Abstract
Non–small cell lung cancers (NSCLCs) that harbor mutations within the epidermal growth factor receptor (EGFR) gene are sensitive to the tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib. Unfortunately, all patients treated with these drugs will acquire resistance, most commonly as a result of a secondary mutation within EGFR (T790M). Because both drugs were developed to target wild-type EGFR, we hypothesized that current dosing schedules were not optimized for mutant EGFR or to prevent resistance. To investigate this further, we developed isogenic TKI-sensitive and TKI-resistant pairs of cell lines that mimic the behavior of human tumors. We determined that the drug-sensitive and drug-resistant EGFR-mutant cells exhibited differential growth kinetics, with the drug-resistant cells showing slower growth. We incorporated these data into evolutionary mathematical cancer models with constraints derived from clinical data sets. This modeling predicted alternative therapeutic strategies that could prolong the clinical benefit of TKIs against EGFR-mutant NSCLCs by delaying the development of resistance.
INTRODUCTION
Gefitinib (Iressa) and erlotinib (Tarceva) are first-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) that were designed as adenosine triphosphate (ATP) mimetics to block wild-type receptor activity. While being developed, these drugs were serendipitously found to be most clinically effective against those non–small cell lung cancers (NSCLCs) that harbor mutations in exons encoding the kinase domain of EGFR (1–3). Common alterations include small in-frame deletions in exon 19 (19 dels) and a point mutation within exon 21 (L858R), both of which lead to sustained activity of the kinase (4–6). More than 70% of patients with EGFR-mutant tumors treated prospectively with either TKI show tumor volume decreases, with an overall median survival of ~30 months (7–9).
Unfortunately, lung tumors in all patients eventually develop acquired resistance (7, 10). The most common mechanism of resistance is a second site mutation within exon 20 of EGFR (T790M), observed in ~50% of cases (11, 12). This change leads to altered binding of the drug within the ATP pocket (13).
Currently, targeted therapeutic options for T790M-harboring NSCLCs are limited. Second-generation EGFR TKIs [for example, HKI-272 (neratinib) and BIBW-2992 (afatinib)] are more potent than gefitinib and erlotinib against EGFR T790M (14, 15). However, because they inhibit drug-sensitive mutants at lower doses than they inhibit the T790M mutant, they still select for T790M-harboring clones in models of acquired resistance in vitro (14). Their antitumor activity in patients with acquired resistance to gefitinib and erlotinib has been disappointing (16, 17).
We hypothesized that, because clinically available EGFR TKIs were developed against wild-type EGFR, current empiric dosing regimens were not optimally designed to inhibit the EGFR mutants in NSCLC nor to minimize the development of drug resistance. Here, we have identified differences in the growth kinetics of TKI-sensitive and TKI-resistant (T790M-containing) isogenic NSCLC cells. We incorporated these findings, along with patient data, into evolutionary cancer models (18) to generate mathematical models predictive of tumor behavior. This approach identified several strategies to improve the treatment of EGFR-mutant NSCLC before and after the emergence of T790M-mediated acquired resistance.
RESULTS
Derivation of EGFR-mutant TKI-resistant lung adenocarcinoma cells
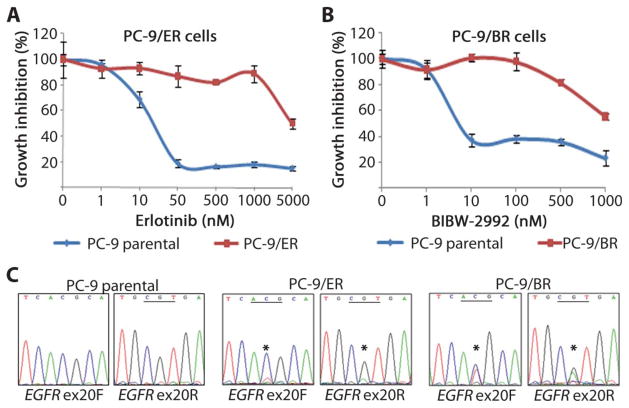
To determine the physical characteristics of TKI-sensitive and TKI-resistant cells, we derived in vitro cellular models of T790M-mediated resistance using EGFR-mutant TKI-sensitive PC-9 cells (19 del), well-established TKI dose-escalation protocols (14, 19, 20), and either a reversible quinazoline (erlotinib) or an irreversible quinazoline (BIBW-2992) that binds covalently to C797 in EGFR. After 120 days in culture, PC-9 cells resistant to erlotinib and BIBW-2992 emerged that grew in drug concentrations ~100-fold (5 μM) and ~1000-fold (500 nM) the initial IC50 (median inhibitory concentration), respectively, of the parental cells (Fig. 1, A and B). On comparative genomic hybridization arrays, the EGFR locus appeared further amplified in erlotinib-resistant (ER) and BIBW-2992–resistant (BR) cells compared to parental cells (fig. S1, A and B). Fluorescence in situ hybridization (FISH) analyses indicated that EGFR alleles were not amplified on double-minute chromosomes, as reported in other studies (21) (fig. S1C). The resistant cells had no evidence of MET amplification, another mechanism of acquired resistance to EGFR TKIs (fig. S1, A and B) (20, 22). No other obvious amplifications or deletions were found.
Fig. 1.
Derivation and characterization of TKI-resistant cells. (A and B) PC-9 erlotinib-resistant cells (PC-9/ER) (panel A) and PC-9 BIBW-2992–resistant cells (PC-9/BR) (panel B) were derived after ~120 days of culture with increasing concentrations of drug. Growth inhibition assays show that these cells are resistant to respective TKIs compared to the parental cells. (C) Direct dideoxynucleotide sequencing chromatograms from EGFR exon 20 (ex20) show the presence of the T790M mutation (*ACG→ATG) in the PC-9/ER and PC-9/BR cells but not in parental cells. F, forward; R, reverse directions.
DNA from polyclonal PC-9/ER and PC-9/BR cells harbored the T790M allele plus the primary drug-sensitizing exon 19 del (Fig. 1C). No other mutations were found within any coding exons of EGFR. Analysis of cloned PC-9/BR complementary DNA (cDNA) products generated by reverse transcription–polymerase chain reaction (RT-PCR) showed that the T790M mutation was in cis with the primary drug-sensitive mutation. Signaling within the EGFR pathway was minimally affected by erlotinib in the PC-9/ER cells or by BIBW-2992 in the PC-9/BR cells (Fig. 2C and fig. S1D). Phospho-receptor tyrosine kinase arrays showed grossly similar profiles for PC-9/ER and PC-9/BR cells (fig. S1E).
Fig. 2.
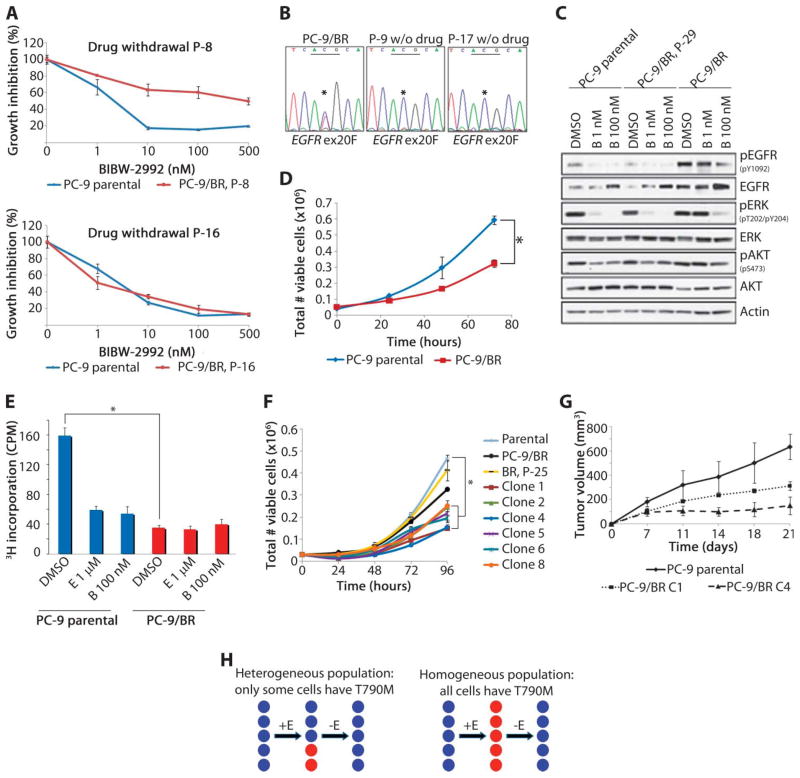
Growth characteristics of TKI-sensitive and TKI-resistant cells. (A) Polyclonal PC-9/BR cells cultured without BIBW-2992 for 8 and 16 passages (P-8 and P-16, respectively) regained intermediate and complete sensitivity to BIBW-2992, respectively. (B) Sequencing of EGFR exon 20 (ex20) showed a decrease in the T790M allele that correlates with restored TKI sensitivity. Genomic DNA was extracted from cells after 9 and 17 passages (P-9 and P-17, respectively) without drug (*ACG→ATG). (C) Parental and late-passage resistant cells (P-29) show decreased phosphorylation of EGFR and its downstream targets in the presence of BIBW-2992, whereas signaling in the PC-9/BR cells remained intact. Cells were treated with vehicle (DMSO) or BIBW-2992 (B) for 3 hours. (D) T790M-containing PC-9/BR cells proliferated more slowly than parental cells over 72 hours in the absence of inhibitor. Graphs represent the average of triplicate wells ± SD. *P < 0.01. (E) [3H]Thymidine incorporation confirms the slower proliferation rate of the PC-9/BR cells compared to parental cells. Cells were treated with DMSO, erlotinib (E), or BIBW-2992 (B) for 24 hours. Data are expressed as counts per million (CPM) relative to each other. (F) Cell counts for PC-9 parental cells, BR (polyclonal), BR late-passage (P-25), and T790M-containing BR clones (1, 2, 4, 5, 6, 8) show that the clones grow more slowly than parental and P-25 cells. (G) PC-9 parental and PC-9/BR cells (clones 1 and 4) were injected subcutaneously into nude mice, and tumor growth in the absence of drug was monitored over time. The slower growth pattern of T790M-harboring PC-9/BR clones 1 and 4 is maintained in vivo. Data are average tumor volumes (n = 3 per group) ± SEM. (H) At the onset of acquired resistance, an EGFR-mutant tumor (blue) develops T790M in a small proportion of cells (red) after exposure to erlotinib (E; left). Upon withdrawal of drug, previously growth-arrested TKI-sensitive cells repopulate the tumor. Alternatively (right), all cells contain some level of T790M at progression. Upon discontinuation of the inhibitor, all cells revert back to parental genotype.
Restoration of drug sensitivity after EGFR TKI withdrawal
We cultured resistant polyclonal cells in the absence of drug. After eight passages, PC-9/BR cells regained partial sensitivity to BIBW-2992 (Fig. 2A, upper panel). By 16 passages, drug sensitivity was restored to parental levels (Fig. 2A, lower panel). Loss of resistance corresponded with a decrease in the proportion of the T790M allele (Fig. 2B). Consistent with these data, lysates from parental cells and late-passage PC-9/BR–resistant cells treated with BIBW-2992 showed significantly reduced phosphorylation of EGFR and its downstream targets, extracellular signal–regulated kinase (ERK) and AKT, whereas lysates from resistant cells maintained in the presence of TKI and treated with the same concentrations of drug did not (Fig. 2C).
To extend our observations, we examined other EGFR-mutant isogenic pairs of drug-sensitive and drug-resistant lung cancer cell lines. Like PC-9 cells, HCC827/ER cells (19 del + T790M) regained TKI sensitivity after multiple passages in the absence of inhibitor and lost the T790M allele (fig. S2, A and B). By contrast, H3255 (L858R) cells, which also acquired T790M in response to continuous TKI exposure, neither regained TKI sensitivity nor lost the T790M after multiple passages in the absence of inhibitor, and they grew at the same rate as the parental line (fig. S3). Thus, two of three lines studied displayed resensitization. We characterized representative PC-9/BR cells in more detail.
Growth of parental cells and EGFR-mutant cells with T790M
To investigate the growth properties of drug-sensitive and drug-resistant cells, we counted the total number of viable cells in culture after plating each cell cohort in the absence of drug. To avoid contact inhibition as a confounding factor, cultures were not allowed to reach confluence. The parental cultures always had more cells (Fig. 2D) and showed more DNA synthesis (Fig. 2E) than did PC-9/BR cells. On average, parental cells doubled ~1.22 times faster than T790M-containing resistant cells. PC-9/BR cells withdrawn from drug for 25 passages (P-25) displayed parental growth kinetics (Fig. 2F). PC-9/ER cells harboring T790M followed the same slower growth pattern as PC-9/BR cells (fig. S2C).
To confirm these observations, we established single-cell clones from both PC-9/BR and PC-9/ER cell lines. Six T790M-containing clones were derived in the presence of BIBW-2992 from the PC-9/BR cells. All clones displayed slower growth kinetics compared to parental cells or polyclonal resistant cells that had been passaged 25 times without drug. PC-9/BR clones also grew more slowly than the resistant polyclonal population maintained under selective pressure (Fig. 2F). Four T790M-harboring clones were derived in the presence of erlotinib from PC-9/ER cells and showed analogous characteristics (fig. S2, D and F). The growth properties of parental cells and PC-9/BR–resistant clones were also maintained in vivo (Fig. 2G). We saw no differences in cell cycle profiles, rates of apoptosis, or rates of senescence between parental and resistant cells. Notably, the T790M-harboring cells described here are distinct from the recently described subpopulation of EGFR-mutant cancer cells lacking T790M that transiently exhibit a distinct phenotype characterized by the engagement of insulin-like growth factor 1 receptor (IGF-1R) activity, hypersensitivity to histone deacetylase (HDAC) inhibition, altered chromatin, and an intrinsic ability to tolerate drug exposure (23).
Collectively, these results can be explained by two scenarios (Fig. 2H). First, tumor cell populations with acquired resistance are composed of a heterogeneous mixture of cells, some of which harbor the T790M allele. After drug withdrawal, previously growth-arrested but faster-growing TKI-sensitive cells repopulate the tumor, and the population of cells displays resensitization. In the second possibility, tumor cell populations with acquired resistance are composed of a homogeneous group of resistant cells; after drug withdrawal, every cell loses the T790M allele and the population becomes resensitized. Several lines of evidence support the former possibility. Polyclonal populations of resistant cells “lose” the T790M allele after passages in vitro in the absence of inhibitor. By contrast, single-cell clones harboring T790M retain the allele even after multiple passages in the absence of drug in vitro (fig. S2, E and F) and as xenografts (up to 78 days) (Fig. 2G and fig. S2G). Retention of the T790M allele in the single-cell clones, both in vitro and in vivo, suggests that acquisition of that allele is not reversible within individual cells, making the second scenario less likely.
Effect of the percentage of T790M clones on the sensitivity and growth of a population of mixed tumor cells
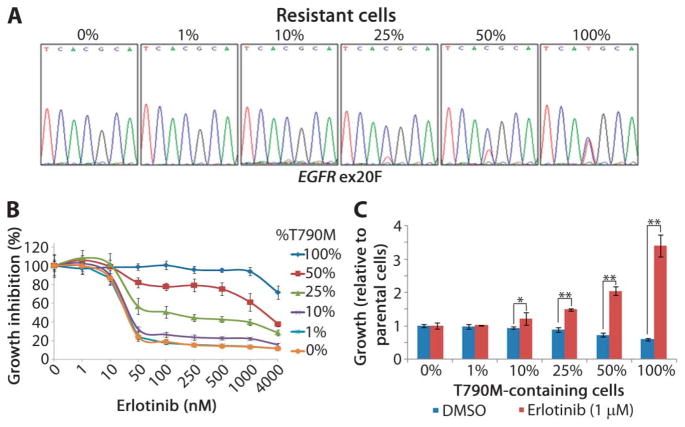
Biopsies of growing lesions in patients do not provide information on whether a tumor consists of a heterogeneous mix of sensitive and resistant cells or a homogeneous mass of only resistant cells (Fig. 2H), because which cells harbor the T790M mutation cannot be ascertained directly. To estimate the proportion of T790M-containing cells within a population necessary for the entire population to display resistance, we performed reconstitution experiments. T790M-harboring clonal cells (PC-9/BR clone 1) were mixed with TKI-sensitive parental cells at known percentages (Fig. 3A), and cell mixture drug sensitivity was measured by growth inhibition assays. Populations with small percentages of resistant cells (1 and 10%) displayed similar sensitivity to erlotinib as parental cells (0%), whereas sensitivity was reduced when T790M clones made up >25% of the population (Fig. 3B). These data can explain why patients whose tumors harbor low levels of T790M can still undergo an objective radiographic response to EGFR TKI treatment (24, 25).
Fig. 3.
Reconstitution experiments to study T790M-mediated resistance. (A) T790M-containing PC-9 cells (BR, clone 1) were spiked into parental PC-9 cells at various proportions. The increased proportion of the T790M allele (*ACG→ATG) is evident from representative direct sequencing chromatograms of EGFR exon 20. (B) Mixed populations of cells were treated with increasing concentrations of erlotinib for 72 hours, at which point growth inhibition was measured. (C) Cell populations with varying proportions of T790M-containing cells were grown in the presence of DMSO or erlotinib (1 μM) to mimic various states of a TKI-resistant heterogeneous solid tumor. Total cell number was determined after 72 hours and graphed as the percent growth compared to parental cells (0%) ± SD. *P < 0.05; **P < 0.01.
We next estimated the percent of resistant cells needed in a population of cells to display tumor growth. Cell mixtures were treated with dimethyl sulfoxide (DMSO) or 1 μM erlotinib for 72 hours (Fig. 3C). The addition of erlotinib did not alter the growth of cell populations with low proportions of resistant cells (1%). However, compared to DMSO-treated cells, populations containing greater than 10% T790M-positive cells proliferated faster than parental cells (0%) in the presence of drug. These findings are further consistent with the notion that tumor cell populations with acquired resistance to EGFR TKIs can be composed of heterogeneous tumor cell mixtures.
Biological properties of sensitive and resistant cells compared to EGFR-mutant NSCLC in human patients
The T790M substitution confers synergistic kinase activity and transformation potential when combined with drug-sensitive EGFR mutations (5, 14). We had therefore expected that resistant clones harboring the T790M allele would display a growth advantage compared to parental drug-sensitive cells. Our results to the contrary prompted us to examine the clinical course of patients with EGFR-mutant NSCLC to verify that our preclinical findings reflected the human disease.
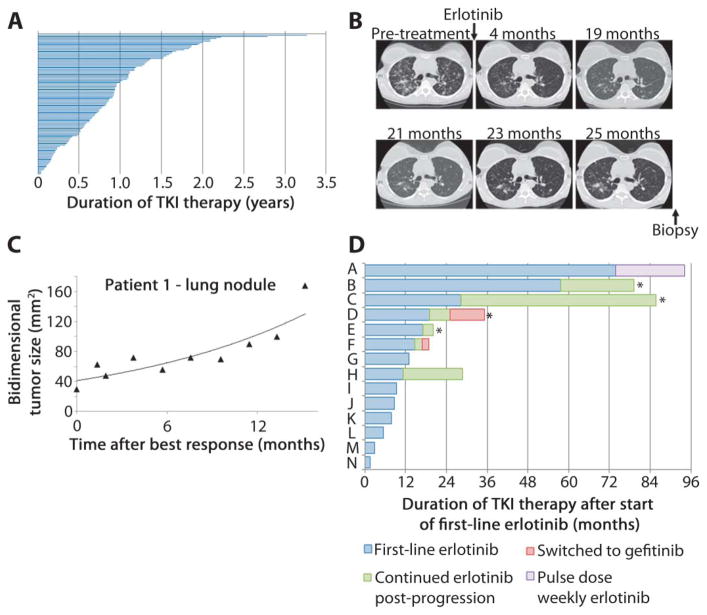
First, we asked what percentage of patients with EGFR-mutant tumors could remain on long-term EGFR TKI therapy. We analyzed unpublished data from patients enrolled in NEJ002, a prospective trial for patients with untreated metastatic EGFR-mutant tumors (7), to evaluate the range of time on TKI therapy. Although the median duration on gefitinib was 0.83 years, the range was as high as 3.3 years (Fig. 4A). Patients (32, 4, and 1%) were on drug for 1, 2, and 3 years, respectively. These data are consistent with the notion that some EGFR-mutant tumors display indolent progression.
Fig. 4.
Indolent progression of T790M-harboring patient tumors. (A) A subset of patients with EGFR-mutant tumors (n = 114) treated with first-line gefitinib (7) displayed prolonged responses to treatment. The average time on gefitinib before progression was 0.9 years. (B) Serial computed tomography scans from a patient with an EGFR-mutant tumor (ex 19 del) [images from (11)]. (C) Serial bidimensional measurements taken from the time of best response for the patient in panel B illustrate the slow rate of progression in this lesion. (D) Patients receiving first-line erlotinib as part of a phase II trial. Four of 14 patients (28%; patients B, C, D, and H) were continued on treatment with single-agent TKI (erlotinib or gefitinib) for >6 months after RECIST progression. Asterisk denotes the presence of T790M.
Second, we examined the prospective clinical course of patients with EGFR-mutant tumors and T790M-mediated acquired resistance. We extracted unpublished data from a clinical trial in which a cohort of 14 patients was treated prospectively with erlotinib (26). There were four patients whose tumors had a documented T790M mutation at the time of disease progression, two of whom had measurable disease amenable to analysis. Both patients displayed slow growth of the T790M-harboring lesion (Fig. 4, B and C, and fig. S4). In the first case, the patient was biopsied when she was deemed to have progression of disease after 25 months (11). Analysis of previous computed tomography (CT) scans indicated that the tumor began to grow at least 6 months before the biopsy and that it grew slowly from the time of maximal response (Fig. 4C). The second patient showed similar findings (fig. S4A). By comparison, a similar analysis of two patients with EGFR wild-type tumors that progressed after receiving benefit from first-line chemotherapy (27) showed that both displayed rapid tumor growth from the time of maximal response to the time when criteria for progressive disease were met (fig. S4B). Notably, the median time to progression on chemotherapy is ~4 months in unselected NSCLC but more than 9 months on erlotinib for EGFR-mutant tumors (10, 28).
Third, using unpublished data from the same prospective erlotinib study that was used for image analysis (26), we asked what proportion of patients displayed progression of disease on erlotinib but continued a TKI as a result of indolent tumor growth (Fig. 4, B and C, and fig. S4A). Among 14 patients, 4 patients continued on single-agent TKI for at least 6 months beyond disease progression, because they were relatively asymptomatic (Fig. 4D). Three of the four patients had biopsies within 2 months of coming off study, and all three (patients B, C, and D) harbored the T790M mutation. T790M was also detected in the re-biopsy specimen from one patient despite the addition of chemotherapy to continuous erlotinib treatment.
Fourth, we examined the frequency of the T790M allele by using 454 sequencing of DNA extracted from 16 untreated early-stage resected EGFR-mutant NSCLCs (Table 1). From mutant allele dilution experiments, the limit of detection of the 454 method was ~0.2%. We did not detect the T790M allele in any of the resected specimens or in parental PC-9 cell DNA. These data are consistent with our evolutionary modeling results (see below) and demonstrate that in the absence of TKI selection, the T790M allele is either absent or highly infrequent (less than 1 in 500).
Table 1.
454 sequencing of EGFR exon 20. About 100,000 454 reads per sample were generated from PCR products generated with EGFR exon 20 (ex20)–spanning primers. All tumors were from treatment-naïve patients. TKI-R-1 and TKI-R-2 were run as positive control samples. 1°mutn, primary EGFR mutation, as assessed by direct sequencing.
| Sample | Stage | 1°mutn | 454 ex20 T790M (%) |
|---|---|---|---|
| Lung TKI-R-1 | IV | 19 DEL | Y (59%) |
| Lung TKI-R-2 | IV | L858R | Y (1.07%) |
| H3255 | Cell Line | L858R | N |
| PC-9 | Cell Line | 19 DEL | N |
| 130T | IA | 19 DEL | N |
| 169T | IA | 19 DEL | N |
| 230T | IA | 19 DEL | N |
| 474T | IA | 19 DEL | N |
| 631T | IA | 19 DEL | N |
| 20T | IB | 19 DEL | N |
| 734T | IB | 19 DEL | N |
| 739T | IB | 19 DEL | N |
| 388T | IIIA | 19 DEL | N |
| 3T | IA | L858R | N |
| 5T | IA | L858R | N |
| 485T | IA | L858R | N |
| 570T | IB | L858R | N |
| 685T | IB | L858R | N |
| 25T | IB | L858R | N |
| 166T | IB | L858R | N |
Finally, numerous published reports support our preclinical data: (i) EGFR-mutant tumors can “flare” after patients stop EGFR TKI treatment (29); (ii) serial biopsies over the course of treatment demonstrate a decrease in prevalence of the T790M allele during the period off therapy (30); (iii) EGFR-mutant cancers that recur after stopping adjuvant TKI do not harbor the T790M mutation, suggesting a growth disadvantage to these clones (31); (iv) EGFR-mutant tumors with documented progression can re-respond to an EGFR TKI after a hiatus off TKI therapy (30, 31); (v) patients with EGFR-mutant tumors and T790M-mediated acquired resistance paradoxically have a better survival than those with acquired resistance and no T790M (32, 33); and (vi) ultra-sensitive locked nucleic acid technology (LNA-PCR; limit of detection ~0.1%) was unable to detect T790M in TKI-naïve samples, half of which harbored the mutation upon progression (33). Collectively, these data all support the notion that tumors with acquired resistance to gefitinib or erlotinib may be composed of mixed populations of cells, and continued TKI selection is needed to promote the outgrowth of slower-growing T790M-mutant cells.
Evolutionary cancer modeling applied to EGFR-mutant NSCLC
Having established the clinical relevance of our preclinical models, we applied evolutionary cancer modeling, involving detailed mathematical descriptions of our tumor cell populations in vitro, to design optimized dosing strategies for EGFR-mutant NSCLC. We incorporated pharmacokinetic data from human clinical trials with erlotinib to ensure that the drug doses proposed were clinically achievable in humans (34, 35).
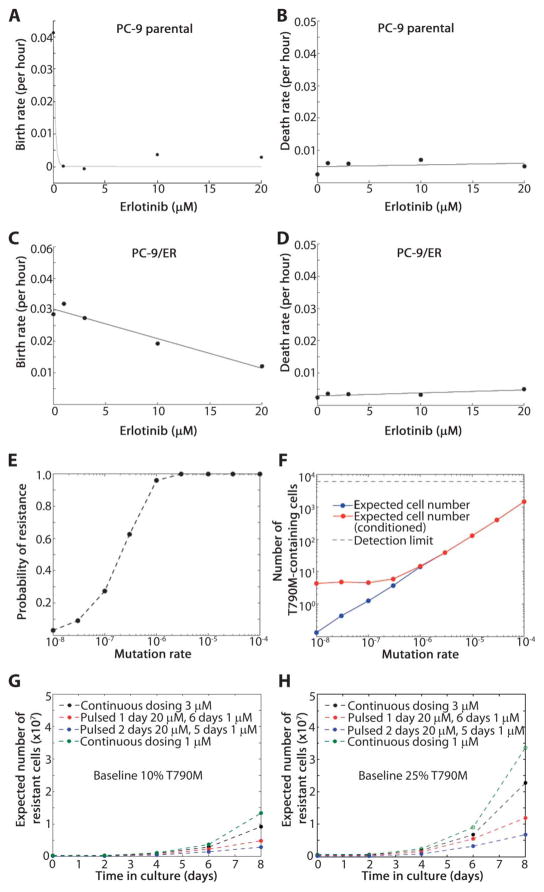
First, we tested whether this approach could be used to determine the potential frequency of T790M-containing clones within a population of untreated PC-9 cells. We measured the growth and death rates of PC-9 and PC-9/ER cells (see Materials and Methods) cultured in the presence of various doses of erlotinib (fig. S5) and modeled the drug-sensitive and drug-resistant cell populations as a multitype binary branching process (36). Application of the experimentally determined estimates of viable cells as well as apoptosis in the presence and absence of drug generated fitted curves describing the birth and death rates of both cell populations as a function of the concentration of erlotinib (Fig. 5, A to D). These curves were used to estimate the number of resistant cells present in a population of 3 million cells [an estimate of the number of cells in a ~1-cm tumor (37)] that initiated from a single cell harboring only a drug-sensitive EGFR mutation and that grew in the absence of drug. The probability that at least one resistant cell exists in the 3 million cell population (Fig. 5E) ranged from 3% (for a mutation rate of 10−8 per TKI-sensitive cell division) to 100% (for mutation rates above 10−6). We also estimated the number of T790M-containing cells expected when the population reaches 3 million cells for a range of mutation rates (10−4 to 10−8) (Fig. 5F); we determined the expected number of cells both averaged over all cases and averaged over the subset of cases in which at least one resistant cell was present. By the standard estimate of mutation rates per base pair per cell division (10−8 to 10−7) (38, 39), cells with T790M in the final population would be about 1 cell out of 3 million. These data were consistent with our 454 deep sequencing results (Table 1), which showed that T790M was rare (<0.2%) in untreated early-stage EGFR-mutant tumors.
Fig. 5.
Evolutionary cancer modeling predictions to delay the development of resistance. (A) PC-9 cell birth rate at erlotinib concentrations of 0, 1, 3, 10, and 20 μM. (B) PC-9 cell death rate as a function of increasing erlotinib concentration. (C) PC-9/ER cell birth rate in the presence of erlotinib (0, 1, 3, 10, and 20 μM). (D) PC-9/ER cell death rates as a function of increasing erlotinib concentration. (E) Probability of preexisting T790M-harboring cells in a population of 3 million cells initiating from one cell harboring just a drug-sensitive EGFR mutation that grew in the absence of drug for a range of mutation rates (10−4 to 10−8 per cell division). (F) Expected number of resistant cells present in the population, both averaged over all cases and averaged only over the subset of cases, where at least one resistant cell is present. (G) An initial population of 750,000 cells, 10% of which harbor T790M, treated with continuous low-dose erlotinib (1 and 3 μM) selects for the emergence of T790M-harboring cells (green and black lines). The addition of one or two high-dose erlotinib “pulses” (20 μM) followed by 1 μM for the remaining days of a 7-day cycle decreases the expected number of resistant cells (red and blue lines). (H) Analogous results as in panel (G) starting with an initial population with 25% T790M-harboring cells.
Second, we used mathematical modeling to predict how long it would take to restore drug sensitivity in the PC-9/BR polyclonal resistant cell population, based on our cell growth and death rates. Using the observed doubling times for sensitive cells and resistant cells of ~19 hours and ~23 to 25 hours, respectively, we estimated that after drug withdrawal, about 35 to 40 days would be needed for a population of cells with 87.5% resistant cells to have only 1% resistant cells. These data fit well with the observation that between 8 and 16 passages (with 1 passage every 3 days) were required to restore full sensitivity after drug withdrawal (Fig. 2). Collectively, these data demonstrate that evolutionary cancer modeling could accurately describe and predict our biologically observed phenomena and were consistent with the presence of a mixed cell population in resistant tumors (Fig. 2H).
Modeling-predicted delay of resistance by high-dose pulses combined with a continuous low dose of TKI
We then used mathematical modeling to predict the relative effects of alternative dosing schedules on the development of resistance. Under the previously mentioned pharmacokinetic constraints, we hypothesized that intermittent high-dose pulses of erlotinib (20 μM) in conjunction with a continuous low-dose administration (1 μM) could be a tolerated dosing schedule to delay the establishment of large resistant cell populations. Using the cell growth and death rates calculated in the presence of 1 and 20 μM erlotinib, we determined the number of PC-9/ER cells expected under various pulsed continuous treatment schedules, starting from an initial population of 750,000 cells with 10 and 25% of the population initially containing T790M (Fig. 5C). Modeling predicted that the use of continuous low-dose treatment with simultaneous high-dose pulsed administration of erlotinib should delay the acquisition of T790M-mediated resistance (Fig. 5, G and H).
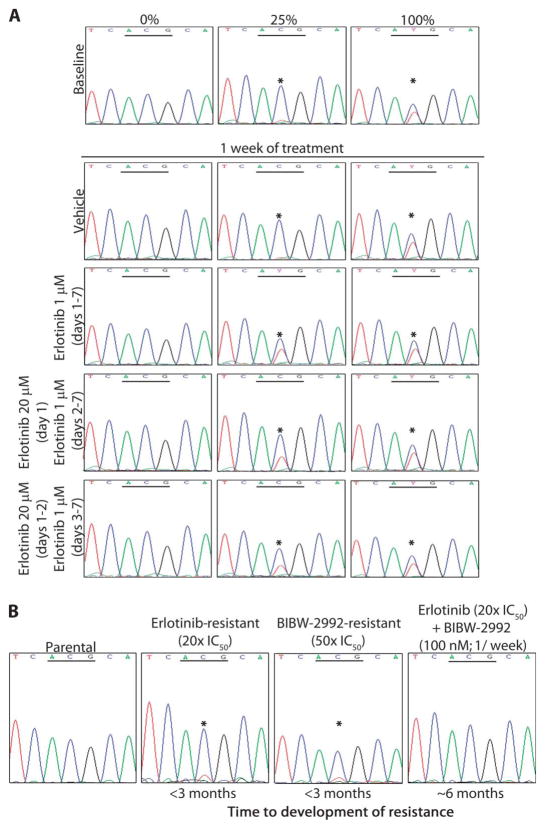
To corroborate the mathematical predictions, we applied the calculated dosing schedules to our cell lines. Using T790M-harboring clones mixed with parental cells, we treated cell populations with 0, 25, or 100% resistant cells with erlotinib at the indicated doses for 7 days (one cycle, as modeled in Fig. 5H). As expected, T790M mutations were not selected for in the absence of drug in the population with 25% resistant cells but were enriched for when 1 μM erlotinib was administered daily (Fig. 6A). Addition of one or two high-dose pulses of erlotinib (20 μM) together with daily low-dose erlotinib (1 μM) also selected for T790M-harboring cells in the mixed population. However, the frequency of the mutant allele was lower than with daily dosing (Fig. 6A), consistent with our mathematical predictions (Fig. 5G) and with the notion that this regimen could delay the acquisition of T790M-mediated resistance.
Fig. 6.
Effect of pulsed high-dose TKI treatment on the number of T790M-harboring cells. (A) Using a T790M-harboring clone (PC-9/BR, c1), we mixed cell populations to have 0, 25, or 100% resistant cells. The baseline panel shows forward sequence tracings from exon 20 of EGFR (the underlined codon encodes T790). Cell populations were then treated with erlotinib at the indicated doses for 7 days. (B) Chromatograms display exon 20 forward sequences of EGFR from PC-9 cells treated with different drugs (*ACG→ATG).
To circumvent the limitations of erlotinib dosing (maximum of 20 μM) and to apply our predictions to a long-term model, we substituted BIBW-2992, which is more potent against the T790M mutant, with an IC50 of about 100 nM (15). This concentration of drug can be achieved in humans at the standard dose of 40 mg daily (40). We used the same continuous dose escalation protocol as we used for erlotinib and BIBW-2992 to select for T790M-mediated resistance in PC-9 cells. Whereas T790M was detectable with low concentrations of BIBW-2992 and erlotinib alone, the combination of high-dose weekly BIBW-2992 plus continuous erlotinib did not select for this mode of resistance (Fig. 6B). Furthermore, the total time for the development of complete resistance (100- to 1000-fold the initial IC50) was twice as long for cells treated with the pulsed dosing regimen. The mechanism of resistance in these cells remains under investigation. These data confirm the modeling predictions and show that resistance can be delayed with combination of a pulsed high-dose potent TKI and continuous low doses of erlotinib.
Modeling-predicted tumor cell control after the emergence of EGFR T790M by continuation of TKI therapy
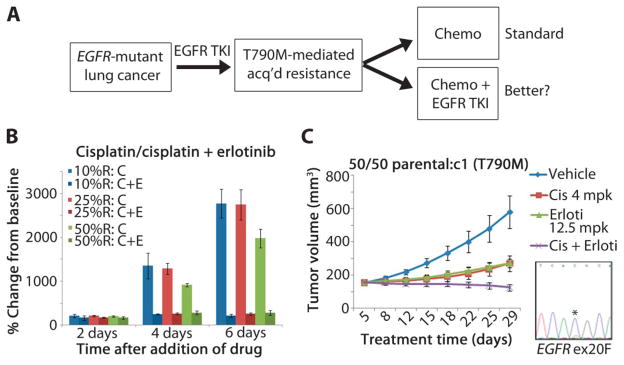
We next used our models to predict better treatment strategies for patients whose tumor cell populations have developed T790M-mediated resistance. In standard oncology practice, progression while on a specific therapy leads to cessation of that therapy and initiation of a new treatment. However, our data suggest that resistant tumors may be composed of a heterogeneous mix of TKI-sensitive and TKI-resistant cells (Fig. 3) and that stopping TKI therapy may permit expansion of the faster-growing TKI-sensitive cells. We modeled this scenario in vitro by diluting PC-9 T790M-containing clonal cells in parental cells at various concentrations, as described above. Cell populations were treated with physiologically achievable doses of chemotherapy alone or chemotherapy plus erlotinib, and cell numbers were counted every 48 hours (Fig. 7A). We used two chemotherapeutic agents with activity in lung cancer: a platinum-based drug, cisplatin (41), and an antifolate, pralatrexate (42). The latter was used rather than the chemically related pemetrexed, because pralatrexate is more stable after reconstitution.
Fig. 7.
Effect of continuation of TKI therapy with chemotherapy on heterogeneous TKI-resistant tumors. (A) Schematic outline of treatment options for patients with EGFR-mutant disease. (B) PC-9/BR c1–resistant cells were diluted in parental cells at various concentrations (see Fig. 3) and treated with chemotherapy (cisplatin, 500 nM) or chemotherapy plus erlotinib (3 μM). In all cases, the TKI-chemotherapy combination was more efficacious at inhibiting cell growth. (C) Athymic nude mice with established tumors (50:50 mixture of PC-9 parental and BR c1 cells) were administered vehicle, cisplatin (4 mg/kg), erlotinib (12.5 mg/kg), or cisplatin plus erlotinib. At the start of treatment, T790M-containing cells made up ~25% of the population, as measured by direct sequencing (bottom right). Tumor volumes were graphed as averages (n = 5 per group) ± SEM. mpk, mg/kg. * indicates residue of interest.
Cells treated with cisplatin (500 nM) (28) grew slower in the presence of erlotinib, both in vitro and in vivo (Fig. 7, B and C). Similar in vitro results were obtained with pralatrexate (100 μM) (43) (fig. S6). Collectively, these data suggest that patients may benefit from continued treatment with an EGFR TKI, even after developing T790M-mediated progression of disease.
DISCUSSION
All patients with metastatic EGFR mutant–harboring lung adenocarcinomas will eventually develop acquired resistance if treated with the EGFR TKIs gefitinib and erlotinib. In ~50% of cases, tumor cells harbor a second mutation in the EGFR kinase domain (T790M), which alters a gatekeeper residue within the ATP-binding pocket. Because existing treatment schedules were established empirically with drugs developed against wild-type EGFR, we hypothesized that evolutionary cancer modeling could be used to develop more optimal dosing strategies against the mutant receptors in NSCLC. We combined in vitro cell culture experiments, multiple clinically relevant data sets, and mathematical modeling to describe tumor behavior. We then used the models to predict dosing strategies that were validated in vitro and in vivo. These dosing regimens will need to be further validated in clinical trials with patients with EGFR-mutant lung cancer. We propose the use of high-dose pulsed once-weekly BIBW-2992 with daily low-dose erlotinib to delay the emergence of T790M-mediated resistance. PC-9 cells treated with this regimen required twice as long to develop resistance and did not show selection for T790M mutations. In patients, the combination of two EGFR TKIs could lead to overlapping toxicities involving rash and diarrhea. Thus, in a phase IB dose-safety trial, we would recommend a more tolerable strategy, with lower doses of erlotinib still known to be effective against EGFR-mutant tumors (25 or 50 mg daily, orally) (44). For BIBW-2992, we would suggest starting at 40 mg once a week and escalating to the maximum tolerated dose, aiming to achieve as high a concentration of the drug in patients as possible.
We determined that tumors with acquired resistance likely harbor mixed populations of drug-sensitive and drug-resistant cells with differential growth rates. To treat these tumors, we would propose continuing EGFR TKI suppression with chemotherapy beyond T790M-mediated progression for maximal disease control, based on the benefits of this approach in both in vitro and in vivo models. Such practice would be analogous to what is done for HER2-positive breast cancer patients who continue receiving the anti-HER2 antibody trastuzumab even after the development of progressive disease (45).
Our findings raise a paradox involving the T790M mutation. Surrogate kinase assays in Sf9 transfectants and transformation assays in fibroblasts showed that the addition of the T790M mutant to a drug-sensitive mutant confers synergistic oncogenic activity (5, 46). Yet, our preclinical data demonstrate that acquisition of the T790M mutation is associated with a growth disadvantage in the absence of TKI selection. One explanation is that the oncogenic activity of double-mutant EGFRs can be toxic to lung adenocarcinoma cells when the protein is expressed at a certain level. This hypothesis is supported by our own observations that transfectants with lower levels of the double-mutant receptor are spontaneously selected for over time (fig. S7A). Other lung cancer cells expressing a different gatekeeper mutation also display a growth disadvantage (fig. S7B). How double-mutant EGFR signaling leads to slower growth rates remains an area of investigation.
Although our preclinical data are supported by many data sets of patients with resectable early-stage to metastatic late-stage EGFR-mutant tumors, the slower growth rate of T790M-harboring cells in our in vitro models may not be representative of all T790M-positive cells in human patients. Consistent with this, one of the three isogenic pairs of EGFR drug-sensitive/drug-resistant cell lines that we developed did not display resensitization after drug withdrawal. Other factors, such as fibroblast growth factor receptor (FGFR), IGF-1R, and nuclear factor κB (NFκB) signaling, may also modulate responses to EGFR TKIs (47–49).
The characteristics of T790M-harboring NSCLC may be broadly applicable to other tumor types. For example, in gastrointestinal stromal tumors (GISTs) harboring imatinib-sensitive activating mutations in KIT, “flares” have been observed after imatinib cessation upon progression (50), and a recent clinical study showed that patients with documented imatinib resistance can re-respond to imatinib after a period off TKI therapy (51). Furthermore, an analogous ABL gatekeeper mutation (T315I) observed in imatinib-resistant chronic myelogenous leukemia (CML) cells decreases in abundance after imatinib therapy is stopped (52).
We predict that in patients with acquired resistance whose disease begins to accelerate rapidly, genetic alterations other than just the presence of the EGFR T790M mutation may play a role in tumor progression. That is, a third hit could enable escape from a slower growth phenotype and contribute to accelerated disease progression. Candidate third-hit genetic alterations remain to be identified.
In summary, evolutionary cancer modeling coupled with an understanding of the unique biological properties of TKI-sensitive and TKI-resistant cells has allowed us to propose optimized dosing schedules for the treatment of EGFR-mutant lung cancer. This approach could be more generally applied toward the optimization of dosing strategies of other targeted therapies used against oncogene-driven cancers.
MATERIALS AND METHODS
Patient samples and data
Tumor specimens were obtained with patients’ consent under Memorial Sloan-Kettering Cancer Center (MSKCC) Institutional Review Board–approved protocols. Samples were frozen and stored at −80°C in institutional tumor banks.
CT scans from four patients treated prospectively with erlotinib (26) who then developed T790M-containing tumors were available for analysis (11). Two patients had a complete response with no residual measurable disease, leaving two patients evaluable. The presence of T790M was confirmed in rebiopsy samples from the remaining two patients after progression of disease, as determined by Response Evaluation Criteria in Solid Tumors (RECIST) (53). These cases were re-reviewed for characteristics of indolent progression. Serial bi-dimensional measurements of reference lesions were performed by a radiologist (M.S.G.) from the time of best response.
Growth assays
Growth inhibition was measured with CellTiter Blue Reagent (Promega) as per the manufacturer’s instructions using cells plated in triplicate at a density of 4000 cells per well. Fluorescence was measured on a SpectraMax fluorometer. Growth inhibition was calculated as percentage of vehicle-treated wells ± SD.
For PC-9 cell counting assays, 20,000 to 40,000 cells per well were plated in six-well plates. Cell counting was performed in triplicate with an automated ViCell counter (Beckman Coulter) or a Coulter Counter. H322M cells were plated at a density of 100,000 per well and counted in quadruplicate with a Z2 Coulter Counter (Beckman Coulter). Cells were not allowed to reach >70% confluence at the final time point. Statistical significance was determined with the Student’s two-tailed t test. For all assays conducted in the presence of drug, fresh TKI was added every 72 hours.
For reconstitution experiments, PC-9/BR C1 cells were mixed with parental cells at the indicated concentrations before plating. DNA was isolated in parallel from each dilution for PCR-based EGFR exon 20 sequencing to confirm mutant peak levels. All experiments were conducted at least two independent times.
Mathematical modeling
PC-9 and PC-9/ER populations were modeled as a two-type stochastic birth and death process. In the context of our model, each PC-9 or PC-9/ER cell waits an exponentially distributed amount of time to divide or die; this waiting time is governed by the cell birth and death rates. During PC-9 cell replication, a cell harboring the T790M mutation arises with a given probability (the mutation rate). Sensitive and resistant cells have distinct growth and death rates that vary depending on the drug concentration; these parameters were experimentally determined. We estimated the net (birth minus death) growth rate by fitting the mathematical model to cell counts at 48, 60, and 72 hours in the presence and absence of drug. Death rates were estimated from annexin V/propidium iodide fluorescence-activated cell sorting (FACS) counts; we considered double-positive cells to make up the dead cell population. We accounted for cell clearance by assuming that 50% of dead cells are cleared every 12 hours; this assumption was made to account for the degradation of dead cells in the cell culture over time. Measurements were performed at drug concentrations of 0, 1, 3, 10, and 20 μM erlotinib (Fig. 5A).
To calculate the expected frequency of T790M alleles in a population of 3 million cells that initiated with a single cell harboring only a drug-sensitive EGFR mutation and growing in the absence of drug, we used the growth and death rates obtained in the absence of drug (0 μM data point). We performed 100,000 Monte Carlo simulations of the model ending when the total population reaches the desired size (3 million cells). We recorded the number of resistant cells present at the final time point. For details, see (18, 54).
To compare the relative effects of various dosing strategies on the development of resistance, we used analytical formulas describing the expected resistant cell population size under time-dependent dosing strategies. These calculations are based on the generating function for an inhomogeneous two-type birth and death process; birth and death rates of the sensitive and resistant cell population at each drug concentration are informed by data shown in Fig. 5A [see (18, 55)].
Xenograft studies
Cells (5 × 106 to 10 × 106) were injected with Matrigel into the hind flanks of 8-week-old athymic (nu/nu) female mice (Harlan). When tumors reached ~150 mm3, animals were randomized to receive vehicle alone, cisplatin (4 mg/kg twice per week, intraperitoneally), erlotinib (12.5 mg/kg daily, orally), or a combination of both erlotinib and cisplatin. Tumor volume was calculated as L × W2 × π/6 and recorded twice per week. All animals were housed in pathogen-free facilities and provided with abundant food and water under guidelines approved by the MSKCC Institutional Animal Care and Use Committee and Research Animal Resource Center.
Supplementary Material
Acknowledgments
We thank Y. Lin for assistance with PCRs, R. Maki for helpful discussions, and C. Arteaga and G. Riely for critically reviewing the manuscript.
Funding: Supported by NIH/National Cancer Institute (NCI) grants R01-CA121210 (W.P.), P01-CA129243 (M.G.K. and W.P.), and U54-CA143798 (F.M. and W.P.). The MSKCC Genomics Core is supported by an NCI Cancer Center Support Grant award to MSKCC (P30-CA008748). W.P. received additional support from Vanderbilt’s SPORE in Lung Cancer grant (CA90949) and the Vanderbilt-Ingram Cancer Center Core grant (P30-CA68485). R.K.T. is supported by the German Ministry of Science and Education as part of the NGFNplus program (grant 01GS08100), Max Planck Society (M.I.F.A.NEUR8061), Deutsche Forschungsgemeinschaft through SFB832 (TP6), Deutsche Krebshilfe (grant 107954), and Fritz-Thyssen-Stiftung (grant 10.08.2.175).
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/3/90/90ra59/DC1
Materials and Methods
Fig. S1. Further characterization of PC-9 TKI-resistant cell lines.
Fig. S2. Characterization of EGFR-mutant TKI-resistant cells.
Fig. S3. Characterization of H3255 TKI-resistant cells.
Fig. S4. Rate of progression in T790M-harboring EGFR-mutant lung cancer.
Fig. S5. Derivation of mathematical parameters.
Fig. S6. Continuation of TKI therapy in cell populations with T790M-harboring clones leads to better tumor cell control.
Fig. S7. Toxicity of “gatekeeper” mutations in other cell models.
References
Author contributions: J.C. and W.P. conceived the project and wrote the initial draft of the manuscript. J.C., J.F., K.H., K.O., R.S., L.W., K.R.A., M.L.S., and E.d.S. performed the experiments. J.C., J.F., G.R.O., M.A., N.D.S., A.V., M.S.G., M.G.K., M.L., V.A.M., F.M., and W.P. analyzed the data. A.I. contributed unpublished patient data sets. All authors commented on the final version of the manuscript.
Competing interests: R.K.T. has received consulting and lecture fees from Sequenom, Sanofi-Aventis, Merck, Roche, Infinity, Boehringer-Ingelheim, AstraZeneca, Johnson & Johnson, and Atlas-Biolabs, and research support from Novartis and AstraZeneca. V.A.M. has consulted for Boehringer-Ingelheim, Genentech, and Roche. M.G.K. has consulted for Boehringer-Ingelheim and Allos Therapeutics. A.I. has consulted for AstraZeneca and Chugai. W.P. has consulted for MolecularMD and AstraZeneca. Rights to a patent application for EGFR T790M testing were licensed on behalf of V.A.M., W.P., and others to MolecularMD. The patent application has been filed by MSKCC.
REFERENCES AND NOTES
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulloy R, Ferrand A, Kim Y, Sordella R, Bell DW, Haber DA, Anderson KS, Settleman J. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67:2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 6.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 7.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T. North-East Japan Study Group, Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, Shimizu E, Saito H, Toyooka S, Nakagawa K, Fukuoka M. West Japan Oncology Group, Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sánchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M. Spanish Lung Cancer Group, Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 10.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 13.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godin-Heymann N, Ulkus L, Brannigan BW, McDermott U, Lamb J, Maheswaran S, Settleman J, Haber DA. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 15.Bean J, Riely GJ, Balak M, Marks JL, Ladanyi M, Miller VA, Pao W. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14:7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, Caicun Z, Oberdick M, Shahidi M, Yang CH. Phase IIB/III double-blind randomized trial of afatinib (BIBW 2992, an irreversible inhibitor of EGFR/HER1 and HER2) + best supportive care (BSC) versus placebo + BSC in patients with NSCLC failing 1–2 lines of chemotherapy and erlotinib or gefitinib (LUX-Lung 1) Ann Oncol. 2010;21:LBA1. [Google Scholar]
- 17.Sequist LV, Besse B, Lynch TJ, Miller VA, Wong KK, Gitlitz B, Eaton K, Zacharchuk C, Freyman A, Powell C, Ananthakrishnan R, Quinn S, Soria JC. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: Results of a phase II trial in patients with advanced non–small-cell lung cancer. J Clin Oncol. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 18.Foo J, Michor F. Evolution of resistance to targeted anti-cancer therapies during continuous and pulsed administration strategies. PLoS Comput Biol. 2009;5:e1000557. doi: 10.1371/journal.pcbi.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T, Takigawa N, Takata M, Kiura K, Tanimoto M. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non–small cell lung cancer cell line. Cancer Res. 2007;67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 20.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Jänne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 21.Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, Lifshits E, Brown A, Lee C, Christensen JG, Kwiatkowski DJ, Engelman JA, Jänne PA. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mok TS. Living with imperfection. J Clin Oncol. 2010;28:191–192. doi: 10.1200/JCO.2009.25.8574. [DOI] [PubMed] [Google Scholar]
- 25.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller VA, Riely GJ, Zakowski MF, Li AR, Patel JD, Heelan RT, Kris MG, Sandler AB, Carbone DP, Tsao A, Herbst RS, Heller G, Ladanyi M, Pao W, Johnson DH. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol. 2008;26:1472–1478. doi: 10.1200/JCO.2007.13.0062. [DOI] [PubMed] [Google Scholar]
- 27.Miller VA, O’Connor P, Soh C, Kabbinavar F for the ATLAS Investigators. A randomized, double-blind, placebo-controlled, phase IIIb trial (ATLAS) comparing bevacizumab (B) therapy with or without erlotinib (E) after completion of chemotherapy with B for first-line treatment of locally advanced, recurrent, or metastatic non-small cell lung cancer (NSCLC) J Clin Oncol. 2009;27:LBA8002. doi: 10.1200/JCO.2012.47.3983. [DOI] [PubMed] [Google Scholar]
- 28.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Eastern Cooperative Oncology Group, Comparison of four chemotherapy regimens for advanced non–small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 29.Riely GJ, Kris MG, Zhao B, Akhurst T, Milton DT, Moore E, Tyson L, Pao W, Rizvi NA, Schwartz LH, Miller VA. Prospective assessment of discontinuation and reinitiation of erlotinib or gefitinib in patients with acquired resistance to erlotinib or gefitinib followed by the addition of everolimus. Clin Cancer Res. 2007;13:5150–5155. doi: 10.1158/1078-0432.CCR-07-0560. [DOI] [PubMed] [Google Scholar]
- 30.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, Heist RS, Temel J, Christensen JG, Wain JC, Lynch TJ, Vernovsky K, Mark EJ, Lanuti M, Iafrate AJ, Mino-Kenudson M, Engelman JA. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oxnard GR, Janjigian YY, Arcila ME, Kris MG, Ladanyi M, Azzoli CJ, Miller VA. Maintained sensitivity to EGFR tyrosine kinase inhibitors (TKIs) in EGFR-mutant lung cancers which recur after adjuvant TKI. abstract presented at the American Society of Clinical Oncology Annual Meeting; Chicago, IL. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oxnard GR, Arcila ME, Sima CS, Riely GJ, Chmielecki J, Kris MG, Pao W, Ladanyi M, Miller VA. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, Zakowski MF, Kris MG, Pao W, Miller VA, Ladanyi M. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milton DT, Azzoli CG, Heelan RT, Venkatraman E, Gomez JE, Kris MG, Krug LM, Pao W, Rizvi NA, Dunne M, Miller VA. A phase I/II study of weekly high-dose erlotinib in previously treated patients with nonsmall cell lung cancer. Cancer. 2006;107:1034–1041. doi: 10.1002/cncr.22088. [DOI] [PubMed] [Google Scholar]
- 35.Hidalgo M, Siu LL, Nemunaitis J, Rizzo J, Hammond LA, Takimoto C, Eckhardt SG, Tolcher A, Britten CD, Denis L, Ferrante K, Von Hoff DD, Silberman S, Rowinsky EK. Phase I and pharmacologic study of OSI-774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J Clin Oncol. 2001;19:3267–3279. doi: 10.1200/JCO.2001.19.13.3267. [DOI] [PubMed] [Google Scholar]
- 36.Parzen E. Stochastic Processes. Society for Industrial and Applied Mathematics; Philadelphia, PA: 1999. [Google Scholar]
- 37.Detterbeck FC, Gibson CJ. Turning gray: The natural history of lung cancer over time. J Thorac Oncol. 2008;3:781–792. doi: 10.1097/JTO.0b013e31817c9230. [DOI] [PubMed] [Google Scholar]
- 38.Seshadri R, Kutlaca RJ, Trainor K, Matthews C, Morley AA. Mutation rate of normal and malignant human lymphocytes. Cancer Res. 1987;47:407–409. [PubMed] [Google Scholar]
- 39.Oller AR, Rastogi P, Morgenthaler S, Thilly WG. A statistical model to estimate variance in long term-low dose mutation assays: Testing of the model in a human lymphoblastoid mutation assay. Mutat Res. 1989;216:149–161. doi: 10.1016/0165-1161(89)90001-0. [DOI] [PubMed] [Google Scholar]
- 40.Eskens FA, Mom CH, Planting AS, Gietema JA, Amelsberg A, Huisman H, van Doorn L, Burger H, Stopfer P, Verweij J, de Vries EG. A phase I dose escalation study of BIBW 2992, an irreversible dual inhibitor of epidermal growth factor receptor 1 (EGFR) and 2 (HER2) tyrosine kinase in a 2-week on, 2-week off schedule in patients with advanced solid tumours. Br J Cancer. 2008;98:80–85. doi: 10.1038/sj.bjc.6604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gralla RJ, Casper ES, Kelsen DP, Braun DW, Jr, Dukeman ME, Martini N, Young CW, Golbey RB. Cisplatin and vindesine combination chemotherapy for advanced carcinoma of the lung: A randomized trial investigating two dosage schedules. Ann Intern Med. 1981;95:414–420. doi: 10.7326/0003-4819-95-4-414. [DOI] [PubMed] [Google Scholar]
- 42.Krug LM, Ng KK, Kris MG, Miller VA, Tong W, Heelan RT, Leon L, Leung D, Kelly J, Grant SC, Sirotnak FM. Phase I and pharmacokinetic study of 10-propargyl-10-deazaaminopterin, a new antifolate. Clin Cancer Res. 2000;6:3493–3498. [PubMed] [Google Scholar]
- 43.Azzoli CG, Krug LM, Gomez J, Miller VA, Kris MG, Ginsberg MS, Henry R, Jones J, Tyson L, Dunne M, Pizzo B, Farmer A, Venkatraman E, Steffen R, Sirotnak FM. A phase 1 study of pralatrexate in combination with paclitaxel or docetaxel in patients with advanced solid tumors. Clin Cancer Res. 2007;13:2692–2698. doi: 10.1158/1078-0432.CCR-06-1754. [DOI] [PubMed] [Google Scholar]
- 44.Yeo WL, Riely GJ, Yeap BY, Lau MW, Warner JL, Bodio K, Huberman MS, Kris MG, Tenen DG, Pao W, Kobayashi S, Costa DB. Erlotinib at a dose of 25 mg daily for non-small cell lung cancers with EGFR mutations. J Thorac Oncol. 2010;5:1048–1053. doi: 10.1097/JTO.0b013e3181dd1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blackwell KL, Burstein HJ, Storniolo AM, Rugo H, Sledge G, Koehler M, Ellis C, Casey M, Vukelja S, Bischoff J, Baselga J, O’Shaughnessy J. Randomized study of lapatinib alone or in combination with trastuzumab in women with ErbB2-positive, trastuzumab-refractory metastatic breast cancer. J Clin Oncol. 2010;28:1124–1130. doi: 10.1200/JCO.2008.21.4437. [DOI] [PubMed] [Google Scholar]
- 46.Godin-Heymann N, Bryant I, Rivera MN, Ulkus L, Bell DW, Riese DJ, II, Settleman J, Haber DA. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res. 2007;67:7319–7326. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, Moonsamy P, Dahlman K, Miller VA, Costa C, Hannon G, Sawyers CL. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong Y, Yao E, Shen R, Goel A, Arcila M, Teruya-Feldstein J, Zakowski MF, Frankel S, Peifer M, Thomas RK, Ladanyi M, Pao W. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507) PLoS One. 2009;4:e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ware KE, Marshall ME, Heasley LR, Marek L, Hinz TK, Hercule P, Helfrich BA, Doebele RC, Heasley LE. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One. 2010;5:e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van den Abbeele AD, Badawi RD, Manola J, Morgan JA, Desai J, Kazanovicz A, St Armand M, Baum C, Demetri GD. Effects of cessation of imatinib mesylate (IM) therapy in patients (pts) with IM-refractory gastrointestinal stromal tumors (GIST) as visualized by FDG-PET scanning. J Clin Oncol. 2004;22:3012. [Google Scholar]
- 51.Fumagalli E, Coco P, Morosi C, Dileo P, Bertulli R, Gronchi A, Casali PG. Rechallenge with imatinib in GIST patients resistant to second or third line therapy. abstract presented at the 15th Connective Tissue Oncology Society Meeting; Miami Beach, FL. 2009. [Google Scholar]
- 52.Hanfstein B, Müller MC, Kreil S, Ernst T, Schenk T, Lorentz C, Schwindel U, Leitner A, Hehlmann R, Hochhaus A. Dynamics of mutant BCR-ABL-positive clones after cessation of tyrosine kinase inhibitor therapy. Haematologica. 2011;96:360–366. doi: 10.3324/haematol.2010.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 54.Iwasa Y, Nowak MA, Michor F. Evolution of resistance during clonal expansion. Genetics. 2006;172:2557–2566. doi: 10.1534/genetics.105.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foo J, Michor F. Evolution of resistance to anti-cancer therapy during general dosing schedules. J Theor Biol. 2010;263:179–188. doi: 10.1016/j.jtbi.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.