Abstract
Repeat proteins contain tandem arrays of small structural motifs. As a consequence of this architecture, they adopt non-globular, extended structures that present large, highly specific surfaces for ligand binding. Here we discuss recent advances toward understanding the functional role of this unique modular architecture. We showcase specific examples of natural repeat proteins interacting with diverse ligands and also present examples of designed repeat protein–ligand interactions.
Introduction
Repeat proteins consist of tandem arrays of a small structural motif. Here we focus on six different families (Table 1), highlighting examples that illustrate general themes of repeat protein function. In the past two years there have been significant advances both in our understanding of the functioning of natural repeat proteins and in our ability to engineer novel repeat proteins. Here we discuss features of repeat protein function that are intimately dependent upon their unique structure. Repeat proteins are, by their very nature, extended structures, and thus have a larger surface area to volume ratio than typical globular proteins. They are therefore particularly well suited to mediate protein–protein interactions and to organize multiple proteins into functional complexes. Moreover, the modular structure allows for different sets of repeats to be used to bind to different ligands.
Table 1.
Repeat protein families and their characteristics
| Repeat protein family | Number of amino acids in a repeat | Structural motif of a repeat | Range of numbers of repeats for natural proteins* most common number |
|---|---|---|---|
| HEAT | 37–47 | Two α-helices (A & B) | 3–36 |
| TPR | 34 | Helix-turn-helix (A & B) | 3–16, 3* |
| Armadillo | 42 | Three α-helices (H1, H2, H3) | 6–15, 12* |
| Ank | 30 | Helix-helix-loop (or β-hairpin) (H1, H2) | 4–24, 6* |
| LRR | 20–29 | β-strand-loop-helix | Up to 28 |
| WD40 | 40–60 | Four-stranded (a–d) antiparallel β-sheet | 3–16, 7*–8* |
Names for protein families originate from: HEAT, Huntingtin; Elongation factor 3; A subunit of PP2A; lipid kinase TOR; TPR, tetratricopeptide repeat; Armadillo, the appearance of embryos that are mutant for the Drosophila segment polarity gene armadillo; Ank, ankyrin-like repeat; LRR, leucine rich repeat; WD40 (also known as WD or beta-transducin repeats) amino acid motifs, often terminating in a Trp-Asp (WD) dipeptide.
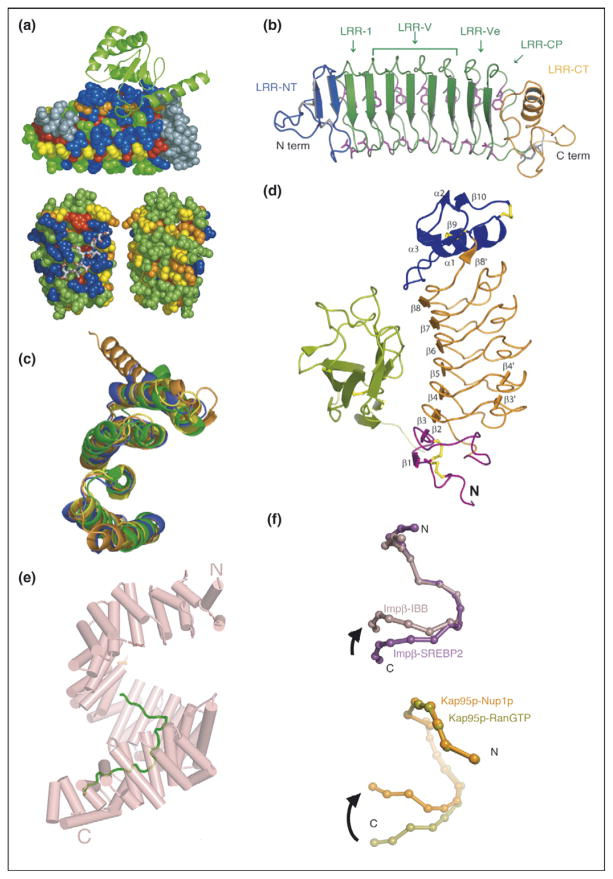
For several classes of repeat proteins, including Ank, TPR, and LRR (Table 1), analyses of the amino acid variability at different positions within a single repeat have revealed an interesting feature—the residues that comprise the ligand binding site are significantly more variable than other positions on the protein surface (Figure 1a) [1–3]. Further analysis of these hypervariable positions, in examples where structures are available, reveals that they are in direct contact with ligand. This finding is consistent with the idea that the repeat protein provides a constant framework that can be ‘decorated’ with functional residues. In fact, sequence ‘hypervariation’ alone can be used to predict the ligand binding sites. This observation is exactly analogous to that of Wu and Kabat, who first identified the complementarity determining regions (CDR) of antibodies on the basis of sequence hypervariability [4].
Figure 1.
Repeat proteins as conserved scaffolds. (a) Sequence variability analysis repeat proteins. Upper panel: variability analysis of 15 497 Ank repeats. The 4-Ank domain from GABPβ1 (spheres) is shown bound to the ligand GABPα (ribbons) (PBD ID: 1awc). Coloring is from most variable = blue to least variable = red. The binding surface is clearly more blue than the rest of the proteins’ surface. Lower panel: sequence variability analysis of 6887 TPR repeats mapped onto the co-crystal structures of HOP-TPR1/Hsc70 peptide (PBD ID: 1elw) with the TPR domain rendered in spheres and the ligands in sticks. Two views from 180° rotation of the molecule are shown. The concave, ligand binding surface, left, is clearly more variable (blue) than the convex, solvent exposed surfaces (green, yellow), right [1,3]. (b) Crystal structure of hagfish variable lymphocyte receptor (VLR-A). The VLR-A protein comprises 8 LLR modules. The constant N-terminal (LRR-NT) and C-terminal (LRR-CT) modules are shown in blue and orange, respectively. The variable LRR modules (LRR-V) that can be present in variable numbers are colored in green [2••]. (c) Alignment of TPR domains. Structural alignment of TPR domains unliganded and peptide-bound: CTPR3 (PDB ID 1NA0) [35] in blue, TPR domain (residues 19-177) of PP5 in orange (PDB ID: 1a17) [36], and TPR2A (PDB ID: 1elr) in green and TPR1 (PDB ID: 1elw) in yellow, domains of Hop from the co-crystal structures with their peptide ligands, C-terminal peptides of Hsp90 and Hsp70, respectively [37]. The structures are all completely superimposable, with backbone RMSD values that vary from 1.1 to 1.9 Å for different pairwise structural alignments [5]. (d) Structure of Slit2 D2 bound to Robo1 Ig1. Ig1 is in green; Slit2 D2 N-terminal and C-terminal caps are in purple and blue, respectively; LRRs 1–6 are in orange; and the disulfide bridges are in yellow [6]. (e) Structure of HEAT domain of karyopherin Kapβ2 bound to NLS substrate (PDB ID: 1ot8). The helices are represented as cylinders and structurally disordered loops as dashed lines. The extended NLS sequence of the cargo protein hnRNPA1 is shown in green. A1-NLS peptide binds in an extended conformation on the concave surface of the C-terminal arch of the karyopherin superhelix [10••]. (f) Schematic illustration of the hinge motion between subgroups of HEAT repeats. (Top) Schematic illustration of the hinge movement between HEAT repeat subdomains H1–H12 and H14–H19 in Impβ and Kap95p upon ligand binding. IMM-bound Impβ (pink) and SREBP2-bound Impβ (purple) are superimposed at HEAT repeats 5–10, with a sphere representing the center of each HEAT repeat. (Bottom) Schematic illustration of the hinge movement between HEAT repeat subdomains H1–H14 and H15–H19. RanGTP-bound Kap95p (light green) and Nup1p-bound Kap95p (orange) are superimposed at HEAT repeats 6–13, with a sphere representing the center of each HEAT repeat [10••].
An additional mechanism by which to enhance binding diversity is seen in the variable lymphocyte receptors (VLR) of the adaptive immune system of jawless vertebrates [2••]. VLRs are composed of LRRs, with a constant N-terminal and C-terminal LRRs flanking an array of one to eight variable central LRRs. The central LRRs are responsible for antigen binding. Germ-line rearrangements insert different numbers of central LRRs as a unique means by which to increase the diversity of ligand recognition (Figure 1b).
No conformational changes upon ligand binding
A common theme in repeat protein structure is that both the individual repeats, and their positions relative to each other, are the same regardless of the protein in which they occur. Moreover, upon ligand binding there is typically little, if any, conformational change. The structures of different repeat proteins, with and without ligand bound, have been compared with RMSD between the ligand-bound and ligand-free structures as little as 1.0 Å. An example for TPR proteins is shown in Figure 1c [5]. In other cases, the same protein with and without ligand bound has been compared, with RMSD differences as little as 0.4 Å and 0.43 Å for LRR and WD40 examples, respectively [6,7].
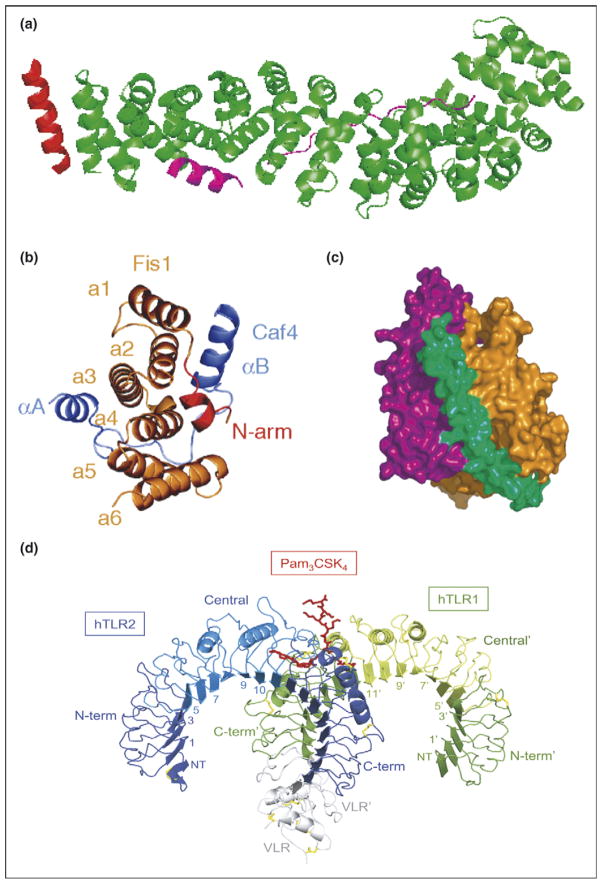
The structure of β-catenin, a protein containing 12 armadillo repeats, in ternary complex with BCL9 and Tcf4 clearly shows that the armadillo domain is virtually identical to the previously reported structure of this domain alone (Figure 4a) [8]. Slit proteins 1–3 each have an N-terminal domain composed of four tandem LRR domains (D 1–4), each of which contains multiple LRR. The second LRR domain of the Slit protein always interacts with the first Ig domain of the Robo protein (Figure 1d). This complex participates in development of bilateral symmetry in both insects and vertebrates. A comparison of the structure of the Slit2 D2-Robo1 Ig1 complex with that of the uncomplexed domains shows that no structural changes occur upon binding. Interestingly, in this example, complex formation involves an interface that consists of two distinct regions—the first region encompassing repeats 1 and 2 is predominantly electrostatic in nature, whereas the second, encompassing repeats 4, 5, and 6, is predominantly hydrophobic [6]. One can speculate about the modularity of recognition being taken even further in this case.
Figure 4.
Typical and atypical modes of binding. (a) Overall structure of the β-catenin/BCL9/Tcf-4 complex in green, magenta, and red, respectively. Packing of BCL9 against the groove formed between H2 and H3 of the N-terminal capping armadillo repeat to form a helical bundle illustrates atypical binding interface for armadillo repeat proteins [8]. (b) Fis1 TPR-Caf4 complex show a new two surfaces binding mode. Crystal structures of the TPR-like protein Fis1 in complex with Caf4 protein (PDB ID: 2pqr). The Fis1 N-terminal arm (red) remains packed against the original hydrophobic groove and stabilizes the B helix of Caf4 (purple), which packs against a second hydrophobic groove on the concave surface of Fis1. In addition to the B helix packed against the concave Fis1 surface, Caf4 (purple) uses the A helix and the intervening loop to bind the convex Fis1 surface [18]. (c) Molecular surface representation of CSL–Notch–Mastermind ternary complex. CSL (orange) and Notch (purple) interact to present composite surface for binding of Mastermind (green). (d) Overall structure of the human TLR1–TLR2–Pam3CSK4 complex. The TLR1 fragments and the TLR2 fragments are shown schematically in green and blue, respectively. The central domains are colored in light green or light blue, and the Pam3CSK4 lipopeptide in red. Disulfide bridges are represented as yellow lines. Domains belonging to the TLR1 hybrid proteins are labeled with apostrophes [21••].
Karyopherinβ2 (Kapβ2), a protein involved in transporting other proteins into the nucleus, contains about 20 tandem HEAT repeats divided into three major segments: HEAT repeats 1–8 form a Ran binding site [9], whereas HEAT repeats 9–13 and 14–18 are substrate binding sites (Figure 1e) [10••]. The structures of the HEAT repeats, in either complex with different NLS (nuclear localization signal) sequences [11,12], or without ligand bound, are essentially identical [10••]. The interesting conformational change that does occur in this protein upon substrate binding is a rotation of the sets of HEAT repeats relative to each other, as rigid elements, about a flexible hinge between HEAT repeats 13 and 14 (Figure 1f) [10••]. Similar behavior is seen in the importin-β family [10••].
Repeat proteins bind extended ligands
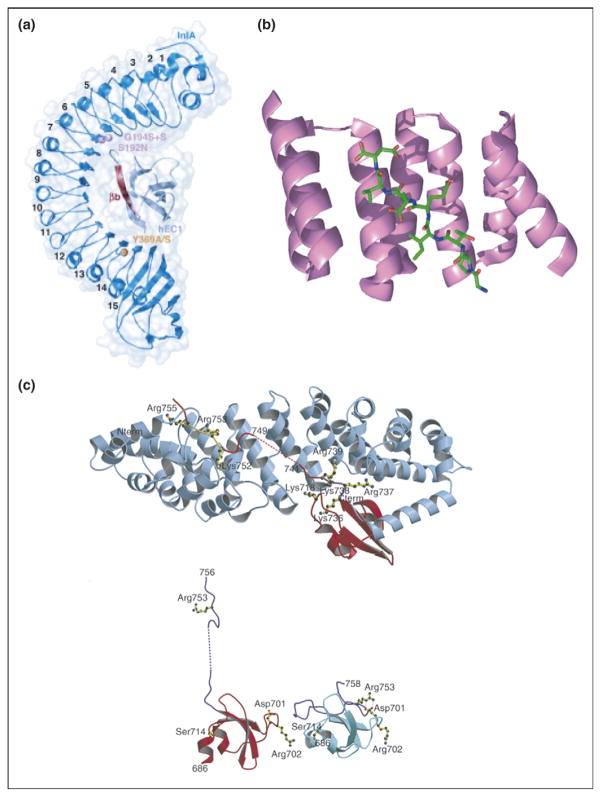
Employing multiple repeats to form an extended surface area of interaction with an extended ligand is an efficient route to tight binding. A common theme is that the repeat protein interacts with an extended peptide (Figure 2b) or element of secondary structure in the target protein, allowing maximal surface area of contact per amino acid. For example, the eight LRRs of the protein internalin interact with a long, extended, β-strand of E-cadherin (Figure 2a) [13••]. Similarly, 12 consecutive HEAT repeats of karyopherin β2 make extensive contacts with the extended NLS peptides (Figure 1e) [10••]. A particularly dramatic example is seen in the interaction of human importin α5 with the C-terminal domain of influenza virus polymerase PB2, where the last 20 residues of PB2 actually unfold into an extended conformation for interaction with the 10 armadillo repeats of importin (Figure 2c) [14••].
Figure 2.
Repeat proteins bind to extended ligands. (a) Overall structure of internalin (InlA)/N-terminal domain of human E-cadherin (hEC1) complex shown to be in dark and light blue, respectively. The eight LRRs of InlA interact with a long, extended, β-strand of hEC1 [13••]. (b) Crystal structure of TPR1 domain of HOP in complex with the extended C-terminal peptide of Hsp70 bound in the concave face of the TPR domain. TPR1 domain is shown as a ribbon (purple) and the 8-mer C-terminal peptide of Hsp70 is shown as sticks (green) [37]. (c) (Top) Ribbon diagram showing C-terminal domain of influenza virus polymerase PB2 subunit (red) bound to human importin α5 (blue), comprising 10 armadillo repeats. (Bottom) Comparison of the PB2 domain structure in complexed (red) and free solution state (cyan) demonstrates unfolding of residues 736–759 (purple) upon binding to importin α5 [14••].
Repeat proteins as multi-protein complex organizers
The extended, modular nature of repeat proteins also allows for different regions of the repeat protein to be used to interact with different ligands, thus bringing them together into a functional complex. Such repeat protein mediated multi-protein organization can occur in different ways.
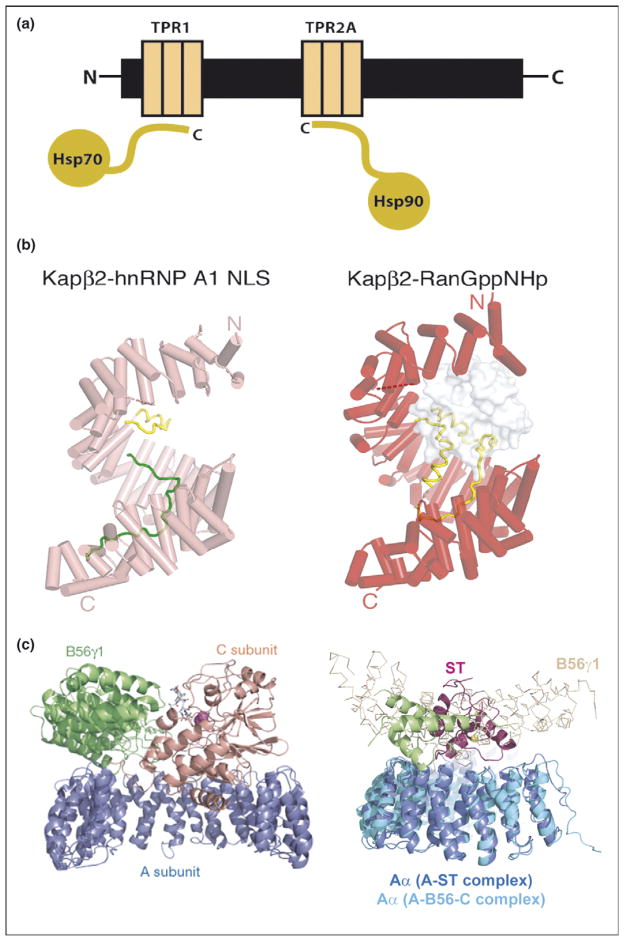
In Hsp organizing protein (HOP), for example, there are two discrete sets of TPR modules, one of which binds to Hsp70 and the other to Hsp90, bringing the chaperones together into a functional complex (Figure 3a) [15].
Figure 3.
Repeat proteins as multi-protein complex organizers. (a) Schematic representation of Hsp90/Hsp70 organizing protein (HOP) showing how two independent TPR domains serve to assemble the multi-chaperone complex. The figure shows the interactions between the TPR1 and TPR2A domain of HOP with the C-terminal sequences of Hsp70 and Hsp90, respectively. (b) Heat repeats of karyopherins present different binding surfaces for multi-protein binding. Structure of karyopherin Kapβ2 bound to NLS substrate (PDB ID: 1ot8) (left panel) and bound to the regulatory protein Ran (PDB ID: 1qbk) (right panel). The helices are represented as cylinders (pink in susbtrate-bound structure and red in the Ran-bound structure); structurally disordered loops are shown as dashed lines and HEAT repeat 8 (H8) loops are in yellow. The substrate hnRNP A1-NLS is shown as a green ribbon and the Ran protein as a surface representation in gray [10••]. (c) Protein phosphatase 2A (PP2A) HEAT repeats as scaffolding subunit. Left panel: Ribbon representation of the heterotrimeric PP2A holoenzyme crystal structure. The scaffolding subunit A colored in blue binds the regulatory subunit B56γ1 (green) and the catalytic subunit C (orange). Right panel: SV 40 Small T antigen competes with the regulatory subunit B. Structural superimposition of the A-ST complex (PDB ID: 2pf4) and the A-B56-C PP2A holoenzyme complex (PDB ID: 2iae). The two complexes are superimposed using A subunit HEAT repeats 2–10. The J and unique domain of ST are colored green and pink, respectively. The Cα trace of B56γ1 regulatory subunit are shown in light orange. It is clear that ST and B56γ1 bind to the same sites on PP2A A subunit [16••,17••].
HEAT repeats are used to assemble multi-protein complexes in proteins that function in nucleocytoplasmatic transport in a very different fashion. The 20–40 HEAT repeats in karyopherin form a superhelix, the external convex surface is involved in nucleoporin binding whereas the inner, concave face interacts with the NLS of cargo proteins (Figure 3b, left) [10••,11,12]. The concave face also presents a binding site for the regulatory protein Ran-GTP (repeats 1–8) (Figure 3b, right) [9].
Another example of using different sets of HEAT repeats for binding different proteins is seen in protein phosphatase 2A (PP2A). PP2As are heterotrimeric proteins in which a ‘scaffolding’ A subunit (with 15 HEAT repeats) binds to both the regulatory ‘B’ subunit and the catalytic ‘C’ subunit with different sets of HEAT repeats (Figure 3c, left) [16••]. Both AC and ABC versions of the complex exist, demonstrating the independence of binding to different sets of repeats within the HEAT domain. Interestingly, it has recently been shown that SV40 small T antigen perturbs the functioning of PP2A by competing with the regulatory B subunit for binding to HEAT repeats 3–7 (Figure 3c, right) [17••].
Atypical binding modes
In the majority of cases, when binding involves multiple repeats, each repeat contributes to the binding interface with the same structural element. For example in armadillo repeats a concave binding interface is lined with H3 helices, in ankyrin repeats a concave binding surface is formed by H1 helices, in LRR proteins parallel β strands create a concave curved β sheet, and in TPRs the concave binding surface is formed by the A-helices (Figure 2b, Table 1). Here we discuss exceptions to these ‘typical’ binding interfaces.
In the β-catenin/BCL9/Tcf4 complex, β-catenin binds to Tcf4 via typical H3-mediated interactions with six armadillo repeats. The interaction with BCL9, however, is atypical with helical BCL9 packing between H2 and H3 of the N-terminal capping armadillo repeat to form a helical bundle (Figure 4a) [8].
The TPR-like protein Fis1 forms complexes with either Mdv1 protein or Caf4 protein [18]. Surprisingly, in both complexes an N-terminal α-helix of Fis1 occupies the typical hydrophobic groove on the concave surface of its own TPR domain and an α-helix of the target protein packs in a second hydrophobic groove on this concave face. However, there is an additional atypical interaction with Caf4 in which a second α-helix of this target protein binds in the convex face of the TPR domain (Figure 4b).
Another variation is that a repeat protein may interact with one protein to create a composite surface for the binding of a third protein. For example, in the CSL–Notch–Mastermind ternary complex, Mastermind simultaneously interacts with Notch1 (Ank repeats three through seven) and CSL. Neither Notch nor CSL undergoes a significant conformational change upon complex formation, indicating that Mastermind recognizes the composite surface derived from both proteins rather than an allosterically induced binding site on either CSL or Notch1 alone (Figure 4c) [19•,20•].
Mammalian innate immune system proteins TLR1 and TLR2 heterodimerize by simultaneously binding to a lipopetide ligand. Such dimerization has been proposed to be a key activation event. TLR1 interacts with the ligand via the ‘typical’ concave face, whereas TLR2 interacts with the lipopeptide via the atypical convex face (Figure 4d) [21••].
Beyond protein–protein interactions
Repeat proteins are continually being shown to bind an ever-increasing repertoire of ligands. Contrary to well known ‘specialized’ folds, such as KH domains [22], which only bind nucleic acids, in repeat proteins we see that exactly the same repeat and fold can bind many different types of ligands. Probably the best described examples are the 16–28 LRR extracellular domains of the Toll-like receptors (TLR) of the mammalian innate immune system that bind proteins, lipoproteins, lipopeptides, and nucleic acids [23•,24]. HEAT repeats are also extremely versatile binding modules. They mostly mediate protein–protein interactions, but there are also examples in which they bind nucleic acids, such as the autoantigen protein Ro, which binds a variety of misfolded RNAs and small non-coding RNAs (Y RNAs) [25,26].
New repeat proteins by selection and design
Following the above discussion, it is not surprising that repeat proteins are appealing targets for protein design. They bind a variety of ligands using scaffolds built from a simple, short, repeated module. Moreover, multiple sequence alignments and structural characterizations allow a clear delineation of structurally and functionally important residues. Several exciting new Ankyrin and TPR repeat protein-based designs have been presented in the past two years. Two complementary strategies have been employed: The first has been to introduce novel binding specificities onto existing repeat scaffolds [27••], the second has been to create new scaffolds onto which known binding sites are grafted [28].
Ank repeats have been used as scaffolds in selections to identify proteins with novel binding specificities. Different numbers of a consensus Ank module, in which six putative binding residues per repeat are randomized, are assembled into large libraries, whose diversity can be increased still further by mutagenic PCR. The small number of Ank repeats in the library, which bind to the specific target are selected in vitro, by ribosome display [27••,29••]. This strategy has yielded novel Ank proteins that bind to new targets, for example the extracellular domain of HER2, the Multi-drug Exporter AcrB, and Caspase-2 with high affinity [29••,30,31]. Interestingly, many mutations that result from the mutagenic PCR step and that enhance binding are at so called framework positions, rather than binding positions [29••]—a result whose significance is not yet understood.
A TPR module, which binds the C-terminal peptide of Hsp90, has been designed by grafting Hsp90-binding residues from natural TPRs onto a consensus TPR scaffold. This designed protein binds Hsp90 with greater affinity and specificity than natural Hsp90 co-chaperones [32••]. Significantly, introduction of this designed protein into breast cancer cells inhibits Hsp90 activity, presumably by out-competing the interaction of Hsp90 with its natural co-chaperones (Figure 3a). The consequence of inhibiting Hsp90 is that Hsp90-dependent proteins, such as HER2, are not folded but are degraded, resulting in cancer cell death [32••].
Future directions
Armadillo repeats bind extended peptide ligands via helix H3: one dipeptide of the ligand per armadillo repeat. Such modular recognition is appealing from the design standpoint because it suggests that one can develop ‘dipeptide recognizing’ modules that can then be combined in any desired combination. Although preliminary results from this approach are encouraging [33], this strategy awaits the full realization.
A consensus sequence for a WD40 repeat has been reported, and protein that contains between 4 and 10 such tandem repeats have been created. Because members of the WD40 propeller fold are characterized by a distinctive diversity of sequence and can support a wide spectrum of biological functions, the authors of this report speculate that such motifs may be the first choice for future functional design. [34••].
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Magliery TJ, Regan L. Beyond consensus: statistical free energies reveal hidden interactions in the design of a TPR motif. J Mol Biol. 2004;343:731–745. doi: 10.1016/j.jmb.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 2••.Kim HM, Oh SC, Lim KJ, Kasamatsu J, Heo JY, Park BS, Lee H, Yoo OJ, Kasahara M, Lee JO. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007;282:6726–6732. doi: 10.1074/jbc.M608471200. Inserting multiple LRR repeats from a germline repertoire to accomplish diversity. [DOI] [PubMed] [Google Scholar]
- 3.Magliery TJ, Regan L. Sequence variation in ligand binding sites in proteins. BMC Bioinformatics. 2005;6:240. doi: 10.1186/1471-2105-6-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson G, Wu TT. Kabat database and its applications: 30 years after the first variability plot. Nucleic Acids Res. 2000;28:214–218. doi: 10.1093/nar/28.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortajarena AL, Regan L. Ligand binding by TPR domains. Protein Sci. 2006;15:1193–1198. doi: 10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morlot C, Thielens NM, Ravelli RB, Hemrika W, Romijn RA, Gros P, Cusack S, McCarthy AA. Structural insights into the Slit-Robo complex. Proc Natl Acad Sci U S A. 2007;104:14923–14928. doi: 10.1073/pnas.0705310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 8.Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu W. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 10••.Cansizoglu AE, Chook YM. Conformational heterogeneity of karyopherin beta2 is segmental. Structure. 2007;15:1431–1441. doi: 10.1016/j.str.2007.09.009. Study showing bending between rigid sets of repeats. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM. Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Biol. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BJ, Cansizoglu AE, Süel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by Karyopherinβ2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Wollert T, Heinz DW, Schubert WD. Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin. Proc Natl Acad Sci U S A. 2007;104:13960–13965. doi: 10.1073/pnas.0702199104. An interesting study that describes ligand binding to the extended surface of the repeat protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. Dramatic example of ligand unfolding to bind to the repeat protein in extended conformation. [DOI] [PubMed] [Google Scholar]
- 15.Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 16••.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. See annotation to [17••] [DOI] [PubMed] [Google Scholar]
- 17••.Cho US, Morrone S, Sablina AA, Arroyo JD, Hahn WC, Xu W. Structural basis of PP2A inhibition by small t antigen. PLoS Biol. 2007;5:e202. doi: 10.1371/journal.pbio.0050202. Study that characterizes the use of repeats in multi-protein complex assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci U S A. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. Back to back studies [20•] showing formation of composite surfaces for ligand binding. [DOI] [PubMed] [Google Scholar]
- 20•.Wilson JJ, Kovall RA. Crystal structure of the CSL–Notch–Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. Back to back studies [19•] showing formation of composite surfaces for ligand binding. [DOI] [PubMed] [Google Scholar]
- 21••.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1–TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. Structural basis of inate immune response. [DOI] [PubMed] [Google Scholar]
- 22.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275(11):2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 23•.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. Recent review on structure, function, and ligand binding of LRRS in Toll receptors. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of Toll-Like Receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nat Struct Biol. 2006;13:1002–1009. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- 27••.Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grutter MG, Pluckthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. Method for design and in vitro selection of novel repeat protein binders. [DOI] [PubMed] [Google Scholar]
- 28.Cortajarena AL, Kajander T, Pan W, Cocco MJ, Regan L. Protein design to understand peptide ligand recognition by tetratricopeptide repeat proteins. PEDS. 2004;17:399–409. doi: 10.1093/protein/gzh047. [DOI] [PubMed] [Google Scholar]
- 29••.Zahnd C, Wyler E, Schwenk JM, Steiner D, Lawrence MC, McKern NM, Pecorari F, Ward CW, Joos TO, Pluckthun A. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol. 2007;369:1015–1028. doi: 10.1016/j.jmb.2007.03.028. Study that describes novel high affinity binder developed by selection. [DOI] [PubMed] [Google Scholar]
- 30.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz HK, Capitani G, Baici A, Pluckthun A, et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: specificity, structure, and inhibition mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 32••.Cortajarena AL, Yi F, Regan L. Designed TPR modules as novel anticancer agents. ACS Chem Biol. 2008;3:161–166. doi: 10.1021/cb700260z. Study that describes designed repeat protein that is active in vivo. [DOI] [PubMed] [Google Scholar]
- 33.Parmeggiani F, Pellarin R, Larsen AP, Varadamsetty G, Stumpp MT, Zerbe O, Caflisch A, Pluckthun A. Designed armadillo repeat proteins as general peptide-binding scaffolds: consensus design and computational optimization of the hydrophobic core. J Mol Biol. 2008;376:1282–1304. doi: 10.1016/j.jmb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 34••.Nikkhah M, Jawad-Alami Z, Demydchuk M, Ribbons D, Paoli M. Engineering of beta-propeller protein scaffolds by multiple gene duplication and fusion of an idealized WD repeat. Biomol Eng. 2006;23:185–194. doi: 10.1016/j.bioeng.2006.02.002. This is perhaps the future scaffold of choice for functional design. [DOI] [PubMed] [Google Scholar]
- 35.Main ERG, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 36.Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]