Abstract
Barley (Hordeum vulgare L.) leaves were used to isolate and characterize the chloroplast NAD(P)H dehydrogenase complex. The stroma fraction and the thylakoid fraction solubilized with sodium deoxycholate were analyzed by native polyacrylamide gel electrophoresis, and the enzymes detected with NADH and nitroblue tetrazolium were electroeluted. The enzymes electroeluted from band S from the stroma fraction and from bands T1 (ET1) and T2 from the thylakoid fraction solubilized with sodium deoxycholate had ferredoxin-NADP oxidoreductase (FNR; EC 1.18.1.2) and NAD(P)H-FeCN oxidoreductase (NAD[P]H-FeCNR) activities. Their NADPH-FeCNR activities were inhibited by 2′-monophosphoadenosine-5′-diphosphoribose and by enzyme incubation with p-chloromercuriphenylsulfonic acid (p-CMPS), NADPH, and p-CMPS plus NADPH. They presented Michaelis constant NADPH values that were similar to those of FNRs from several sources. Their NADH-FeCNR activities, however, were not inhibited by 2′-monophosphoadenosine-5′-diphosphoribose but were weakly inhibited by enzyme incubation with NADH, p-CMPS, and p-CMPS plus NADH. We found that only ET1 contained two polypeptides of 29 and 35 kD, which reacted with the antibodies raised against the mitochondrial complex I TYKY subunit and the chloroplast ndhA gene product, respectively. However, all three enzymes contained two polypeptides of 35 and 53 kD, which reacted with the antibodies raised against barley FNR and the NADH-binding 51-kD polypeptide of the mitochondrial complex I, respectively. The results suggest that ET1 is the FNR-containing thylakoidal NAD(P)H dehydrogenase complex.
The plastid genomes of higher plants contain 11 reading frames (ndhA–K), which show significant homologies with genes encoding subunits of the mitochondrial NADH-ubiquinone oxidoreductase (complex I; Sugiura, 1992; Berger et al., 1993b). Seven chloroplast genes are homologous to mitochondrial genes coding subunits of complex I, and the other four chloroplast genes are homologous to nuclear genes coding the subunits of the NADH dehydrogenase (Matsubayashi et al., 1987; Shimada and Sugiura, 1991). The ndh genes have been shown to be transcribed (Matsubayashi et al., 1987; Schantz and Bogorad, 1988; Steinmüller et al., 1989), and the polypeptide products of six of them, NdhA (Martin et al., 1996), NdhB and NdhJ (Guedeney et al., 1996), NdhH (Berger et al., 1993b), NdhI (Wu et al., 1989; Sazanov et al., 1996), and NdhK (Nixon et al., 1989; Sazanov et al., 1996), have been found in thylakoid membranes. Moreover, NdhH (Berger et al., 1993b) and NdhK (Nixon et al., 1989) are localized in stroma thylakoids.
It has been suggested that ndh genes encode subunits of a chloroplastic NAD(P)H dehydrogenase complex (Ohyama et al., 1988) that might be involved in chlororespiration, a chloroplastic respiratory process mainly studied in the unicellular green alga Chlamydomonas reinhardtii (Peltier et al., 1987; Bennoun, 1994). By using inhibitors such as rotenone, Godde and Trebst (1980) concluded that the thylakoid membranes of C. reinhardtii have a bound, respiratory-like NADH dehydrogenase that is specific for plastoquinone. This enzyme was partially purified and characterized (Godde, 1982). Until recently, direct proof for the thylakoid NADH dehydrogenase complex was lacking, but we have demonstrated the existence of an NADH dehydrogenase complex from barley (Hordeum vulgare L.) thylakoids (Cuello et al., 1995a). This has been separated by native PAGE and its polypeptidic composition analyzed (Quiles et al., 1996). Additionally, Sazanov et al. (1996) have detected and partially characterized an NADH dehydrogenase complex from pea thylakoids.
Recent evidence from leaves of potato has suggested an association of ndhB and ndhJ gene products with FNR as components of a chloroplastic NAD(P)H dehydrogenase complex (Guedeney et al., 1996). The polypeptide of FNR was recognized by an antibody against FNR (Guedeney et al., 1996). Scherer et al. (1988) showed that, in the cyanobacterium Anabaena variabilis, NADPH oxidation coupled to respiratory electron transport is performed by a thylakoid-associated FNR. However, Sazanov et al. (1996) reported that the chloroplast NADH dehydrogenase complex does not contain FNR.
The dark reduction of plastoquinone by NADPH has been demonstrated in broken chloroplasts (Mills et al., 1979), leaf discs (Harris and Heber, 1993), and intact leaves (Groom et al., 1993), supporting the idea that a chlororespiratory pathway, similar to that thought to exist in algae, is present in higher plants. Evidence of the existence of chlororespiratory activity in higher plants has also been presented (Garab et al., 1989; Singh et al., 1992; Gruszecki et al., 1994). Such a process might be important during leaf senescence (Cuello et al., 1995b). However, in the light the NADH dehydrogenase complex might participate in a cyclic electron flow around PSI (Bendall and Manasse, 1995). This has been proven in cyanobacteria (Mi et al., 1995) and it also seems to occur in higher plants (Mano et al., 1995).
In the study presented in this paper we isolated the chloroplast NADH dehydrogenase complex with the aim of studying its relation to FNR. To do this, SF and TFS were isolated from barley leaf chloroplasts. These were analyzed by native PAGE and the separated enzymes, detected with NADH and NBT, were electroeluted for further study. FNR was shown to be associated with the thylakoid NADH dehydrogenase complex on the basis of the presence of FNR activity, properties of their NAD(P)H-FeCNR activities, and the presence of polypeptide subunits reacting with antibodies raised against FNR, the chloroplast NdhA polypeptide, and the 51-kD and TYKY polypeptides from bovine complex I.
MATERIALS AND METHODS
Barley (Hordeum vulgare L. cv Hassan) was grown at 24°C under an 18-h photoperiod of white light, approximately 15 W m−2 (12.7 W m−2 PAR), as described previously (Cuello et al., 1987). All experiments were carried out with the oldest leaf of 14-d-old plants (about 1.42 mg chlorophyll g−1) and the base and tip were discarded.
Isolation and Fractionation of Chloroplasts and Solubilization of NAD(P)H Dehydrogenase from Thylakoids
For the isolation of chloroplasts, leaf segments were homogenized at 0 to 5°C in a Waring blender operating at the lowest speed for 12 s in 6 volumes of a freshly prepared buffer (E) that contained 0.35 m Suc, 25 mm Na-Hepes, 2 mm Na2-EDTA, 2 mm ascorbic acid, 4 mm DTT, and 1 mm PMSF (pH 7.6). The homogenate was strained through four layers of muslin and centrifuged at 200g for 5 min. The supernatant was centrifuged at 2500g for 10 min and the pellet was washed with buffer E. The chloroplasts were finally resuspended and fractionated into SF and thylakoids as described previously (Quiles et al., 1996). As described elsewhere (Cuello et al., 1995a), the preparation of chloroplasts was essentially free from mitochondria and other cellular components. The protein containing NAD(P)H-FeCNR activity was solubilized from thylakoids by adding 20% (w/v) sodium deoxycholate as described previously (Quiles et al., 1996), and the TFS was obtained.
Gel Electrophoresis and Zymograms
SDS-PAGE was carried out in a linear gradient of 10 to 20% polyacrylamide (2.5% bis-acrylamide, O'Farrel, 1975) as previously described (Quiles et al., 1996). Protein samples were denatured by the addition of 3.5% (w/v) SDS and 5% (v/v) mercaptoethanol and heating for 10 min at 100°C before being subjected to SDS-PAGE.
Native PAGE was carried out at 5°C in the same way as SDS-PAGE in a linear gradient of 5 to 22% of polyacrylamide, with the exception that the gels contained Triton X-100 instead of SDS (Kuonen et al., 1986). Samples were applied either in a single continuous band across the gel, which was subsequently sliced, or in individual wells formed in 3% acrylamide.
NAD(P)H-NBTR activities were detected by incubation of gel slices for 30 min at 30°C in darkness in 50 mm KH2PO4 (pH 8.0), 1 mm Na2-EDTA, 0.2 mm NADH or NADPH, and 0.5 mg mL−1 NBT. No color developed in controls without NAD(P)H.
Proteins and molecular mass markers were detected after electrophoresis by staining with Coomassie blue. Prestained SDS-PAGE standards (Bio-Rad) were used for immunoblot analysis.
Electroelution
NADH-NBTR-activity-staining bands were excised from the native gel, and the enzymes were electroeluted from gels in 25 mm Tris, 192 mm Gly, and 0.1% (w/v) Triton X-100 buffer (pH 8.3) in an electro-eluter unit (Bio-Rad) according to the protocol provided by the manufacturer.
Enzyme Assays
NAD(P)H dehydrogenase activities were determined with FeCN (kinetic assays) and NBT (zymogram detection) as the electron acceptors.
NAD(P)H-FeCNR activities were determined at 25°C, as described by Galante and Hatefi (1979), by continuous measurement of the decreases in A340 caused by oxidation of NAD(P)H, in a spectrophotometer (ATI, Cambridge, UK). Unless otherwise stated, the reaction mixture, with a final volume of 1 mL, included 50 mm KH2PO4 (pH 8.0), 1 mm Na2-EDTA, 0.15 mm NADH or NADPH, 1 mm FeCN, and enzyme sample. Usually, the decrease in absorbance was recorded for the first 1 to 2 min after the addition of the enzyme sample, and the linear decrease was determined between 10 and 70 s of the reaction. As indicated, prior to the enzymatic reaction, the enzyme was incubated with p-CMPS and/or NAD(P)H at 30°C for various times. Control values obtained from media without enzyme preparation were subtracted. Control rates without FeCN were zero. The effects of electron-transport inhibitors were also examined as indicated below. Rotenone and antimycin A were added in a small volume (5 μL) from ethanol solutions. Ethanol (5 μL added to 1 mL of the reaction mixture) did not produce any inhibition by itself.
FNR activity was assayed by measuring Fd-dependent Cyt c reduction in the presence of NADPH, as described by Morigasaki et al. (1990). Control values (ranging between 5 and 15% of the values obtained with complete reaction mixture) obtained without Fd were subtracted.
One unit of activity is defined as the amount of enzyme preparation that oxidizes 1 μmol of substrate (NADH or NADPH) or reduces 1 μmol of substrate (Cyt c) per minute under the conditions of reaction. Specific activities are expressed per milligram of protein. The extinction coefficients of 6.22 at 340 nm and 19 mm−1 cm−1 at 550 nm were used to calculate NAD(P)H oxidation and Cyt c reduction rates, respectively.
Km values were calculated from rate measurements with the Enzfitter program (Elsevier-Biosoft, Cambridge, UK).
Immunoblot Analysis
After SDS-PAGE, polypeptides were electroblotted from gels to PVDF membranes (Immobilon-P, Millipore) in 48 mm Tris, 39 mm Gly, and 0.0375% (w/v) SDS buffer (pH 9.2) containing 20% (v/v) methanol in a NovaBlot electrophoretic transfer unit (Pharmacia) at 0.8 mA cm−2 of the gel for 1 h and 15 min. Following transfer, the PVDF membrane was washed for 5 min in TBS (25 mm Tris-HCl and 150 mm NaCl, pH 7.4). The nonspecific-binding sites on PVDF membranes were blocked by incubation with 5% (w/v) nonfat dry milk in TBS at 37°C for 1 h. Antibodies diluted 1,000-fold (anti-NdhA, anti-51 kD, and anti-TYKY) or 10,000-fold (anti-FNR) in TBS containing 1% (w/v) nonfat dry milk and 0.3% (v/v) Tween 20 (TBS-T buffer) were incubated with sheets of membrane for 3 h at 23°C. The sheets were washed four times in the above buffer and incubated for 1 h with anti-rabbit IgG-alkaline phosphatase conjugate (Sigma) diluted 30,000-fold in the same buffer. The membranes were washed with TBS-T buffer and then with TBS-T buffer containing 2 m NaCl and rinsed with TBS, and the color was developed by using 5-bromo-4-chloro-3-indolyl phosphate/NBT (Sigma) as the substrate.
The antibodies raised against the bovine complex I subunits 51 and 23.5 kD (TYKY), barley NdhA polypeptide, and barley FNR were kindly donated, respectively, by Drs. J.E. Walker and J.M. Skehel (Medical Research Council, Cambridge, UK), Drs. M. Martín and B. Sabater (University of Alcalá de Henares, Spain), and Dr. H. Scheller (Royal Veterinary and Agricultural University, Copenhagen, Denmark).
Determination of Protein
Protein was quantified by the method of Lowry et al. (1951) after precipitation with 10% (w/v) TCA.
RESULTS
Electrophoresis, Electroelution, and Characterization of the Enzymes
Purification of the NADH dehydrogenase complex from the thylakoid membrane is complicated because of its low abundance (the complex represents approximately 0.2% of total thylakoid membrane protein, Sazanov et al., [1996]) and to its possible instability (well known for complex I from other systems, Friedrich et al., [1995]).
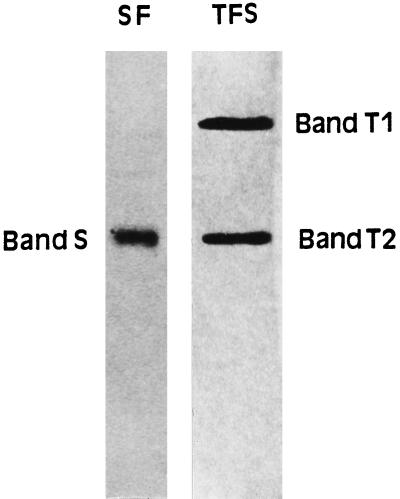
As previously reported (Quiles et al., 1996), the native PAGE of the chloroplast fractions produces three intense bands after NADH-NBT gel staining: one from SF (band S) and two from TFS (bands T1 and T2; Fig. 1). TFS also gave rise to a third band containing NADH-NBTR activity on the 3% constant polyacrylamide gel, with a generally more variable and lower intensity than bands T1 and T2 but with an identical polypeptidic pattern to that of band T1 (Quiles et al., 1996). For this reason, the third thylakoid band was not further characterized. Figure 1 shows the results of a typical electrophoretic analysis using the starting samples (SF or TFS). However, with the aim of electroeluting the enzymes from the gel bands for further characterizations, each sample was applied as a single continuous band across the gel.
Figure 1.
Electrophoretic separation of chloroplast NADH dehydrogenase. Zymograms were revealed with NADH and NBT after native PAGE (5–22% linear polyacrylamide gel gradient). The starting samples contained 0.2 and 0.5 units of NADH-FeCNR for the SF and TFS enzymes, respectively.
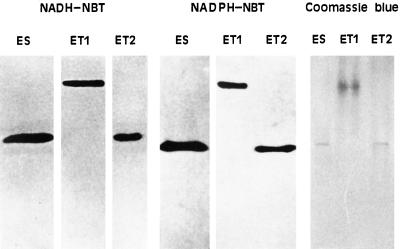
To study the purity of ES, ET1, and ET2, these enzymes were analyzed by native PAGE and subsequent staining with NADH-NBT, NADPH-NBT, and Coomassie blue. Figure 2 shows the results obtained by depositing in each well the corresponding electroeluate (after concentration to 200 μL) obtained from one (for NAD[P]H-NBTR activity assays) or two (for Coomassie blue staining) continuous bands. Each enzyme produced a single colored band with each of the staining reactions and with electrophoretic mobility similar to that observed in Figure 1. These results show that the electroeluted enzymes, which also had NADPH-NBTR activity (Fig. 2), were relatively pure and that the electrophoresis and electroelution processes did not significantly alter the enzyme structure.
Figure 2.
Native PAGE of the enzymes ES, ET1, and ET2 electroeluted from the gels shown in Figure 1. Zymograms were revealed with NADH-NBT and NADPH-NBT and the gels were stained with Coomassie blue after native PAGE (5–22% linear polyacrylamide gel gradient). For further explanation, see the text.
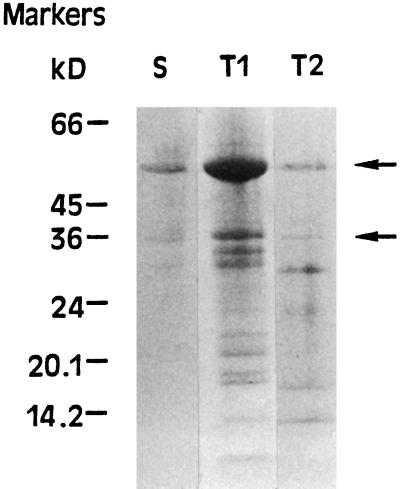
Figure 3 shows the SDS-PAGE peptide profiles of bands S, T1, and T2 indicated in Figure 1, as visualized by staining with Coomassie blue. As previously reported (Quiles et al., 1996), band T1 contains at least nine polypeptides that range from 12 to 53 kD (Fig. 3). Moreover, the molecular masses of six of them agreed with those deduced from six chloroplast ndh genes (Quiles et al., 1996). All enzymes contained, among others, a main polypeptide of 53 kD and another of approximately 35 kD (Fig. 3), which probably correspond to the NADH-binding subunit and FNR, respectively (see below). Silver staining of the gel after Coomassie blue staining did not increase the intensity or the number of protein bands.
Figure 3.
SDS-PAGE profiles of the chloroplast NADH dehydrogenase of bands S, T1, and T2 shown in Figure 1. The polypeptides were visualized by Coomassie blue staining. Migration of molecular mass markers is indicated on the left. The arrows indicate the polypeptides cited in the text.
Table I shows the recoveries and specific NAD(P)H-FeCNR and FNR activities of the electroeluted enzymes ES, ET1, and ET2, as well as the NADPH-FeCNR/FNR and NADPH-FeCNR/NADH-FeCNR activity ratios. The low recoveries may be explained by inactivation of the enzyme during the isolation process and because many enzymes in SF and TFS (different from ES, ET1, and ET2) that have NAD(P)H-NBTR activity (Quiles et al., 1996), and presumably NAD(P)H-FeCNR activity, were not electroeluted. Therefore, these were not quantified. It is also possible that the enzymes ES, ET1, and ET2 were incompletely electroeluted from the native gels.
Table I.
Recovery of protein, NAD(P)H-FeCNR, and FNR activities and activity ratios of the enzymes ES, ET1, and ET2 electroeluted from bands in Figure 1
| Parameter | SF | TFS | ES | ET1 | ET2 |
|---|---|---|---|---|---|
| Protein (mg) | 1.87 ± 0.28 | 9.35 ± 1.22 | 0.04 ± 0.008 | 0.07 ± 0.009 | 0.06 ± 0.007 |
| Yield (%) | 100 | 100 | 2.14 | 0.75 | 0.64 |
| Enzymatic activity | |||||
| NADH-FeCNR | |||||
| Specific (units mg−1 protein) | 0.49 ± 0.06 | 0.23 ± 0.01 | 0.49 ± 0.09 | 0.34 ± 0.07 | 0.53 ± 0.12 |
| Total (units) | 0.92 | 2.15 | 0.02 | 0.02 | 0.03 |
| Yield (%) | 100 | 100 | 2.17 | 0.93 | 1.40 |
| NADPH-FeCNR | |||||
| Specific (units mg−1 protein) | 3.10 ± 0.38 | 1.08 ± 0.25 | 3.62 ± 0.67 | 1.57 ± 0.41 | 1.40 ± 0.21 |
| Total (units) | 5.80 | 10.10 | 0.14 | 0.11 | 0.08 |
| Yield (%) | 100 | 100 | 2.41 | 1.09 | 0.79 |
| FNR | |||||
| Specific (units mg−1 protein) | nda | 0.20 ± 0.01 | 0.47 ± 0.10 | 0.16 ± 0.01 | 0.17 ± 0.03 |
| Total (units) | nd | 1.87 | 0.02 | 0.01 | 0.01 |
| Yield (%) | nd | 100 | nd | 0.53 | 0.53 |
| Specific activity ratio | |||||
| NADPH-FeCNR/FNR | nd | 5.40 | 7.70 | 9.81 | 8.24 |
| NADPH-FeCNR/NADH-FeCNR | 6.33 | 4.70 | 7.39b | 4.62 | 2.64 |
For comparison, the values for starting SF and TFS are shown. The values are means ± se from 6 to 11 independent experiments.
nd, Not determined.
Variable values ranging between 2.41 and 11.44.
As for recoveries in the chromatographic purification of barley thylakoid NADH dehydrogenase on Q or hydroxylapatite columns, we obtained higher values (5–6%; Quiles et al., 1996) than those in Table I but values similar to those obtained in the purification of the NADH dehydrogenase from the cyanobacterium Anabaena variabilis on a Mono-Q column (Alpes et al., 1989) and of the mitochondrial complex I from broad bean on a hydroxylapatite column (Leterme and Boutry, 1993). However, the enzyme complex was dissociated during the chromatographic purification and only one NADH dehydrogenase of small size was detected by native PAGE (Quiles et al., 1996).
The three enzymes that we analyzed showed FNR activity, although for enzymes ET1 and ET2 the specific activities were lower (0.16 and 0.17 unit mg−1 protein, respectively) than that of the starting sample (0.20 unit mg−1 protein; Table I). FNR activity was not determined in SF, because this fraction showed a rapid reduction of Cyt c in a reaction that was independent of added Fd. Consequently, the change in specific FNR activity and the recovery of FNR with regard to SF are unknown in the case of ES. The NADPH-FeCNR/FNR activity ratios of ES, ET1, and ET2 showed constant values in the different assays of each enzyme (results not shown), with the values for the three enzymes being similar (7.70, 9.81 and 8.24, respectively; Table I). However, although relatively constant in the different assays of both ET1 and ET2, the values of the NADPH-FeCNR/NADH-FeCNR activity ratios were different for each of the three enzymes, with those of ET1 and ES being almost double and triple, respectively, that of ET2. The values of the NADPH-FeCNR/NADH-FeCNR activity ratio for many different ES samples varied considerably, from 2.41 to 11.44 (Table I).
Direct and double-reciprocal plots of the effects of NADPH concentration on the NADPH-FeCNR activity of the electroeluted enzymes (not shown) indicated that the enzymes exhibited Michaelis-Menten kinetics, and the Km values for NADPH, calculated with the Enzfitter program, were 55, 35, and 62 μm for ES, ET1, and ET2, respectively (means obtained from two to three independent experiments and with no individual values differing by more than 20% from the mean value).
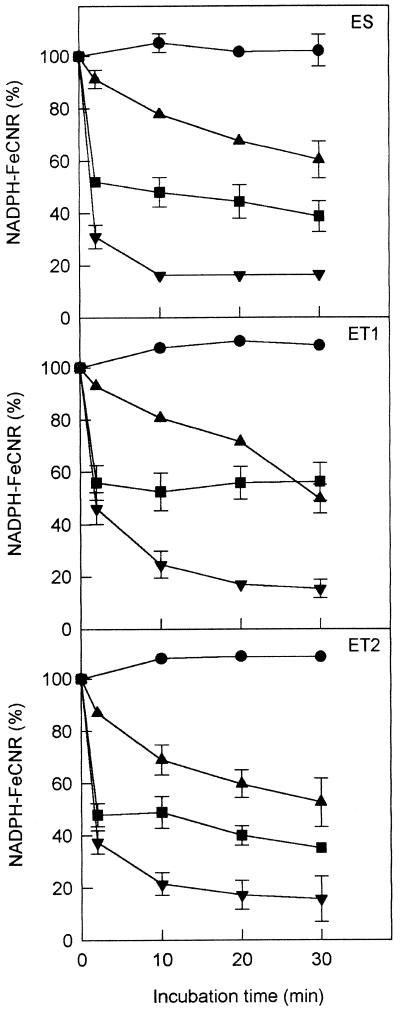
Figure 4 shows the effects on the NADPH-FeCNR activities of ES, ET1, and ET2 when each enzyme was incubated separately with 0.5 mm NADPH, 0.05 mm p-CMPS, or 0.5 mm NADPH plus 0.05 mm p-CMPS at 30°C for variable times. Except for quantitative differences, ES, ET1, and ET2 behaved similarly. The inhibition by p-CMPS increased progressively with the enzyme incubation time, the activity decreasing to between 50 and 60% at 30 min. The inhibition by NADPH was very rapid: after 2 min of incubation the activity decreased to 48 to 56% and remained at this level (ET1) or increased slightly (ES and ET2) at 30 min. The results of incubation with NADPH plus p-CMPS showed in all cases the cooperative effect of both compounds (Fig. 4).
Figure 4.
Effects of NADPH, p-CMPS, and NADPH plus p-CMPS on the NADPH-FeCNR activity of the enzymes ES, ET1, and ET2. The enzymes were incubated at 30°C for variable times with 0.5 mm NADPH (▪), 0.05 mm p-CMPS (▴), or 0.5 mm NADPH plus 0.05 mm p-CMPS (▾) prior to the activity assays. Control assays incubating the enzymes with distilled water (•) were also made. The values indicate activity percentages in relation to nontreated samples (Table I) and are means of three to five independent assays. Only ses ≥ 3.5 are represented.
Results contrasting with those depicted in Figure 4 were obtained when the effects of incubating the three enzymes with NADH, p-CMPS, and NADH plus p-CMPS on their respective NADH-FeCNR activities were studied at the same concentrations and temperature. Table II shows the results for 20 min of incubation. NADH alone had no effect on ET1, slightly activated ET2, and slightly inhibited ES. Moreover, p-CMPS alone had a comparatively slight inhibitory effect (the activity diminished to approximately 90% in ES, ET1, and ET2), in contrast to its effect on the NADPH-FeCNR activity (which diminished to 60 to 70% at 20 min, Fig. 4). This slight inhibition of the NADH-FeCNR activity decreased slightly (ES) or was maintained (ET1 and ET2) when the enzyme was incubated with NADH plus p-CMPS (Table II).
Table II.
Effects of NADH, p-CMPS, and NADH plus p-CMPS on the NADH-FeCNR activity of the enzymes ES, ET1, and ET2
| Enzyme | NADH-FeCNR Activity
|
||
|---|---|---|---|
| NADH | p-CMPS | NADH + p-CMPS | |
| % | |||
| ES | 92.0 ± 5.0 | 88.5 ± 4.5 | 77.5 ± 13.0 |
| ET1 | 99.5 ± 5.5 | 92.0 ± 6.5 | 87.0 ± 4.5 |
| ET2 | 114.5 ± 4.5 | 89.5 ± 5.0 | 92.0 ± 4.0 |
The enzymes were incubated at 30°C for 20 min with 0.5 mM NADH, 0.05 mM p-CMPS, or 0.5 mM NADH plus 0.05 mM p-CMPS prior to the activity assays. The values indicate activity percentages in relation to nontreated samples (see Table I) and are means ± se of three to five independent assays.
The effects of 5 μm antimycin A, 1.5 mm PADR, and 20 μm rotenone on the NAD(P)H-FeCNR activities of ES, ET1, and ET2 are shown in Table III. Antimycin A has been reported to act as an inhibitor of the Fd-dependent reduction of plastoquinone in cyclic electron flow (Mano et al., 1995). PADR is a specific inhibitor of FNR (Avron, 1981), whereas rotenone is an inhibitor of complex I of mammalian mitochondria. Antimycin A had no effect on the NADPH-FeCNR activity of any of the enzymes (Table III), whereas in the case of the NADH-FeCNR activity, it had no (ES) or a slight (ET2) or moderate (ET1) inhibitory effect. Rotenone (20 μm) had no effect on the NAD(P)H-FeCNR activities of any of the three enzymes (Table III). However, 40 μm rotenone showed no effect on the NADPH-FeCNR activities of ES, ET1, or ET2 but strongly activated their NADH-FeCNR activities (not shown). Finally, 1.5 mm PADR showed a strong inhibitory effect on the NADPH-FeCNR activities (which decreased to 45 and 42% with ES and ET1, respectively), whereas it had no effect on the NADH-FeCNR activities (which remained at the control values for the same enzymes; Table III).
Table III.
Effects of various inhibitors on the NAD(P)H-FeCNR activities of the enzymes ES, ET1, and ET2
| Inhibitor | NADH-FeCNR
|
NADPH-FeCNR
|
||||
|---|---|---|---|---|---|---|
| ES | ET1 | ET2 | ES | ET1 | ET2 | |
| % | ||||||
| Antimycin A (5 μm) | 99.0 ± 12.6 | 71.0 ± 3.5 | 86.0 ± 6.3 | 100.5 ± 1.2 | 98.6 ± 1.0 | 99.2 ± 1.3 |
| PADR (1.5 mm) | 103.5 | 96.0 | nda | 45.0 | 42.0 | nd |
| Rotenone (20 μm) | 100.0 ± 0.0 | 97.5 ± 13.0 | 100.0 ± 8.1 | 104.0 ± 4.0 | 102.0 ± 2.0 | 102.5 ± 2.4 |
The values indicate activity percentages in relation to nontreated samples (see Table I), and for antimycin and rotenone are means ± se of three to five independent assays, and for PADR are means of two independent assays in which no individual value deviates more than 6% from the mean.
nd, Not determined.
Immunoblot Analysis
The antibodies raised against FNR, NdhA, and the 51- and 23.5-kD subunits (or TYKY as characterized by its first four amino acids according to the method of Walker, 1992) of the bovine complex I were used to investigate the presence of the corresponding polypeptides in the chloroplastic enzymes ES, ET1, and ET2. After SDS-PAGE, carried out with 10 μg of protein of each of the denatured enzymes, the polypeptides were transferred onto PVDF membrane and used for immunoblot analysis. As a control, we also assayed TFS in such quantities that 20 μg of protein was applied. Controls in which the primary and/or secondary (alkaline phosphatase conjugate) antibodies were omitted during incubation did not reveal any bands (not shown).
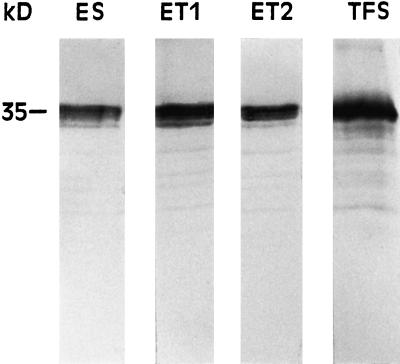
Figure 5 shows immunoblots of ES, ET1, ET2, and TFS proteins using antibody against barley FNR. In all of the enzymes and, as expected, in TFS a polypeptide of approximately 35 kD reacted strongly with the anti-FNR antibody. The molecular mass of this polypeptide is in agreement with that previously reported for barley FNR (Andersen et al., 1992). Other weaker bands below 35 kD were also recognized by the anti-FNR antibody (Fig. 5). These bands were probably degradation products of FNR, a phenomenon previously reported for other polypeptides (Martin et al., 1996).
Figure 5.
Immunoblot analysis of the polypeptides of the enzymes ES, ET1, and ET2 using an anti-FNR antibody. After excision of the NADH-NBT staining gel bands and electroelution, 10 μg of protein of each enzyme was subjected to SDS-PAGE prior to immunoblot assays. The analysis of starting TFS (20 μg of protein applied to the lane) is shown as a control. For details, see the text.
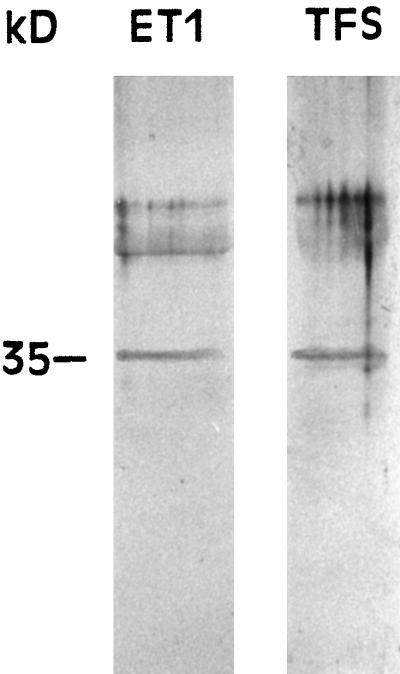
With regard to the existence of the ndhA gene product in the electroeluted enzymes, immunoblot assays showed that antibody raised against barley NdhA reacts with another 35-kD polypeptide of ET1 and TFS (Fig. 6), although no 35-kD antigen was detected in ES and ET2 (results not shown). Two additional bands above 35 kD were also recognized by the anti-NdhA antibody (Fig. 6). The nature of these bands, which might be due to the association of NdhA with other polypeptides or to unspecific binding of the antibody, remains undetermined.
Figure 6.
Immunoblot analysis of the polypeptides of the enzyme ET1 using an anti-NdhA antibody. After excision of band T1 and electroelution, 10 μg of protein of the enzyme was subjected to SDS-PAGE prior to immunoblot assay. The analysis of starting TFS (20 μg of protein applied to the lane) is shown as a control.
The chloroplast 35-kD polypeptide reacting with anti-NdhA antibody has been found in thylakoid membranes, but not in stroma, of barley chloroplasts by Martin et al. (1996). Hence, this 35-kD polypeptide must be the protein product (NdhA) of the plastid-encoded ndhA gene.
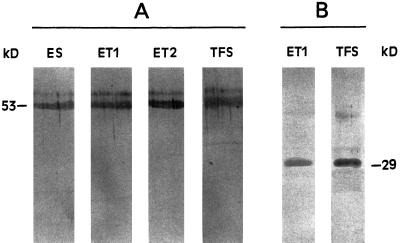
To study the presence in the chloroplast of the polypeptides homologous to subunits of bovine mitochondrial complex I, the proteins of TFS and electroeluted enzymes were analyzed by immunoblotting using antibodies raised against the 51-kD and TYKY subunits of the bovine complex I (Fig. 7). A band at approximately 53 kD was recognized by the anti-51-kD antibody in ES, ET1, and ET2 (Fig. 7A). The antibody against the TYKY subunit reacted with a 29-kD polypeptide only in ET1 (Fig. 7B); no polypeptide was detected in ES and ET2 (not shown). As expected, both antibodies recognized the corresponding polypeptide in TFS (Fig. 7).
Figure 7.
Immunoblot analysis of the polypeptides of the enzymes ES, ET1, and ET2 using antibodies against the bovine complex I subunits 51 (A) and 23.5 kD (TYKY; B). After excision of bands and electroelution, 10 μg of protein of each enzyme was subjected to SDS-PAGE prior to immunoblot assays. The analyses of starting TFS with anti-51-kD and anti-TYKY antibodies (20 μg of protein applied to each lane) are shown as controls.
The peripheral 51-kD subunit of complex I contains the site for binding NADH. However, this subunit and the respective gene have not been identified in chloroplasts (Friedrich et al., 1995). Our immunoblot results with the anti-51-kD antibody demonstrated the existence in chloroplasts of a polypeptide homologous to the NADH-binding subunit in mitochondria.
DISCUSSION
We isolated three NADH dehydrogenase enzymes (ES, ET1, and ET2) from barley chloroplasts by native PAGE and electroelution. Electrophoresis of the enzymes electroeluted from the electrophoretic bands obtained starting with SF and TFS (Fig. 1) showed that they were pure (Fig. 2). Moreover, the fact that the single bands of Figure 2 showed the same electrophoretic mobility as the enzymes with NADH-NBTR activity from SF and TFS (Fig. 1) indicates that, in our conditions at least, the complexes neither decay nor lose subunits during the separation process, as is also the case in the prokaryotic complex I (Yagi et al., 1988). Although the S, T1, and T2 bands (Fig. 1) also have NADPH-NBTR activity (Quiles et al., 1996), we revealed them with NADH and NBT prior to electroelution of their respective enzymes, unlike the chloroplastic NAD(P)H dehydrogenase complex analyzed by Guedeney et al. (1996), which was revealed with NADPH and NBT.
All three enzymes, ES, ET1, and ET2, show FNR activity (Table I). Their constant NADPH-FeCNR/FNR ratios and the results of the above-described characterizations indicate that their NADPH-FeCNR activities are due to the diaphorase activity of FNR (Forti, 1977). The Km values for NADPH of the NADPH-FeCNR activity (55, 35, and 62 μm for ES, ET1, and ET2, respectively) are similar to those found for the same activity of FNR: 66 μm in A. variabilis (Sancho et al., 1988) and 31 μm in spinach (Forti and Sturani, 1968). Furthermore, the sensitivity of FNR to inhibition by p-CMPS is strongly enhanced if the enzyme is incubated with this inhibitor in the presence of NADPH (Forti and Sturani, 1968). This also occurred during the incubation of ES, ET1, and ET2 with NADPH plus p-CMPS (Fig. 4). Also, a brief incubation of ES, ET1, and ET2 with NADPH alone inhibits their NADPH-FeCNR activities (Fig. 4), as was described for the incubation of FNR with NADPH (Forti and Sturani, 1968). Our results agree with those obtained by Sancho et al. (1988), who incubated FNR of A. variabilis with NADPH and/or p-hydroxymercuribenzoate.
The previous results are in contrast to those obtained when studying the effect on NADH-FeCNR activity of incubating the three enzymes with NADH, p-CMPS, and NADH plus p-CMPS (Table II). NADH had no significant inhibitory effect on any of them and it did not increase the small inhibitory effect of p-CMPS. A small inhibitory effect was found by Alpes et al. (1989) after application of p-CMPS to the NADH dehydrogenase enzyme from thylakoids of A. variabilis.
The inactivation by PADR of the NADPH-FeCNR activity of the enzymes ES and ET1 (Table III) lends additional support to the idea that this activity is due to FNR. In contrast, the lack of effect of PADR on the NADH-FeCNR activity (Table III) and the variable NADPH-FeCNR/NADH-FeCNR ratios for the enzymes (Table I) suggest that the NADH-FeCNR activity is caused by a part of the enzymatic complex other than FNR. Moreover, NADH is a very poor electron donor for FNR (Sancho et al., 1988; Cartagena et al., 1994). The variable NADPH-FeCNR/NADH-FeCNR ratios might be due to differential inactivations of FNR and that part of the enzymatic complex that shows NADH-FeCNR activity, to different stoichiometries of FNR and the NADH-binding polypeptide, and/or to differential effects on either of the above activities of other parts of the chloroplast NADH dehydrogenase complex present or absent in the ES, ET1, and ET2 enzymes. However, 20 μm rotenone has no effect on the NADH-FeCNR activity of ES, ET1, and ET2 (Table III) or on the chloroplast total NAD(P)H-FeCNR activity (Cuello et al., 1995a). This is not surprising, since the catalytic activity of purified mitochondrial complex I from red beetroot is completely unaffected by rotenone (Soole et al., 1992; Rasmusson et al., 1994) until the enzyme is reconstituted into phosphatidylcholine vesicles. Furthermore, the inhibition of the NADH dehydrogenase from Chlamydomonas reinhardtii. thylakoids by 20 μm rotenone was lower than 10%, and the activity was even stimulated by rotenone after (NH4)2SO4 fractionation of the enzyme (Godde, 1982). The fact that antimycin A had no or little inhibitory effect on the NAD(P)H-FeCNR activities of the enzymes (Table III) may indicate that the thylakoid NADH dehydrogenase associated with FNR is involved in antimycin A-insensitive cyclic electron transport, a pathway suggested by Hosler and Yocum (1985).
The immunoblots of ES, ET1, and ET2 proteins using the anti-FNR antibody revealed the suspected presence of FNR in the three enzymes (Fig. 5). This coincides with the results of Guedeney et al. (1996), who found FNR associated with chloroplastic NAD(P)H dehydrogenase complex from potato. Our results, however, contrast with those of Sazanov et al. (1996), who did not find FNR in the chloroplast NADH dehydrogenase complex from pea, although it may be that the purified enzyme used by these authors did not contain NADPH-FeCNR activity because the FNR had been released from the complex through the use of dodecyl-β-d-maltoside. The immunoblots with the anti-NdhA antibody demonstrated that NdhA, the 35-kD protein product of the plastid-encoded ndhA gene, is present only in ET1 (Fig. 6). Likewise, the antigen corresponding to the anti-TYKY antibody was found only in ET1 (Fig. 7B), whereas the chloroplast ndhI gene encodes a homolog to the 23.5-kD (TYKY) subunit (Dupuis et al., 1991; Friedrich et al., 1995).
An important result is that the three enzymes studied contain a 53-kD polypeptide that is recognized by the antibody raised against the 51-kD subunit of the NADH-oxidizing subcomplex of mitochondrial complex I. Eleven subunits are known to be encoded by the 11 chloroplast ndh genes, and this 53-kD polypeptide could be one of the three remaining unknown subunits necessary for the chloroplast NADH dehydrogenase complex (Friedrich et al., 1995). By analogy with the complex I from other sources, the 53-kD polypeptide might be a peripheral subunit, which binds to NADH, and could be codified in the nucleus (Friedrich et al., 1995). In agreement with the previous results, Peltier and Schmidt (1991) found a major subunit of 51 kD in thylakoid-bound NADH-plastoquinone oxidoreductase from C. reinhardtii, and Alpes et al. (1989) described a 52-kD polypeptide in the thylakoid-bound NADH dehydrogenase from A. variabilis.
Many available data suggest that ET1 is the chloroplast NADH dehydrogenase complex (ET2 and ES would be subcomplexes) that contains subunits of the three subcomplexes into which the cyanobacterial NAD(P)H dehydrogenase complex can be fractionated (Berger et al., 1993a). ET1 contains NdhA, which is an intrinsic protein of the membrane belonging to the hydrophobic subcomplex (Friedrich et al., 1995). It also contains a peripheral or predominantly peripheral subunit homologous to TYKY (NdhI; Friedrich et al., 1995), belonging to another subcomplex (solubilized from membranes of Synechocystis sp. PCC6803 by Berger et al., 1993a). Finally, ET1 contains the 53-kD polypeptide homologous to the 51-kD polypeptide that binds to NADH and that belongs to the peripheral subcomplex (Friedrich et al., 1995). In our case, ES might be the peripheral subcomplex associated with FNR. Thus, the chloroplast NAD(P)H dehydrogenase complex would have two doors available for electrons to pass through, FNR (the most important because of the abundance of NADPH in chloroplasts) for the electrons of NADPH, and the 53-kD polypeptide for those of NADH. The use of NADPH does not seem to depend on Fd, because it does not affect the NADPH-FeCNR activity of any of the three enzymes analyzed (results not shown). However, it should be indicated that NdhI shows homology to Fds and has been called frxB, or Fd-like protein, although no function has been attributed to it (Dupuis et al., 1991).
The association of FNR with relatively small polypeptides has been described in previous papers. It has been reported as being bound to a 17.5-kD (probably NdhJ) polypeptide in spinach thylakoids (Matthijs et al., 1986), to a 10.8-kD polypeptide in barley thylakoids (Andersen et al., 1992), and to a “binding protein” of thylakoids in A. variabilis (Scherer et al., 1988). However, Fd and FNR have been shown to form a very stable electrostatic complex in a 1:1 ratio, irrespective of their redox states (Zanetti and Curti, 1981).
However, although the data presented in our work provide environmental evidence for the binding of FNR to a pyridine nucleotide dehydrogenase complex, further evidence, such as a constant molar ratio of FNR to ndh gene products during the complex purification, is necessary to confirm such binding.
It is probable that the chloroplast complex associated with FNR is the complex involved in the entry of electrons from a soluble stromal pool of NAD(P)H into the plastoquinone pool (Guedeney et al., 1996). During illumination this complex, rather than the as-yet-unidentified Fd-plastoquinone reductase proposed by Bendall and Manasse (1995) and Scheller (1996), might participate in a cyclic electron pathway around PSI. It has been suggested that the Cyt b-559 of low-redox potential participates in the electron transfer to the plastoquinone during antimycin A-sensitive cyclic electron transport (Miyake et al., 1995). It is noteworthy that a PSI gene is included in one of the four transcriptional units encoding the plastidial complex (Shimada and Sugiura, 1991).
Alternatively, the chloroplast complex may be involved in chlororespiration, a process that has been extensively studied in unicellular green algae (Bennoun, 1982, 1994; Peltier et al., 1987) but less so in higher plants (Garab et al., 1989; Gruszecki et al., 1994). The breakdown of carbohydrates in chloroplasts in the dark occurs by glycolysis and the pentose phosphate pathway, producing NADPH (Singh et al., 1992), which would be reoxidized by the chloroplast NAD(P)H dehydrogenase.
ACKNOWLEDGMENTS
We thank Drs. J.E. Walker and J.M. Skehel (Medical Research Council, Cambridge, UK), Drs. M. Martín and B. Sabater (University of Alcalá de Henares, Spain), and Dr. H.V. Scheller (Royal Veterinary and Agricultural University, Copenhagen, Denmark) for supplying the antibodies.
Abbreviations:
- ES
enzyme electroeluted from band S
- ET1
ET2, enzyme electroeluted from bands T1 and T2, respectively
- FNR
Fd-NADP oxidoreductase
- NAD(P)H-FeCNR
NAD(P)H-FeCN oxidoreductase
- NAD(P)H-NBTR
NAD(P)H-NBT oxidoreductase
- NBT
nitroblue tetrazolium
- PADR
2′-monophosphoadenosine-5′-diphosphoribose
- p-CMPS
p-chloromercuriphenylsulfonic acid
- SF
stroma fraction
- TFS
thylakoid fraction solubilized with sodium deoxycholate
Footnotes
This work was supported by the Spanish Dirección General de Investigación Científica y Técnica (grant no. PB94-1141).
LITERATURE CITED
- Alpes I, Scherer S, Böger P. The respiratory NADH dehydrogenase of the cyanobacterium Anabaena variabilis: purification and characterization. Biochim Biophys Acta. 1989;973:41–46. [Google Scholar]
- Andersen B, Scheller HV, Moller BL. The PSI-E subunit of photosystem I binds ferredoxin:NADP oxidoreductase. FEBS Lett. 1992;311:169–173. doi: 10.1016/0014-5793(92)81391-x. [DOI] [PubMed] [Google Scholar]
- Avron M (1981) Photosynthetic electron transport and photophosphorylation. In MD Hatch, NK Boardman, eds, The Biochemistry of Plants, Vol 8. Academic Press, New York, pp 163–191
- Bendall DS, Manasse RS. Cyclic photophosphorylation and electron transport. Biochim Biophys Acta. 1995;1229:23–38. [Google Scholar]
- Bennoun P. Evidence for a respiratory chain in the chloroplast. Proc Natl Acad Sci USA. 1982;79:4352–4356. doi: 10.1073/pnas.79.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennoun P. Chlororespiration revisited: mitochondrial-plastid interactions in Chlamydomonas. Biochim Biophys Acta. 1994;1186:59–66. [Google Scholar]
- Berger S, Ellersiek U, Kinzelt D, Steinmüller K. Immunopurification of a subcomplex of the NAD(P)H-plastoquinone-oxidoreductase from the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 1993a;326:246–250. doi: 10.1016/0014-5793(93)81800-f. [DOI] [PubMed] [Google Scholar]
- Berger S, Ellersiek U, Westhoff P, Steinmüller K. Studies on the expression of NDH-H, a subunit of the NAD(P)H-plastoquinone oxidoreductase of higher-plant chloroplasts. Planta. 1993b;190:25–31. [Google Scholar]
- Cartagena E, Bes MT, Gomez-Moreno C, Peleato ML. Purification and characterization of ferredoxin-NADP+ reductase from the green alga Chlorella fusca. Physiol Plant. 1994;91:645–650. [Google Scholar]
- Cuello J, Quiles MJ, Albacete ME, Sabater B. Properties of a large complex with NADH dehydrogenase activity from barley thylakoids. Plant Cell Physiol. 1995a;36:265–271. [Google Scholar]
- Cuello J, Quiles MJ, Rosauro J, Sabater B. Effects of growth regulators and light on chloroplasts NAD(P)H dehydrogenase activities of senescent barley leaves. Plant Growth Regul. 1995b;17:225–232. [Google Scholar]
- Cuello J, Quiles MJ, Sabater B. Control by phytochrome of the synthesis of protein related to senescence in chloroplasts of barley (Hordeum vulgare) Physiol Plant. 1987;71:341–344. [Google Scholar]
- Dupuis A, Skehel JM, Walker JE. A homologue of a nuclear-coded iron-sulfur protein subunit of bovine mitochondrial complex I is encoded in chloroplast genomes. Biochemistry. 1991;30:2954–2960. doi: 10.1021/bi00225a032. [DOI] [PubMed] [Google Scholar]
- Forti G. Flavoproteins. In: Trebst A, Avron M, editors. Encyclopedia of Plant Physiology, New Series, Vol 5. Berlin: Springer; 1977. pp. 222–226. [Google Scholar]
- Forti G, Sturani E. On the structure and function of reduced nicotinamide adenine dinucleotide phosphate-cytochrome f reductase of spinach chloroplasts. Eur J Biochem. 1968;3:461–472. doi: 10.1111/j.1432-1033.1967.tb19553.x. [DOI] [PubMed] [Google Scholar]
- Friedrich T, Steinmüller K, Weiss H. The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett. 1995;367:107–111. doi: 10.1016/0014-5793(95)00548-n. [DOI] [PubMed] [Google Scholar]
- Galante YM, Hatefi Y. Purification and molecular and enzymic properties of mitochondrial NADH dehydrogenase. Arch Biochem Biophys. 1979;192:559–568. doi: 10.1016/0003-9861(79)90126-7. [DOI] [PubMed] [Google Scholar]
- Garab G, Lajkó F, Mustárdy L, Márton L. Respiratory control over photosynthetic electron transport in chloroplasts of higher-plant cells: evidence for chlororespiration. Planta. 1989;179:349–358. doi: 10.1007/BF00391080. [DOI] [PubMed] [Google Scholar]
- Godde D. Evidence for a membrane bound NADH-plastoquinone-oxidoreductase in Chlamydomonas reinhardtii CW-15. Arch Microbiol. 1982;131:197–202. [Google Scholar]
- Godde D, Trebst A. NADH as electron donor for the photosynthetic membrane of Chlamydomonas reinhardtii. Arch Microbiol. 1980;127:245–252. [Google Scholar]
- Groom QJ, Kramer DM, Crofts AR, Ort DR. The non-photochemical reduction of plastoquinone in leaves. Photosynth Res. 1993;36:205–215. doi: 10.1007/BF00033039. [DOI] [PubMed] [Google Scholar]
- Gruszecki WI, Bader KP, Schmid GH. Light-induced oxygen uptake in tobacco chloroplasts explained in terms of chlororespiratory activity. Biochim Biophys Acta. 1994;1188:335–338. [Google Scholar]
- Guedeney G, Corneille S, Cuiné S, Peltier G. Evidence for an association of ndh B, ndh J gene products and ferredoxin-NADP-reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett. 1996;378:277–280. doi: 10.1016/0014-5793(95)01473-x. [DOI] [PubMed] [Google Scholar]
- Harris GC, Heber U. Effects of anaerobiosis on chlorophyll fluorescence yield in spinach (Spinacia oleracea) leaf discs. Plant Physiol. 1993;101:1169–1173. doi: 10.1104/pp.101.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosler JP, Yocum CF. Evidence for two cyclic photophosphorylation reactions concurrent with ferredoxin-catalyzed non-cyclic electron transport. Biochim Biophys Acta. 1985;808:21–31. [Google Scholar]
- Kuonen DR, Roberts PJ, Cottingham IR. Purification and analysis of mitochondrial membrane proteins on nondenaturing gradient polyacrylamide gels. Anal Biochem. 1986;153:221–226. doi: 10.1016/0003-2697(86)90084-9. [DOI] [PubMed] [Google Scholar]
- Leterme S, Boutry M. Purification and preliminary characterization of mitochondrial complex I (NADH:ubiquinone reductase) from broad bean (Vicia faba L.) Plant Physiol. 1993;102:435–443. doi: 10.1104/pp.102.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mano J, Miyake C, Schreiber U, Asada K. Photoactivation of the electron flow from NADPH to plastoquinone in spinach chloroplasts. Plant Cell Physiol. 1995;36:1589–1598. [Google Scholar]
- Martin M, Casano LM, Sabater B. Identification of the product of ndh A gene as a thylakoid protein synthesized in response to photooxidative treatment. Plant Cell Physiol. 1996;37:293–298. doi: 10.1093/oxfordjournals.pcp.a028945. [DOI] [PubMed] [Google Scholar]
- Matsubayashi T, Wakasugi T, Shinozaki K, Yamaguchi-Shinozaki K, Zaita N, Hidaka T, Meng BY, Ohto C, Tanaka M, Kato A and others. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol Gen Genet. 1987;210:385–393. doi: 10.1007/BF00327187. [DOI] [PubMed] [Google Scholar]
- Matthijs HCP, Coughlan SJ, Hind G. Removal of ferredoxin:NADP+ oxidoreductase from thylakoid membranes, rebinding to depleted membranes, and identification of the binding site. J Biol Chem. 1986;261:12154–12158. [PubMed] [Google Scholar]
- Mi H, Endo T, Ogawa T, Asada K. Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 1995;36:661–668. [Google Scholar]
- Mills JD, Mitchell PD, Barber J. The cyclic electron transport pathway in chloroplasts. Reduction of plastoquinone by reduced nicotinamide adenine dinucleotide phosphate in the dark. Photobiochem Photobiophys. 1979;1:3–9. [Google Scholar]
- Miyake C, Schreiber U, Asada K. Ferredoxin-dependent and antimycin A-sensitive reduction of cytochrome b-559 by far-red light in maize thylakoids; participation of a Menadiol-reducible cytochrome b-559 in cyclic electron flow. Plant Cell Physiol. 1995;36:743–748. [Google Scholar]
- Morigasaki S, Takata K, Suzuki T, Wada K. Purification and characterization of a ferredoxin-NADP+ oxidoreductase-like enzyme from radish root tissues. Plant Physiol. 1990;93:896–901. doi: 10.1104/pp.93.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Gounaris K, Coomber SA, Hunter CN, Dyer TA, Barber J. psbG is not a photosystem two gene but may be an ndh gene. J Biol Chem. 1989;264:14129–14135. [PubMed] [Google Scholar]
- O'Farrel PM. High resolution two-dimensional electrophoresis of protein. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Kohchi T, Sano T, Yamada Y. Newly identified groups of genes in chloroplasts. Trends Biochem Sci. 1988;13:19–22. doi: 10.1016/0968-0004(88)90013-8. [DOI] [PubMed] [Google Scholar]
- Peltier G, Ravenel J, Verméglio A. Inhibition of a respiratory activity by short saturating flashes in Chlamydomonas: evidence for a chlororespiration. Biochim Biophys Acta. 1987;893:83–90. [Google Scholar]
- Peltier G, Schmidt GW. Chlororespiration: an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1991;88:4791–4795. doi: 10.1073/pnas.88.11.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles MJ, Albacete ME, Sabater B, Cuello J. Isolation and partial characterization of the NADH dehydrogenase complex from barley chloroplast thylakoids. Plant Cell Physiol. 1996;37:1134–1142. [Google Scholar]
- Rasmusson AG, Mendel-Hartvig J, Moller IM, Wiskich JT. Isolation of the rotenone-sensitive NADH-ubiquinone reductase (complex I) from red beet mitochondria. Physiol Plant. 1994;90:607–615. [Google Scholar]
- Sancho J, Peleato ML, Gomez-Moreno C, Edmondson DE. Arch Biochem Biophys. 1988;260:200–207. doi: 10.1016/0003-9861(88)90441-9. [DOI] [PubMed] [Google Scholar]
- Sazanov LA, Burrows P, Nixon PJ. Detection and characterization of a complex I-like NADH-specific dehydrogenase from pea thylakoids. Biochem Soc Trans. 1996;24:739–743. doi: 10.1042/bst0240739. [DOI] [PubMed] [Google Scholar]
- Schantz R, Bogorad L. Maize chloroplast genes ndhD, ndhE, and psaC. Sequences, transcripts and transcript pools. Plant Mol Biol. 1988;11:239–247. doi: 10.1007/BF00027381. [DOI] [PubMed] [Google Scholar]
- Scheller HV. In vitro cyclic electron transport in barley thylakoids follows two independent pathways. Plant Physiol. 1996;110:187–194. doi: 10.1104/pp.110.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S, Alpes I, Sadowski H, Böger P. Ferredoxin-NADP+ oxidoreductase is the respiratory NADPH dehydrogenase of the cyanobacterium Anabaena variabilis. Arch Biochem Biophys. 1988;267:228–235. doi: 10.1016/0003-9861(88)90027-6. [DOI] [PubMed] [Google Scholar]
- Shimada H, Sugiura M. Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 1991;19:983–995. doi: 10.1093/nar/19.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Chen C, Gibbs M. Characterization of an electron transport pathway associated with glucose and fructose respiration in the intact chloroplasts of Chlamydomonas reinhardtii and spinach. Plant Physiol. 1992;100:327–333. doi: 10.1104/pp.100.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soole KL, Dry IB, Wiskich JT. Partial purification and characterization of complex I, NADH:ubiquinone reductase, from the inner membrane of beetroot mitochondria. Plant Physiol. 1992;98:588–594. doi: 10.1104/pp.98.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmüller K, Ley AC, Steinmetz AA, Sayre RT, Bogorad L. Characterization of the ndhC-psbG-ORF157/159 operon of maize plastid DNA and of the cyanobacterium Synechocystis sp. PCC6803. Mol Gen Genet. 1989;216:60–69. doi: 10.1007/BF00332231. [DOI] [PubMed] [Google Scholar]
- Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- Walker JE. The NADH-ubiquinone oxidoreductase of respiratory chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- Wu M, Nie ZQ, Yang J. The 18-kD protein that binds to the chloroplast DNA replicative origin is an iron-sulfur protein related to a subunit of NADH dehydrogenase. Plant Cell. 1989;1:551–557. doi: 10.1105/tpc.1.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Hon-nami K, Ohnishi T. Purification and characterization of two types of NADH-quinone reductase from Thermus thermophilus HB-8. Biochemistry. 1988;27:2008–2013. doi: 10.1021/bi00406a030. [DOI] [PubMed] [Google Scholar]
- Zanetti G, Curti B. Interactions between ferredoxin-NADP+ reductase and ferredoxin at different reduction levels of the two proteins. FEBS Lett. 1981;129:201–204. doi: 10.1016/0014-5793(81)80165-2. [DOI] [PubMed] [Google Scholar]