Abstract
Cholesterol is so essential to the proper function of mammalian cell membranes that even strikingly small inborn errors in cholesterol synthesis can be devastating. Here we combine molecular dynamics simulations with small angle x-ray diffraction experiments to compare mixed sterol/DMPC membranes over a wide range of sterol compositions for two types of sterols: cholesterol and its immediate metabolic precursor 7DHC, that differs from cholesterol by one double bond. We find that while most membrane properties are only slightly affected by the replacement of one sterol by the other, the tilt degree of freedom, as gauged by the tilt modulus, is significantly larger for cholesterol than for 7DHC over a large range of concentrations. In silico mutations of one sterol into the other further support these findings. Moreover, bending rigidities calculated from simulations and estimated in experiments show that cholesterol stiffens membranes to a larger extent than 7DHC. We discuss the possible mechanistic link between sterol tilt and the way it impacts the membrane mechanical properties, and comment on how this link may shed light on the way replacement of cholesterol by 7DHC leads to disease.
Keywords: cholesterol metabolism, 7DHC, SLOS, cholesterol tilt modulus, membrane bending rigidity
Introduction
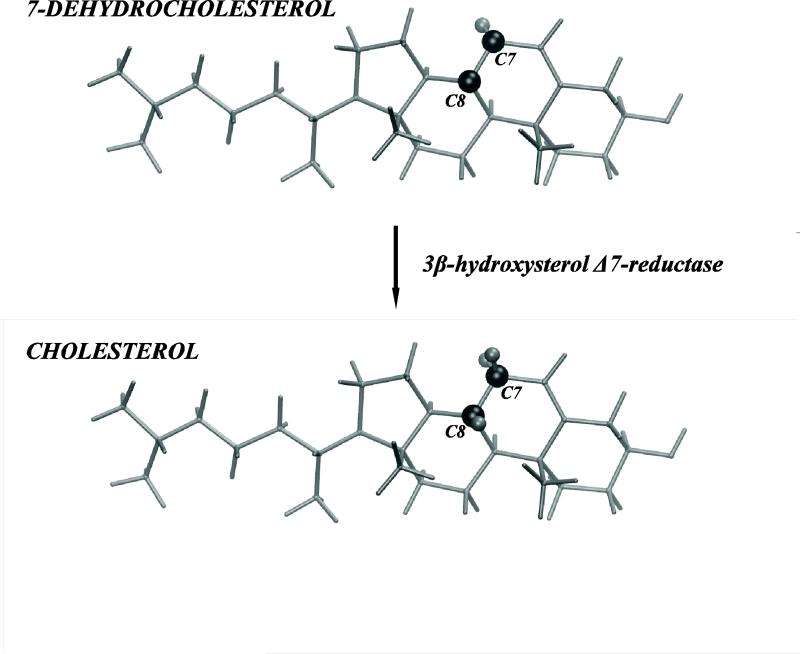
Long recognized as vital to proper cellular membrane function, cholesterol is unjustly vilified. While cholesterol can be ingested, it can also be endogenously synthesized by all animal cells in a series of enzymatic steps allowing to maintain homeostasis. Changes in the physiological balance of cholesterol have been linked to various pathologies ranging from coronary heart disease to genetic metabolic disorders that involve the biochemical pathways of cholesterol synthesis (1). A striking example is the Smith-Lemli-Opitz syndrome (SLOS) caused by an inborn deficient activity of 3β-hydroxysterol Δ7-reductase (DHCR7), the enzyme responsible for the final step in cholesterol synthesis from 7-dehydrocholesterol (7DHC) to cholesterol, see Fig.1. This DHCR7 deficiency is responsible for the accumulation of 7DHC and reduced levels of cholesterol in patients with SLOS, leading to multiple congenital malformations. Puzzlingly, these dire consequences are due to just one double bond present in 7DHC but not in cholesterol, Fig. 1.
Figure 1.
3β-hydroxysterol Δ7-reductase (DHCR7) is the enzyme responsible for the final step in cholesterol synthesis from 7-dehydrocholesterol (7DHC). In this enzymatic reaction, one double bond between C7 and C8 atoms in 7DHC is saturated (transformed into a single bond in cholesterol). The schematic drawing highlights (dark shade) C7 and C8 carbons and their associated hydrogen atoms on cholesterol and 7DHC.
While SLOS pathology has been well characterized, the underlying pathophysiological processes responsible for these manifestations are unknown and are likely due to multiple mechanisms. First, replacing cholesterol (Chol) with 7DHC is expected to influence many biomolecules to which cholesterol is a precursor, including hormones, neurosteroids, oxysterols, and bile acids (1). A complimenting proposed mechanism implicates changes in the material properties due to cholesterol substitution by 7DHC as effectors of membrane function as well as the ability of membrane proteins to organize and function properly (1-3). This impact to lipid membranes could potentially modify their function as a barrier to extracellular solutes as well as their ability to properly modulate membrane protein function important to microdomain formation, signal transduction, and membrane trafficking (4-6).
Support of specific mechanisms has been hampered by the lack of conclusive evidence for changes in membrane structure by substitution with 7DHC. In favor of changes in material properties, it was found that cholesterol is more effective at stiffening saturated DMPC membranes (2, 7). In addition, changes in shape and size of sterol-containing membrane domains (8-11) as well as altered membrane organization (detected by x-ray diffraction), and membrane fluidity (as determined by fluorescence anisotropy) (3) were found in 7DHC containing membranes. In contrast, other studies have shown that domain formation can be either even more pronounced with 7DHC (12), or remain unaltered. Overall it seems that cholesterol to 7DHC substitution only subtly modulates many membrane material properties, making it harder to assess the mechanistic role of this modification.
Recent molecular dynamics (MD) simulations have been instrumental in beginning to unravel the molecular alterations resulting from 7DHC substitution. In model membranes of the disaturated dioleoyl-phosphocholine (DOPC) [18:1 18:1] lipid, there were no significant differences in membrane ordering by the different sterols. However, model membranes composed of saturated dipalmitoylphosphocholine (DPPC) [16:0 16:0] lipids were shown to change structure due to cholesterol substitution with 7DHC, as witnessed for example in chain order parameters, sterol location in the membrane, and lateral pressure profiles (13, 14). These differences were traced to the looser packing near the modified part of the 7DHC sterol ring. Interestingly, sterol tilt was found to be an important measure of sterol's ordering capabilities. It remains an open question how this important degree of freedom is altered due to changes in sterol concentration in membranes, and how these lead to changes in the material properties of membranes, such as lipid rigidity, as well as to the membrane ability to serve as a proper solvating environment to proteins embedded within it.
To assess the impact of cholesterol to 7DHC substitution on membrane material properties, we have performed MD simulation of sterol/dimyristoylphosphatidylcholine (DMPC) [14:0 14:0] mixtures over a wide range of sterol:lipid ratios. Our aim was to link material properties to the structural changes in the lipid membranes upon cholesterol-7DHC substitution. We find only small differences in most membrane properties upon substitution to 7DHC at all concentrations studied, as verified by our x-ray scattering experiments. We have previously shown that changes in the preferred sterol tilt in membranes can be assessed using the tilt modulus (15), a thermodynamic property quantifying the work required to change sterol's orientation in a membrane. We show here that this measure is significantly different for 7DHC and cholesterol and that these differences persist over a wide range of sterol compositions. By using mutation substitutions in silico, we were able to estimate the differences in sterol free energies, showing that these differences are strongly encoded in the structure of a single sterol molecule embedded within a membrane of other (similar or dissimilar) sterol molecules. These in silico mutations reaffirm the lower free energy of 7DHC in membranes relative to Chol. Using spectral analysis of membrane fluctuations, we find that membranes with Chol are much stiffer than membranes containing 7DHC. Finally, we show that the work required to orient two sterols at a non parallel (splayed) configuration is strongly linked to membrane rigidity. We comment on the possible importance of these findings to the function of biologically relevant membranes.
MATERIALS AND METHODS
Sample Preparation
DMPC was purchased from Avanti Polar Lipids (Birmingham, AL), cholesterol and 7DHC were from Sigma-Aldrich (St. Louis, MO). All lipids were used without further purification. Lipid stock solutions were prepared by dissolving weighted amounts of lipid powder in chloroform (pro analysis quality). DMPC/Chol samples at 20 and 40% molar ratios and DMPC/7DHC samples at 0, 5, 10, 20, 30, and 40% molar ratios were obtained by mixing appropriate amounts of the stock solutions. Organic solvent was removed by placing the samples first under a gentle stream of N2 and then under vacuum for approximately 8 hours. Dry lipid films were suspended in excess of 18 MΩ/cm water (lipid concentration: 5 wt. %). 7DHC containing samples were further carefully flushed with Ar in order to avoid oxidation. Finally, samples were kept at 35°C for 4 hours and intermittently vortex mixed to achieve fully hydrated multilamellar vesicles (MLVs).
Small-angle X-ray Diffraction (SAXD)
SAXD experiments were performed at the Austrian SAXS beamline (Elettra, Sincrotrone Trieste, Italy) using 8 keV photons and a mar300 (Marresearch, Norderstedt, Germany) image plate detector. Samples were filled into reusable quartz-glass capillaries and kept in a brass sample holder connected to a circulating water bath. All experiments were performed at T=35 °C at an exposure time of 3 minutes. Silver behenate was used for angular calibration and the program FIT2D was used for angular integration of the 2D diffraction patterns.
Although, due to problems with the background acquisition, we were not able to apply a full q-range analysis to derive the structural membrane parameters as published earlier (16), improved data resolution as compared to earlier experiments (e.g. (16)) enabled us to discern at least four diffraction orders, even for pure DMPC samples. This was enough to calculate electron density profiles using the integrated intensities of the peaks (16, 17). Therefore, electron densities were calculated using the standard Fourier synthesis
| (1) |
where αh = ± 1 is the phase, d the lamellar repeat distance and Fh the form factor of the hth diffraction order determined from Lorentzian fits to the Bragg peaks as , (q = 4π / λ sin θ).
Estimates of bending rigidities from SAXD experiments
In order to estimate the bending rigidity KC of cholesterol containing DMPC bilayers in comparison to 7DHC/DMPC mixtures, we fitted the first diffraction order using a modified Caillé theory structure factor (16, 18). This yields the fluctuation parameter
| (2) |
In the above expression, B represents the bulk modulus describing interactions between bilayers (see e.g., (19), for a recent review), and kB denotes Boltzmann's constant.
Molecular Dynamics (MD) Simulations
MD simulations were performed on hydrated DMPC bilayers containing 0, 5, 10, 20, 30, 40 and 50% sterol (either Chol or 7DHC) number fractions, and at a temperature of T=35°C. We chose DMPC as the lipid component because it is particularly amenable to physico-chemical studies due to its relatively low gel-to-fluid transition temperature (~23.5°C) (20). The specific simulation parameters were chosen based on available experimental (2, 8, 21-26) and computational data (13, 15, 27-30) on cognate systems.
The simulations were conducted on two differently sized bilayer patches. As listed in Table 1, we studied DMPC/sterol complexes consisting of 400 lipids in total and at various lipid/sterol ratios (designated as SIM1), and larger DMPC/sterol bilayers, containing 1600 lipids and at 0 and 30% sterol concentrations (SIM2).
Table 1.
Lipid complexes studied with MD simulations
| DMPC/Sterol | NDMPC | NSTEROL | Nw | Time, ns |
|---|---|---|---|---|
| 0%a | 400 | 0 | 14261 | 50 |
| 0%b | 1600 | 0 | 51280 | 20 |
| 5%a | 380 | 20 | 12883 | 50 |
| 10%a | 360 | 40 | 13613 | 50 |
| 20%a | 320 | 80 | 13483 | 50 |
| 30%a | 280 | 120 | 12971 | 50 |
| 30%b | 1120 | 480 | 52228 | 20 |
| 40%a | 240 | 160 | 13416 | 50 |
| 50%a | 200 | 200 | 13013 | 50 |
Simulations were conducted using NAMD 2.7b1 and CHARMM 36 force field parameters for lipids (SIM1).
Simulations were conducted using Gromacs 3.3.1 and Gromos 43A1-S3 parameter set for lipids (SIM2). NDMPC, NSTEROL, Nw denote number of DMPC, sterol (either Cholesterol or 7DHC), and water molecules in mixtures respectively.
For SIM1 simulations DMPC/Chol bilayers containing 400 molecules with appropriate Chol compositions were first built with the CHARMM-GUI Membrane Builder web tool (31). Briefly, CHARMM-GUI utilizes a structural library containing 2000 different conformations for each lipid molecule to build, assemble, and solvate a membrane. In the process, care is taken to resolve bad lipid/lipid contacts, by using a replacement method. The final assembled structures were additionally checked for any remaining bad contacts, such as between cholesterol rings and other types of lipids (31). DMPC/7DHC mixtures were then built by replacing Chol molecules with 7DHC in assembled DMPC/Chol bilayers.
After initial energy minimization of 5400 steps, SIM1 complexes were simulated using the CHARMM36 force field for lipids (32) and propagated for 50 ns using the NAMD modeling package (ver 2.7b1) (33). Only the last 20ns trajectory segments, that were deemed as converged, were used for further analysis (see below). The simulations were conducted in the constant temperature and pressure (NPT) ensemble with semi-isotropic pressure coupling and utilizing Particle-Mesh-Ewald (PME) for long-range electrostatics (34). The Nose-Hoover Langevin piston method (35, 36) was used to control the target 1atm pressure with the Langevin piston period set to 100 fs and Langevin piston decay set to 50 fs. All MD simulations were performed with rigid bonds allowing a 2 fs time step.
For simulations of large membrane patches (SIM2 simulations), we chose to use the Gromacs 3.3.1 software (37-40) and the united-atom representations for lipids. This choice was based on two specific reasons: first, the simulation of 1600-lipid size system, required for proper determination of membrane rigidities, is significantly less demanding computationally if performed with united-atom representations compared to all-atom; and second, the improved united-atom forcefield 43A1-S3 we used here, has been recently validated and used by us and others (15, 27, 41, 42) for different lipid systems, including DMPC/cholesterol membranes (27), to consistently yield good agreement with the experimental data. In addition, comparisons of the effects of different sterols (Chol and 7DHC) on the structural and mechanical properties of phospholipid membranes (see below) as derived using different force-field potentials provides an important additional consistency check to our model predictions (see Table S1 in the Supplemental Data), and shows that these go beyond differences in specific forcefields.
Pure DMPC and DMPC/Chol bilayers with 30% Chol containing 1600 molecules were constructed from previously equilibrated 400-lipid DMPC/Chol membrane patches that were further MD simulated for 40ns (15, 27). These original membrane structures were replicating in the bilayer plane (x and y directions). To construct the corresponding DMPC/7DHC mixture with 30% 7DHC, we first replaced Chols with 7DHC in the original united-atom 30% DMPC/Chol membrane (15, 27). This 400-lipid 30% DMPC/7DHC mixture was then equilibrated and simulated (for ~30ns) using Gromacs 3.3.1 with the 43A1-S3 forcefield, before it was replicated and expanded in the x and y directions. The final 1600-lipid size united-atom 30% DMPC/7DHC bilayer was then simulated for an additional 20ns using Gromacs 3.3.1 and 43A1-S3 forcefield. Because the structural properties derived for these large membranes and the bilayers from SIM1 simulations were similar, in this manuscript we compare DMPC/Chol and DMPC/7DHC membranes structurally from SIM1 trajectories, and use SIM2 simulations specifically for the spectral analysis of membrane undulations leading to calculations of bending rigidities (see below).
For all Gromacs simulations, we used the LINCS algorithm to constrain all bond lengths (43) allowing for 2fs time steps. Periodic boundary conditions were applied in all three dimensions, and long-range electrostatics were calculated using the PME algorithm (34). A cutoff of 18Å was employed for van der Waals interactions. The systems were simulated in the NPT ensemble. A constant pressure of 1 atm was maintained using the Parrinello-Rahman semi-isotropic pressure coupling scheme (44, 45), and constant temperature was maintained using the Noose-Hoover temperature coupling method (46).
Parameterization of 7DHC
Forcefield potentials for 7DHC representation used in SIM1 simulations were derived from all-atom CHARMM parameters for cholesterol (32) using the general CHARMM36 force field parameter set (47). The united-atom force-field for 7DHC used in our SIM2 simulations was obtained from the united-atom force-field of cholesterol molecule (15, 27, 41, 42). Because in 7DHC the C7 and C8 carbons form a double bond (Fig. 1), the united atom types and non-bonded C6 and C12 parameters for these two carbons had to be modified. To derive the relevant bonded interactions, and in particular for the torsion dihedral C5=C6-C7=C8, ab initio level calculations on the B3LYP/6-31g** theory level were performed on 7DHC as a whole as well as smaller molecular compounds, using 1,3-cyclohexadiene and cis,cis-2,4-hexadiene as model molecules (see Figure S1 in the Supplemental Data).
Calculation of sterol tilt modulus from MD Simulations
We have previously described the procedure for calculating the cholesterol tilt modulus χ from MD simulations (15, 27, 28). Here, we use the same framework to obtain χ for Chol and 7DHC in the different systems. Briefly, the sterol tilt angle, θ, was defined in the range [0°; 180°] as the polar angle between the vector joining C3-C17, commonly used to describe the sterol ring plane orientation, and the bilayer normal (15, 27). In this definition, θ=0 represents a sterol orientation where the ring plane is parallel to the bilayer normal z axis. Then, the normalized probability density P(θ) of the sterol tilt angle with respect to bilayer normal was constructed for each simulated system. The P(θ) distribution was obtained by creating a sterol orientational angle histogram in the angular range [0°; 90°]. To calculate the sterol tilt modulus, a quadratic fit was performed to an expression of the form –kBT ln[P(θ) / sinθ] over the [8°; 20°] interval, a range chosen to represent the best-sampled angular region and at the same time to limit the fit to low θ regime where the quadratic expansion of the free energy with respect to tilt is valid (see Results)(15, 27, 48). Finally, χ was determined from the coefficient that corresponded to the best fit.
Calculation of bilayer bending modulus from MD Simulations
To investigate the effect of Chol and 7DHC on the bending modulus, KC, of DMPC-sterol membranes, we used the Helfrich continuum representation for lipid bilayer undulations u(x,y) (49), and performed spectral analysis of undulations (50-53) of large DMPC and DMPC/sterol bilayers as derived from our SIM2 simulation trajectories. According to the Helfrich description, the Hamiltonian of the fluctuating membrane can be expanded to lower order terms in u(x,y) (neglecting Gaussian curvature contribution which remains constant for bilayer shape fluctuations) (51):
| (3) |
Here the two terms represent bending and surface tension contributions, with their respective KC and γ constants. Analysis of the bending rigidity from the bilayer undulations is most conveniently performed in Fourier space using the expansion (see Ref. (50-53), for example):
| (4) |
Where and denote two-dimensional real space and reciprocal space vectors, respectively. Assuming a tensionless membrane, the spectral amplitude profile <u2(q)> for the small-q modes can be obtained from the equipartition theorem (50-53) as: (kBT / ABOX) × KC × q-4, where ABOX represents the lateral area of the simulation box.
In addition, as has been detailed elsewhere (51-53), local protrusion modes in <u2(q)> can also be observed in high q-regime. Governed by the microscopic protrusion tension, γP, such modes can be approximated by (kBT / ABOX) × γP-1 × q-2. Thus, the full spectrum of the <u2(q)> amplitudes is expected to contain the two q-4 and q-2 regimes, corresponding to the small and high wavevector modes respectively.
The converged last 14ns segments of SIM2 trajectories (see Results) were analyzed for wavevector-dependent undulatory modes (53) by representing the position of each bilayer leaflet using the vertical z-coordinates of lipid backbone C13 carbon atoms. Resulting traces for two monolayers where then subjected to interpolation with spline functions of the fifth order to yield smooth leaflet shapes. Application of a two-dimensional Fourier transform (50) to the smoothed monolayer shapes yielded undulatory spectral amplitudes, which were subsequently time-averaged and converted into histograms according to wavevector magnitude, q. The calculated Fourier amplitudes <u2(q)> presented here for all simulated bilayers were scaled by the lateral area of the respective simulated systems, ABOX, and the values for KC were obtained from the fitting coefficients to the q-4 function (50, 54) (see also Results).
To test the sensitivity of our results towards the splining order used for shape interpolation as well as on trajectory sampling, we repeated the spectral analysis for the first and the last 7ns intervals of the entire 14ns trajectory segment, and used fourth and sixth order spline functions as alternatives to shape smoothing. All these different calculations resulted in a standard deviation of ~2kBT in KC. This fact is reflected in the reported uncertainties we associate with the derived KC values.
For completeness, we also calculated the KC modulus from SIM1 trajectories, but using a somewhat different approach. Thus, for each model SIM1 bilayer, we first derived the area compressibility KA from the extent of area fluctuations, and then obtained KC using the polymer brush theory as KC=KA(2Dc)2/24, where Dc represents the hydrophobic thickness of the bilayer (see, for example reference (55) and references therein). As highlighted in Table S1 (see Supplemental Data) listing results for 30% DMPC/sterol mixtures, the relative change in KC upon Chol-to-7DHC substitution calculated from the SIM2 (using spectral analysis) and SIM1 simulations agree reasonably well, but the absolute values for the bending rigidities obtained from the smaller SIM1 bilayers are much higher than anticipated (see Results). This discrepancy should not be surprising and can be explained by the suppression of the long wavelength undulations in the small SIM1 membranes (see above). To conclude, this analysis confirms the importance of including such undulatory modes for accurate calculations of the bending rigidity, and therefore the need for simulating large bilayers as extensively discuss previously, see e.g., ref (56).
Free energy perturbation (FEP) calculations
Free Energy Perturbation (FEP) calculations were carried out on selected systems from SIM1 trajectories to simulate in silico mutation of cholesterol into 7DHC (Chol → 7DHC) or of 7DHC into cholesterol (7DHC → Chol). Specifically, to perform these mutations we identified Chol and 7DHC molecules at different tilt angles with respect to the bilayer normal axis (see Table 2 and Results) in frames from the final, equilibrated parts of the corresponding 5% and 30% DMPC/sterol mixtures from SIM1 simulations. Then, restraining the molecule's orientation, Chol → 7DHC or 7DHC → Chol FEP transformations were performed on each selected sterol molecule separately with NAMD 2.7b1 and using the same simulations parameters as described for SIM1 simulations above.
Table 2.
Cholesterol → 7DHC in silico mutations in 30% DMPC/Cholesterol bilayera.
| CHOL # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| θ,° | 10.29 | 10.58 | 10.43 | 10.73 | 33.65 | 33.02 | 32.93 | 31.67 |
| Δμ, kcal/mol | -2.420 ± 0.195 | -2.724 ± 0.465 | ||||||
The tilt angles with respect to the membrane normal for 8 Cholesterol molecules in 30% DMPC/Cholesterol mixture from SIM1 trajectory chosen for the mutations are shown along with the corresponding free energies for mutations in “upright” and “tilted” cholesterols separately. The error bars in the free energy values were calculated as standard deviations over FEP calculations performed in forward/backward directions and over 1200ps and 2400ps mutation time intervals for Chols 1-4 and 5-8 respectively.
For each of the FEP computations, the coupling parameter λ varied from 0 to 1 by increments of 0.05 and a soft-core potential was employed according to the better-adapted sampling strategy (57, 58). Since 7DHC structurally differs from Chol due to the double bond between C7 and C8 carbons (see Fig. 1), we used a hybrid topology to annihilate/create two hydrogen atoms covalently attached to C7 and C8 in Chol.
During FEP, the mutated sterol molecule was restrained to its orientation relative to the membrane normal direction using the tiltCon collective variable in NAMD. The total time for a single mutation was either 1200 ps or 2400 ps (60ps or 120ps per iteration, respectively) and both forward and backward FEP transformations were employed on each construct. These different conditions resulted in a transformation free energy error of |Δμ| < 0.6 kcal/mol (see Figure S2 in the Supplemental Data and Tables 2-4 in the main text).
Table 4.
Cholesterol → 7DHC and 7DHC → Cholesterol in silico mutations in 5% DMPC/Cholesterol and DMPC/7DHC bilayers, respectivelya.
| STEROL # | CHOL → 7DHC 1 | 7DHC → CHOL 1 | CHOL → 7DHC 2 | 7DHC → CHOL 2 |
|---|---|---|---|---|
| θ,° | 9.03 | 9.94 | 37.12 | 33.92 |
| Δμ, kcal/mol | -2.222 ± 0.6 | 2.085 ± 0.436 | -2.477 ± 0.308 | 1.773 ± 0.584 |
The tilt angles with respect to the membrane normal for 4 sterol molecules in 5% DMPC/Chol (CHOL → 7DHC 1, CHOL → 7DHC 2) or 5% DMPC/7DHC (7DHC → CHOL 1, 7DHC → CHOL 2) mixtures from SIM1 simulations chosen for the mutations are shown along with the corresponding free energies for mutations. The error bars in the free energy values were calculated as standard deviations over FEP calculations performed in forward/backward directions and over 1200ps and 2400ps mutation time intervals.
RESULTS
Structural comparison of DMPC/Cholesterol and DMPC/7DHC membranes
We begin by reporting on several structural properties of DMPC/sterol membranes obtained from our SAXD experiments and MD simulations, and discuss how these differ for the two sterols, Chol and 7DHC, at different concentration. We used 2 sets of simulations, SIM1 (400 lipids, CHARMM forcefield) and SIM2 (1600lipids, GROMOS forcefield), both for model verification and to gain access to bending rigidities, as well as other membrane parameters, as detailed in the Materials and Methods. The convergence of all simulated systems was confirmed by the constant simulation box dimensions (within fluctuation) over the last 20ns or 14ns for SIM1 and SIM2 simulations, respectively (Figures S3 and S4 in the Supplemental Data).
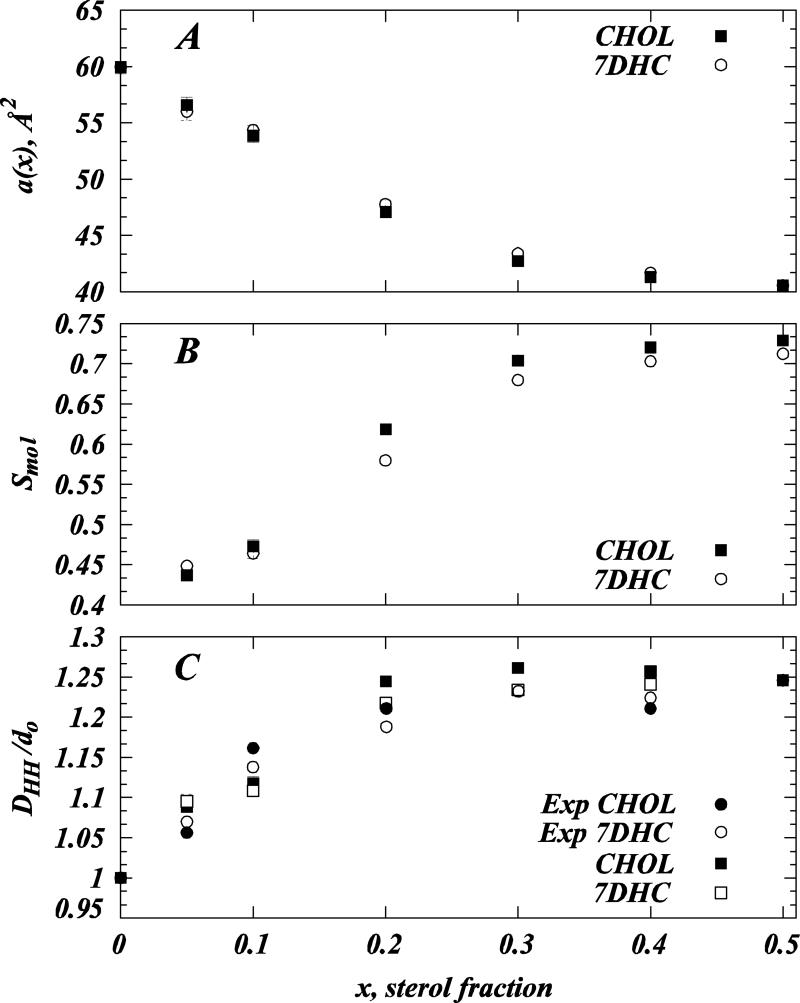
Figure 2A presents the equilibrium values for the areas per lipid head-group a(x) for DMPC/sterol membranes calculated from the converged 20ns trajectory segment of SIM1 simulations. For each sterol mole fraction x, the area per molecule was obtained as: a(x) = 2ABOX / (NDMPC+ NCHOL). We find strong non-linear decrease in molecular area with increasing Chol or 7DHC content, indicative of the previously described sterol condensation on phospholipids, Fig. 2A (15, 27, 59, 60). Furthermore, a(x) values are similar for the two sterols over the entire concentration range we have explored (0-50% sterol).
Figure 2.
Membrane molecular parameters compared for cholesterol and DMPC at different concentrations and to SAXD experiments. (A) Area per lipid a(x) as a function of sterol fraction for DMPC/Cholesterol (closed symbols) and DMPC/7DHC (open symbols) mixtures. (B) Average molecular order parameter Smol for DMPC lipid tails as a function of sterol fraction for DMPC/Cholesterol (closed symbols) and DMPC/7DHC (open symbols) mixtures. (C) Comparison of experimental (circles) and simulation (squares) peak-to-peak DHH distances for different DMPC/Chol (closed symbols) and DMPC/7DHC (open symbols) mixtures calculated from the corresponding electron density profiles. DHH values presented are normalized by the thickness d0 of the pure DMPC bilayer in experiment and simulations. SAXD data for 5 and 10 mol% DMPC/Chol mixtures are from earlier studies (21, 27). Note that error bars for some data points in all three panels are comparable to the symbol size used to plot this graphs and thus are hardly visible. All the MD data presented is from the analysis of SIM1 simulations (see Table 1).
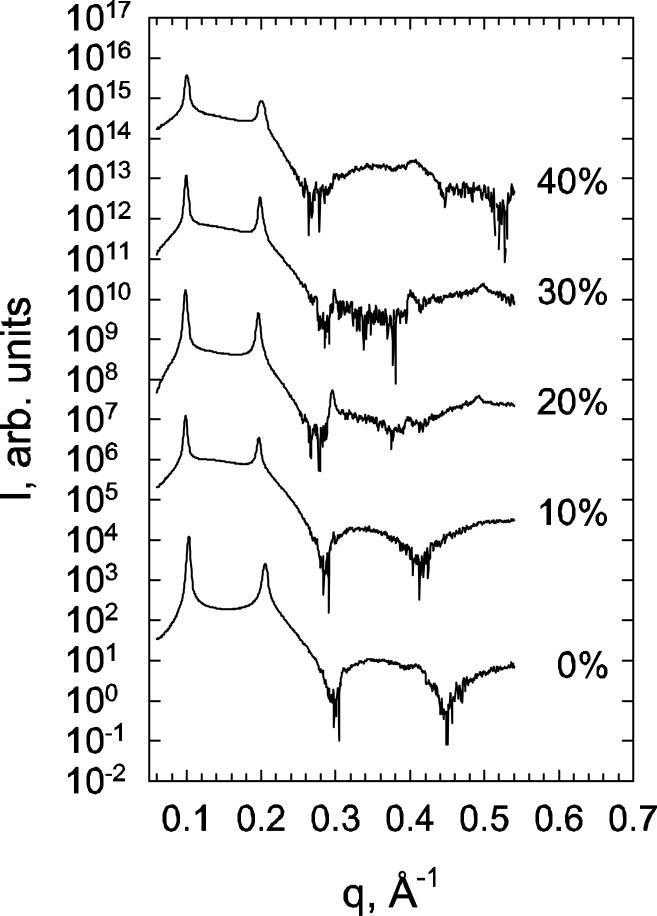
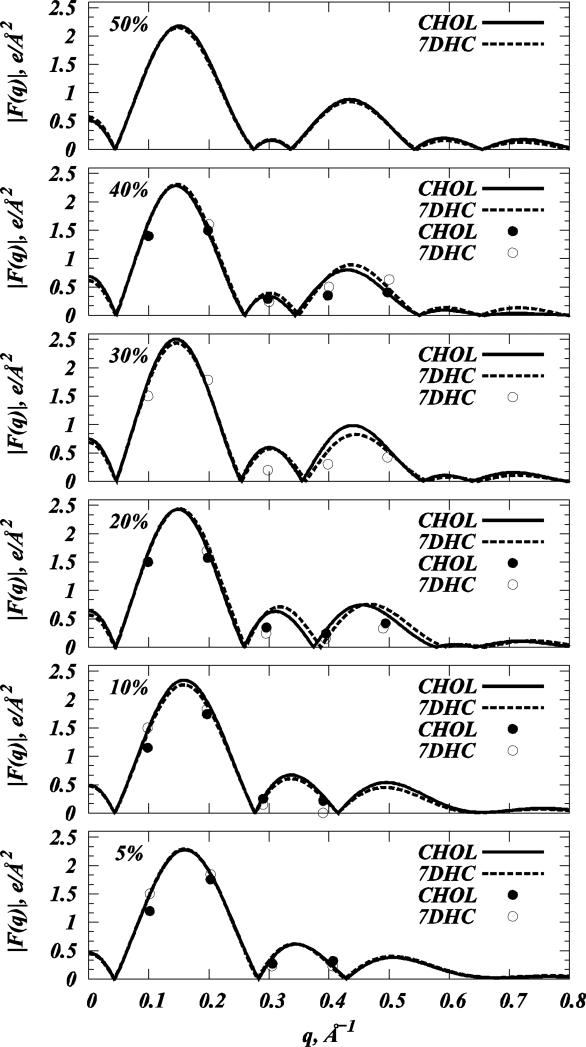
To further quantify the structural effects of Chol and 7DHC on DMPC bilayers, we evaluated SAXD scattering patterns, experimental and simulated form factors and electron densities, as well as lipid order parameters from SIM1 trajectories. Figure 3 shows the radially integrated SAXD scattering patterns for various DMPC/7DHC mixtures, and Figure 4 compares the form factors Fh derived from these scattering intensities to the simulated form factors F(q) (see Tables S1 and S2 in the Supplemental Data for Fh and corresponding d-spacing values). To obtain F(q) from MD simulations, we used the standard integration of the symmetric electron density profile for each model membrane (see for example (15, 27, 32, 42, 61, 62)).
Figure 3.
SAXD integrated intensity patterns for selected DMPC/7DHC mixtures. Numbers (right) to the data correspond to 7DHC concentration in mol%. SAXD data for 5 and 10 mol% DMPC/Chol mixtures are from earlier studies (21, 27).
Figure 4.
Comparison of the form factors for various model DMPC/Cholesterol and DMPC/7DHC bilayers from SIM1 simulations (solid lines - DMPC/Cholesterol, dashed lines - DMPC/7DHC) and SAXD experiments (closed symbols - DMPC/Cholesterol, open symbols - DMPC/7DHC).
The form factor plots in Fig. 4 illustrate a good agreement between experiment and simulations, and together with the SAXD traces in Fig. 3, show the expected trends (15, 21, 27), for both Chol and 7DHC. Thus, upon increasing sterol concentration, the maxima in form factors shift towards lower q values, giving rise to additional lobes in the high q-range. These features are usually indicative of bilayer thickening, and suggest increased ordering within the membrane, as we further discuss below. However, the profiles shown in Fig. 4 also reveal that F(q)-s overlap for DMPC/Chol and DMPC/7DHC mixtures at 5, 10, and 50% sterol concentrations, suggesting structurally similar organizations in these mixtures. For 20, 30 and 40% systems, the second and third lobes in the form factors appear to be somewhat shifted towards the lower q wave-vector range of in DMPC/Chol bilayers compared to DMPC/7DHC, indicating that cholesterol-containing membranes at these compositions are somewhat thicker, and that lipid hydrocarbon tails order more strongly.
The trends observed in the form factor profiles are confirmed by our analysis of lipid chain order parameters and electron densities from simulations. Specifically, Figure 2B illustrates the average molecular order parameters Smol for DMPC lipid tails in SIM1 simulations. Smol values are derived using the relation Smol = 2 |<SCD>| (13, 14), where <SCD> represents chain- and time-averaged deuterium order parameter defined at each carbonyl atom in the DMPC sn-1 and sn-2 chains as (63): SCD = ½ <3cos2β-1>, β denoting the angle between the C-H bond in the carbonyl group and the normal to the bilayer slab. Although the process of DMPC tail ordering with increasing sterol content (13, 15, 27, 30, 64, 65) saturates at similar levels of Chol and 7DHC, cholesterol orders slightly more with respect to 7DHC in the 20%-40% composition range (see Smol profiles in Fig. 2B). We detect the largest difference in lipid tail order ΔSmol ≈0.04 between Chol and 7DHC mixtures at 20% sterol concentration. This ΔSmol value is consistent with that reported in MD studies of saturated dipalmitoyl-phosphatidylcholine (DPPC) lipid at 20% Chol or 7DHC (13, 14, 29, 30).
In the SAXD electron density profiles, DHH distances can be closely assigned to an average separation between phosphate atoms on lipids from opposite leaflets, and thus provides a measure of bilayer thickness (22). Figure 2C presents head-head (peak-to-peak) distances, DHH, in SIM1 bilayers (scaled by the thickness of the pure DMPC bilayer derived from SIM1 simulations, d0) versus sterol content in different DMPC/Chol and DMPC/7DHC mixtures. For comparison, the panel also shows the same measure calculated from our SAXD experiments (scaled by the thickness of the pure DMPC bilayer derived from the experiment). We find good agreement between the MD simulations and the SAXD data (see Fig. 2C), and only small differences in DMPC/Chol versus DMPC/7DHC bilayer thicknesses, the largest difference in DHH appearing at 20% sterol concentration, consistent with the Smol data in Fig. 2B.
To summarize the results presented so far, our MD and SAXD data indicate that at 5, 10, and 50% concentrations the two sterols have a similar effect on DMPC lipid organization. But in the 20-40% composition range, cholesterol appears to affect DMPC tail ordering and membrane thickening somewhat stronger compared to 7DHC. These results are in line with experimental (6, 8, 9) and computational (13, 14, 29, 30, 66) studies that have established that the 7DHC effects on saturated lipids are weaker than those of cholesterol in the 20-40% sterol concentration range. The computational studies from different groups (13, 14, 29, 30, 66) as well as our own (15, 27) have further established that an important factor in sterol's ability to order lipid membranes is the extent of sterol tilt. Therefore, we have pursued the orientation of cholesterol and 7DHC in our SIM1 simulations over the range of sterol concentrations. The findings from this analysis are presented next.
Preferred orientation of cholesterol and 7DHC in DMPC membranes
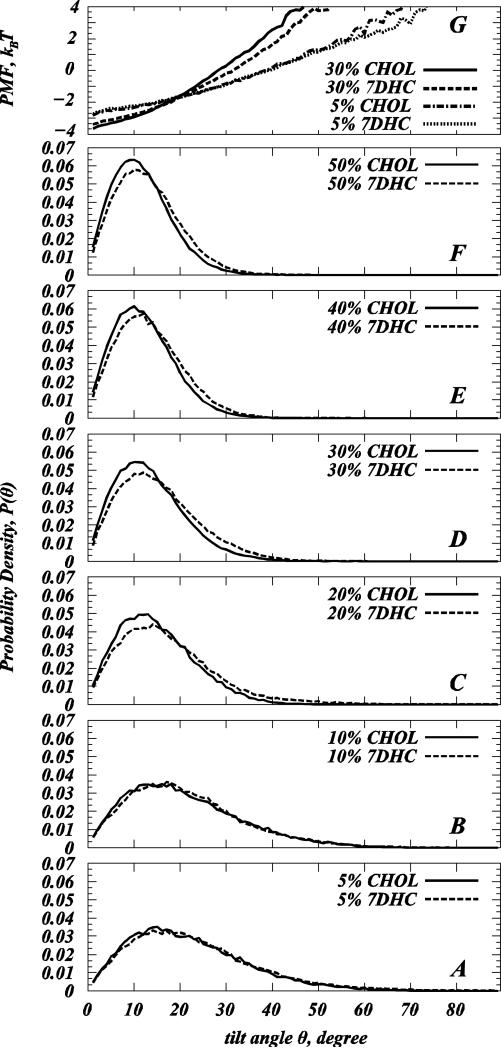
We compared normalized probability distributions P(θ) of Chol and 7DHC tilt angles θ with respect to the normal to the membrane interface (see Materials and Methods) in SIM1 simulations, panels A-F of Figure 5. Comparative analysis of P(θ) distributions for each sterol at different concentrations reveals that increasing Chol or 7DHC content results in stronger sterol alignment along the bilayer normal, as the peaks in the P(θ) plots for both Chol and 7DHC shift towards lower θ angles with sterol levels.
Figure 5.
(A-F) Normalized probability distributions P(θ) of sterol tilt angle θ in model DMPC/Chol (closed symbols) and DMPC/7DHC (dashed lines) bilayers from SIM1 simulations. The tilt angle is defined as the angle between the vector C3-C17 describing sterol ring plane orientation and the bilayer normal (see Materials and Methods). (G) Potential of Mean Force (PMF) plots from the distributions in panels A and D (see text for details).
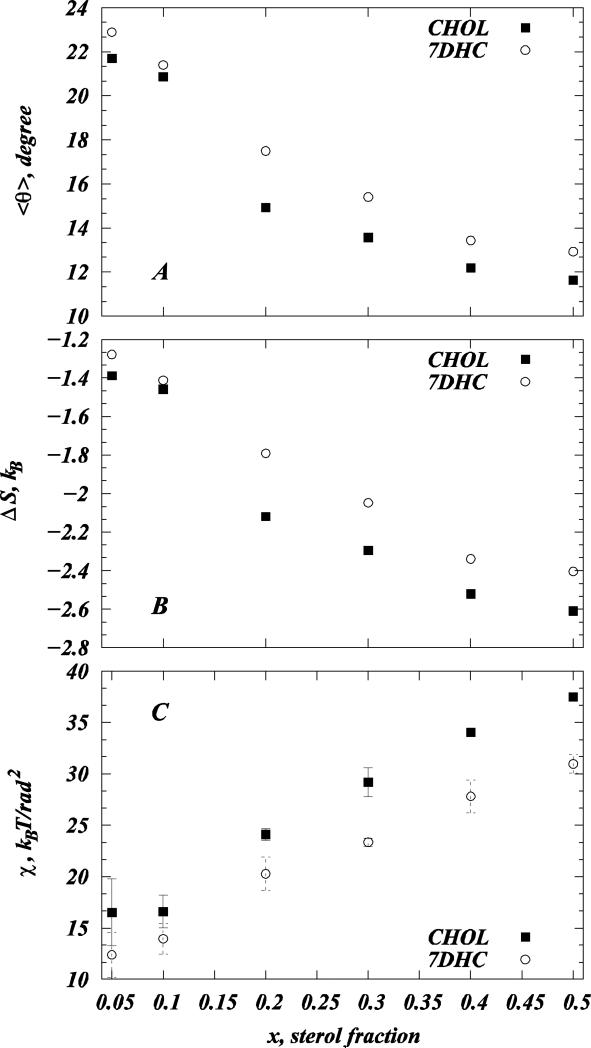
Using P(θ) for each sterol composition, we also calculated the average sterol tilt angle <θ> as (15, 27): . We find that the average tilt angle <θ> for both sterols decreases with sterol concentration. The comparison of <θ> values for the two sterols in different mixtures is presented in Figure 6A. Furthermore, consistent with earlier MD studies from our (15, 27) and other groups (30, 67), trends in <θ> and membrane thickness with increasing sterol composition (Fig. 2C) are strongly anticorrelated, emphasizing the important mechanistic role of sterol tilt in ordering lipid bilayers (30). As noted elsewhere (15, 27), in principle, the average cholesterol tilt angle can be defined in several ways, and the definition employed here may differ from the ones used in other MD studies (30, 62, 68-71).
Figure 6.
(A) Average sterol tilt angle <θ> for model DMPC/Chol (solid symbols) and DMPC/7DHC (open symbols) membranes from SIM1 simulations as a function of sterol fraction. (B) Losses in sterol orientational entropy with increasing sterol concentration for DMPC/Chol (solid symbols) and DMPC/7DHC (open symbols) membranes from SIM1 simulations. (C) Tilt modulus χ for cholesterol and 7DHC as a function of sterol composition for model DMPC/Chol (solid symbols) and DMPC/7DHC (open symbols) membranes from SIM1 simulations. For each system, χ was obtained by the fitting quadratic function to the PMF plots in Figure 3G in the [8°; 20°] angular range of θ. The error bars represent standard deviations from the similar fits obtained over [8°; 12°], [12°; 16°], and [16°; 20°] angular intervals.
Comparing P(θ) plots, Fig. 5A-F, for the different sterols at the same concentrations, indicates that at sterol levels of 20% and higher, Chol orients differently than 7DHC in DMPC membrane. Specifically, while 7DHC in the 20-50% concentration range exhibits a stronger tendency for larger (20-40°) θ angles compared to cholesterol, the probability of finding Chol molecules at smaller (<20°) tilt angles is higher compared to 7DHC. These inferences are in agreement with the MD studies of saturated DPPC lipid mixtures with Chol or its metabolic precursors (including 7DHC) with 20% sterol (14) where, of all the sterols studied, cholesterol was found to attain the most “upright” orientation in DPPC bilayer.
Overall, over the entire range of sterol compositions, the average tilt <θ> of cholesterol molecules is lower than that of 7DHC in our SIM1 simulations (Fig. 6A, see also Table S1). Stated differently, at a given concentration, the cone angle (72) for 7DHC is larger than that of cholesterol. This result agrees with other MD studies that compared cholesterol and 7DHC orientation in different lipids and at 20% sterol content (13, 14, 30). Interestingly, the largest difference between Chol and 7DHC average tilts in our simulations is observed in the 20-30% sterol concentration range, in line with the Smol and DHH data, that also show the largest differences in 20-30% composition interval, see Figs. 2B-C.
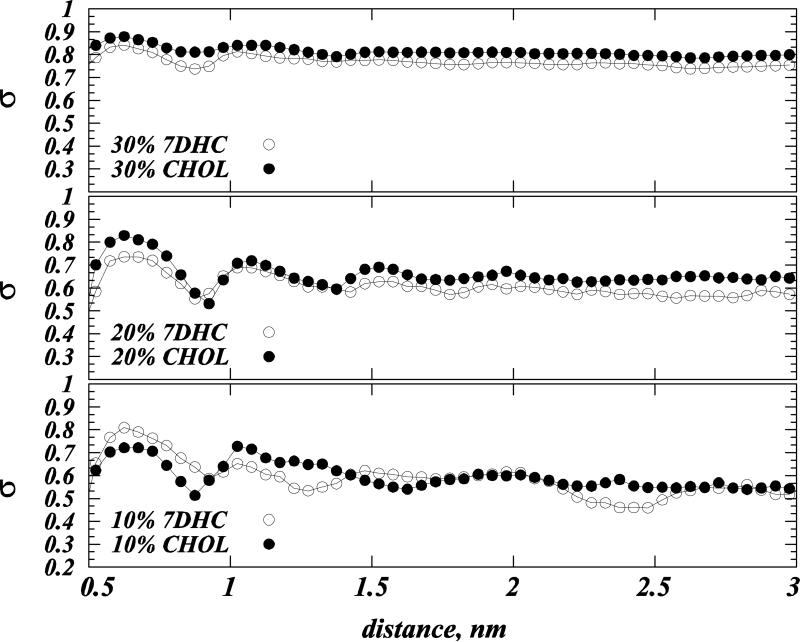
Trends in P(θ) distributions in Fig. 5A-F are indicative of a nematic-like ordering (73) of Chol and 7DHC in DMPC membranes, whereby sterols align along the bilayer normal (15, 27). Such alignment can be better quantified by measuring sterol orientational order parameter σ (15, 27, 28, 74) defined as: σ=(1/2)(3cos2α-1), where α denotes the angle between the vectors defining the ring planes of two sterols (see Materials and Methods). If sterol molecules are perfectly aligned σ=1, while for the orientationally isotropic mixture σ=0.
The variation of σ with the distance between cholesterols, for Chol and 7DHC in 10, 20 and 30% mixtures, indicates that with increasing sterol content, the orientational ordering of both Chol and 7DHC molecules increases, and that this high order persists over large distances, Figure 7. In addition, comparing σ profiles for the two sterols in 20 and 30% mixtures reveals stronger alignment tendencies for cholesterol, as σ values are larger for Chol compared to 7DHC. However, for lower compositions (10%), the σ plots for Chol and 7DHC overlap over the entire range of pair distances, indicating similar long-range orientational pattern for the two sterols at these concentrations.
Figure 7.
Orientational order parameter σ (see text) as a function of distance between cholesterol (solid symbols) and 7DHC (open symbols) molecules. Results are for model 10% (lower panel), 20% (middle panel) and 30% (upper panel) sterol mixtures derived from SIM1 simulations.
Losses in orientational entropy upon sterol alignment are larger for cholesterol than for 7DHC
We have recently found that the nematic-like ordering of sterol molecules found for higher cholesterol content can be accompanied by losses in sterol orientational entropy ΔS=ΔS(x) (15, 27, 28) defined as (see (15, 27) and references therein). Note that ΔS, as defined here, vanishes if sterol molecules are randomly oriented, but deviates from zero for anisotropic orientations of sterols, reflecting losses in orientational entropy (15, 27). To compare these losses in orientational entropy for Chol versus 7DHC, we plot ΔS(x) as a function of sterol composition in our SIM1 simulations, Figure 6B.
Indeed, ΔS is negative for both Chol and 7DHC mixtures (Fig. 6B) and decreases for both sterols with concentration x. However, consistent with cholesterol's stronger alignment tendencies (Figs. 5A-F, 6A, and 7), ΔS is lower for Chol compared to 7DHC for the entire range of sterol compositions we explored. Perhaps not too surprisingly, we find the trends in ΔS and in the average sterol tilt <θ> with increasing sterol concentrations to be strongly correlated, compare Fig.6A and 6B.
Energy cost for substituting Chol with 7DHC is higher for sterol molecules that are more upright: in silico mutations
To further quantify differences in the alignment tendency for Chol and 7DHC, as observed in Figs. 5 and 6A-B, we used free energy perturbation (FEP) calculations to perform in silico mutations on selected, differently oriented, Chol and 7DHC molecules in SIM1 membranes (see Materials and Methods). In particular, from the final trajectory frames of 30% DMPC/Chol and 30% DMPC/7DHC systems, we selected for mutations four sterols in each membrane at low tilt angles (θ ≈ 10°), and four at high tilt angles (θ ≈ 30°) (see Tables 2-3). Subsequently, we performed eight individual Chol → 7DHC and eight 7DHC → Chol mutations in 30% DMPC/Chol and 30% DMPC/7DHC bilayers respectively. In addition, we selected one sterol at low tilt angle and one at high tilt angle in 5% DMPC/Chol and 5% DMPC/7DHC bilayers (Table 4), and performed two additional Chol → 7DHC and two 7DHC → Chol mutations in 5% DMPC/Chol and 5% DMPC/7DHC mixtures correspondingly. For all FEP calculations, we restrained the orientations of mutated sterols, so that our analysis quantified specifically Δμ(θ), the free energy difference associated with substituting one sterol with the other at a particular specified sterol tilt angle θ.
Table 3.
7DHC → Cholesterol in silico mutations in 30% DMPC/7DHC bilayera.
| 7DHC # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| θ,° | 12.09 | 10.98 | 11.15 | 11.66 | 31.13 | 30.83 | 34.29 | 31.06 |
| Δμ, kcal/mol | 2.508 ± 0.556 | 2.65 ± 0.423 | ||||||
The tilt angles with respect to the membrane normal for 8 7DHC molecules in 30% DMPC/7DHC mixture from SIM1 trajectory chosen for the mutations are shown along with the corresponding free energies for mutations in “straight” and “tilted” 7DHC separately. The error bars in the free energy values were calculated as standard deviations over FEP calculations performed in forward/backward directions and over 1200ps and 2400ps mutation time intervals for 7DHCs 1-4 and 5-8 respectively.
The results of the whole set of in silico mutations we performed (Tables 2-4) reveals that irrespective of sterol orientation or sterol composition, Chol → 7DHC transformation is energetically favorable, whereas 7DHC → Chol mutation is unfavorable. Thus, we find free energy gains upon Chol → 7DHC substitutions and free energy losses upon 7DHC → Chol transformations. Although the Δμ free energies reported in Tables 2-4 are similar in magnitude for all tested transformations, we note some subtle differences. In particular, Chol → 7DHC substitution in 30% DMPC/Chol bilayer (Table 2) is somewhat more favorable (by ~0.5 kBT) for cholesterols that are in a tilted orientation than for ones in relatively upright conformation, suggesting that the Chol → 7DHC transformation is energetically more favorable for sterols that are in the tilted conformation. Stated differently, the chemical potential of two tilted sterols is more disparate than when they are standing up (75). 7DHC → Chol mutations in 30% DMPC/7DHC membrane are energetically more favorable for sterols in the “upright” orientation (Table 3), but the differences in Δμ between the transformations in the upright vs. tilted sterol orientations in 30% DMPC/7DHC bilayer are even smaller (~0.25 kBT) than those in 30% DMPC/Chol membrane (Table 2). This is probably due to the “soft” nature of 7DHC environment surrounding the mutated molecule (see below), that maintains its composition even as 7DHC mutates into Chol, when the two sterols are orientated similarly, comparing Δμ values for 5% (Table 4) versus 30% DMPC/sterol (Tables 2-3) mixtures suggests that Chol → 7DHC transformations are energetically more favorable in 30% than in 5% sterol systems, whereas conversely, 7DHC → Chol mutations bare lower energy cost in 5% mixtures than in 30% systems.
As we discuss below, these trends in Δμ result from an interplay between differences in the volumes accessible to Chols versus 7DHCs at different orientations and in membranes of different cholesterol content. These properties, in turn, directly affect the sterol's orientational entropy.
Cholesterol is characterized by higher tilt modulus than 7DHC
The free energies measured in the FEP calculations can be directly compared with the potential of mean force (PMF) that quantifies the free energy associated with tilting a sterol molecule under different membrane conditions. Specifically, we have recently introduced a PMF that is based on the P(θ) distributions as: PMF(θ) = –kBT ln[P(θ) / sinθ], where sinθ is the normalization factor describing the probability density of sterol tilt angle in the ideal system of non-interacting sterol molecules (see (15, 27) for details). With that ΔPMF (θ2 – θ1) = PMF(θ1) – PMF(θ2) can be interpreted as the work required to tilt a sterol molecule from an angle θ1 to an angle θ2.
We find close agreement between PMF(θ) values and the free energies calculated from in silico mutations. For comparison, PMF(θ) for 5 and 30% mixtures from SIM1 simulations are presented in Figure 5G, and these are used to compare with Tables 2-4. For example, from Fig. 5G we find that ΔPMF(30° –10°) for 30% DMPC/Chol and 30% DMPC/7DHC mixtures are different by 0.57 kBT, a value consistent with the free energy difference of 0.5 kBT that we found for the Chol → 7DHC mutations performed on Chols at 30° and 10° tilt angles in 30% DMPC/Chol membranes, Table 2.
The PMF description, in addition, provides a framework to calculate the sterol tilt modulus χ, an important quantitative empirical measure of the force associated with tilting a sterol molecule (15, 27). χ represents the proportionality coefficient for the second order term in the series expansion around θ=0 angle of the sterol desolvation free energy, ΔG ≈ ΔG0 + (1/ 2)χθ2 (15, 27, 48). Thus, χ can be conveniently obtained from the quadratic fit to the low angular range of the PMF(θ) function, as described in ref (15, 27). Following the protocol we have previously developed (15, 27), we calculated here χ values for cholesterol and 7DHC molecules at different sterol concentrations, taking for each system the best fits of the quadratic function to the PMF(θ) profiles in the [8°; 20°] θ angle interval (see Materials and Methods). In fact, using the derived values of χ, it is possible to reconstruct the whole tilt probability distribution, and it is therefore more general than the average tilt angle alone. We find that the functional approximation for P(θ) well fits the distribution in simulation even outside of the region of the original fit (See Figure S5 in the Supporting Data).
Consistent with our previous studies on DMPC/Chol (15, 27), we find that χ for both Chol and 7DHC is constant (within statistical uncertainty) for sterol concentrations less than 10%, but rises sharply upon sterol addition beyond that, due to the onset of a sterol-induced spatial liquid ordered fluidity state, see Fig. 6C. At concentrations of 20% and higher, the χ profiles illustrate substantial difference in the values of the tilt modulus for the two sterols, Fig 6C. Thus, for concentrations above 20% sterol content, χCHOL is larger than χ7DHC. Furthermore, these differences between the tilt modulus for Chol and 7DHC appear to progressively increase with sterol content. Larger χ values generally signify a higher free energy penalty for sterol molecules to be inserted at a tilted orientation into a lipid membrane relative to the upright configuration. Therefore, the results in Fig. 6C indicate that with 20% and higher sterol content it costs less free energy to introduce an additional 7DHC in a tilted orientation into DMPC/7DHC mixtures than to add a cholesterol molecule in the same tilted conformation into DMPC/Chol membranes.
Having established that the different orientation profiles of Chol and 7DHC in DMPC membranes result in different values for Chol and 7DHC tilt modulus, we next hypothesized that the different χCHOL and χ7DHC values we calculated may have a direct effect on the material properties, such as the bending rigidity, KC, of the cholesterol- and 7DHC-containing simulated lipid membranes (see Materials and Methods). It has been suggested experimentally (2, 23) that cholesterol and 7DHC alter KC of phospholipid membranes to a different extent. To establish the possible link between the tilt modulus χ and the bending modulus KC, we conducted a set of MD simulations on large 1600-lipid DMPC/sterol membranes (SIM2 simulations, Table 1) at 0 and 30% sterol compositions, and calculated KC values for different mixtures by performing spectral analysis for membrane undulations on these SIM2 trajectories (see Materials and Methods). These results are discussed next.
Differences in the tilt modulus correlate with the mechanical properties of DMPC/Cholesterol and DMPC/7DHC membranes
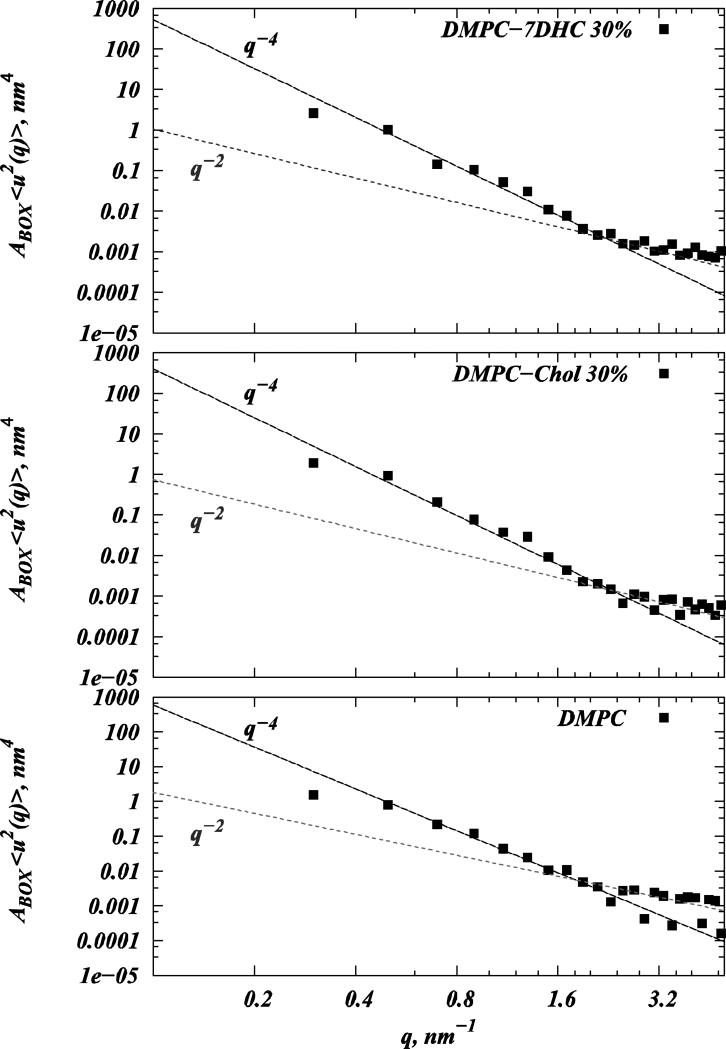
Figure 8 shows results from the spectral analysis of undulations conducted on trajectories from large-scale SIM2 simulations. As expected (see Materials and Methods), the spectrum contains two regimes well-fitted by q-4 and q-2 power laws. From the best fits, we can estimate the bending rigidities for pure DMPC, 30% DMPC/7DHC, and 30% DMPC/Chol systems to be 16±2 kBT, 19±2 kBT, and 28±2 kBT, respectively. Providing good agreement with experimentally measured KC for DMPC bilayers (13±4 kBT (22), 20±4 kBT (2)), our data suggest that at 30% sterol concentration, DMPC/Chol membranes are ≈1.47 times more rigid compared to DMPC/7DHC bilayers. This number is in excellent agreement with experimentally derived estimates of 1.48 for this ratio, reported in Refs. (2, 23).
Figure 8.
Spectral analysis of bilayer undulations carried out on pure DMPC (lower panel), 30% DMPC/Cholesterol (middle panel), and 30% DMPC/7DHC (upper panel) mixtures from SIM2 simulations. Logarithmic plots on the three panels depict calculated 〈u2(q)〉 Fourier amplitudes multiplied by the simulation box size ABOX (black symbols) as a function of wave vector q for different systems. Also shown are q-4 and q-2 fits to the undulatory and protrusion regimes of the spectral data. Bending rigidities KC for different membranes were obtained from the coefficients of the best fits to the q-4 function.
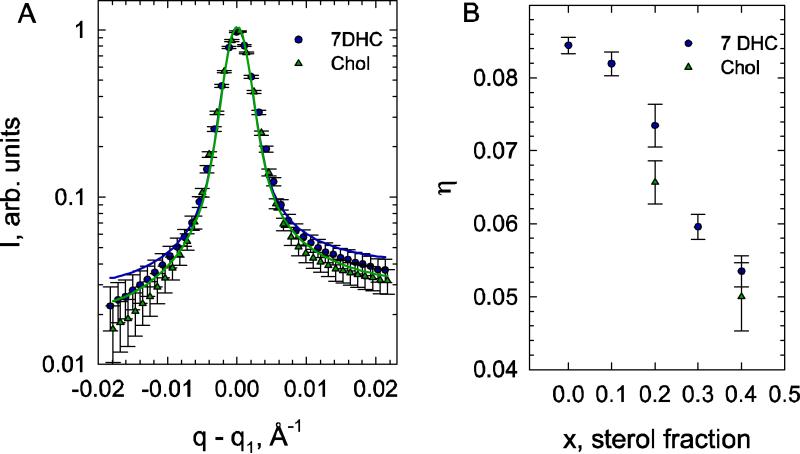
To provide additional experimental support to the impact on membrane bending rigidity that we find for the two sterols, we further tested whether similar effects on bending rigidity can be found also from SAXD experiments at 20% sterol concentrations. Using the modified Caillé theory, we can estimate KC for 20% DMPC/Chol and DMPC/7DHC mixtures from our SAXD experiments by analyzing the first order diffraction peaks (see Materials and Methods).
We found a general decrease of bending fluctuations with increased 7DHC content (Figure 9B), as previously reported also for cholesterol, sigmasterol, and sitosterol (21) indicating a significant increase of membrane stiffness (22, 76). Comparing the bending fluctuation data of DMPC/7DHC with that of DMPC/Chol mixtures at equivalent molar ratios, we found that 7DHC is less effective in reducing bending fluctuations than cholesterol, as evident by the comparison of the different sterols at 20% content, data in Fig. 9A, where the diffuse part of the Bragg wings is significantly more expressed for DMPC/7DHC than for DMPC/Chol. Using theory, these differences can be shown to indicate that cholesterol containing bilayers exhibit less bending fluctuations than DMPC/7DHC (18).
Figure 9.
Effect of sterols on bending fluctuations. (A) The fits to the normalized first quasi-Bragg peaks of DMPC/7DHC and DMPC/Chol (20 mol% sterol content). (B) Comparison of the fluctuation parameters of 7DHC and cholesterol containing DMPC bilayers.
Assuming that the bulk modulus B of interactions between adjacent membranes is the same for DMPC/7DHC and DMPC/Chol, we estimated the difference in bending rigidities using Eq. (2) as
| (6) |
We therefore estimate that 20% DMPC/7DHC membranes are 1.27 times softer than 20% DMPC/Chol membranes. Taken together with the KC values we obtained for 30% mixtures and the tilt modulus profiles in Fig. 6C, our analysis establishes a link between the sterol tilt modulus and bilayer bending rigidity. Thus, in the 20 to 30% sterol concentration range, where χCHOL becomes progressively larger than χ7DHC, DMPC/Chol membranes become gradually more rigid compared to DMPC/7DHC membrane (KC ratios grow from ~30% to ~50%).
DISCUSSION
To trace differences in the impact of two sterols on lipid membranes: cholesterol and its metabolic precursor 7DHC, we have followed mixtures of these sterols with DMPC lipids. Whereas there have been numerous reports in the literature on the effect of Chol and other sterols on ordering of lipid membranes, as we discuss below, this work offers two novel aspects. First, we illustrate that one can discuss the differential condensing effect of sterols on phospholipids in terms of the organization of membrane sterols; this should be complementary to following changes in measures of lipid order. Second, our results offer a quantitative description of the important energetic and entropic factors directly related to the mechanistic properties of lipid/sterol membranes.
Our MD results complemented by SAXD experiments indicate only minor differences in many structural properties of DMPC/Chol and DMPC/7DHC membranes. All structural characteristics we have measured for these bilayers, including membrane form factors, areas per lipid, membrane thickness, and lipid tail order parameters, are similar for DMPC/Chol and DMPC/7DHC mixtures, both at low (< 10%) and high (>40%) sterol compositions. At 20% and 30% sterol concentrations, DMPC/Chol membranes appear somewhat more ordered (ΔSmol ≈ 0.04) than DMPC/7DHC bilayers, but even at these intermediate compositions the structural differences between the two mixtures are rather small, in line with other structural studies on these and similar mixtures (13, 14, 66).
One important difference in organization of DMPC/Chol and DMPC/7DHC membranes that we measure is a significantly altered extent of tilt for the two sterols. These findings are in agreement with earlier reports (see for example (13, 14, 30, 66)). Thus, although upon increasing sterol concentration both Chol and 7DHC tend to align along the membrane normal, the preferred orientations of the two sterols reflect that Chols are stabilized in a more upright orientation within DMPC bilayers compared to 7DHC molecules.
The different tilting of the two sterols is a direct consequence of the free energy losses associated with tilting sterol molecules away from the bilayer normal. Indeed, we find that in sterol concentrations of 20% and higher, the tilt modulus χCHOL for cholesterol is substantially larger (by 5-8 kBT/rad2) than the corresponding tilt modulus χ7DHC for 7DHC. As a consequence, in this composition range, we find that DMPC/Chol membranes become progressively stiffer (by ~20%) than DMPC/7DHC membrane.
Because our data suggests that the different elastic properties of DMPC/Chol and DMPC/7DHC bilayers stems from the different orientations of the two sterols in DMPC membranes, it is important to establish a quantitative molecular mechanistic description for the differences observed in Chol and 7DHC arrangement in phospholipid bilayers. In the following we develop this description and discuss possible implications that the distinct tilt of Chol and 7DHC may have on the organization and function of membrane inserted proteins.
Preferred orientation of Cholesterol and 7DHC is determined by the interplay between the sterol orientational entropy and excluded volume effects
What drives cholesterol and 7DHC to assume distinct preferred orientations inside DMPC bilayers? From molecular structural considerations, namely, because cholesterol molecules possess two hydrogen atoms (covalently bound to carbons C7 and C8, Fig 1) that are not present in 7DHC, it is reasonable to suspect that the organization of Chol and 7DHC will be at least in part regulated by the volumes that are accessible to the two sterols in a lipid membrane. In fact, comparative MD studies of cholesterol, desmosterol, or 7DHC in DPPC membranes (14) have shown that DPPC lipids are expected to be more loosely packed around the C7=C8 double bond region in 7DHC system than they are around the C7-C8 bond segment in Chol containing bilayers. These findings imply that there may be a larger “free” volume available to 7DHC molecules inside phospholipid membranes than there is to cholesterols.
We have previously quantified the effect of the free volume on cholesterol's tendency to align with increasing concentration (15, 27). In particular, we found that the preferred cholesterol configuration in a lipid bilayer is dictated by the interplay between energy and entropy components. Thus, pair-wise interactions between Chol molecules and between Chol and lipids will tend to preferentially exclude cholesterol molecules from each other, because Chol-Chol pair interactions are in general less favorable energetically compared to Chol-lipid interactions (see (15, 27) and references therein). In addition to this free energy contribution, and with analogy to simple hard-rod molecules, each sterol molecule also possesses degrees of freedom associated with orientational and translational entropy of the molecule inside the membrane (15). The greater the sterol tilt, the higher the orientational entropy, but higher too is the excluded volume (see Figs. 6A-B). Thus, in addition to the enthalpic contributions, the tendency for sterols to align with increasing concentration can also be due to the sterol's attempt to minimize excluded volume (or gain “free” volume) and hence maximize translational entropy at the expense of orientational entropy (27).
Taken together, the preferred orientation of Chol and 7DHC in a lipid membrane should be expected to be largely dictated by the different interplay of orientational and translational entropy factors that is in turn regulated by the different extent of free volume available to Chol and 7DHC inside the membrane. Our FEP calculations support this mechanistic picture (see Tables 2-4). In particular, irrespective of sterol tilt or composition, the positive and negative free energy changes due to Chol →7DHC and 7DHC→Chol transformations correspondingly can be a consequence of the reduced effective excluded volume interactions (gain in free volume release) upon Chol→7DHC transformation and of the free volume loss upon 7DHC→Chol substitution. Furthermore, our findings that Chol → 7DHC mutations are accompanied by lower energy cost in 30% DMPC/Chol than in 5% DMPC/Chol membranes is consistent with a mechanism whereby sterols gain free volume when they align, as they do in mixtures of high (≥20%) sterol content.
Interestingly, in 5% DMPC/sterol mixtures, both Chol→7DHC and 7DHC→Chol transformations bare a lower energy cost if the substituted sterol is held in a tilted conformation compared to an upright orientation. This is because at low compositions orientational entropy dominates the free energy of sterol insertion into a lipid membrane (Fig. 6B) at the rather small expense of the translational entropy, and thus of the excluded volume effects (see also (15, 27)). Taken together, our quantitative FEP analysis suggests that the preferred orientation of Chol and 7DHC inside a phospholipid membrane will be stabilized by the interplay between the orientational entropy of the sterol that prefers a “lying down” conformation, and the free volume effects, as well as additional free energy contributions that favor “standing up”.
Mechanistic link between sterol tilt modulus and membrane rigidity
How can the tilt modulus of sterol molecules impact membrane rigidity? To make first mechanistic links between these properties at the molecular level, it is useful to consider another related property, the modulus describing the tilting of one sterol molecule with respect to another, χ12, which we term the pair tilt modulus (PTM). When a membrane undergoes bending deformations, sterol molecules are forced to rearrange in the membrane, and on average assume a more splayed configuration with respect to one another. One contribution to the bending free energy must be associated with this tilting degree of freedom of one molecule with respect to the other. If it becomes harder to tilt sterols with respect to one another, a stiffening of the membrane should ensue.
To gain access to this property, we repeated the procedure for deriving the tilt modulus χ for a single sterol from P(θ) (Figs. 5 and 6C) only this time considering the normalized density P12(α) that describes the probability of finding two sterol molecules at some angle α between them (see Figure S7 in the Supporting Data). By determining P12(α) for sterols that are closer than 1 nm to each other (this includes molecule pairs within the first peak in the orientational correlation function of sterols, Fig. 7) we are able to derive approximate estimates to the free energy penalty for tilting one sterol molecule with respect to the other. Thus, we find that, for cholesterol, the PTM χ12 can be rather high, 25 kBT/rad2 at 20% Chol, compared to 14 kBT/rad2 for 7DHC at the same mole fraction.
To connect the PTM χ12 to the bilayer bending rigidity, consider a highly simplified model where two neighboring sterol molecules 1 nm apart are set in a membrane cylindrically deformed with small curvature 1/R, so that the tilt angle α between sterols is, α ≈ sinθ12 = 1/R. The energy associated with this tilt is given by ETILT = (1/2)χ12(1/ R)2, or and per cholesterol and 7DHC, when using the respective values we find for χ12. On the other hand, using the Helfrich formulation for the bending energy (see Materials and Methods), the contribution per sterol molecule occupying an area of asterol is given by EH = (1/ 2)KC (1/ R)2 asterol. Estimating asterol from the lateral dimensions of our simulated membranes to be approximately 2.35 nm2, and using the bending modulus values calculated for DMPC/Chol and DMPC/7DHC membranes (see Results), we obtain and .
These simple estimations provide two important mechanistic insights: i) trends in the PTM χ12 and bending modulus KC appear to be correlated in that for stiffer (DMPC/Chol) membranes the pair tilt modulus is higher; and ii) the tilting of one sterol with respect to the other, quantified here by the PTM, accounts for a significant (~30-40%) portion of the total bending free energy increase in the two sterol/DMPC mixtures with respect to the pure DMPC bilayer. These finding suggests that bending rigidity due to changes in sterol can be traced to the easier or more difficult splaying of two cholesterol molecules with respect to each other. With that, our framework establishes the pair tilt modulus χ12 as a convenient measure for estimating differences in membrane stiffness in sterol-containing membranes.
While further studies are required to decode the full mechanism of cholesterol action on membrane rigidity, it would be interesting to determine how these properties also affect membrane function. Specifically, if the tilt modulus is much different in cholesterol versus 7DHC membranes, it is reasonable to expect this tilt degree of freedom to impact also the way that proteins fold and associate within the membrane environment. The excluded volume and aligning forces of cholesterol are already known to affect the structure and function of membrane proteins (see for example, (28)). It would be interesting to determine whether the same structural differences between sterols that impact membrane mechanical properties also determine the ability of membranes to serve as environments in which proteins can fold and function properly.
Supplementary Material
ACKNOWLEDGEMENTS
GK would like to thank Jerome Henin for his advice on setting up the free energy perturbation analysis with collective variables in NAMD. We also acknowledge helpful discussions with Harel Weinstein. DH acknowledges support from the Israel Science Foundation (grants 1011/07, 1012/07). GK is supported by NIH grant U54 GM087519. Computational resources of the David A. Cofrin Center for Biomedical Information in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine are gratefully acknowledged.
Footnotes
SUPPLEMENTAL DATA
Two tables and six figures are presented in the Supplemental Data.
REFERENCES
- 1.Porter FD, Herman GE. J Lipid Res. 52:6–34. doi: 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gondre-Lewis MC, Petrache HI, Wassif CA, Harries D, Parsegian A, Porter FD, Loh YP. J Cell Sci. 2006;119:1876–1885. doi: 10.1242/jcs.02906. [DOI] [PubMed] [Google Scholar]
- 3.Tulenko TN, Boeze-Battaglia K, Mason RP, Tint GS, Steiner RD, Connor WE, Labelle EF. J Lipid Res. 2006;47:134–143. doi: 10.1194/jlr.M500306-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay A, Paila YD, Jafurulla M, Chaudhuri A, Singh P, Murty MR, Vairamani M. Biochem Biophys Res Commun. 2007;363:800–805. doi: 10.1016/j.bbrc.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 5.Paila YD, Murty MR, Vairamani M, Chattopadhyay A. Biochim Biophys Acta. 2008;1778:1508–1516. doi: 10.1016/j.bbamem.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Shrivastava S, Paila YD, Dutta A, Chattopadhyay A. Biochemistry. 2008;47:5668–5677. doi: 10.1021/bi8001677. [DOI] [PubMed] [Google Scholar]
- 7.Petrache HI, Harries DP, Parsegian VA. Macromol. Symp. 2005;219:39–50. [Google Scholar]
- 8.Staneva G, Chachaty C, Wolf C, Quinn PJ. Journal of Lipid Research. 2010;51:1810–1822. doi: 10.1194/jlr.M003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berring EE, Borrenpohl K, Fliesler SJ, Serfis AB. Chem Phys Lipids. 2005;136:1–12. doi: 10.1016/j.chemphyslip.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Rebolj K, Ulrih NP, Macek P, Sepcic K. Biochim Biophys Acta. 2006;1758:1662–1670. doi: 10.1016/j.bbamem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Wolf C, Chachaty C. Biophys Chem. 2000;84:269–279. doi: 10.1016/s0301-4622(00)00135-6. [DOI] [PubMed] [Google Scholar]
- 12.Xu X, Bittman R, Duportail G, Heissler D, Vilcheze C, London E. J Biol Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]
- 13.Rog T, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M. Biochim Biophys Acta. 2009;1788:97–121. doi: 10.1016/j.bbamem.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 14.Rog T, Vattulainen I, Jansen M, Ikonen E, Karttunen M. J Chem Phys. 2008;129:154508. doi: 10.1063/1.2996296. [DOI] [PubMed] [Google Scholar]
- 15.Khelashvili G, Pabst G, Harries D. J Phys Chem B. 114:7524–7534. doi: 10.1021/jp101889k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pabst G, Rappolt M, Amenitsch H, Laggner P. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2000;62:4000–4009. doi: 10.1103/physreve.62.4000. [DOI] [PubMed] [Google Scholar]
- 17.Rappolt M. Journal of Applied Physics. 2010;101:7. [Google Scholar]
- 18.Zhang R, Suter RM, Nagle JF. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1994;50:5047–5060. doi: 10.1103/physreve.50.5047. [DOI] [PubMed] [Google Scholar]
- 19.Pabst G, Kucerka N, Nieh MP, Rheinstadter MC, Katsaras J. Chem Phys Lipids. 163:460–479. doi: 10.1016/j.chemphyslip.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Almeida PFF, Vaz WLC, Thompson TE. Biochemistry. 1992;31:6739–6747. doi: 10.1021/bi00144a013. [DOI] [PubMed] [Google Scholar]
- 21.Hodzic A, Rappolt M, Amenitsch H, Laggner P, Pabst G. Biophys J. 2008;94:3935–3944. doi: 10.1529/biophysj.107.123224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan JJ, Mills TT, Tristram-Nagle S, Nagle JF. Physical Review Letters. 2008;100 doi: 10.1103/PhysRevLett.100.198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrache HI, Harries DP, Parsegian VA. Macromol. Symp. 2005;219:39–50. [Google Scholar]
- 24.Bernsdorff C, Winter R. Journal of Physical Chemistry B. 2003;107:10658–10664. [Google Scholar]
- 25.Shrivastava S, Paila YD, Dutta A, Chattopadhyay A. Biochemistry. 2008;47:5668–5677. doi: 10.1021/bi8001677. [DOI] [PubMed] [Google Scholar]
- 26.Keller RK, Arnold TP, Fliesler SJ. Journal of Lipid Research. 2004;45:347–355. doi: 10.1194/jlr.M300232-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khelashvili G, Pabst G, Harries D, Weinstein H. Biophysical Society Meeting; San Francisco, CA, USA. 2010. p. 80a. [Google Scholar]
- 28.Khelashvili G, Mondal S, Andersen OS, Weinstein H. Journal of Physical Chemistry B. 2010;114:12046–12057. doi: 10.1021/jp106373r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ollila OHS, Rog T, Karttunen M, Vattulainen I. Journal of Structural Biology. 2007;159:311–323. doi: 10.1016/j.jsb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Aittoniemi J, Rog T, Niemela P, Pasenkiewicz-Gierula M, Karttunen M, Vattulainen I. J Phys Chem B. 2006;110:25562–25564. doi: 10.1021/jp064931u. [DOI] [PubMed] [Google Scholar]
- 31.Jo S, Lim JB, Klauda JB, Im W. Biophysical Journal. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Journal of Physical Chemistry B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- 35.Martyna GJ, Tobias DJ, Klein ML. Journal of Chemical Physics. 1994;101:4177–4189. [Google Scholar]
- 36.Feller SE, Zhang YH, Pastor RW, Brooks BR. Journal of Chemical Physics. 1995;103:4613–4621. [Google Scholar]
- 37.Hess B, Kutzner C, van der Spoel D, Lindahl E. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 38.Lindahl E, Hess B, van der Spoel D. Journal of Molecular Modeling. 2001;7:306–317. [Google Scholar]
- 39.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 40.Berendsen HJC, Vanderspoel D, Vandrunen R. Computer Physics Communications. 1995;91:43–56. [Google Scholar]
- 41.Chiu SW, Pandit SA, Scott HL, Jakobsson E. J Phys Chem B. 2009 doi: 10.1021/jp807056c. [DOI] [PubMed] [Google Scholar]
- 42.Pandit SA, Chiu SW, Jakobsson E, Grama A, Scott HL. Langmuir. 2008;24:6858–6865. doi: 10.1021/la8004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. Journal of Computational Chemistry. 1997;18:1463–1472. [Google Scholar]
- 44.Nose S, Klein ML. Journal of Chemical Physics. 1983;78:6928–6939. [Google Scholar]
- 45.Parrinello M, Rahman A. Journal of Applied Physics. 1981;52:7182–7190. [Google Scholar]
- 46.Evans DJ, Holian BL. Journal of Chemical Physics. 1985;83:4069–4074. [Google Scholar]
- 47.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. Journal of Computational Chemistry. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kessel A, Ben-Tal N, May S. Biophysical Journal. 2001;81:643–658. doi: 10.1016/S0006-3495(01)75729-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helfrich W. Z Naturforsch C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 50.Olsen BN, Schlesinger PH, Baker NA. Journal of the American Chemical Society. 2009;131:4854–4865. doi: 10.1021/ja8095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brandt EG, Braun AR, Sachs JN, Nagle JF, Edholm O. Biophysical Journal. 2011;100:8. doi: 10.1016/j.bpj.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hofsass C, Lindahl E, Edholm O. Biophys J. 2003;84:2192–2206. doi: 10.1016/S0006-3495(03)75025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindahl E, Edholm O. Biophys J. 2000;79:426–433. doi: 10.1016/S0006-3495(00)76304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiu SW, Vasudevan S, Jakobsson E, Mashl RJ, Scott HL. Biophys J. 2003;85:3624–3635. doi: 10.1016/S0006-3495(03)74780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Biophys J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waheed Q, Edholm O. Biophys J. 2009;97:2754–2760. doi: 10.1016/j.bpj.2009.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beutler TC, Mark AE, Vanschaik RC, Gerber PR, Vangunsteren WF. Chemical Physics Letters. 1994;222:529–539. [Google Scholar]
- 58.Zacharias M, Straatsma TP, Mccammon JA. Journal of Chemical Physics. 1994;100:9025–9031. [Google Scholar]
- 59.Hung WC, Lee MT, Chen FY, Huang HW. Biophysical Journal. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Meyer F, Smit B. Proc Natl Acad Sci U S A. 2009;106:3654–3658. doi: 10.1073/pnas.0809959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klauda JB, Kucerka N, Brooks BR, Pastor RW, Nagle JF. Biophysical Journal. 2006;90:2796–2807. doi: 10.1529/biophysj.105.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kucerka N, Perlmutter JD, Pan J, Tristram-Nagle S, Katsaras J, Sachs JN. Biophysical Journal. 2008;95:2792–2805. doi: 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wassall SR, McCabe MA, Wassall CD, Adlof RO, Feller SE. J Phys Chem B. 114:11474–11483. doi: 10.1021/jp105068g. [DOI] [PubMed] [Google Scholar]
- 64.Aittoniemi J, Niemela PS, Hyvonen MT, Karttunen M, Vattulainen I. Biophys J. 2007;92:1125–1137. doi: 10.1529/biophysj.106.088427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smondyrev AM, Berkowitz ML. Biophysical Journal. 2001;80:1649–1658. doi: 10.1016/S0006-3495(01)76137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samuli Ollila OH, Rog T, Karttunen M, Vattulainen I. J Struct Biol. 2007;159:311–323. doi: 10.1016/j.jsb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Alwarawrah M, Dai J, Huang J. J Phys Chem B. 114:7516–7523. doi: 10.1021/jp101415g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pasenkiewicz-Gierula M, Rog T, Kitamura K, Kusumi A. Biophysical Journal. 2000;78:1376–1389. doi: 10.1016/S0006-3495(00)76691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Czub J, Baginski M. Biophysical Journal. 2006;90:2368–2382. doi: 10.1529/biophysj.105.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandit SA, Bostick D, Berkowitz ML. Biophys J. 2004;86:1345–1356. doi: 10.1016/S0006-3495(04)74206-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rog T, Pasenkiewicz-Gierula M. Biophysical Journal. 2001;81:2190–2202. doi: 10.1016/S0006-3495(01)75867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lipari G, Szabo A. Biophys J. 1980;30:489–506. doi: 10.1016/S0006-3495(80)85109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Gennes PG, Prost J. The physics of liquid crystals. Clarendon Press; Oxford: 1993. [Google Scholar]
- 74.Kramer D, Benshaul A, Chen ZY, Gelbart WM. Journal of Chemical Physics. 1992;96:2236–2252. [Google Scholar]
- 75.de Joannis J, Coppock PS, Yin F, Mori M, Zamorano A, Kindt JT. J Am Chem Soc. 133:3625–3634. doi: 10.1021/ja110425s. [DOI] [PubMed] [Google Scholar]
- 76.Pan J, Tristram-Nagle S, Nagle JF. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.