Abstract
Context
Clinical and experimental studies have suggested a link between S100 gene expression and neoplastic disorders, however, the molecular mechanisms of this association are not well understood. The aim of this review was to conduct a comprehensive literature search in order to understand the possible underlying molecular mechanisms of this association. We also discuss their application as diagnostic and prognostic markers in colorectal and hepatocellular carcinoma.
Evidence Acquisitions
We searched Pubmed (NLM) and Web of Science (ISI Web of Knowledge).
Results
S100 genes display a complex expression pattern in colorectal and hepatocel lular carcinoma. They are expressed in tumor and/or tumor stroma cells, and they exert both pro- and antitumorigenic actions. In view of this complexity, it becomes clear that S100 proteins might act as both friend and foe. The biological role of the S100 genes is predicted to depend on the relative contributions of the different cell types at specific stages of tumor progression.
Conclusions
Further research is required in order to uncover the functional role of S100 genes in tumorigenesis. Answers to this issue are needed before we can more fully understand the clinical relevance of S100 protein expression within epithelial tumors, with regard to their potential applicability as biomarkers for diagnosis and therapy decisions.
Keywords: Biological Markers; Cell Transformation, Neoplastic; Matrix Metalloproteinases
1. Context
Over the last decade a number of S100 genes have been found to be differentially expressed in cancer cells, by comparative and functional genomics. However, the molecular mechanisms by which they promote tumorigenesis and progression to malignancy remain unclear. In general, S100 overexpression is coupled with; poor tumor differentiation, aggressiveness, advanced stage, and metastatic growth. S100 expression has therefore been considered to be a negative prognosticator for patients´ survival. On the other hand, recent studies have also demonstrated that S100 genes can act as tumor suppressors in some cancer entities. The aim of this review was to conduct a comprehensive literature search to better understand the putative cellular functions of S100 proteins in the context of tumorigenesis. We discuss their complex expression patterns in tumor and stromal cells as well as their pro- and antitumorigenic actions. Furthermore, we present evidence for their application as diagnostic and prognostic markers in colorectal and hepatocellular carcinoma.
2. Evidence Acquisition
Pubmed (NLM) and Web of Science (ISI Web of Knowledge) were searched with key words ‘S100 genes’, ‘colorectal carcinoma ‘, ‘hepatocellular carcinoma’, and ‘inflammation associated tumorigenesis’, in the past 10 years.
3. Results
We were able to find 161 research and review articles relevant to this topic, either directly or indirectly. From the information given in these papers, we drew out the following aspects.
3.1. The S100 Protein Family
The S100 protein family is a multigenic group of nonubiquitous cytoplasmic EF-hand Ca2+-binding proteins, sharing significant structural similarities at both genomic and protein levels. They are differentially expressed in a wide variety of cell types (1) and have been reported to be involved in the regulation of inflammatory responses (2), as well as in the metastasis development of several cancers (3). The S100 protein family comprises 24 known human members each coded by a separate gene. At least 19 of these gene are located on chromosome 1q21. This S100 gene cluster is close to the epidermal differentiation complex (4) as well as to a psoriasis susceptibility region, the PSORS4 locus (5, 6). An additional important indication for their involvement in neoplastic disorders is that the S100 gene cluster is found near a break-point region on human chromosome 1q21 which, if affected, is responsible for a number of genetic abnormalities related to cancer (7-11). S100 proteins are p53 (12-14), NF-κB (15-19) or AP-1/Fos (20-25) target genes, thus playing an important role in inflammation-associated carcinogenesis. Although the function of S100 proteins in cancer cells is still unknown, the specific expression patterns of these proteins are a valuable prognostic tool. Several general articles on S100 proteins have been published recently (26, 27), but in this chapter we will only include details of those functions which have an impact on cancer.
S100 proteins are complex in their actions as they have both intracellular and extracellular functions. In resting cells, S100 proteins are localized intracellular. However, upon cellular activation or exposure to proinflammatory cytokines, they are either specifically released (mainly from cells of the myeloid lineage) or constitutively secreted from epithelial-like cells and tumor cells. Upon their release into the extracellular environment, they exert regulatory effects on several different cell types. S100 proteins can signal this by binding to the receptor for advanced glycation end products (RAGE), toll-like receptors (TLRs), or other receptors.
In addition, S100 proteins interact with multiple molecular targets both in a calcium-dependent and independent manner. Thereby, they regulate multiple cellular pathways that play key roles in tumor progression and metastasis.
3.1.1. Binding With Cytoskeletal Proteins Thereby Increasing Cell Migration
One molecular mechanism by which S100 proteins exert their protumorigenic effect depends on their interaction with cytoskeletal proteins resulting in an enhancement of cell migration. The number of cytoskeletal proteins includes tropomyosin (28, 29), nonmuscle myosin (30-38), actin (39, 40), and tubulin (41-43).
3.1.2. Binding With DNA Binding Factors Thereby Increasing Cell Migration
S100 proteins also modulate cell migration due to their transcriptional activity, either by direct DNA binding, or by interacting with other DNA-binding proteins. For example, S100A4 negatively regulates the expression of Ecadherin (44, 45) or reduces the expression of occludin, which is a tight junction protein (46), thus loosening epithelial cell integrity. Similarly, the overexpression of S100P leads to changes in the expression levels of several cytoskeletal proteins, including cytokeratins 8, 18, and 19. A cellular consequence of this change is a disorganization of the actin cytoskeleton network and changes in the phosphorylation status of the actin regulatory protein, cofilin (47).
3.1.3. Interaction With p53
The tumor suppressor p53 protein is a homotetrameric transcription factor that regulates several cellular processes. In response to stress (ie, DNA strand break), p53 prevents tumorigenic transformation through the induction of cell cycle arrest or apoptosis. The crucial role that this protein plays is reflected in the fact that more than 50% of human cancers contain mutations in this gene (48). In unstressed cells, the level of p53 tumor suppressor is low, however, upon stress challenge; p53 is activated through posttranslational modifications that increase its stability. The regulation of protein stability is one of the most effective mechanisms for controlling the function of p53. Key to this process is MDM2, an E3 ligase that targets p53 for ubiquitination. Several S100 proteins such as S100B (49, 50), S100A1, S100A2 (50, 51), S100A4 (46, 51) , S100A6, and S100A14 (52, 53) have been shown to interact with MDM2 (49). Direct protein-protein interaction thereby promoting its degradation, has been demonstrated in S100B (54) and S100A4 (55). Both processes result in the loss of p53-dependent tumor suppression activities. The consequences are intriguing, connecting the metastasispromoting activities of S100 proteins to the large set of important p53-mediated functions, with broad potential importance in cancer development and metastasis. In addition, S100 proteins also appear to interact with two other members of the p53 family, p63 and p73 (56). It has been discussed that the target preference of each individual S100 protein, their specific expression pattern and their individual calcium dependency, as well as the splicing variation of the p53 family, has to be considered when studying the biological effects of S100 proteins in their biological context. It is noteworthy, that potential p53 binding sites have been identified in the promoter of several S100 genes (see above), indicating that the metastasis- promoting properties of S100 proteins are not as clear-cut as has previously been suggested, this is due to their interaction with p53-dependent apoptosis. This fact may explain why some members of the S100 family are markedly down-regulated in malignant cells, in comparison to normal cells. In addition, it has been reported that S100A4 enhances p53-dependent apoptosis while S100A2 promotes p53 transcriptional activity (57).
3.1.4. S100 Proteins: Role in Degradation of Extracellular Matrix and Metastasis
Degradation of the extracellular matrix is another important factor affecting the motility of cancer cells. Herein, S100 proteins have been ascribed to contribute by various molecular mechanisms. For example, intracellular S100A14 promotes cell motility and invasiveness by regulating the expression and function of matrix metalloproteinase (MMP)-2 in a p53-dependent manner (52, 58). P53 is a transrepressor of MMP2 gene expression. Intracellular S100A14 affects p53 transactivity and stability, resulting in an enhancement of MMP2 gene expression. Extracellular S100A4 binds to RAGE and thereby induces the upregulation of MMP-13 (59, 60), MMP-2 (58), and MMP-9 gene expression (61), allowing cell invasion and thus promoting metastasis formation. S100A8/A9 overexpression also caused the upregulation of MMP-9 in HaCaT keratinocytes (62). Here, MMP9 gene induction depends on NF-κB activation, and intracellular S100A8/A9 was shown to promote epithelial NADPH oxidases and subsequently NF-κB activation. S100P induces the expression of cathepsin D, an aspartyl protease, that takes part in the proteolytic degradation of the extracellular matrix, hence it increases the invasive potential of the tumor (47).
3.1.5. Intra- and Extracellular Localization With Opposite Effects on Cell Growth
Contradictory findings have been reported for the effect of S100 proteins on cell growth. Beside p53 (see above), another checkpoint for cells, either to set about repairing themselves or to commit suicide through apoptosis, is p21/WAF1 also known as cyclin-dependent kinase (CDK) inhibitor 1 or CDK-interacting protein 1. The p21/WAF1 protein functions as a regulator of cell cycle progression at G1 via inhibition of cyclin-CDK2 or -CDK1 activity. The expression of this gene is tightly controlled by p53, through which this protein mediates the p53-dependent cell cycle G1 phase arrest, in response to a variety of stress stimuli. Accumulation of S100A11 in the nucleus has been observed in normal human keratinocytes by high calcium concentrations or TGF-ß exposure, where it induced p21/ WAF1 (63, 64). In contrast, S100A11 has been shown to be overexpressed in many human cancers. This apparent discrepancy has been resolved by the finding that the production and secretion of S100A11 are markedly enhanced in human squamous cancer cells. Extracellular S100A11 binds to RAGE thereby enhancing the production of epidermal growth factor family proteins and this results in growth stimulation (65).
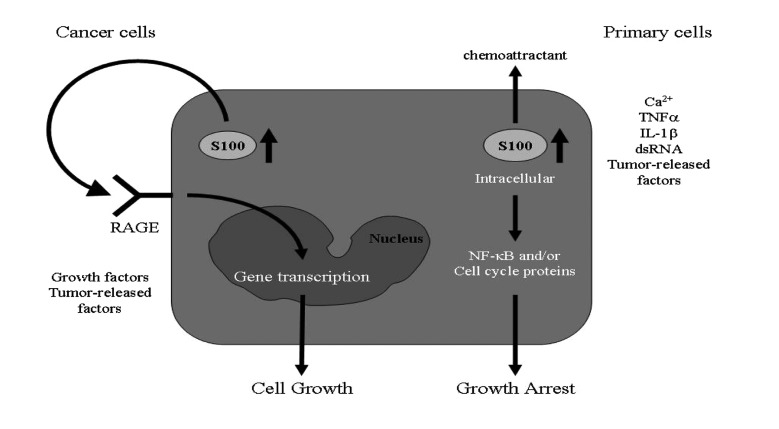
In human hepatocellular carcinoma cells (HCC), TGF-ß has been demonstrated to induce growth suppression (66). S100A11 has also been shown to be involved since TGF-ß induces S100A11 gene expression and translocation into the nucleus, where it interacts with p21/Waf1. In addition, S100A8/A9 appears to have both growth-stimulatory and anti-proliferative properties. Extracellular S100A8/ A9 stimulates the proliferation of normal human keratinocytes (67), while intracellular S100A8/A9 functions as a mediator for growth suppression (62). Thus, S100A8/A9 plays an ambivalent role with respect to the growth regulation of keratinocytes, and the biological effects of S100 proteins mediated by intracellular versus extracellular actions, have to be carefully elucidated (Figure 1).
Figure 1. In Resting Epithelial Cells, S100 Proteins are Primarily Localized in the Cytoplasm.
3.1.6. Extracellular S100 Proteins: Interaction With RAGE
Another important feature is that S100 proteins form heteromers, as well as higher order complexes, and these complexes display different affinities to target proteins depending on their oligomerization state. This has been demonstrated for p53 and RAGE (56, 68). Moreover, extracellular S100 proteins bind to various receptors and they also bind to different epitopes on individual receptors. RAGE has been shown to transduce the extracellular effects of S100B, S100A4, S100A6, S100A8/A9, S100A11, S100A12, S100A13, and S100P. For S100B and S100A6, it has been shown that they not only interact with distinct RAGE immunoglobulin domains, but they also exert opposite effects on cell survival. At similar concentrations, S100B increased cellular proliferation, whereas S100A6 triggered apoptosis. In addition, both S100 proteins induced the formation of reactive oxygen species; however, S100B recruited phosphatidylinositol 3-kinase/AKT and NF-κB, whereas S100A6 activated JNK. These data indicate the complexity of S100/RAGE cellular signaling (69).
Another layer of complexity is attributed to the fact that some S100 proteins exert both protumorigenic and antitumorigenic effects. The prosurvival activity of S100B depends on its binding to RAGE, RAGE-induced activation of MEK–ERK1/2–NF-κB pathway, and the up-regulation of anti-apoptotic factor bcl-2 (70). On the other hand, high concentrations of S100B induce apoptotic cell death (70). Remarkably, the bimodal function of S100B is mediated by RAGE engagement. In contrast, S100A8/A9 also exerts both growth-stimulating and apoptosis-inducing activities; however, these cellular effects are mediated by distinct receptors. S100A8/A9 exerts its apoptosis-inducing activity through the mitochondrial pathway since overexpression of the mitochondrial transmembrane deficient form of the proapoptotic BNIP3 (ΔTM-BNIP3) partially reversed the cytotoxicity of S100A8/A9 (71, 72). Further features are a drop in the mitochondrial membrane potential; Bak and BNIP3 activation, selective translocation of Smac/Diablo and Omi/HtrA2, and decreased Drp1 expression. RAGE gene knockdown and RAGE blocking experiments have demonstrated that the apoptosisinducing activity of S100A8/A9 was not mediated by RAGE ligation. However, the growth-promoting activity obvious at low micro molar concentrations was mediated by RAGE (71). In another study, the first evidence which was provided is that S100A8/A9 induces both apoptosis and autophagy. S100A8/A9-induced cell death involves BNIP3 and an increase in reactive oxygen species (ROS) production by the mitochondria, subsequently this is followed by mitochondrial damage and lysosomal activation. These data indicate that ROS is the critical factor that integrates S100-induced mitochondrial and lysosomal death pathways (73).
3.2. S100 Gene Expression in Tumor Stroma Cells
Comparative and functional genomics revealed a number of S100 proteins to be differentially expressed in cancer cells. Some of these studies are hampered by the fact that there has not been sufficient discrimination between the expression within the tumor or the tumor stroma cells. Thus, although differential expression has been noted in tumors, the cellular role of S100 proteins in tumorigenesis needs to be defined in the context of their cell-type specific expression pattern. Tumors are heterogeneous organs containing myofibroblasts, endothelial cells, inflammatory cells, vascular cells and other cells (see below), in addition to malignant cells. These tumor-companion cells are intermingled in the tumor-associated stroma that comprises most of the tumor mass in many carcinomas. The stromal microenvironment of tumors is involved in the suppression of immune control, in the promotion of neoangiogenesis, in the proliferation and invasion of carcinoma cells. A dynamically molecular crosstalk takes place between the cancer and stroma cells, mostly in the form of growth factor signaling, and this contributes to niche formation for the metastasis. The relevance of the reactive stroma that surround the cancer cells, is highlighted by the fact that the molecular properties of tumor stroma are predictive of disease outcomes (74). Research, aiming to establish the roles of the tumor-associated stroma in facilitating the spread of carcinoma cells into distant organs, has provided an abundance of data and greater knowledge of the biology of metastatic carcinoma and associated stromal cells (75). This has stimulated further advances in the development of novel therapeutic approaches targeting tumor metastasis. Thus, understanding the roles of S100 proteins in cancer presumes the cell-type(s) in which the S100 proteins are expressed and released, and the molecular mechanisms by which their gene expression is regulated. Finally, the investigation of the cellular responses and the underlying mechanisms, whereby these proteins mediate their biological effects, may offer new therapeutic strategies to prevent, treat, and predict cancer.
3.2.1. Fibrocytes
CD34+ fibrocytes are constitutive elements of the connective tissue, where they play a role in matrix synthesis and tumor-associated stromal remodeling. In colorectal adenocarcinoma (CRC), normal colonic stroma harbor CD34+ fibrocytes, whereas this cell population is absent from the stroma of invasive adenocarcinoma (76). They are a distinct population of leukocytes with the characteristics of fibroblasts. They are capable of producing matrix proteins, such as fibroblast collagen type 1 and 3 (77, 78), and they are fully functional in their presentation of antigens to naive T-cells. Fibrocytes play a prominent role in fibrosis, and they can be discriminated from fibroblasts in fibrotic lesions by a set of specific markers, including S100A8/A9 (79). To date, there has been no data available concerning the cellular role of S100A8/A9, or other S100 proteins in fibrocytes in the context of tumorigenesis.
3.2.2. Dendritic Cells
The clinical significance of tumor-infiltrating dendritic cells (DCs) has been reported in a variety of human solid tumors, as shown by the correlations found between the presence of tumor-infiltrating dendritic cells and the clinical prognosis (80-83). S100A4 (84), S100A6 (84), S100A8 (85), S100A9 (85), S100A12 (86, 87) and S100B (87) have been found in DCs. A significant role in DCs has been shown for S100A8/A9 (85). S100A8/A9 expression is found in both myeloid DCs and plasmacytoid DCs. DCs of S100a9-/- mice promote higher T cell proliferation compared to wild-type DC in mixed lymphocyte reactions, and they express more DC cell surface markers, (CD205, IA) and co-stimulatory molecules (CD40, CD86) compared to wild-type cells after lipopolysaccharide (LPS) stimulation (88). In addition, S100A9 regulates B7 molecule expression and reduces antigen presentation by DCs and subsequent T-cell priming. The absence of S100A9 markedly increases T-cell activation and exacerbates allograft rejection (89).
Consistent with these results, DC differentiation is blocked by S100A9 overexpression (86). The same study showed that up-regulation of S100A8 and S100A9 by tumor- derived factors might represent one major mechanisms by which abnormal DC differentiation is established in cancer (90-93).
3.2.3. Myeloid-Derived Suppressor Cells
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of immune cells that accumulate in tumor-bearing hosts, as well as in response to inflammation. In mice, these cells are defined by the surface expression of CD11b and Gr-1 for granulocytic MDSCs or CD11b, Gr 1, CD115, and F4/80 for monocytic MDSCs, respectively (86). To date, only two members of the S100 protein family, S100A8 and S100A9, have been detected in MDSCs. Interestingly, mice lacking S100A9 showed significantly reduced tumor incidence, growth and metastasis, reduced chemokine levels, and reduced infiltration of CD11b+ Gr1+ cells within tumors and pre-metastatic organs. Studies using bone marrow chimeric mice revealed that S100A8/ A9 expression on myeloid cells is essential for the development of colon tumors (94). MDSCs synthesize and secrete S100A8/A9, and the binding of S100A8/A9 not only induces MDSCs via RAGE activation, but it also promotes MDSC migration (95). Furthermore, S100A8/A9+ MDSCs impair tumor immunity by inhibiting T and NK cell activation, by polarizing immunity toward tumor-promoting type 2 phenotypes, and by inhibiting DC differentiation. These responses are considered to be responsible for the limited effectiveness or failure of cancer vaccines and other immunotherapies (85). Tumor cells and Mac-1+myeloid cells utilize a common pathway for their migration to the lung (96). S100A8/A9 also exerts chemotactic activity for Mac-1+ myeloid cells. After preparation of the target tissue to accept the malignant cells, tumor cells mimic Mac-1+ myeloid cells in response to S100A8/A9 chemotactic signaling and they migrate to the lung. These findings suggest that S100A8/A9 is an attractive target for the development of future strategies to counteract the tumor metastasizing to certain organs. In accordance with this suggestion, the suppression of Mac-1+ myeloid cells was shown to enhance tumor response to radiation (97). As mentioned above, S100A4 plays an important role in tumor metastasis, due to its effects on tumor cell migration. Recently, it has been shown that S100A4 is expressed in macrophage chemotaxis (98). S100A4 deficiency affected chemotactic motility of bone marrow macrophages in vitro and impaired recruitment of macrophages to sites of inflammation in vivo. In addition, S100A4-/- bone marrow macrophages formed unstable protrusions, over assembled myosin-IIA, and exhibited altered colony-stimulating factor-1 receptor signaling. This study established S100A4 as a regulator of physiological macrophage motility. It is becoming increasingly clear that S100 proteins have distinct functions in different cell types which are involved in metastasis development of several cancers. Further studies are required to elucidate whether the effects of S100 proteins are mediated by intracellular or extracellular mechanisms and whether they modulate cellular responses in tumor stroma cells and/or tumor cells.
3.3. S100 Gene Expression in Tumor Cells
The family of S100 Ca2+-binding proteins is closely related to several tumors, including those of the colon and the liver. Table 1 summarizes the expression of S100 genes in different stages of tumor progression within these two tumor entities.
Table 1. S100 Expression in Different Stages of Tumor Progression.
| S100 Expression | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Colorectal Carcinoma (119) | A1 (153) | A2 (153) | A3 (153) | A4 (99, 154) | A5 (153) | A6 (107, 109, 153) | A7 | A8 (109) | A9 (109) | A10 | A11 (109, 155) | A14 (156) | B (112) | P (133-135, 140) | G |
| Normal epithelium | expression at very low level | expression at very low level | expression at very low level | expression at very low level | protein expression verified by IHC and/or WB | protein expression verified by IHC and/ or WB | N.D. | protein expression verified by IHC and/or WB | protein expression verified by IHC and/or WB | N.D. | protein expression verified by IHC and/or WB | protein expression verified by IHC and/ or WB | protein expression verified by IHC and/or WB | expression at very low level | N.D. |
| Preneoplastic adenomas | expression at very low level | N.D. | N.D. | protein expression verified by IHC and/or WB | reduced/ lost expression compared to normal tissue | protein expression verified by IHC and/ or WB | N.D. | protein expression verified by IHC and/or WB | stage-dependent overexpression | N.D. | protein expression verified by IHC and/or WB | protein expression verified by IHC and/ or WB | protein expression verified by IHC and/or WB | protein expression verified by IHC and/or WB | N.D. |
| Neoplastic primaries | reduced/ lost expression compared to normal tissue | N.D. | N.D. | overexpression compared to normal tissue | reduced/ lost expression compared to normal tissue | stage-dependent overexpression | N.D. | stagedependent overexpression | stage-dependent overexpression | protein expression verified by IHC and/or WB | stage-dependent overexpression | reduced/ lost expression compared to normal tissue | stagedependent overexpression | stagedependent overexpression | N.D. |
| Metastatic lesions | reduced/ lost expression compared to normal tissue | N.D. | N.D. | stagedependent overexpression | reduced/ lost expression compared to normal tissue | stage-dependent overexpression | N.D. | stagedependent overexpression | stage-dependent overexpression | N.D. | protein expression verified by IHC and/or WB | expression at very low level | stagedependent overexpression | stagedependent overexpression | N.D. |
| Hepatocellular carcinoma | A1 (157) | A2 | A3 | A4 (145) | A5 | A6 (142) | A7 | A8 (17) | A9 (17) | A10 (158) | A11 (159) | A14 (156) | B | P (160) | G |
| Normal epithelium | N.E. | N.D. | N.D. | expression at very low level | N.D. | N.E. | N.D. | expression at very low level | protein expression verified by IHC and/or WB | expression at very low level | expression at very low level | protein expression verified by IHC and/ or WB | N.D | N.E. | N.D. |
| Preneoplastic lesions | N.D. | N.D. | N.D. | expression at very low level | N.D. | protein expression verified by IHC and/ or WB | N.D. | protein expression verified by IHC and/or WB | protein expression verified by IHC and/or WB | N.D. | N.D. | N.D. | N.D. | protein expression verified by IHC and/or WB | N.D. |
| Neoplastic primaries | N.D. | N.D. | N.D. | stagedependent overexpression | N.D. | stage-dependent overexpression | N.D. | overexpression compared to normal tissue | overexpression compared to normal tissue | protein expression verified by IHC and/or WB | N.D. | N.D | N.D. | stagedependent overexpression | N.D. |
| Metastatic lesions | N.D | N.D. | N.D. | stagedependent overexpression | N.D | stage-dependent overexpression | N.D. | overexpression compared to normal tissue | overexpression compared to normal tissue | overexpression compared to normal tissue (161) | overexpression compared to normal tissue (161) | N.D. | N.D. | stagedependent overexpression | N.D. |
Abbreviations: IHC, immunohistochemistry; N.D., no data available; N.E., no expression; WB, Western blotting
3.3.1. S100 Proteins as Biomarkers for Diagnosis and Staging of Colorectal Carcinoma?
CRC development is the result of accumulating genetic and epigenetic alterations that transform normal epithelial cells into invasive adenocarcinomas. This event may either be triggered by exogenous stimuli (eg, high alcohol consumption, smoking and/or high body weight), or chronic intestinal inflammation. Besides these initiators, CRC may also arise in the context of hereditary origins. Despite improved screening and treatment strategies, CRC remains the third leading cause of cancer-related deaths in the world. Therefore, identifying novel biomarkers for the early diagnosis of CRC which will help to improve patients´ outcomes is essential. Examining S100 protein expression has gained a great deal of attention, since these proteins have been implicated in tumor development, metastasis and, in some cases, even chemoresistance. In addition to tests based on analyzing blood or serum samples, S100 proteins may potentially be used as additional markers for tagging patients bearing pre-malignant adenomas of the colorectum. Analysis may ideally be performed on biopsies obtained following a colonoscopy. This may also be conducive in the development of an improved screening protocol. S100A4 (metastasin) is a target gene of the Wnt/ß-catenin pathway, which is constitutively active in most CRC cases. Overexpression was shown to be associated with lymph node metastasis, advanced TNM stage, and poor outcomes (ie, increased recurrence and decreased overall survival), but not Duke subtypes of CRC (99-101). Thus, it was suggested that S100A4 may be necessary for dissemination to distant metastatic sites rather than for local tumor invasion, and it could possibly be a good biomarker for metastasis and prognosis (102). Boye et al. have identified differences in CRC prognosis in terms of cellular localization. Nuclear S100A4 expression was a negative predictor of metastasis- free and overall survival, whereas cytoplasmic S100A4 was not associated with patient outcomes (103). On the contrary, circulating S100A4 is not suitable as an ideal tumor marker, since red blood cells and mononuclear cells also contain S100A4. This can lead to false positive plasma levels in hemolytic samples (104). Whether S100A4 is a useful biomarker or not for CRC has to be carefully re-evaluated. More consistency exists for S100A14, whose expression is inversely correlated with S100A4. Epithelial S100A14 expression was found to be associated with lower invasiveness, metastatic potential and better differentiation, compared to S100A4+ cases (105). Interestingly, reduced expression of S100A14 tends to co-occur with increased S100A4 expression in CRC cell lines and tissue specimens. Therefore, S100A14 is a useful prognostic marker for CRC patients (105). Likewise, S100A6 (calcyclin) might function as a valuable biomarker for the early diagnosing of colonic transformation. It was the first S100 protein to be specifically related to the state of cellular proliferation. S100A6 has two EF-hand motifs. Although S100A6 protein levels can be found in normal colonic mucosa, their expression is dramatically increased during the course of malignant transformation (106). S100A6 is preferentially expressed in the G1 phase of the cell cycle. It has been implicated in regulating cell growth and proliferation via interaction with various ligands, eg, Ca2+/phospholipid-binding proteins of the annexin family and glyceraldehyde-3-phosphate dehydrogenase (107, 108). Besides these quantitative changes, Stulik et al. have also identified differential isoforms in normal and malignant colonic mucosa (107). Clinically, S100A6 expression is significantly correlated with two parameters; lymphatic permeation and Duke’s tumor status (107). S100A8 and S100A9 proteins, usually occurring as the heterodimeric complex, calprotectin, are changed significantly at the early stages of cancer. Plasma levels of S100A8 and S100A9 increase appreciably in patients with colorectal adenomas and established CRCs (109). Therefore, these proteins proteins might, in combination with other serum markers, serve as candidates for serological biomarkers for a CRC diagnosis (110). However, they have not proved effective as a candidate stool marker for detecting CRC. In a clinical study of 77 CRC patients, levels of fecal S100A9 were significantly higher than in the controls, but S100A8 did not show any difference. With regard to sensitivity and specificity, there was no benefit in analyzing S100A9, compared to the conventional fecal occult blood test (111).
Serum S100B protein concentration in stage II-IV melanoma is a reliable prognostic marker. However, no data exists for circulating S100B levels in CRC patients. A recent study found that overexpression within tumors is a more important predictor than merely a conventional pathological variable (112). As determined by immunohistochemistry, S100B overexpression, together with the presence of vascular and perineural invasion, as well as a high postoperative CEA level, was found to be an independent predictor of postoperative early relapse (112). On the contrary, positive S100B protein expression was only found in 6% of non-early relapsed CRC patients. Thus, determining the S100B expression status would probably help to identify patients with a high-risk for early relapse. These patients could then be monitored more intensively and treated accordingly. S100P expression is not restricted to neoplastic cells, but it is also detectable in various normal cell types (eg, enterocytes and epithelial cells). It is therefore not expected to be useful for diagnostic and therapeutic applications (113). Nevertheless, a recent study proposed the likelihood of predicting CRCassociated hepatic metastasis by examining S100P-expression (114). This conclusion follows the observation of increased expression levels during disease progression. Further studies are needed to show if S100P determination will be clinically useful.
3.3.2. S100 Expression and Chronic Inflammation – a Direct Link Towards Tumor Progression?
Several S100 protein family members are differentially expressed in inflammatory bowel diseases, ie, ulcerative colitis and Crohn’s disease, and these have been implicated in inflammation-induced colorectal carcinogenesis (115). Brentnall et al. detected increased expression profiles of S100P, S100A6, S100A11, and S100H during the course of malignant transformation (116). On the other hand, S100A8, S100A9, S100A10, and S100A4 displayed heterogeneous expression patterns in ulcerative colitis patients showing neoplasia (116). Another study proposed that S100A8/A9 expression is an early step in neoplastic transformation during inflammation-associated carcinogenesis. Accordingly, immunohistochemical studies identified S100A8 and A9 expressing macrophages and polymorphonuclear leukocytes along the invasive margin of CRC specimens (109). In normal and chronically inflamed colon mucosa, diffuse staining is usually ob served. Whether this can be seen as an immunological defense mechanism, helping local tumor growth control by chemotactically attracting immune cells to the local microenvironment, or rather forces tumor invasion and metastasis by keeping activated immune cells away from the central tumor burden, is largely unknown. However, the fact that (cytotoxic) T cells can usually be largely found at the invasive margins of CRCs, but only rarely within tumors, hints towards the latter hypothesis. As stated earlier, S100 proteins interact functionally with RAGE. The S100–RAGE signaling pathway plays an important role in linking inflammation and cancer, as well as in tumor cell survival and malignant progression (117). RAGE-deficient tumors are characterized by accelerated apoptosis, reduced NF-κB activation and significantly impaired proliferation (118). Further studies are needed to understand the significance of S100–RAGE interaction in inflammation-induced carcinogenesis in order to assess its appropriateness as a potential target for therapeutic strategies.
3.3.3. S100 Protein Expression in Primary CRCs, Local and Distant Metastases as Well as Possible Implications for Therapeutic Approaches
Local and in particular, distant metastases, are the principal causes of tumor-related complications and death. A common site of metastases derived from the colon following its spread into regional lymph nodes, is the liver. In addition, the presence or absence of lymph node metastasis determines the particular treatment regimen. Metastases are involved in tumor initiation and promotion, uncontrolled proliferation, angiogenesis, invasion, intra- and extravasation, as well as colony formation at the liver site (119). In recent years, several S100 proteins have gained interest because of their involvement in CRC-associated liver metastasis. Despite the well known contribution of S100A4 in mediating CRC progression and driving metastasis, the exact mechanism of how it interacts with other intracellular proteins and exerts pro-metastastic effects remains elusive. Two other S100 family members, namely S100A2 and B were shown to interact with the tumor-suppressor p53 at the transcriptional level and down-regulate wildtype p53 (50). This may drive tumorigenesis. Thus, S100A4 was thought to interact physically and in a regulatory manner with p53, too. However, recent in vitro data indicate that although S100A4 and p53 are present in the same cells, no reciprocal regulatory mechanisms exist (55, 101). Irrespective of the intracellular p53 status, S100A4 levels increase parallel to the number of epithelial cells in the S phase (107). In this regard, calcimycin, a calcium ionophore and transcriptional inhibitor of S100A4, restricts cell motility in CRC cells and inhibits metastasis formation in the intrasplenic HCT116 xenograft mouse model (120). The very same research group also identified that niclosamide, which is an established antihelminthic drug, may act as a potential therapy against S100A4-mediated metastatic colon tumors (121). In addition, a very recent study provided evidence for the suppression of cell growth, migration and invasiveness by specifically silencing S100A4. Metastasis- related genes MMP9, MMP10 and CDH11 were found to be down-regulated after S100A4 suppression, thus indicating a specific association of S100A4 with multiple molecules (122). Likewise, chemosensitization towards methotrexate was achieved by silencing S100A4 in HT-29 CRC cells, which normally do not respond to this cytostatic drug (123). Comparable results were obtained for other chemotherapeutics, suggesting that S100A4 expression can be considered as a biomarker of resistance (124). By 1998, Nakamura and Takenaga had already described the functional mechanisms underlying S100A4 expression in CRCs. They found that hypomethylation of the second intron triggers its expression (125). This supports the emerging concept that hypomethylation may play a role in the up-regulation of genes during later stages of tumorigenesis. Co-expression of S100A6 and S100A11, allows for the discrimination between; primary CRC, liver metastasis, and primary HCC, as well as between CRC and HCC (119). S100A6 and A11 regulate intracellular activities such as; cell growth and motility, cell-cycle progression, transcription, and cell differentiation. Both have been found in epithelial tumors and are linked to metastasis (106). Of particular interest is the finding that S100A6, but not S100A11, is a potential candidate to distinguish between CRC-derived liver metastasis and primary HCC, which is currently a complicated procedure (119). However, S100A11 has just been identified as a valuable marker for regional lymph node metastasis. Immunohistochemical S100A11 expression is significantly correlated with the nodal status, but not tumor depth or grading (126). No studies have so far analyzed the biological functions of S100A6 and/or A11 on chemoresponsiveness.
S100A10 is a plasminogen receptor and this has been suggested as being a promoter of angiogenesis and tumor metastasis. S100 deficiency is associated with early cessation of tumor growth in vivo and this is thought to be due to reduced recruitment of tumor-promoting macrophages to the tumor site (127). Despite lacking firm clinical data, it has been shown that S100A10 silencing decreases plasminogen-dependent invasiveness in a CRC cell line (128). Of particular interest from a therapeutic point of view, is the finding that intracellular S100A10 protein expression levels correlate with sensitivity of CRC cells to oxaliplatin, but not 5-FU or irinotecan (129, 130). Oxaliplatin, a third-generation platinum compound, is commonly used as a combination therapy (=FOLFOX) for CRC patients. However, approximately half of these patients will be unlikely to benefit from this treatment regimen due to chemoresistance (129). Assessing the S100A10 status could thus facilitate prediction of individual responsiveness towards platinum-based chemotherapies. Comparably little is known about the functional contribution of S100B to CRC progression and metastasis. S100B binds to different oligomeric forms of p53 and regulates its activity. S100B binding to p53 disables its biological function as a tumor suppressor and probably drives carcinogenesis. These activities are mediated via interactions with target proteins such as; annexins, cytosolic phospholipase A2, the Ca2+ release channel of the sarcoplasmic reticulum, and myosin (131). In Taiwanese CRC patients, liver metastasis is correlated with S100B overexpression (132).
Besides pancreatic and breast cancer, S100P, originally isolated from a placenta, is overexpressed in tumors of the colorectum. Here, elevated S100P levels have been found in epithelial cells (113, 133). S100P is linked to immortalization, malignancy, hormone-independency, chemoresistance and poor clinical outcomes (134). S100P expression, among other genes, seems to be essential for CRC liver metastasis (114). In a similar vein, S100P expression correlates with two other metastasis-inducing proteins, S100A4 and osteopontin. Recently, Chandramouli et al. shed light on the regulatory mechanisms of this pro-tumorigenic molecule. They showed that S100P is significantly up-regulated by prostaglandin E2 (PGE2) via the EP4 receptor, which is overexpressed in epithelial CRC cells. Hence, pro-tumorigenic PGE2 stimulation activates downstream signals (ie, ERK phosphorylation and NF-κB activation) and this influences morphology, colony growth, tumor cell motility, and chemoresponsiveness (135).
Elevated NF-κB activity is linked to increased chemoresistance (136, 137). Bertram et al. observed doxorubicin resistance in S100P-expressing CRC cells (138), while S100P absence or knockdown inhibits CRC cell growth, migration and invasion in vitro, as well as tumor growth and liver metastasis in a xenograft nude mouse model in vivo (139, 140). S100P regulation by the PGE2/EP4 receptor signaling pathway may therefore constitute a feedback regulatory mechanism by which tumor cells perpetuate growth and migration during carcinogenesis.
3.4. Hepatocellular Carcinoma
Tumors in the liver arise due to; chronic liver injury, inflammation, and hepatocyte proliferation, and this can be provoked by several causes such as; chronic hepatitis B and C viral infections, chronic alcohol consumption, and aflatoxin B1–contaminated food (141). Screening for HCC has so far not improved the prognosis of this condition. Hence, novel markers for early disease detection and therapy decision making are warranted. Here, we discuss the current value on S100 protein detection within HCC.
3.4.1. S100 Proteins as Biomarkers for Hepatocellular Carcinoma Diagnosis and Staging?
S100A6 is very specific marker for malignancy, since expression is absent in normal, non-cancerous liver tissue. This signal transduction molecule forces cell proliferation, differentiation, migration, and cytoskeletal dynamics. S100A6 expression, particularly when associated with osteopontin, another pro-metastatic protein and high a-fetoprotein levels, are associated with poor tumor differentiation. It may therefore be a promising diagnostic marker and therapeutic target for HCC (142). Comparing S100A9 expression in 70 HCC with non-carcinomatous hepatocytes and bile duct epithelia revealed exclusive immunoreactivity in tumor cells. These findings hint towards a neo-expression in differentiated malignant hepatocytes, which is related to deprived tumor differentiation. However, no correlation exists with regard to vascular invasion (143). Therefore, S100A9 may be involved in initial tumor formation rather than in metastasis.
3.4.2. Detecting S100 Proteins in HCC Cell Lines and Primary as Well as Metastatic Lesions With Possible Implications for Therapeutic Approaches
Very few studies have so far analyzed S100P expression in HCC, despite its well-known association with a variety of neoplastic diseases. Liver cancer cell lines express higher S100P levels than normal non-malignant hepatocytes, which frequently lack S100P. In an in vitro study, Kim et al. showed that endogenous S100P silencing decreases HCC cell growth. The anti-mitogenic effect could be partially attributed to cell cycle deregulation and apoptosis induction. Nevertheless, as opposed to CRCs, silencing of endogenous S100P by siRNA did not alter phosphorylation or protein expression levels of the S100-downstream molecules ERK and NF-κB (110). The functional role of S100P on liver carcinogenesis, therefore, still remains elusive. In situ, S100P expression gradually increases from dysplasia, precancerous lesions, to HCC. Hence, S100P, which is known to act as an aggressiveness factor in HCC cells, may be conducive for HCC formation or progression. S100A6 contributes to cell proliferation and invasion in different ways. These include activin A inhibition and transcription factor 3 activation. This can affect MMP functions (eg, MMP-2) and cytoskeletal reorganization in conjunction with cathepsin D (142). Besides, cellular S100A6 expression is inducible by NF-κB (p65) following TNF-α stimulation (18).
S100A4 expression is associated with epithelial mesenchymal transition (EMT), a tumor-cell invasion process that results in the loss of epithelial characteristics and the acquisition of mesenchymal features. In HCC, S100A4 expression, together with other EMT-related proteins, is synonymous with metastasis as well as decreased diseasefree and overall survival (144). Hence, S100A4-expressing tumor cells have a much stronger metastatic ability than their S100A4-negative counterparts (145). In cholangiocarcinoma cell lines, S100A4-silencing leads to reduced motility, invasiveness, and MMP-9 secretion in vitro. Up to now, S100A4 function has remained largely unknown, as it is still uncertain if S100A4 is just a biomarker of cellular aggressiveness or represents a functional target for therapeutic interventions. S100A8 and S100A9 are NF-κB target genes in HCC cells during inflammation-associated liver carcinogenesis. While both proteins are seldom found in normal liver tissues, their expression increases with malignant transformation and facilitates disease progression. The underlying molecular mechanism has been described as due to activation of ROS-dependent signaling pathways and protection from TNF-a induced cell death (17). Possible consequences for the clinics are still pending.
S100A11 has an EF-hand domain and mediates growth inhibition in normal fibroblasts through p21/Waf1 activation. In an experimental study, comparable effects were described for human HCC cells, in which proliferation was blocked following TGF-ß treatment (66). TGF-ß is an important physiological inducer of apoptosis in HCC cells that exerts its various effects via two transmembrane serine/threonine kinases. This may hint towards another role of S100A11, not only its function as a pro-tumorigenic molecule, but also as a growth inhibitor. These findings fit well with the ambiguous and varying role S100A11 plays between different tumor entities.
3.4.3. Latent Chronic Liver Inflammation and the Relevance of S100 Proteins?
Chronic inflammation represents a high risk factor for the development of various cancers, and S100 proteins have been implicated in many aspects of the interaction between tumor, stromal, and immune cells contributing to the formation of an inflammatory tumor microenvironment (146). Apparently, liver cancer is not related to chronic inflammation; however, persistent viral or bacterial inflammation has been demonstrated to initiate or trigger HCC. The prevalence of chronic hepatitis B (circular DNA virus) and C (positive-strand RNA virus) infection is directly linked to HCC. While HBV integrates into the host genome and has reverse transcriptase activity, HCV does not. Common to both viruses is an immune-mediated inflammatory response that either clears the infection or slowly destroys the liver. Viruses are recognized by the hosts’ immune system as pathogen-associated molecular patterns via TLRs (e.g. TLR 3 and 7). This activates a signal cascade, leading to NF-κB activation, which finally links inflammation and immunity to cancer development and progression. Despite the relevance of S100 proteins in inflammation and cancer, these proteins have rarely been studied in virus-driven HCC. HCV-associated carcinogenesis arises as a consequence of complicating cirrhosis. The process of liver fibrosis following a HCV infection may be accompained by EMT of the bile duct epithelial cells. In situ as well as experimentally, EMT is induced by TGF-ß1, transforming normal HCV infected cells into invasive cells. Moreover, TGF-ß1 induces S100A4 expression upon LPS stimulation of human intrahepatic biliary epithelial cells (147) and elevated levels of S100A4-positive cells are found in all forms of chronic liver disease, including chronic HCV infection, alcoholic liver disease, non-fatty liver disease, hereditary hemochromatosis, and cryptogenic cirrhosis (148). Hence, S100A4 expression is directly linked to inflammation-induced carcinogenesis.
In the case of HBV, HCC is habitually found in non-cirrhotic livers. S100A10 inhibits the HBV Pols’ DNA polymerase activity (149). Besides this, serum S100B seems to be a sensitive marker of endothelial and tissue damage in chronic HCV hepatitis (150). Interestingly, while S100A8/A9 is actively involved in chronic bowel inflammation, this complex is supposed to exert the opposite function in the liver by indirectly suppressing nitrogen oxide overproduction from activated neutrophils and/or macrophages. In a rat model of acute LPS-induced inflammation, the activity of proinflammatory cytokines could be neutralized by intraperitoneal injections (151). Hence, S100A8/A9 is rather more involved in liver regeneration than in promoting inflammation. Further indirect evidence is given by the observation that S100A8/A9 gene expression is induced in virus-infected keratinocytes via TLR3 ligation (152).
4. Conclusions and Future Perspectives
The family of S100 Ca2+-binding proteins is closely related to several tumors including those of the colon and liver. In general, S100 overexpression is coupled with poor tumor differentiation, aggressiveness, advanced stage, and metastatic growth. Therefore, S100 expression is considered to be a negative prognosticator for patient’s survival. However, recent data has suggested that this conclusion is too short-sighted and S100 proteins may therefore be regarded as playing both positive and negative roles. Further research is therefore required in order to discriminate the complex expression patterns of S100 proteins in tumor and stromal cells, as well as in their pro-tumorigenic and anti-tumorigenic actions. Answers to these issues are needed, before we can more fully understand the clinical relevance of S100 protein expression within epithelial tumors with regard to their potential applicability as biomarkers, for diagnosis and therapy decisions.
Acknowledgments
None declared.
Footnotes
Implication for health policy/practice/research/medical education: Clinical and experimental studies have suggested a linkage between S100 gene expression and neoplastic disorders, however, the molecular mechanisms of this association are not well understood. In this review, we discuss their application as diagnostic and prognostic markers in colorectal and hepatocellular carcinoma.
Please cite this paper as: Maletzki C, Bodammer P, Breitrück A, Kerkhoff C. S100 Proteins as Diagnostic and Prognostic Markers in Colorectal and Hepatocellular Carcinoma. Hepat Mon. 2012; 12(10 HCC): e7240. DOI: 10.5812/hepatmon.7240
Authors’ Contribution: All authors took part in reviewing the literature, writing the manuscript, and revising it for final publication.
Financial Disclosure: None declared.
Funding/Support: These studies were supported in part by “Deutsche Forschungsgemeinschaft”, projects KE 820/6-1 and KE 820/2-4 (CK).
References
- 1.Schafer BW, Heizmann CW. The S100 family of EF-hand calciumbinding proteins: functions and pathology. Trends Biochem Sci. 1996;21(4):134–40. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 2.Nacken W, Roth J, Sorg C, Kerkhoff C. S100A9/S100A8: Myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc Res Tech. 2003;60(6):569–80. doi: 10.1002/jemt.10299. [DOI] [PubMed] [Google Scholar]
- 3.Ghavami S, Chitayat S, Hashemi M, Eshraghi M, Chazin WJ, Halayko AJ, et al. S100A8/A9: a Janus-faced molecule in cancer therapy and tumorgenesis. Eur J Pharmacol. 2009;625(1-3):73–83. doi: 10.1016/j.ejphar.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 4.Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex ("epidermal differentiation complex") on human chromosome 1q21. J Invest Dermatol. 1996;106(5):989–92. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- 5.Hardas BD, Zhao X, Zhang J, Longqing X, Stoll S, Elder JT. Assignment of psoriasin to human chromosomal band 1q21: coordinate overexpression of clustered genes in psoriasis. J Invest Dermatol. 1996;106(4):753–8. doi: 10.1111/1523-1747.ep12345807. [DOI] [PubMed] [Google Scholar]
- 6.Semprini S, Capon F, Tacconelli A, Giardina E, Orecchia A, Mingarelli R, et al. Evidence for differential S100 gene over-expression in psoriatic patients from genetically heterogeneous pedigrees. Hum Genet. 2002;111(4-5):310–3. doi: 10.1007/s00439-002-0812-5. [DOI] [PubMed] [Google Scholar]
- 7.Emberley ED, Murphy LC, Watson PH. S100A7 and the progression of breast cancer. Breast Cancer Res. 2004;6(4):153–9. doi: 10.1186/bcr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson H, Petersson S, Enerback C. Cluster analysis of S100 gene expression and genes correlating to psoriasin (S100A7) expression at different stages of breast cancer development. Int J Oncol. 2005;27(6):1473–81. [PubMed] [Google Scholar]
- 9.Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46(3):256–69. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 10.Luthra MG, Ajani JA, Izzo J, Ensor J, Wu TT, Rashid A, et al. Decreased expression of gene cluster at chromosome 1q21 defines molecular subgroups of chemoradiotherapy response in esophageal cancers. Clin Cancer Res. 2007;13(3):912–9. doi: 10.1158/1078-0432.CCR-06-1577. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Li X, Dong GL, Zhang HW, Chen DL, Du JJ, et al. In silico analysis and verification of S100 gene expression in gastric cancer. BMC Cancer. 2008;8:261. doi: 10.1186/1471-2407-8-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lapi E, Iovino A, Fontemaggi G, Soliera AR, Iacovelli S, Sacchi A, et al. S100A2 gene is a direct transcriptional target of p53 homologues during keratinocyte differentiation. Oncogene. 2006;25(26):3628–37. doi: 10.1038/sj.onc.1209401. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Chen H, Ding F, Zhang Y, Luo A, Wang M, et al. A novel p53 target gene, S100A9, induces p53-dependent cellular apoptosis and mediates the p53 apoptosis pathway. Biochem J. 2009;422(2):363–72. doi: 10.1042/BJ20090465. [DOI] [PubMed] [Google Scholar]
- 14.Pustylnyak VO, Lisachev PD, Shtark MB, Epstein OI. Regulation of S100B gene in rat hippocampal CA1 area during long term potentiation. Brain Res. 2011;1394:33–92. doi: 10.1016/j.brainres.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Wu F, Chakravarti S. Differential expression of inflammatory and fibrogenic genes and their regulation by NF-kappaB inhibition in a mouse model of chronic colitis. J Immunol. 2007;179(10):6988–7000. doi: 10.4049/jimmunol.179.10.6988. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y, Liu J, Xu Z, Sun K, Fu W. HLA-B gene participates in the NF kappaB signal pathway partly by regulating S100A8 in the laryngeal carcinoma cell line Hep2. Oncol Rep. 2008;19(6):1453–9. [PubMed] [Google Scholar]
- 17.Nemeth J, Stein I, Haag D, Riehl A, Longerich T, Horwitz E, et al. S100A8 and S100A9 are novel nuclear factor kappa B target genes during malignant progression of murine and human liver carcinogenesis. Hepatology. 2009;50(4):1251–62. doi: 10.1002/hep.23099. [DOI] [PubMed] [Google Scholar]
- 18.Joo JH, Kim JW, Lee Y, Yoon SY, Kim JH, Paik SG, et al. Involvement of NF-kappaB in the regulation of S100A6 gene expression in human hepatoblastoma cell line HepG2. Biochem Biophys Res Commun. 2003;307(2):274–80. doi: 10.1016/S0006-291X(03)01199-9. [DOI] [PubMed] [Google Scholar]
- 19.Tsoporis JN, Izhar S, Parker TG. Expression of S100A6 in cardiac myocytes limits apoptosis induced by tumor necrosis factoralpha. J Biol Chem. 2008;283(44):30174–83. doi: 10.1074/jbc.M805318200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bottini F, Mazzocco K, Abbondi T, Tonini GP. Identification of an AP-1-like sequence in the promoter region of calcyclin, a S-100-like gene. Enhancement of binding during retinoic acid-induced neuroblastoma cell differentiation. Neurosci Lett. 1994;181(1-2):35–8. doi: 10.1016/0304-3940(94)90554-1. [DOI] [PubMed] [Google Scholar]
- 21.Cohn MA, Hjelmso I, Wu LC, Guldberg P, Lukanidin EM, Tulchinsky EM. Characterization of Sp1, AP-1, CBF and KRC binding sites and minisatellite DNA as functional elements of the metastasis associated mts1/S100A4 gene intronic enhancer. Nucleic Acids Res. 2001;29(16):3335–46. doi: 10.1093/nar/29.16.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pawliczak R, Cowan MJ, Huang X, Nanavaty UB, Alsaaty S, Logun C, et al. p11 expression in human bronchial epithelial cells is increased by nitric oxide in a cGMP-dependent pathway involving protein kinase G activation. J Biol Chem. 2001;276(48):44613–21. doi: 10.1074/jbc.M104993200. [DOI] [PubMed] [Google Scholar]
- 23.Huang X, Pawliczak R, Yao XL, Madara P, Alsaaty S, Shelhamer JH, et al. Characterization of the human p11 promoter sequence. Gene. 2003;310:133–42. doi: 10.1016/S0378-1119(03)00529-8. [DOI] [PubMed] [Google Scholar]
- 24.Gebhardt C, Breitenbach U, Tuckermann JP, Dittrich BT, Richter KH, Angel P. Calgranulins S100A8 and S100A9 are negatively regulated by glucocorticoids in a c-Fos-dependent manner and overexpressed throughout skin carcinogenesis. Oncogene. 2002;21(27):4266–76. doi: 10.1038/sj.onc.1205521. [DOI] [PubMed] [Google Scholar]
- 25.Zenz R, Eferl R, Kenner L, Florin L, Hummerich L, Mehic D, et al. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature. 2005;437(7057):369–75. doi: 10.1038/nature03963. [DOI] [PubMed] [Google Scholar]
- 26.Averill MM, Kerkhoff C, Bornfeldt KE. S100A8 and S100A9 in cardiovascular biology and disease. Arterioscler Thromb Vasc Biol. 2012;32(2):223–9. doi: 10.1161/ATVBAHA.111.236927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerkhoff c, Voss A, Scholzen TE, Averill MM, Zänker KS, Bornfeldt KE. Novel Insights into the Role of S100A8/A9 in Skin Biology. Exp Derm. 2012 doi: 10.1111/j.1600-0625.2012.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Fernig DG, Rudland PS, Sparks A, Wilkinson MC, Barraclough R. Binding to intracellular targets of the metastasis inducing protein, S100A4 (p9Ka). Biochem Biophys Res Commun. 2001;286(5):1212–7. doi: 10.1006/bbrc.2001.5517. [DOI] [PubMed] [Google Scholar]
- 29.Breen EC, Tang K. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. J Cell Biochem. 2003;88(4):848–54. doi: 10.1002/jcb.10398. [DOI] [PubMed] [Google Scholar]
- 30.Kriajevska MV, Cardenas MN, Grigorian MS, Ambartsumian NS, Georgiev GP, Lukanidin EM. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J Biol Chem. 1994;269(31):19679–82. [PubMed] [Google Scholar]
- 31.Kriajevska M, Bronstein IB, Scott DJ, Tarabykina S, Fischer-Larsen M, Issinger O, et al. Metastasis-associated protein Mts1 (S100A4) inhibits CK2-mediated phosphorylation and self-assembly of the heavy chain of nonmuscle myosin. Biochim Biophys Acta. 2000;1498(2-3):252–63. doi: 10.1016/S0167-4889(00)00100-2. [DOI] [PubMed] [Google Scholar]
- 32.Kim EJ, Helfman DM. Characterization of the metastasis-associated protein, S100A4. Roles of calcium binding and dimerization in cellular localization and interaction with myosin. J Biol Chem. 2003;278(32):30063–73. doi: 10.1074/jbc.M304909200. [DOI] [PubMed] [Google Scholar]
- 33.Li ZH, Spektor A, Varlamova O, Bresnick AR. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42(48):14258–66. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- 34.Malashkevich VN, Varney KM, Garrett SC, Wilder PT, Knight D, Charpentier TH, et al. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47(18):5111–26. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.House RP, Pozzuto M, Patel P, Dulyaninova NG, Li ZH, Zencheck WD, et al. Two functional S100A4 monomers are necessary for regulating nonmuscle myosin-IIA and HCT116 cell invasion. Biochemistry. 2011;50(32):6920–32. doi: 10.1021/bi200498q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elliott PR, Irvine AF, Jung HS, Tozawa K, Pastok MW, Picone R, et al. Asymmetric mode of Ca(2)(+)-S100A4 interaction with nonmuscle myosin IIA generates nanomolar affinity required for filament remodeling. Structure. 2012;20(4):654–66. doi: 10.1016/j.str.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao XQ, Naka M, Muneyuki M, Tanaka T. Ca(2+)-dependent inhibition of actin-activated myosin ATPase activity by S100C (S100A11), a novel member of the S100 protein family. Biochem Biophys Res Commun. 2000;267(1):77–9. doi: 10.1006/bbrc.1999.1918. [DOI] [PubMed] [Google Scholar]
- 38.Du M, Wang G, Ismail TM, Gross S, Fernig DG, Barraclough R, et al. S100P dissociates myosin IIA filaments and focal adhesion sites to reduce cell adhesion and enhance cell migration. J Biol Chem. 2012;287(19):15330–44. doi: 10.1074/jbc.M112.349787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider G, Nieznanski K, Jozwiak J, Slomnicki LP, Redowicz MJ, Filipek A. Tubulin binding protein, CacyBP/SIP, induces actin polymerization and may link actin and tubulin cytoskeletons. Biochim Biophys Acta. 2010;1803(11):1308–17. doi: 10.1016/j.bbamcr.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Jung MJ, Murzik U, Wehder L, Hemmerich P, Melle C. Regulation of cellular actin architecture by S100A10. Exp Cell Res. 2010;316(7):1234–40. doi: 10.1016/j.yexcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Vogl T, Ludwig S, Goebeler M, Strey A, Thorey IS, Reichelt R, et al. MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood. 2004;104(13):4260–8. doi: 10.1182/blood-2004-02-0446. [DOI] [PubMed] [Google Scholar]
- 42.Davey GE, Murmann P, Hoechli M, Tanaka T, Heizmann CW. Calcium-dependent translocation of S100A11 requires tubulin filaments. Biochim Biophys Acta. 2000;1498(2-3):220–32. doi: 10.1016/S0167-4889(00)00098-7. [DOI] [PubMed] [Google Scholar]
- 43.Broome AM, Eckert RL. Microtubule-dependent redistribution of a cytoplasmic cornified envelope precursor. J Invest Dermatol. 2004;122(1):29–38. doi: 10.1046/j.0022-202X.2003.22105.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang HY, Zheng XZ, Wang XH, Xuan XY, Wang F, Li SS. S100A4 mediated cell invasion and metastasis of esophageal squamous cell carcinoma via the regulation of MMP-2 and E-cadherin activity. Mol Biol Rep. 2012;39(1):199–208. doi: 10.1007/s11033-011-0726-1. [DOI] [PubMed] [Google Scholar]
- 45.Moriyama-Kita M, Endo Y, Yonemura Y, Heizmann CW, Miyamori H, Sato H, et al. S100A4 regulates E-cadherin expression in oral squamous cell carcinoma. Cancer Lett. 2005;230(2):211–8. doi: 10.1016/j.canlet.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 46.Nishioku T, Furusho K, Tomita A, Ohishi H, Dohgu S, Shuto H, et al. Potential role for S100A4 in the disruption of the blood-brain barrier in collagen-induced arthritic mice, an animal model of rheumatoid arthritis. Neuroscience. 2011;189:286–92. doi: 10.1016/j.neuroscience.2011.05.044. [DOI] [PubMed] [Google Scholar]
- 47.Whiteman HJ, Weeks ME, Dowen SE, Barry S, Timms JF, Lemoine NR, et al. The role of S100P in the invasion of pancreatic cancer cells is mediated through cytoskeletal changes and regulation of cathepsin D. Cancer Res. 2007;67(18):8633–42. doi: 10.1158/0008-5472.CAN-07-0545. [DOI] [PubMed] [Google Scholar]
- 48.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. Nat Rev Cancer. 2002;2(8) doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 49.van Dieck J, Lum JK, Teufel DP, Fersht AR. S100 proteins interact with the N-terminal domain of MDM2. FEBS Lett. 2010;584(15):3269–74. doi: 10.1016/j.febslet.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 50.Lin J, Yang Q, Wilder PT, Carrier F, Weber DJ. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J Biol Chem. 2010;285(35):27487–98. doi: 10.1074/jbc.M110.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grigorian M, Andresen S, Tulchinsky E, Kriajevska M, Carlberg C, Kruse C, et al. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: functional consequences of their interaction. J Biol Chem. 2001;276(25):22699–708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- 52.Chen H, Yuan Y, Zhang C, Luo A, Ding F, Ma J, et al. Involvement of S100A14 protein in cell invasion by affecting expression and function of matrix metalloproteinase (MMP)-2 via p53-dependent transcriptional regulation. J Biol Chem. 2012;287(21):17109–19. doi: 10.1074/jbc.M111.326975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sapkota D, Costea DE, Blo M, Bruland O, Lorens JB, Vasstrand EN, et al. S100A14 inhibits proliferation of oral carcinoma derived cells through G1-arrest. Oral Oncol. 2012;48(3):219–25. doi: 10.1016/j.oraloncology.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Wilder PT, Charpentier TH, Liriano MA, Gianni K, Varney KM, Pozharski E, et al. In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction. Int J High Throughput Screen. 2010;2010(1):109–26. doi: 10.2147/IJHTS.S8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berge G, Maelandsmo GM. Evaluation of potential interactions between the metastasis-associated protein S100A4 and the tumor suppressor protein p53. Amino Acids. 2011;41(4):863–73. doi: 10.1007/s00726-010-0497-3. [DOI] [PubMed] [Google Scholar]
- 56.van Dieck J, Brandt T, Teufel DP, Veprintsev DB, Joerger AC, Fersht AR. Molecular basis of S100 proteins interacting with the p53 homologs p63 and p73. Oncogene. 2010;29(14):2024–35. doi: 10.1038/onc.2009.490. [DOI] [PubMed] [Google Scholar]
- 57.Mueller A, Schafer BW, Ferrari S, Weibel M, Makek M, Hochli M, et al. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J Biol Chem. 2005;280(32):29186–93. doi: 10.1074/jbc.M505000200. [DOI] [PubMed] [Google Scholar]
- 58.Mathisen B, Lindstad RI, Hansen J, El-Gewely SA, Maelandsmo GM, Hovig E, et al. S100A4 regulates membrane induced activation of matrix metalloproteinase-2 in osteosarcoma cells. Clin Exp Metastasis. 2003;20(8):701–11. doi: 10.1023/B:CLIN.0000006819.21361.03. [DOI] [PubMed] [Google Scholar]
- 59.Yammani RR, Carlson CS, Bresnick AR, Loeser RF. Increase in production of matrix metalloproteinase 13 by human articular chondrocytes due to stimulation with S100A4: Role of the receptor for advanced glycation end products. Arthritis Rheum. 2006;54(9):2901–11. doi: 10.1002/art.22042. [DOI] [PubMed] [Google Scholar]
- 60.van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem. 2009;284(20):13804–11. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleem M, Kweon MH, Johnson JJ, Adhami VM, Elcheva I, Khan N, et al. S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc Natl Acad Sci U S A. 2006;103(40):14825–30. doi: 10.1073/pnas.0606747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voss A, Bode G, Sopalla C, Benedyk M, Varga G, Bohm M, et al. Expression of S100A8/A9 in HaCaT keratinocytes alters the rate of cell proliferation and differentiation. FEBS Lett. 2011;585(2):440–6. doi: 10.1016/j.febslet.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 63.Sakaguchi M, Miyazaki M, Takaishi M, Sakaguchi Y, Makino E, Kataoka N, et al. S100C/A11 is a key mediator of Ca(2+)-induced growth inhibition of human epidermal keratinocytes. J Cell Biol. 2003;163(4):825–35. doi: 10.1083/jcb.200304017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gorsler T, Murzik U, Ulbricht T, Hentschel J, Hemmerich P, Melle C. DNA damage-induced translocation of S100A11 into the nucleus regulates cell proliferation. BMC Cell Biol. 2010;11:100. doi: 10.1186/1471-2121-11-100. 21167017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakaguchi M, Sonegawa H, Murata H, Kitazoe M, Futami J, Kataoka K , et al. S100A11, an dual mediator for growth regulation of human keratinocytes. Mol Biol Cell. 2008;19(1):78–85. doi: 10.1091/mbc.E07-07-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyazaki M, Sakaguchi M, Akiyama I, Sakaguchi Y, Nagamori S, Huh NH. Involvement of interferon regulatory factor 1 and S100C/A11 in growth inhibition by transforming growth factor beta 1 in human hepatocellular carcinoma cells. Cancer Res. 2004;64(12):4155–61. doi: 10.1158/0008-5472.CAN-03-2750. [DOI] [PubMed] [Google Scholar]
- 67.Nukui T, Ehama R, Sakaguchi M, Sonegawa H, Katagiri C, Hibino T, et al. S100A8/A9, a key mediator for positive feedback growth stimulation of normal human keratinocytes. J Cell Biochem. 2008;104(2):453–64. doi: 10.1002/jcb.21639. [DOI] [PubMed] [Google Scholar]
- 68.Fritz G, Botelho HM, Morozova-Roche LA, Gomes CM. Natural and amyloid self-assembly of S100 proteins: structural basis of functional diversity. FEBS J. 2010;277(22):4578–90. doi: 10.1111/j.1742-4658.2010.07887.x. [DOI] [PubMed] [Google Scholar]
- 69.Leclerc E, Fritz G, Weibel M, Heizmann CW, Galichet A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J Biol Chem. 2007;282(43):31317–31. doi: 10.1074/jbc.M703951200. [DOI] [PubMed] [Google Scholar]
- 70.Huttunen HJ, Kuja-Panula J, Sorci G, Agneletti AL, Donato R, Rauvala H. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J Biol Chem. 2000;275(51):40096–105. doi: 10.1074/jbc.M006993200. [DOI] [PubMed] [Google Scholar]
- 71.Ghavami S, Kerkhoff C, Los M, Hashemi M, Sorg C, Karami-Tehrani F. Mechanism of apoptosis induced by S100A8/A9 in colon cancer cell lines: the role of ROS and the effect of metal ions. J Leukoc Biol. 2004;76(1):169–75. doi: 10.1189/jlb.0903435. [DOI] [PubMed] [Google Scholar]
- 72.Ghavami S, Kerkhoff C, Chazin WJ, Kadkhoda K, Xiao W, Zuse A , et al. S100A8/9 induces cell death via a novel, RAGE-independent pathway that involves selective release of Smac/DIABLO and Omi/HtrA2. Biochim Biophys Acta. 2008;1783(2):297–311. doi: 10.1016/j.bbamcr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 73.Ghavami S, Eshragi M, Ande SR, Chazin WJ, Klonisch T, Halayko AJ, et al. S100A8/A9 induces autophagy and apoptosis via ROS mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res. 2010;20(3):314–31. doi: 10.1038/cr.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 75.Horimoto Y, Takahashi Y, Polanska U, Orimo A. Emerging roles of the tumor-associated stroma in promoting tumor metastasis. Cell Adh Migr. 2012;6(3) doi: 10.4161/cam.20631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barth PJ, Ebrahimsade S, Ramaswamy A, Moll R. CD34+ fibrocytes in invasive ductal carcinoma, ductal carcinoma in situ, and benign breast lesions. Virchows Arch. 2002;440(3):298–303. doi: 10.1007/s004280100530. [DOI] [PubMed] [Google Scholar]
- 77.Chesney J, Bucala R. Peripheral blood fibrocytes: novel fibroblas tlike cells that present antigen and mediate tissue repair. Biochem Soc Trans. 1997;25(2):520–4. doi: 10.1042/bst0250520. [DOI] [PubMed] [Google Scholar]
- 78.Metz CN. Fibrocytes: a unique cell population implicated in wound healing. Cell Mol Life Sci. 2003;60(7):1342–50. doi: 10.1007/s00018-003-2328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pilling D, Fan T, Huang D, Kaul B, Gomer RH. Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PLoS One. 2009;4(10):e7475. doi: 10.1371/journal.pone.0007475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai F, Liu L, Che G, Yu N, Pu Q, Zhang S, et al. The number and microlocalization of tumor-associated immune cells are associated with patient's survival time in non-small cell lung cancer. BMC Cancer. 2010;10:220. doi: 10.1186/1471-2407-10-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sandel MH, Dadabayev AR, Menon AG, Morreau H, Melief CJ, Offringa R, et al. Prognostic value of tumor-infiltrating dendritic cells in colorectal cancer: role of maturation status and intratumoral localization. Clin Cancer Res. 2005;11(7):2576–82. doi: 10.1158/1078-0432.CCR-04-1448. [DOI] [PubMed] [Google Scholar]
- 82.Nagorsen D, Voigt S, Berg E, Stein H, Thiel E, Loddenkemper C. Tumor-infiltrating macrophages and dendritic cells in human colorectal cancer: relation to local regulatory T cells, systemic T cell response against tumor-associated antigens and survival. J Transl Med. 2007;5:62. doi: 10.1186/1479-5876-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boomershine CS, Chamberlain A, Kendall P, Afshar-Sharif AR, Huang H, Washington MK, et al. Autoimmune pancreatitis results from loss of TGFbeta signalling in S100A4-positive dendritic cells. Gut. 2009;58(9):1267–74. doi: 10.1136/gut.2008.170779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clarke LE, Helm KF, Hennessy J, Bruggeman RD, Clarke JT. Dermal dendritic cells in psoriasis, nummular dermatitis, and normal appearing skin. J Am Acad Dermatol. 2012;66(1):98–105. doi: 10.1016/j.jaad.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 85.Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205(10):2235–49. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, Johnson-Huang LM, Nograles KE, White TR, et al. Identification of TNF-related apoptosis-inducing ligand and other molecules that distinguish inflammatory from resident dendritic cells in patients with psoriasis. J Allergy Clin Immunol. 2010;125(6):1261–8 e9. doi: 10.1016/j.jaci.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morbini P, Villa C, Campo I, Zorzetto M, Inghilleri S, Luisetti M. The receptor for advanced glycation end products and its ligands: a new inflammatory pathway in lung disease? Mod Pathol. 2006;19(11):1437–45. doi: 10.1038/modpathol.3800661. [DOI] [PubMed] [Google Scholar]
- 88.Averill MM, Barnhart S, Becker L, Li X, Heinecke JW, Leboeuf RC, et al. S100A9 differentially modifies phenotypic states of neutrophils, macrophages, and dendritic cells: implications for atherosclerosis and adipose tissue inflammation. Circulation. 2011;123(11):1216–26. doi: 10.1161/CIRCULATIONAHA.110.985523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimizu K, Libby P, Rocha VZ, Folco EJ, Shubiki R, Grabie N, et al. Loss of myeloid related protein-8/14 exacerbates cardiac allograft rejection. Circulation. 2011;124(25):2920–32. doi: 10.1161/CIRCULATIONAHA.110.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Almand B, Resser JR, Lindman B, Nadaf S, Clark JI, Kwon ED, et al. Clinical significance of defective dendritic cell differentiation in cancer. Clin Cancer Res. 2000;6(5):1755–66. [PubMed] [Google Scholar]
- 91.Wojas K, Tabarkiewicz J, Jankiewicz M, Rolinski J. Dendritic cells in peripheral blood of patients with breast and lung cancer--a pilot study. Folia Histochem Cytobiol. 2004;42(1):45–8. [PubMed] [Google Scholar]
- 92.Della Bella S, Gennaro M, Vaccari M, Ferraris C, Nicola S, Riva A, et al. Altered maturation of peripheral blood dendritic cells in patients with breast cancer. Br J Cancer. 2003;89(8):1463–72. doi: 10.1038/sj.bjc.6601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoffmann TK, Muller-Berghaus J, Ferris RL, Johnson JT, Storkus WJ, Whiteside TL. Alterations in the frequency of dendritic cell subsets in the peripheral circulation of patients with squamous cell carcinomas of the head and neck. Clin Cancer Res. 2002;8(6):1787–93. [PubMed] [Google Scholar]
- 94.Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9(2):133–48. doi: 10.1158/1541-7786.MCR-10-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sinha P, Okoro C, Foell D, Freeze HH, Ostrand-Rosenberg S, Srikrishna G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181(7):4666–75. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8(12):1369–75. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 97.Ahn GO, Tseng D, Liao CH, Dorie MJ, Czechowicz A, Brown JM. Inhibition of Mac-1 (CD11b/CD18) enhances tumor response to radiation by reducing myeloid cell recruitment. Proc Natl Acad Sci U S A. 2010;107(18):8363–8. doi: 10.1073/pnas.0911378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li ZH, Dulyaninova NG, House RP, Almo SC, Bresnick AR. S100A4 regulates macrophage chemotaxis. Mol Biol Cell. 2010;21(15):2598–610. doi: 10.1091/mbc.E09-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor S, Herrington S, Prime W, Rudland PS, Barraclough R. S100A4 (p9Ka) protein in colon carcinoma and liver metastases: association with carcinoma cells and T-lymphocytes. Br J Cancer. 2002;86(3):409–16. doi: 10.1038/sj.bjc.6600071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kwak JM, Lee HJ, Kim SH, Kim HK, Mok YJ, Park YT, et al. Expression of protein S100A4 is a predictor of recurrence in colorectal cancer. World J Gastroenterol. 2010;16(31):3897–904. doi: 10.3748/wjg.v16.i31.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berge G, Costea DE, Berg M, Rasmussen H, Grotterod I, Lothe RA, et al. Coexpression and nuclear colocalization of metastasispromoting protein S100A4 and p53 without mutual regulation in colorectal carcinoma. Amino Acids. 2011;41(4):875–84. doi: 10.1007/s00726-010-0514-6. [DOI] [PubMed] [Google Scholar]
- 102.Huang LY, Xu Y, Cai GX, Guan ZQ, Sheng WQ, Lu HF, et al. S100A4 over-expression underlies lymph node metastasis and poor prognosis in colorectal cancer. World J Gastroenterol. 2011;17(1):69–78. doi: 10.3748/wjg.v17.i1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boye K, Nesland JM, Sandstad B, Maelandsmo GM, Flatmark K. Nuclear S100A4 is a novel prognostic marker in colorectal cancer. Eur J Cancer. 2010;46(16):2919–25. doi: 10.1016/j.ejca.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 104.Flatmark K, Maelandsmo GM, Mikalsen SO, Nustad K, Varaas T, Rasmussen H, et al. Immunofluorometric assay for the metastasis-related protein S100A4: release of S100A4 from normal blood cells prohibits the use of S100A4 as a tumor marker in plasma and serum. Tumour Biol. 2004;25(1-2):31–40. doi: 10.1159/000077721. [DOI] [PubMed] [Google Scholar]
- 105.Wang HY, Zhang JY, Cui JT, Tan XH, Li WM, Gu J, et al. Expression status of S100A14 and S100A4 correlates with metastatic potential and clinical outcome in colorectal cancer after surgery. Oncol Rep. 2010;23(1):45–52. [PubMed] [Google Scholar]
- 106.Stulik J, Koupilova K, Osterreicher J, Knizek J, Macela A, Bures J, et al. Protein abundance alterations in matched sets of macroscopically normal colon mucosa and colorectal carcinoma. Electrophoresis. 1999;20(18):3638–46. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3638::AID-ELPS3638>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 107.Stulik J, Osterreicher J, Koupilova K, Knizek J, Bures J, Jandik P, et al. Differential expression of the Ca2+ binding S100A6 protein in normal, preneoplastic and neoplastic colon mucosa. Eur J Cancer. 2000;36(8):1050–9. doi: 10.1016/S0959-8049(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 108.Zeng FY, Gerke V, Gabius HJ. Identification of annexin II, annexin VI and glyceraldehyde-3-phosphate dehydrogenase as calcyclinbinding proteins in bovine heart. Int J Biochem. 1993;25(7):1019–27. doi: 10.1016/0020-711X(93)90116-V. [DOI] [PubMed] [Google Scholar]
- 109.Stulik J, Osterreicher J, Koupilova K, Knizek , Macela A, Bures J, et al. The analysis of S100A9 and S100A8 expression in matched sets of macroscopically normal colon mucosa and colorectal carcinoma: the S100A9 and S100A8 positive cells underlie and invade tumor mass. Electrophoresis. 1999;20(4-5):1047–54. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<1047::AID-ELPS1047>3.3.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 110.Kim JK, Jung KH, Noh JH, Eun JW, Bae HJ, Xie HJ, et al. Targeted disruption of S100P suppresses tumor cell growth by down-regulation of cyclin D1 and CDK2 in human hepatocellular carcinoma. Int J Oncol. 2009;35(6):1257–64. [PubMed] [Google Scholar]
- 111.Yoo BC, Shin YK, Lim SB, Hong SH, Jeong SY, Park JG. Evaluation of calgranulin B in stools from the patients with colorectal cancer. Dis Colon Rectum. 2008;51(11):1703–9. doi: 10.1007/s10350-008-9381-6. [DOI] [PubMed] [Google Scholar]
- 112.Hwang CC, Chai HT, Chen HW, Tsai HL, Lu CY, Yu FJ, et al. S100B protein expressions as an independent predictor of early relapse in UICC stages II and III colon cancer patients after curative resection. Ann Surg Oncol. 2011;18(1):139–45. doi: 10.1245/s10434-010-1209-7. [DOI] [PubMed] [Google Scholar]
- 113.Parkkila S, Pan PW, Ward A, Gibadulinova A, Oveckova I, Pastorekova S, et al. The calcium-binding protein S100P in normal and malignant human tissues. BMC Clin Pathol. 2008;8:2. doi: 10.1186/1472-6890-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ding Q, Chang CJ, Xie X, Xia W, Yang JY, Wang SC, et al. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J Clin Invest. 2011;121(11):4526–36. doi: 10.1172/JCI45008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10(5):445–56. doi: 10.1093/hmg/10.5.445. [DOI] [PubMed] [Google Scholar]
- 116.Brentnall TA, Pan S, Bronner MP, Crispin DA, Mirzaei H, Cooke K, et al. Proteins That Underlie Neoplastic Progression of Ulcerative Colitis. Proteomics Clin Appl. 2009;3(11):1326. doi: 10.1002/prca.200900061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gebhardt C, Nemeth J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72(11):1622–31. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 118.Sedaghat F, Notopoulos A. S100 protein family and its application in clinical practice. Hippokratia. 2008;12(4):198–204. [PMC free article] [PubMed] [Google Scholar]
- 119.Melle C, Ernst G, Schimmel B, Bleul A, von Eggeling F. Colon-derived liver metastasis, colorectal carcinoma, and hepatocellular carcinoma can be discriminated by the Ca(2+)-binding proteins S100A6 and S100A11. PLoS One. 2008;3(12):e3767. doi: 10.1371/journal.pone.0003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sack U, Walther W, Scudiero D, Selby M, Aumann J, Lemos C, et al. S100A4-induced cell motility and metastasis is restricted by the Wnt/beta-catenin pathway inhibitor calcimycin in colon cancer cells. Mol Biol Cell. 2011;22(18):3344–54. doi: 10.1091/mbc.E10-09-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sack U, Walther W, Scudiero D, Selby M, Kobelt D, Lemm M, et al. Novel effect of antihelminthic Niclosamide on S100A4-mediated metastatic progression in colon cancer. J Natl Cancer Inst. 2011;103(13):1018–36. doi: 10.1093/jnci/djr190. [DOI] [PubMed] [Google Scholar]
- 122.Huang L, Xu Y, Cai G, Guan Z, Cai S. Downregulation of S100A4 expression by RNA interference suppresses cell growth and invasion in human colorectal cancer cells. Oncol Rep. 2012;27(4):917–22. doi: 10.3892/or.2011.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mencia N, Selga E, Rico I, de Almagro MC, Villalobos X, Ramirez S, et al. Overexpression of S100A4 in human cancer cell lines resistant to methotrexate. BMC Cancer. 2010;10:250. doi: 10.1186/1471-2407-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pedersen KB, Andersen K, Fodstad O, Maelandsmo GM. Sensitization of interferon-gamma induced apoptosis in human osteosarcoma cells by extracellular S100A4. BMC Cancer. 2004;4:52. doi: 10.1186/1471-2407-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakamura N, Takenaga K. Hypomethylation of the metastasis associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin Exp Metastasis. 1998;16(5):471–9. doi: 10.1023/A:1006589626307. [DOI] [PubMed] [Google Scholar]
- 126.Meding S, Balluff B, Elsner M, Schone C, Rauser S, Nitsche U, et al. Tissue-based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J Pathol. 2012 doi: 10.1002/path.4021. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 127.Phipps KD, Surette AP, O'Connell PA, Waisman DM. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer Res. 2011;71(21):6676–83. doi: 10.1158/0008-5472.CAN-11-1748. [DOI] [PubMed] [Google Scholar]
- 128.Zhang L, Fogg DK, Waisman DM. RNA interference-mediated silencing of the S100A10 gene attenuates plasmin generation and invasiveness of Colo 222 colorectal cancer cells. J Biol Chem. 2004;279(3):2053–62. doi: 10.1074/jbc.M310357200. [DOI] [PubMed] [Google Scholar]
- 129.Suzuki S, Yamayoshi Y, Nishimuta A, Tanigawara Y. S100A10 protein expression is associated with oxaliplatin sensitivity in human colorectal cancer cells. Proteome Sci. 2011;9:76. doi: 10.1186/1477-5956-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fenouille N, Grosso S, Yunchao S, Mary D, Pontier-Bres R, Imbert V, et al. Calpain 2-dependent IkappaBalpha degradation mediates CPT-11 secondary resistance in colorectal cancer xenografts. J Pathol. 2012;227(1):118–29. doi: 10.1002/path.3034. [DOI] [PubMed] [Google Scholar]
- 131.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci U S A. 2005;102(13):4735-40. 2005;102(13):4735–40. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang MY, Wang HM, Chang HJ, Hsiao CP, Wang JY, Lin SR. Overexpression of S100B, TM4SF4, and OLFM4 genes is correlated with liver metastasis in Taiwanese colorectal cancer patients. DNA Cell Biol. 2012;31(1):43–9. doi: 10.1089/dna.2011.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Arumugam T, Logsdon CD. S100P: a novel therapeutic target for cancer. Amino Acids. 2011;41(4):893–9. doi: 10.1007/s00726-010-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gibadulinova A, Tothova V, Pastorek J, Pastorekova S. Transcriptional regulation and functional implication of S100P in cancer. Amino Acids. 2011;41(4):885–92. doi: 10.1007/s00726-010-0495-5. [DOI] [PubMed] [Google Scholar]
- 135.Fuentes MK, Nigavekar SS, Arumugam T, Logsdon CD, Schmidt AM, Park JC, et al. RAGE activation by S100P in colon cancer stimulates growth, migration, and cell signaling pathways. Dis Colon Rectum. 2007;50(8):1230–40. doi: 10.1007/s10350-006-0850-5. [DOI] [PubMed] [Google Scholar]
- 136.Arlt A, Vorndamm J, Muerkoster S, Yu H, Schmidt WE, Folsch UR, et al. Autocrine production of interleukin 1beta confers constitutive nuclear factor kappaB activity and chemoresistance in pancreatic carcinoma cell lines. Cancer Res. 2002;62(3):910–6. [PubMed] [Google Scholar]
- 137.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221–7. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 138.Bertram J, Palfner K, Hiddemann W, Kneba M. Elevated expression of S100P, CAPL and MAGE 3 in doxorubicin-resistant cell lines: comparison of mRNA differential display reverse transcription-polymerase chain reaction and subtractive suppressive hybridization for the analysis of differential gene expression. Anticancer Drugs. 1998;9(4):311–7. doi: 10.1097/00001813-199804000-00004. [DOI] [PubMed] [Google Scholar]
- 139.Jiang L, Lai YK, Zhang J, Wang H, Lin MC, He ML, et al. Targeting S100P inhibits colon cancer growth and metastasis by Lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17(7-8):709–16. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chandramouli A, Mercado-Pimentel ME, Hutchinson A, Gibadulinova A, Olson ER, Dickinson S, et al. The induction of S100p expression by the Prostaglandin E(2) (PGE(2))/EP4 receptor signaling pathway in colon cancer cells. Cancer Biol Ther. 2010;10(10):1056–66. doi: 10.4161/cbt.10.10.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15(2):223–43, vii-x. doi: 10.1016/j.cld.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hua Z, Chen J, Sun B, Zhao G, Zhang Y, Fong Y, et al. Specific expression of osteopontin and S100A6 in hepatocellular carcinoma. Surgery. 2011;149(6):783–91. doi: 10.1016/j.surg.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 143.Arai K, Yamada T, Nozawa R. Immunohistochemical investigation of migration inhibitory factor-related protein (MRP)-14 expression in hepatocellular carcinoma. Med Oncol. 2000;17(3):183. doi: 10.1007/BF02780526. [DOI] [PubMed] [Google Scholar]
- 144.Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE, et al. Human hepatocellular carcinomas with “Stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 2011;54(5):1707–17. doi: 10.1002/hep.24559. [DOI] [PubMed] [Google Scholar]
- 145.Cui JF, Liu YK, Zhang LJ, Shen HL, Song HY, Dai Z, et al. Identification of metastasis candidate proteins among HCC cell lines by comparative proteome and biological function analysis of S100A4 in metastasis in vitro. Proteomics. 2006;6(22):5953–61. doi: 10.1002/pmic.200500460. [DOI] [PubMed] [Google Scholar]
- 146.Lukanidin E, Sleeman JP. Building the niche: the role of the S100 proteins in metastatic growth. Semin Cancer Biol. 2012;22(3):216–25. doi: 10.1016/j.semcancer.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 147.Zhao L, Yang R, Cheng L, Wang M, Jiang Y, Wang S. LPS-induced epithelial-mesenchymal transition of intrahepatic biliary epithelial cells. J Surg Res. 2011;171(2):819–25. doi: 10.1016/j.jss.2010.04.059. [DOI] [PubMed] [Google Scholar]
- 148.Osterreicher CH, Penz-Osterreicher M, Grivennikov SI, Guma M, Koltsova EK, Datz C, et al. Fibroblast-specific protein 1 identifies an inflammatory subpopulation of macrophages in the liver. Proc Natl Acad Sci U S A. 2011;108(1):308–13. doi: 10.1073/pnas.1017547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Choi J, Chang JS, Song MS, Ahn BY, Park Y, Lim DS, et al. Association of hepatitis B virus polymerase with promyelocytic leukemia nuclear bodies mediated by the S100 family protein p11. Biochem Biophys Res Commun. 2003;305(4):1049–56. doi: 10.1016/S0006-291X(03)00881-7. [DOI] [PubMed] [Google Scholar]
- 150.Falasca K, Mancino P, Ucciferri C, Dalessandro M, Zingariello P, Lattanzio FM, et al. Inflammatory cytokines and S-100b protein in patients with hepatitis C infection and cryoglobulinemias. Clin Invest Med. 2007;30(5):E167–76. doi: 10.25011/cim.v30i5.2892. [DOI] [PubMed] [Google Scholar]
- 151.Ikemoto M, Murayama H, Itoh H, Totani M, Fujita M. Intrinsic function of S100A8/A9 complex as an anti-inflammatory protein in liver injury induced by lipopolysaccharide in rats. Clin Chim Acta. 2007;376(1-2):197–204. doi: 10.1016/j.cca.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 152.Voss A, Gescher K, Hensel A, Nacken W, Zanker KS, Kerkhoff C. Double-stranded RNA induces S100 gene expression by a cycloheximide-sensitive factor. FEBS Lett. 2012;586(2):196–203. doi: 10.1016/j.febslet.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 153.Bronckart Y, Decaestecker C, Nagy N, Harper L, Schafer BW, Salmon I, et al. Development and progression of malignancy in human colon tissues are correlated with expression of specific Ca(2+)-binding S100 proteins. Histol Histopathol. 2001;16(3):707–12. doi: 10.14670/HH-16.707. [DOI] [PubMed] [Google Scholar]
- 154.Takenaga K, Nakanishi H, Wada K, Suzuki M, Matsuzaki O, Matsuura A, et al. Increased expression of S100A4, a metastasis-associated gene, in human colorectal adenocarcinomas. Clin Cancer Res. 1997;3(12 Pt 1):2309–16. [PubMed] [Google Scholar]
- 155.Wang G, Wang X, Wang S, Song H, Sun H, Yuan W, et al. Colorectal cancer progression correlates with upregulation of S100A11 expression in tumor tissues. Int J Colorectal Dis. 2008;23(7):675–82. doi: 10.1007/s00384-008-0464-6. [DOI] [PubMed] [Google Scholar]
- 156.Pietas A, Schluns K, Marenholz I, Schafer BW, Heizmann CW, Petersen I. Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics. 2002;79(4):513–22. doi: 10.1006/geno.2002.6744. [DOI] [PubMed] [Google Scholar]
- 157.Zimmer DB, Chessher J, Wilson GL, Zimmer WE. S100A1 and S100B expression and target proteins in type I diabetes. Endocrinology. 1997;138(12):5176–83. doi: 10.1210/en.138.12.5176. [DOI] [PubMed] [Google Scholar]
- 158.Kittaka N, Takemasa I, Takeda Y, Marubashi S, Nagano H, Umeshita K, et al. Molecular mapping of human hepatocellular carcinoma provides deeper biological insight from genomic data. Eur J Cancer. 2008;44(6):885–97. doi: 10.1016/j.ejca.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 159.Inada H, Naka M, Tanaka T, Davey GE, Heizmann CW. Human S100A11 exhibits differential steady-state RNA levels in various tissues and a distinct subcellular localization. Biochem Biophys Res Commun. 1999;263(1):135–8. doi: 10.1006/bbrc.1999.1319. [DOI] [PubMed] [Google Scholar]
- 160.Kim HJ, Kang HJ, Lee H, Lee ST, Yu MH, Kim H, et al. Identification of S100A8 and S100A9 as serological markers for colorectal cancer. J Proteome Res. 2009;8(3):1368–79. doi: 10.1021/pr8007573. [DOI] [PubMed] [Google Scholar]
- 161.Song HY, Liu YK, Feng JT, Cui JF, Dai Z, Zhang LJ, et al. Proteomic analysis on metastasis-associated proteins of human hepatocellular carcinoma tissues. J Cancer Res Clin Oncol. 2006;132(2):92–8. doi: 10.1007/s00432-005-0044-x. [DOI] [PMC free article] [PubMed] [Google Scholar]