Abstract
Mycobacterium tuberculosis is an obligate human respiratory pathogen that encodes approximately ten arsenic repressor (ArsR) family regulatory proteins that allow the organism to respond to a wide range of changes in its immediate microenvironment. How individual ArsR repressors have evolved to respond to selective stimuli is of intrinsic interest. The Ni(II)/Co(II)-specific repressor NmtR and related actinomycete nickel sensors harbor a conserved N-terminal αNH2-Gly2-His3-Gly4 sequence. Here, we present the solution structure of homodimeric apo-NmtR and show that the core of the molecule adopts a typical winged-helix ArsR repressor (α1-α2-α3-αR-β1-β2-α5) “open conformation” that is similar to the related zinc sensor Staphylococcus aureus CzrA, but harboring long, flexible N-terminal (residues 2-16) and C-terminal (residues 109-120) extensions. Ni(II) binding to the regulatory sites induces strong paramagnetic broadening of the α5 helical region and the extreme N-terminal tail to residue 10. Ratiometric pulse chase amidination mass spectrometry reveals that the rate of amidination of the Gly2 α-amino group is strongly attenuated in the Ni(II) complex relative to the apo-state and non-cognate Zn(II) complex. Ni(II) binding also induces dynamic disorder in the μs-ms timescale of key DNA interacting regions that likely contributes to the negative regulation of DNA binding by Ni(II). Molecular dynamics simulations and quantum chemical calculations reveal that NmtR readily accommodates a distal Ni(II) hexacoordination model involving the α-amine and His3 of the N-terminal region and α5 residues Asp91′, His93′, His104 and His107, which collectively define a new metal sensing site configuration in ArsR family regulators.
INTRODUCTION
Transition metal ions including zinc, copper, nickel, cobalt, iron, and manganese are essential for a diversity of biological processes, the essentiality of which derives from their catalytic roles as cofactors in oxidation-reduction, electron transfer and hydrolytic enzymes, to structural roles in biomolecules.1-3 All metal ions are toxic in excess however and as a result, cells encode machinery to precisely control the intracellular total and bioavailable concentrations of cytoplasmic metal ions. All cells including pathogenic organisms have evolved the capacity to quickly respond to chronic and acute changes of microenvironment in which the availability of both biologically essential metal ions and heavy metal pollutants varies.4-6 The central players in this process are the metalloregulatory proteins that regulate the expression of genes encoding metal transporters and trafficking proteins, which collectively manage metal homeostasis and resistance.6,7
Mycobacterium tuberculosis (Mtb) is the causative pathogen of human tuberculosis. Mtb encodes a remarkable diversity of known and putative metal ion transporters, including multiple putative P-type ATPases, ABC transporters, and cation diffusion facilitator (CDF) transporters.8 A large panel of metalloregulatory proteins in Mtb regulate transcription of those transporters. For example, M. tuberculosis CsoR represses transcription of the cso (Cu-sensitive operon) which encodes CsoR itself, a Cu(I)-effluxing P-type ATPase CtpV and a gene of unknown function.9 Mtb also encodes approximately ten arsenic repressor (ArsR) family proteins that regulate the expression of genes encoding for proteins responsible for metal ion detoxification, sequestration and efflux, and perhaps other processes.10-13
Several members of the ArsR family have been extensively studied.5,12 Among them, Staphylococcus aureus Zn(II) sensor CzrA is the most well characterized family repressor with regard to its structural, mechanistic and thermodynamic understanding of allosteric negative regulation of DNA operator binding.14-18 S. aureus CzrA represses the transcription of the cation diffusion facilitator (CDF) CzrB in the absence of metal stress.19,20 The quaternary structure of CzrA adopts a typical winged-helix homodimeric fold21,22 denoted an “α5” ArsR family sensor.23 CzrA binds Zn(II) with negative homotropic cooperativity15,17 at two regulatory tetrahedral sites positioned across the pair of C-terminal α5 helices. A previous site-specific mutagenesis study of CzrA revealed that two metal ligands were necessary to maintain the native coordination geometry and Zn(II)-mediated regulation, while two other ligands were required only for maintaining high metal binding affinity with no or small effects on regulation, in what termed a “division of labor” organization.15 Taken together, it has been suggested that the metal coordination geometry is a fundamental determinant of metal selectivity for metalloregulators.7,14,24. The recent NMR structure of CzrA in the DNA-bound state suggested that metal binding drives a “closed-to-open” conformational change to reduce the affinity of DNA binding, and induced long-range disorder in the allosteric metal sites.16,18
There are two Ni(II) specific ArsR repressors encoded by Mtb.11,25 These are NmtR and KmtR, which repress the transcription of ctpJ and cdf, encoding a P-type ATPase metal transporter and a putative cation diffusion facilitator (CDF) transporter, respectively, at low Ni(II) concentrations. At high Ni(II) concentrations, NmtR and KmtR bind Ni(II) and dissociate from their DNA operators, allowing transcription of ctpJ and cdf, to export Ni(II) and Co(II), respectively, from the cytoplasm.8,11 A previous in vivo study proposed that NmtR binds Ni(II) in an octahedral coordination geometry with four metal ligands in the α5 helix, D91, H93, H104, and H107, and two C-terminal residues H109 and H116.25 We recently proposed an alternative Ni(II) coordination model involving the N-terminal Gly2-His3-Gly4 motif in NmtR (in place of H109 and H116) that specifically drives Ni(II)-binding and allosteric negative regulation of DNA binding on the basis of in vitro metal binding and DNA binding assays of a collection of conservative glutamine substitution mutants.26
Here we present the solution NMR structure of homodimeric NmtR in apo-state. The overall structure reveals a typical core winged-helix fold of ArsR family, but with distinct long flexible N- and C-terminal tails. The binding of paramagnetic Ni(II) induces strong line broadening of the regulatory sites composed of α5 helices as well as the N-terminal tail to residue 10. In contrast, residues in the C-terminal tail beyond 110 (residues 111-120) are unaffected by Ni(II) binding; as such, these findings rule out Ni(II) coordination by His116. Ratiometric pulse chase amidination mass spectrometry27 reveals that the α-amino group of Gly2 is strongly protected from amidination in the Ni(II) bound form, with the non-cognate, tetrahedral Zn(II) complex14 providing little significant protection relative to the apo-state. These and previous findings taken collectively are consistent with a regulatory octahedral Ni(II) complex formed by the α-amine and His3 from the N-terminus and α5 residues Asp91′, His93′, His104 and His107. This chelate structure is supported by a range of molecular dynamics (MD) simulations18 of the non-cognate Zn(II) and cognate Ni(II) complexes.
EXPERIMENTAL PROCEDURES
Protein Purification
Recombinant NmtR was expressed and purified as described previously.26 For the NMR experiments, 15N-labeled or 13C-and 15N-labeled NmtR were expressed in E. coli BL21 (DE3) cells in M9 minimal medium and purified essentially as described previously.16,26 MALDI-TOF mass spectrometry of purified apo-NmtR used in this work reveals 12,699.7 D (12,704.3 D expected for residues 2-120, with the N-terminal Met processed).26
NMR Spectroscopy
For NMR experiments, NmtR was prepared in 10 mM Hepes, pH 7.0, 100 mM NaCl in as 10% 2H2O/90% H2O mixture or 100% 2H2O at protomer protein concentrations ranging from 0.35-0.50 mM. All NMR experiments were performed on a Varian DDR 600 or 800 MHz spectrometer fitted with cryogenic probe systems at 310K in the METACyt Biomolecular NMR Laboratory. NMR spectra were referenced to external DSS. NMR data processing and analysis were performed using NMRPipe28 and NMRView.29 Backbone resonance assignments were obtained using standard triple-resonance NMR data set as described previously.26 Assignments of side chain resonances were made using three-dimensional (3D) HCCH-COSY30, H(CCO)NH-TOCSY31,32 and C(CO)NH-TOCSY32 experiments. Distance restraints were derived from 3D 15N-edited NOESY-HSQC (τm = 100 ms) and 13C-edited NOESY-HSQC (τm = 80 ms) spectra. Backbone amide residual dipolar couplings, 1DNH, were obtained from the difference in the scalar coupling (1J) measured in stretched polyacrylamide gels (4%) using 2D IPAP-HSQC spectroscopy.33
Structure Calculations
NOE crosspeak assignments and initial NOE constraints were obtained with CYANA 2.134 and CANDID.35 All NOE constraints were manually confirmed during CYANA calculations. Hydrogen bonds for the helical regions of the NmtR were added to facilitate the automated assignment of additional NOE restraints by CANDID. Chemical shift-based restraints from TALOS+36 were included only for secondary structural regions.
The initial 100 symmetric homodimer structures were calculated by a simulated annealing protocol with Xplor-NIH37 and were further refined with the addition of non-crystallographic symmetry (NCS) and RDC restraints. A total of 54 intersubunit NOE restraints were cross-validated during the structure calculation procedure by randomly removing one-third of these constraints and recalculating the global precision of the structures.16 Iterative refinement and editing of the distance restraints based on the NOESY spectra to remove incorrect and ambiguous assignments reduced the number of restraints. The final 19 structures with the lowest energy were chosen for analysis and were deposited in the PDB (accession code 2LKP).
Simulations of NmtRs
Molecular dynamics simulations of the apo-, Zn(II)-bound and Ni(II)-bound allosteric forms of NmtR were performed using the AMBER 11 suite of programs38,39 utilizing the ff99SBildn40 force-field. The protonation states for charged amino acids not involved in metal binding were obtained from the H++ protonation state server.41 The metal binding residues in the α5 helix (Asp91, His93, His104′ and His107′) were represented in their metal binding protonation states adopted from CzrA in simulations of Zn•NmtR and Ni•NmtR.22 A protonation state model similar to that employed in our previous simulations of apo-CzrA featuring a positively charged His104 and a negatively charged Asp91 was found to be the most likely candidate for apo-NmtR.18
A common solvation and equilibration scheme was implemented to prepare the allosteric forms of NmtR for production MD simulations. The protein was immersed in a periodically replicated truncated octahedral box containing explicit SPC/E water molecules42,43 that provided an 8 Å water shell around it. In these simulations, a range of 11578 to 26933 water molecules was used to solvate the protein. The solvated protein was next net-neutralized using explicit Na+ or Cl− counter-ions.44 A single Cl− ion was required to neutralize the apo allosteric form of the protein, while two additional Cl− ions were required to neutralize the metal-ions bound forms of the protein. The systems were prepared for production simulations using a well defined equilibration protocol. First, a series of restrained-minimizations was performed in which constituent parts of the system were gradually relaxed over multiple stages to avoid artifacts and steric clashes. In the first phase of minimization, the water molecules and counter-ions were minimized while the protein was kept restrained. Employing a multi-stage approach, increasingly larger parts of the protein were gradually relaxed. In the first stages of minimization, protein hydrogen atom positions were optimized followed by a minimization including side-chain groups. In the next stage of energy minimization only the backbone Cα atoms were kept restrained, and in the final step the entire protein structure was minimized along with the solvent molecules and counter-ions. The phases of minimizations were performed for 10000, 10000, 20000, 40000 and 40000 steps respectively, unless convergence was achieved earlier. The minimized system was then slowly heated to 300K for a canonical ensemble (NVT) over 200 ps, while maintaining weak restraints on all protein heavy atoms. Finally, all restraints were removed and the system was further equilibrated for 1 ns at 300 K for an isobaric and isothermal ensemble (NPT). MD trajectories were propagated from this well-equilibrated structure at 300K for an NPT ensemble. The temperature of the simulated system was maintained using Langevin Dynamics with a collision frequency of 1 ps−1, and the SHAKE algorithm was employed to constrain heavy atom bonds to hydrogen atoms.42,45,46 Long-range electrostatics were calculated using the particle mesh Ewald method.47 A time-step of 2 fs was employed and snapshots from the trajectories were collected for analysis after every 1000 steps.
Modeling the Zn(II) Binding Site of NmtR
Starting from a solvated and well equilibrated structure of apo-NmtR, constrained MD and energy-minimization methods were used to bring the possible metal binding residues to within a reasonable distance for Zn(II) binding. A Zn(II) ion was placed at each metal binding site using the Maestro GUI.48 Both metal binding regions [D91, H93, H104, H107 and Zn(II))] were treated using DFT based QM/MM methods49 (M06-L/LACVP* for QM and OPLS-2005 force field for MM residues)50-52 53 as implemented in the Qsite QM/MM program.50
To further validate the calculated binding site of Zn(II)-bound NmtR and to capture the effect of Zn(II) binding on the entire protein structure, 1 ns of QM/MM MD simulations were performed on well solvated and equilibrated Zn•NmtR and Zn2•NmtR structures at the SCC-DFTB level of theory54,55 using the AMBER suite of programs. A detailed description of these QM/MM and QM/MM MD calculations and their successful implementation have been provided elsewhere.18,56,57 The bond distances and relative orientations of coordinating residues in this α5 site of NmtR is similar to that observed in CzrA.18
Modeling of the Ni(II) binding site
We performed model system calculations at the DFT level of theory to validate the proposed Ni(II) coordination complex in NmtR. These calculations included the Ni(II) ion coordinated to Gly2, His3 and a reduced representation of Asp91, His93, His104 and His107 residues. We used the Gaussian09 electronic structure program58 to perform these calculations with the M06-L functional51 paired with the valence double zeta polarized 6-31G* basis set for C and H atoms, the 6-31+G* basis set for O and N atoms, and the LANL2TZ effective core potentials for Ni(II).59-62 The calculated Ni(II) bond lengths from these calculations were similar to those obtained from XAS experiments.14 We then performed steered-MD (SMD) calculations in an effort to determine whether the N-terminal amino acids (Gly2 and His3) could access the distal and proximal binding models of Ni(II) coordination (Fig. S2). In these calculations, Gly2 and His3 were “reeled in” from distances of tens of angstroms into the proximal and distal Ni(II) binding sites over hundreds of picoseconds. The tetra-coordinated metal ion was represented using our previously developed “bonded model” parameters for a DHHH binding motif using the MTK++ program.63,64 Preliminary simulations performed using a generalized Born implicit solvent model65,66 indicated that introducing Gly2 and His3 at the distal Ni(II) coordination sites caused large changes in the structure of the protein. Analogous simulations in which Gly2 and His3 were introduced into the proximal Ni(II) coordination sphere failed to elicit a similar conformational change suggesting that the distal binding mode was the correct representation. To create a model construct for Ni(II)-bound NmtR and validate the distal binding mode, multiple SMD simulations were performed in which Gly2 and His3 were introduced into the distal metal binding site in explicit solvent. These simulations were initiated from explicitly solvated and well-equilibrated structures of tetra-coordinated Ni2•NmtR, and were performed over tens of nanoseconds.
Ratiometric Pulse-Chase Amidination Mass spectrometry (rPAm-MS)
These mass-coded experiments were carried out essentially as described previously27 in which a pulse of S-methyl thioacetimidate (SMTA) was added for time, t, followed by chase with and excess of S-methyl thiopropionimidate (SMTP) (Δ=14 amu), quenched and digested with trypsin and the N-terminal heptapeptide (2GHGVEGR8) quantified by high resolution matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. The expected molecular weights of the acetamidinated peptide 2-8 is 752.4 D while that of the propionamidinated peptide 2-8 is 766.4 D. 20 μM NmtR monomer (containing 20 μM total amine) was reacted in either the apo-, Zn(II)-bound (2.5 monomer mol•equiv Zn) or Ni(II)-bound (1.5 monomer mol•equiv Ni) states and mixed with 100-fold (apo-NmtR, Zn-NmtR) or 500-fold (Ni-NmtR) monomer molar excess of SMTA, followed by 2000-fold (for apo, Zn) or 10,000-fold (Ni) monomer molar excess of SMTP. Quantitation of the intensities (I) of the acetimidated (A) and propionimidated (P) peptides as a function of time t as carried out as previously described27 with mol fraction propionimidated (ΘP) calculated from ΘP=IP/(IA+IP). Each data set was fitted to a single exponential decay, ΘP = Aoe−kt to obtain k and divided by [SMTA] in the pulse to obtain the second-order rate constant (MSMTA−1 min−1).
RESULTS AND DISCUSSION
Solution Structure of Apo-NmtR
Uniformly 15N, 13C-labeled apo-NmtR that conforms to the sequence in Fig. S1 (residues 2-120, with the N-terminal Met processed)26 was subjected to detailed multidimensional NMR analysis using standard methods to obtain nearly complete backbone (Cα, Hα, C’, N, HN) and ≈90% of the side chain assignable protons assigned. The structure was calculated on the basis of 509 intraprotomer and 54 interprotomer 1H-1H NOE restraints and 49 1DNH RDCs per protomer (Table 1). The final ensemble of nineteen structures shows excellent statistics and structural convergence with a final pairwise heavy atom rmsd of 1.47 ± 0.42 Å (Table 1).
Table 1. NMR Restraints and Structural Statistics for the apo-NmtR Homodimera.
| NMR restraints | |
| Total NOE distance restraints | 563 |
| Intrasubunit | 509 |
| Intersubunit | 54 |
| Hydrogen bonds | 96 |
| Total dihedral angle restraints | 142 |
| RDC restraints | 46 |
| Structural statistics (19 structures) | |
| Violations | |
| Number of distance restraints > 0.5 Å | 0 |
| Number of dihedral angle restraints > 5° | 0 |
| Rms deviation from experiments | |
| Distance (Å) | 0.065 ± 0.006 |
| Dihedral angle (°) | 0.464 ± 0.013 |
| Rms deviation from idealized geometry | |
| Bonds (Å) | 0.004 ± 0.000 |
| Angles (°) | 0.643 ± 0.136 |
| Impropers (°) | 0.390 ± 0.020 |
| Ramachandran analysis (%) (all residues) | |
| Most favored region | 81.8 |
| Additionally allowed region | 13.1 |
| Generously allowed region | 4.3 |
| Disallowed region | 0.7 |
| Average pairwise RMS deviations (Å) (residues 17-108)b | |
| Backbone heavy atoms | 1.47 ± 0.42 |
| All heavy atoms | 2.17 ± 0.39 |
Resonance assignments and other NMR data for apo-NmtR have been deposited in the BioMagResBank as accession number 18003, with structure bundle apo-NmtR deposited in the Protein Data Bank under accession code 2LKP.
Coordinate precision was calculated by comparing individual structures in the bundle with the coordinates of the average structure. Only residues 17-108 for each subunit was used in this analysis.
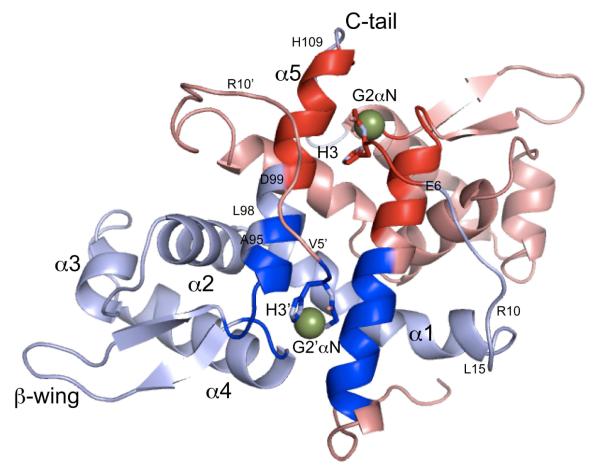
A ribbon representation of a global superposition of the 19 models is shown in Figure 1, compared to the average structure, from the DNA binding interface (Fig. 1A) and the allosteric sites (Fig. 1B). The core of apo-NmtR adopts the typical ArsR repressor (α1-α2-α3-α4-β1-β2-α5) fold encompassing residues 17-108 as previously observed in Mtb CmtR,67 S. aureus CadC,68 Xylella fastidiosa BigR13 and in the zinc sensors S. aureus CzrA16,22 and Synechococcus SmtB,22,69 but in this case is flanked by long unstructured, highly mobile N-terminal (residues 2-16) and C-terminal (residues 109-120) “tails”.26 These tails emerge on opposite sides of the molecule with the N-terminal tail of one protomer closest to the C-terminal tail of the other protomer (Fig. 1). Helix α4 is the DNA-recognition helix, also denoted αR. The β-wing tip is structurally less well-defined relative to the core of the molecule, as was previously found in solution for Zn(II)-bound CzrA.16 There is no evidence from these data that NmtR harbors an additional N-terminal αhelix like that present in the α3N Cd/Pb sensor CadC.23,68
Figure 1.
Ribbon representation of the solution structure of apo-NmtR. 19 lowest energy structures are shown (2LKP) in yellow (residues 2-16), indigo (residues 17-107) and grey (residues 108-120) superimposed on the average structure (residues 16-107) shaded magenta. (A) View from the α1/αR DNA binding interface; (B) View from the allosteric α5 helices.
S. aureus CzrA is strongly related to M. tuberculosis NmtR (≈30% identity; ≈60% similarity) and detailed comparative studies of this pair of sensors has long served as a model system with which to investigate the evolution of metal selectivity in ArsR family repressors.5,6,14,70 In fact, CzrA can be considered NmtR without the “tails” and indeed both sensors harbor a standard “α5-type” metal binding Asp1-x-His2-x10-His3-x2-His/Glu4 motif (where x is any amino acid) (Fig. S1).12,71 In CzrA, this sequence is found in the α5 helix, and the two symmetry-related, interprotomer zinc sensing sites are defined by tetrahedral coordination to Asp1-His2-His3′-His4′ and Asp1′-His2′-His3-His4 sites (Fig. S2). CzrA is selective for Zn(II) and shows no response to Ni(II); in contrast, NmtR is selective for Ni(II)/Co(II) and shows an attenuated (in vitro) or no response (in vivo) to Zn(II).6,14,25 Although our structure establishes that the core folds of NmtR and CzrA are similar, a global superposition of the core apo-NmtR structure bundle with the average structure (Fig. 2A) reveals detectable differences to that of the bundle superimposed on the crystallographic structure of apo-CzrA (Fig. 2B). These differences extend beyond the uncertainty of the coordinates of the structure bundle, and derive at least partly from distinct protomer packing (≈1.6 Å Cα rmsd) (Fig. 2B). In any case, apo-NmtR, like apo-CzrA, clearly adopts an “open” dimer conformation, in which the interprotomer Ser61 Cα-Ser61′ Cα distance is quite large, ≈48 Å, a distance comparable to the largest analogous (Ser54-Ser54′ Cα) distance observed in molecular dynamics simulations of allosterically inhibited Zn2 CzrA.18 This form of apo-NmtR is clearly not pre-organized to bind DNA without a significant change to a more “closed” conformation, perhaps like previously observed for apo-CzrA.16
Figure 2.
Ribbon representation of the folded core of apo-NmtR (residues 17-107). The bundle of 19 structures (yellow) are globally superimposed on the average structure (magenta) (A) and on the crystallographic structure of S. aureus CzrA (shaded marine blue, pdb 1R1U). The subunit pairwise Cα rmsd (using residues 9-101 or 9-102 in CzrA, residues 17-107 for NmtR) is 1.62 and 1.69 Å, while the dimer pairwise Cα rmsd is 2.53 Å. αR, DNA recognition helix α4.
Paramagnetic Broadening Induced by Ni(II) Directly implicates the N-terminal Tail and the C-terminal α5 helix as Providing Ligand Donor Atoms to the Ni(II)
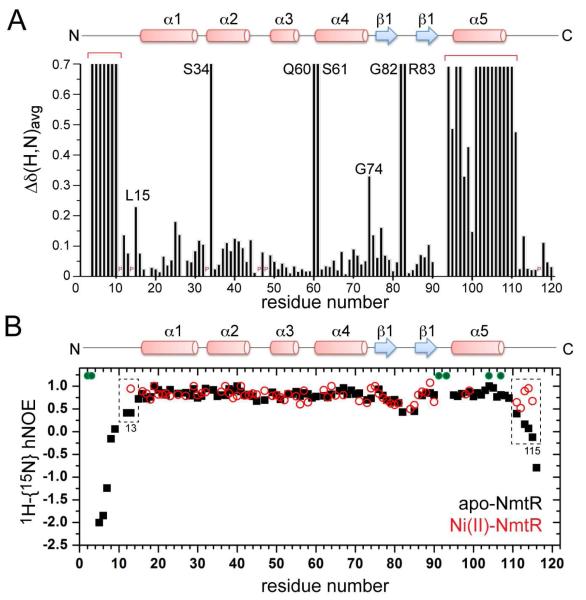
Addition of Ni(II) to apo-NmtR induces significant changes in the 15N-TROSY spectrum, including a number of shifts superimposed on a significant reduction in the number of crosspeaks (Fig. S3). Ni(II) is expected to broaden resonances within ≈10 Å of the paramagnetic, high-spin Ni(II)72,73 consistent with a six-coordinate, distorted octahedral geometry determined previously.14 These spectral perturbations are shown in a bar chart (Fig. 3A) and schematically on a ribbon representation of the solution structural model from three different views (Fig. 4). What is clearly seen is that Ni(II) binding extensively broadens resonances at the extreme N-terminus (from residues 4-10; residues 2 and 3 are unassigned in the apo-state26) as well as residues at both the N-terminal and C-terminal regions of the α5 helix (residues 91-93 are exchange broadened in apo-NmtR26), and extending to only L110, after which point there is little broadening observable.
Figure 3.
Perturbations of 15N-1H HSQC spectra of Ni(II) binding to apo-NmtR. (A) Chemical shift changes as a function of residue number. Resonances not observed in the “diamagnetic” spectral window in the Ni(II) complex are arbitrarily assigned a Δδ(H,N)avg value of 0.7 and those close to the Ni(II) are clustered by the red bracket. Perturbations for other residues for which a resonance crosspeak is observed are calculated from Δδ(H,N)avg = sqrt[(ΔδHN)2 + (ΔδN/5)2]. Resonances of interest are highlighted and a secondary structure schematic is shown at the top of the figure. P, Pro residue. Residues 91-93 are conformationally exchange broadened in the apo-state26 and thus no data are shown for these residues. (B) Plot of the 1H-{15N} heteronuclear NOE vs. residues number is shown for apo-NmtR26 (black) and Ni(II)-NmtR (red open circles). Regions of significant difference between the two states are boxed, and proposed Ni(II) ligands are indicated by the filled green circles.
Figure 4.
Schematic representation of the spectral perturbations of Ni(II) binding to apo-NmtR painted on the ribbon representation of the average structure of apo-NmtR from three different views: (A) bottom, DNA binding interface; (B) “top” or allosteric site view; and (C) side view. All residues for which backbone amide correlations are paramagnetically broadened/shifted are shaded gold, while residues 91-93 (conformationally broadened in apo-NmtR) are highlighted in orange. Green, backbone amides conformationally broadened in Ni(II) NmtR far from the metal site (see Fig. 3A) with L15 (panel B) and G74 (panel C) highlighted due to significant perturbations in chemical shift. In panel (B), D114 (shaded cyan) defines the penultimate C-terminal residue that is ordered on Ni(II) binding (see also Fig. S5, Supporting Information). D and P define the proximal and distal α5 metal sites, respectively, relative to the N-terminus labeled N at bottom right. See also Fig. S2, Supporting information.
These spectral features are consistent with the conclusions reached earlier on the basis of thermodynamic measurements of the KNi and allosteric coupling free energy, ΔGcNi and suggest that the Ni(II) binds to four residues analogous to the tetrahedral zinc complex of CzrA (D91, H93, H104 and H107) and the two residues derived from the extreme N-terminus, the α-amino group and the side chain of His3.26 Although these studies do not distinguish between proximal (P) and distal (D) Ni(II) binding site models (see Fig. 4B; Fig. S2), it is interesting to note that there is complete broadening of residues 4-10 and L15 just N-terminal to the α1 helix is strongly shifted on Ni(II) binding. Given the relatively short-range (≈10 Å) paramagnetic sphere of influence of the bound Ni(II), only in the distal binding mode would a substantial region of the N-terminal tail pass close enough to one or the other Ni(II) ion to result in paramagnetic broadening (see Fig. 4B). Thus, these data favor a model in which the Ni(II) forms an octahedral complex via coordination by G2, H3, D91′, H93′, H104 and H107 and its symmetry-related pair (Fig. S2 of the Supporting Information) (see below).
Due to the unfavorable spectral characteristics of the Ni(II)-complex, we were unable to obtain a structural model of the Ni(II)-complex. Inspection of the 1H-{15N} heteronuclear NOE reveals that Ni(II) binding stabilizes both the N-terminal (to residue 13) and the C-terminal (to residue 115) tail regions compared to apo-NmtR, such that these regions now tumble on the ps-ns timescale with the rest of the protein (Fig. 3B). The structural origin of this conformational ordering is unknown but for the vast majority of residues, Ni(II)-induced conformational changes are quaternary structural in nature, given the high degree of correspondence of 13Cα chemical shifts (where they are observable) in the apo- and Ni(II)-bound states (Fig. S4).
We also acquired NMR data for the non-cognate Zn(II)-bound NmtR and Fig. S3 shows an overlay of 15N TROSY spectra the apo-, Ni(II)- and Zn(II)-bound complexes. Inspection of these spectra suggest that the conformational change induced by Zn(II) is distinct from that of Ni(II), but we could do little else with the Zn(II)-NmtR sample given the poor solubility of this metallated derivative. X-ray absorption spectroscopy reveals a tetrahedral Zn(II) complex and QM/MM calculations18 reveal that a first coordination complex formed by the α5 residues only (Asp91, His93, His104′, His107′) can be modeled that is fully consistent with the data (Fig. 5A). Thus, these data and previous studies26 argue that Zn(II) is a weaker allosteric regulator than Ni(II) because it does not recruit the extreme N-terminus into the first metal coordination shell and thus is incapable of fully switching NmtR to an allosterically inhibited conformation.70
Figure 5.
Results of quantum-mechanical molecular-mechanical molecular dynamics (QM/MM MD) calculations of the non-cognate Zn(II) complex (A) and steered molecular dynamics simulations of the cognate Ni(II) complex (B) in NmtR.14 The image shown in panel (B) rotated ≈90° along the vertical relative to panel (A). Metal-ligand distances varied from calculation to calculation and were found to range from 1.95 Å to 2.20 Å in our models. For the model shown, the Ni(II)-ligand distances are Gly2 α-amino N: 1.96 Å, His3 Nδ1: 2.09 Å, Asp91 Oδ1: 1.96 Å, His104 Nδ1: 2.14 Å and His107 Nε2: 2.11 Å. Residue identities are indicated with prime nomenclature indicative of the opposite protomer. See Experimental Procedures for full details.
In striking contrast, an identical series of calculations reveal that it is indeed possible to recruit two ligands from the N-terminus to create an pseudooctahedral complex with good stereochemistry around the Ni(II) ion (Fig. 5B, see also Fig. S5). In this complex, the Nδ1 atoms of His3 and His104 are the axial ligands to the octahedrally coordinated Ni(II) ion, with the α-NH2, Asp91′, His93′ (Nδ1) and His107 (Nε2) lying roughly in a plane. Multiple SMD calculations suggest that in this coordination model, the N-terminus reaches over the “top” of the α5 helices to coordinate to the distal α5 metal site (Fig. 6). In models derived from these calculations, the Nε2 face of His104 points towards the backbone carbonyl groups of Leu73′ and Gly74′, and in some instances specifically forms a hydrogen bond with the backbone carbonyl group of Gly74. Such a hydrogen bonding interaction could explain the perturbations in the 15N-1H HSQC spectra observed at Gly74 in response to Ni(II) binding (Fig. 3A) and suggests that a CzrA-like hydrogen-bonding pathway18 may play a role in the mechanism of allosteric regulation. The structure shown (Fig. 6) is in excellent agreement with the paramagnetic broadening observed in the NMR spectra of Ni(II) NmtR in detail (Fig. 3A), except for residues 6-10 in the N-terminal region. Although these resonances are experimentally broadened beyond detection (Fig. 3A), in this model these residues are just outside of the zone of paramagnetic influence of the Ni(II) ions (≤13 Å) (Fig. 6). It is important to recognize that the precise path that each N-terminal tail takes is not well-defined by these simulations. However, these SMD models suggest that a number of favorable electrostatic interactions between charged residues in the N-terminal region and α5 helices of NmtR may stabilize the distal binding model. On the other hand, if these resides do indeed lie outside of the Ni(II) broadening zone, then conformational exchange might be contributing to dynamic disorder in this region. Additional studies are required to distinguish between these two possibilities.
Figure 6.
Ribbon diagram of a MD-derived model of the Ni(II)-bound NmtR with each N-terminus coordinating to the distal Ni(II) site, such the N-terminal tails cross the C-terminal α5 helices.. Each protomer is shaded light blue and salmon and Ni(II) ions are shaded green. The Gly2 α-NH2 group and His3 Nδ1 are shown coordinated to the Ni(II); other coordinating side chains (D91, H93, H104 and H107) are not shown for clarity. A zone of 10 Å around each Ni(II) ion is shaded blue and red, respectively; this is the region where one expects to see complete paramagnetic broadening on amide crosspeaks (compare to Fig. 3A). The agreement between theory and experiment is excellent, with the exception of residues 6-10 (see text for details).
Ratiometric Pulsed Amidination Mass Spectrometry
To obtain additional evidence of the participation of the N-terminal α-amino group in Ni(II) coordination, we took advantage of the fact that NmtR contains no lysine residues, with the only primary amine the N-terminal α-NH2 group (see Fig. S1). We reasoned that the reactivity of the α-amine to an amine-modifying reagent would be greatly attenuated in Ni(II)-bound state vs. the apo-state or the Zn(II)-bound state, which adopts a tetrahedral coordination geometry (Fig. 5A).14,26,70 A ratiometric pulsed amidination mass spectrometry method was used to obtain second order rate constants for amidination of the α-amino group in these allosteric states.27 Here, NmtR is reacted with a pulse of S-methyl thioacetimidate (SMTA) for time, t, followed by chase with an excess of S-methyl thiopropionimidate (SMTP) (Δ=14 amu), with the reaction quenched and protein subsequently digested with trypsin. Tryptic peptides are then subjected to high resolution MALDI-TOF mass spectrometry and peak areas of the amidinated (SMTA) and propionimidated (SMTP) N-terminal heptapeptide (2GHGVEGR8) quantified by integration. The raw mass spectrometry of this region of the mass spectrum is shown in Fig. 7, with the data fit to a single exponential to extract the pseudo-first order rate constant for amidination (k) (Fig. 8) and second order rate constants compiled in Table 2. These data reveal that the α-amino group is highly reactive in the apo-state (≈94 M−1 min−1) and is at most 2-fold protected when bound to non-cognate metal ion Zn(II) (≈50 M−1 min−1). In strong contrast, Ni(II) affords 100-fold protection relative to apo-NmtR (≈1 M−1 min−1). These data taken collectively strongly support Ni(II) coordination model shown (Fig. 5B) in which the α-amino group of Gly2 forms a direct coordination bond to the Ni(II) ion via a multidentate interaction with other donor atoms in the N-terminal region, e.g. His3,26 as found in other proteins involved in Ni(II) sensing and trafficking.74-77
Figure 7.
A series of MALDI-TOF spectra of tryptic peptide 2-8 are shown as a function of pulse amidination time (t) in apo-NmtR (A), Zn(II)-NmtR (B) and Ni(II)-NmtR (C). The isotope distribution of two different modified peptides are labeled as A (singly-acetamidinated) (752.34 D observed; 752.40 D expected) and P (singly-propionamidinated) (766.34 D observed; 766.40 D expected) with observed m/z shown.
Figure 8.
Mol fraction of propionamidinated α-amino group, ΘP, plotted as a function of pulse amidination time (t) in apo-NmtR (A), Zn(II)-NmtR (B) and Ni(II)-NmtR (C). Pseudo first order rate constants were determined by fitting the data with single exponential decay Θ = Ae−kt with parameters compiled in Table 2.
Table 2.
Second-order Amidination Rate constants Obtained for the α-amino group of various NmtRsa
| NmtR | Second order rate constant (MSMTA−1 min−1) |
Amplitude (Ao) |
|---|---|---|
| Apo | 93.5 ± 9.8 | 0.96 ± 0.05 |
| Zn(II) | 49.2 ± 10.1 | 0.76 ± 0.02 |
| Ni(II) | 0.96 ± 0.43 | 0.82 ± 0.02 |
Derived from the data shown in Figs. 7-8 by fitting ΘP vs. pulse time to a single exponential function with no offset.
Dynamical changes in NmtR on Ni(II) binding and implications for DNA operator binding
Although the paramagnetic zone of influence broadens beyond detection those residues within ≈10 Å of the bound Ni(II) ions,73 there are other changes in the 15N-1H HSQC spectrum of the Ni(II) bound NmtR that are likely reporting on changes in conformational dynamics that are important for Ni(II)-mediated negative regulation of DNA binding (Fig. 3A). In particular, inspection of the DNA binding interfacial region reveals several 15N-1H correlations that are significantly exchange broadened beyond detection upon Ni(II) binding, including those corresponding to S34 near the N-terminus of α2, Q60-S61 at the N-terminus of the α4 recognition helix, and G82-R83 at the tip of the β-wing (Fig. 4; Fig. S6). These residues are conserved in at least 8/9 of the candidate NmtRs (see Fig. S1) and correspond to N27, Q53-S54 and G75-Q76 in the related zinc sensor S. aureus CzrA.14 Q53E and S54A substitutions in CzrA show 4800- and 105-fold reductions in DNA binding affinity16 and inspection of the molecular dynamics-based model of the apo-CzrA-DNA operator complex18 reveals that all five residues define key contact regions across the entire protein-DNA interface (Fig. S7). Although the structure of the apo-NmtR-DNA complex is not yet available, these residues likely define the interfacial region of the NmtR-DNA complex as well, given the very high conservation of the residues in the DNA recognition α4 helices and the near identity of the DNA operators to which CzrA and NmtR bind (NmtO: 5′-TGAACATATGATCA; CzrO: 5′-TGAACAAATATTCA).16 If so, these data suggest that Ni(II)-induced dynamic disorder in the μs-ms timescale of key DNA interacting regions contributes to the negative regulation of DNA binding.
The amide group crosspeak corresponding to G74 in the turn between the α4 and β1 strands is also significantly broadened and shifted on Ni(II) binding (Fig. 4C). G74 corresponds to His67 in CzrA, the backbone carbonyl group of which accepts an intersubunit hydrogen bond from the nonliganding side chain of H97′ (H104′) to stabilize the allosterically inhibited low DNA binding affinity conformation.18,22 This suggests the possibility that this key interprotomer H104′-G74 linkage plays an important role in allosteric coupling by Ni(II) in NmtR as well, which explains in part why Zn(II) drives some negative regulation of DNA binding in NmtR even without coordination to the extreme N-terminus.26
CONCLUSIONS
In this work, we present the solution structure of the Ni/Co-specific ArsR-family regulator NmtR from M. tuberculosis and propose a Ni(II) coordination model that is consistent with previous thermodynamic experiments,26 x-ray absorption spectroscopy,14 chemical modification and Ni(II)-induced paramagnetic broadening measurements. Molecular dynamics simulations and quantum chemical calculations reveal such a coordination model can be readily accommodated by the structure of NmtR. It is unknown as yet precisely how distal Ni(II) site coordination by the extreme N-terminus drives a structural change in the dimer that mediates allosteric negative regulation of nmt DNA operator-promoter binding, except to say that loss of N-terminal coordination changes the coordination number and the rank order of allosteric effectors from Ni(II)>Zn(II) to Zn(II)>Ni(II).26 Efforts are underway to obtain a high resolution structural model of the Ni(II)-NmtR complex.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Dejian Ma, Indiana University, for his assistance in acquisition of the NMR data and Alfredo Guerra, Indiana University, for assistance with figures. We gratefully acknowledge the University of Florida High-Performance Computing Center for their support.
Funding
This work was supported by grants from the NIH to D. P. G. (GM042569) and K.M.M. (GM044974 and GM066859).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Supplementary Figures S1-S6 are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Berg JM, Godwin HA. Lessons from zinc-binding peptides. Annu Rev Biophys Biomol Struct. 1997;26:357–371. doi: 10.1146/annurev.biophys.26.1.357. [DOI] [PubMed] [Google Scholar]
- (2).Seneque O, Bonnet E, Joumas FL, Latour JM. Cooperative metal binding and helical folding in model peptides of treble-clef zinc fingers. Chemistry. 2009;15:4798–4810. doi: 10.1002/chem.200900147. [DOI] [PubMed] [Google Scholar]
- (3).Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- (4).Giedroc DP, Arunkumar AI. Metal sensor proteins: Nature’s metalloregulated allosteric switches. Dalton Trans. 2007;29:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- (5).Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chen PR, He C. Selective recognition of metal ions by metalloregulatory proteins. Curr Opin Chem Biol. 2008;12:214–221. doi: 10.1016/j.cbpa.2007.12.010. [DOI] [PubMed] [Google Scholar]
- (8).Agranoff D, Krishna S. Metal ion transport and regulation in Mycobacterium tuberculosis. Front Biosci. 2004;9:2996–3006. doi: 10.2741/1454. [DOI] [PubMed] [Google Scholar]
- (9).Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- (10).Busenlehner LS, Pennella MA, Giedroc DP. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 2003;27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- (11).Campbell DR, Chapman KE, Waldron KJ, Tottey S, Kendall S, Cavallaro G, Andreini C, Hinds J, Stoker NG, Robinson NJ, Cavet JS. Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. J Biol Chem. 2007;282:32298–32310. doi: 10.1074/jbc.M703451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Osman D, Cavet JS. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep. 2010;27:668–680. doi: 10.1039/b906682a. [DOI] [PubMed] [Google Scholar]
- (13).Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, Leme AF, Murakami MT, Benedetti CE. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem. 2011;286:26148–26157. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Structural elements of metal selectivity in metal sensor proteins. Proc Natl Acad Sci U S A. 2003;100:3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pennella MA, Arunkumar AI, Giedroc DP. Individual metal ligands play distinct functional roles in the zinc sensor Staphylococcus aureus CzrA. J Mol Biol. 2006;356:1124–1136. doi: 10.1016/j.jmb.2005.12.019. [DOI] [PubMed] [Google Scholar]
- (16).Arunkumar AI, Campanello GC, Giedroc DP. Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc Natl Acad Sci U S A. 2009;106:18177–18182. doi: 10.1073/pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009;131:17860–17870. doi: 10.1021/ja906131b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chakravorty DK, Wang B, Lee CW, Giedroc DP, Merz KM. Simulations of allosteric motions in the zinc sensor CzrA. J Am Chem Soc. 2012;134:3367–3376. doi: 10.1021/ja208047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kuroda M, Hayashi H, Ohta T. Chromosome-determined zinc-responsible operon czr in Staphylococcus aureus strain 912. Microbiol Immunol. 1999;43:115–125. doi: 10.1111/j.1348-0421.1999.tb02382.x. [DOI] [PubMed] [Google Scholar]
- (20).Singh VK, Xiong A, Usgaard TR, Chakrabarti S, Deora R, Misra TK, Jayaswal RK. ZntR is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of Staphylococcus aureus. Mol Microbiol. 1999;33:200–207. doi: 10.1046/j.1365-2958.1999.01466.x. [DOI] [PubMed] [Google Scholar]
- (21).Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- (22).Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- (23).Busenlehner LS, Weng TC, Penner-Hahn JE, Giedroc DP. Elucidation of primary (α3N) and vestigial (α5) heavy metal-binding sites in Staphylococcus aureus pI258 CadC: Evolutionary implications for metal ion selectivity of ArsR/SmtB metal sensor proteins. J. Mol. Biol. 2002;319:685–701. doi: 10.1016/S0022-2836(02)00299-1. [DOI] [PubMed] [Google Scholar]
- (24).Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- (25).Cavet JS, Meng W, Pennella MA, Appelhoff RJ, Giedroc DP, Robinson NJ. A nickel-cobalt-sensing ArsR-SmtB family repressor. Contributions of cytosol and effector binding sites to metal selectivity. J Biol Chem. 2002;277:38441–38448. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- (26).Reyes-Caballero H, Lee CW, Giedroc DP. Mycobacterium tuberculosis NmtR harbors a nickel sensing site with parallels to Escherichia coli RcnR. Biochemistry. 2011;50:7941–7952. doi: 10.1021/bi200737a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chang FM, Lauber MA, Running WE, Reilly JP, Giedroc DP. Ratiometric pulse-chase amidination mass spectrometry as a probe of biomolecular complex formation. Anal Chem. 2011;83:9092–9099. doi: 10.1021/ac202154r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- (29).Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;5:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- (30).Bax A, Clore GM, Gronenborn AM. 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 1990;88:425–431. [Google Scholar]
- (31).Montelione GT, Lyons BA, Emerson SD, Tashiro M. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J Am Chem Soc. 1992;114:10974–10975. [Google Scholar]
- (32).Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. 1993;101:114–119. [Google Scholar]
- (33).Ding K, Gronenborn AM. Sensitivity-enhanced 2D IPAP, TROSY-anti-TROSY, and E.COSY experiments: Alternatives for measuring dipolar 15N-1HN couplings. J Magn Reson. 2003;163:208–214. doi: 10.1016/s1090-7807(03)00081-8. [DOI] [PubMed] [Google Scholar]
- (34).Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- (35).Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- (36).Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Mag Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- (38).Case DA, Darden TA, T E, Cheatham I, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 11. University of California; San Francisco: 2010. [Google Scholar]
- (39).Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins-Struct Funct Bioinform. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pK(a)s and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Allen MP, Tildesley DJ. Computer Simulations of Liquids. Clarendon Press; Oxford: 1987. [Google Scholar]
- (43).van der Spoel D, van Maaren PJ, Berendsen HJC. A systematic study of water models for molecular simulation: Derivation of water models optimized for use with a reaction field. J Chem Phys. 1998;108:10220–10230. [Google Scholar]
- (44).Joung IS, Cheatham TE., 3rd. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Adelman SA, Doll JD. Generalized Langevin equation approach for atom-solid-surface scattering - General formulation for classical scattering off harmonic solids. J Chem Phys. 1976;64:2375–2388. [Google Scholar]
- (46).Doll JD, Dion DR. Generalized Langevin equation approach for atom-solid-surface scattering - Numerical techniques for Gaussian generalized Langevin dynamics. J Chem Phys. 1976;65:3762–3766. [Google Scholar]
- (47).York DM, Darden TA, Pedersen LG. The effect of long-range electrostatic interactions in simulations of macromolecular crystals - a Comparison of the Ewald and truncated list methods. J Chem Phys. 1993;99:8345–8348. [Google Scholar]
- (48).Tools M-DI. Maestro-Desmond Interoperability Tools. Version 3.0 ed Schrödinger; New York, NY: 2011. [Google Scholar]
- (49).Hartsough DS, Merz KM. Dynamic force field models: Molecular dynamics simulations of human carbonic anhydrase II using a quantum mechanical/molecular mechanical coupled potential. J. Phys. Chem. 1995;99:11266–11275. [Google Scholar]
- (50).Qsite. 5.7 ed Schrodinger, I. N. Y.; NY: 2011. [Google Scholar]
- (51).Zhao Y, Truhlar DG. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Acc. 2008;120:215–241. [Google Scholar]
- (52).Cramer CJ, Truhlar DG. Density functional theory for transition metals and transition metal chemistry. Phys Chem Chem Phys. 2009;11:10757–10816. doi: 10.1039/b907148b. [DOI] [PubMed] [Google Scholar]
- (53).Jorgensen WLT-R,J. Proc Natl Acad Sci USA. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Seabra GD, Walker RC, Elstner M, Case DA, Roitberg AE. Implementation of the SCC-DFTB method for hybrid QM/MM simulations within the amber molecular dynamics package. J Phys Chem A. 2007;111:5655–5664. doi: 10.1021/jp070071l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Gaus M, Cui QA, Elstner M. DFTB3: Extension of the self-consistent-charge density-functional tight-binding method (SCC-DFTB) J Chem Theory Computation. 2011;7:931–948. doi: 10.1021/ct100684s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Chakravorty DK, Wang B, Ucisik MN, Merz KM. Insight into the cation-pi interaction at the metal binding site of the copper metallochaperone CusF. J Am Chem Soc. 2011;133:19330–19333. doi: 10.1021/ja208662z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Yang Y, Wang B, Ucisik MN, Cui G, Fierke CA, Merz KM. Insights into the mechanistic dichotomy of the protein farnesyltransferase peptide substrates CVIM and CVLS. J Am Chem Soc. 2012;134:820–823. doi: 10.1021/ja209650h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JAJ, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamao C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople J. Gaussian 09. A.01 ed Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- (59).Petersson GA, Bennett A, Tensfeldt TG, Allaham MA, Shirley WA, Mantzaris J. A Complete Basis Set Model Chemistry .1. The Total Energies of Closed-Shell Atoms and Hydrides of the 1st-Row Elements. J Chem Phys. 1988;89:2193–2218. [Google Scholar]
- (60).Petersson GA, Allaham MA. A Complete Basis Set Model Chemistry .2. Open-Shell Systems and the Total Energies of the 1st-Row Atoms. J Chem Phys. 1991;94:6081–6090. [Google Scholar]
- (61).Wadt WR, Hay PJ. Abinitio effective core potentials for molecular calculations - potentials for main group elements Na to Bi. J Chem Phys. 1985;82:284–298. [Google Scholar]
- (62).Roy LE, Hay PJ, Martin RL. Revised basis sets for the LANL effective core potentials. J. Chem Theory Computation. 2008;4:1029–1031. doi: 10.1021/ct8000409. [DOI] [PubMed] [Google Scholar]
- (63).Peters MB, Yang Y, Wang B, Fusti-Molnar L, Weaver MN, Merz KM. Structural survey of zinc-containing proteins and development of the zinc AMBER force field (ZAFF) J Chem Theory Computation. 2010;6:2935–2947. doi: 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Yang Y, Chakravorty DK, Merz KM., Jr. Finding a needle in the haystack: computational modeling of Mg2+ binding in the active site of protein farnesyltransferase. Biochemistry. 2010;49:9658–9666. doi: 10.1021/bi1008358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- (66).Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free-energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- (67).Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Cavet JS, Dennison C, Graham AI, Harvie DR, Robinson NJ. NMR structural analysis of cadmium sensing by winged helix repressor CmtR. J Biol Chem. 2007;282:30181–30188. doi: 10.1074/jbc.M701119200. [DOI] [PubMed] [Google Scholar]
- (68).Ye J, Kandegedara A, Martin P, Rosen BP. Crystal structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J Bacteriol. 2005;187:4214–4221. doi: 10.1128/JB.187.12.4214-4221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Cook WJ, Kar SR, Taylor KB, Hall LM. Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins. J Mol Biol. 1998;275:337–346. doi: 10.1006/jmbi.1997.1443. [DOI] [PubMed] [Google Scholar]
- (70).Guerra AJ, Giedroc DP. Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch Biochem Biophy. 2011;519:210–222. doi: 10.1016/j.abb.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).VanZile ML, Chen X, Giedroc DP. Structural characterization of distinct alpha3N and alpha5 metal sites in the cyanobacterial zinc sensor SmtB. Biochemistry. 2002;41:9765–9775. doi: 10.1021/bi0201771. [DOI] [PubMed] [Google Scholar]
- (72).Pochapsky TC, Pochapsky SS, Ju T, Hoefler C, Liang J. A refined model for the structure of acireductone dioxygenase from Klebsiella ATCC 8724 incorporating residual dipolar couplings. J Biomol NMR. 2006;34:117–127. doi: 10.1007/s10858-005-5735-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Pochapsky SS, Sunshine JC, Pochapsky TC. Completing the circuit: direct-observe 13C,15N double-quantum spectroscopy permits sequential resonance assignments near a paramagnetic center in acireductone dioxygenase. J Am Chem Soc. 2008;130:2156–2157. doi: 10.1021/ja710187x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J Am Chem Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Nickel superoxide dismutase structure and mechanism. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- (76).Sankararamakrishnan R, Verma S, Kumar S. ATCUN-like metal-binding motifs in proteins: identification and characterization by crystal structure and sequence analysis. Proteins. 2005;58:211–221. doi: 10.1002/prot.20265. [DOI] [PubMed] [Google Scholar]
- (77).Chung KC, Cao L, Dias AV, Pickering IJ, George GN, Zamble DB. A high-affinity metal-binding peptide from Escherichia coli HypB. J Am Chem Soc. 2008;130:14056–14057. doi: 10.1021/ja8055003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.