Background: The loss of McpC has been shown to reduce chemotaxis to 19 of the 20 amino acids.
Results: McpC can directly bind 11 amino acids and indirectly sense four others.
Conclusion: McpC can sense a variety of amino acids by using two discrete mechanisms.
Significance: We elucidate the mechanisms by which a single receptor can sense a wide variety of ligands.
Keywords: Bacillus, Bacteria, Chemotaxis, Ligand-binding Protein, Receptors
Abstract
Bacillus subtilis can perform chemotaxis toward all 20 l-amino acids normally found in proteins. Loss of a single chemoreceptor, McpC, was previously found to reduce chemotaxis to 19 of these amino acids. In this study, we investigated the amino acid-sensing mechanism of McpC. We show that McpC alone can support chemotaxis to 17 of these amino acids to varying degrees. Eleven amino acids were found to directly bind the amino-terminal sensing domain of McpC in vitro. Sequence analysis indicates that the McpC sensing domain exhibits a dual Per-Arnt-Sim (PAS) domain structure. Using this structure as a guide, we were able to isolate mutants that suggest that four amino acids (arginine, glutamine, lysine, and methionine) are sensed by an indirect mechanism. We identified four candidate binding lipoproteins associated with amino acid transporters that may function in indirect sensing: ArtP, GlnH, MetQ, and YckB. ArtP was found to bind arginine and lysine; GlnH, glutamine; MetQ, methionine; and YckB, tryptophan. In addition, we found that ArtP, MetQ, and YckB bind the sensing domain of McpC, suggesting that the three participate in the indirect sensing of arginine, lysine, methionine, and possibly tryptophan as well. Taken together, these results further our understanding of amino acid chemotaxis in B. subtilis and gain insight into how a single chemoreceptor is able to sense many amino acids.
Introduction
The Gram-positive bacterium Bacillus subtilis can perform chemotaxis toward all 20 l-amino acids normally found in proteins (1). These amino acids are sensed by at least two transmembrane chemoreceptors, McpB and McpC, which contain an extracellular sensing domain and a cytoplasmic signaling domain (2, 3). These chemoreceptors form ternary complexes with the CheA histidine kinase and the CheW and CheV scaffolding proteins (4). The binding of amino acids to these chemoreceptors increases the rate of CheA autophosphorylation (2, 5). The phosphoryl group is then passed to the CheY response regulator protein, which in its phosphorylated form can bind to the cytoplasmic face of the flagellar motor and induce a chemotactic response (6).
McpB is the sole chemoreceptor for asparagine, and it additionally supports chemotaxis toward aspartate, glutamine, and histidine (2). Previous work has demonstrated that the sensing domain of McpB contains a dual Per-Arnt-Sim (PAS)3 domain architecture and that asparagine binds in the upper PAS domain (7). This architecture is found in other chemoreceptors, including B. subtilis McpC and Vibrio cholerae McpN, as well as numerous other sensor histidine kinases (7, 8). McpC is the sole receptor for proline and was previously found to support chemotaxis to all other amino acids except asparagine (3). In particular, an mcpC-null mutant showed reduced chemotaxis to 19 amino acids. These results suggest that McpC senses 19 amino acids. The goal of this study is to investigate amino acid sensing by McpC. In particular, we sought to determine whether a single receptor can indeed sense 19 different amino acids and, if so, to begin to elucidate the mechanisms involved.
Previous work from Escherichia coli demonstrated that chemotaxis sensing may be direct, where the chemoreceptor directly binds a ligand, or indirect, where a second protein is involved. A well studied example of indirect sensing is the maltose-binding protein from E. coli involved in maltose chemotaxis (9–11). This periplasmic protein binds maltose and interacts with the sensing domain of the Tar chemoreceptor (10). Given the diversity of amino acid ligands for McpC, an indirect sensing mechanism is likely involved as well. However, the sensing domains of the E. coli chemoreceptors exhibit a different structure than those of the B. subtilis McpB and McpC chemoreceptors (7). An example of indirect sensing by a dual PAS domain sensor comes from LuxQ, a sensor histidine kinase involved in quorum sensing in Vibrio harveyi (12). LuxQ indirectly senses AI-2 through its interactions with LuxP, a periplasmic binding protein. LuxP directly binds the small molecule AI-2 and can then activate the LuxQ sensor kinase. Both PAS domains of LuxQ are involved in the interaction with LuxP, and there are numerous residues along the LuxQ/LuxP interface that are crucial for this interaction (12).
In this work we demonstrate that McpC alone can support chemotaxis to 17 amino acids. Of these, McpC directly binds 11 amino acids. Using the structure of LuxQ bound to LuxP, we were able to identify mutants that suggest four amino acids are sensed by an indirect mechanism. Moreover, we were able to show three binding lipoproteins that interact with the sensing domain of McpC and a fourth that may as well. We also found that these four lipoproteins respectively bind these four amino acids, suggesting that they enable indirect sensing by McpC.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
All B. subtilis strains are derived from the chemotactic strain (che+) OI1085 (13). All cloning and plasmid propagations were performed in E. coli strain TG1 (Amersham Biosciences). Recombinant protein overexpression was done in E. coli strain BL-21 (Amersham Biosciences). Plasmids used for protein purification were expressed in pGEX-6P-2 (GE Healthcare).
All B. subtilis strains were created by using QuikChange (Stratagene) mutagenesis on pAIN750 variants that contained the full-length mcpC gene (14). These plasmids were then transformed into the amyE locus of the Δ10mcp strain (OI3545) and selected for spectinomycin resistance. Expression of McpC was confirmed by Western blotting, and its levels were found to be equal to the wild type (data not shown).
The GST fusion pGEX-6P-2 plasmids were made by amplifying the amino terminus from the mcpC gene (residues 33–276) or the binding protein genes without the transmembrane anchor (artP residues 34–322, glnH residues 48–297, metQ residues 33–317, yckB residues 47–314) from OI1085 genomic DNA by PCR. The primers used were engineered to create a 5′ EcoRI and 3′ NotI site. This fragment was then ligated into pGEX-6P-2.
Capillary Assay
The capillary assay was performed as described previously to quantitatively measure the chemotactic ability of various strains (15, 16). Briefly, cells were grown overnight at 30 °C on TBAB plates (1% tryptone, 0.3% beef extract, 0.5% NaCl, 1.5% agar). The cells were then scraped from the plate and resuspended to A600 nm = 0.03 in 5-ml capillary assay minimal medium (50 mm K3PO4, pH 7.0, 1.2 mm MgCl2, 0.14 mm CaCl2, 1 mm (NH4)2SO4, 0.01 mm MnCl2, 20 mm sorbitol, and 0.02% tryptone, supplemented with 50 μg/ml histidine, methionine, and tryptophan). The cultures were grown to A600 nm = 0.4 at 37 °C and 250 rpm shaking, after which 50 μl of the 5% glycerol, 0.5 m sodium lactate solution was added, and the cells were incubated a further 15 min. The cells were then washed three times with chemotaxis buffer (10 mm K3PO4, pH 7.0, 0.14 mm CaCl2, 0.3 mm (NH4)2SO4, 0.1 mm EDTA, 5 mm sodium lactate, 0.05% (v/v) glycerol) and diluted to A600 nm = 0.001. The resuspended cells were aliquoted into 0.3-ml ponds on a temperature-controlled plate at 37 °C, and closed-end capillary tubes filled with appropriate chemoattractant were inserted and incubated for 30 min. Cells in the capillaries were harvested and transferred to 0.5 ml of top agar (1% tryptone, 0.8% NaCl, 0.8% agar, 0.5 mm EDTA) and plated onto TBr (1% tryptone, 0.5% NaCl, 1.5% agar) plates. These plates were incubated at 37 °C for 16 h at which point colonies were counted to derive the data. Experiments were performed in duplicate and on 2 different days to assure reproducibility.
Protein Purification
To purify the GST fusion proteins, cells containing pGEX-6P-2 with the assorted chemotaxis proteins cloned in the multiple cloning site were grown in 6 liters of LB medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) supplemented with 100 μg/ml ampicillin at 37 °C and with 250 rpm shaking until A600 nm = 0.8. Expression was then induced by addition of 1 mm isopropyl β-d-1-thiogalactopyranoside, and the culture was grown at 25 °C with 250 rpm shaking for 12 h. Cells were collected by centrifugation at 7000 × g for 10 min. The cell pellet was resuspended in 3 ml of TBS (50 mm Tris, pH 7.5, 150 mm NaCl) + 1% Triton X-100 + 1 mm dithiothreitol (DTT) for every 1-g cell pellet. The cells were then disrupted by sonication (5 × 10-s pulse), and the cellular debris was removed by centrifugation at 10,000 × g for 11 min. Lysate was further centrifuged for 1 h at 40,000 × g. Finally for further clarification, the lysate was passed through a 0.2-μm filter to remove any other aggregates or insoluble particles.
The cell lysate was then passed through a 5-ml GSTrap column (GE Healthcare) and washed with at least 5 bed volumes of TBS. The fusion proteins were eluted from the column with 20 ml of GEB (50 mm Tris, pH 8.0, 10 mm glutathione). If necessary, the GST tag was removed by digestion with 100 units of PreScission protease for 12 h at 4 °C. This solution was again passed over the GSTrap column to remove the GST and protease. The pure Che protein flow-through was collected and stored in TKMDmod (50 mm Tris, pH 8.0, 50 mm KCl, 5 mm MgCl2) buffer at −80 °C.
Isothermal Titration Calorimetry (ITC)
All ITC measurements were carried out using a VP-ITC titration calorimeter (MicroCal) at 25 °C, and data were extracted and processed using the Origin software package. During the titration reaction, the 1.4-ml reaction cell was constantly stirred at 300 rpm. A total of 28 injections, all 10 μl in volume, were performed. Prior to analysis, the pH of all solutions used was checked and adjusted so that every solution had an identical pH the day of the reaction (the pH of the solutions was between 7.2 and 7.3). All solutions were also degassed for 15 min prior to the experiment.
GST Pulldowns
The pulldowns were performed as described previously (17). Briefly, 100 μl of 30 μm GST-McpCn was added to 50 μl of prewashed (with 400 μl of TBS-X (TBS + 1% Triton X-100)) glutathione beads (GE Healthcare) in a Handee spin cup column (Pierce) and incubated for 10 min. The beads were washed with 800 μl of TBS-X, and 100 μl of 150 μm of purified binding protein was added and incubated for 10 min. The beads were again washed with 800 μl of TBS-X, and the protein was eluted with 75 μl of GEB. 25 μl of 4× SDS solubilizer was added, and the samples were run on SDS-polyacrylamide gels and visualized with Coomassie stain. A GST control was also run, as was a nonspecific GST-tagged protein control, and no binding of the binding protein was observed (data not shown).
RESULTS
McpC Alone Can Support Chemotaxis to 17 Amino Acids
B. subtilis can perform chemotaxis toward all 20 l-amino acids normally found in proteins. Previous work suggested that a single receptor, McpC, supports taxis to 19 of these amino acids; an mcpC mutant exhibited reduced chemotaxis to all amino acids except asparagine (Table 1, column 2). One possible interpretation of these results is that McpC alone is capable of sensing 19 amino acids. The other would be that McpC may somehow enable one of the other nine chemoreceptors present in B. subtilis to sense these amino acids. We first sought to examine whether McpC alone is capable of sensing these amino acids by expressing it in a strain lacking the other nine chemoreceptors (Δ10 mcp amyE::mcpC, strain OI4007) (14). We then employed the capillary assay to test for chemotaxis toward all 20 amino acids using this strain.
TABLE 1.
ITC results and capillary assays of all 20 amino acids
| Amino acid | mcpC mutanta | Capillary assay accumulationb | KDc | ITC KD | McpC E115A mutant | McpC K195A mutant |
|---|---|---|---|---|---|---|
| % reduced | μm | μm | % reduced | % reduced | ||
| Cys | 100 | 28,175 ± 5,408 | 10 | NAd | >99 | 20 |
| Pro | 99 | 29,119 ± 4,845 | 1 | 14 | >99 | 8 |
| Thr | 99 | 30,120 ± 6,375 | 5.6 | 21 | 97 | 11 |
| Gly | 99 | 13,081 ± 6,440 | 320 | 699 | 94 | 51 |
| Ser | 99 | 9,395 ± 816 | 32 | 90 | 95 | 9 |
| Lysh | 99 | 15,500 ± 5,348 | 1,000 | NBe | 50 | 88 |
| Val | 98 | 15,653 ± 6,372 | 32 | 154 | 96 | 60 |
| Argh | 98 | 12,887 ± 1,574 | 100 | NBe | <1 | 95 |
| Ala | 92 | 12,225 ± 3,766 | 0.32 | 18 | 97 | 4 |
| Tyr | 92 | 2,378 ± 701 | 320 | 360 | 95 | <1 |
| Glu | 89 | 3,892 ± 2,515 | 32,000 | NBe | 98 | 54 |
| Ile | 87 | 3,355 ± 219 | 100 | 1,000 | 97 | 67 |
| Meth | 79 | 8,650 ± 2,036 | 100 | NBe | 62 | 91 |
| Trp | 78 | 254 ± 42 | 3,200 | 3,700 | <1 | 86 |
| Phe | 72 | 8,713 ± 1,061 | 320 | 492 | 95 | 77 |
| Leu | 70 | 8,713 ± 1,061 | 32 | 72 | 97 | 80 |
| Asp | 35 | 3,355 ± 1,061 | 10,000 | NBe | 97 | 51 |
| Glnh | 34 | 1,475 ± 369 | 32 | NBe | 55 | 95 |
| His | 27 | 26 ± 12 | 3,200 | 320f | NAg | NAg |
| Asn | 0 | 0 ± 35 | 56 | NBe | NAg | NAg |
a As previously determined experimentally (3).
b Strain OI4007 (Δ10 mcp amyE::mcpC). All amino acids were tested at 0.01 m, except for Glu and Asp, which were tested at 0.1 m.
c As previously determined experimentally (26).
d Could not be tested.
e No binding under the conditions tested.
f Exothermic binding.
g Accumulation too low.
h Chemotaxis mediated by an amino acid binding protein.
The results from the capillary assay show that McpC alone is able to support chemotaxis to 17 amino acids (Table 1, column 3). Accumulation to tryptophan and histidine was very low, suggesting chemotaxis to these two attractants might require an additional chemoreceptor. In the case of histidine, McpB is the likely chemoreceptor involved (3). However, in the case of tryptophan, the additional chemoreceptor is not known.
The Sensing Domain of McpC Binds 11 Amino Acids Directly
We next sought to test whether these 17 amino acids are sensed directly by McpC. The amino-terminal sensing domain of McpC (McpCn, residues 33–276) was purified, and then ITC was used to determine which amino acids it bound in vitro. Previous work using ITC showed that the sensing domain of McpB binds aparagine in vitro with affinities comparable with those inferred from in vivo experiments (7).
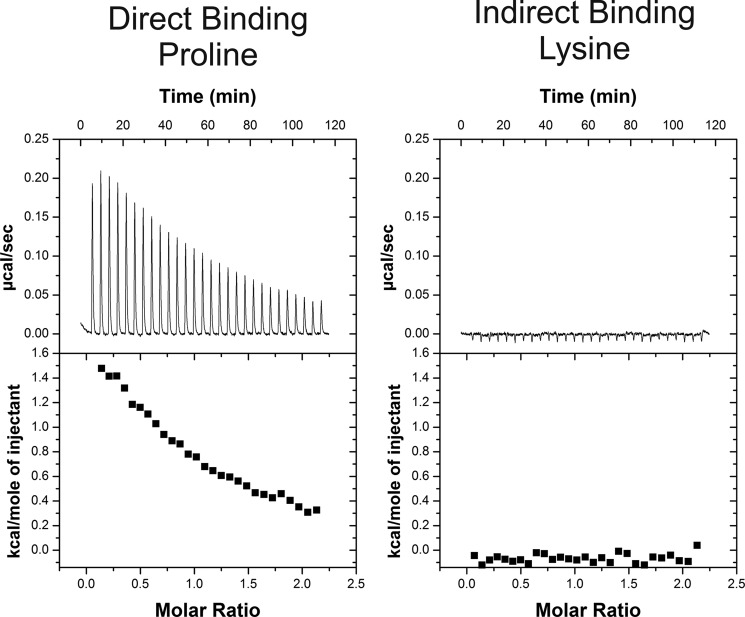
The ITC results show that the sensing domain of McpC directly binds 12 amino acids in vitro, and the binding is endothermic for 11 of them (Fig. 1 and Table 1, column 5). The in vitro affinities derived from the binding curves generally agree with the previously reported in vivo affinities determined using a specialized capillary assay (Table 1, column 4). The exceptions are proline, alanine, and isoleucine, where the in vitro affinities are at least 10-fold lower than the in vivo one. McpC clearly binds proline and alanine because in vitro binding was strong (KD = 14 and 18 μm, respectively). In the case of isoleucine, in vitro binding was much weaker (KD = 1 mm), although still sufficiently strong to suggest that McpC directly binds it as well. The other exception was histidine, which exhibited exothermic binding, unlike the endothermic change observed with the other 11 amino acids. McpC alone does not support chemotaxis to histidine. In addition, it appears to be a shared ligand with McpB so we did not explore it further.
FIGURE 1.
ITC binding isotherms for the purified amino-terminal sensing domain of McpC reveals direct binding to many amino acids. Proline binds directly to McpC, whereas lysine does not bind. The ITC results for all 20 amino acids are summarized in Table 1.
Of the 20 amino acids tested, 7 did not bind the sensing domain of McpC, and 1, cysteine, could not be tested because DTT, necessary to prevent disulfide bond formation, interferes with ITC analysis. Aside from asparagine, the amino acids that did not bind in vitro are lysine, arginine, glutamate, methionine, aspartate, and glutamine. Both glutamate and aspartate have weak in vivo affinity for the chemoreceptors (Table 1, column 4); likely, in vitro binding is too weak to be detected by ITC. The simplest explanation for the remaining four (lysine, arginine, methionine, and glutamine) is that McpC senses them by an indirect mechanism, most likely one involving ancillary proteins.
Identification of Mutants That Separate Direct from Indirect Sensing
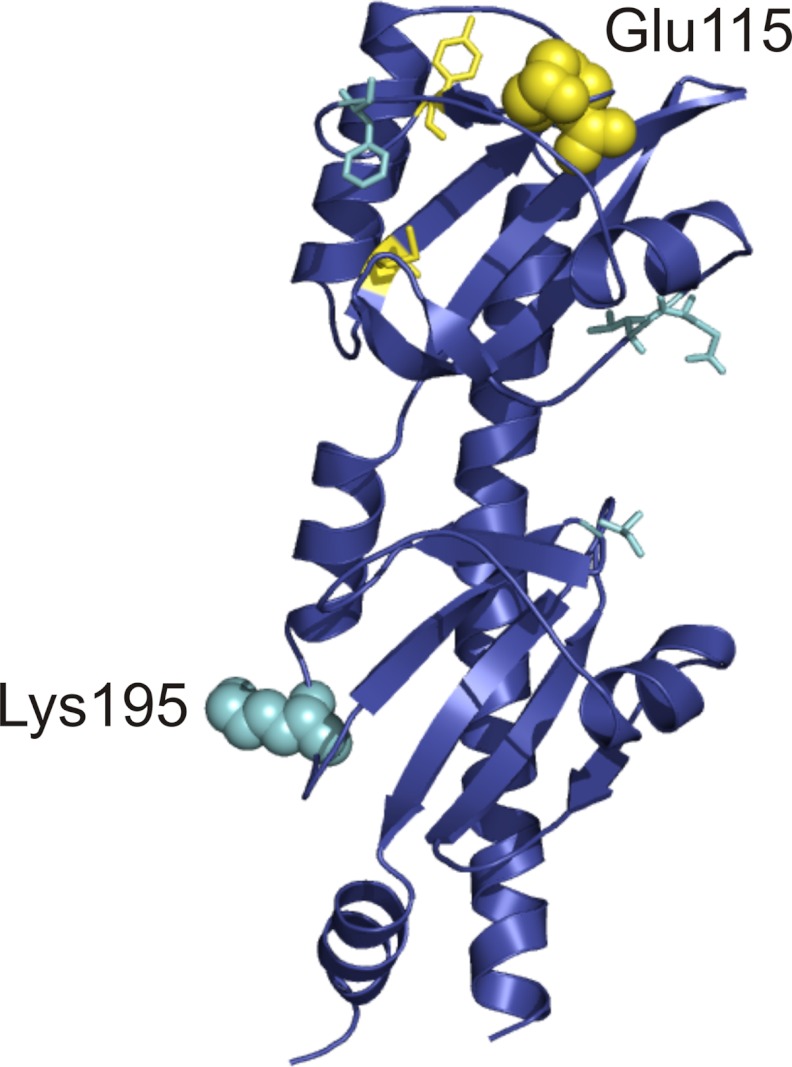
The sensing domains of B. subtilis McpB and V. cholerae McpN consist of two dual PAS domains; the ligand binds in the upper PAS domain. Homology modeling predicts that the sensing domain of McpC also adopts the same dual PAS domain structure (Fig. 2). Given the structural similarity among the sensing domains of these three chemoreceptors, we expect that the upper PAS domain of McpC is also the site of direct binding. Using the homology model, we identified residues on McpC that might play a role in direct ligand binding in a putative upper PAS domain binding pocket (Fig. 2). Indirect binding, on the other hand, likely involves a second binding protein. Based on the structure of LuxQ and LuxP, we would expect that this binding protein interacts with both PAS domains of McpC. Previous work has shown that certain residues on LuxQ are vital for its interaction with LuxP (12). We were able to identify similar residues on McpC that might play a role in its interaction with a putative binding protein (Fig. 2). Initially, we made alanine substitutions in these putative binding pocket and binding protein interface residues in a strain expressing the mutated McpC as the sole chemoreceptor.
FIGURE 2.
Identification of residues involved in ligand sensing on the extracellular amino-terminal domain of the McpC chemoreceptor. Putative residues that were tested for direct binding (yellow) and indirect binding (cyan) are highlighted on a structural model of the McpC sensing domain. The two residues chosen for further study, Glu-115 and Lys-195, are shown in a space-filling representation for clarity.
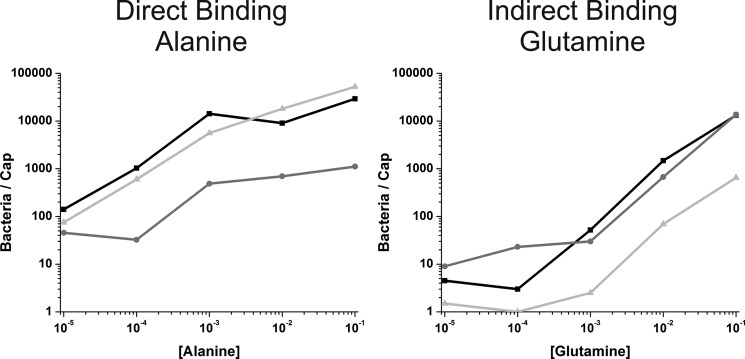
By screening these numerous mutants using capillary assays (data not shown), we were able to identify one binding pocket mutant and one binding protein interface mutant. The first point mutant, E115A (Δ10 mcp amyE::mcpC E115A), is located in the binding pocket of the upper PAS domain. It exhibited reduced chemotaxis to all amino acids tested as determined using the capillary assay. However, the reduction was less for lysine, arginine, methionine, and glutamine (Table 1, column 6; Fig. 3). The second point mutant, K195A (Δ10 mcp amyE::mcpC K195A), is located in the lower PAS domain along the putative binding protein interface and is distal to the upper PAS domain binding pocket. This mutant also exhibited varying levels of reduced chemotaxis to all amino acids tested (Table 1, column 7; Fig. 3). However, the reduction was greater for lysine, arginine, methionine, and glutamine. When the reductions for both mutants are plotted on the same figure, these four amino acids clearly segregate from the others for the two mutants (supplemental Fig. S1).
FIGURE 3.
Capillary assays toward alanine and glutamine show bacterial accumulation over a range of attractant concentrations. The wild-type McpC chemoreceptor is shown in black (Δ10 mcp amyE::mcpC), the E115A mutant (Δ10 mcp amyE::mcpC E115A) is shown in dark gray, and the K195A mutant (Δ10 mcp amyE::mcpC K195A) is shown in light gray. Accumulation to alanine is indicative of direct ligand binding, whereas the results for glutamine are an example of indirect ligand binding. The results of capillary assays to all 20 amino acids are summarized in Table 1.
These results are entirely consistent with the ITC results if we discount glutamate and aspartate due to their high apparent KD values. Moreover, when we locate these two residues on the structural model for McpC sensing domain, they are entirely consistent with our hypothesis derived from structural analysis. Residue 115 is located in the upper PAS domain in the putative ligand binding pocket, and residue 195 is located in the lower PAS domain on the external face of the protein (Fig. 2). Taken together, these results suggest that McpC directly senses the binding of at least 11 amino acids and indirectly senses the binding of 4 amino acids.
Identification of the Ancillary Proteins Involved in Indirect Sensing
What is the indirect sensing mechanism for McpC? Based on results from E. coli chemotaxis and the LuxQ/LuxP interaction, ancillary proteins are expected to directly bind these four amino acids and interact with McpC. The B. subtilis genome contains >40 binding lipoproteins associated with ATP-binding cassette (ABC) transporters that are potential candidates. Eight of these proteins are known or have been hypothesized to be involved in amino acid transport (18, 19). Of these eight, four appear to be involved in cysteine uptake: TcyA, TcyJ, TcyK, and YxeM (20, 21). The other four are: ArtP (arginine), GlnH (glutamine), MetQ (methionine), and YckB (unknown) (22–24). These four were chosen for further study. Three of them are responsible for the same amino acids that do not directly bind McpC; and the fourth, YckB, is annotated as a binding lipoprotein associated with an amino acid transporter (18). These proteins all contain a hydrophobic amino-terminal region, presumably to anchor these proteins to the membrane.
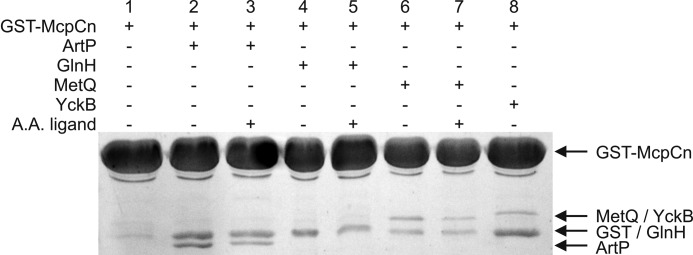
To test whether these four binding proteins interact with the sensing domain of McpC, we purified them without the anchor region and then used the glutathione S-transferase (GST) pulldown assay to test for binding. Briefly, the McpC sensing domain tagged to GST was bound to glutathione beads and incubated with the binding protein, both in the presence or absence of their cognate amino acid. For instance, ArtP was tested in the presence or absence of 1 mm arginine. The beads were then washed twice and eluted using glutathione.
These results showed that McpC binds to ArtP, MetQ, and YckB (Fig. 4). The results for GlnH are ambiguous as GST, the likely product of degradation due to a labile linker, runs at the same molecular mass. Additionally, we found that the presence of amino acid did not appear to have any significant effect on this binding. The ability of the McpC sensing domain to bind three of the four binding proteins tested suggests that an indirect binding mechanism is indeed plausible for chemotaxis toward lysine, arginine, glutamine, and perhaps methionine as well. As a control, we also tested whether the McpC sensing domain binds CheC and BSA and found that it did not (supplemental Fig. S2), indicating that the interaction with the binding lipoproteins is specific.
FIGURE 4.
GST pulldown assays show an interaction between the amino-terminal sensing domain of McpC and four different ABC transport-binding proteins. GST-McpCn was immobilized on glutathione beads, incubated with a binding protein (in the absence of presence of its cognate amino acid ligand), washed, and eluted with glutathione.
We also tested whether the K195A mutant sensing domain was able to bind ArtP, MetQ, and YckB and found that it still did. These results are not surprising as multiple residues on McpC are predicted to interact with the binding lipoproteins. However, the affinity appears to be somewhat less for the K195A mutant than for the wild-type sensing domain (supplemental Fig. S3). Likely, the K195A mutation affects the allosteric coupling between receptor and binding lipoproteins, including by reducing the affinity between the two.
Finally, we tested whether these proteins bind amino acids using ITC. We found that ArtP bound arginine tightly (KD = 43 nm) and lysine with a much lower affinity (KD = 657 μm). GlnH bound glutamine (KD = 120 μm) and MetQ bound methionine (KD = 3.8 μm). We additionally found that YckB bound tryptophan very weakly (KD = 2020 μm); however, McpC alone does not support chemotaxis to this amino acid. Taken together, these results show that ABC transport-binding lipoproteins bind both the McpC chemoreceptor and an amino acid ligand, demonstrating that indirect sensing by McpC likely involves ABC transport-binding lipoproteins.
DISCUSSION
McpC is a nearly universal amino acid chemoreceptor, alone capable of supporting chemotaxis to 17 amino acids in B. subtilis. Eleven of these amino acids were found to directly bind the sensing domain of McpC as determined using in vitro ITC analysis. As the in vitro affinities were similar to those inferred from in vivo analysis, we conclude that all are directly sensed by McpC. Mutational analysis further revealed that four amino acids (arginine, glutamine, lysine, and methionine) are likely sensed by an indirect mechanism involving ancillary proteins. We were able to identify and characterize three lipoproteins (ArtP, GlnH, and MetQ) associated with amino acid transporters that may function as ancillary proteins involved in the indirect sensing of arginine, glutamine, lysine, and methionine. Although our results show that these proteins provide a sufficient mechanism for indirect sensing, we were unable to demonstrate that they are necessary as null mutations to these binding lipoproteins appear to affect chemotaxis toward all amino acids (data not shown).
How is the sensing domain of McpC capable of directly binding 11 different amino acids? Clearly, high resolution structures of the sensing domain of McpC in the apo and bound form for all 11 amino acids are needed to definitively elucidate the binding mechanism. However, based on previous work, we can speculate on possible mechanisms. In particular, previous work has demonstrated that ligand binds in the upper PAS domain in a dual PAS domain sensor (8). Moreover, the mutational analysis from this study supports this conclusion. If true, then the binding pocket of the McpC sensing domain is fairly flexible and likely requires multiple contacting residues. In the crystal structure of the sensing domain of the McpN chemoreceptor from V. cholerae bound to alanine, at least eight residues contact this ligand (Protein Data Bank code 3C8C). Assuming that McpC binds ligand in a similar fashion, multiple residues would be available to contact these 11 amino acids, which suggests that multiple ligands can be accommodated in the same binding pocket. Furthermore, the 11 amino acids identified in this study are all relatively small in size and uncharged, so all could feasibly physically fit in the same binding pocket.
Our results suggest that four amino acids (lysine, arginine, methionine, and glutamine) are sensed by McpC using an indirect binding mechanism involving ABC transport-binding lipoproteins. Lysine and arginine are both charged ligands, which might make them poor ligands for the McpC binding pocket. Glutamate and aspartate are also charged, and perhaps the weak affinity (Table 1) also reflects poor binding in the McpC binding pocket. The ArtP-binding lipoprotein was able to bind both lysine and arginine and is likely responsible for the ability of McpC to sense both of these ligands. As for methionine and glutamine, an indirect sensing mechanism may enable the cells to couple chemotaxis to transport as recently proposed by Neumann and co-workers (25). Methionine is a sulfur-containing amino acid, and perhaps sulfur is limiting for B. subtilis. Cysteine, the other sulfur-containing amino acid, is sensed directly by McpC. However, B. subtilis possesses four binding lipoproteins specific for cysteine, suggesting that it may be more important for sulfur scavenging. Alternatively, methionine may be expensive for the cell to synthesize, as it requires both cobalamin and a folic acid derivative. Glutamine may be an important source of nitrogen for the B. subtilis. This amino acid is also a ligand for the McpB receptor and actually binds McpB in vitro (data not shown), so it appears that glutamine can be sensed indirectly by McpC and directly by McpB.
The protein binding interface of the dual PAS domain may be conserved among diverse sensor histidine kinases and chemoreceptors. Our results show that residues along the protein binding interface of LuxQ are conserved in McpC, in particular the lysine at residue 195. This suggests that these two sensors employ similar mechanisms for binding accessory proteins. Moreover, the signaling mechanisms are similar. We found that the amino acid ligand does not alter the interaction between the McpC sensing domain and binding lipoprotein, implying that the two proteins interact without ligand present. The same is seen with LuxQ and LuxP, where two are bound in the absence of ligand; activation is induced when AI-2 binds LuxP (12). Presumably the same occurs with McpC and the binding lipoproteins. Finally, we were not able to examine whether the presence of bound binding protein might affect the affinity of McpC for ligands binding in the upper PAS domain, although this is certainly an intriguing area for future work.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM054365 (to G. W. O and C. V. R.).

This article contains supplemental Figs. S1–S3.
- PAS
- Per-Arnt-Sim
- ABC
- ATP-binding cassette
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Ordal G. W., Gibson K. J. (1977) Chemotaxis toward amino acids by Bacillus subtilis. J. Bacteriol. 129, 151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanlon D. W., Ordal G. W. (1994) Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J. Biol. Chem. 269, 14038–14046 [PubMed] [Google Scholar]
- 3. Müller J., Schiel S., Ordal G. W., Saxild H. H. (1997) Functional and genetic characterization of mcpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology 143, 3231–3240 [DOI] [PubMed] [Google Scholar]
- 4. Karatan E., Saulmon M. M., Bunn M. W., Ordal G. W. (2001) Phosphorylation of the response regulator CheV is required for adaptation to attractants during Bacillus subtilis chemotaxis. J. Biol. Chem. 276, 43618–43626 [DOI] [PubMed] [Google Scholar]
- 5. Garrity L. F., Ordal G. W. (1997) Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology 143, 2945–2951 [DOI] [PubMed] [Google Scholar]
- 6. Szurmant H., Bunn M. W., Cannistraro V. J., Ordal G. W. (2003) Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J. Biol. Chem. 278, 48611–48616 [DOI] [PubMed] [Google Scholar]
- 7. Glekas G. D., Foster R. M., Cates J. R., Estrella J. A., Wawrzyniak M. J., Rao C. V., Ordal G. W. (2010) A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J. Biol. Chem. 285, 1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z., Hendrickson W. A. (2010) Structural characterization of the predominant family of histidine kinase sensor domains. J. Mol. Biol. 400, 335–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hazelbauer G. L. (1975) Maltose chemoreceptor of Escherichia coli. J. Bacteriol. 122, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kossmann M., Wolff C., Manson M. D. (1988) Maltose chemoreceptor of Escherichia coli: interaction of maltose-binding protein and the Tar signal transducer. J. Bacteriol. 170, 4516–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y., Gardina P. J., Kuebler A. S., Kang H. S., Christopher J. A., Manson M. D. (1999) Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl. Acad. Sci. U.S.A. 96, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neiditch M. B., Federle M. J., Pompeani A. J., Kelly R. C., Swem D. L., Jeffrey P. D., Bassler B. L., Hughson F. M. (2006) Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ullah A. H., Ordal G. W. (1981) In vivo and in vitro chemotactic methylation in Bacillus subtilis. J. Bacteriol. 145, 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kristich C. J., Ordal G. W. (2002) Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J. Biol. Chem. 277, 25356–25362 [DOI] [PubMed] [Google Scholar]
- 15. Adler J. (1973) A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74, 77–91 [DOI] [PubMed] [Google Scholar]
- 16. Zimmer M. A., Szurmant H., Saulmon M. M., Collins M. A., Bant J. S., Ordal G. W. (2002) The role of heterologous receptors in McpB-mediated signalling in Bacillus subtilis chemotaxis. Mol. Microbiol. 45, 555–568 [DOI] [PubMed] [Google Scholar]
- 17. Chao X., Muff T. J., Park S. Y., Zhang S., Pollard A. M., Ordal G. W., Bilwes A. M., Crane B. R. (2006) A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 124, 561–571 [DOI] [PubMed] [Google Scholar]
- 18. Quentin Y., Fichant G., Denizot F. (1999) Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287, 467–484 [DOI] [PubMed] [Google Scholar]
- 19. Bunai K., Ariga M., Inoue T., Nozaki M., Ogane S., Kakeshita H., Nemoto T., Nakanishi H., Yamane K. (2004) Profiling and comprehensive expression analysis of ABC transporter solute-binding proteins of Bacillus subtilis membrane based on a proteomic approach. Electrophoresis 25, 141–155 [DOI] [PubMed] [Google Scholar]
- 20. Auger S., Danchin A., Martin-Verstraete I. (2002) Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184, 5179–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burguière P., Auger S., Hullo M. F., Danchin A., Martin-Verstraete I. (2004) Three different systems participate in l-cystine uptake in Bacillus subtilis. J. Bacteriol. 186, 4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sekowska A., Robin S., Daudin J. J., Hénaut A., Danchin A. (2001) Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2, RESEARCH0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu L., Welker N. E. (1991) Cloning and characterization of a glutamine transport operon of Bacillus stearothermophilus NUB36: effect of temperature on regulation of transcription. J. Bacteriol. 173, 4877–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hullo M. F., Auger S., Dassa E., Danchin A., Martin-Verstraete I. (2004) The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, d- and l-methionine. Res. Microbiol. 155, 80–86 [DOI] [PubMed] [Google Scholar]
- 25. Neumann S., Hansen C. H., Wingreen N. S., Sourjik V. (2010) Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J. 29, 3484–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ordal G. W., Villani D. P., Gibson K. J. (1977) Amino acid chemoreceptors of Bacillus subtilis. J. Bacteriol. 129, 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.