Summary
In Drosophila oocytes, after the completion of recombination, meiotic chromosomes form a compact cluster called the karyosome within the nucleus, and later assemble spindle microtubules without centrosomes. Although these oocyte-specific phenomena are also observed in humans, their molecular basis is not well understood. Here, we report essential roles for the conserved kinase SRPK in both karyosome formation and spindle microtubule assembly in oocytes. We have identified a female-sterile srpk mutant through a cytological screen for karyosome defects. Unlike most karyosome mutants, the karyosome defect is independent of the meiotic recombination checkpoint. Heterochromatin clustering found within the wild-type karyosome is disrupted in the mutant. Strikingly, a loss of SRPK severely prevents microtubule assembly for acentrosomal spindles in mature oocytes. Subsequently, bi-orientation and segregation of meiotic chromosomes are also defective. Therefore, this study demonstrates new roles of this conserved kinase in two independent meiotic steps specific to oocytes.
Key words: Drosophila, SRPK, Kinase, Meiosis, Mitosis, Oocyte, Spindle
Introduction
Meiosis consists of recombination followed by two rounds of chromosome segregation. Oocytes have a very long intervening period between recombination and chromosome segregation: 10–40 years in humans and 2–3 days in Drosophila. During this period, Drosophila meiotic chromosomes cluster together to form a spherical body, called the karyosome, within the enlarged oocyte nucleus (King, 1970). Similar clustering of chromosomes is also observed in the growing phase of mouse and human oocytes (Parfenov et al., 1989). This conservation suggests an important role for chromosome clustering, but the precise biological significance is not yet established. In mice, the clustering of meiotic chromosomes is known to correlate with the developmental competency of the oocytes (Zuccotti et al., 1998; Zuccotti et al., 2002). In Drosophila, we proposed that clustering of meiotic chromosomes facilitates the formation of one unified spindle from multiple chromosomes (Cullen et al., 2005).
Despite being widespread across animals, only limited information is available on the molecular mechanism of chromosome clustering in the oocyte nucleus. In Drosophila, we previously found that the conserved kinase NHK-1 directly regulates karyosome formation through phosphorylation of BAF, a linker between the nuclear envelope and chromatin (Lancaster et al., 2007). Furthermore, when DNA double strand breaks (DSBs) are not repaired, the meiotic recombination checkpoint (hereafter referred to as the meiotic checkpoint) prevents karyosome formation through suppression of NHK-1 (Lancaster et al., 2010).
Once oocytes are mature, the nuclear envelope breaks down and the meiotic spindle is assembled. In many animals including humans and Drosophila, oocytes do not contain centrosomes, the major microtubule organising centres in mitotic cells (McKim and Hawley, 1995). Instead, chromosomes play a central role in spindle formation in oocytes. In Xenopus egg extracts, the Ran-GTP gradient is essential for spindle assembly around chromosomes (Carazo-Salas et al., 1999; Kalab et al., 2002). In addition, several other proteins were also shown to be essential for spindle microtubule assembly in Xenopus extracts (Gruss et al., 2001; Tournebize et al., 2000; Sampath et al., 2004; Wilde and Zheng, 1999). On the other hand, spindle microtubule assembly in living oocytes is far from being fully understood. For example, in mouse and Drosophila oocytes, the spindle can be formed around chromosomes even if the Ran-GTP gradient is disrupted (Dumont et al., 2007; Cesario and McKim, 2011). In Drosophila, no mutants have been identified which severely prevent spindle microtubule assembly.
Here we report a mutation in the conserved kinase SRPK which disrupts both karyosome formation and spindle microtubule assembly in Drosophila oocytes. The karyosome defect in the mutant is independent of the meiotic checkpoint. Remarkably, spindle microtubule assembly is compromised much more severely than in any other mutants so far reported. This study demonstrates crucial roles of this kinase in these two independent steps in meiosis.
Results and Discussion
A screen for mutants with defective karyosomes identified the conserved kinase SRPK
To gain insight into the molecular mechanism of karyosome formation, we screened a collection of mutants for abnormal karyosome morphology. The genomes were first chemically mutagenised and germline clones of mutations on the right arm of the second chromosome were generated in females using the FRT/ovoD system (Fig. 1A) (Vogt et al., 2006). About 100 mutants produced eggs that failed to develop at all. These mutants were cytologically screened for abnormal karyosome morphology in germline clones. This report focuses on one mutant (2R-129-09) identified in this screen. The mutant is sterile in females and males, but is fully viable without obvious morphological defects, including ones typically associated with mitotic defects such as rough eyes or missing bristles.
Fig. 1.
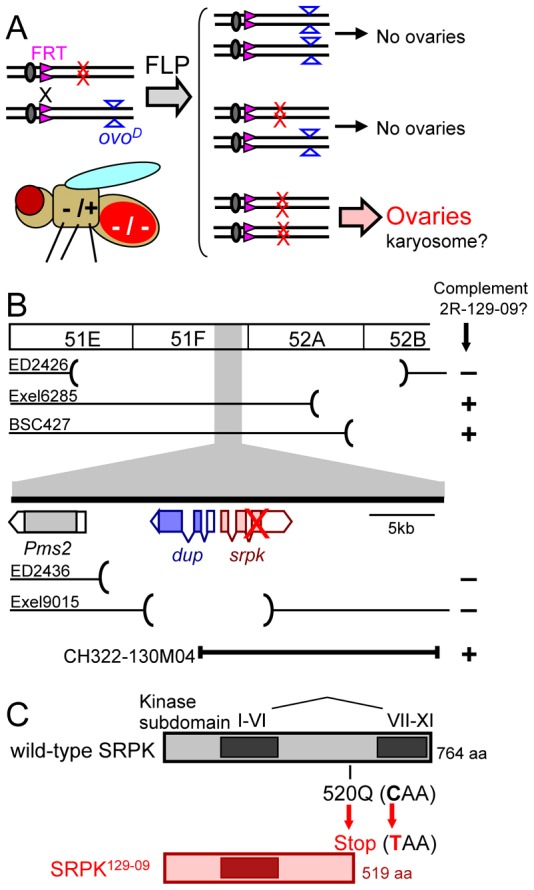
Identification of a mutation in the gene for the conserved kinase SRPK. (A) Generation of female germline clones to screen for karyosome mutants. (B) Deficiency mapping of the female sterile mutation in 2R-129-09. The deleted genomic regions of deficiencies are indicated by the absence of lines and parentheses. The transgene (CH322-130M04) rescued the mutation. (C) A nonsense srpk mutation was indentifed in 2R-129-09 (the position marked by the red cross in B).
To identify the mutation that causes the karyosome defect in 2R-129-09, we first mapped the mutation using a series of chromosomal deletions (known as deficiencies). Two genes (srpk and dup) were disrupted by all of the three overlapping deficiencies, which did not rescue the sterility (Fig. 1B). Sequencing of the genomic region from the mutant and the parental line identified one nucleotide difference, which represents a nonsense mutation in the srpk gene. A transgene of a wild-type srpk genomic region fully rescued the sterility and the cytological defects in the mutant (Fig. 1B; supplementary material Fig. S1). This demonstrated that the mutation in srpk is the cause of the sterility and cytological defects in the mutant.
The srpk gene encodes SRPK (SR Protein Kinase), which is highly conserved and belongs to a distinct subfamily of serine/threonine kinases. The unique feature of this subfamily is a large spacer region within the kinase domain. The nonsense mutation in 2R-129-09 results in a truncated protein which lacks half of the kinase domain including subdomains VII-XI essential for kinase activity (Fig. 1C). Consistent with our results, female sterile mutations in the srpk gene (also called cuaba) have also been identified previously through a screen for abnormal oocyte polarity (Barbosa et al., 2007). However, karyosome defects have not been reported before.
SRPK is crucial for karyosome formation
To gain insight into the role of SRPK in karyosome formation, we analysed the karyosome defect in the srpk mutant in detail. In wild-type oocytes, meiotic chromosomes are clustered together to form a compact spherical karyosome. In srpk129-09/Df mutant oocytes, the spherical karyosome morphology was frequently disrupted (79% at stage 6; Fig. 2A,B; supplementary material Fig. S2). The karyosome was often deformed with one or more lobes extended from a mass of chromosomes. In other cases, chromosomes appeared to be separated into multiple masses. Sometimes chromosomes appeared to be extensively attached to the nuclear envelope. The chromatin morphology of nurse and follicle cells was not affected (supplementary material Fig. S3).
Fig. 2.
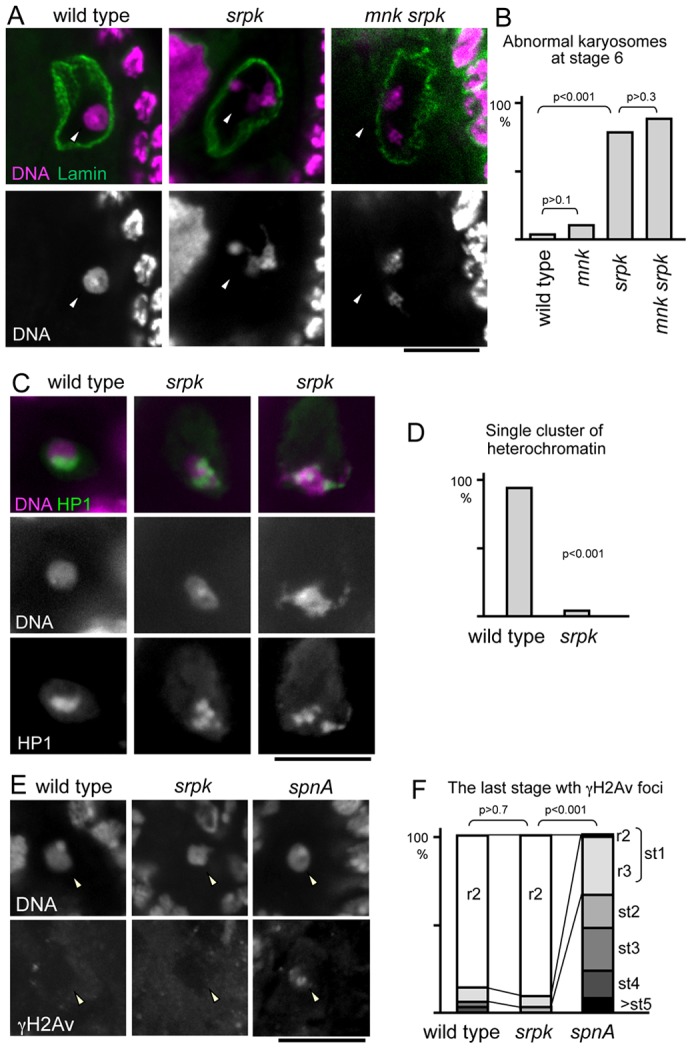
The karyosome defect in the srpk mutant is not caused by the meiotic checkpoint. (A) Karyosomes in wild-type, srpk129-09 and mnkp6 srpk129-09 oocytes. The arrowheads indicate the position of the karyosome in the oocyte nucleus. (B) The frequency of abnormal karyosomes in stage-6 oocytes (n≥110). (C) Heterochromatin within the karyosome is visualised with an anti-HP1 antibody. (D) The frequency of karyosomes with a single heterochromatin cluster at stage 3 or 4. (E) γ-H2Av signals in a stage-3 or -4 wild-type, srpk and spnA oocyte nucleus. (F) The last oogenesis stage when γ-H2Av foci were observed in each ovariole. st, stage; r, region. n≥33. Scale bars: 10 µm.
In wild-type oocytes, the karyosome is first established at oogenesis stage 3. In mutant oocytes, karyosome defects were first observed from stage 3 and progressively worsened during oogenesis (supplementary material Fig. S2). Therefore, SRPK is crucial for both establishment and maintenance of the karyosome.
Peri-centromeric heterochromatin is clustered within wild-type karyosomes (Fig. 2C,D) (Dernburg et al., 1996). Immunostaining of Heterochromatin Protein 1 (HP1) showed that heterochromatin clustering was disrupted in a large majority of mutant karyosomes, including ones with relatively normal morphology (Fig. 2C,D).
The karyosome defect in the srpk mutant is independent of the meiotic checkpoint
Karyosome defects can be induced by activation of the meiotic checkpoint pathway. The meiotic checkpoint detects DSBs caused by a failure in DNA repair or in rasiRNA processing which suppresses retrotransposition (Ghabrial et al., 1998; Klattenhoff et al., 2007). Indeed, the defects in many karyosome mutants have been shown to be caused by the meiotic checkpoint, while the karyosome defect of an nhk-1 mutant is not caused by the meiotic checkpoint (Lancaster et al., 2010).
To monitor the existence of unrepaired DSBs, oocytes were immunostained for γ-H2Av (γ-H2AX in mammals), which accumulates on DSBs (Fig. 2E). In wild type, the DSBs induced during meiotic recombination were repaired by oogenesis stage 2, and γ-H2Av foci were restricted to stage 1 (Mehrotra and McKim, 2006). In a mutant of spnA (the homologue of the rad51 DNA repair gene), γ-H2Av foci persist well after stage 2, as previously reported (Staeva-Vieira et al., 2003). In the srpk mutant, γ-H2Av foci were restricted to stage 1 (Fig. 2F). This indicates that DSBs were repaired with normal timing in the srpk mutant.
To conclusively determine whether the karyosome defect in the srpk mutant is due to the meiotic checkpoint, the meiotic checkpoint was inactivated by a mutation in mnk (also known as chk2, which encodes a conserved kinase required for the meiotic checkpoint and the DNA damage checkpoint). The mnk mutation has been shown to rescue karyosome defects in mutants defective in DNA repair or rasiRNA processing (Abdu et al., 2002; Klattenhoff et al., 2007). Immunostaining of double mutants between srpk129-09/Df and mnkp6 showed that the mnk mutation failed to rescue the karyosome defect in the srpk mutant (Fig. 2A,B). This demonstrates that the karyosome defect in the srpk mutant is not caused by the meiotic checkpoint.
SRPK is essential for the assembly of spindle microtubules in mature oocytes
To identify further roles of SRPK in meiosis, we examined meiotic spindles in mature oocytes (stage-14 oocytes) by immunostaining. A mature wild-type oocyte contains a single meiotic spindle arrested in metaphase I. In oocytes, the meiotic spindle forms without centrosomes. However, the molecular requirements of microtubule assembly in oocytes remain a mystery, as no mutants have been identified that prevent microtubule assembly in oocytes.
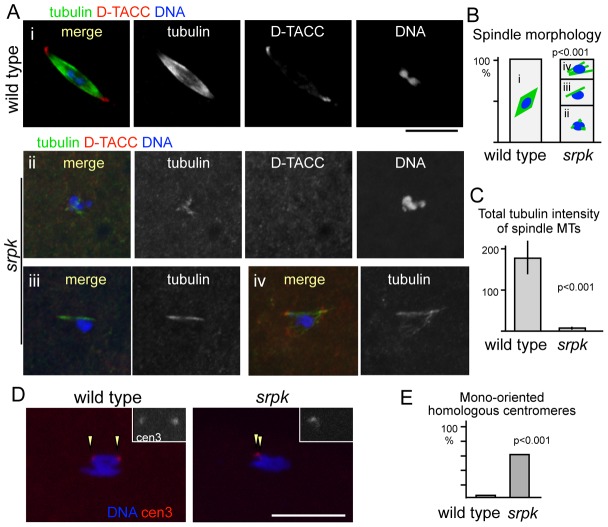
To test the role of SRPK in spindle assembly, mature oocytes which arrest in metaphase I were immunostained for α-tubulin, the pole protein D-TACC and DNA. In a mature wild-type oocyte, a single bipolar spindle was associated with a cluster of meiotic chromosomes (Fig. 3A). In contrast, meiotic spindles were severely defective in mature oocytes of the srpk mutant (Fig. 3A,B). In all mutant oocytes, only residual microtubules were observed and a proper bipolar spindle structure did not form. Frequently (13 out of 30 oocytes), no or only a few short stubs of microtubules were associated with the chromosomes. Often (10) one or a few longer microtubule bundles were observed. In the other cases (7), a partially organised structure was formed but was very short, weak, multipolar and/or loose.
Fig. 3.
Spindle microtubule assembly is severely compromised in the srpk mutant. (A) Metaphase I spindles in mature oocytes. (i) Robust bipolar spindles; (ii) no or short microtubules; (iii) one or a few microtubule bundles; and (iv) other abnormal spindles, including multipolar spindles and/or spindles with loose and/or weak microtubules. (B) Spindle morphology in wild-type and srpk oocytes. (C) Total spindle microtubule intensity (means±s.e.m.) in mature oocytes from wild-type and srpk mutant flies. (D) FISH using the pericentromeric dodeca satellite on chromosome 3 as a probe. Arrowheads indicate the positions of the centromere 3. The inserts show FISH signals alone in a higher magnification. (E) Frequency of mono-oriented homologous centromeres. Scale bars: 10 µm.
To objectively compare the amount of spindle microtubules assembled around chromosomes, the total fluorescent intensity of tubulin signals above the background was quantified in wild type and the mutant. The total spindle microtubule signal in the srpk mutant was dramatically decreased to less than 4% of the wild-type level (P<0.001, Wilcoxon test) (Fig. 3C).
Chromosomes were misaligned, but their condensation states appeared to be normal in the mutant. In some cases (5 out of 30), more than one cluster of chromosomes was observed in combination with the above defects (supplementary material Fig. S4). Distinct foci of the pole protein D-TACC were sometimes observed at the end of microtubule bundles (Fig. 3A, iv). Immunostaining of the central spindle protein Subito (the homologue of Mklp2, a kinesin-6) (Jang et al., 2005) revealed its association with spindle microtubules near chromosomes in the mutant, although the signal was weak, as expected from poorly assembled microtubules (supplementary material Fig. S5). Taken together, these observations suggest a specific defect in spindle microtubule assembly.
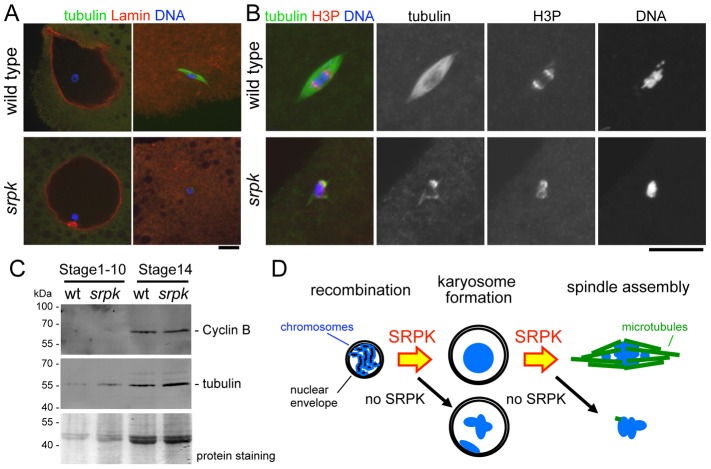
To test whether mutant oocytes had properly matured, we first examined disassembly of the nuclear envelope protein Lamin by immunostaining. Lamin was properly disassembled in the mutant as in wild type (Fig. 4A). To further confirm oocyte maturation, we immunostained phosphorylated Histone 3 (at serine 10), a commonly used M-phase marker. Histone 3 was phosphorylated in the srpk mutant as in wild type, despite poorly assembled spindle microtubules (Fig. 4B). Furthermore, immunoblotting showed both wild-type and the mutant stage-14 oocytes have a high level of Cyclin B, an activating subunit of Maturation Promoting Factor (Fig. 4C). These results argue strongly against the possibility that severe reduction of spindle microtubules is due to a failure to maturate or maintain M-phase properly.
Fig. 4.
srpk oocytes matured correctly. (A) Lamin localisation in oocytes at stage 14 (right) and earlier (left). (B) Phosphorylation of H3 at serine 10 in the wild-type and mutant mature oocytes. (C) Cyclin B accumulates both in wild-type and mutant stage-14 oocytes, but not in early-stage egg chambers. (D) SRPK is required for two essential steps in meiotic progression in oocytes. Scale bars: 10 µm.
RNA interference of srpk in Drosophila cultured cells did not reveal spindle defects in mitosis (supplementary material Fig. S6; Bettencourt-Dias et al., 2004), suggesting a specific role for SRPK in meiosis. The spindle defect seen in the srpk mutant oocytes is very penetrant and the most severe among all mutants reported so far. These results demonstrate that SRPK is essential for spindle microtubule assembly in oocytes.
SRPK is required for centromere bi-orientation and chromosome separation in oocytes
We further investigated the consequence of the spindle defect in the srpk mutant. To establish whether bi-orientation of homologous centromeres is disrupted in the srpk mutant, we carried out fluorescent in situ hybridisation (FISH) using pericentromeric dodeca satellite DNA located on chromosome 3. In mature wild-type oocytes, two signals were located at the ends of the chromosome mass (Fig. 3D). These signals represent homologous centromeres pulled from two opposite poles. Each of the signals represents two tightly attached sister-centromeres. In most mature mutant oocytes, homologous centromeres were located close together and failed to achieve bi-orientation (Fig. 3D,E).
To examine meiotic progression beyond metaphase I, activated eggs were immunostained. In wild type, various stages of development were found, including a pair of tandem meiosis II spindles which shared a spindle pole (supplementary material Fig. S7A). In contrast, most srpk mutant oocytes (22 out of 42) contained figures that may correspond to products of failed chromosome segregation (supplementary material Fig. S7A, a cluster of chromosomes or a large nucleus). Few mutant oocytes contained tandem meiosis II spindles or progressed into mitotic divisions (supplementary material Fig. S7B). This confirms that meiotic chromosome segregation is defective in the mutant.
SRPK is required for karyosome formation and spindle microtubule assembly
Our study showed that a loss of SRPK results in defects in both karyosome formation and spindle microtubule assembly (Fig. 4D). We do not think that the spindle assembly defect is simply due to a general karyosome defect, as this spindle defect is unique among karyosome mutants isolated in the same screen. How does the loss of SRPK lead to these meiotic defects? Known substrates of SRPK or its homologues include the splicing factors SR proteins, the nuclear envelope-chromatin linker Lamin B Receptor (LBR), and the sperm chromatin protein protamine (Giannakouros et al., 2011). It is possible that under-phosphorylation of SR proteins may lead to the reduction of proteins required for karyosome formation or spindle assembly. However, the effect must be specific, as our preliminary results show no global changes in the protein expression pattern. As SR proteins are also known to interact with chromatin and HP1 (Loomis et al., 2009), the effect may be independent of splicing. It is also possible that phosphorylation of LBR is required for the release of chromosomes from the nuclear envelope during karyosome formation.
Interestingly, the fission yeast homologue of SRPK, Dsk1, was isolated as a high copy suppressor of a mitotic defect of a dis1 mutation (Takeuchi and Yanagida, 1993). dis1 encodes one of two homologues of Msps/XMAP215, a crucial microtubule assembly factor (Ohkura et al., 2001). Dsk1 is more active in mitosis (similar to mammalian SRPK1) and hyperphosphorylated in mitosis, suggesting a mitotic role (Takeuchi and Yanagida, 1993). The mechanism of the suppression of dis1 is still unknown, but Dsk1 may activate other microtubule assembly proteins.
In this study we have shown that the conserved kinase SRPK is involved in two critical steps in meiotic progression in oocytes. Importantly this is the first protein whose absence virtually eliminates spindle microtubule assembly in oocytes. Identification of critical regulators and substrates of this conserved kinase would lead to an understanding of the molecular regulation of these critical but poorly understood meiotic processes.
Materials and Methods
Drosophila genetics and cytology
Standard fly techniques were followed (Ashburner et al., 2005). All stocks were grown at 25°C in standard cornmeal media. Details of mutations and chromosome aberrations can be found in Lindsley and Zimm (Lindsley and Zimm, 1992) or at Flybase (http://flybase.org; McQuilton et al., 2012). Female germline clones were induced in hs-FLP122/w; FRT ovoD/2R-129-09 (FRT G13 c px sp. *) by heat shocking (at 37°C for 45 minutes) each day for 3 days in larval stages. mnkp6 (Abdu et al., 2002) was a kind gift from W. Theurkauf (University of Massachusetts). The srpk129-09 phenotype was examined over a deficiency, Df(2R)ED2436. w1118 or the parental chromosome over Df(2R)ED2436 were used as wild-type controls. Immunostaining and FISH were carried out according to Lancaster et al. (Lancaster et al., 2007), Cullen and Ohkura (Cullen and Ohkura, 2001) and Meireles et al. (Meireles et al., 2009). Chi-square test was used for statistical analysis unless stated otherwise.
Identification of the female sterile mutation in 2R-129-09
To map the mutation in the line 2R-129-09, a series of deficiencies on the right arm of the second chromosome were crossed with the mutant line and the lethality and female sterility of transheterozygotes were tested for each cross. Two overlapping deficiencies, Df(2R)ED2426 and Df(2R)ED2436, were found to be female sterile over 2R-129-09. After confirming the karyosome defects in the transheterozygotes, further deficiencies around the regions were tested as above. 2R-129-09 over Df(2R)ED2426, Df(2R)ED2436 or Df(2R)Exel9015 was fully viable and female sterile, and showed karyosome defects identical to female germline clones of 2R-129-09. 2R-129-09 over Df(2R)Exel6285, or Df(2R)ED2436 over the parental chromosome (FRT G13 c px sp) were fertile and did not show any karyosome defects. These results indicate that a mutation responsible for female sterility and the karyosome defect is located in the genes (dup and srpk) uncovered by Df(2R)ED2426. The entire genomic region spanning dup and srpk was amplified from 2R-129-09 over Df(2R)ED2426 and the parental chromosome over the same deficiency as a series of overlapped segments by PCR using Taq polymerase (Roche), and sequenced using Big Dye terminator (Applied Biosystems) by the Edinburgh SBS sequence facility (Gene pool). The details of PCR and sequence primers used for this study are available upon request.
To perform a rescue experiment, the BAC plasmid, CH322-130M04, in attB-P[acman]-CmR-BW (Venken et al., 2009) carrying the wild-type srpk gene was obtained from BACPAC Resource Centre (BPRC) at Oakland, USA. This plasmid was used to generate transgenic flies by Genetic Service Inc. (Sudbury, USA) using the docking site VK33 at 65B2. The srpk/Df(2R)Exel9015 flies possessing the transgene CH322-130M04 (Pac[SRPK]) were tested for male and female fertility, and a karyosome and spindle phenotype, together with siblings without the transgene.
Cytological and immunological techniques
Immunostaining of ovaries, mature non-activated oocytes and activated oocytes was carried out according to Lancaster et al. (Lancaster et al., 2007), Cullen and Ohkura (Cullen and Ohkura, 2001) and Cullen et al. (Cullen et al., 2005), respectively. Fluorescent in situ hybridisation (FISH) of ovaries was carried out according to Meireles et al. (Meireles et al., 2009) using an oligonucleotide corresponding to the dodeca satellite as a probe. The oligonucleotide (cccgtactggtcccgtactggtcccgtactcggtcccgtactcggt) was 3′ end-labelled by the following method. A hundred pmol of the oligonucleotide was incubated at 37°C for 1 hour with 30 units of terminal deoxynucleotidyl transferase (Promega), 2 nmole of Alexa546-conjugated dUTP (Invitrogen) and 16 nmole of unlabelled dTTP (Promega) in 20 µl of the transferase buffer (Promega). After incubation at 70°C for 10 minutes, un-incorporated dTTP was removed by passing through a G25 Mini Quick spin column (Roche). 4 µl of this labelled oligonucleuotide was hybridised with ovaries in 40 µl of the hybridisation buffer at 30°C overnight.
Immunostained samples were examined by an Axioimager (Zeiss) attached to a confocal laser scanning head, LSM5 Exciter (Zeiss). Either one Z-plane or maximum intensity Z projections of selected planes have been shown in the figures after contrast and brightness were adjusted uniformly across the field using Photoshop (Adobe). No features were added or removed in the process.
The total tubulin intensity of the spindle was estimated using the following formula. Two areas (1,2) were drawn on the maximum intensity projection made from z-series of images taken using the same gain (500). Area 1 includes the spindle and surrounding region, and area 2 includes mainly the spindle. The result was then taken as: [I2-(I1-I2)N2/(N1-N2)]/L, where I and N are the total pixel intensity and pixel number in the specified area, respectively, and L is the laser power.
Immunoblotting of egg chambers were carried out using fluorescent secondary antibodies (Li-Cor) and detected by an Odyssey scanner (Li-Cor). Ovaries were first dissected in methanol to prevent protein degradation, and then ones at stages 1–10 and 14 were identified according to morphology.
The primary antibodies used in this study include antibodies against α-tubulin (mouse monoclonal DM1A; 1:500; Sigma), Lamin (mouse monoclonal ADL67.10; 1∶100; Developmental Studies Hybridoma Bank, or mouse monoclonal ADL84; 1∶200; kind gift from P. Fisher, SUNY), γ-H2Av (rabbit polyclonal; 1∶25; Lancaster et al., 2010), phosphorylated H3 at serine10 (rabbit polyclonal; 1∶500; Millipore), Cyclin B (mouse monoclonal F2F4; 1∶100; Developmental Studies Hybridoma Bank), D-TACC (rabbit polyclonal D-TACC-CTD; 1∶1000; this study), Subito (rat polyclonal antibody; 1∶250; this study). The polyclonal rabbit D-TACC-CTD antibody was made against D-TACC-CTD expressed in E. coli from a construct kindly given by J. Raff, Oxford (Gergely et al., 2000). The polyclonal rat Subito antibody was made against GST-Subito expressed in E. coli. The secondary antibodies were purchased from Jackson Immunologicals or Molecular Probes.
RNA interference
RNA interference and immunostaining was carried out according to Dzhindzhev et al. (Dzhindzhev et al., 2005). dsRNA was produced from wild-type genomic DNA using a pair of primers (5′-CGACTCACTATAGGGAGATTGGCTGTGCTGGGATTT-3′, 5′-CGACTCACTATAGGGAGAGCCGTTCTGCACCTCATT-3′). dsRNA corresponding to E. coli β-lactamase gene was used as a control. The cells were fixed and immunostained after three and five days' incubation with dsRNA.
Acknowledgments
We thank the Bloomington Stock Center, Exelixis, BACPAC Resources Center, Developmental Studies Hybridoma Bank, J. Raff and W. Theurkauf for providing reagents. We also thank Heather Syred, Sally Beard, Manuel Breuer and Elvira Nikalayevich for invaluable assistance.
Footnotes
Funding
This study was funded by the Wellcome Trust [grant numbers 081849, 087448, 092076]. N. V. was supported by the Boehringer Ingelheim Fonds and the Max Planck Society. Deposited in PMC for immediate release.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.107979/-/DC1
References
- Abdu U., Brodsky M., Schüpbach T. (2002). Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12, 1645–1651. 10.1016/S0960-9822(02)01165-X [DOI] [PubMed] [Google Scholar]
- Ashburner M., Golic K. G., Hawley R. S. (2005). Drosophila: A Laboratory Handbook, New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Barbosa V., Kimm N., Lehmann R. (2007). A maternal screen for genes regulating Drosophila oocyte polarity uncovers new steps in meiotic progression. Genetics 176, 1967–1977. 10.1534/genetics.106.069575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt–Dias M., Giet R., Sinka R., Mazumdar A., Lock W. G., Balloux F., Zafiropoulos P. J., Yamaguchi S., Winter S., Carthew R. W.et al. (2004). Genome-wide survey of protein kinases required for cell cycle progression. Nature 432, 980–987. 10.1038/nature03160 [DOI] [PubMed] [Google Scholar]
- Carazo–Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. (1999). Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181. 10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Cesario J., McKim K. S. (2011). RanGTP is required for meiotic spindle organization and the initiation of embryonic development in Drosophila. J. Cell Sci. 124, 3797–3810. 10.1242/jcs.084855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C. F., Ohkura H. (2001). Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3, 637–642. 10.1038/35083025 [DOI] [PubMed] [Google Scholar]
- Cullen C. F., Brittle A. L., Ito T., Ohkura H. (2005). The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J. Cell Biol. 171, 593–602. 10.1083/jcb.200508127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., Sedat J. W., Hawley R. S. (1996). Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86, 135–146. 10.1016/S0092-8674(00)80084-7 [DOI] [PubMed] [Google Scholar]
- Dumont J., Petri S., Pellegrin F., Terret M. E., Bohnsack M. T., Rassinier P., Georget V., Kalab P., Gruss O. J., Verlhac M. H. (2007). A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell Biol. 176, 295–305. 10.1083/jcb.200605199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N. S., Rogers S. L., Vale R. D., Ohkura H. (2005). Distinct mechanisms govern the localisation of Drosophila CLIP-190 to unattached kinetochores and microtubule plus-ends. J. Cell Sci. 118, 3781–3790. 10.1242/jcs.02504 [DOI] [PubMed] [Google Scholar]
- Gergely F., Kidd D., Jeffers K., Wakefield J. G., Raff J. W. (2000). D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 19, 241–252. 10.1093/emboj/19.2.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A., Ray R. P., Schüpbach T. (1998). okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12, 2711–2723. 10.1101/gad.12.17.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakouros T., Nikolakaki E., Mylonis I., Georgatsou E. (2011). Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J. 278, 570–586. 10.1111/j.1742-4658.2010.07987.x [DOI] [PubMed] [Google Scholar]
- Gruss O. J., Carazo–Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83–93. 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Jang J. K., Rahman T., McKim K. S. (2005). The kinesinlike protein Subito contributes to central spindle assembly and organization of the meiotic spindle in Drosophila oocytes. Mol. Biol. Cell 16, 4684–4694. 10.1091/mbc.E04-11-0964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. (2002). Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452–2456. 10.1126/science.1068798 [DOI] [PubMed] [Google Scholar]
- King R. C. (1970). Ovarian Development in Drosophila melanogaster. New York, NY: Academic Press. [Google Scholar]
- Klattenhoff C., Bratu D. P., McGinnis–Schultz N., Koppetsch B. S., Cook H. A., Theurkauf W. E. (2007). Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55. 10.1016/j.devcel.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Lancaster O. M., Cullen C. F., Ohkura H. (2007). NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J. Cell Biol. 179, 817–824. 10.1083/jcb.200706067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster O. M., Breuer M., Cullen C. F., Ito T., Ohkura H. (2010). The meiotic recombination checkpoint suppresses NHK-1 kinase to prevent reorganisation of the oocyte nucleus in Drosophila. PLoS Genet. 6, e1001179 10.1371/journal.pgen.1001179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Zimm G. G. (1992). The Genome of Drosophila melanogaster. San Diego, CA: Academic Press. [Google Scholar]
- Loomis R. J., Naoe Y., Parker J. B., Savic V., Bozovsky M. R., Macfarlan T., Manley J. L., Chakravarti D. (2009). Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell 33, 450–461. 10.1016/j.molcel.2009.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Hawley R. S. (1995). Chromosomal control of meiotic cell division. Science 270, 1595–1601. 10.1126/science.270.5242.1595 [DOI] [PubMed] [Google Scholar]
- McQuilton P., St Pierre S. E., Thurmond J.FlyBase Consortium (2012). FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 40, D706–D714. 10.1093/nar/gkr1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., McKim K. S. (2006). Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2, e200 10.1371/journal.pgen.0020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles A. M., Fisher K. H., Colombié N., Wakefield J. G., Ohkura H. (2009). Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184, 777–784. 10.1083/jcb.200811102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Garcia M. A., Toda T. (2001). Dis1/TOG universal microtubule adaptors - one MAP for all? J. Cell Sci. 114, 3805–3812. [DOI] [PubMed] [Google Scholar]
- Parfenov V., Potchukalina G., Dudina L., Kostyuchek D., Gruzova M.1989). Human antral follicles: oocyte nucleus and the karyosphere formation (electron microscopic and autoradiographic data). Gamete Res. 22219–231. 10.1002/mrd.1120220209 [DOI] [PubMed] [Google Scholar]
- Sampath S. C., Ohi R., Leismann O., Salic A., Pozniakovski A., Funabiki H. (2004). The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118, 187–202. 10.1016/j.cell.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Staeva–Vieira E., Yoo S., Lehmann R. (2003). An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22, 5863–5874. 10.1093/emboj/cdg564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Yanagida M. (1993). A mitotic role for a novel fission yeast protein kinase dsk1 with cell cycle stage dependent phosphorylation and localization. Mol. Biol. Cell 4, 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., Ashford A. J., Rybina S., Pozniakovsky A., Mayer T. U., Walczak C. E., Karsenti E., Hyman A. A. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., Spokony R., Wan K. H., Koriabine M., de Jong P. J., White K. P., Bellen H. J., Hoskins R. A. (2009). Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434. 10.1038/nmeth.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N., Koch I., Schwarz H., Schnorrer F., Nüsslein–Volhard C. (2006). The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963–3972. 10.1242/dev.02570 [DOI] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. (1999). Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359–1362. 10.1126/science.284.5418.1359 [DOI] [PubMed] [Google Scholar]
- Zuccotti M., Giorgi Rossi P., Martinez A., Garagna S., Forabosco A., Redi C. A. (1998). Meiotic and developmental competence of mouse antral oocytes. Biol. Reprod. 58, 700–704. 10.1095/biolreprod58.3.700 [DOI] [PubMed] [Google Scholar]
- Zuccotti M., Ponce R. H., Boiani M., Guizzardi S., Govoni P., Scandroglio R., Garagna S., Redi C. A. (2002). The analysis of chromatin organisation allows selection of mouse antral oocytes competent for development to blastocyst. Zygote 10, 73–78. 10.1017/S0967199402002101 [DOI] [PubMed] [Google Scholar]