Abstract
An integrated software package, Compartment Model Kinetic Analysis Tool (COMKAT), is presented in this report.
Methods
COMKAT is an open-source software package with many functions for incorporating pharmacokinetic analysis in molecular imaging research and has both command-line and graphical user interfaces.
Results
With COMKAT, users may load and display images, draw regions of interest, load input functions, select kinetic models from a predefined list, or create a novel model and perform parameter estimation, all without having to write any computer code. For image analysis, COMKAT image tool supports multiple image file formats, including the Digital Imaging and Communications in Medicine (DICOM) standard. Image contrast, zoom, reslicing, display color table, and frame summation can be adjusted in COMKAT image tool. It also displays and automatically registers images from 2 modalities. Parametric imaging capability is provided and can be combined with the distributed computing support to enhance computation speeds. For users without MATLAB licenses, a compiled, executable version of COMKAT is available, although it currently has only a subset of the full COMKAT capability. Both the compiled and the noncompiled versions of COMKAT are free for academic research use. Extensive documentation, examples, and COMKAT itself are available on its wiki-based Web site, http://comkat.case.edu. Users are encouraged to contribute, sharing their experience, examples, and extensions of COMKAT.
Conclusion
With integrated functionality specifically designed for imaging and kinetic modeling analysis, COMKAT can be used as a software environment for molecular imaging and pharmacokinetic analysis.
Keywords: kinetic modeling, imaging software, pharmacokinetics, COMKAT
In molecular imaging, kinetic or compartment models can be used to describe the pharmacokinetics of tracers and quantify physiology. For example, the 2-compartment kinetic model of 18F-FDG has been well studied and extensively used for measuring the glucose metabolic rate (1,2). However, one challenge for applying kinetic modeling in image quantification is the lack of software designed specifically for kinetic modeling analysis in molecular imaging research. Quite a few sophisticated kinetic modeling software packages are publicly available, including RFit (3), Pk-Fit (4), and SAAM II (5). Their drawback nevertheless is that they are not specifically designed for image analysis and therefore lack the functionality for image processing. On the other hand, many software packages provided by scanner vendors have image-processing but not kinetic-modeling capabilities. Similarly, most publicly available software packages for biomedical image processing, such as AMIDE (6) and ImageJ (7), do not provide functionality for kinetic modeling analysis. The only alternative solutions with imaging and modeling capability of which we are aware include commercial products such as PMOD (8), which requires licensing fees, or Internet-based applications such as KIS (9). However, there has been no software package specifically designed for both modeling and imaging, publicly available with open-source distribution, and capable of specifying new kinetic models with graphical user interfaces (GUIs). To address this unmet need, we developed Compartment Model Kinetic Analysis Tool (COMKAT) (10) as an integrated software environment for model-based analysis and physiologic interpretation of biomedical image data.
First developed as a toolbox for kinetic modeling and published in 2001 (10), COMKAT has been a powerful package for kinetic modeling, with many unique features including intuitive command-line functions for building and solving kinetic models, parameter estimation for fitting experimental data, and a GUI-based model creation tool. Since 2001, COMKAT has been significantly improved to make it more useful to a larger user base. Distributed as a free, open-source software package for academic research use, COMKAT has undergone major improvements in performance, user interfaces, capabilities, and documentation.
MATERIALS AND METHODS
Framework of COMKAT
The functions and GUIs of COMKAT can be categorized into several components, including the main COMKAT GUI, the COMKAT image tool, the COMKAT input function GUI, COMKAT command-line functions, and miscellaneous analytic tools. The integration of and relationship between these components is diagrammed in Figure 1. The command-line and GUI are complementary, with the former providing underlying support for the latter and users having the flexibility to apply both interfaces. The COMKAT GUI is linked to the COMKAT input function GUI and the COMKAT image tool for users to specify input functions and image data. Model solving and parameter estimation in the COMKAT GUI is performed by calling the COMKAT command-line functions behind the scenes. A brief summary of COMKAT functionality is provided in Table 1.
FIGURE 1.
Framework of COMKAT software. COMKAT GUI serves as the front-end GUI of software. It calls several other GUIs of COMKAT to import data and kinetic models. Behind scenes, COMKAT GUI calls COMKAT command-line functions to calculate model output and estimate parameters.
TABLE 1.
Summary and Comparison of Functionalities of COMKAT Distributions
| Function | COMKAT on MATLAB | Compiled COMKAT application |
|---|---|---|
| COMKAT GUI | ||
| Loading of input functions from files | Yes | Yes |
| Simulation of model output | Yes | Yes |
| Creation of new kinetic models | Yes | No |
| Parameter estimation | Yes | Yes |
| Loading of tissue time–activity curves from files | Yes | Yes |
| Loading of tissue time–activity curves from COMKAT image tool |
Yes | Yes |
| Calculation of parametric images | Yes | Yes |
| Distributed computing for parametric imaging* | Yes | No |
| COMKAT image tool | ||
| Support for multiple image formats (Table 2) | Yes | Yes |
| Image display and contrast adjustments | Yes | Yes |
| Frame summation | Yes | Yes |
| Spatial filtering | Yes | Yes |
| Drawing of ROIs or volumes of interest | Yes | Yes |
| Image coregistration | Yes | Yes |
| Image translation and rotation | Yes | Yes |
| Image reslicing in arbitrary orientations | Yes | Yes |
| MATLAB scripting with COMKAT command-line functions |
Yes | No |
| Available for Windows, Linux, and MacOS X† | Yes | Yes |
| COMKAT licensing | Free for academic research use |
Free for academic research use |
| MATLAB licensing | Requires MATLAB installation and licenses |
Requires MATLAB Compiler Runtime (no licensing fees) |
Requires MATLAB licenses for MATLAB Distributed Computing Server and Parallel Computing Toolbox.
We have successfully tested COMKAT on the following platforms: Windows XP and Vista in both 32-bit and 64-bit; MacOS X 10.5.1 and later; Linux Ubuntu in both 32-bit and 64-bit.
COMKAT Command-Line Functions
The COMKAT command-line functions, as previously reported (10), enable users to specify kinetic models, solve for model outputs, and estimate parameters. Considerable improvement has been made to the COMKAT command-line functions since 2001. First, to speed up solving of the differential equations, a “mex” function—a compiled, C-language function with MATLAB (The MathWorks, Inc.) interface (11)—has been created. This function uses CVODES (12) to solve the differential equations with several strategies (e.g., caching the results of intermediate calculations, reusing memory, and reducing the number of function calls back into MATLAB) to enhance efficiency. This is especially important when parameter estimation is required because the model is solved using many different values of the parameters. Second, COMKAT now supports several different built-in kinetic rules (e.g., diffusion, receptor–ligand saturable binding, and Michaelis–Menten). In addition, the user can define customized kinetic rules for concentration- or time-dependent rate constants. Third, COMKAT now supports several weighting mechanisms such as penalized weighted least-squares criteria for fitting data under different noise models (13).
COMKAT GUI
The COMKAT GUI serves as the front-end interface for users who wish to use COMKAT without typing commands. In the COMKAT GUI, an input function and a kinetic model are required. Experimental data are required when data are to be fit by a model. Users may specify input functions with the COMKAT input function GUI. It is assumed that a user has the appropriate knowledge for the task at hand, such as using a well-defined model versus determining the appropriate model for a new radiopharmaceutical. To specify the kinetic model, users can either choose from a list of preprogrammed model templates or create their own kinetic model in a GUI. Once a model and input function have been specified, the COMKAT GUI solves for the model output immediately using the default initial guesses for the parameter values. Users may then adjust the values of the initial guess, and the plot is updated instantaneously.
Parameter estimation or data fitting is performed similarly. Users must specify the experimental data either from regional time–activity curves from the COMKAT image tool or from previously calculated curves that have been stored in files. Supported file formats are listed in Table 2. The COMKAT GUI automatically converts the time–activity curves to the built-in data units as µCi/mL for activity concentration and minutes for time. For the values of each parameter, the COMKAT GUI supports separate initial guesses and bounds for each region of interest (ROI). Users may then initiate fitting, in which case COMKAT sequences through each ROI and estimates its parameter values by minimizing the difference between model output and the data. Fitted curves are then plotted on the output panel so that users can visually assess the fit. Goodness of fit can also be evaluated by sum-of-square errors and runs tests within COMKAT GUI. If desired, the user may adjust initial guesses or bounds and redo the estimation until a satisfactory fit is achieved.
TABLE 2.
Data Formats Supported by COMKAT GUI and COMKAT Image Tool
| Data type | Supported file formats in COMKAT |
|---|---|
| Input function | Text files (.txt), comma-separated-value files (.csv), Excel spreadsheets (.xls), MATLAB binary files (.mat) |
| Output function (time–activity curves) | Text files (.txt), comma-separated-value files (.csv), Excel spreadsheets (.xls), MATLAB binary files (.mat) |
| Image files | DICOM (part 10), NIFTI, Analyze, Siemens microPET (ASIPro), Siemens ECAT Exact (ECAT7), Philips Allegro and Gemini (ImageIO), Bruker Biospin |
Input Function GUI
Input functions are essential for kinetic modeling, and therefore there are provisions for handling them within the software. In COMKAT, there is an input function GUI specifically developed for loading input functions from data stored in a variety of file formats as listed in Table 2. In addition to the activities of blood samples, users also may specify the radionuclide, specific activity, hematocrit, and fraction of blood activity attributed to unmetabolized radiopharmaceutical in plasma. Within the GUI, users may adjust the input function, including the temporal offsets. The COMKAT input function GUI can also correct for spillover and partial-volume effects in an image-derived input function for small animals based on methods described by Fang and Muzic (14).
COMKAT Image Tool
The COMKAT image tool is the front-end GUI for image processing. Image-reading functions are written for a number of file formats as listed in Table 2. With the images loaded and displayed, users may adjust contrast, color lookup table, and zoom for better visualization. Users may also navigate spatially and temporally on the 3 orthogonal views sampled from the volume, including translations and rotations. For multimodality images, the COMKAT image tool is capable of loading and displaying 2 different image datasets as superimposed slices. The COMKAT image tool automatically processes the information about image orientation, resolution, and pixel spacing for both datasets so that they will be displayed with the same magnification and, if relevant information is available, the same positioning of the subject. Otherwise, an automatic image registration method is provided that uses the mutual-information similarity criterion (15,16) to help users coregister the image volumes.
Once the tissue or organ of interest is displayed, a user can draw ROIs on any view, slice, or frame on the images. For each ROI, the COMKAT image tool calculates the average pixel value and automatically converts that to the calibrated value in, for example, activity concentration or Hounsfield units. Users may also create volumes of interest in COMKAT image tool by drawing multiple ROIs and grouping them. The COMKAT image tool allows users to specify multiple ROIs or volumes of interest for the same dataset, and the associated time–activity curves will be returned back to the COMKAT GUI for model analysis. Optionally, the time–activity curves may be saved in text or spreadsheet-compatible files. The user-defined ROIs and volumes of interest themselves can be stored as files and later loaded into the COMKAT image tool for application to the same or other image data.
Distributed-Computing Functionality
COMKAT GUI is capable of pixelwise estimation of parameters. The GUI for computing parametric images uses the kinetic model specified in the COMKAT GUI, prompts users for estimation settings, and determines the parameter values for each pixel. For each pixel, its estimated parameter values are used to construct a new image volume, which is called a parametric image. A new volume will be saved for each parameter after the computation is completed. For example, with 18F-FDG there may be K1, k2, k3, and k4 images.
Because generation of parametric images is a pixelwise operation, it is especially computation-intensive. One method to accelerate this operation, when processing of pixels may be done independently, is to divide the calculation into subsets and assign each to a worker process that may be run on a selected core, central processing unit (CPU), or computer. In the case of distributed computing in MATLAB, each worker is a separate MATLAB process that can receive data, process the operation, and return results to the main MATLAB program. Therefore, the parametric imaging function in the COMKAT GUI is implemented to support distributed computing to enable users to make use of computational resources spanning from 2 cores in the CPU of a notebook computer to multiple cores of multiple-CPU computers in a cluster. The MATLAB function “parfor” under the Parallel Computing Toolbox is used in the COMKAT GUI to distribute the pixelwise computation over workers. Once the computation on all workers is complete, results are returned back to the user’s computer and automatically consolidated.
Validation and Performance Evaluation for COMKAT
COMKAT is validated both for its command-line functions and for the COMKAT GUI. This is particularly useful to confirm that COMKAT is functioning correctly after installation. The accuracy of COMKAT command-line functions has been verified in the previous publication (10) and further examined with more models under the new COMKAT validation suite. This suite solves for the model output using COMKAT and an independent reference method that is analytic, when possible. At completion, a hyper-text-markup-language report is created and displayed on the screen. The report includes plots of COMKAT and reference solutions obtained on the user’s computer and the same obtained on the developer’s computers. An example validation report based on 18F-FDG can be found in the supplemental materials (available online only at http://jnm.snmjournals.org). The performance of COMKAT command-line functions is evaluated by its speed for solving model equations for the output and sensitivity equations (10). This compiled solver is compared with the MATLAB built-in ordinary-differential-equation solver ode15s (17). Both solvers are used to solve the 18F-FDG model under the same model configuration. Performance is measured according to the mean time and SD, of 500 repetitions, to solve the model using a dual-core Core2 (Intel) desktop computer operating at 2.4 GHz.
The COMKAT GUI is tested with input functions and image data from small-animal PET studies to evaluate whether results from the COMKAT GUI equal those from scripts implemented with COMKAT command-line functions. The animal study protocol was approved by the Institutional Animal Care and Use Committee of Case Western Reserve University. To evaluate the speed and accuracy of COMKAT GUI, we used data from 1 female 236-g Sprague–Dawley rat. It was injected intravenously with 30.7 MBq of 18F-FDG and scanned with a microPET R4 system (Siemens) (18). Detailed data acquisition was described elsewhere (14), and this test dataset can be downloaded from the COMKAT Web site. A standard 2-compartment 18F-FDG kinetic model was used to generate model-predicted output and to fit experimental data (1). Initial values for k1 to k4 are 0.1, 0.2, 0.5, and 0.0069 min−1. Lower and upper bounds were set to zero and one, respectively. Parameters were first estimated for a brain ROI and a myocardium ROI with the COMKAT GUI. Then, the time–activity curves of these 2 ROIs were fitted for parameter estimation again with a MATLAB script that uses COMKAT command-line functions. Results from the COMKAT GUI and a script were compared.
Some of the major functions were tested for speed in the COMKAT GUI and COMKAT image tool. For the COMKAT GUI, speed is evaluated as the time required for initializing the GUI, loading a kinetic model from the templates, solving and plotting model output, and estimating parameters. Average time required is calculated from 10 repetitions. In the COMKAT image tool, speed is evaluated for loading the test dataset of small-animal PET images, summing frames, and refreshing the image each time the display is adjusted.
The performance advantage of distributed computing is evaluated for parametric imaging by comparing the time required for local computation versus that required for distributed computing in estimating 18F-FDG kinetic rate constants pixelwise in volume for a small-animal PET study of 44 frames, each consisting of 128 × 128 × 63 pixels. Either local computation is performed using 1 worker locally, or distributed computing is performed using 2, 4, 8, 16, or 31 workers on a mini cluster. The mini cluster consists of 4 PowerEdge 1950 III servers (Dell) running Windows Server 2003 (Microsoft). Each server has 2 quad-core Xeon (Intel) 2.5-GHz CPUs (32 cores total) and 9 GB of random-access memory. The speed-up ratio is calculated as follows:
where n denotes the number of workers (1 per core) used in a specific test in the distributed computing mode. Each test of a specific number of workers and local processing was executed 5 times, and the result was averaged.
RESULTS
Implementation and Validation
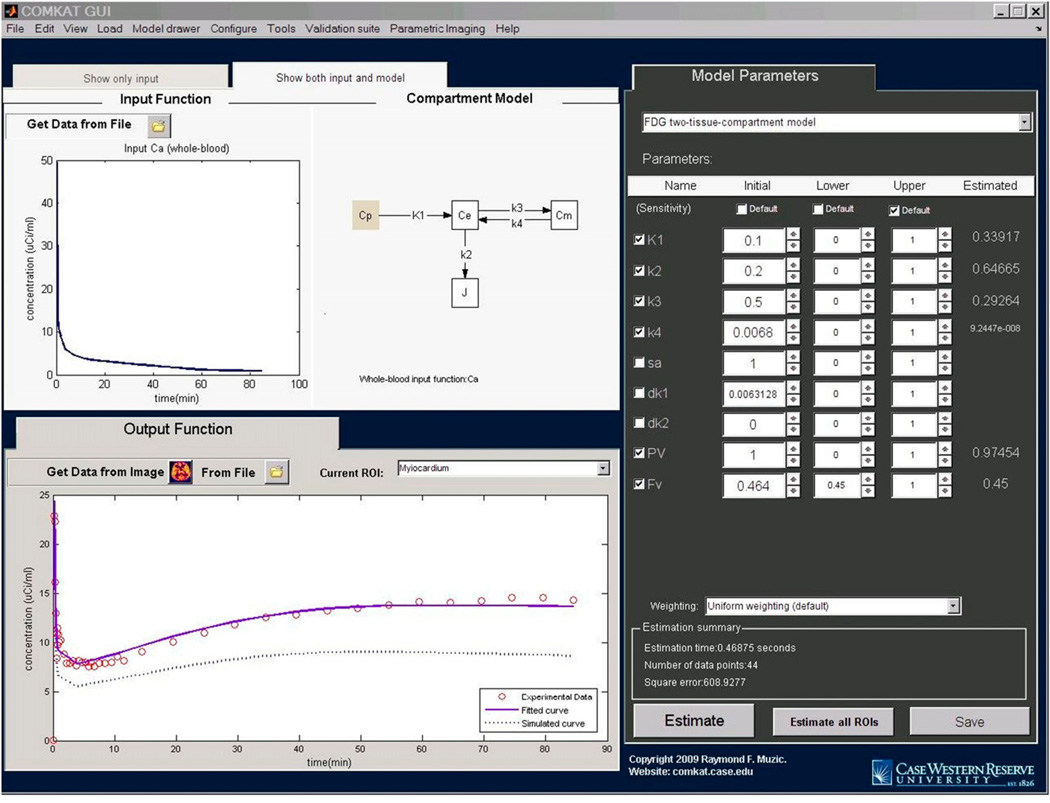
Supplemental Figure 1 shows a screen snapshot of the COMKAT GUI window before data are loaded or a model is specified. The input function data were loaded with the COMKAT input function GUI as shown in Supplemental Figure 2. The COMKAT input function GUI then returned the blood and plasma time–activity curves, in the form of first-order piecewise polynomial (i.e., linear interpolation), back to the COMKAT GUI, wherein the 2-compartment 18F-FDG model was selected from a drop-down list. Model-simulated output was automatically calculated—using default values for the model parameters—and plotted on the output function panel. Subsequently, the values of the parameters may be changed interactively and the plot is updated instantaneously. To specify experimental data for parameter estimation, COMKAT image tool was opened from the COMKAT GUI. Figure 2 shows the appearance of COMKAT image tool after loading and displaying a set of microPET R4 image data. Two ROIs were drawn on the brain and myocardium as shown. Corresponding time–activity curves were automatically calculated from the images and returned back to the COMKAT GUI. After the tissue time–activity curves were returned back to the COMKAT GUI, parameters for both ROIs were estimated as shown in Figure 3. These 2 ROIs were processed sequentially and independently, with the estimation results stored and displayed separately. Optionally, these results may be stored as a report in a spreadsheet file and can be used for further analysis.
FIGURE 2.
Appearance of COMKAT image tool when small-animal PET images are loaded. With COMKAT image tool, users may adjust the image display and draw ROIs or volumes of interest. In this figure, ROIs are drawn. On the sagittal view, an ROI is drawn on the brain. On the axial view, an ROI is drawn on the myocardium. Perpendicular views show ROIs in cross-section. Corresponding time–activity curves are shown on the plot. Once users finish drawing ROIs and click “Return to main GUI” button, COMKAT image tool returns ROI information and time–activity curves back to COMKAT GUI. Displayed time–activity curve on the bottom right panel is associated with the myocardial ROI.
FIGURE 3.
COMKAT GUI with input function and experimental data specified. On screen, both model and data are loaded. Users may click buttons next to parameter values or enter their values, and model output curves are immediately updated. Once the user is satisfied with the initial guess, the computer will optimize values to fit data when the “Estimate” button is clicked. Parameter values will be estimated, and the fitted curve will be plotted on output function figure. Values for rate constants are specified in units of min−1.
The accuracy of COMKAT command-line functions and of the COMKAT GUI was validated separately. Command-line functions were validated with the COMKAT validation suite. The model output and sensitivity functions were calculated using COMKAT and a reference method. As expected, the different solutions agreed closely, with a maximum difference of 0.01%. After the command-line functions were validated, parameter estimation results obtained from the COMKAT GUI were compared with those obtained using command-line functions. Parameter estimation results were identical between the GUI-based and command-line implementation.
Performance Evaluation
The speed evaluation of the major COMKAT command-line functions, COMKAT GUI, and COMKAT image tool is summarized in Table 3. The speed of COMKAT command-line functions was evaluated for solving the 18F-FDG model output with both the compiled and the built-in solvers. The model output was solved by a compiled solver in 3.2 ± 0.1 ms. This was approximately 40 times faster than with the noncompiled ode15s solver (0.12 ± 0.01 s).
TABLE 3.
Summary of Computation Speed for Major Functions in COMKAT
| COMKAT command-line functions, output solving |
COMKAT GUI |
COMKAT image tool |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | Compiled solver |
Built-in solver (ode15s) |
Initialization | Model loading |
Output solving and plotting |
Parameter estimation |
Reading and displaying images |
Frame summation |
Refreshing display |
| Mean | 3.2 | 124.1 | 519.6 | 615.6 | 154.2 | 595.2 | 8,146.0 | 2,044.0 | 120.7 |
| SD | 0.1 | 9.4 | 9.9 | 102.1 | 0.8 | 1.5 | 1,004.3 | 47.0 | 26.7 |
Values are time, in milliseconds. Computation is based on the 18F-FDG kinetic model of small-animal PET rat study data. For COMKAT command-line functions, the model was solved 500 times and mean and SD calculated. For GUIs, 10 repetitions were used.
Average times required to complete some major functions of the COMKAT GUI and COMKAT image tool were measured. Loading of the approximately 700-MB image (44 frames, 128 × 128 × 63 pixels) was found to be the most time-consuming, requiring 8.1 ± 1.0 s. Summing all 44 frames in volume took approximately 2 s. All other tested major functions of the COMKAT GUI and COMKAT image tool took less than 0.7 s. Solving models and refreshing plots was especially fast, averaging 0.15 ± 0.00 s.
The performance of distributed computing in the pixelwise parameter estimation for parametric images was evaluated. Results of the computation for parametric images were summarized in Table 4. The speed-up ratio was approximately linear with respect to the number of workers, with r = 0.99. Maximum acceleration was 25 times faster when 31 workers were used, reducing the time required for computing a complete volume of parametric imaging from approximately 150 min to approximately 6 min.
TABLE 4.
Summary of Computation Speed for Parametric Imaging
| Number of workers |
||||||
|---|---|---|---|---|---|---|
| Parameter | Local | 2.0 | 4.0 | 8.0 | 16.0 | 31.0 |
| Time, mean (min) | 149.49 | 85.48 | 44.14 | 22.79 | 11.38 | 5.97 |
| Time, SD (min) | 0.15 | 0.12 | 0.12 | 0.08 | 0.02 | 0.01 |
| Speed-up ratio | 1.75 | 3.39 | 6.56 | 13.14 | 25.05 | |
| Efficiency (%) | 87.45 | 84.68 | 81.99 | 82.12 | 80.82 | |
Mean time required for computation was calculated from 5 repetitions of computing a parametric image for the whole volume. Local computation was used as a reference for calculating speed-up ratios by estimating parameters pixelwise on 1 core of 1 CPU without distributed computing. Efficiency was calculated as (speed-up ratio/number of workers). With regression analysis, linear relationship was found in speed-up ratio (y) vs. number of workers (x) as y = 0.812x, with r = 0.99.
Software Distribution
COMKAT was packaged for users to download from the Web site (http://comkat.case.edu) as an open-source project with free licenses for academic research use. COMKAT has been supported and tested on multiple operating systems including Windows XP and Vista (Microsoft Corp.) (32- and 64-bit), Linux (Ubuntu 32- and 64-bit), and MacOS X (Apple Inc.). The COMKAT GUI was compiled with the MATLAB compiler into a standalone executable file that also contained the compiled COMKAT image tool and the COMKAT input function GUI. This compiled COMKAT distribution does not require users to have MATLAB installed; however, it does require users to install The MathWorks’ MATLAB Compiler Runtime (MCR), which is redistributable without licensing fees. The compiled version of COMKAT supports most of the full functionality of the standard version of COMKAT as detailed in Table 1.
DISCUSSION
In molecular imaging, numerous reports have proven the value of using kinetic modeling to absolutely quantify physiology (1,19–21). However, lack of software that is specifically designed for applying kinetic modeling to image data has been an impediment for researchers and particularly those without a computer programming background. This report describes the software package COMKAT, which we believe will be useful to many researchers for their needs in image quantification. COMKAT has several unique features. First, COMKAT may be downloaded without licensing costs for academic, nonprofit research applications. Users incur costs for MATLAB licensing fees only if they do not already have MATLAB or if they require functionality not available in the compiled version of COMKAT. Second, COMKAT has been developed as an open-source project to maximize transparency, extensibility, and collaboration. For example, users may trace the program to diagnose potential errors or add new functionality to COMKAT for their specific needs in image quantification. Experienced users may modify the source code of COMKAT, such as modifying the convergence criteria for kinetic models in the COMKAT GUI, or may even create their own GUI supported by the COMKAT command-line functions. Third, COMKAT is developed as a user-friendly application with various GUIs. Users who do not have any experience in MATLAB programming can refer to our documentation about the COMKAT GUI and quickly learn how to apply COMKAT for their data analysis. As for data compatibility, COMKAT already supports various image and data formats, including DICOM, and can be extended by users to support additional formats. Finally, COMKAT allows users to easily add novel kinetic models either with a GUI or with command-line functions. For users who wish to evaluate different kinetic models, COMKAT can help streamline and simplify model development, output solving, and analysis for sensitivity functions.
COMKAT has also been shown to provide a significant improvement in speed, compared with early releases in kinetic modeling. In a previous report of COMKAT performance, solving an 18F-FDG model for output and sensitivity functions with a noncompiled ode15s solver took 1.14 ± 0.01 s (10). Using the same benchmarking program with the current versions of COMKAT and ode15s, the computation time is now 0.12 ± 0.01 s. The approximately 10-fold performance improvement is attributed mainly to improvements in computer hardware. When this improvement is combined with the approximately 40-fold (Table 2) advantage of the compiled solver over ode15s that is independent of hardware gains, the new version of COMKAT is 350-fold faster than the 2001 version of COMKAT (10). This improvement is important because during the iterations within parameter estimation, model output and sensitivity functions will be solved many times before the convergence. Furthermore, the extremely fast speed for solving the model (3.2 ± 0.1 ms) resolves potential concerns about solution efficiency in the MATLAB environment.
Based on the speed improvement from this compiled solver, the COMKAT GUI can solve for model output and refresh the plot within 0.2 s each time a parameter value is changed. In fact, because our results show that the time required for model solving is on the order of milliseconds, most of the 0.2 s is actually spent on updating the graphics. This fast computation speed in GUI helps users by allowing them to quickly simulate model output or make appropriate initial guesses for parameter estimation. Other major functions of the COMKAT GUI and COMKAT image tools have also been evaluated. For example, loading image data is intrinsically a time-consuming computation. With the COMKAT image tool, only about 8 s are required to load a 700-MB 4-dimensional dataset. Display adjustment is also done quickly, in less than 0.2 s. In summary, the speed in COMKAT for both command-line and GUI functions is fast and will help users to increase throughput in image quantification.
Another unique and useful feature of COMKAT is the capability to generate parametric images with support for distributed computing. This report shows that distributed computing can be applied to accelerate pixelwise estimation by using the computation power of a cluster system. When 31 workers were used, the COMKAT GUI achieved approximately 25-fold acceleration and reduced the time requirement from 149.49 ± 0.15 to 5.97 ± 0.01 min. This represents an efficiency of 25/31, or about 80%. Because the speed-up ratio has an approximately linear dependence on the number of workers up to 31 as shown in Table 4, further speed enhancement can be expected if more workers are used.
COMKAT is an ongoing project, and we will continue to maintain and enhance it to the best of our ability. COMKAT will be expanded by adding more kinetic models, supporting more imaging modalities such as dynamic contrast-enhanced MRI (22), and improving the functionality available to users without MATLAB licenses. Documentation continues to be added to the COMKAT Web site, especially for examples of specific research applications. We hope to see more users benefit from using COMKAT as the software environment for molecular imaging research.
CONCLUSION
COMKAT is a software package capable of analyzing molecular images integrated with kinetic modeling methods. COMKAT has numerous useful features, including seamless connection between imaging and modeling GUIs, support for user-defined kinetic models and experimental data, parameter estimation, parametric imaging functionality, and no licensing fees for academic research use. COMKAT is suitable for a wide spectrum of data analysis tasks, including quantifying physiology, kinetic model development, image processing, and data simulation. To download COMKAT, please visit http://comkat.case.edu.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by NIH grants R33-CA-101073 and R24-CA-110943.
REFERENCES
- 1.Huang SC, Phelps ME, Hoffman EJ, Sideris K, Selin CJ, Kuhl DE. Noninvasive determination of local cerebral metabolic rate of glucose in man. Am J Physiol. 1980;238:E69–E82. doi: 10.1152/ajpendo.1980.238.1.E69. [DOI] [PubMed] [Google Scholar]
- 2.Phelps ME, Huang SC, Hoffman EJ, Selin C, Sokoloff L, Kuhl DE. Tomographic measurement of local cerebral glucose metabolic rate in humans with (F-18)2-fluoro-2-deoxy-D-glucose: validation of method. Ann Neurol. 1979;6:371–388. doi: 10.1002/ana.410060502. [DOI] [PubMed] [Google Scholar]
- 3.Huesman RH, Knittel BL, Mazoyer BM, et al. Notes on RFIT: A Program for Fitting Compartmental Models to Region-of-Interest Dynamic Emission Tomographic Data. Berkeley, CA: Lawrence Berkeley Laboratory; 1993. Report LBL-37621. [Google Scholar]
- 4.Farenc C, Fabreguette JR, Bressolle F. Pk-fit: a pharmacokinetic/pharmacodynamic and statistical data analysis software. Comput Biomed Res. 2000;33:315–329. doi: 10.1006/cbmr.2000.1548. [DOI] [PubMed] [Google Scholar]
- 5.Barrett PH, Bell BM, Cobelli C, et al. SAAM II: simulation, analysis, and modeling software for tracer and pharmacokinetic studies. Metabolism. 1998;47:484–492. doi: 10.1016/s0026-0495(98)90064-6. [DOI] [PubMed] [Google Scholar]
- 6.Loening AM, Gambhir SS. AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging. 2003;2:131–137. doi: 10.1162/15353500200303133. [DOI] [PubMed] [Google Scholar]
- 7.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–43. [Google Scholar]
- 8.Burger C, Buck A. Requirements and implementation of a flexible kinetic modeling tool. J Nucl Med. 1997;38:1818–1823. [PubMed] [Google Scholar]
- 9.Huang SC, Truong D, Wu HM, et al. An Internet-based “kinetic imaging system” (KIS) for microPET. Mol Imaging Biol. 2005;7:330–341. doi: 10.1007/s11307-005-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muzic RF, Jr, Cornelius S. COMKAT: compartment model kinetic analysis tool. J Nucl Med. 2001;42:636–645. [PubMed] [Google Scholar]
- 11.Matlab: The Language of Technical Computing. Natick, MA: The MathWorks, Inc; 1996. [Google Scholar]
- 12.Hindmarsh AC, Serban R. User Documentation for CVODES, an ODE Solver with Sensitivity Analysis Capabilities. Livermore, CA: Lawrence Livermore National Laboratory; 2002. p. 189. [Google Scholar]
- 13.Muzic RF, Jr, Christian BT. Evaluation of objective functions for estimation of kinetic parameters. Med Phys. 2006;33:342–353. doi: 10.1118/1.2135907. [DOI] [PubMed] [Google Scholar]
- 14.Fang YH, Muzic RF., Jr Spillover and partial-volume correction for image-derived input functions for small-animal 18F-FDG PET studies. J Nucl Med. 2008;49:606–614. doi: 10.2967/jnumed.107.047613. [DOI] [PubMed] [Google Scholar]
- 15.Fei B, Wheaton A, Lee Z, Duerk JL, Wilson DL. Automatic MR volume registration and its evaluation for the pelvis and prostate. Phys Med Biol. 2002;47:823–838. doi: 10.1088/0031-9155/47/5/309. [DOI] [PubMed] [Google Scholar]
- 16.Pluim JP, Maintz JB, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 17.Shampine LF, Reichelt MW. The MATLAB ODE suite. SIAM J Sci Comput. 1997;18:1–22. [Google Scholar]
- 18.Knoess C, Siegel S, Smith A, et al. Performance evaluation of the microPET R4 PET scanner for rodents. Eur J Nucl Med Mol Imaging. 2003;30:737–747. doi: 10.1007/s00259-002-1052-6. [DOI] [PubMed] [Google Scholar]
- 19.Muzik O. Validation of nitrogen-13-ammonia tracer kinetic model for quantification of myocardial blood flow using PET. J Nucl Med. 1993;34:83–91. [PubMed] [Google Scholar]
- 20.Acton PD, Zhuang H, Alavi A. Quantification in PET. Radiol Clin North Am. 2004;42:1055–1062. doi: 10.1016/j.rcl.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Price JC, Klunk WE, Lopresti BJ, et al. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh compound-B. J Cereb Blood Flow Metab. 2005;25:1528–1547. doi: 10.1038/sj.jcbfm.9600146. [DOI] [PubMed] [Google Scholar]
- 22.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.