SUMMARY
The Wilms’ tumor 1 protein WT1 is a transcriptional regulator that is involved in cell growth and differentiation. The transcriptional corepressor BASP1 interacts with WT1 and converts WT1 from a transcriptional activator to a repressor. Here, we demonstrate that the N-terminal myristoylation of BASP1 is required in order to elicit transcriptional repression at WT1 target genes. We show that myristoylated BASP1 binds to nuclear PIP2, which leads to the recruitment of PIP2 to the promoter regions of WT1-dependent target genes. BASP1’s myristoylation and association with PIP2 are required for the interaction of BASP1 with HDAC1, which mediates the recruitment of HDAC1 to the promoter and elicits transcriptional repression. Our findings uncover a role for myristoylation in transcription, as well as a critical function for PIP2 in gene-specific transcriptional repression through the recruitment of histone deacetylase.
INTRODUCTION
The Wilms’ tumor 1 protein WT1 plays a central role in development and in both pediatric and adult cancers (Rivera and Haber, 2005; Hohenstein and Hastie, 2006; Huff, 2011). WT1 is a transcriptional regulator that can either activate or repress transcription (Roberts, 2005). We identified BASP1 (NAP-22, CAP-23) as a WT1-binding protein that mediates its transcriptional repression activity (McKay et al., 1999; Carpenter et al., 2004). Several previous studies showed that BASP1 is stoichiometrically N-terminally myristoylated in several tissues (e.g., neuronal, kidney, testis, and lymphoid) and cell lines, and specifically associates with phosphatidylinositol 4,5-bisphosphate (PIP2) at the cell membrane (Maekawa et al., 1994; Mosevitsky et al., 1997; Takasaki et al., 1999; Laux et al., 2000; Terashita et al., 2002; Mosevitsky, 2005; Epand, 2008; see Figure 1A for summary). The interaction of BASP1 with PIP2 requires both the myristoylation motif and the N-terminal region of BASP1, which contains a conserved serine (residue 6) that can be phosphorylated by protein kinase C (PKC) in vivo (Maekawa et al., 1994; Mosevitsky et al., 1997; Kashihara et al., 2000; Mosevitsky, 2005). Phosphorylation of this residue disrupts the interaction of BASP1 with lipids (Peitzsch and McLaughlin, 1993; Mosevitsky et al., 1997; Mosevitsky, 2005).
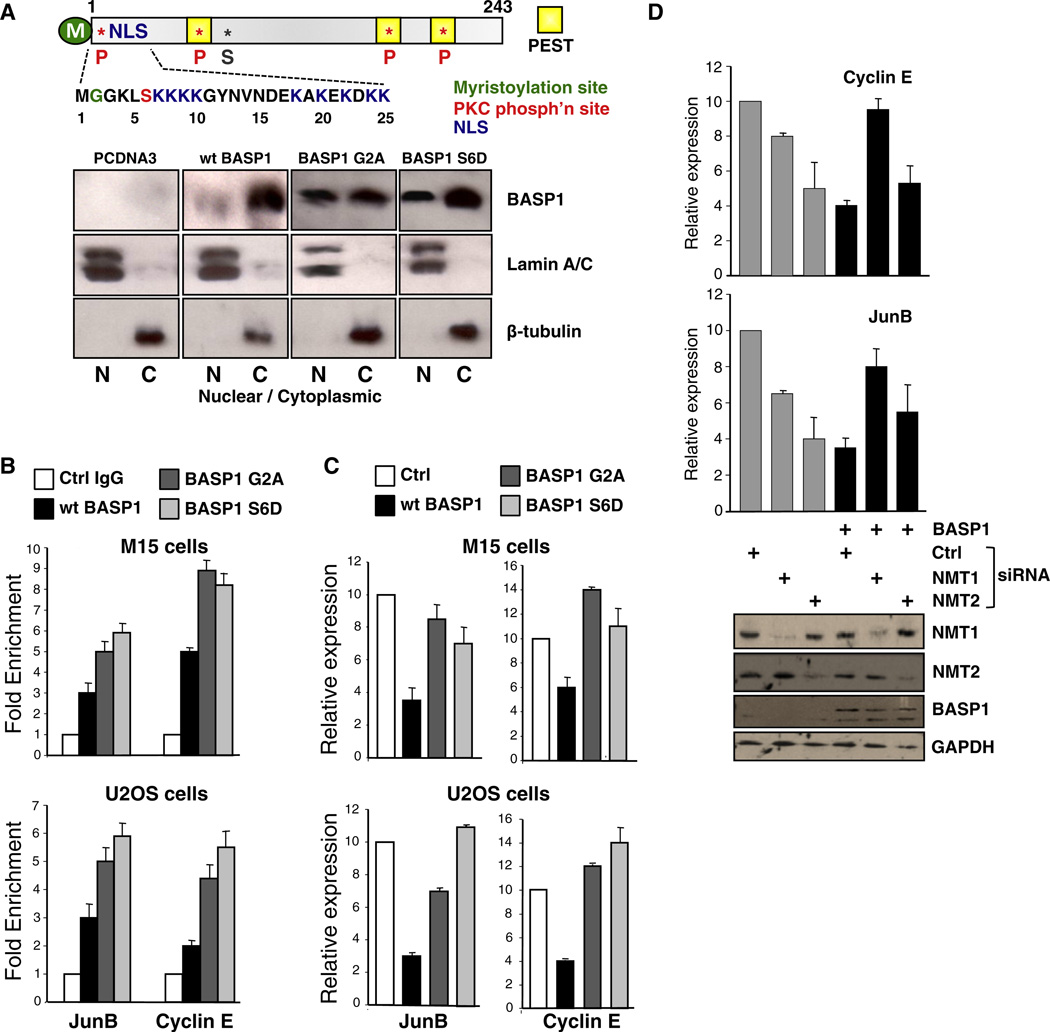
Figure 1. The Myristoylation of BASP1 Is Required for Its Transcriptional Repressor Activity.
(A) Schematic of BASP1. The N-terminal 20 residues and myristoylated G2 are shown below, and the PKC site at S6, and NLS and PEST sequences are indicated. pCDNA3 alone or driving the expression of wild-type BASP1, BASP1 G2A, or BASP1 S6D were transfected into M15 cells, and 48 hr later the nuclear (N) and cytoplasmic (C) fractions were immunoblotted with the antibodies indicated. Lamin and β-tubulin are nuclear and cytoplasmic controls. M, myristoylation site; NLS, nuclear localization sequence; P, phosphorylation site; S, sumoylation site.
(B) M15 cells (top) or U2OS cells (bottom) were transfected as in (A), and ChIP was performed with anti-BASP1 antibodies or control IgG. Primers to amplify the WT1-binding regions of the JunB and Cyclin E promoters were used in quantitative PCR (qPCR) to determine fold enrichment relative to a noncoding region. Error bars are the standard deviation from the mean (SDM) of three independent experiments.
(C) M15 cells (top) or U2OS cells (bottom) were transfected as in (A) and 48 hr later, RNA and cDNA were prepared. qPCR was performed to detect GAPDH, JunB, and Cyclin E mRNA. The data are presented relative to GAPDH mRNA and error bars denote the SDM of three independent experiments.
(D) M15 cells were transfected with pCDNA3 or pCDNA3 driving expression of BASP1 along with control, NMT1, or NMT2 siRNA. cDNA was prepared 48 hr later, and qPCR was performed to detect GAPDH, Cyclin E, and JunB mRNA. The graphs represent Cyclin E (top) and JunB (bottom) expression relative to GAPDH (error bars are the SDM of three independent experiments). Below, simultaneously prepared whole-cell extracts were immunoblotted with the antibodies indicated.
See also Figure S1.
BASP1 is present in the nucleus, mediated by a functional bipartite nuclear localization sequence (NLS) located between residues 7 and 25, and localizes to the promoters of several WT1 target genes (Carpenter et al., 2004; Green et al., 2009; Essafi et al., 2011; Goodfellow et al., 2011; see Figure 1A for summary). BASP1 also regulates other transcription factors, and can inhibit cellular transformation induced by v-myc and block the regulation of myc target genes (Hartl et al., 2009). Moreover, BASP1 is downregulated in myc-transformed cells and in a significant proportion of hepatocellular carcinomas and leukemias through silencing of the BASP1 gene by methylation (Yeoh et al., 2002; Moribe et al., 2008; Hartl et al., 2009).
The mechanisms by which BASP1 acts as a transcriptional corepressor are not known. In this study, we demonstrate that the N-terminal myristoylation of BASP1 and the capacity of BASP1 to interact with PIP2 are critical for its transcriptional corepressor function with WT1. The BASP1-PIP2 interaction promotes the recruitment of HDAC1 to the gene promoter region, which then leads to transcriptional repression. Our findings uncover a role for myristoylation in transcription and a mode of gene-specific transcriptional repression through nuclear lipids.
RESULTS AND DISCUSSION
Myristoylation of BASP1 Is Required for Its Transcriptional Corepressor Function
BASP1 is myristoylated at its N terminus, which promotes its interaction with PIP2 at the cell membrane (reviewed in Mosevitsky, 2005; Epand, 2008; see Figure 1A for summary). Mutation of BASP1 glycine-2 to alanine (G2A) completely abrogates myristoylation, disrupts the interaction with PIP2, and leads to membrane detachment (Takasaki et al., 1999; Terashita et al., 2002; Epand, 2008; Korshunova et al., 2008). Phosphorylation of BASP1 at serine-6 also causes the release of BASP1 from lipids by creating an unfavorable electrostatic environment close to the myristoyl motif (Peitzsch and McLaughlin, 1993; Mosevitsky et al., 1997). Thus, the phosphomimetic mutant BASP1 derivative S6D also leads to membrane detachment (Mosevitsky, 2005). We hypothesized that a BASP1 mutant derivative that prevents myristoylation (G2A), or a mimic of phosphorylation of BASP1 (serine-6 [S6D]) would lead to increased nuclear localization of BASP1. Mouse embryonic kidney M15 cells were transfected with pcDNA3 or pcDNA3, driving the expression of wild-type BASP1, BASP1 G2A, or BASP1 S6D. An analysis of nuclear and cytoplasmic extracts demonstrated that both the prevention of BASP1 myristoylation (G2A) and the use of the phosphomimetic S6D led to an increase of the proportion of BASP1 in the nucleus (Figure 1A).
WT1 and BASP1 coimmunoprecipitate from nuclear extracts of both M15 (Carpenter et al., 2004) and U2OS cells (Figure S1). Therefore, we performed a chromatin immunoprecipitation (ChIP) analysis to analyze the association of wild-type BASP1, BASP1 G2A, or BASP1 S6D with the WT1-binding regions of the JunB and Cyclin E promoters in either M15 cells or U2OS cells. Consistent with the enhanced nuclear localization of BASP1 G2A and BASP1 S6D compared with wild-type BASP1, there was an increase in promoter localization of the mutant BASP1 derivatives (Figure 1B). Surprisingly, however, when we analyzed the expression of the endogenous JunB and Cyclin E genes in either M15 cells or U2OS cells, we found that BASP1 G2A and BASP1 S6D were both significantly defective in transcriptional repression compared with wild-type BASP1 (Figure 1C). These results suggest that myristoylation of BASP1 and the capacity for PIP2 binding are required for BASP1’s function as a WT1 transcriptional corepressor.
We sought to confirm and extend these findings by blocking BASP1 myristoylation through ablation of N-myristoyltransferase (NMT) activity. Mammalian cells express two isoforms of NMT, termed NMT1 and NMT2. M15 cells were transfected with either pcDNA3 or the same vector, driving expression of BASP1 along with either a control small interfering RNA (siRNA) or siRNA directed to NMT1 or NMT2. Western blotting confirmed successful NMT1 and NMT2 ablation by their respective siRNAs (Figure 1D). As before, expression of wild-type BASP1 repressed transcription of both the Cyclin E and JunB genes. However, when expression of NMT1, but not NMT2, was ablated, the effect of BASP1 expression on both genes was reduced. These data independently confirm that myristoylation of BASP1 is required for its transcriptional corepressor activity.
BASP1 Associates with Nuclear PIP2
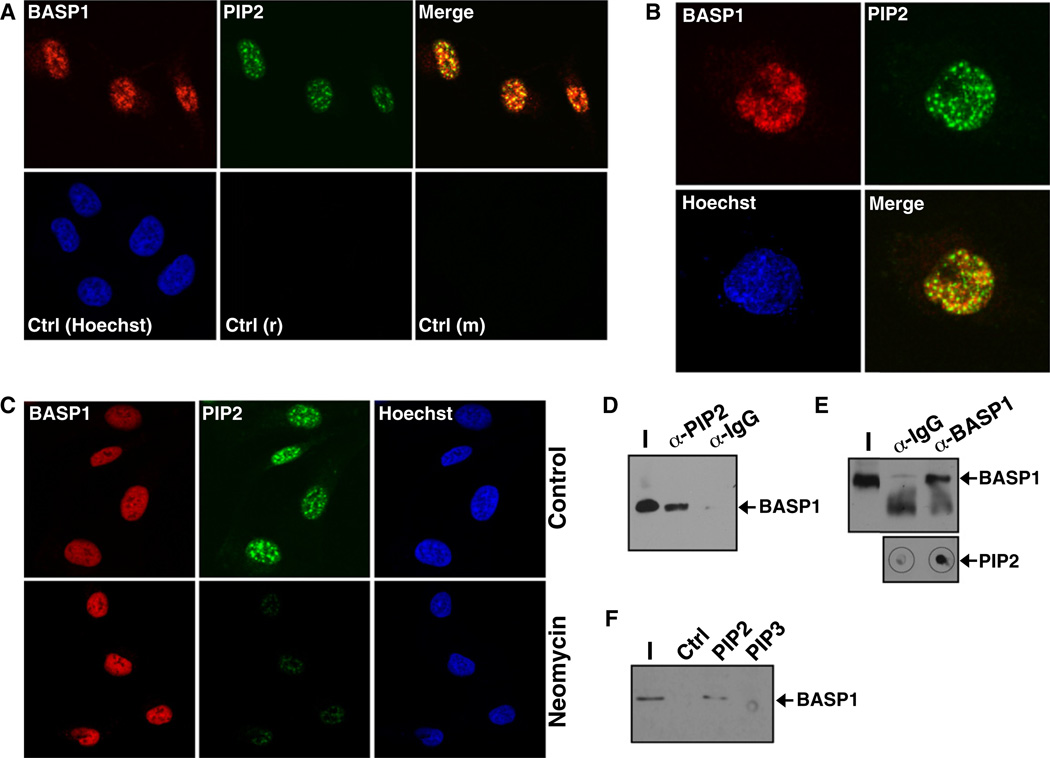
It is well established that various phospholipids, including PIP2, are present in the cell nucleus (Albi and Viola Magni, 2004; Barlow et al., 2010; Keune et al., 2011). We therefore sought to determine whether endogenous BASP1 colocalizes with PIP2 in the nucleus by using coimmunofluorescence. We used anti-PIP2 antibodies in a protocol that was previously optimized to specifically detect detergent-resistant PIP2 in the nucleus (Osborne et al., 2001; Mortier et al., 2005). Both PIP2 and BASP1 were detected in the nucleus of U2OS cells (Figure 2A), and confocal microscopy revealed significant nuclear colocalization (Figure 2B). To confirm that the anti-PIP2 antibodies were recognizing PIP2, we treated the U2OS cells with neomycin (2 mg/ml) for 12 hr prior to immunofluorescence with anti-PIP2 antibodies. Neomycin chelates PIP2 and was previously shown to block its interaction with the same antibodies used in our experiments (Osborne et al., 2001). Treatment of U2OS cells with neomycin significantly reduced the nuclear signal with the anti-PIP2 antibodies, confirming that they specifically recognize phosphoinositides (Figure 2C). Comparable results were obtained when we analyzed BASP1 and PIP2 localization in MCF7 cells (Figure S2). To provide further evidence that BASP1 interacts with nuclear PIP2, we performed immunoprecipitation with HeLa cell nuclear extracts. Anti-PIP2 immunoprecipitates contained BASP1 (Figure 2D), and dot-blotting confirmed that the anti-BASP1 immunoprecipitates contained PIP2 (Figure 2E). We sought to confirm these results by direct binding of BASP1 from nuclear extracts to control agarose beads, or agarose beads containing PIP2, or phosphatidylinositol 3,4,5 triphosphate (PIP3; Figure 2F). Nuclear BASP1 was retained by the PIP2 beads but not by the PIP3 beads. Taken together, the data in Figure 2 demonstrate that BASP1 interacts with nuclear PIP2.
Figure 2. BASP1 Associates with Nuclear PIP2.
(A) U2OS cells were subjected to coimmunofluorescence with rabbit BASP1 antibodies and mouse PIP2 antibodies. Control anti-mouse (m) and anti-rabbit (r) antibodies are shown in the bottom row.
(B) As in (A) except that confocal microscopy was used to visualize nuclear BASP1 and PIP2.
(C) U2OS cells were mock-treated or treated with 2 mg/ml neomycin for 24 hr prior to immunofluorescence with either BASP1 or PIP2 antibodies.
(D) HeLa nuclear extract was subjected to immunoprecipitation with control IgG or PIP2 antibodies, and the samples were immunoblotted with BASP1 antibodies.
(E) HeLa nuclear extract was subjected to immunoprecipitation with control IgG or BASP1 antibodies, and the samples were immunoblotted with BASP1 antibodies or dot-blotted with PIP2 antibodies.
(F) Nuclear extract derived from K562 cells stably transfected to express wild-type BASP1 was incubated with control agarose beads or beads containing equal amounts of either PIP2 or PIP3. After the beads were washed, the bound products were subjected to immunoblotting with BASP1 antibodies. I, input nuclear extract.
See also Figure S2.
BASP1-Dependent Recruitment of PIP2 to the Promoter of WT1 Target Genes
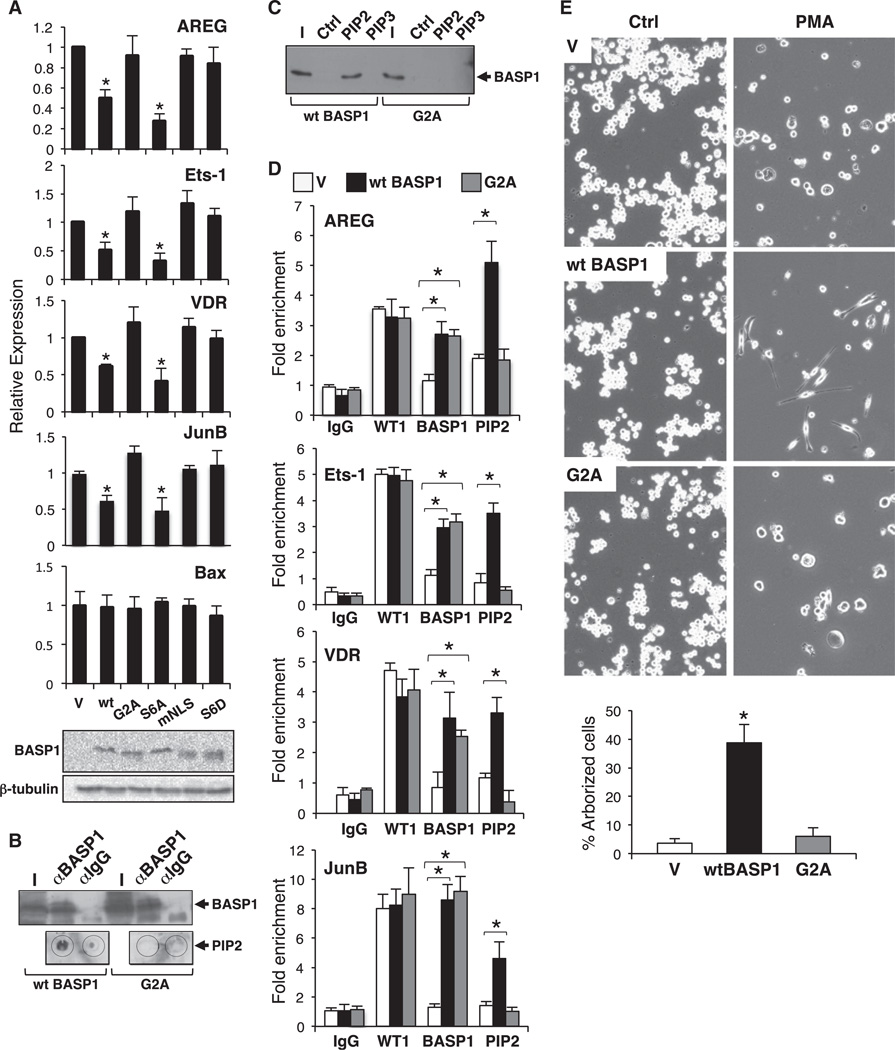
We next explored the functional role that PIP2 plays in BASP1-dependent transcriptional repression. K562 cells express wild-type WT1 but do not express BASP1, and thus the endogenous WT1 acts primarily as a transcriptional activator (Goodfellow et al., 2011). Introduction of BASP1 into K562 cells leads to the repression of WT1 target genes via a WT1-dependent mechanism that requires the recruitment of BASP1 to the gene promoter (Goodfellow et al., 2011). We therefore generated several K562 cell line derivatives that express either wild-type BASP1 or the BASP1 mutant derivatives aG2A (which prevents myristoylation), S6A (which prevents phosphorylation of BASP1 at Ser-6), S6D (which mimics phosphorylation of Ser-6), and mNLS (K7A:K9A, which abolishes nuclear localization of BASP1, as we previously demonstrated; Goodfellow et al., 2011). We then prepared cDNA from the cell lines and measured the expression of the WT1 target genes amphiregulin (AREG), Ets-1, Vitamin D receptor (VDR), and JunB, along with Bax as a control non-WT1 target gene (Figure 3A). As we reported in a recent study (Goodfellow et al., 2011), expression of wild-type BASP1 in K562 cells resulted in reduced expression of AREG, Ets-1, VDR, and JunB, but not Bax. In addition, as we also noted in that study, disruption of the nuclear localization signal of BASP1 (mNLS) ablated BASP1 corepressor function. Consistent with the data in Figure 1, expression of BASP1 G2A failed to repress expression of any of the WT1 target genes. BASP1 S6A, which retains PIP2-binding capacity, repressed transcription of the WT1 target genes. As before (Figure 1), BASP1 S6D failed to act as a transcriptional repressor.
Figure 3. BASP1 Recruits PIP2 to Promoter-Bound WT1.
(A) Vector control K562 cells (V) or K562 cell line derivatives expressing wild-type BASP1 (wt) or the mutant derivatives G2A, S6A, mNLS (K7A:K9A), and S6D were used to prepare cDNA. Expression of the WT1 target genes AREG, Ets-1, VDR, JunB, and the non-WT1 target gene Bax was analyzed in comparison with GAPDH. Errorbars are the SDM of three independent experiments; *p < 0.05 by Student’s t test. Immunoblotting confirmed that the BASP1 derivatives are equivalently expressed.
(B) Immunoprecipitation of extracts derived from K562 expressing wild-type BASP1 or BASP1 G2A was performed with BASP1 antibodies, and the immunoprecipitates were either immunoblotted with BASP1 antibodies (above) or dot-blotted with PIP2 antibodies.
(C) Nuclear extract derived from K562 cells expressing either wild-type BASP1 or the G2A mutant derivative were incubated with control, PIP2, or PIP3 agarose beads. Stably bound products were subjected to immunoblotting with BASP1 antibodies.
(D) ChIP was performed with vector control K562 cells (V) or K562 cells expressing wild-type BASP1 or the mutant BASP1 derivative G2A with control IgG, WT1, BASP1, and PIP2 antibodies. The data are presented as fold enrichment at the AREG, Ets-1, VDR, and JunB promoter regions compared with a control genome region. Error bars are the SDM of three independent experiments; *p < 0.05 by Student’s t test.
(E) Vector control K562 cells (V) or K562 cells expressing either wild-type BASP1 (wt BASP1) or the mutant derivatives G2A were mock-treated (Ctrl) or treated with 100 nM PMA and imaged 72 hr later. Below, quantitation of the percentage of arborized cells is shown (error bars are the SDM of three independent experiments; *p < 0.05 by Student’s t test).
To determine whether the transcriptional effects we observed above were PIP2 dependent, we immunoprecipitated wild-type BASP1 or BASP1 G2A from K562 cell extracts and performed a dot-blot analysis with anti-PIP2 antibodies. Although both wild-type BASP1 and BASP1 G2A were equivalently immunoprecipitated, PIP2 was only detected in the precipitate from cells expressing wild-type BASP1 (Figure 3B). Moreover, whereas wild-type BASP1 from nuclear extract was retained by PIP2-agarose beads (but not PIP3-agarose beads) in pull-down assays (as in Figure 1F), BASP1 G2A was not (Figure 3C). These results confirm that myristoylation of BASP1 is required for its interaction with PIP2, and that this interaction is specific to PIP2.
We next performed ChIP assays with vector control K562 cells and K562 cells expressing wild-type BASP1 or BASP1 G2A (Figure 3D). WT1 occupied the AREG, Ets-1, VDR, and JunB promoter regions in both vector control K562 cells and BASP1-expressing K562 cells. In addition, K562 cells expressing BASP1 G2A also retained recruitment of WT1 at the promoters. ChIP analysis with anti-PIP2 antibodies revealed that PIP2 showed enhanced recruitment to all four promoters in K562 cells expressing wild-type BASP1 when compared with the vector control K562 cells. Thus, BASP1-PIP2 complexes are recruited along with WT1 to the promoters. Moreover, although BASP1 G2A is present at the promoters along with WT1, it failed to recruit PIP2 above the level observed in the vector control K562 cells. Taken together, these data demonstrate that PIP2-bound BASP1 is recruited to the promoter of a WT1 target gene, and that myristoylation of BASP1 is required for inclusion of PIP2 in the complex.
We previously showed that WT1 and BASP1 transcriptionally cooperate to divert the phorbol ester-induced differentiation program of K562 cells to an arborized cell type with neuronal characteristics (Goodfellow et al., 2011). We therefore treated control K562 cells or K562 cells stably expressing either wild-type wildtype BASP1 or BASP1 G2A with phorbol myristyl acetate (PMA) and allowed the cells to differentiate over 3 days. As before, K562 cells expressing wild-type BASP1 exhibited an arborized phenotype (Figure 3E). Consistent with the lack of WT1 corepressor activity, BASP1 G2A failed to support significant arborization of K562 cells.
Myristoylation-Dependent Association of BASP1 with Histone Deacetylase
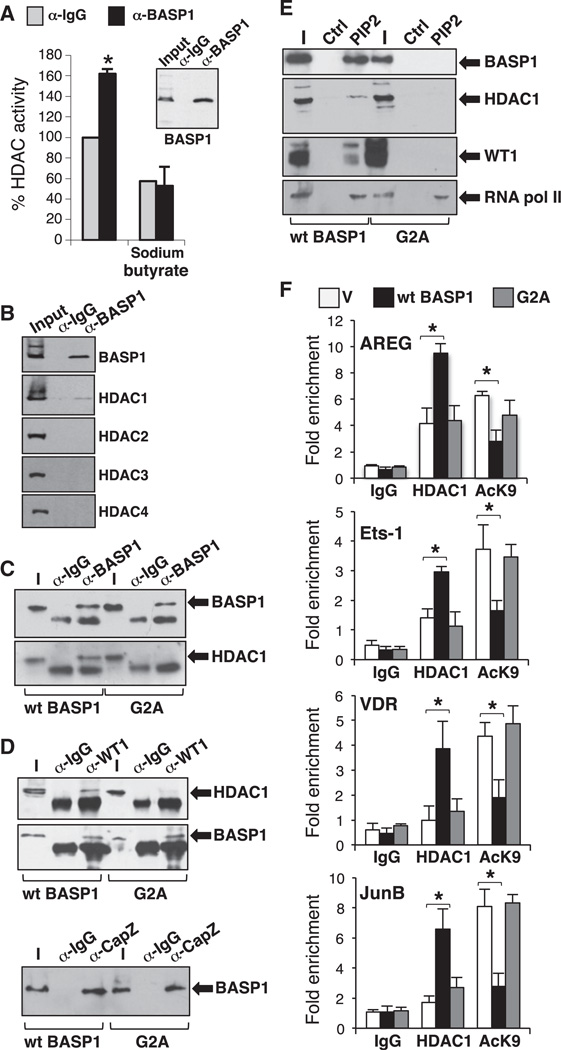
BASP1 has been associated with localized histone modification at the Wnt4 gene promoter (Essafi et al., 2011). We therefore sought to determine whether BASP1 uses histone deacetylases as part of its transcriptional corepressor function. HeLa cell nuclear extract was subjected to immunoprecipitation with anti-BASP1 antibodies, and the presence of histone deacetylase activity was determined (Figure 4A). BASP1 immunoprecipitates contained significantly more histone deacetylase activity than control immunoprecipitates, and this activity was inhibited by sodium butyrate, which predominantly inhibits class 1 and 2 HDACs. Immunoblotting of BASP1 immunoprecipitates with a battery of HDAC antibodies revealed that BASP1 specifically associated with HDAC1 (Figure 4B). To determine whether this BASP1-HDAC1 association required the BASP1-PIP2 interaction, we immunoprecipitated BASP1 from nuclear extracts derived from K562 cells that express either wild-type BASP1 or BASP1 G2A (which is defective in myristoylation). Whereas the immunoprecipitates of wild-type BASP1 contained HDAC1, the immunoprecipitates of BASP1 G2A did not (Figure 4C). These results indicate that the BASP1-PIP2 interaction is required for the association of BASP1 with HDAC1.
Figure 4. Myristoylated BASP1 Recruits Functional HDAC1 to the Promoter.
(A) BASP1 antibodies or control IgG were used in immunoprecipitation with HeLa nuclear extracts and subjected to histone deacetylase assay. Error bars are the SDM of three independent experiments; *p < 0.05 by Student’s t test.
(B) BASP1 was immunoprecipitated as in (A) and immunoblotted with the HDAC antibodies indicated.
(C) BASP1 was immunoprecipitated from K562 cells expressing wild-type BASP1 or BASP1 G2A, and the samples were immunoblotted with BASP1 (top) or HDAC1 antibodies. α-IgG are control antibodies.
(D) As in (C) except that anti-WT1 antibodies were used for immunoprecipitation, and HDAC1 and BASP1 were detected by immunoblotting (above) or anti-CapZ antibodies were used for immunoprecipitation followed by immunoblotting with BASP1 antibodies.
(E) Nuclear extracts derived from K562 cell expressing either wild-type BASP1 or BASP1 G2A were incubated with control agarose beads or beads containing PIP2. The bound fraction was immunoblotted with the antibodies indicated.
(F) ChIP was performed with vector control K562 cells or K562 cells expressing wild-type BASP1 (wt BASP1) or the mutant BASP1 derivative G2A with control IgG, HDAC1, or acetyl H3K9 antibodies. The data are presented as fold enrichment at the AREG, Ets-1, VDR, and JunB promoter regions compared with a control genome region. Error bars are the SDM of three independent experiments; *p < 0.05 by Student’s t test.
Our results suggest that BASP1 provides a link between WT1 and HDAC1, and that this would require the myristoylation of BASP1. In support of this hypothesis, we found that anti-WT1 antibodies specifically coimmunoprecipitate HDAC1 from K562 cells stably expressing wild-type BASP1, but not from K562 cells expressing BASP1 G2A (Figure 4D). In contrast, and consistent with the fact that BASP1 G2A localizes to the promoter regions of WT1-responsive genes (Figure 3D), we found that anti-WT1 antibodies coimmunoprecipitated both wild-type BASP1 and BASP1 G2A (Figure 4D). The actin-capping protein CapZ was previously shown to bind to BASP1 independently of myristoylation status (Odagaki et al., 2009), and consistent with this we found that CapZ antibodies coimmunoprecipitated both wild-type BASP1 and BASP1 G2A (Figure 4D, lower panel). Taken together, the data in Figure 4D suggest that WT1 interacts with HDAC1 through myristoylation of BASP1 and that BASP1 G2A is specifically defective in interacting with HDAC1.
We next determined whether WT1 and HDAC1 interact with PIP2 via myristoylated BASP1. HDAC1 and WT1 were recruited to PIP2-agarose beads only from K562 cell extracts containing wild-type BASP1 and not BASP1 G2A (Figure 4E). RNA polymerase II (RNA pol II) was included as a control protein that binds to PIP2 independently of BASP1. These results suggest the formation of a complex among WT1, BASP1, and HDAC1 that is dependent on the myristoylation of BASP1 and the inclusion of PIP2 in the complex.
We next performed ChIP assays to determine whether the myristoylation of BASP1 is required for the recruitment of HDAC1 to the promoters of the AREG, Ets-1, VDR and JunB genes, and whether this in turn affects histone H3K9 acetylation status at each promoter. Vector control K562 cells or K562 cell line derivatives expressing either wild-type BASP1 or BASP1 G2A were subjected to ChIP with control antibodies, anti-HDAC1, or anti-acetyl-H3K9 antibodies. Compared with the vector control cells, cells expressing wild-type BASP1 showed an increase in HDAC1 recruitment to all four promoters and a concomitant reduction in H3K9 acetylation (Figure 4F). In contrast, cells expressing BASP1 G2A did not show enhanced HDAC1 recruitment compared with control cells, and also did not result in a significant reduction in histone H3K9 acetylation. Thus, the PIP2-dependent association of BASP1 with HDAC1 leads to a repressive chromatin state at the AREG, Ets-1, VDR, and JunB promoters.
It is well established that PIP2 is present in the nucleus, where it has been shown to associate with RNA pol II and to regulate the activity of ATP-dependent chromatin-remodeling activities (Zhao et al., 1998; Osborne et al., 2001; Rando et al., 2003; Shen et al., 2003; Steger et al., 2003). Our findings demonstrate that PIP2 associates with transcription factors at the promoter to direct a gene-specific role in transcriptional regulation.
How the BASP1-PIP2 interaction drives association with HDAC1 is not clear. However, other phospholipids have been shown to regulate protein complexes that possess HDAC activity (Gozani et al., 2003; Hait et al., 2009; Han and Emr, 2011). Therefore, PIP2 may be directly involved in the interaction with HDAC1 rather than acting exclusively on BASP1 conformation. Our findings suggest that myristoylation provides an interface between nuclear lipids and the chromatin-remodeling machinery. Because BASP1 can also regulate the activity of other transcription factors, such as myc, this myristoylation- and PIP2-dependent mechanism of transcriptional repression will likely also apply to other DNA-binding transcriptional regulators.
EXPERIMENTAL PROCEDURES
Tissue Culture, Immunofluorescence, Plasmids, and Transfection
BASP1 mutant derivatives were generated using the QuikChange mutagenesis kit (QIAGEN). M15 and U2OS cells were cultured in Dulbecco’s modified Eagle’s medium, and K562 cells were cultured in RPMI medium. Stable K562 cell line derivatives were generated as described previously (Goodfellow et al., 2011). Cells were transfected with Lipofectamine 2000 (Invitrogen) or Effectene (QIAGEN). Immunofluorescence was performed as previously described (Carpenter et al., 2004). BASP1 antibodies were described previously (Carpenter et al., 2004). Antibodies anti-WT1 (C-19), anti-GAPDH (6C5), anti-CapZ (C-7), and anti-Lamin A/C (N-18) were obtained from Santa Cruz Biotechnology. Anti-NMT1 (ab84666), anti-NMT2 (ab102473), anti-H3K9Ac (ab10812), anti-PIP2 (ab11039), anti-pol II (ab5408), anti-β-tubulin (ab6160), and HDAC1 (ab19485) were obtained from Abcam. HDAC1-4 antibodies (sampler kit 9928) were obtained from Cell Signaling.
Protein, mRNA, and ChIP Analysis
Nuclear/cytoplasmic extracts were prepared as described previously (Hartkamp et al., 2010). Total RNA was prepared using the QIAGEN RNeasy kit, and cDNA was prepared using the Bio-Rad cDNA synthesis kit. ChIP assays were performed as described previously (Hartkamp et al., 2010). Histone deacetylase activity was assayed with the use of a kit from Millipore. siRNAs to mouse NMT1 and NMT2 were obtained from QIAGEN. PIP2 and PIP3 agarose beads were purchased from Echelon Biosciences, and binding assays were performed as previously described (Lewis et al., 2011). For the primers used in this study, see Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Keith Brown, Karim Malik, and Chris Paraskeva for comments on the manuscript and Alan Segal for help with the confocal microscopy. H.A.C. was funded by a studentship from the Biotechnology and Biological Sciences Research Council. This work was funded by the National Institute of General Medical Sciences (1R01GM098609 to S.G.E.R.), Cancer Research UK (C1356/A6630 to S.G.E.R.), the National Science Foundation (NSF0917893 to K.F.M.), and the Association for International Cancer Research and Well-come Trust (WT061207MA to P.S.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and two figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.08.005.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
REFERENCES
- Albi E, Viola Magni MP. The role of intranuclear lipids. Biol. Cell. 2004;96:657–667. doi: 10.1016/j.biolcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2010;20:25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B, Hill KJ, Charalambous M, Wagner KJ, Lahiri D, James DI, Andersen JS, Schumacher V, Royer-Pokora B, Mann M, et al. BASP1 is a transcriptional cosuppressor for the Wilms’ tumor suppressor protein WT1. Mol. Cell. Biol. 2004;24:537–549. doi: 10.1128/MCB.24.2.537-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM. Proteins and cholesterol-rich domains. Biochim. Biophys. Acta. 2008;1778:1576–1582. doi: 10.1016/j.bbamem.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Essafi A, Webb A, Berry RL, Slight J, Burn SF, Spraggon L, Velecela V, Martinez-Estrada OM, Wiltshire JH, Roberts SGE, et al. A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev. Cell. 2011;21:559–574. doi: 10.1016/j.devcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow SJ, Rebello MR, Toska E, Zeef LA, Rudd SG, Medler KF, Roberts SGE. WT1 and its transcriptional cofactor BASP1 redirect the differentiation pathway of an established blood cell line. Biochem. J. 2011;435:113–125. doi: 10.1042/BJ20101734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Green LM, Wagner KJ, Campbell HA, Addison K, Roberts SGE. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res. 2009;37:431–440. doi: 10.1093/nar/gkn955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BK, Emr SD. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkamp J, Carpenter B, Roberts SGE. The Wilms’ tumor suppressor protein WT1 is processed by the serine protease HtrA2/Omi. Mol. Cell. 2010;37:159–171. doi: 10.1016/j.molcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M, Nist A, Khan MI, Valovka T, Bister K. Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1) Proc. Natl. Acad. Sci. USA. 2009;106:5604–5609. doi: 10.1073/pnas.0812101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenstein P, Hastie ND. The many facets of the Wilms’ tumour gene, WT1. Hum. Mol. Genet. 2006;15(Spec No 2):R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- Huff V. Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat. Rev. Cancer. 2011;11:111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashihara M, Miyata S, Kumanogoh H, Funatsu N, Matsunaga W, Kiyohara T, Sokawa Y, Maekawa S. Changes in the localization of NAP-22, a calmodulin binding membrane protein, during the development of neuronal polarity. Neurosci. Res. 2000;37:315–325. doi: 10.1016/s0168-0102(00)00132-2. [DOI] [PubMed] [Google Scholar]
- Keune W, Bultsma Y, Sommer L, Jones D, Divecha N. Phosphoinositide signalling in the nucleus. Adv. Enzyme Regul. 2011;51:91–99. doi: 10.1016/j.advenzreg.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Korshunova I, Caroni P, Kolkova K, Berezin V, Bock E, Walmod PS. Characterization of BASP1-mediated neurite outgrowth. J. Neurosci. Res. 2008;86:2201–2213. doi: 10.1002/jnr.21678. [DOI] [PubMed] [Google Scholar]
- Laux T, Fukami K, Thelen M, Golub T, Frey D, Caroni P. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol. 2000;149:1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AE, Sommer L, Arntzen MØ, Strahm Y, Morrice NA, Divecha N, D’Santos CS. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003376. M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay LM, Carpenter B, Roberts SGE. Regulation of the Wilms’ tumour suppressor protein transcriptional activation domain. Oncogene. 1999;18:6546–6554. doi: 10.1038/sj.onc.1203046. [DOI] [PubMed] [Google Scholar]
- Maekawa S, Murofushi H, Nakamura S. Inhibitory effect of calmodulin on phosphorylation of NAP-22 with protein kinase C. J. Biol. Chem. 1994;269:19462–19465. [PubMed] [Google Scholar]
- Moribe T, Iizuka N, Miura T, Stark M, Tamatsukuri S, Ishitsuka H, Hamamoto Y, Sakamoto K, Tamesa T, Oka M. Identification of novel aberrant methylation of BASP1 and SRD5A2 for early diagnosis of hepatocellular carcinoma by genome-wide search. Int. J. Oncol. 2008;33:949–958. [PubMed] [Google Scholar]
- Mortier E, Wuytens G, Leenaerts I, Hannes F, Heung MY, Degeest G, David G, Zimmermann P. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 2005;24:2556–2565. doi: 10.1038/sj.emboj.7600722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosevitsky MI. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 2005;245:245–325. doi: 10.1016/S0074-7696(05)45007-X. [DOI] [PubMed] [Google Scholar]
- Mosevitsky MI, Capony JP, Skladchikova GYu, Novitskaya VA, Plekhanov AYu, Zakharov VV. The BASP1 family of myristoylated proteins abundant in axonal termini. Primary structure analysis and physicochemical properties. Biochimie. 1997;79:373–384. doi: 10.1016/s0300-9084(97)80032-6. [DOI] [PubMed] [Google Scholar]
- Odagaki S, Kumanogoh H, Nakamura S, Maekawa S. Biochemical interaction of an actin-capping protein, CapZ, with NAP-22. J. Neurosci. Res. 2009;87:1980–1985. doi: 10.1002/jnr.22040. [DOI] [PubMed] [Google Scholar]
- Osborne SL, Thomas CL, Gschmeissner S, Schiavo G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 2001;114:2501–2511. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chi TH, Crabtree GR. Second messenger control of chromatin remodeling. Nat. Struct. Biol. 2003;10:81–83. doi: 10.1038/nsb0203-81. [DOI] [PubMed] [Google Scholar]
- Rivera MN, Haber DA. Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. Nat. Rev. Cancer. 2005;5:699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- Roberts SGE. Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki A, Hayashi N, Matsubara M, Yamauchi E, Taniguchi H. Identification of the calmodulin-binding domain of neuron-specific protein kinase C substrate protein CAP-22/NAP-22. Direct involvement of protein myristoylation in calmodulin-target protein interaction. J. Biol. Chem. 1999;274:11848–11853. doi: 10.1074/jbc.274.17.11848. [DOI] [PubMed] [Google Scholar]
- Terashita A, Funatsu N, Umeda M, Shimada Y, Ohno-Iwashita Y, Epand RM, Maekawa S. Lipid binding activity of a neuron-specific protein NAP-22 studied in vivo and in vitro. J. Neurosci. Res. 2002;70:172–179. doi: 10.1002/jnr.10407. [DOI] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.