Abstract
Migrating birds fly thousand miles and more, often without visual cues and in treacherous winds, yet keep direction. They employ for this purpose, apparently, as a powerful navigational tool the photoreceptor protein cryptochrome to sense the geomagnetic field. The unique biological function of cryptochrome supposedly arises from a photoactivation reaction involving radical pair formation through electron transfer. Radical pairs, indeed, can act as a magnetic compass; however, the cryptochrome photo-reaction pathway is not fully resolved yet. To reveal this pathway and underlying photochemical mechanisms we carried out a combination of quantum chemical calculations and molecular dynamics simulations on plant (Arabidopsis thaliana) cryptochrome. The results demonstrate that after photoexcitation a radical pair forms becomes stabilized through proton transfer, and decays back to the protein’s resting state on timescales allowing the protein, in principle, to act as a radical pair-based magnetic sensor. We briefly relate our findings on Arabidopsis thaliana cryptochrome to photo-reaction pathways in animal cryptochromes.
Introduction
Cryptochromes are flavoprotein photoreceptors originally identified in the plant Arabidopsis thaliana,1 where they play a key role in growth and development.2,3 Subsequently discovered in prokaryotes, archaea, and eukaryotes,3 cryptochromes were shown to be involved in circadian rhythms2,4,5 and are proposed to function as light-dependent magnetoreceptors in insects and migratory birds apparently leading to visual perception of the magnetic field6–15. The unique biological role of cryptochromes in insect and animal magnetosensing arises due to photoactivation of a flavin-pigment bound by the protein: exposure to blue light results in a transient one-electron reduction of flavin, which leads to the formation of a spin-entangled pair of radicals (molecules with a single unpaired electron), the so-called radical pair. Radical pair reactions are well known to exhibit a sensitivity to weak magnetic fields.16–19
Indeed, evidence has been provided that cryptochrome enables the fruit fly, Drosophila mela-nogaster, to sense magnetic fields.20,21 According to a widely accepted theory fields as weak as the geomagnetic field (0.5 Gauss strength) affects the entanglement of electron spins in a radical pair photoreaction; the entanglement, in turn, affects the cryptochrome signaling state lifetime.11–13 The radical pair reaction furnishes a mechanism by which the geomagnetic field can be sensed by insects. During the last decade, the hypothesis16,17 that the radical pair mechanism plays a role in animal navigation has been further elaborated theoretically11–14,22 and experimentally.18,19,23
Cryprochromes in plants control growth and development of seedlings3,24 with the signaling state being governed by a semireduced flavin cofactor. A report on cryptochrome mediated magnetic field effects on plant growth24 had been contested.25 The formation of the signaling state in plant cryptochromes involves photoinduced electron transfer to flavin from a tryptophan triad (W400, W377, W234) that bridges the space between flavin and the protein surface26–28 as depicted in Fig. 1a. The triad is conserved in the primary sequence of different cryptochromes and is also found, together with the flavin cofactor, in a related family of light-activated DNA repair enzymes, the photolyases. The exact steps of the tryptophan triad→flavin electron transfer are still debated, in particular, in regard to the interpretation of spectroscopic observations.22
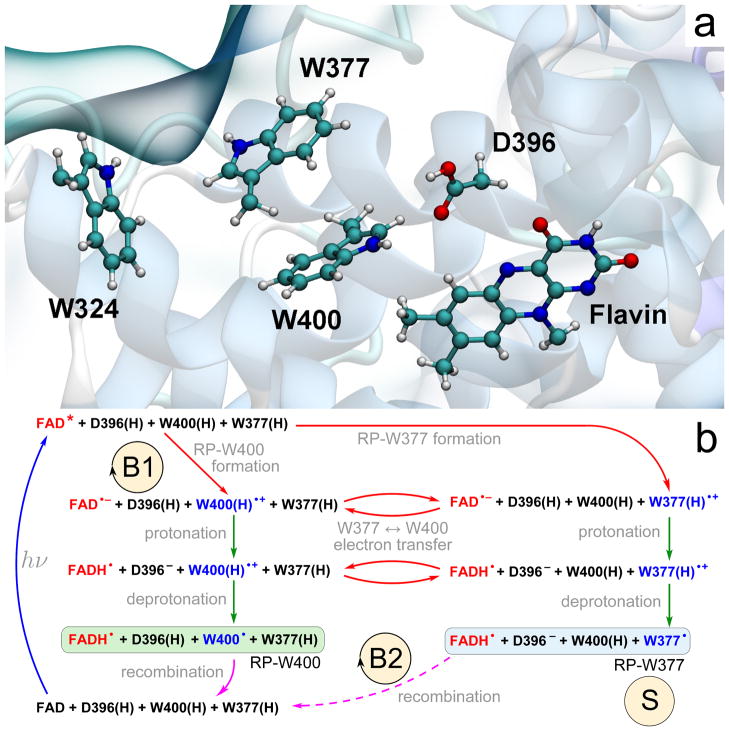
Figure 1. Electron and proton-transfer reactions in cryptochrome.
(a) Flavin cofactor, the tryptophan triad W400, W377, W324 and the D396 residue forming the active site of crypto-chrome-1 from Arabidopsis thaliana.34 (b) Schematic representation of the photoactivation reaction. Cryptochrome photoactivation is triggered by blue-light photoexcitation of the FAD cofactor (blue arrow) initially present in the oxidized state. Excited flavin, FAD*, receives an electron from one of the nearby tryptophans (red arrows); either W400(H) (RP-W400) or W377(H) (RP-W377). Electron transfer from tryptophan leads to formation of an ionic FAD•− + W(H)•+ radical pair, which is then transformed into a stable neutral FADH• + W• radical pair state through proton exchange with the nearby D396 (two green arrows). RP-W400 and RP-W377 interconvert through a W400(H)↔W377(H) electron transfer process (red arrows). In contrast to RP-W377, the neutral radical pair RP-W400 recombines back to the initial state through coupled electron-proton transfer (solid purple arrow). The RP-W377 state is stabilized through W377(H)•+ deprotonation into solution and, therefore, returns to the initial state only on a very long time scale (dashed purple arrow). The reaction cycle B1 and the reaction B2 are primary candidates for establishing magne-toreception;12,13 the state labeled S is a primary candidate for being the signaling state (for details see text).
In recent years, quantum chemistry and molecular dynamics (MD) studies provided crucial contributions towards identification of molecular mechanisms underlying flavin-based photochemistry. MD simulations allow detailed characterization of protein dynamics,29 while quantum chemistry calculations describe chemical transformations in the protein,30,31 i.e., of the processes experimentally observed through transient absorption spectroscopy methods.26,32,33
The present theoretical analysis focuses on plant cryptochrome-1 from Arabidopsis thaliana. Plant cryptochrome is particularly attractive for such analysis, since its atomic level structure is known34 and since electron transfer kinetics data derived from transient absorption spectra are available.28,33,35–37 Using first principles (so-called ab initio) quantum chemistry as well as classical all-atom MD simulations we investigate plant cryptochrome photoactivation, answering how radical pairs form, become stabilized, and decay. Our approach lays the foundation for future studies of magnetoreceptive properties of cryptochrome, as it accounts for the intermediate states in the protein, the transitions between which, in principle, can be magnetic field sensitive.
Methods
Calculations were performed using a variety of theoretical methods, described in Supplementary Information (SI). Quantum-chemical calculations involving cryptochrome active site models were carried out using the CASSCF and XMCQDPT238 methods available in the Firefly39 program, which is partially based on the GAMESS (US)40 source code. The MS-CASPT241 method was employed for the QM/MM calculations with the AMBER94 force field,42 using the TINKER43 and MOLCAS44 programs. Molecular dynamics simulations were performed using NAMD 2.845 with the CHARMM22 force field.46,47 All images and a video were rendered with VMD.48
Electron transfer from tryptophan W400
A model of the cryptochrome active site, shown in Fig. S1a of SI, comprising flavin, D396, W400 and several amino acid residues (R362, D390 and S251) surrounding the flavin, was used to study flavin photoexcitation and RP-W400 state formation. For this purpose, energies and optimized geometries of the model system were calculated in the ground and excited electronic states using CASSCF and perturbation theory-based XMC-QDPT238 methods (see SI for detail). The total energies of the model system calculated using the XMCQDPT2 method are summarized in Tab. S2 of SI, while the energy diagram in Fig. 2a shows the corresponding relative energies; the results of the CASSCF calculations are summarized in Tab. S1 and Fig. S3 in SI.
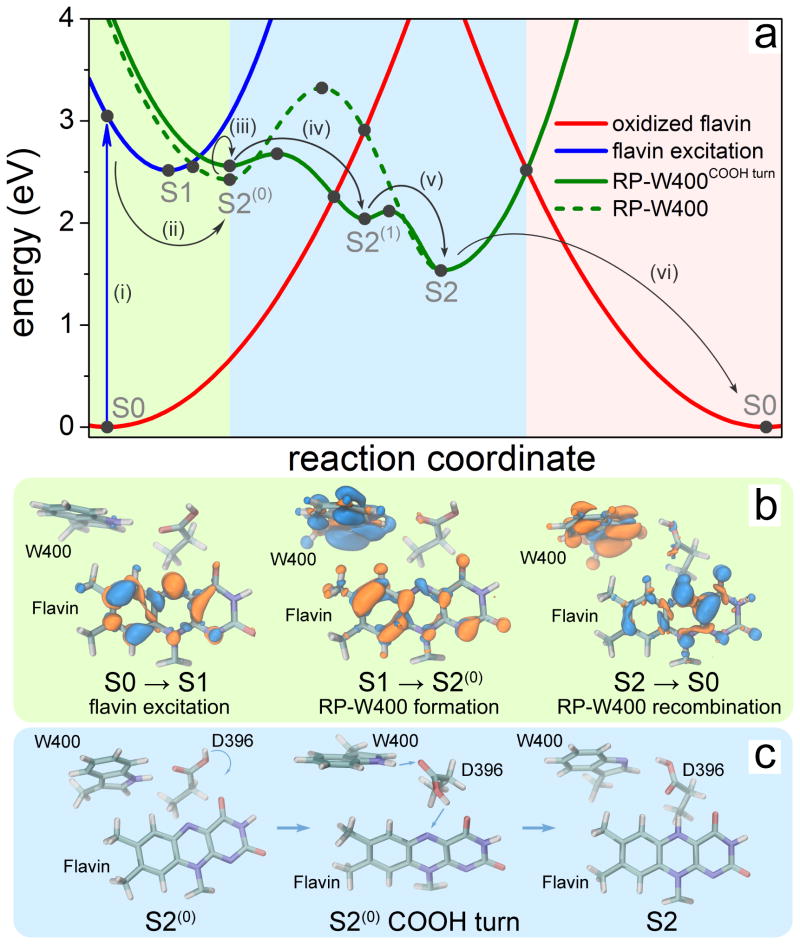
Figure 2. Characterization of the cryptochrome photoreaction involving flavin, W400 and D396.
(a) Calculated potential energy profiles of the key electronic states describing cryptochrome photoactivation. The energy of oxidized flavin is shown in red, of excited flavin in blue, and of the radical pair state RP-W400 in green. Filled circles represent computed energies while lines show schematic potential energy surfaces. The colored background distinguishes electron transfer step (light green), proton transfer steps (light blue) and coupled electron-proton transfer step (pink). Reaction steps (i-vi) refer to explanations in the text. (b) Change of electron density due to flavin photoexcitation (S0→S1), photoinduced radical pair formation (S1→S2(0)) and recombination (S2→S0). The initial distribution of electron density is shown in blue, while the final distribution is shown in orange. (c) Rearrangement of the COOH- group of the D396(H) residue catalyzing the protonation of flavin by the W400(H)•+ radical through formation of a D396− intermediate (minimum S2(1) in (a)).
Electron transfer from tryptophan W377
In order to describe involvement of W377 and formation of the RP-W377 state, a model system of the cryptochrome active site consisting of flavin, D396, W377 and W400 was described quantum-chemically. Six electronic states were included in the calculation. The energies of these states were determined using the XMCQDPT2 method and are summarized in Tab. S4 in SI; the results of the CASSCF calculations are summarized in Tab. S3 and Fig. S5 in SI. The active site model containing W377 does not include side chains of amino acids surrounding the flavin. Thus, the excitation energies computed for this model are slightly different from the energies shown in Fig. 2a; the differences are not relevant for the conclusions of the paper.
Stabilization of the W377 radical
To examine W377-solvent interaction, the cryptochrome active site model was extended by adding three water molecules which form hydrogen bonds with each other and with W377 on the cryptochrome surface as illustrated in Fig. 4b (see also Fig. S1c in SI). The energy of the relevant states, calculated at the S3(1) minimum, are shown in Fig. 4a, labeled 3H2O, and are listed in Tab. S3 (CASSCF calculation) and Tab. S4 (XMCQDPT2 calculation) in SI, respectively.
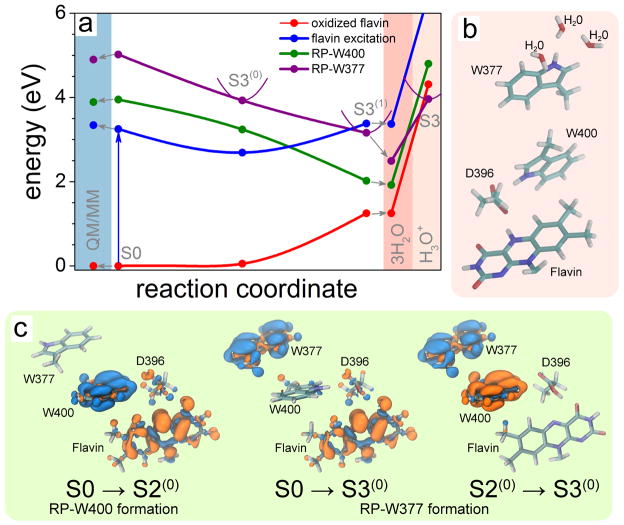
Figure 4. Electron transfer through tryptophan diad.
(a) Calculated potential energy pro-files for the flavin oxidized state (red), flavin excited state (blue), radical pair state RP-W400 (green) and radical pair state RP-W377 (purple). Filled circles represent the computed energies while lines show a schematic profile of the potential energy surfaces. The colored background highlights the results of QM/MM calculations (light blue), and calculations that account for the presence of three water molecules around W377 (pink). (b) Active site model containing three water molecules in the W377 vicinity. (c) Change of electron density due to electron transfer from W400(H) to flavin (S0→S2(0)), from W377(H) to flavin (S0→S3(0)) and from W377(H) to W400(H)•+ (S2(0) →S3(0)). The initial distribution of electron density is shown in blue; the final electron density is shown in orange.
Protein environment and flavin optical spectrum
A QM/MM calculation was employed to study the effect of the protein environment on the electronic excitation energies of cryptochrome. The QM system included the lumiflavin moiety of the FAD chromophore as well as the side chains of residues W377, D396 and W400 (see Fig. S2); the remaining atoms of the moieties were included in the MM region. The absolute energies obtained in the QM/MM calculation using the CASPT2 method are summarized in Tab. S5 in SI.
Results
Electron and proton transfers in cryptochrome
The active site of plant cryptochrome containing the flavin moiety of the flavin adenine dinucleotide (FAD) chromophore, the tryptophan triad (W400, W377, W324) and an aspartic acid in close proximity to the flavin is shown in Fig. 1a. The tryptophan triad is a conduit for electron transfer, triggered by flavin photoexcitation, permitting the three electron transfer steps W400→FAD*, W377→W400 and W324→W377.3 W377 and W324, being both surface exposed, can stabilize their oxidized state through proton release into solvent. For the sake of simplicity the present study considers only W377 in this role and describes only the two tryptophans, W400 and W377, as being engaged jointly with flavin and D396 in electron and proton transfer.
Figure 1b illustrates schematically the photoactivation reaction studied, which consists of a series of electron and proton transfers. After photon absorption, the excited FAD* receives an electron from the nearby W400, leading to formation of the FAD•− + W400(H)•+ radical pair (RP-W400 state). Alternatively, W377 donates an electron to the excited flavin, leading to formation of the FAD•− + W377(H)•+ radical pair (RP-W377 state). The interconversion of RP-W400 into RP-W377 has been widely discussed in previous studies.3,26,27
In plant cryptochrome, a protonated aspartic acid D396(H) is neighboring the flavin moiety of FAD and acts as a proton donor to the flavin FAD•− radical.33,37 Subsequently, the formed D396− anion attracts a proton from the cation W400(H)•+ radical, leading to formation of a neutral FADH• + W400• radical pair. This step is specific for RP-W400 and is not possible for the RP-W377 state since D396(H) is not in contact with W377. The RP-W377 state, in turn, is stabilized through tryptophan deprotonation into the solvent. Radical recombination, i.e., electron back-transfer flavin→W400, should occur readily for the RP-W400 state, but not for the neutral RP-W377 state, where release and diffusion of a proton into the bulk solvent makes the recombination reaction unfavorable.13
The reaction cycle denoted B1 in Fig. 1b, involving the steps FAD + W400(H)→ FAD* + W400(H) → FAD•− + W400(H)•+ →FADH• + W400• → FAD + W400(H), is a primary candidate for establishing magnetoreception, controlling the population of the signaling state represented by the FADH• + W377• denoted as state S in Fig. 1b.12 An alternative, or additional, magnetosensitive reaction could control the decay of the signalling state FADH• + W377• back to FAD + W377(H) through FADH• + W377• → FAD + W377(H), denoted as cycle B2 in Fig. 1b, the so-called dark reaction.13
Electron transfer from tryptophan W400
The electronic states of the active site with the lowest energies are shown in Fig. 2a. Flavin photoexcitation (step (i) in Fig. 2a) is a single-electron transition from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO) with a calculated vertical energy of 3.05 eV, which is in a good agreement with the measured FAD→FAD• excitation energy of 2.8 eV.2,28,33 The change of the total electron density due to flavin excitation is shown in the left panel of Fig. 2b. For the initial (S0) structure of the cryptochrome active site the energy of the radical pair RP-W400 state is higher than the energy of the flavin photoexcitation. Here the radical pair is described by a single-electron transition from the HOMO of W400 to the LUMO of flavin.
Relaxation of the active site after flavin photoexcitation results in a structural rearrangement, leading to the energy minimum S1 on the potential energy surface of the flavin excited state. From this minimum the the FAD•− + W400(H)•+ radical pair is formed readily and corresponds to the energy minimum S2(0) in Fig. 2a. Our calculation yields a very small energy of 0.03 eV for the S1→S2(0) transition barrier such that the FAD•− + W400(H)•+ radical pair should be formed on an extremely fast time scale (step (ii) in Fig. 2a), in agreement with experimental observations35 that do not discern any significant fluorescence after cryptochrome excitation. The change of the total electron density due to the photo-induced electron transfer process is shown in the middle panel of Fig. 2b, which presents the formation of the charge-separated state in the cryptochrome active site.
Structural rearrangement in cryptochrome
Structural rearrangement subsequent to FAD•− + W400(H)•+ radical pair formation was described through MD simulations. The formation of this radical pair was seen to trigger within cryptochrome a slight, but crucial reorientation of the D396(H) residue as illustrated in Fig. 2c and Fig. 3. Prior to radical pair formation, the COOH-group of D396(H) forms a hydrogen bond with the neutral W400(H) residue with a bond length, d(HW400-OD396), fluctuating around 3 Å as seen in Fig. 3a; the O-HD396 group of D396(H) forms a hydrogen bond with the backbone oxygen atom of the M381 residue (data not shown) as also observed in the cryptochrome crystal structure.34 We note that a hydrogen bond between the hydrogen atom of the COOH- group and the N5 atom of the flavin group is not possible as is evident from the d(HD396-N5flavin) distance fluctuating around 6 Å as shown in the bottom plot of Fig. 3a, due to steric constraints in the cryptochrome active site.
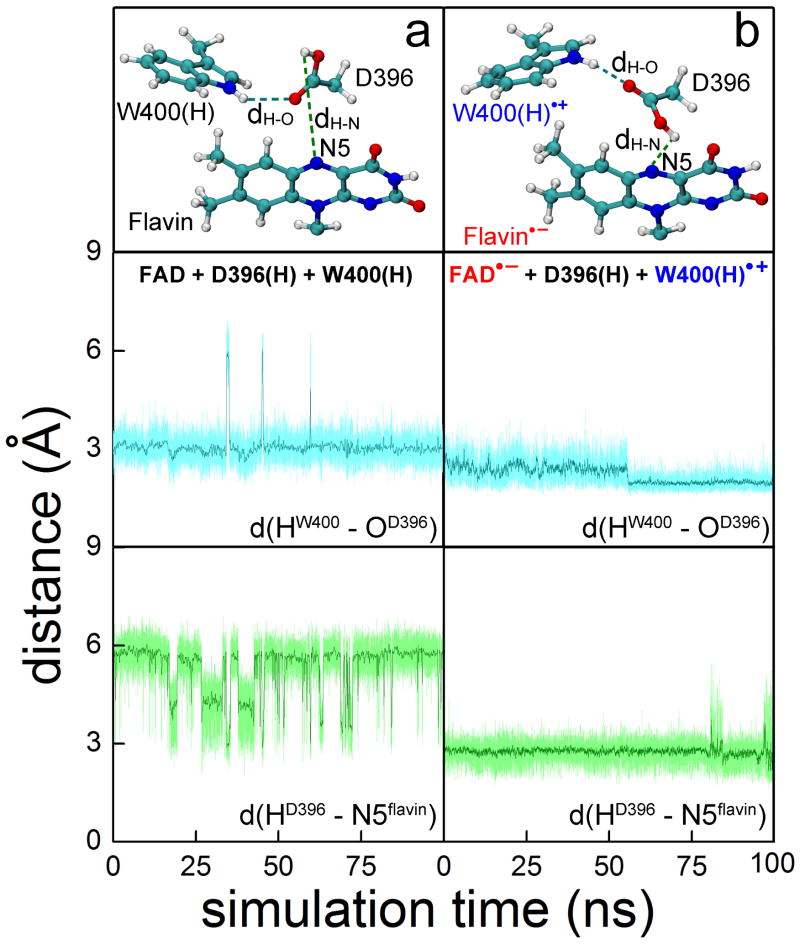
Figure 3. Rearrangement of D396(H) in the cryptochrome active site.
(Top) Relative orientations of flavin, D396(H) and W400(H) residues obtained through all-atom MD simulations of Arabidopsis thaliana cryptochrome in water for (a) cryptochrome with oxidized flavin, i.e., FAD + W400(H), and (b) cryptochrome in the radical pair state FAD•− + W400(H)•+. (Middle, Bottom) Time dependence of the hydrogen bond lengths d(HW400-OD396), labeled dH−O at top, and d(HD396-N5flavin), labeled dH−O at top, calculated for the two different redox states shown at top.
Once flavin gains a negative charge and W400 becomes positively charged, i.e., once the FAD•− + W400(H)•+ radical pair is formed, D396(H) is seen in a second MD simulation to become involved in a remarkable structural transformation. Figure 3b shows that in the radical-pair state two stable hydrogen bonds connecting flavin FAD•−, D396(H) and W400(H)•+ exist, resulting within a nanosecond in the d(HW400-OD396) and d(HD396-N5flavin) distances assuming average values of 2.0 Å and 2.7 Å, respectively. This rearrangement of D396(H) permits FAD•− protonation and W400(H)•+ deprotonation. A similar rearrangement of D396(H) is seen in a MD simulation for negatively-charged FAD•− and neutral W400(H) as shown in Fig. S4 in SI.
Proton transfer and stabilization of the radical pair
The D396 rotation, shown in Fig. 2c, initiates W400(H)•+ →FAD•− proton transfer between aspartic acid and flavin. Alternatively, proton transfer could be initiated by W400(H)•+, leading to a D396(H2)+ intermediate. However, as Fig. 2a shows, the transition energy barrier (dashed green line) would be 0.9 eV in this case, suggesting a fairly long-lived FAD•− + W400(H)•+ radical pair state in cryptochrome.
The proton transfer initiated by D396(H) involves spontaneous rearrangement of the COOH group identified in the MD simulation that first increases the energy of the S2(0) minimum by 0.14 eV (step (iii) in Fig. 2a), but then through protonation of FAD•− decreases the energy, leading to the S2(1) minimum (step (iv)). This state, involving the neutral FADH• radical and the D396− anion, lowers its energy further through W400(H)•+ → D396− proton transfer (step (v)) leading to formation of the minimum S2 and involving radicals FADH• and W400• together with D396(H). The corresponding active site configuration is shown in the right panel of Fig. 2c. The two protonation steps (iv) and (v), involved in the process FAD•− + W400(H)•+ →FADH• + W400•, have energy barriers of about 0.1 eV, which are overcome readily.
A key result of our study is that the S2(1) and S2 minima are found on the ground potential energy surface of the system. Thus, upon flavin protonation, the ground state changes its electronic character from oxidized flavin to FADH•. Return to the FAD state in a S2→S0 back-reaction is possible through proton-coupled electron transfer transforming FADH• + W400• to the initial FAD + W400(H) state. The S2 minimum lies energetically about 1.5 eV above the S0 minimum; the two minima are separated by a 0.72 eV energy barrier as shown in Fig. 2a (step (vi)). We emphasize that the back-reaction (vi) is an important aspect of cryptochrome’s photoactivation process, as it actually prevents the protein from signaling, i.e., reduces the signaling level.
A movie in SI illustrates the formation of the neutral RP-W400 state. The process consists of the five steps as indicated in Fig. 2a: (i) flavin photoexcitation; (ii) electron transfer from W400(H) to FAD*; (iii) rearrangement of the D396(H) side chain; and two consecutive proton transfers from D396(H) to FAD•− (iv) and from W400(H)•+ to D396− (v).
Electron transfer from tryptophan W377
The S2→S0 back reaction prevents cryptochrome from reaching the millisecond-lived putative signaling state28,35 and denoted S in Fig. 1b. However, cryptochrome signaling can take place if the distance separation between the two radicals increases, namely through W377→W400 electron transfer; indeed, this transfer has often be postulated.3
Figure 4 shows the four electronic states that are relevant for the formation of the FADH• + W377• radical pair, namely oxidized flavin, flavin electronic excitation, RP-W400 and RP-W377. The RP-W377 state corresponds to a single-electron promotion from the HOMO of W377 to the LUMO of flavin. The change in electron density accompanying this promotion is shown in the middle panel of Fig. 4c. The S3(0) minimum in Fig. 4a corresponds to the FAD•− + W377(H)•+ radical pair, whereas the S3(1) minimum is associated with the neutral FADH• radical and anionic D396−. Thus, the S3(0) and S3(1) minima are analogous to the S2(0) and S2(1) minima for RP-W400 discussed above. The higher energy of the RP-W377 state, as compared to the RP-W400 state, is due to the wider separation of W377 and flavin.49 For W377→W400 electron transfer to appear feasible, factors not yet included in the present model need to be taken into account, in particular interactions with solvent near the protein surface. W377→W400 electron transfer is feasible as long as the potential energy surfaces of the RP-W400 and RP-W377 states cross and the energy minimum of RP-W377 becomes the electronic ground state. The condition can be met since the W377 radical is solvent exposed and, therefore, can transfer a proton to the solvent.
Stabilization of the W377 radical
To investigate the role of solvent in stabilizing the RP-W377 state we added three water molecules to our description as specified in Methods. The added water stabilized the RP-W377 state by 0.67 eV, as shown in Fig. 4a. Adding more water would likely lower the energy further, but an energy decrease far enough to render RP-W377 the new ground state would rather require deprotonation of W377(H)•+ into the solvent. We investigated, therefore, in the respective optimized geometry a deprotonated W377• radical and a protonated water trimer (H3O+ + 2×H2O). Figure 4a (right most part) shows that in this case the energy minimum S3 of RP-W377, indeed, becomes the ground state of the system, but only at a significantly higher energy than the analogous S2 minimum in Fig. 2a. Our results suggest that RP-W377 forms through electron transfer to a protonated W377 radical state coupled to deprotonation. An important difference in the active site geometry, characterized either through S2 or S3 minima, is the D396 residue, which is neutral for the S2 configuration, while it is anionic in the S3 case. The D396 residue becomes D396− for the S3 minimum, because the W377→W400 electron transfer is coupled to D396(H)→W400 proton transfer. Thus, the S3 minimum corresponds to the configuration in which W400− has become protonated through D396(H) and W377(H)•+ has released a proton.
Protein environment and flavin optical spectrum
The protein environment may influence the electronic excitation energies of cryptochrome. Therefore, a study of the environmental effect on cryptochrome photoactivation was performed using a QM/MM model in which flavin, D396, W400 and W377 side chains were treated quantum mechanically and the rest of the protein and the surrounding water were described with the AMBER94 force field (see Fig. S2 in SI). Figure 4a (left) presents the calculated excitation energies (denoted as QM/MM) obtained for the optimized geometry of cryptochrome. The result shows only insignificant influence of the protein environment on the excitation energies, validating the results of the quantum mechanical calculations of cryptochrome active site models described above.
Conclusion
The present study provides a detailed view of Arabidopsis thaliana cryptochrome photoactivation through quantum chemistry calculations and MD simulations. The results are clearly indicative of an ultrafast photoinduced radical pair formation. Remarkably, plant and animal cryptochromes differ greatly in regard to their activation from other flavin-containing photoreceptors, e.g., BLUF and LOV.30,50–52 Apparently, ultrafast formation of a long-lived radical pair is unique to the cryptochromes and should contribute critically to the proteins’ biological function. Magnetoreception, perhaps, is the most prominent function for which radical pair formation in cryptochrome plays a key role.11,12,15,26
Our study explains the observed35 ultrafast quenching of flavin fluorescence by photoinduced electron transfer from the neighboring W400(H) side group. Unexpected is the finding of a concomitant proton transfer reaction leading to formation of a neutral FADH• + W400• radical pair and involving the D396 residue that initiates the transfer through a two step process. This transfer exhibits a low activation barrier such that the anionic FAD•− state should be short-lived. In contrast, the neutral radical FADH• is fairly stable, as the calculations show that the back-reaction exhibits a large energy barrier (0.7 eV) seen in Fig. 2a. The S2→S0 back-reaction (vi), which would actually preclude cryptochrome activation, can be readily prevented by W377→W400 electron transfer. Our calculations suggest a scenario in which this electron transfer ultimately leads to proton transfer from W377 into the protein environment, trapping the FADH• state, namely, the putative signaling state of cryptochrome. Possibly, W377 deprotonation requires an external proton acceptor; W377→W400 electron transfer may be stabilized energetically also by an external redox agent or by amino acid side groups.
The suggested mechanism provides a basis for an interpretation of spectroscopic data recorded for plant cryptochromes. Recently the time constant for the initial photoinduced electron transfer from tryptophan to flavin had been reported to be less than 100 ns.33 A subsequent spectral change described by a 1.7 μs time constant was assigned to protonation of flavin by D396(H).33 Such slow protonation is surprising considering the close proximity of D396(H) to the anionic flavin. Our calculations suggest, instead, that flavin becomes protonated on the nanosecond time scale, shortly after radical pair formation, with a subsequent W377→W400 electron transfer, expected to be slow, occurring on the μs timescale. Thus, the experimentally observed 1.7 μs process may be attributed to deprotonation of D396(H) and protonation of W400•.
The signaling state of plant cryptochrome is associated with the FADH• state of the flavin cofactor. Time-resolved spectroscopy studies observed decay of this state over a millisecond.28,35 Calculations suggest that further chemical transformations involving the cryptochrome active site are needed and possible to stabilize FADH• for a millisecond. The key role is played, likely, by coupled W377→W400 electron transfer and D396 deprotonation.
Cryptochrome provides the pathway for radical pair formation and also the molecular environment shielding the radical pair that may act as a magnetoreceptor; the radical pair must retain to a significant degree spin entanglement, i.e., partners cannot be exchanged, and the radical pair reaction must proceed within about a microsecond, a time too short to involve other molecular partners, except ones that may form a tight complex with cryptochrome, but there is no evidence for such complex.
In animal cryptochromes, the aspartic acid D396 is replaced by a cysteine. This replacement changes the entire electron-transfer mechanism, because in the absence of a proton donor a stable FADH• + W400• radical pair cannot be formed and anionic FAD•− should be formed instead. FAD•− may serve the same signalling purpose as FADH• + D396− does in plant cryptochrome, i.e., may trigger a similar structural rearrangement once cryptochrome is activated. However, differing electron transfer dynamics in plant and animal cryptochromes can impact the magnetoreceptive responsiveness and sensitivity of cryptochromes.
Supplementary Material
Acknowledgments
The authors thank Johan Strümpfer and Vita Solovyeva for stimulating discussions. This work has been supported, in part, by National Science Foundation grants NSF MCB-0744057 and NSF PHY0822613 as well as by National Institutes of Health grant P41-RR005969. K.S. thanks the Alexander von Humboldt Foundation for support and I.S. acknowledges support as a Beckman Fellow. T.D. thanks the MPG Minerva-program.
Footnotes
Supplementary Information Available
In Supplementary Information (SI) we present details of theoretical methods used for the calculations. Additionally, Supplementary Tables 1–6, Supplementary Figures 1–5, Supplementary Files and Supplementary Movie are also available. This information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Ahmad M, Cashmore AR. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 2.Cashmore AR, Jarillo JA, Wu YJ, Liu D. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 3.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 4.Partch CL, Sancar A. Meth Enzym. 2005;393:726–745. doi: 10.1016/S0076-6879(05)93038-3. [DOI] [PubMed] [Google Scholar]
- 5.Yoshii T, Ahmad M, Helfrich-Foerster C. PLoS Biology. 2009;7:813–819. doi: 10.1371/journal.pbio.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zapka M, Heyers D, Hein CM, Engels S, Schneider NL, Hans J, Weiler S, Dreyer D, Kishkinev D, Wild JM, Mouritsen H. Nature. 2009;461:1274–1277. doi: 10.1038/nature08528. [DOI] [PubMed] [Google Scholar]
- 7.Mouritsen H, Janssen-Bienhold U, Liedvogel M, Feenders G, Stalleicken J, Dirks P, Weiler R. Proc Natl Acad Sci USA. 2004;101:14294–14299. doi: 10.1073/pnas.0405968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liedvogel M, Feenders G, Wada K, Troje N, Jarvis E, Mouritsen H. Eur J Neurosci. 2007;25:1166–1173. doi: 10.1111/j.1460-9568.2007.05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyers D, Manns M, Luksch H, Güntürkün O, Mouritsen H. PLoS ONE. 2007;2:e937. doi: 10.1371/journal.pone.0000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liedvogel M, Mouritsen H. J R Soc Interface. 2010;7:S147–S162. doi: 10.1098/rsif.2009.0411.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritz T, Adem S, Schulten K. Biophys J. 2000;78:707–718. doi: 10.1016/S0006-3495(00)76629-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solov’yov IA, Chandler D, Schulten K. Biophys J. 2007;92:2711–2726. doi: 10.1529/biophysj.106.097139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solov’yov IA, Schulten K. Biophys J. 2009;96:4804–4813. doi: 10.1016/j.bpj.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solov’yov IA, Mouritsen H, Schulten K. Biophys J. 2010;99:40–49. doi: 10.1016/j.bpj.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liedvogel M, Maeda K, Henbest K, Schleicher E, Simon T, Timmel CR, Hore P, Mouritsen H. PLoS ONE. 2007;2:e1106. doi: 10.1371/journal.pone.0001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulten K, Swenberg CE, Weller A. Z Phys Chem. 1978;NF111:1–5. [Google Scholar]
- 17.Schulten K, Staerk H, Weller A, Werner HJ, Nickel B. Z Phys Chem. 1976;NF101:371–390. [Google Scholar]
- 18.O’Dea AR, Curtis AF, Green NJB, Timmel CR, Hore P. J Phys Chem A. 2005;109:869–973. doi: 10.1021/jp0456943. [DOI] [PubMed] [Google Scholar]
- 19.Maeda K, Henbest KB, Cintolesi F, Kuprov I, Rodgers CT, Liddell PA, Gust D, Timmel CR, Hore PJ. Nature. 2008;453:387–390. doi: 10.1038/nature06834. [DOI] [PubMed] [Google Scholar]
- 20.Foley LE, Gegear RJ, Reppert SM. Nat Commun. 2011;2:168. doi: 10.1038/ncomms1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gegear RJ, Foley LE, Casselman A, Reppert SM. Nature. 2010;463:804–807. doi: 10.1038/nature08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solov’yov IA, Schulten K. J Phys Chem B. 2012;116:1089–1099. doi: 10.1021/jp209508y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodgers CT, Hore PJ. Proc Natl Acad Sci USA. 2009;106:353–360. doi: 10.1073/pnas.0711968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad M, Galland P, Ritz T, Wiltschko R, Wiltschko W. Planta. 2007;225:615–624. doi: 10.1007/s00425-006-0383-0. [DOI] [PubMed] [Google Scholar]
- 25.Harris SR, Henbest KB, Maeda K, Pannell JR, Timmel CR, Hore PJ, Okamoto H. J R Soc Interface. 2009:1193–1205. doi: 10.1098/rsif.2008.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda K, Robinson AJ, Henbest KB, Hogben HJ, Biskup T, Ahmad M, Schle-icher E, Weber S, Timmel CR, Hore PJ. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1118959109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biskup T, Schleicher E, Okafuji A, Link G, Hitomi K, Getzoff ED, Weber S. Angew Chem Int Ed Engl. 2009;48:404–407. doi: 10.1002/anie.200803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giovani B, Byrdin M, Ahmad M, Brettel K. Nat Struct Biol. 2003;10:489–490. doi: 10.1038/nsb933. [DOI] [PubMed] [Google Scholar]
- 29.Song SH, Freddolino PL, Nash AI, Carroll EC, Schulten K, Gardner KH, Larsen DS. Biochem. 2011;50:2411–2423. doi: 10.1021/bi200198x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udvarhelyi A, Domratcheva T. Photochem Photobiol. 2011;87:554–563. doi: 10.1111/j.1751-1097.2010.00884.x. [DOI] [PubMed] [Google Scholar]
- 31.Domratcheva T. J Am Chem Soc. 2011;133:18172–18182. doi: 10.1021/ja203964d. [DOI] [PubMed] [Google Scholar]
- 32.Henbest KB, Maeda K, Hore PJ, Joshi M, Bacher A, Bittl R, Weber S, Tim-mel CR, Schleicher E. Proc Natl Acad Sci USA. 2008;105:14395–14399. doi: 10.1073/pnas.0803620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langenbacher T, Immeln D, Dick B, Kottke T. J Am Chem Soc. 2009;131:14274–14280. doi: 10.1021/ja901628y. [DOI] [PubMed] [Google Scholar]
- 34.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisen-hofer J. Proc Natl Acad Sci USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeugner A, Byrdin M, Bouly JP, Bakrim N, Giovani B, Brettel K, Ahmad M. J Biol Chem. 2005;280:19437–19440. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 36.Immeln D, Pokorny R, Herman E, Moldt J, Batschauer A, Kottke T. J Phys Chem B. 2010;114:17155–17161. doi: 10.1021/jp1076388. [DOI] [PubMed] [Google Scholar]
- 37.Kottke T, Batschauer A, Ahmad M, Heberle J. Biochem. 2006;45:2472–2479. doi: 10.1021/bi051964b. [DOI] [PubMed] [Google Scholar]
- 38.Granovsky AA. J Chem Phys. 2011;134(1–14):214113. doi: 10.1063/1.3596699. [DOI] [PubMed] [Google Scholar]
- 39.Granovsky AA. Firefly version 7.1.G. http://classic.chem.msu.su/gran/firefly/index.html.
- 40.Schmidt M, Baldridge K, Boatz J, Elbert S, Gordon M, Jensen J, Koseki S, Mat-sunaga N, Nguyen K, Su S, Windus T, Dupuis M, Montgomery J. J Comp Chem. 1993;14:1347–1363. [Google Scholar]
- 41.Finley J, Malmqvist P-Å, Roos BO, Serrano-Andrés L. Chem Phys Lett. 1998;288:299–306. [Google Scholar]
- 42.Cornell W, Cieplak P, Bayly C, Gould I, Merz K, Ferguson D, Spellmeyer D, Fox T, Caldwell J, Kollman P. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 43.Ponder JW. TINKER, Software Tools for Molecular Design, version 4.2. Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine; St. Louis, MO: 2004. The most updated version for the TINKER program can be obtained from the Internet at http://dasher.wustl.edu/tinker. [Google Scholar]
- 44.Aquilante F, De Vico L, Ferré N, Ghigo G, Malmqvist P, Neogrády P, Pedersen TB, Pitoňák M, Reiher M, Roos BO, Serrano-Andrés L, Urban M, Veryazov V, Lindh R. J Comp Chem. 2010;31:224–247. doi: 10.1002/jcc.21318. [DOI] [PubMed] [Google Scholar]
- 45.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacKerell AD, Jr, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 47.MacKerell AD, Jr, Feig M, Brooks CL., III J Comp Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 48.Humphrey W, Dalke A, Schulten K. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 49.Izmaylov AF, Tully JC, Frisch MJ. J Phys Chem A. 2009;113:12276–12284. doi: 10.1021/jp900357f. [DOI] [PubMed] [Google Scholar]
- 50.Dittrich M, Freddolino PL, Schulten K. J Phys Chem B. 2005;109:13006–13013. doi: 10.1021/jp050943o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salzmann S, Silva-Junior MR, Thiel W, Marian CM. J Phys Chem B. 2009;113:15610–15618. doi: 10.1021/jp905599k. [DOI] [PubMed] [Google Scholar]
- 52.Domratcheva T, Fedorov R, Schlichting I. J Chem Theory Comput. 2006;2:1565–1574. doi: 10.1021/ct0600114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.