fluorimetry with Sypro Orange (Sigma Aldrich, St. Louis, MO) and a real-time PCR machine, iCycler (Bio-Rad, Hercules, CA). Sypro Orange is a dye that binds hydrophobic sites on proteins, and thus its fluorescence emission intensity increases upon protein denaturation [27,28]. The advantage of this assay is that small volumes and low protein concentrations can be utilized. Here, 1–2 μg of WT or RAPA protein were incubated with 2× Sypro Orange in a 20 μL reaction volume in 250 mM KCl, 10 mM KH2PO4, pH 7.5, 5% glycerol, 0.5 mM EDTA, 0.01% Igepal, then subjected to thermal denaturation. The temperature increased 0.5 °C every 15 sec from 20–70 °C and fluorescence emission was monitored using the iCycler melting curve software. Midpoints of unfolding transition (Tm) were determined from the thermal unfolding curves. Briefly, the background subtracted fluorescence values were converted into fraction denatured as a function of temperature and normalized by the van't Hoffs analysis [35]. The reported Tm is an average of two measurements from two independent purifications.
4.4 Oligonucleotide substrates
Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA), and sequences are listed in Table I. Substrates were radiolabeled at the 5′ end with [γ-32P] ATP (Perkin Elmer Life Sciences, Waltham, MA) and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). MicroSpin G-25 columns (GE Healthcare, Piscataway, NJ) were used to remove unincorporated [γ-32P] ATP. For those substrates that required duplex formation, the partially complementary oligonucleotide was annealed in 10 mM Tris-HCl pH 8.0, 1 mM EDTA and 50 mM NaCl at a molar ratio 1:2 of labeled to unlabeled oligonucleotide. To promote duplex formation the oligonucleotides were heated to 95°C for 10 minutes then allowed to slow cool to room temperature.
4.5 Strand annealing
The indicated amounts of each RECQL4 protein were incubated with single stranded radiolabeled T1 substrate (0.5 nM) and unlabeled complementary oligonucleotide B1 (1 nM) substrate for 20 minutes at 37 °C in a reaction volume of 10 μl containing 30 mM Tris pH 7.4, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 10% glycerol, and where indicated 5 mM ATP. Reactions were stopped by addition of 10 μl of 3× native stop dye (50 mM EDTA, 40% glycerol, 0.9% SDS, 0.05% bromophenol blue, and 0.05% xylene cyanol). The percent of annealed complementary oligonucleotide substrate generated by the RECQL4 proteins was plotted against protein concentration. The experiments were done at least three times and the average and standard deviation are shown.
4.6 Helicase assays
The indicated amounts of each RECQL4 protein were incubated with forked duplex substrate, T1/B1 (0.5 nM, Table I) for 20 minutes at 37 °C in a reaction volume of 10 μl containing 30 mM Tris pH 7.4, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 10% glycerol, and 5 mM ATP. Helicase reactions were stopped by addition of 10 μl of 3× native stop dye. For those reactions that required excess single stranded DNA, cold T1, 12.5 nM, was added, and all other reaction conditions remained the same. The reaction products were separated by electrophoresis on 10% native polyacrylamide gel, exposed to a PhosphorImager screen (GE Healthcare, Piscataway, NJ), then imaged with a Typhoon scanner (GE Healthcare, Piscataway, NJ). ImageQuant version 5.2 (GE Healthcare, Piscataway, NJ) was employed to analyze the images. The percent of the single stranded substrate that was liberated by the RECQL4 proteins was plotted against protein concentrations. The experiments were done four times, and the average and standard deviation for the four experiments are shown.
4.7 ssDNA-stimulated ATPase assay
Briefly, the RECQL4 proteins were incubated with 8 pmol of a 61 nucleotide ssDNA oligonucleotide (61-mer, Table I) and 1 μCi [γ-32P] ATP (PerkinElmer Life Sciences, Waltham, MA) in 10 μl helicase assay buffer with 50 μM cold ATP for 1 h at 37 °C. Reactions were stopped by addition of 5 μl of 0.5 M EDTA and placed on ice. Two microliters from each reaction were spotted onto a cellulose polyethyleneimine thin-layer chromatography sheet (TLC sheets, JT-Baker, Phillipburg, NJ), and resolved using a solution of 0.8 M LiCl and 1 M formic acid. The chromatography sheet was exposed to a PhosphorImager screen (GE Healthcare, Piscataway, NJ), and the percent of ATP hydrolyzed was reported. The reactions were done in triplicate, and the average and standard deviation are shown.
Highlights
-
“
RAPADILINO syndrome is associated with mutations in RECQL4. (60 characters including spaces)
-
“
We characterize RAPADILINO RECQL4's catalytic activities relative to WT RECQL4. (82 characters including spaces)
-
“
We show that RAPADILINO RECQL4 lacks helicase and ssDNA-dependent ATPase activity.
-
“
RAPADILINO and RTS patients may develop cancer due to loss of helicase activity. (84 characters including spaces)
Acknowledgements
We would like to thank Dr. Avik Ghosh and Martin Jensen for their assistance with the purification of WT RECQL4 and for editing the manuscript. We would like to thank Robert Brosh for critically reading the manuscript. We would like to thank Patrick Sung (Yale, CT) for the pGEX-RECQL4-His bacterial expression vector and Dr. Sharon Plon (Baylor College of Medicine, TX) for the pGFP-Δ exon 7 RECQL4 mammalian expression vector. This work was supported by the intramural research program of the National Institute on Aging, NIH, Z01 AG000726-20.
Abbreviations
- WT
wild type
- RAPA
RAPADILINO
- RTS
Rothmund-Thomson syndrome
- BGS
Baller-Gerold syndrome
- RQC
RecQ Helicase Conserved domain
- HRDC
Helicase and RNase D C-terminal Domain
- bp
base pair
- EMSA
electrophoretic mobility shift assay
- ssDNA
single stranded DNA
- dsDNA
double stranded DNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest The authors declare no conflicts of interest.
References
- [1].Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophys. Chem. 2010;149:67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- [3].Wang LL, Gannavarapu A, Kozinetz CA, Levy ML, Lewis RA, Chintagumpala MM, Ruiz-Maldanado R, Contreras-Ruiz J, Cunniff C, Erickson RP, Lev D, Rogers M, Zackai EH, Plon SE. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J. Natl. Cancer Inst. 2003;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- [4].Siitonen HA, Sotkasiira J, Biervliet M, Benmansour A, Capri Y, Cormier-Daire V, Crandall B, Hannula-Jouppi K, Hennekam R, Herzog D, Keymolen K, Lipsanen-Nyman M, Miny P, Plon SE, Riedl S, Sarkar A, Vargas FR, Verloes A, Wang LL, Kaariainen H, Kestila M. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Larizza L, Roversi G, Volpi L. Rothmund-Thomson syndrome. Orphanet. J. Rare. Dis. 2010;5:2. doi: 10.1186/1750-1172-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kaariainen H, Ryoppy S, Norio R. RAPADILINO syndrome with radial and patellar aplasia/hypoplasia as main manifestations. Am. J. Med. Genet. 1989;33:346–351. doi: 10.1002/ajmg.1320330312. [DOI] [PubMed] [Google Scholar]
- [7].Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria), Cancer Epidemiol. Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- [8].Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- [9].Burks LM, Yin J, Plon SE. Nuclear import and retention domains in the amino terminus of RECQL4. Gene. 2007;391:26–38. doi: 10.1016/j.gene.2006.11.019. [DOI] [PubMed] [Google Scholar]
- [10].Singh DK, Karmakar P, Aamann M, Schurman SH, May A, Croteau DL, Burks L, Plon SE, Bohr VA. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell. 2010;9:358–371. doi: 10.1111/j.1474-9726.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- [12].Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Suzuki T, Kohno T, Ishimi Y. DNA helicase activity in purified human RECQL4 protein. J. Biochem. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- [14].Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst) 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yin J, Kwon YT, Varshavsky A, Wang W. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 2004;13:2421–2430. doi: 10.1093/hmg/ddh269. [DOI] [PubMed] [Google Scholar]
- [16].Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- [17].Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp. Cell Res. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- [18].Croteau DL, Rossi ML, Canugovi C, Tian J, Sykora P, Ramamoorthy M, Wang Z, Singh DK, Akbari M, Kasiviswanathan R, Copeland WC, Bohr VA. RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell. 2012 doi: 10.1111/j.1474-9726.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].De S, Kumari J, Mudgal R, Modi P, Gupta S, Futami K, Goto H, Lindor NM, Furuichi Y, Mohanty D, Sengupta S. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J. Cell Sci. 2012 doi: 10.1242/jcs.101501. [DOI] [PubMed] [Google Scholar]
- [20].Dietschy T, Shevelev I, Pena-Diaz J, Huhn D, Kuenzle S, Mak R, Miah MF, Hess D, Fey M, Hottiger MO, Janscak P, Stagljar I. p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 2009;122:1258–1267. doi: 10.1242/jcs.037747. [DOI] [PubMed] [Google Scholar]
- [21].Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, III, Croteau DL, Bohr VA. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum. Mol. Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kumata Y, Tada S, Yamanada Y, Tsuyama T, Kobayashi T, Dong YP, Ikegami K, Murofushi H, Seki M, Enomoto T. Possible involvement of RecQL4 in the repair of double-strand DNA breaks in Xenopus egg extracts. Biochim. Biophys. Acta. 2007;1773:556–564. doi: 10.1016/j.bbamcr.2007.01.005. [DOI] [PubMed] [Google Scholar]
- [23].Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- [24].Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr., Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics. 1999;61:268–276. doi: 10.1006/geno.1999.5959. [DOI] [PubMed] [Google Scholar]
- [27].Goldberg DS, Bishop SM, Shah AU, Sathish HA. Formulation development of therapeutic monoclonal antibodies using high-throughput fluorescence and static light scattering techniques: Role of conformational and colloidal stability. J. Pharm. Sci. 2010 doi: 10.1002/jps.22371. [DOI] [PubMed] [Google Scholar]
- [28].Layton CJ, Hellinga HW. Thermodynamic analysis of ligand-induced changes in protein thermal unfolding applied to high-throughput determination of ligand affinities with extrinsic fluorescent dyes. Biochemistry. 2010;49:10831–10841. doi: 10.1021/bi101414z. [DOI] [PubMed] [Google Scholar]
- [29].Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- [31].Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- [32].Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008;47:10247–10254. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Capp C, Wu J, Hsieh TS. RecQ4: the second replicative helicase? Crit Rev. Biochem. Mol. Biol. 2010;45:233–242. doi: 10.3109/10409231003786086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen CF, Brill SJ. An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. EMBO J. 2010;29:1713–1725. doi: 10.1038/emboj.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]