Abstract
A proteomic approach was taken to study how fish respond to changes in calorie availability, with the longer-term goal of understanding the evolution of lipid metabolism in vertebrates. Zebrafish (Danio rerio) were fed either high (3 rations/day) or low (1 ration/7 days) calorie diets for 5 weeks and liver proteins extracted for proteomic analyses. Proteins were separated on two-dimensional electrophoresis gels and homologous spots compared between treatments to determine which proteins were up-regulated with high-calorie diet. Fifty-five spots were excised from the gel and analyzed via LC–ESI MS/MS, which resulted in the identification of 69 unique proteins (via multiple peptides). Twenty-nine of these proteins were differentially expressed between treatments. Differentially expressed proteins were mapped to Gene Ontology (GO) terms, and these terms compared to the entire zebrafish GO annotation set by Fisher's exact test. The most significant GO terms associated with high-calorie diet are related to a decrease in oxygen-binding activity in the high-calorie treatment. This response is consistent with a well-characterized response in obese humans, indicating there may be a link between lipid storage and hypoxia sensitivity in vertebrates.
Keywords: Danio rerio, Diet, Gene ontology, Heme proteins, Obesity, Oxygen binding activity
1. Introduction
Calorie availability pivotally affects metabolic rate, reproductive fitness, growth, and survival in fish (Tocher, 2003). Calories in the form of fatty acids are the most significant source of ATP for many species of fish. Accordingly, fish manipulate storage and mobilization of fatty acids as part of their natural history. Striped bass increase intracellular lipid droplets 13-fold in red muscle during cold acclimation (Egginton and Sidell, 1989) and salmon increase serum cholesterol without eating during spawning migrations (Farrell and Munt, 1983). The expression of lipid metabolism genes is regulated by free long-chain fatty acids and their metabolic by-products, however the definitive mechanism by which they do so remains elusive (Duplus et al., 2000). Characterizing the molecular signaling behind these changes in lipid metabolism has traditionally been approached by looking at candidate proteins, (e.g. fatty acid binding proteins (FABPs; Londraville and Sidell,1995) organelle function (peroxisomes; Crockett and Sidell, 1993), and enzymatic indicators of fatty acid flux (Crockett et al., 1999). However, these specific indicators are only part of a poorly characterized integrated system through which cells respond to changes in calorie availability.
Studying the effects of a single protein in isolation is limited in explanatory power for many reasons, one of which is uncertainty about how the rest of the proteome is responding. Proteomics can quantitatively estimate changes in tissue protein expression on a large scale (Abbott, 1999). By using protein extracted from tissue, proteomics documents those proteins that are actually expressed (vs. those potentially expressed as in microarray studies, e.g. (Hogstrand et al., 2002; Paulson et al., 2003). Also, by separating large numbers of proteins simultaneously, researchers can examine relative differences in expression among pathways.
In this study, we employed proteomics to gain further insight into how fish respond to changes in calorie availability. Our long-term goal is to characterize the evolution of lipid metabolism in vertebrates. As the most basal vertebrate class, fish represent a valuable reference for comparison to mammalian lipid metabolism. Using proteomics, we documented expression differences in fish under increased-calorie diet. We chose to use zebrafish (Danio rerio) due to the near-complete knowledge of their genome (www.sanger.ac.uk), which greatly facilitates protein identification. The liver proteome was chosen because it is a central lipid-storage organ in fish (Sheridan, 1988; Sheridan, 1994) and because zebrafish on high-calorie diets increase liver lipid storage (Schlegel and Stainier, 2006).
Our a priori hypothesis was that proteins in lipid metabolism pathways would be the predominant up-regulated proteins in livers of fish on a high-calorie diet. However, the largest signal comes from changes in oxygen-binding heme proteins. This is consistent with a well-established response in humans (Le Guenno et al., 2007), but is not a pathway we were likely to pursue had it not been suggested by proteomics. This illustrates the power and utility of this approach for questions in comparative biology.
2. Materials and methods
2.1. Animals
Wild-type, age-matched adult D. rerio (Cypriniformes, Cyprinidae) were purchased from Scientific Hatcheries Inc. (Huntington Beach, CA, USA). Zebrafish were sorted by gender and treatment in separate 38-liter aquaria (26 fish/tank) with filtered, dechlorinated tap water at a constant 28.5 °C. Water quality was monitored for ammonia levels and temperature daily. Fish were fed Scientific Hatcheries (Huntington Beach, CA) 3-pigment mash fed 3×/day (high-calorie) or 1×/week (low-calorie) at ~1% of body weight per feeding via automatic feeders for 5 weeks. Diet composition was: protein 52%, carbohydrate 19%, fat 10%, ash 10%, moisture 8%, and fiber 1% (pers. comm. Silver Cup Fish Feed, Murray, UT, USA). In the high-calorie treatment, this left a minimal excess of food, ensuring fish ate to satiety (excess food and feces removed daily). After completion of the experiment, treatment animals were euthanized by overdose of clove oil.
Total length, weight, and body condition (100*mass/length3) were recorded from all individuals (Table 1). Whole-body total fat mass was determined from a subset of each population via petroleum ether extraction (Folch et al., 1957) and total protein by Bradford assay (Bradford, 1976). Morphometric data were compared between high and low-calorie treatments using a non-parametric Wilcoxin sign-rank test. Because females on the low-calorie treatment were smaller, livers had to be pooled for the proteomics experiments, thus reducing the sample size available for fat mass and total protein determination. This left a relatively small sample size in the low-calorie group, which was not normally distributed and thus would violate the assumption of a parametric test. From the remaining individuals, liver was harvested and immediately frozen in liquid nitrogen.
Table 1.
Whole-body diet effects
| Sex | Treatment | Lipid % dry weighta | Protein % dry weight | Initial total weight (g) | Final total weight (g) | Final condition factorb |
|---|---|---|---|---|---|---|
| Female | High-calorie | 30.6 ± 2.5c* (14) | 34.8 ± 2.9 (12) | 0.85 ± 0.04 (26) | 0.90 ± 0.04(26)c*** | 1.48 ± 0.05c* (24) |
| Female | Low-calorie | 16.0 ± 6.7 (3) | 24.0 ± 2.7 (3) | 0.62 ± 0.03 (25) | 1.11 ± 0.03 (25) | |
| Male | High-calorie | 26.7 ± 5.4 (11) | 38.0 ± 14.7 (13) | 0.71 ± 0.03 (25) | 0.64 ± 0.23 (25)c*** | 1.08 ± 0.03c* (26) |
| Male | Low-calorie | 16.4 ± 2.7 (7) | 34.4 ± 4.7 (8) | 0.45 ± 0.02 (26) | 0.88 ± 0.02 (26) |
Variables measured on a whole-body scale for zebrafish on the two diets.
Extracted weight of lipid expressed as a percentage of lyophilized total weight of the fish.
Condition factor was defined as 100 times the mass divided by the cube of the length.
Indicates a significant difference between high and low-calorie treatment as detected by the Wilcoxon sign-rank test (*p<0.05, ***p<0.001). Sample size indicated in parentheses. All errors are standard error.
2.2. 2-D electrophoresis
Only females were characterized for protein expression changes with diet due to their greater magnitude change in body composition under to 2 diet regimes, and due to their significantly larger livers (final female liver weight 2–3×> male). A 10% (w/v) homogenate of each zebrafish liver was made in 50 mM Tris (pH 7.8) via sonication (5 s at 5 W). Total protein (1 mg/first dimension strip, as estimated by Bradford assay) was extracted using a commercial kit (Perfect-FOCUS, GenoTech, St. Louis, MO, USA).
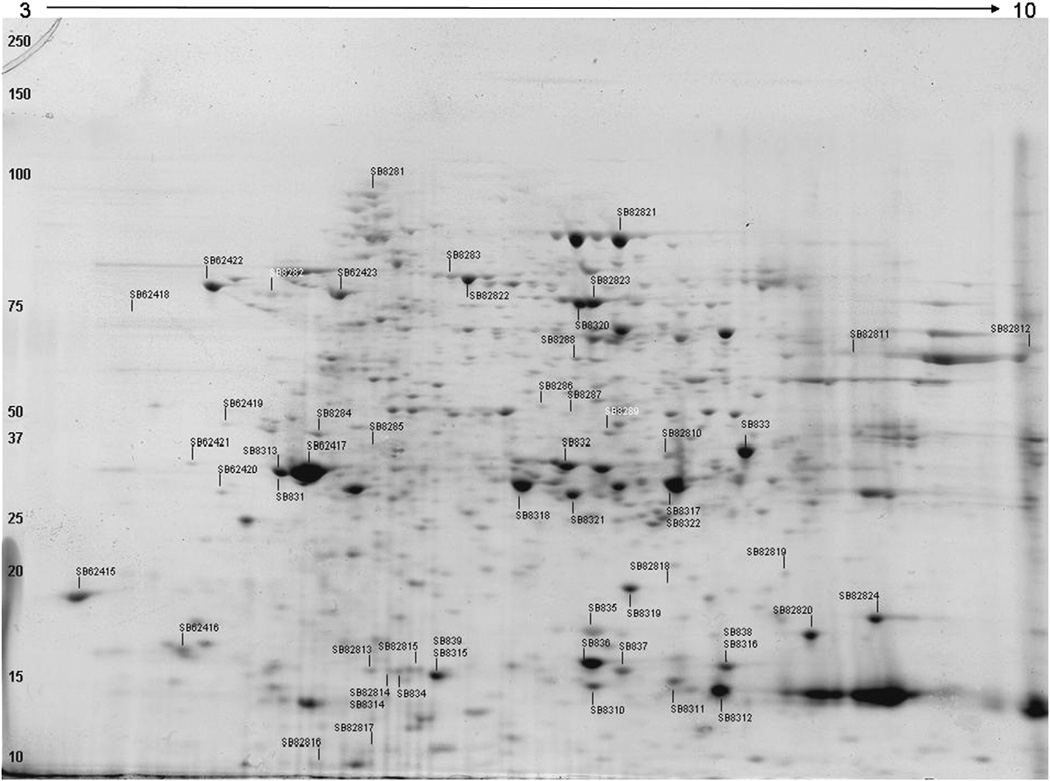
Extracted protein pellets were resuspended in a 2-D suspension buffer consisting of 7 M urea, 2 M thiourea, 1% (v/v) Triton X-100, 100 mM DTT, 1% (v/v) ampholytes,1% (w/v) CHAPS,1% (v/v) of protease inhibitor cocktail (Sigma), and a trace of bromophenol blue. First-dimension focusing was done on commercial immobilized pH gradient strips (BioRad IEF strips, pH 3–10) with active overnight rehydration 50 V overnight (BioRad Protein IEF Cell), followed by focusing to an accumulation of 10,000 V h (gradient of 0–1000 V for approximately 2.5 h). Following focusing, strips were equilibrated in 50 mM Tris, 6 M urea, 30% (v/v) glycerol, and 1% (w/v) SDS, and 3.3 mM DTT, followed by a second equilibration was done in 50 mM Tris, 6 M urea, 35% v/v glycerol, and 1% (w/v) SDS, and 12 mM iodoacetamide. Equilibrated strips were run on 15% polyacrylamide gels (BioRad Criterion). Gels were fixed in 50% ETOH, 10% acetic acid and stained with a colloidal Coomassie blue stain (GelCode Blue, Pierce, Rockford, IL, USA) (Fig. 1).
Fig. 1.
Representative 2-DE electrophoresis map of zebrafish liver (female, low-calorie treatment), visualized with Coomassie brilliant blue. The pH gradient is listed at the top and MW (kDA) on the left. All labeled spots were characterized by LC–ESI MS/MS; multiple numbers on the same spot indicated multiple analyses from different fish. Labels without obvious spots are only visible in high-calorie treatments.
Stained gels were scanned with a transmittance scanner (Epson 2450, 3.3 Dynamic range) and analyzed with Melanie software (version 4.0, Genebio, Geneva, Switzerland). This software can quantify spot volume (area*density) and normalize values relative to total staining intensity, correcting for gel-to-gel variation in total protein loaded. Spot selecting algorithms matched proteins among replicate gels from established landmarks, accounting for local distortions in electrophoretic migration and pairing them to a master gel.
2.3. Mass spectrometry
Spots selected for mass spectrometry analyses were excised from the gel and digested in-gel using the protocol detailed in (Goel et al., 2005). Briefly, spots were destained, dehydrated in acetonitrile, washed in 0.1 M ammonium bicarbonate, and again dehydrated in acetonitrile before drying in a vacuum centrifuge. Trypsin (Mass Spec grade, Promega) was prepared in 50 mM ammonium bicarbonate to a concentration of 20 ng/µL and enough volume added to just rehydrate the gel pieces (typically 10–20 µL). Digestion proceeded overnight at room temperature. Digested peptides were extracted from the gel piece in 50% acetonitrile/5% formic acid, concentrated in a vacuum centrifuge, and resuspended in 1% acetic acid for injection into the HPLC.
Digests were analyzed on either a LCQ Deca quadrupole ion trap or a LTQ linear ion trap mass spectrometer (ThermoFisher, San Jose, CA, USA) utilizing an operator packed 8 cm × 75 µm id Phenomenex Jupiter C18 reversed-phase capillary chromatography column. The individual extracts were injected and the peptides eluted from the column by an acetonitrile/0.05 M acetic acid gradient at a flow rate of ~200 nL/min. The microelectrospray ion source was operated at 2.5 kV. Digests were analyzed using the data dependent multitask capability of the instrument, acquiring full scan mass spectra to determine peptide molecular weights and collisionally induced dissociation (CID) product ion spectra to determine amino acid sequence in successive instrument scans.
The data were analyzed by using all CID spectra collected in the experiment to search the NCBI non-redundant database with the search program Mascot (www.matrixscience.com). Parameters were set to monoisotopic, peptide tolerance ± 3 Da, MS/MS tolerance ± 1.5 Da, fixed modifications: carbamidomethyl (C-term) and variable modifications Pyro-glu (N-term Q) and oxidation (M). Putative matches (Probability Mowse scores> 100, corresponding to p<0.05 that match is a random event) verified by manual interpretation. If the search program was unable to identify the protein based on these spectra, then the CID spectra were interpreted manually by a published strategy (Kinter and Sherman, 2000) to determine the amino acid sequence. This sequence was then used to search sequence databases. The great majority (83%) of identifications were made by ≥3 peptides covering ≥10% of total amino acid sequence. All spectra were searched against a chicken + zebrafish RefSeq database (NCBI) compiled 12/07.
2.4. Analysis
Relative pixel volume ((spot area*density)/total pixel density) for all spots on each gel was estimated by Melanie software, and compared across and within groups for matched spots (spots were matched by the software and matched sets verified visually). Both ‘landmark’ spots (prominent spots on the gel used for orientation) and putative differentially expressed spots were selected for MS identification. Spots were identified as putatively differentially expressed if: 1) the same spot was found in at least 2 gels per treatment and its mean relative pixel volume varied by > 200% between treatments, and variability within a treatment was <60% (standard deviation as a percent of the mean), or if 2) the spot was present in multiple gels of one treatment and absent in the other.
All differentially expressed proteins (Table 3) were matched to Gene Ontology (GO) terms using Blast2Go software (Conesa et al., 2005). To determine which GO terms were expressed as an effect of treatment, we used Fisher's exact test to determine whether GO terms from differentially expressed proteins represented a non-random subset of all mapped GO terms for zebrafish. Analyses were done separately for differentially expressed proteins (between groups) defined one of four ways:
relative pixel volume significantly different via Student's t-test
mean relative pixel volume ≥2-fold different
presence in one group and absent in another
categories 1–3 combined.
Table 3.
Gene ontology terms associated with differentially expressed proteins
| A. | |
|---|---|
| GO ID and descriptiona | p-valueb |
| GO:0030731 : guanidinoacetate N-methyltransferase activity | 2.30E-04 |
| GO:0008199 : ferric iron binding | 0.004823142 |
| GO:0008757 : S-adenosylmethionine-dependent methyltransferase activity | 0.010995826 |
| GO:0005319 : lipid transporter activity | 0.016455839 |
| GO:0016853 : isomerase activity | 0.042049128 |
| GO:0008168 : methyltransferase activity | 0.043158809 |
| GO:0016741 : transferase activity, transferring one-carbon groups | 0.044489012 |
| GO:0003735 : structural constituent of ribosome | 0.053097825 |
| GO:0008289 : lipid binding | 0.062514577 |
| GO:0005506 : iron ion binding | 0.090085189 |
| GO:0004252 : serine-type endopeptidase activity | 0.098139 |
| B. | |
|---|---|
| GO ID and description c | p-valueb |
| GO:0004364 : glutathione transferase activity | 0.00551 |
| GO:0003779 : actin binding | 0.00585 |
| GO:0008092 : cytoskeletal protein binding | 0.008973 |
| GO:0004175 : endopeptidase activity | 0.01043 |
| GO:0005344 : oxygen transporter activity | 0.010536 |
| GO:0019825 : oxygen binding | 0.015539 |
| GO:0004298 : threonine endopeptidase activity | 0.015993 |
| GO:0004252 : serine-type endopeptidase activity | 0.017129 |
| GO:0016859 : cis–trans isomerase activity | 0.018258 |
| GO:0003755 : peptidyl-prolyl cis–trans isomerase activity | 0.018258 |
| GO:0016765 : transferase activity, transferring alkyl or aryl (other than methyl) groups | 0.019615 |
| GO:0008236 : serine-type peptidase activity | 0.0203 |
| GO:0017171 : serine hydrolase activity | 0.0203 |
| GO:0008236 : serine-type peptidase activity | 0.0203 |
| GO:0004089 : carbonate dehydratase activity | 0.020971 |
| GO:0008233 : peptidase activity | 0.024492 |
| GO:0003824 : catalytic activity | 0.03972 |
| GO:0016836 : hydro-lyase activity | 0.039765 |
| GO:0016835 : carbon-oxygen lyase activity | 0.043307 |
| GO:0016787 : hydrolase activity | 0.068755 |
| GO:0016853 : isomerase activity | 0.082339 |
| C. | |
|---|---|
| GO ID and description d | p-valueb |
| GO:0005344 : oxygen transporter activity | 5.02E-08 |
| GO:0019825 : oxygen binding | 1.69E-07 |
| GO:0046906 : tetrapyrrole binding | 1.33E-04 |
| GO:0020037 : heme binding | 1.33E-04 |
| GO:0005506 : iron ion binding | 2.96E-04 |
| GO:0004069 : aspartate transaminase activity | 0.001035 |
| GO:0004422 : hypoxanthine phosphoribosyltransferase activity | 0.00138 |
| GO:0008943 : glyceraldehyde-3-phosphate dehydrogenase activity | 0.002758 |
| GO:0004365 : glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity | 0.002758 |
| GO:0048037 : cofactor binding | 0.005614 |
| GO:0004550 : nucleoside diphosphate kinase activity | 0.00654 |
| GO:0004550 : nucleoside diphosphate kinase activity | 0.00654 |
| GO:0016620 : oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | 0.009283 |
| GO:0016776 : phosphotransferase activity, phosphate group as acceptor | 0.010993 |
| GO:0016903 : oxidoreductase activity, acting on the aldehyde or oxo group of donors | 0.013042 |
| GO:0008483 : transaminase activity | 0.013383 |
| GO:0022892 : substrate-specific transporter activity | 0.013863 |
| GO:0051287 : NAD binding | 0.015769 |
| GO:0016769 : transferase activity, transferring nitrogenous groups | 0.018149 |
| GO:0019205 : nucleobase, nucleoside, nucleotide kinase activity | 0.022894 |
| GO:0016763 : transferase activity, transferring pentosyl groups | 0.02357 |
| GO:0005215 : transporter activity | 0.028768 |
| GO:0030170 : pyridoxal phosphate binding | 0.029973 |
| GO:0019842 : vitamin binding | 0.061757 |
| GO:0050662 : coenzyme binding | 0.076978 |
| D. | |
|---|---|
| GO IDe | p-valueb |
| GO:0005344 : oxygen transporter activity | 6.79E-09 |
| GO:0019825 : oxygen binding | 3.53E-08 |
| GO:0005506 : iron ion binding | 4.54E-05 |
| GO:0046906 : tetrapyrrole binding | 2.38E-04 |
| GO:0020037 : heme binding | 2.38E-04 |
| GO:0030731 : guanidinoacetate N-methyltransferase activity | 9.97E-04 |
| GO:0004069 : aspartate transaminase activity | 0.002989 |
| GO:0004422 : hypoxanthine phosphoribosyltransferase activity | 0.003983 |
| GO:0008943 : glyceraldehyde-3-phosphate dehydrogenase activity | 0.007951 |
| GO:0004365 : glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity | 0.007951 |
| GO:0004252 : serine-type endopeptidase activity | 0.009592 |
| GO:0004364 : glutathione transferase activity | 0.011903 |
| GO:0008236 : serine-type peptidase activity | 0.012238 |
| GO:0017171 : serine hydrolase activity | 0.012238 |
| GO:0008236 : serine-type peptidase activity | 0.012238 |
| GO:0005215 : transporter activity | 0.01401 |
| GO:0016853 : isomerase activity | 0.014701 |
| GO:0022892 : substrate-specific transporter activity | 0.017835 |
| GO:0004175 : endopeptidase activity | 0.018272 |
| GO:0004550 : nucleoside diphosphate kinase activity | 0.018783 |
| GO:0004550 : nucleoside diphosphate kinase activity | 0.018783 |
| GO:0008199 : ferric iron binding | 0.020741 |
| GO:0003779 : actin binding | 0.026343 |
| GO:0016620 : oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | 0.02659 |
| GO:0003824 : catalytic activity | 0.02974 |
| GO:0008289 : lipid binding | 0.031318 |
| GO:0016776 : phosphotransferase activity, phosphate group as acceptor | 0.031439 |
| GO:0004298 : threonine endopeptidase activity | 0.034337 |
| GO:0016903 : oxidoreductase activity, acting on the aldehyde or oxo group of donors | 0.037227 |
| GO:0008483 : transaminase activity | 0.038189 |
| GO:0016859 : cis–trans isomerse activity | 0.039149 |
| GO:0003755 : peptidyl-prolyl cis–trans isomerse activity | 0.039149 |
| GO:0008092 : cytoskeletal protein binding | 0.039528 |
| GO:0016765 : transferase activity, transferring alkyl or aryl (other than methyl) groups | 0.042025 |
| GO:0048037 : cofactor binding | 0.04391 |
| GO:0051287 : NAD binding | 0.044893 |
| GO:0004089 : carbonate dehydratase activity | 0.044893 |
| GO:0008757 : S-adenosylmethionine-dependent methyltransferse activity | 0.0468 |
Gene ontology terms and the probability that the GO term is identified randomly for differentially expressed proteins, defined 3 different ways (A,B,C).
A) Gene ontology terms associated with proteins differentially expressed via Student's t-test (see Table 2).
B) Gene ontology terms associated with proteins differentially expressed via presence in low-calorie and absence in high-calorie (see Table 2).
C) Gene ontology terms associated with proteins differentially expressed via ≥2-fold difference in mean pixel volume between treatments (see Table 2).
D) Gene ontology terms associated with proteins differentially expressed via all methods combined (t-test, ≥2-fold difference, presence/absence; see Table 2).
Probability value for Fisher's exact test-comparing number of GO terms nested within each GO term with total GO terms in the category for the entire zebrafish reference set. Terms that are significant for Fisher's exact test after Bonferroni correction are highlighted.
GO terms were judged significant if they exceeded the Bonferroni-corrected probability value (0.05/77 GO groups = 0.00065).
3. Results and discussion
Few proteomics studies exist for fishes, probably due to both the relative novelty of the technique and the relative paucity of fish genomic information compared to mammals. The earliest efforts at proteome screening in fish documented changes in protein expression patterns without the ability to identify individual proteins (Kültz and Somero, 1996). Later, proteins could be identified via mass spectrometry, but typically few proteins were positively identified (Martin et al., 2001), and recently large numbers of fish proteins can be identified with high confidence (Martin et al., 2003, 2007; Bosworth et al., 2005; Lee et al., 2006). Martin et al. (2003) also studied changes in the liver proteome of rainbow trout fed control vs. soy-based diets (Martin et al., 2003). Of the 19 proteins positively identified, only apolipoprotein A-1 and aldolase are common between the Martin study and the current study. We also detected a significant change in the expression of apolipoprotein A-1 (Table 2), but not aldolase with diet treatment. Based on these two studies, liver apolipoprotein A-1 expression seems to be sensitive to dietary manipulation in fish. Alternatively, apolipoproteinmay simply be a highly expressed protein that is easily identified and one which responds to a variety of physiological perturbations (Anderson, 2005).
Table 2.
Identification parameters for zebrafish liver protein bands
| Sample # | ID | Acc #a | MWb | pIc | Pr.Sc.d | # Pepe | % Covf | N Lowg | N Highg | AVG Lh | AVG Hh | Pi |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A) Proteins found in one sample | ||||||||||||
| SB8-3-1 | Zgc:92590 | 54035518 | 28 | 4.9 | 628 | 8 | 47 | 12 | 7 | 0.23 | 0.27 | 0.654 |
| SB8-3-2 | Hypoxanthine phosphoribosyltransferase | 47085697 | 24.9 | 6.1 | 402 | 6 | 30 | 7 | 4 | 0.13 | 0.06 | 0.206 |
| Zgc:92745 | 51010935 | 29.4 | 6.2 | 467 | 7 | 30 | ||||||
| SB8-3-3 | Zgc:101897 | 55250043 | 27.8 | 8.1 | 374 | 6 | 25 | 9 | 1 | 0.13 | 0.11 | NA |
| SB8-3-4 | FABP 7 | 18858657 | 14.9 | 5.4 | 612 | 8 | 71 | 8 | 5 | 0.11 | 0.09 | 0.565 |
| TPA:Transgelin 2 | 68364340 | 25 | 9.6 | 131 | 2 | 9 | ||||||
| SB8-3-5 | CuZn SOD | 56790262 | 16.1 | 6.1 | 435 | 4 | 42 | 13 | 5 | 0.23 | 0.23 | 0.593 |
| SB8-3-7 | 40S ribosomal protein S12 | 41152459 | 14.8 | 6.8 | 253 | 4 | 42 | 11 | 5 | 0.13 | 0.07 | 0.045 |
| SB8-3-9 | Retinol binding protein 2b | 50540292 | 15.7 | 6.7 | 323 | 5 | 38 | 1 | 0 | NA | NA | NA |
| SB8-3-10 | FABP 6 | 50344806 | 14.4 | 6.6 | 138 | 2 | 22 | 14 | 5 | 0.11 | 0.07 | 0.151 |
| SB8-3-11 | Myoglobin | 41053652 | 15.6 | 6.7 | 494 | 5 | 48 | 11 | 4 | 0.12 | 0.05 | 0.256 |
| Glyceraldehyde 3-phosphate dehydrogenase | 47085833 | 36.5 | 6.6 | 198 | 4 | 13 | ||||||
| SB8-3-13 | TPA:anionic trypsin II precursor | 68383831 | 27.8 | 4.8 | 795 | 9 | 54 | 10 | 7 | 0.23 | 0.27 | 0.707 |
| SB8-3-14 | TPA:tyrosine3 monooxygenase/tryptophan monooxygenase activation protein | 68436321 | 200.9 | 8.9 | 226 | 4 | 16 | 8 | 5 | 0.11 | 0.08 | 0.565 |
| SB8-3-17 | Chymotrypsinogen B1 | 47086795 | 28.9 | 7.0 | 384 | 6 | 28 | 8 | 2 | 0.32 | 0.21 | 0.386 |
| SB8-3-19 | Malate dehydrogenase Ia | 41053939 | 33.7 | 8.1 | 175 | 2 | 7 | 12 | 5 | 0.23 | 0.23 | 0.890 |
| TPA: methylmalonyl CoA mutase | 68382837 | 27.3 | 5.8 | 133 | 2 | 10 | ||||||
| SB8-3-21 | Guanidinoacetate N-methyltransferase | 45387893 | 26.9 | 65.8 | 222 | 5 | 23 | 9 | 4 | 0.22 | 0.05 | 0.011 |
| SB8-3-22 | Hypothetical protein LOC445032 | 51010935 | 29.4 | 6.1 | 255 | 5 | 21 | 9 | 2 | 0.32 | 0.21 | 0.229 |
| SB8-28-1 | Myosin, heavy polypeptide 11, smooth muscle | 66773050 | 228.6 | 5.4 | 393 | 9 | 5 | 9 | 0 | 0.07 | 0 | NA |
| SB8-28-2 | TPA:mucin-like protein | 68417777 | 17.8 | 5.4 | 73 | 1 | 3 | 5 | 2 | 0.06 | 0.09 | 0.775 |
| SB8-28-3 | cb825 protein | 27881963 | 55 | 6.3 | 698 | 11 | 26 | 7 | 1 | 0.07 | 0.03 | NA |
| SB8-28-4 | Heat shock protein 8 | 38649356 | 71 | 5.3 | 260 | 5 | 10 | 11 | 4 | 0.09 | 0.07 | 0.256 |
| SB8-28-5 | Prohibitin | 41152028 | 29.7 | 5.3 | 93 | 1 | 6 | 4 | 2 | 0.02 | 0.04 | 0.582 |
| SB8-28-6 | No ID | 9 | 1 | 0.05 | 0.03 | NA | ||||||
| SB8-28-7 | Ela1 protein | 70887623 | 31.6 | 6.1 | 196 | 3 | 16 | 4 | 0 | 0.02 | 0 | NA |
| SB8-28-8 | No ID | 6 | 2 | 0.03 | 0.02 | 0.161 | ||||||
| SB8-28-9 | NAD(P)H dehydrogenase | 45387531 | 31.2 | 6.3 | 514 | 7 | 30 | 5 | 2 | 0.03 | 0.02 | 0.194 |
| Carbonyl reductase-1 | 35902843 | 30.4 | 6.9 | 307 | 5 | 20 | ||||||
| SB8-28-10 | Zgc:92726 | 50369319 | 29 | 6.2 | 242 | 5 | 17 | 5 | 0 | 0.07 | 0 | NA |
| Hydroxyacyl glutathione hydrolase | 37681823 | 29 | 6.6 | 285 | 5 | 21 | ||||||
| SB8-28-12 | Glutamate oxaloacetate transaminase 2 | 41053395 | 47.8 | 8.9 | 240 | 4 | 11 | 6 | 5 | 0.08 | 0.19 | 0.089 |
| aA1 globin | 1477577 | 15 | 7.8 | 143 | 3 | 26 | ||||||
| SB8-28-13 | Type I cytokeratin (Danio rerio) | 18858423 | 46 | 5.3 | 417 | 7 | 13 | 8 | 5 | 0.11 | 0.09 | 0.565 |
| Nucleolar protein family A | 41055780 | 22.9 | 10.9 | 67 | 1 | 4 | ||||||
| SB8-28-14 | Thioredoxin domain containing 17 | 51011001 | 14.4 | 5.4 | 114 | 3 | 30 | 2 | 3 | 0.09 | 0.08 | 0.833 |
| SB8-28-15 | Profilin 2 | 56118638 | 15.1 | 6.1 | 296 | 4 | 35 | 4 | 1 | 0.09 | 0.19 | NA |
| SB8-28-16 | Zgc:110343 | 60552803 | 22.2 | 6.4 | 187 | 3 | 18 | 6 | 0 | 0.08 | 0 | NA |
| Enoyl CoA hydratase | 52218912 | 31.5 | 8.8 | 150 | 3 | 9 | ||||||
| SB8-28-17 | Histone 2 | 73611922 | 25.1 | 11.2 | 197 | 3 | 13 | 3 | 2 | 0.06 | 0.07 | 0.860 |
| SB8-28-18 | Galectin 9 | 41152379 | 35.1 | 8.5 | 227 | 2 | 9 | 3 | 3 | 0.01 | 0.03 | 0.173 |
| SB8-28-19 | KH domain RNA binding protein | 46195759 | 40.3 | 8.3 | 154 | 5 | 9 | 2 | 0 | 0.01 | 0 | NA |
| SB8-28-20 | Nme2 protein | 33416409 | 17 | 7.7 | 288 | 8 | 73 | 8 | 4 | 0.17 | 0.05 | 0.167 |
| Ba1 globin | 18858329 | 16.6 | 7.7 | 98 | 1 | 8 | ||||||
| SB8-28-22 | Cb825 protein | 27881963 | 55 | 6.3 | 1247 | 17 | 36 | 10 | 2 | 0.14 | 0.03 | 0.007 |
| SB8-28-24 | Peptidoylprolyl isomerase A | 41152400 | 17.7 | 8.3 | 352 | 5 | 21 | 7 | 1 | 0.16 | 0.03 | NA |
| BA2 globin | 53749219 | 16.6 | 7.7 | 302 | 4 | 34 | ||||||
| SB6-24-15 | Translationally-controlled tumor protein | 37700237 | 19 | 4.5 | 1187 | 20 | 88 | j | ||||
| SB6-24-16 | Eukaryotic translation initiation factor 5A | 47085971 | 17 | 5.1 | 791 | 13 | 61 | j | ||||
| Eukaryotic translation initiation factor 5A | 55742597 | 17 | 5.1 | 715 | 13 | 62 | ||||||
| Signal sequence receptor delta | 50344818 | 18.4 | 5.2 | 275 | 5 | 27 | ||||||
| SMT3 suppresssor of mif 2 homolog 3 | 55742575 | 10.7 | 5.3 | 189 | 4 | 36 | ||||||
| SB6-24-18 | Reticulacalbin 3 | 50344972 | 37 | 4.5 | 467 | 11 | 28 | j | ||||
| SB6-24-19 | Eukaryotic translation elongation factor 1 2 | 41053941 | 24.5 | 4.5 | 521 | 10 | 44 | j | ||||
| Proteasome subunit 5 | 45387823 | 26.6 | 4.7 | 211 | 3 | 16 | ||||||
| Predicted: zgc:101730 | 68439615 | 25.9 | 4.7 | 171 | 4 | 22 | ||||||
| SB6-24-20 | TPA:Wu:fb64f03 | 68384010 | 29.4 | 4.8 | 1440 | 23 | 68 | j | ||||
| SB6-24-22 | HSP60 | 31044489 | 61.4 | 5.5 | 2965 | 51 | 94 | j | ||||
| SB6-24-23 | Sb:cb825 protein | 4538389 | 54.7 | 6.3 | 1420 | 24 | 33 | j | ||||
| B) Proteins found in multiple samples | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SB8-3-1 | Carbonic anhydrase | 41351397 | 29 | 7.1 | 211 | 2 | 20 | 12 | 7 | 0.23 | 0.27 | 0.654 |
| SB8-3-3 | 708 | 12 | 65 | 9 | 1 | 0.13 | 0.06 | 0.206 | ||||
| SB8-3-4 | FABP7 | 18858657 | 14.9 | 5.4 | 612 | 8 | 71 | 8 | 5 | 0.11 | 0.09 | 0.565 |
| SB8-3-12 | 190 | 2 | 21 | 11 | 3 | 0.28 | 0.35 | 0.789 | ||||
| SB8-3-14 | 666 | 8 | 71 | 8 | 5 | 0.11 | 0.08 | 0.565 | ||||
| SB8-28-14 | 345 | 4 | 38 | 2 | 3 | 0.09 | 0.08 | 0.833 | ||||
| SB8-28-15 | 245 | 3 | 31 | 4 | 1 | 0.09 | 0.19 | NA | ||||
| SB8-3-4 | Profilin 2 | 56118638 | 15.1 | 6.0 | 176 | 3 | 24 | 8 | 5 | 0.11 | 0.09 | 0.565 |
| SB8-3-8 | 270 | 3 | 22 | 10 | 6 | 0.13 | 0.17 | 0.674 | ||||
| SB8-3-16 | 660 | 3 | 43 | 10 | 6 | 0.13 | 0.17 | 0.674 | ||||
| SB8-28-15 | 245 | 3 | 31 | 4 | 1 | 0.09 | 0.19 | NA | ||||
| SB8-3-4 | Retinol binding protein 2a, cellular | 23308503 | 15.9 | 6.1 | 157 | 2 | 22 | 8 | 5 | 0.11 | 0.09 | 0.565 |
| SB8-3-6 | 457 | 8 | 57 | 13 | 5 | 0.33 | 0.24 | 0.326 | ||||
| SB8-3-8 | Galectin 1 | 41055788 | 13.5 | 6.4 | 166 | 3 | 31 | 10 | 6 | 0.13 | 0.17 | 0.674 |
| SB8-3-10 | 97 | 2 | 21 | 14 | 5 | 0.11 | 0.07 | 0.151 | ||||
| SB8-3-12 | 289 | 6 | 62 | 11 | 3 | 0.28 | 0.35 | 0.789 | ||||
| SB8-3-9 | FABP3 | 23308625 | 15 | 5.7 | 394 | 6 | 14 | 1 | 0 | NA | NA | NA |
| SB8-3-15 | 440 | 7 | 43 | 12 | 7 | 0.33 | 0.45 | 0.329 | ||||
| SB8-3-8 | TPA:chromodomain helicase DNA binding protein 5 | 68368782 | 51.8 | 8.0 | 112 | 2 | 6 | 10 | 6 | 0.13 | 0.17 | 0.674 |
| SB8-3-10 | 394 | 6 | 14 | 14 | 5 | 0.11 | 0.07 | 0.151 | ||||
| SB8-3-12 | 128 | 2 | 5 | 11 | 3 | 0.28 | 0.35 | 0.789 | ||||
| SB8-3-14 | Enolase 3 | 47551317 | 47.8 | 6.3 | 128 | 2 | 8 | 8 | 5 | 0.11 | 0.08 | 0.565 |
| SB8-3-18 | 186 | 4 | 14 | 9 | 4 | 0.64 | 0.04 | 0.006 | ||||
| SB8-3-20 | 1300 | 18 | 47 | 10 | 5 | 0.33 | 0.19 | 0.082 | ||||
| SB8-28-23 | 1193 | 18 | 52 | 10 | 5 | 0.33 | 0.19 | 0.082 | ||||
| SB8-3-13 | Triosephosphate isomerase 1B | 47271422 | 27.1 | 6.9 | 200 | 2 | 15 | 10 | 7 | 0.23 | 0.27 | 0.707 |
| SB8-3-15 | 292 | 4 | 30 | 12 | 7 | 0.33 | 0.45 | 0.329 | ||||
| SB8-3-17 | 1251 | 17 | 74 | 8 | 2 | 0.32 | 0.21 | 0.386 | ||||
| SB8-3-20 | 406 | 6 | 37 | 10 | 5 | 0.33 | 0.19 | 0.082 | ||||
| SB8-3-22 | 997 | 13 | 59 | 9 | 2 | 0.32 | 0.21 | 0.229 | ||||
| SB8-3-15 | Aldolase b | 42734425 | 39.7 | 8.5 | 199 | 3 | 11 | 12 | 7 | 0.33 | 0.45 | 0.829 |
| SB8-3-17 | 872 | 12 | 31 | 8 | 2 | 0.32 | 0.21 | 0.386 | ||||
| SB8-3-22 | 288 | 5 | 10 | 9 | 2 | 0.32 | 0.21 | 0.229 | ||||
| SB8-28-11 | 553 | 8 | 25 | j | ||||||||
| SB8-3-18 | TPA:apolipoprotein A-I | 68443519 | 30.1 | 6.0 | 650 | 12 | 38 | 9 | 4 | 0.64 | 0.04 | 0.006 |
| SB8-3-19 | 568 | 10 | 35 | 12 | 5 | 0.23 | 0.23 | 0.890 | ||||
| SB8-3-21 | 546 | 6 | 36 | 9 | 4 | 0.22 | 0.05 | 0.011 | ||||
| SB6-24-17 | 1221 | 24 | 60 | j | ||||||||
| SB6-24-21 | 1677 | 29 | 79 | j | ||||||||
| SB8-3-21 | Hypothetical protein LOC445032 | 51010935 | 28.4 | 6.1 | 147 | 2 | 8 | 9 | 4 | 0.22 | 0.05 | 0.011 |
| SB8-3-22 | 344 | 7 | 25 | 9 | 2 | 0.32 | 0.21 | 0.229 | ||||
| SB8-3-22 | Chymotrypsinogen B1 | 47086795 | 28.9 | 7.0 | 309 | 5 | 26 | 9 | 2 | 0.32 | 0.21 | 0.229 |
| SB8-28-10 | 148 | 2 | 13 | 5 | 0 | 0.07 | 0 | NA | ||||
| SB8-3-11 | Ba1 globin | 18858329 | 16.6 | 7.7 | 645 | 8 | 66 | 11 | 4 | 0.12 | 0.05 | 0.256 |
| SB8-28-20 | 98 | 1 | 8 | 8 | 4 | 0.17 | 0.05 | 0.167 | ||||
| SB8-28-5 | Transferrin A | 62632717 | 58.4 | 5.6 | 142 | 2 | 6 | 4 | 2 | 0.02 | 0.04 | 0.582 |
| SB8-28-21 | 1113 | 16 | 40 | 11 | 2 | 0.24 | 0.08 | 0.028 | ||||
Electrophoresis and mass spectrometry analyses variables for all analyzed spots.
Accession number.
Molecular mass, in kDa.
Predicted isoelectric point.
Standard protein score (ion score cut-off = 25).
Number of peptides matching the identification.
Percent of the complete primary sequence covered by matching peptides.
Sample size for spot in the low and high treatment groups.
Mean relative pixel volume for low (L) and high (H) treatment groups.
Probability score from Student's t-test comparing relative pixel volume for that spot between groups. Spots that are significantly different between treatments are shaded in medium grey. Spots that have a mean relative expression difference ≥2-fold are shaded in black. Spots that are primarily expressed only in one treatment are shaded in light grey.
Samples SB6-24-15 through SB6-24-23 were from initial electrophoresis optimization experiments with zebrafish liver, and were not derived from fish in the feeding experiment. Therefore, only their identification (and not relative abundance with treatment) is listed. These proteins were not included in the GO analyses.
Five spots (SB3-3-7, SB8-3-18, SB8-3-21, SB8-28-21, SB8-28-22), collectively containing 6 unique proteins were expressed at a significantly lower relative value in the high-calorie treatment (Table 2, outlined in black). Five additional spots (SB8-3-2, SB8-3-11, SB8-28-12, SB8-28-18, SB8-28-20), containing 9 proteins, had a mean expression difference of ≥2-fold (Table 2, medium grey). Finally, 9 spots (SB8-3-3, SB8-28-1, SB8-28-3, SB8-28-6, SB8-28-7, SB8-28-10, SB8-28-15, SB8-28-16, SB8-28-24), containing 15 proteins, were primarily only expressed in the low-calorie treatment (Table 2, light grey). In fact, in nearly all cases where there is an apparent difference in expression between groups (whether by t-test, fold-difference, or presence/absence), expression is higher in the low-calorie vs. high-calorie group (the exceptions are GOT2/A1 globin, profilin2/FABP7, and Galectin 9). Further, the sum total of quantifiable spots is significantly higher in the low-calorie group (650 ± 51 vs. 411 ± 62, p <.05). Therefore, high-calorie diet lowers protein expression diversity in the zebrafish liver (alternatively, the diversity of proteins expressed is not changed, but expression level is reduced below that detected by Coomassie blue).
With the goal of determining which cellular functions respond to the high-calorie treatment, we mapped Gene Ontology (GO) terms to all differentially expressed proteins, and then tested whether these functions represented a non-random sampling of the entire zebrafish GO set via Fisher's exact test (Table 3). Proteins differentially expressed via t-test are associated with guanidinoacetate N-methyl-transferase (GAMT) and ferric-iron binding activity (Table 3A). GAMT is essential for creatine synthesis (knockout mice for GMAT have no measurable creatine (Kan et al., 2004), and GAMT deficiency is associated with creatine deficiency syndrome in humans (Torremans et al., 2005). Reduced GMAT expression in the high-calorie diet may result in altered ATP-buffering capacity, as seen in humans with steatosis (Perseghin et al., 2008). Ferric-iron binding activity is consistent with other GO functions that indicate changes in heme protein metabolism (see below). The decrease in glutathione transferase expression (Table 3B) we interpret as increased exposure to lipid peroxide radicals, as increased lipid deposition typically decreases the titer of glutathione transferases (Grimsrud et al., 2007). Expression changes mapped to the GO term of decreased actin-binding activity in the high-calorie treatment (Table 3B) are consistent with a general rearrangement of the cytoskeleton to accommodate lipid deposition. Zeidan et al. recently determined that leptin increases phosphorylation of actin-binding proteins, and in doing so stimulates F-actin polymerization (Zeidan et al., 2006; 2007a,b). Because leptin is positively correlated with lipid mass in fish (Johnson et al., 2000), increased lipid deposition could explain decreased actin-binding activity in the high-calorie treatment.
Our most consistent result from the GO analyses is that oxygen binding activity decreases in the high-calorie treatment (Tables 2 and 3). If one adopts a 2-fold difference as a criterion for differential expression (Table 3C), or if one combines all criteria (t-test, fold-difference, and presence absence; Table 3D), the most significant GO terms are all associated with decreases in oxygen binding proteins (oxygen transporter activity and binding, tetrapyrole binding, heme binding, and iron ion binding). This is consistent with a well-established phenomenological link between obesity and iron metabolism in mammals. Genetically obese, ob/ob mice (Kennedy et al., 1986; Failla et al., 1988) and rats (Serfass et al., 1988) have decreased iron concentration in the liver but normal plasma iron. Iron absorption, however, is higher in ob/ob mice vs. lean mice, as is hematocrit (Failla et al., 1988). Circulating iron (as ferritin) is highly correlated with insulin resistance in humans (Tuomainen et al., 1997). Obesity is thought to lead to increased erythropoiesis, with increased iron absorption rate as an adaptation and increased hematocrit as a product (Le Guenno et al., 2007). Some aspects of the link between iron metabolism and diet may be species specific, however, as iron absorption is significantly reduced in rats on a high-fat diet (Boesch-Saadatmandi et al., 2007).
We fed high and low-calorie diets to zebrafish in order to determine the major changes in liver protein expression associated with a change in available calories. The high-calorie diet significantly increased total body lipid in females but not males, and total weight and body condition in both sexes (Table 1). We assume that the increase in total body lipid also results in an increase in liver lipid storage, as liver is the central storage organ for lipid in fish (Sheridan, 1988; Sheridan, 1994), and zebrafish on a high-calorie diet deposit lipid in liver (Schlegel and Stainier, 2006). However, we did not extract lipid from liver separately, and significant lipid deposition likely went to eggs, as high-calorie females were visibly more gravid than low-calorie (data not shown). Therefore, even though we interpret results in light of an assumed increase in liver lipid stores (above), the largest effect on protein expression may interact with other processes (e.g. perfusion or growth).
Proteomics experiments are designed to be hypothesis-generating, rather than explicitly hypothesis-testing. Although the techniques are extremely powerful in their sensitivity and breadth, they present significant statistical and analytical challenges in the post-data generation phase of the study (Aebersold and Mann, 2003). One challenge is determining how one will delineate differentially expressed proteins. In a related study, we determined (with a common sample) that total spots detected among gels varied 4.6% and mean pixel volume for any given spot varied 12.5% (Edmonds, 2005). Therefore, we are confident that when pixel volume varies ≥200% between treatments, that most of that variation is biological vs. experimental. However, fold-difference cutoffs do not account for variation among individuals. The Student's t-test does account for variation among individuals, but the probability corrections needed for ~800 comparisons are often prohibitive. Finally, spots that are present only in one treatment are certainly differentially expressed, at least intuitively. We tested which GO functions are associated with differentially expressed proteins, as defined by the above 3 criteria. Ideally, the conclusions derived from each of these methods should converge, and they do to some extent (Table 3). We could now justify pursuing hypotheses related to any of the highly significant GO functions listed in Table 3. However, the largest signal is that the zebrafish liver proteome responds to a high-calorie diet with a decrease in heme binding proteins. From this we would predict that hematocrit increases in the high-calorie diet as predicted in humans (Le Guenno et al., 2007). This response is also consistent with reduced heme proteins and increased lipid deposition seen in cold-bodied fishes (Londraville and Sidell, 1990; Sidell et al., 1997). Going forward, we will test the hypothesis that lipid deposition and heme protein metabolism are linked in fishes using more statistically tractable techniques (e.g. real-time PCR and immunoblotting).
The fact that we did not find support for our initial hypothesis (that pathways of lipid metabolism would be up-regulated in the high-calorie treatment) is a testament to the power of the proteomics experiment. Rather than referring to proteomics derisively as a ‘fishing expedition’ (Aebersold and Mann, 2003), we view it as an unbiased analytical measurement of how a tissue responds to treatment. We took an admittedly conservative approach to analyzing these data, and with less rigorous analyses we could undoubtedly tease out a response in lipid metabolism among the data. However, we can definitely say that changes in lipid metabolism do not account for the largest share of the cell's protein expression budget when exposed to a high-calorie diet.
Acknowledgements
This work was supported in part by NSF Grant IOS 0328554 to RLL and Dr. Peter Niewiarowski, a University of Akron Faculty Research Grant to RLL, and a grant from the Program in Integrated Bioscience at UA to RLL and ZHD.
Abbreviations
- RefSeq
reference sequence
- NCBI
National Center for Biotechnology Information
- CID
collisionally induced dissociation
- ESI
electrospray ionization
- GO
gene ontology
- LC–ESI MS/MS
liquid chromatography, electrospray ionization tandem mass spectrometry
- kDa
kilodaltons
- GAMT
guanidinoacetate N-methyltransferase
References
- Abbott A. A post-genomic challenge: learning to read patterns of protein synthesis. Nature. 1999;402:715–720. doi: 10.1038/45350. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J. Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesch-Saadatmandi C, Most E, Weigand E. Influence of dietary fat and zinc supplementation on the iron utilization in growing rats. Ann. Nutr. Metab. 2007;51:395–401. doi: 10.1159/000108107. [DOI] [PubMed] [Google Scholar]
- Bosworth CAt, Chou CW, Cole RB, Rees BB. Protein expression patterns in zebrafish skeletal muscle: initial characterization and the effects of hypoxic exposure. Proteomics. 2005;5:1362–1371. doi: 10.1002/pmic.200401002. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Crockett EL, Sidell BD. Peroxisomal á-oxidation is a significant pathway for catabolism of fatty acids in a marine teleost. Am. J. Physiol. 1993;264:R1004–R1009. doi: 10.1152/ajpregu.1993.264.5.R1004. [DOI] [PubMed] [Google Scholar]
- Crockett EL, Londraville RL, Wilkes EE, Popesco MC. Enzymatic capacities for beta-oxidation of fatty fuels are low in the gill of teleost fishes despite presence of fatty acid-binding protein. J. Exp. Zool. 1999;284:276–285. doi: 10.1002/(sici)1097-010x(19990801)284:3<276::aid-jez5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Duplus E, Glorian M, Forest C. Fatty acid regulation of gene transcription. J. Biol. Chem. 2000;275:30749–30752. doi: 10.1074/jbc.R000015200. [DOI] [PubMed] [Google Scholar]
- Edmonds S. Unpublished MS Thesis. Akron, OH USA: Department of Biology, University of Akron; 2005. Proteomic analysis of the effect of parasitism by Uvulifer ambloplitis on bluegill (Lepomis macrochirus) plasma protein. [Google Scholar]
- Egginton S, Sidell BD. Thermal acclimation induces adaptive changes in subcellular structure of fish skeletal muscle. Am. J. Physiol. 1989;256:r1–r9. doi: 10.1152/ajpregu.1989.256.1.R1. [DOI] [PubMed] [Google Scholar]
- Failla ML, Kennedy ML, Chen ML. Iron metabolism in genetically obese (ob/ob) mice. J. Nutr. 1988;118:46–51. doi: 10.1093/jn/118.1.46. [DOI] [PubMed] [Google Scholar]
- Farrell AP, Munt B. Cholesterol levels in the blood of Atlantic salmonids. Comp. Biochem. Physiol. A. 1983;75:239–242. doi: 10.1016/0300-9629(83)90076-2. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley G. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Goel M, Sinkins W, Keightley A, Kinter M, Schilling WP. Proteomic analysis of TRPC5- and TRPC6-binding partners reveals interaction with the plasmalemmal Na(+)/K(+)-ATPase. Pflugers Arch. 2005;451:87–98. doi: 10.1007/s00424-005-1454-y. [DOI] [PubMed] [Google Scholar]
- Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol. Cell Proteomics. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- Hogstrand C, Balesaria S, Glover CN. Application of genomics and proteomics for study of the integrated response to zinc exposure in a non-model fish species, the rainbow trout. Comp. Biochem. Physiol. B. 2002;133:523–535. doi: 10.1016/s1096-4959(02)00125-2. [DOI] [PubMed] [Google Scholar]
- Johnson RM, Johnson TM, Londraville RL. Evidence for leptin expression in fishes. J. Exp. Zool. 2000;286:718–724. doi: 10.1002/(sici)1097-010x(20000601)286:7<718::aid-jez6>3.0.co;2-i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan HE, Renema WK, Isbrandt D, Heerschap A. Phosphorylated guanidinoacetate partly compensates for the lack of phosphocreatine in skeletal muscle of mice lacking guanidinoacetate methyltransferase. J. Physiol. 2004;560:219–229. doi: 10.1113/jphysiol.2004.067926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy ML, Failla ML, Smith JC., Jr Influence of genetic obesity on tissue concentrations of zinc, copper, manganese and iron in mice. J. Nutr. 1986;116:1432–1441. doi: 10.1093/jn/116.8.1432. [DOI] [PubMed] [Google Scholar]
- Kinter M, Sherman N. Protein Sequencing and Identification using Tandem Mass Spectrometry. New York: Wiley-Interscience; 2000. [Google Scholar]
- Kültz D, Somero GN. Differences in protein patterns of gill epithelial cells of the fish Gillichthys mirabilis after osmotic and thermal acclimation. J. Comp. Physiol. B. 1996;166:88–100. doi: 10.1007/BF00301172. [DOI] [PubMed] [Google Scholar]
- Le Guenno G, Chanseaume E, Ruivard M, Morio B, Mazur A. Study of iron metabolism disturbances in an animal model of insulin resistance. Diabetes Res. Clin. Pract. 2007;77:363–370. doi: 10.1016/j.diabres.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Lee J, Valkova N, White M, Kultz D. Proteomic identification of processes and pathways characteristic of osmoregulatory tissues in spiny dogfish shark (Squalus acanthias) Comp. Biochem. Physiol. D. 2006;1:328–343. doi: 10.1016/j.cbd.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Londraville RL, Sidell BD. Ultrastructure of aerobic muscle in Antarctic fishes may contribute to maintenance of diffusive fluxes. J. Exp. Biol. 1990;150:205–220. [Google Scholar]
- Londraville RL, Sidell BD. Purification and characterization of fatty acid-binding protein from aerobic muscle of the Antarctic icefish Chaenocephalus aceratus. J. Exp. Zool. 1995;273:190–203. doi: 10.1002/jez.1402730304. [DOI] [PubMed] [Google Scholar]
- Martin SAM, Cash P, Blaney S, Houlihan D. Proteome analysis of rainbow trout (Oncorhychus mykiss) liver proteins during short term starvation. Fish Physiol. Biochem. 2001;24:259–270. [Google Scholar]
- Martin SA, Vilhelmsson O, Medale F, Watt P, Kaushik S, Houlihan DF. Proteomic sensitivity to dietary manipulations in rainbow trout. Biochim. Biophys. Acta. 2003;1651:17–29. doi: 10.1016/s1570-9639(03)00231-0. [DOI] [PubMed] [Google Scholar]
- Martin SA, Mohanty BP, Cash P, Houlihan DF, Secombes CJ. Proteome analysis of the Atlantic salmon (Salmo salar) cell line SHK-1 following recombinant IFN-gamma stimulation. Proteomics. 2007;7:2275–2286. doi: 10.1002/pmic.200700020. [DOI] [PubMed] [Google Scholar]
- Paulson L, Martin P, Persson A, Nilsson C, Ljung E, Westman-Brinkmalm A, Eriksson P, Blennow K, Davidsson P. Comparative genome- and proteome analysis of cerebran cortex from MK-801-treated rats. J. Neurosci. Res. 2003;71:526–533. doi: 10.1002/jnr.10509. [DOI] [PubMed] [Google Scholar]
- Perseghin G, Lattuada G, De Cobelli F, Esposito A, Belloni E, Ntali G, Ragogna F, Canu T, Scifo P, Del Maschio A, Luzi L. Increased mediastinal fat and impaired left ventricular energy metabolism in young men with newly found fatty liver. Hepatology. 2008;47:51–58. doi: 10.1002/hep.21983. [DOI] [PubMed] [Google Scholar]
- Schlegel A, Stainier DY. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry. 2006;45:15179–15187. doi: 10.1021/bi0619268. [DOI] [PubMed] [Google Scholar]
- Serfass RE, Park KE, Kaplan ML. Developmental changes of selected minerals in Zucker rats. Proc. Soc. Exp. Biol. Med. 1988;189:229–239. doi: 10.3181/00379727-189-42803. [DOI] [PubMed] [Google Scholar]
- Sheridan MA. Lipid dynamics in fish: aspects of absorption, transportation, deposition, and mobilization. Comp. Biochem. Physiol. B. 1988;90:679–690. doi: 10.1016/0305-0491(88)90322-7. [DOI] [PubMed] [Google Scholar]
- Sheridan MA. Regulation of lipid metabolism in poikilothermic vertebrates. Comp. Biochem.Physiol. B. 1994;107:495–508. [Google Scholar]
- Sidell BD, Vayda ME, Small DJ, Moylan TJ, Londraville RL, Yuan ML, Rodnick KJ, Eppley ZA, Costello L. Variable expression of myoglobin among the hemoglobinless Antarctic icefishes. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3420–3424. doi: 10.1073/pnas.94.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocher DG. Metabolism and functions of lipids and fatty acids in teleost fish. Revs. Fish. Sci. 2003;11:107–184. [Google Scholar]
- Torremans A, Marescau B, Possemiers I, Van Dam D, D'Hooge R, Isbrandt D, De Deyn PP. Biochemical and behavioural phenotyping of a mouse model for GAMT deficiency. J. Neurol. Sci. 2005;231:49–55. doi: 10.1016/j.jns.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Tuomainen TP, Nyyssonen K, Salonen R, Tervahauta A, Korpela H, Lakka T, Kaplan GA, Salonen JT. Body iron stores are associated with serum insulin and blood glucose concentrations. Population study in 1,013 eastern Finnish men. Diabetes Care. 1997;20:426–428. doi: 10.2337/diacare.20.3.426. [DOI] [PubMed] [Google Scholar]
- Zeidan A, Javadov S, Karmazyn M. Essential role of Rho/ROCK-dependent processes and actin dynamics in mediating leptin-induced hypertrophy in rat neonatal ventricular myocytes. Cardiovasc. Res. 2006;72:101–111. doi: 10.1016/j.cardiores.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Zeidan A, Javadov S, Chakrabarti S, Karmazyn M. Leptin-induced cardio-myocyte hypertrophy involves selective caveolae and RhoA/ROCK-dependent p38 MAPK translocation to nuclei. Cardiovasc. Res. 2007a;77:64–72. doi: 10.1093/cvr/cvm020. [DOI] [PubMed] [Google Scholar]
- Zeidan A, Paylor B, Steinhoff KJ, Javadov S, Rajapurohitam V, Chakrabarti S, Karmazyn M. Actin cytoskeleton dynamics promotes leptin-induced vascular smooth muscle hypertrophy via RhoA/ROCK- and phosphatidylinositol 3-kinase/protein kinase B-dependent pathways. J. Pharmacol. Exp. Ther. 2007b;322:1110–1116. doi: 10.1124/jpet.107.122440. [DOI] [PubMed] [Google Scholar]