Abstract
Currently, there is extensive information about circulating tumor cells (CTCs) and their prognostic value; however, little is known about other characteristics of these cells. In this prospective study, we assessed the gene transcripts of epithelial-to-mesenchymal transition inducing transcription factors (EMT-TFs) and cancer stem cell features in HER2+ metastatic breast cancer (MBC) patients. Epithelial cells were enriched from peripheral blood mononuclear cells (PBMCs) using antibody-coated anti-CD326 antibody (CD326+) magnetic beads, and the residual CD326− PBMCs were further depleted of leukocytes using anti-CD45 antibody-coated magnetic beads (CD326−CD45−). RNA was extracted from all cell fractions, reverse transcribed to cDNA, and subjected to quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to detect EMT-TFs (TWIST1, SNAIL1, ZEB1, and TG2) as a measure of CTCs undergoing EMT (EMT-CTCs). Additionally, PBMCs were analyzed using multi-parameter flow cytometry for ALDH activity and cancer stem cells (CSCs) that express CD24, CD44, and CD133. Twenty-eight patients were included in this study. At least one EMT-TF mRNA was elevated in the CTCs of 88.2% of patients and in the CD326−CD45− cell fraction of 60.7% of patients. The CD326−CD45− fraction of patients with elevated SNAIL1 and ZEB1 transcripts also had a higher percentage of ALDH+/CD133+ cells in their blood than did patients with normal SNAIL1 and ZEB1 expression (P=0.038). Our data indicate that HER2+ MBC patients have EMT-CTCs. Moreover, an enrichment of cancer stem cells was found in CD326−CD45− cells. Additional studies are needed to determine whether EMT-CTCs and CSCs have prognostic value in HER2+ MBC patients treated with trastuzumab-based therapy.
Keywords: circulating tumor cells, epithelial to mesenchymal transition, stem cells, HER2, CD133, metastatic breast cancer
INTRODUCTION
The presence of circulating tumor cells (CTCs) in the blood of metastatic breast cancer (MBC) patients is an independent prognostic and predictive factor.(1–9) Several techniques have been used to isolate, enumerate, and characterize CTCs, all of which rely on procedures to enrich for epithelial cells that express the epithelial cell adhesion molecule, EpCAM/CD326. The Food and Drug Administration-cleared CellSearch system defines CTCs as EpCAM/CD326+ nucleated cells that coexpress cytoplasmic cytokeratins (CK8, CK18, and CK19) and not the common leukocyte antigen, CD45.(1) Recently, we found that CTCs detected by CellSearch are heterogeneous, with different prognostic values among breast cancer subtypes.(10) In particular, CTCs lack predictive value in patients with HER2-amplified (HER2+) tumors treated with targeted therapy.(11, 12) One possible explanation for this observation is the loss of the expressions of EpCAM (13) on CTCs that may be undergoing epithelial-mesenchymal transition (EMT-CTCs) and the high efficacy of treatment with HER2-targeted therapy eliminating HER2+ CTCs.(11, 14)
Earlier we reported that a majority of disseminated tumor cells express a CSC phenotype(15) and others have suggested that a subset of disseminated tumor cells can be induced to become CSCs.(8, 16) To initiate metastases, disseminated tumor cells with characteristics of CSCs must acquire the ability to intravasate into and survive in the peripheral circulation. To acquire the properties of extravasation, migration, invasion, and metastasis seeding, epithelial cancer cells must undergo multiple biochemical and structural changes that enable them to acquire a mesenchymal cell phenotype. Through oncogenic EMT type 3, cancer cells shed many of their epithelial characteristics, detach from epithelial sheets, and undergo drastic morphologic and phenotypic alterations.(17–19) Loss of functional E-cadherin adhesion protein expression, leading to loss of cell polarity, is considered the hallmark of EMT. Members of the E-cadherin repressor/EMT inducers of the zinc-finger transcription factor SNAIL, Slug, ZEB1, ZEB2, FoxC2, and TWIST have been extensively evaluated.(18) In addition to loss of epithelial characteristics, EMT frequently coincides with the acquisition of motility, invasiveness, cytoskeletal protein changes (vimentin and α-smooth muscle actin expression), altered adhesion receptor expression (switching from E- to N-cadherin), and proteinase secretion (e.g., matrix metalloproteinases).(20) These findings suggest that determining the expressions of epithelial-cell surface markers, such as EpCAM (21), and cytoskeletal proteins, such as CKs (22), is not the ideal strategy for evaluating and characterizing heterogeneous CTCs with a mesenchymal phenotype.
Independent of these findings, studies of neoplastic tissues have demonstrated the presence of self-renewing, stem-like cells known also as CSCs (23) constitute a small minority of neoplastic cells in tumors and are defined operationally by their ability to seed new tumors.(24) During the tumor metastasis process, which is often enabled by EMT,(25) disseminated cancer cells seem to acquire a self-renewal capability, similar to that exhibited by stem cells, to spawn macroscopic metastases. This observation raises the possibility that the EMT process, which enables cancer cell dissemination, imparts a self-renewal capability to disseminating cancer cells. Therefore, researchers have addressed the possible association between EMT and stem cell formation. In many respects, cells that have undergone EMT behave similarly to stem cells isolated from normal or neoplastic cell populations.(26) Moreover, evidence demonstrates that HER2 plays a role in mammary carcinogenesis by regulating the stem cell population (27) and that HER2 overexpression in multiple breast cancer cell lines increases the fraction of ALDH1+ cells, which has a greater ability to invade and form tumors when transplanted into immunodeficient mice.(28) The correlation between breast CSCs and EMT has been shown in vitro and here we will try to investigate how the CTCs transit from primary tumor to enter peripheral blood and traverse to distant organs.
In this prospective translational study, we isolated epithelial and non-epithelial cells from blood of patients with HER2+ MBC and characterized their epithelial differentiation, transcript levels of transcription factors known to induce EMT (EMT-TF), and stem cell features to better understand circulating tumor cell seeding.
PATIENTS AND METHODS
Studied Subjects
This is a prospective translational study (LAB08-0079) conducted in the Department of Hematopathology, in collaboration with the Department of Breast Medical Oncology, The University of Texas MD Anderson Cancer Center in Houston, Texas, USA, and was approved by the institutional review board.
All MBC patients with immunohistochemical analysis (IHC) or fluorescent in situ hybridization (FISH) HER2+ disease, regardless of prior treatment history were eligible for this study. Between August 2009 and March 2010, we recruited 28 MBC patients with HER2+ tumors before starting a new line of therapy and 2 with HER2− tumors and HER2+ CTC.(29) At the same time of patient enrollment, 20 healthy women older than 18 years old (HDs) were recruited as control subjects; all MBC patients and HDs gave informed consent according to the institutional review board-approved protocol.
Clinicopathological Analysis
All tumor specimens were assigned a study identification number that was distinct from the patient’s medical record number. The histological type and grade of invasive disease were coded according to the World Health Organization classification system (30) and modified Black nuclear grading system, respectively.(31) Tumor specimens were analyzed in a Clinical Laboratory Improvement Act certified clinical pathology laboratory for estrogen and progesterone receptor status by IHC staining. Patients with at least one positive hormonal receptor were considered hormone receptor positive. HER-2/neu status was determined using IHC analysis or the PathVysion FISH kit (Abbott Molecular, Des Plaines, IL).(32) For FISH, 30 nuclei were scored per sample, and the number of HER2 and CEP17 signals were recorded. A ratio of HER2 to CEP17 >2.2 was defined as gene amplification; polysomy 17 was defined as ≥3 CEP17 signals per nucleus (average for 30 cells). Specimens with no evidence of staining on IHC analysis or no gene amplification by FISH were considered negative for HER-2/neu. Specimens that had a 3+ stain intensity on IHC analysis or gene amplification by FISH were considered HER2+.
Patients were required to have clinical and radiological evidence of MBC, with measurable or evaluable disease, before initiating therapy. All patients underwent imaging studies, laboratory evaluations, and treatment planning at MD Anderson Cancer Center. Disease status was evaluated every 6–12 weeks using the same imaging techniques, depending on the treatment schedule, until loss to follow-up or death. Response to therapy was evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST) v1.1.(33)
Blood Specimens
Patients provided 30 mL of peripheral blood (PB) in heparin anticoagulant before initiating a new treatment to isolate tumor cells. The blood samples were processed immediately or no later than 6 hours after phlebotomy. Contemporaneously, 7.5 mL of PB were collected in a Veridex CellSave tube for CTC isolation and enumeration using CellSearch (Veridex, LLC, Warren, NJ, USA).
Detection of CTCs by CellSearch
The CellSearch system was used to detect CTCs in 7.5 mL of PB, as previously described.(1) In brief, PB samples were subjected to EpCAM+ cell enrichment with anti-EpCAM-coated ferrous particles. Thereafter, EpCAM-enriched cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and reacted with anti-CD45 antibody to identify leukocytes, and with a cocktail of antibodies against CK-8, -18, or -19.(34) CTCs were defined as EpCAM+ nucleated cells (DAPI-stained nucleus) that lacked surface expression of CD45 but expressed cytoplasmic CK.(34) Samples were considered positive if they had ≥5 CTCs per 7.5 mL of PB.
CTC Detection by Magnetic Bead Separation
PBMCs were isolated from 30 mL of PB using a ficoll-hypaque density gradient, washed twice with sterile phosphate-buffered saline (PBS), and counted. Patient samples with >30 × 106 PBMCs were enriched for epithelial cells (EpCAM/CD326+, step 1) using magnetic bead separation methods, and the residual PBMCs were further depleted of leukocytes (CD326−CD45−, step 2). Patient samples with <30 × 106 PBMCs and HD blood were directly separated into leukocyte-enriched (CD45+) and -depleted (CD45−) cell fractions (step 2). The reason for not performing step 1 separation in samples with <30 × 106 PBMCs was the lower yield of cells of interest from each of the depletion steps, compared with samples that contained >30 × 106 PBMCs. The inability to collect sufficient numbers of cells of interest after each depletion step compromised our ability to extract sufficient concentration of RNA for downstream analysis by RT-PCR. In step 1, the PBMCs were adjusted to a concentration of 107 PBMCs per mL and incubated with 100 μL of magnetic beads coated with anti-CD326 antibody (Miltenyi-Biotec, Auburn, CA) and 100 μL of FcR blocking reagent (Miltenyi-Biotec) for 30 minutes at 4°C. In step 2, the CD326-depleted PBMCs were incubated with 20 μL of magnetic beads coated with anti-CD45 antibody (Miltenyi-Biotec) for 15 minutes at 4°C. Thereafter, the PBMCs were passed through a magnet-filled column on an AutoMACSPro Cell Separator system (Miltenyi-Biotec) using the positive selection protocol (POSSELD program) to enrich for EpCAM-positive cells and the negative selection protocol (DEPLETE program) to enrich for CD45-depleted CTCs. The CD45-depleted (CD45−) fraction underwent an additional run through the magnet-filled column using the MACS DEPLETES program to prevent any residual contamination of CD45+ cells. The mean purity of CD45− fraction was 96.6% (range, 89.4%–99.9%). The efficacy of the separation process was already showed in a previous paper.(13)
RNA Isolation and cDNA Synthesis
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions, and purified using the fully automated QIAcube system (Qiagen) to standardize the RNA isolation procedure. All RNA preparation and handling steps were performed under RNase-free conditions in a laminar flow hood. RNA concentration was determined using a NanoDrop 2000 spectrophotometer (ThermoScientific, Wilmington, DE), and RNA quality was assessed by evaluating the RNA integrity number using RNA 6000 Pico Kit and Agilent’s 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), according to the manufacturer’s instructions. RNA was converted in cDNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems Inc., Foster City, CA).
Cell Lines
The MCF-7, BT-474, SKBR-3, MDA-MB-231 breast cancer cells were obtained from the American Type Culture Collection (Manassas, VA) and grown in DMEM/F-12 culture medium supplemented with 10% fetal bovine serum (Tissue Culture Biologicals, Seal Beach, CA), 1% antibiotics penicillin and streptomycin. The immortalized human mammary epithelial cells HMLE and HMLE-TWIST were kindly provided by Dr. S.A. Mani (coauthor) and were cultured in a 1:1 mixture of MEGM (Lonza) and DME/F12 supplemented with 5% horse serum, 1% antibiotics penicillin and streptomycin, 10 μg/mL insulin, 10 ng/mL, EGF, 0.5 μg/mL hydrocortisone. RNA extracted from 1 × 106 cells was used as control for EMT-TFs (Supplemental Figure 1).(35)
Gene Expression Analysis by Reverse Transcription-Polymerase Chain Reaction
Gene expression was quantified with the TaqMan gene expression assay and RT-PCR on a 7900HT Fast Real-Time PCR System (ABI Applied Biosystems), following the manufacturer’s instructions. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used to normalize gene expression levels, and the relative expression of each gene was calculated using the equation 2−ΔCt, where ΔCt = mean Ctgene − CtGAPDH, or 2−ΔΔCt, where ΔΔCt = (Ctgene − CtGAPDH) patient sample − (Ctgene − CtGAPDH) HD sample. The TaqMan assays ZEB1: Hs00232783_m1, SNAIL1: Hs00195591_m1, TWIST1: Hs00361186_m1, TG2: Hs00190278_m1, GAPDH: Hs02758991_g1, EPCAM: Hs00158980_m1, KRT19: Hs00761767_s1, and HER2: Hs00170433_m1 were purchased from ABI.
Cancer Stem Cell Markers
PBMCs were evaluated for ALDH activity using the Aldefluor assay, according to the manufacturer’s protocol (StemCell Technologies, Vancouver, BC, Canada), as previously described.(15) In brief, 4 × 106 PBMCs were suspended in Aldefluor buffer containing a proprietary ATP-binding cassette transport inhibitor. One-third of the cells were reacted with 5 μL of the ALDH inhibitor, diethylamino-benzaldehyde, to serve as a negative control. Both the test reaction and negative control were incubated for 35 minutes at 37°C in a 5% CO2 atmosphere. Purified anti-CD44 monoclonal antibody (BD Pharmingen, San Diego, CA) was conjugated with Alexa700 using the Zenon antibody labeling kit (Invitrogen, Carlsbad, CA) prior to being reacted with the Aldefluor-labeled cells. In addition, pre-conjugated antibodies to anti-CD24 (PE) and anti-CD45 (PE-Cy7), both from BD Pharmingen (San Diego, CA), and anti-CD326 and anti-CD133 (APC, Miltenyi-Biotec) were used to label cells at ambient temperature, in the dark, for 30 minutes. An additional tube of Aldefluor-labeled cells was stained with the appropriate immunoglobulin isotype-matched controls. The stained cells were washed twice with PBS, and the cell pellet was suspended in 200 μL of PBS prior to being analyzed on a LSR-II flow cytometer capable of discriminating six-color fluorescence emission signals (BD Biosciences, San Jose, CA). Cellular debris was excluded from the analysis on the basis of low-forward light scatter. In the Aldefluor+ epithelial cell population, a subset of CSCs was defined as cells with a CD44+CD24lo and CD133+ phenotype.
Statistical Analyses
Thirty (30) patients with MBC were enrolled in this pilot study that was designed to determine the feasibility of isolating RNA from CTCs for the detection of EMT-TF genes. We excluded the two patients with HER2− tumor from this analysis. Patient characteristics were summarized using the median (range) for continuous variables and frequency (percentage) for categorical variables. Fisher’s exact test was used to assess the association between overexpression of EMT-TFs and other patient characteristics. Although the expression and the data are unlikely to yield sufficient data to relate to clinical outcome, we analyzed overall survival (OS) rate at median follow-up in the 17 patients according to the CTC gene expression profiling. OS duration was defined as the time from baseline blood draw to the date of death. All living patients were censored at the last follow-up date. A P value of <0.05 was deemed statistically significant. All statistical analyses were conducted using SPSS 19 software (IBM, Somers, NY).
To determine whether CTC samples overexpressed gene transcripts, we established the following cut-offs: in the CD326+ cell fraction, because the EpCAM/CD326+ cell fraction was not available in HDs, an EMT-TF and HER2 genes were considered elevated if the 2−ΔCt value of each gene was higher than the mean 2−ΔCt + 3SD in the CD45+ cell fraction. In the CD45− cell fraction, an EMT-TF was considered elevated if 2−ΔΔCt value was equal to or higher than 2. EMT-TF gene expression medians in the CD326+ cell fraction were compared using the Mann-Whitney test.
RESULTS
Patient Characteristics
Twenty-eight patients and 20 HDs were included in the study. Patient characteristics are shown in Table 1. Ten patients received trastuzumab plus chemotherapy (docetaxel or paclitaxel) as first-line treatment for metastatic disease, before study enrollment. Among the 28 patients included in this study, 17 had >30 × 106 PBMCs and underwent separation (step 1) plus depletion (step 2), and 11 patients had fewer (only step 2).
Table 1.
Patient characteristics.
| Characteristic | N (%) |
|---|---|
| Overall | 28 (100) |
|
| |
| Age yr (median, range) | 52 (35–72) |
|
| |
| ER/PR+ | 15 (53.6) |
| ER/PR− | 13 (46.4) |
|
| |
| First-line | 10 (35.7) |
| ≥ Second-line | 18 (64.3) |
|
| |
| CTC 0 | 12 (42.9) |
| CTC 1–4 | 7 (25) |
| CTCs ≥5 | 3 (10.7) |
| CellSearch not performed | 6 (21.4) |
Gene Expression Analysis of EpCAM/CD326+ Cell Fractions
In the EpCAM/CD326+ cell fractions, nine of 17 (52.9%) patients presented with low expression of EpCAM or KRT19 in the CD326+ cell fraction. We combined the EpCAM and KRT19 expressions and found that seven (41.2%) patients had low epithelial gene expression in their CD326+ cell fractions. At least one EMT-TF was elevated in 15 of 17 (88.2%) patients. TWIST1, SNAIL1, and TG2 were elevated in 13, 11, and 11 patients, respectively. ZEB1 was not elevated in any patients. HER2 was elevated in 16 of 17 (94.1%) patients. The median expression levels (2−ΔCT) of TWIST1, SNAIL1, TG2, and HER2 in the EpCAM/CD326+ cell fractions were higher than those in the CD45+ fractions (P <0.05). Figure 1 summarizes the mRNA expression levels of the analyzed genes in the CD326+ and CD45+ cell fractions.
Figure 1.
Gene expression levels of EMT-TFs (TWIST1, SNAIL1, ZEB1, and TG2) and HER2 in the CD45+ and CD326+ cell fractions. The GAPDH gene was used to normalize gene expression levels, and the relative expression of each gene was calculated using the equation 2−ΔCt, where ΔCt = mean Ctgene − CtGAPDH. Samples were considered positive if the gene expression of 2−ΔCt was higher than the mean of that in the CD45+ cell fraction 2−ΔCt + 3SD (cut-off line). The 2−ΔCt medians among fractions were compared using the Mann Whitney test.
Gene Expression Analysis of CD45− Cell Fractions
For TWIST1, SNAIL1, and ZEB1 gene expression, samples were available from 28 patients, respectively. For TG2, and HER2/neu gene expression, samples were available from 26 patients. The TWIST1 gene was not elevated (2−ΔΔCT < 2) in any of the patients; SNAIL1 was elevated in five of 28 (17.9%) patients; ZEB1 was elevated in seven of 28 (25%) patients; TG2 was elevated in eight of 26 (30.8%) patients. At least one EMT-TF gene was elevated in 17 of 28 (60.7%) patients. HER2/neu was elevated in seven of 26 (26.9%) patients. Figure 2 summarizes the results for the CD45− cell fractions.
Figure 2.
Gene expression levels of EMT-TFs (TWIST1, SNAIL1, ZEB1, and TG2) and HER2 in the CD45+ and CD45− cell fractions. The GAPDH gene was used to normalize gene expression levels, and the relative expression of each gene was calculated using the equation 2−ΔΔCt, where ΔΔCt = (Ctgene − CtGAPDH) patient sample − (Ctgene − CtGAPDH) HD sample. Samples were considered positive if the gene expression of 2−ΔΔCt was equal to or higher than 2 (cut-off line).
Association between CTCs Detected by CellSearch and EMT-TF Expression by RT-PCR
CTC counts, as determined using CellSearch, were available for 22 of the 28 patients. Nineteen (86.4%) evaluable patients had <5 CTCs per 7.5 mL of PB, and only three (13.6%) had ≥5 CTCs. Two of the three patients with ≥5 CTCs were about to receive first-line trastuzumab at the time of study enrollment. The CellSearch CTC numbers were correlated with EMT-TF expression levels in the CD326+ (n=10) and CD45− (n=22) cell fractions (Table 2). In 19 patients with <5 CTCs per 7.5 mL of PB, seven of nine (77.7%) presented with an elevated EMT-TF transcript in the CD326+ cell fraction and nine of 19 (47.4%) in the CD45− fraction. In particular, even in patients with no CTCs (n=12), we detected at least one EMT-TF transcript that was elevated in 66.7% of CD326+ cell fractions and in 50% of CD45− cell fractions. All patients with ≥5 CTCs per 7.5 mL of PB had elevated EMT-TFs transcripts in their CD45− cell fractions.
Table 2.
EMT-TFs expression according to CTC count and treatment.
| Subgroups | Data available (N. of patients) | EMT-TF+ CD326+, n/total (%) | EMT-TF+ CD45−, n/total (%) |
|---|---|---|---|
| N. of CTC | |||
| 0 | 12 | 4/6 (66.7) | 6/12 (50) |
| 1–4 | 7 | 3/3 (100) | 3/7 (42.9) |
| ≥ 5 | 3 | 1/1 (100) | 3/3 (100) |
|
| |||
| Trastuzumab naïve (group 1) | 9 | 5/6 (83.3) | 8/9 (88.9) |
| Trastuzumab treated (group 2) | 18 | 10/11 (90.9) | 8/18 (44.4) |
Association between Treatment and EMT-TF Expression
Ten patients (group 1) underwent baseline assessments before starting first-line therapy with trastuzumab plus chemotherapy for metastatic disease. The remaining 18 patients (group 2) had been treated with at least one line of trastuzumab-based therapy plus chemotherapy. As shown in Table 2, we detected at least one elevated EMT-TF transcript in the CD326+ cell fractions of groups 1 and 2 (83.3% vs 90.9% of patients, respectively, p = 0.596). On the other hand, we found at least one elevated EMT-TF transcript in the CD45− cell fraction in eight of nine (88.9%) patients in group 1 but only in eight of the 18 (44.4%) patients in group 2 (p = 0.033).
Association between Cancer Stem Cell and EMT-TFs
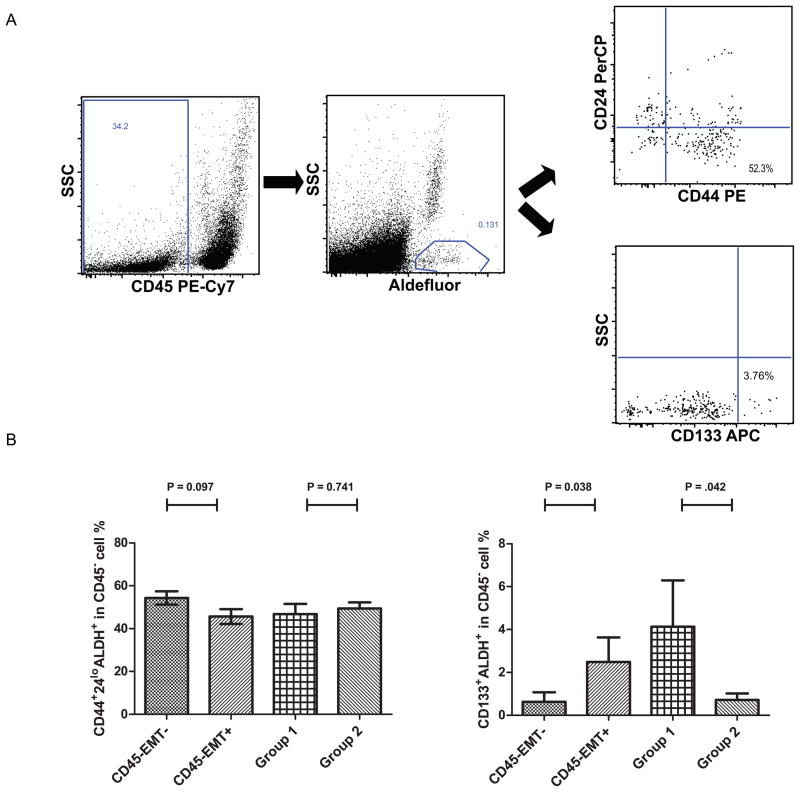
Twenty-eight patients were evaluated for cancer stem cell markers by multiparameter flow cytometry. We identified a cancer stem cell population (Aldefluor+ CD44+CD24lo) in the EpCAM+ (Aldefluor+ CD44+CD24lo/CD326+CD45−) and EpCAM−/CD45− cell fractions (Aldefluor+ CD44+CD24lo/CD326−CD45−) (Figure 3A). A mean of 0.1% of PBMCs were Aldefluor+ (range, 0.008%–0.57%); among these, 4.8% (range, 0.3%–17.2%) were CD326+CD45−and 59% (range, 43%–82%) were CD326−CD45−. A mean of 3% (range, 0.6%–13.5%) of Aldefluor+ cells were CD44+CD24lo/CD326+CD45−, and 48.7% (range, 37.2%–71.9%) of these cells were CD44+CD24lo/CD326− CD45−. After combining the CD44 and CD24 stem cell markers with EMT-TFs and the two groups of treatment, we found no particular association among stem cell markers (Figure 3B). Interestingly, there was an association between CD133 expression and EMT-TFs elevated in CD45− cell fractions. Patients with elevated EMT-TFs (SNAIL1 and ZEB1) in the CD45− cell fraction had a higher expression of CD45−Aldefluor+CD133+ cells in the blood (P =0.038). Moreover, HER2+ trastuzumab-naïve patients had a higher ratio of CD133+ stem cells in the blood than did those who were previously treated with trastuzumab (P =0.042) (Figure 3B).
Figure 3.
(A) Flowcytometric gating strategy to identify the cancer stem cell populations (Aldefluor+ CD44+CD24lo/CD45− and Aldefluor+ CD133+/CD45−). (B) We compared the percentage of stem cell marker-positive cells, Aldefluor+ CD44+CD24lo/CD45− on the left and Aldefluor+ CD133+/CD45− on the right, in the CD45− cell fractions with and without amplified EMT-TF. The groups were compared using the Mann-Whitney test as follows: at least one amplified EMT-TF vs. none, trastuzumab naïve (group 1) vs trastuzumab treated (group 2).
Clinical Outcome and EMT Markers
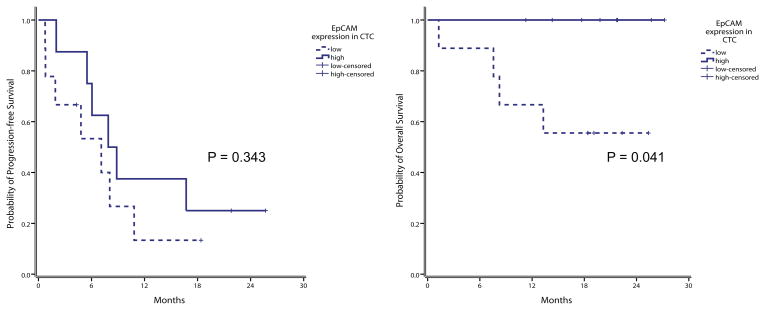
After a median follow-up duration of 19 months, 20 had experienced disease progression and eight of 28 patients have expired. Among the 17 who underwent gene expression profiling in the CD326+ cell fraction, nine patients with low EpCAM transcript expression had lower OS and PFS rates than those of the eight patients with high expression (44.4% of low-expression patients died vs 0% of high-expression patients; p = 0.041; for PFS, 87% vs 75%, respectively, p = 0.343) (Figure 4).
Figure 4.
(A) Progression-free survival (PFS) and (B) overall survival (OS) Kaplan Meier curves for all 17 patients according to EpCAM transcript expression in the CD326+ cell fraction. EpCAM high expression in green (n = 8); EpCAM low expression in blue (n = 9); time was measured from the basal blood draw to the death for OS and to radiologic evidence of disease progression (PFS).
DISCUSSION
In this pivotal translational trial, we characterized CTCs from HER2+ MBC patients, specifically looking for EMT-TFs and CSC features. We successfully isolated CTCs from the PB of MBC patients; these CTCs differed in epithelial differentiation and expressed EMT-TFs. Moreover, we showed that CTCs with EMT features (EMT-CTCs) can be detected even in patients with undetectable CTCs using CellSearch. Specifically, we found TWIST1 and SNAIL1 transcripts were elevated in the CD326+ cell fractions and SNAIL1 and ZEB1 transcripts were elevated in the CD45− cell fractions. These data are in agreement with published reports in which they indicated that a subset of primary breast cancer patients showed a subtype of CTCs that had undergone EMT could not be detected using currently available detection methods for CTCs.(36) Other researchers have found, independently of HER2+ subtype, positive expression of the EMT-related markers vimentin and fibronectin in EpCAM+/CKdim/CD45− cells, which do not meet the established definition of a CTC by CellSearch.(26, 37) The existence of hybrid CTCs with an epithelial/mesenchymal phenotype was observed also in patients with non-small cell lung cancer.(38)
Recent studies (26, 39) have demonstrated an association between EMT and acquisition of CSC characteristics, leading to the hypothesis that cancer cells that undergo EMT are capable of metastasizing through their acquired self-renewal potential, thereby enabling them to spawn the large cell populations that constitute macroscopic metastases. We found that most cells with a CSC phenotype were in the CD45− cell fraction. However, no EMT-TF genes was associated with the putative CSC markers (ALDH+ and CD44+/CD24lo/−). This inconsistency could be explained by the fact that some of patients were heavily pretreated and the heterogeneity of patient population and the small number of samples could explain the absence of significant association between stem cell markers and EMT-TFs. Interestingly, we found an association between ALDH+CD133+ cells and SNAIL1 and ZEB1 transcript expression in the CD45− cell fractions. Moreover the ability of trastuzumab to reduce the percentage of CSCs was demonstrated in a prior report.(28) These findings are consistent with those of Armstrong et al.,(40) that showed in addition to cells expressing both epithelial and mesenchymal markers, an unknown number of CTCs that are more mesenchymal-like and thus are EpCAM−. All these observations lend support to the new metastatic cascade theory postulated by Cheffer et al. in which CTCs are plastic and undergo bidirectional interconversions between stem and non-stem compartments.(41) In our study, CTCs in PB of HER2+ MBC patients is comprised of a heterogeneous population of differentiated epithelial cells that express EpCAM that are readily detected by CellSearch, to a CTC with an EMT/stem cell-like phenotype that does not express EpCAM and hence, cannot be detected by CellSearch. We are cognizant that our study included a small number of patients with only 22 of 28 women analyzed with CellSearch and more studies are needed to understand the biologic characteristics of the metastatic cascade and the possible effect of CSC targeted therapy on clinical outcomes. Another caveat of our study is related to the small quantity of gene material analyzed from a limited number of CTCs. All isolated CTCs were used for the EMT-TF expression and stem cell markers analysis, and no additional assays for protein and DNA assessments were possible.
In conclusion, we demonstrated, for the first time, that a cluster of CTCs with elevated EMT-TFs can be isolated from the PB of HER2+ MBC patients and that this cluster of CTCs is not always detectable using the current EpCAM-enrichment enumeration methods. We found that patients with EMT-TFs in CTCs had more ALDH+CD133+ cancer stem cell-like cells. Considering the small number of patients enrolled in this study, additional studies are needed to determine whether EMT events and CSCs have prognostic value in HER2+ MBC patients treated with targeted therapy.
Supplementary Material
Acknowledgments
We wish to thank Ann M. Sutton from the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editing the manuscript. This study was presented in part at the 2011 ASCO Annual Meeting as a poster presentation by Antonio Giordano.
Grant support: Human Breast Cancer Stem Cell Surrogates, CA138239-02, NIH/NCI (S.A. Mani, M. Cristofanilli, and J.M. Reuben). State of Texas Rare and Aggressive Breast Cancer Research Program, The Morgan Welch Inflammatory Breast Cancer Research Program and Clinic (H. Gao, S. Tin, R.H. Alvarez, V. Valero, N.T. Ueno, M. Cristofanilli, and J.M. Reuben). Assessment of Circulating Breast Cancer Stem Cells To Predict Recurrent Disease, W81XWH-09-1-0031 01, DOD (E. Cohen, and J.M. Reuben). University of Naples Federico II and Second University of Naples, PhD Program in Medical and Surgical Oncology and Clinical Immunology (A. Giordano).
Footnotes
Potential Conflicts of Interest: None.
References
- 1.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 2.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–30. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 3.Dawood S, Broglio K, Valero V, Reuben J, Handy B, Islam R, et al. Circulating tumor cells in metastatic breast cancer: from prognostic stratification to modification of the staging system? Cancer. 2008;113:2422–30. doi: 10.1002/cncr.23852. [DOI] [PubMed] [Google Scholar]
- 4.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–24. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 5.Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, et al. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27:5153–9. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nole F, Munzone E, Zorzino L, Minchella I, Salvatici M, Botteri E, et al. Variation of circulating tumor cell levels during treatment of metastatic breast cancer: prognostic and therapeutic implications. Ann Oncol. 2008;19:891–7. doi: 10.1093/annonc/mdm558. [DOI] [PubMed] [Google Scholar]
- 7.Tewes M, Aktas B, Welt A, Mueller S, Hauch S, Kimmig R, et al. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat. 2009;115:581–90. doi: 10.1007/s10549-008-0143-x. [DOI] [PubMed] [Google Scholar]
- 8.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giordano A, Cristofanilli M. CTCs in metastatic breast cancer. Recent Results Cancer Res. 2012;195:193–201. doi: 10.1007/978-3-642-28160-0_18. [DOI] [PubMed] [Google Scholar]
- 10.Giordano A, Giuliano M, De LM, Eleuteri A, Iorio F, Tagliaferri R, et al. Artificial neural network analysis of circulating tumor cells in metastatic breast cancer patients. Breast Cancer Res Treat. 2011;129:451–8. doi: 10.1007/s10549-011-1645-5. [DOI] [PubMed] [Google Scholar]
- 11.Giuliano M, Giordano A, Jackson S, Hess KR, De GU, Mego M, et al. Circulating tumor cells as prognostic and predictive markers in metastatic breast cancer patients receiving first-line systemic treatment. Breast Cancer Res. 2011;13:R67. doi: 10.1186/bcr2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giordano A, Giuliano M, De LM, Arpino G, Jackson S, Handy BC, et al. Circulating tumor cells in immunohistochemical subtypes of metastatic breast cancer: lack of prediction in HER2-positive disease treated with targeted therapy. Ann Oncol. 2011:mdr434v2-mdr434. doi: 10.1093/annonc/mdr434. [DOI] [PubMed] [Google Scholar]
- 13.Mego M, Mani SA, Lee BN, Li C, Evans KW, Cohen EN, et al. Expression of epithelial-mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int J Cancer. 2011;130:808–16. doi: 10.1002/ijc.26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bidard FC, Mathiot C, Degeorges A, Etienne-Grimaldi MC, Delva R, Pivot X, et al. Clinical value of circulating endothelial cells and circulating tumor cells in metastatic breast cancer patients treated first line with bevacizumab and chemotherapy. Ann Oncol. 2010 doi: 10.1093/annonc/mdq052. [DOI] [PubMed] [Google Scholar]
- 15.Reuben JM, Lee BN, Gao H, Cohen EN, Mego M, Giordano A, et al. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44(+)CD24(lo) cancer stem cell phenotype. Eur J Cancer. 2011;47:1527–36. doi: 10.1016/j.ejca.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mego M, Mani SA, Cristofanilli M. Molecular mechanisms of metastasis in breast cancer-clinical applications. Nat Rev Clin Oncol. 2010;7:693–701. doi: 10.1038/nrclinonc.2010.171. [DOI] [PubMed] [Google Scholar]
- 17.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98:1512–20. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Pluijm G. Epithelial plasticity, cancer stem cells and bone metastasis formation. Bone. 2011;48:37–43. doi: 10.1016/j.bone.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Mego M, Mani SA, Lee BN, Li C, Evans KW, Cohen EN, et al. Expression of epithelial-mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int J Cancer. 2011 doi: 10.1002/ijc.26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gradilone A, Raimondi C, Nicolazzo C, Petracca A, Gandini O, Vincenzi B, et al. Circulating tumor cells lacking cytokeratin in breast cancer: the importance of being mesenchymal. J Cell Mol Med. 2011 doi: 10.1111/j.1582-4934.2011.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 24.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–6. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedard PL, Cardoso F, Piccart-Gebhart MJ. Stemming resistance to HER-2 targeted therapy. J Mammary Gland Biol Neoplasia. 2009;14:55–66. doi: 10.1007/s10911-009-9116-x. [DOI] [PubMed] [Google Scholar]
- 28.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–30. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuben JM, Lee BN, Li C, Gao H, Broglio KR, Valero V, et al. Circulating tumor cells and biomarkers: implications for personalized targeted treatments for metastatic breast cancer. Breast J. 2010;16:327–30. doi: 10.1111/j.1524-4741.2010.00910.x. [DOI] [PubMed] [Google Scholar]
- 30.The World Health Organization Histological Typing of Breast Tumors. The World Organization. Am J Clin Pathol. (2) 1982;78:806–16. doi: 10.1093/ajcp/78.6.806. [DOI] [PubMed] [Google Scholar]
- 31.Black MM, Speer FD. Nuclear structure in cancer tissues. Surg Gynecol Obstet. 1957;105:97–102. [PubMed] [Google Scholar]
- 32.Vranic S, Teruya B, Repertinger S, Ulmer P, Hagenkord J, Gatalica Z. Assessment of HER2 gene status in breast carcinomas with polysomy of chromosome 17. Cancer. 2011;117:48–53. doi: 10.1002/cncr.25580. [DOI] [PubMed] [Google Scholar]
- 33.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1. 1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 35.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, et al. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101:61–6. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasimir-Bauer S, Hoffmann O, Wallwiener D, Kimmig R, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012;14:R15. doi: 10.1186/bcr3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raimondi C, Gradilone A, Naso G, Vincenzi B, Petracca A, Nicolazzo C, et al. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat. 2011;130:449–55. doi: 10.1007/s10549-011-1373-x. [DOI] [PubMed] [Google Scholar]
- 38.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105:1338–41. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting R, Turnbull J, et al. Circulating Tumor Cells from Patients with Advanced Prostate and Breast Cancer Display Both Epithelial and Mesenchymal Markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–5. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.