Background: Cytoplasmic male sterility (CMS) is associated with mitochondrial defects.
Results: Reduction of assembled mitochondrial protein complexes and altered jasmonic acid pathways were observed in the sterile line.
Conclusion: CMS may be related to a mitochondrial complex assembly defect and abnormal jasmonic acid pathway.
Significance: Our discoveries suggest a novel mechanism for CMS.
Keywords: Bioenergetics/Electron Transfer Complex, F1F0-ATPase, Hormones, Mitochondria, Plant, Proteomics, Cytoplasmic Male Sterility, Jasmonic Acid Pathway, Rice
Abstract
It has been suggested that the mitochondrial chimeric gene orfH79 is the cause for abortion of microspores in Honglian cytoplasmic male sterile rice, yet little is known regarding its mechanism of action. In this study, we used a mass spectrometry-based quantitative proteomics strategy to compare the mitochondrial proteome between the sterile line Yuetai A and its fertile near-isogenic line Yuetai B. We discovered a reduced quantity of specific proteins in mitochondrial complexes in Yuetai A compared with Yuetai B, indicating a defect in mitochondrial complex assembly in the sterile line. Western blotting showed that ORFH79 protein and ATP1 protein, an F1 sector component of complex V, are both associated with large protein complexes of similar size. Respiratory complex activity assays and transmission electron microscopy revealed functional and morphological defects in the mitochondria of Yuetai A when compared with Yuetai B. In addition, we identified one sex determination TASSELSEED2-like protein increased in Yuetai A, leading to the discovery of an aberrant variation of the jasmonic acid pathway during the development of microspores.
Introduction
The world's food supply depends on a few high yield crops, such as rice, corn, and wheat. Hybrid seed generation is vital for high yield crop production. Cytoplasmic male sterility (CMS)4 is a maternally inherited phenomenon observed in the higher plant kingdom. CMS leads to pollen abortion but does not affect female fertility and vegetative growth, which form the key technical base for hybrid seed production (1, 2). A number of mitochondrial genes associated with CMS have been identified in the past, but few mechanistic details are known for this phenomenon. Most CMS-associated mitochondrial genes are chimeric genes composed of a fragment of a normal mitochondrial gene, encoding small and low abundance mitochondrial membrane proteins (3).
Based on limited biochemical studies of the molecular mechanisms of CMS, two theories have been proposed (4). The first is the “gain-of-function” theory, which is supported by the finding that T-URF13, an inner mitochondrial membrane protein, may form a pore and interfere with membrane activity in the cytoplasm of maize (5, 6). In addition, ORF138, which forms a complex of over 750 kDa, has been proposed to act as a pore in the inner mitochondrial membrane in a sterile line of rapeseed (7). The second theory, the “loss-of-function” theory, is supported by the characterization of ORF522, which shares a similar N-terminal amino acid sequence with ORFB but competes with ORFB in binding to the ATP synthase (complex V) subunit in sunflowers to reduce the stability and efficiency of this complex (8).
There are three widely distributed CMS rice varieties in China, including “Honglian” (HL), the “Wild Abortive,” and “Chinsurah Boro II/Taichung native 65.” These CMS lines have differences in inheritance, morphology of abortive pollens, and restoration-maintenance relationships (9). HL CMS rice was developed by backcrossing red-awned wild rice (Oryza rufipogon) with the indica variety Lian Tang-Zao in Hainan China in the 1970s. The sterility of Yuetai A (YtA) is maintained by backcrossing with its isogenic fertile line Yuetai B (YtB). Restriction fragment length polymorphism analysis and BAC library screening of the mitochondrial DNA from YtA and YtB revealed the chimeric gene orfH79 located downstream of the atp6 gene in the sterile line (10–12). Transgenic analysis showed that the ORFH79 protein is responsible for CMS in the sterile line (13). However, it remains unclear how the aberrant ORFH79 affects organization and function in mitochondria.
Plant mitochondria not only play a pivotal role in energy production but are also involved in multiple biological processes such as amino acid metabolism, biosynthesis of vitamins and lipids, and programmed cell death (14). Therefore, it is difficult to attribute a precise function to any specific CMS protein in mitochondria, especially when the CMS protein shares limited homology to any known protein. In searching for clues to the mechanism of CMS, past studies have used proteomics to obtain a global view of the composition and spatial distribution of mitochondrial proteins (8, 15, 16). In this study, quantitative proteomics was used to compare the mitochondrial proteome between YtA and its maintainer line YtB. Using blue native-PAGE (BN-PAGE) to separate protein complexes, we also quantitatively compared the protein composition of mitochondrial protein complexes. Compared with YtB, reduced quantities of proteins were detected in mitochondrial complexes in YtA, suggesting a defect in mitochondrial complex assembly. Western blotting indicated that ORFH79 protein location is associated with several large protein complexes, ranging from ∼400 kDa to ∼1.2 MDa. ATP synthase α subunit (ATP1), an F1 sector component of complex V, also belongs to protein complexes of similar size as ORFH79. Respiratory complex activity assays and transmission electron microscopy revealed functional and structural defects in the mitochondria of YtA when compared with YtB. Furthermore, our quantitative proteomic analyses revealed that one sex determination TASSELSEED2-like protein was up-regulated in YtA, leading to the discovery that an aberrant jasmonic acid pathway existed in CMS rice.
EXPERIMENTAL PROCEDURES
Plant Materials
Seeds of Honglian CMS line rice YtA and its corresponding fertile maintainer line YtB were washed in 1% (v/v) bleach for 10 min and rinsed in distilled water. The seedlings were grown in vermiculite trays at 30 °C, with the etiolated seedlings in the dark, and the green seedlings were grown in a 14-h light/8-h dark regime. The seedlings were watered daily and harvested at 10 days for experiments. YtA and YtB were grown in an experimental field of Wuhan University in the summer of 2009. Tassels at specific developmental stages were collected after plants had grown for ∼3 months. Developmental stage of microspores was determined according to the method of Wan et al. (17).
Rice Mitochondrial Isolation
Isolation of rice mitochondria was performed by differential centrifugation followed by continuous Percoll gradients as described by Heazlewood et al. (18). The amount of mitochondrial protein was determined using the modified Lowry protein assay kit (Pierce).
Electrophoretic Techniques
BN-PAGE was performed according to a published method with minor modifications (19). Mitochondrial samples were centrifuged for 10 min at 20,000 × g, and sedimented organelles were resuspended in 100 μl of dodecyl maltoside (DDM) solubilization buffer (750 mm aminocaproic acid, 50 mm BisTris, pH 7.0, 0.5 mm EDTA, 1 mm PMSF, and DDM (2 g per g of protein; Sigma)). Gels consisted of a separating gel (4.5–16 or 4.5–10% (w/v) acrylamide) and a stacking gel (4% (w/v) acrylamide). To separate proteins in the second dimension, BN lanes were cut from gels, equilibrated in standard 1% SDS and 1% 2-mercaptoethanol for 1 h, and laid horizontally on 10% (w/v) acrylamide Tricine/SDS-PAGE separating gels. The gels were stained with Coomassie Blue G-250 after electrophoresis.
Protein Digestion and Dimethyl Labeling
For mitochondrial total protein analysis, a fixed amount of enriched mitochondria (1 mg from YtA or YtB, digested separately) were lysed with buffer containing 25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitor mixtures (Roche Applied Science). Three volumes of 50% acetone, 50% ethanol, 0.1% acetic acid were added to the lysates followed with 2 h of incubation on ice. The protein pellets were resuspended in 8 m urea, 0.2 m Tris, pH 8.0, 4 mm CaCl2 and sonicated for several seconds to fully resuspend the proteins.
Samples were reduced with 10 mm DTT for 1 h and alkylated with 40 mm iodoacetamide in the dark for 30 min. After diluting the urea concentration to 2 m, trypsin (Promega) (1:50; trypsin/mitochondrial proteins) was added, and the digestion was carried out at 37 °C overnight. After digestion, urea and salts were removed by C18 Sep-Pak columns (Waters).
For the stable isotope dimethyl labeling, the digested peptide samples from YtA and YtB were reconstituted separately with 200 μl of CH3COONa, pH 5.9. Eight microliters of CH2O (light labeled) and CD2O (heavy labeled) were added to YtA and YtB samples, respectively. Eight microliters of 0.6 m NaBH3CN was added to both samples and incubated at room temperature for 1 h. To quench the reaction, 32 μl of 1% (v/v) ammonia solution and 16 μl of 5% formic acid were added to both samples on ice. Light labeled YtA and heavy labeled YtB samples were mixed at the ratio of 1:1 and desalted before strong cation exchange (SCX) fractionation.
For the SCX fractionation, the mixed peptides were resuspended in buffer A containing 5 mm KH2PO4, 20% acetonitrile, pH 2.7. SCX was performed on a polysulfoethyl column (2.1 × 50 mm, 5 μm × 200 Å) using a KCl gradient from 0 to 0.5 m to fractionate the peptides. Nine fractions were collected and desalted with C18 ZipTip (Millipore) before MS analysis.
For protein in-gel digestion, BN gel lanes containing complexes above 100 kDa were divided into six fractions. Each fraction was cut into ∼1-mm3 pieces. The gel pieces were washed with 50% acetonitrile, 100 mm NH4HCO3, pH 8.0, three times, reduced with 10 mm DTT, alkylated with 55 mm iodoacetamide in the dark, and digested in-gel with trypsin. The tryptic peptides from YtA and YtB were dimethyl-labeled as mentioned above, and the corresponding fractions were mixed together. Finally, the samples were desalted by C18 Sep-Pak columns (Waters).
LC-MS/MS and Data Analysis
A QSTAR ELITE mass spectrometer (Applied Biosystems) coupled with a nanoflow HPLC system (TempoTM, Applied Biosystems) was used for relative quantitation of mitochondrial proteins from YtA and YtB rice. The LC-MS method was the same as used by Chen et al. (20). Raw data from QSTAR ELITE were analyzed with MASCOT Daemon software (Version 2.2.2, Matrix Science) using a local MASCOT engine. The data were searched against a rice database (Release 6.1, The Institute for Genomic Research, Rice Genome Annotation Project), including mitochondrial and plastid proteins plus ORFH79 (21) with added dimethyl masses. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. Peptide mass tolerance was set as 200 ppm and 0.4 Da. The peptide charge was set to 2+ and 3+, allowing for up to two missed cleavages, and the significance threshold was set at p < 0.05.
After the Mascot search, the rov data obtained from the database search were opened by Mascot Distiller (Version 2.3.2.0) for quantitation. For the quantitation analysis, we set fraction, correlation, and standard error at 0.5, 0.9, and 0.2, respectively. The false-positive rates of peptide spectrum matches were determined by data search against a decoy database. The peptide ratios were calculated as weighted average ratios (ion intensity versus ratio) if several spectra for the same peptide were available. The median of all quantitation data from all peptides was used to normalize the peptide ratios. Protein ratios were also calculated as the geometric mean of the peptide ratios for a protein. Student's t test were performed, and p values were calculated using software SPSS 13.0.
Antibodies
For ORFH79 antibody production, the sequence of orfH79 lacking the 5′-terminal 66-bp region homologous to coxI was amplified by PCR using primers 5′-CCGGAATTCTTCGGTGTCGTTGTAGGA-3′ (the underlined letters show the EcoRI site) and 5′-ACGCGTCGACACCACTGTCCTGTCTTTCTT-3′ (the underlined letters show the SalI site). This PCR fragment was digested with EcoRI and SalI and subcloned into the EcoRI and SalI sites of pET-28a (Merck) for bacterial expression. The recombinant protein was purified by affinity chromatography and then injected into rabbits. The anti-ORFH79 serum was generated by NewEastTM Biosciences.
ATP1 antibodies were a gift from Dr. T. Elthon, and NAD9 antibodies were a gift from Prof. J. M. Grienenberge. PHB2, HSP60, and CYTC antibodies were from Proteintech Group Inc. ORFB antibodies were from Beijing Protein Institute.
Immunoprecipitation and Immunoblotting
Mitochondrial membrane proteins (1 mg) were resuspended at a concentration of 2.5 mg of protein/ml in lysis buffer for BN-PAGE and solubilized with DDM at a protein/detergent ratio of 1:2 (g/g). After centrifugation, the supernatant was used for immunoprecipitation with the co-immunoprecipitation kit (Pierce Lot. 26149) according to the manual.
After BN-PAGE, protein complexes were transferred onto PVDF membranes with electrode buffer (50 mm Tricine, 7.5 mm imidazole, pH 7.0) using a Trans-Blot Semi-Dry Cell (Bio-Rad) for 1 h at 10 V. The Tricine/SDS-PAGE electrode buffer was changed to cathode buffer (300 mm 6-aminocaproic acid, 30 mm Tris, pH 9.2) and anode buffer (300 mm Tris, 100 mm Tricine, pH 8.4).
Respiratory Complex Activity Assays
In-gel activity assay was performed according to the protocol of Sabar et al. (22). After electrophoresis, BN gels were pre-equilibrated in reaction buffer (complex I: 0.2 mm NADH, 0.1 m Tris-HCl, pH 7.4, 10 min; complex IV: 1 mg/ml cytochrome c, 50 mm KH2PO4, pH 7.4, 10 min; complex V: 5 mm ATP, 50 mm glycine, 5 mm MgCl2, 10 mm Tris-HCl, pH 8.4, 1 h) and transferred to fresh reaction buffer with color developing reagents (complex I: 0.2% (w/v) nitro blue tetrazolium; complex IV: 0.1% (w/v) diaminobenzidine; complex V: 0.1% (w/v) Pb(NO3)2). In-solution activity assays were performed using a respiratory complexes activity kit (Genmed Scientifics) according to the manual.
Transmission Electron Microscopy
Leaf squares cut from etiolated seedlings at 10 days were fixed, embedded, cut, and soaked as described by Sosso et al. (23). Microscopy used an H-8100 electron microscope (Hitachi H-8100) at an accelerating voltage of 150 kV.
Quantitation of Jasmonic Acid and Its Biosynthetic Intermediates
Quantitation of the jasmonic acid pathway intermediates was performed by a recently described method (24). Rice seedlings or tassels (0.25 g) were frozen in liquid nitrogen, ground to a fine powder, and extracted with 1.25 ml of 80% (v/v) methanol at 4 °C for 12 h. Then 9,10-dihydrojasmonic acid (50 ng/g), d2-indole-3-acetic acid (50 ng/g), and d6-abscisic acid (50 ng/g) were added to plant samples as internal standards, prior to grinding. After centrifugation (30,000 × g, 4 °C, 20 min), the supernatant was collected and passed through a C18 SPE-cartridge (1 ml, 125 mg) preconditioned with 1 ml of H2O, 1 ml of methanol, and 1 ml of 80% (v/v) methanol. The eluant was pooled and evaporated under a nitrogen gas stream and reconstituted in 250 μl of H2O. The solution was acidified with 30 μl of 0.1 m hydrochloric acid and extracted with ethyl ether (three times, 0.5 ml). The ether phases were combined, dried under nitrogen gas, and reconstituted with 176 μl of acetonitrile. Fifteen microliters of Et3N (20 mm) and 9 μl of 3-bromoactonyltrimethylammonium bromide (20 mm) were added. The derivatization reaction was completed within 30 min at room temperature. The reaction solution was evaporated under a stream of nitrogen gas to dryness, and the residue was dissolved in 200 μl of acetonitrile/water (82:18, v/v). Ten microliters of the solution was injected into an HPLC-ESI-Q-TOF-MS system (Bruker Daltonik GmbH) for analysis. Separations were performed on a reversed-phase/SCX column (C18-SCX, 4:1, 150 × 2.1 mm, 5 μm, Shiseido) at a flow rate of 0.2 ml/min at 30 °C, eluting with 40 mm ammonium acetate and acetonitrile (18:82, v/v).
RESULTS
Mitochondrial Proteome and Protein Complex Analysis
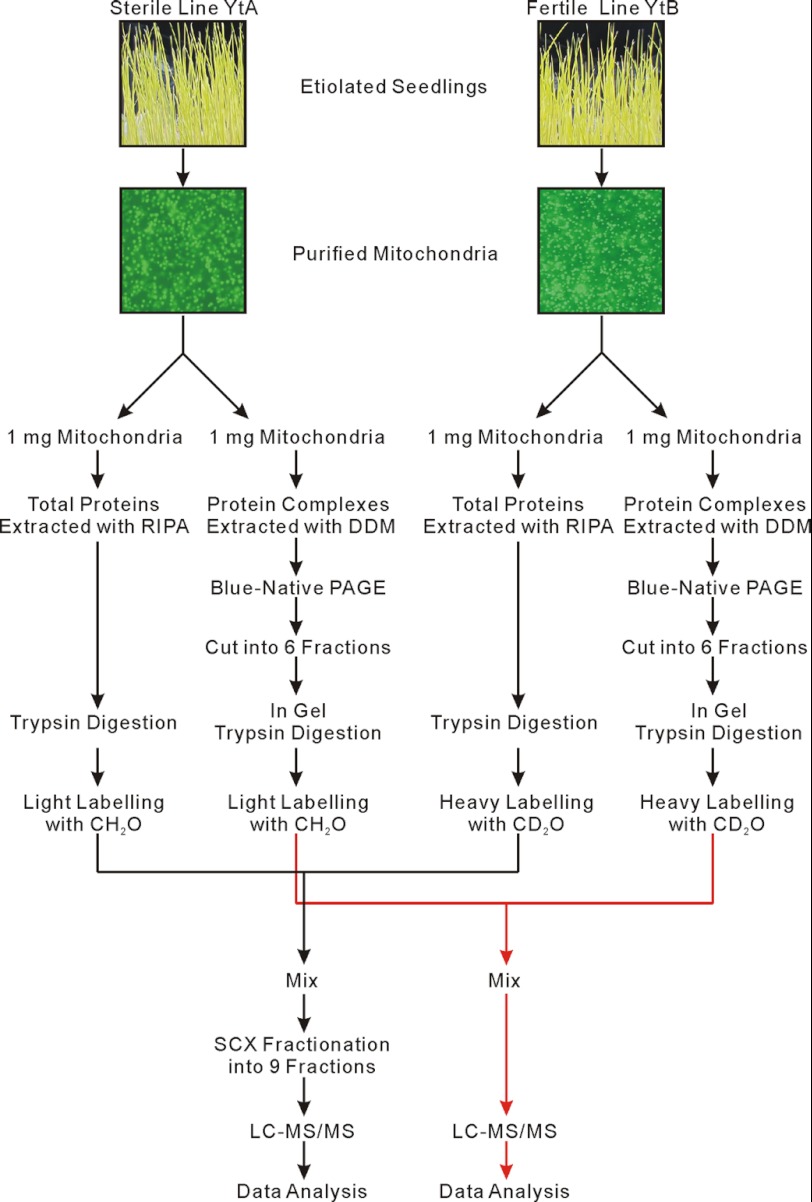
Our proteomics strategy is outlined in Fig. 1. We purified rice mitochondria from YtA and YtB using differential centrifugation and a Percoll gradient. Mitochondrial samples were further separated into two groups. Each group contained 1 mg of a fixed amount of mitochondrial protein. One group was used in quantitative mitochondrial total proteome analysis, which included both mitochondrial membrane proteins and the soluble proteins. The other group containing 1 mg of intact mitochondria was used to analyze mitochondrial protein complexes after BN-PAGE.
FIGURE 1.
Quantitative proteomic analysis work flow.
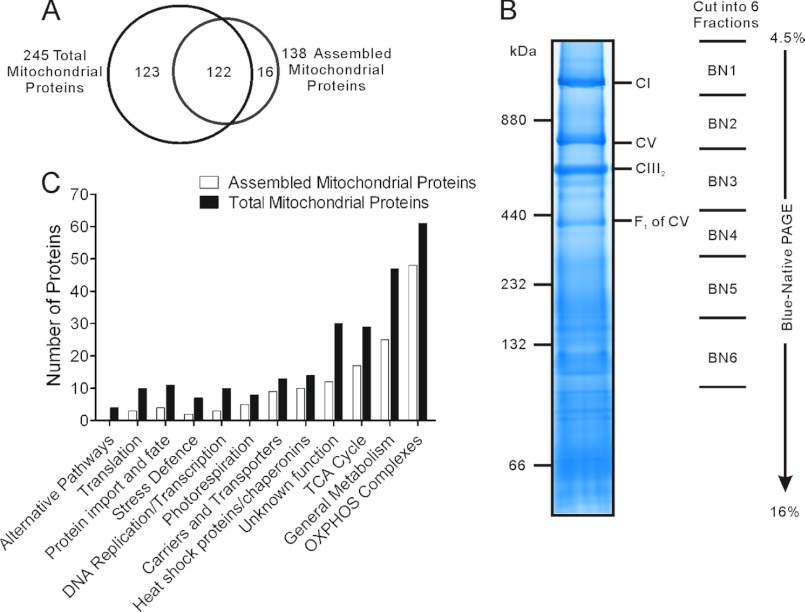
In the total mitochondrial proteome group, nine SCX fractions were collected and labeled with stable isotope dimethyl labels for quantitative LC-MS/MS analysis. After data searching against the rice database from The Institute for Genomic Research, a total of 522 proteins were identified in two biological replicates (supplemental Table S1, with duplicated data alignment shown in supplemental Fig. S1). The false-positive rates of peptide-spectrum matches determined by data search against a decoy database were 0.92 and 1.96%, respectively, for the two biological replicates. Among these 522 proteins, 245 proteins were confirmed as mitochondrial components through comparison with a list of 322 nonredundant rice mitochondrial proteins previously compiled (Fig. 2A; a list of these proteins is shown in supplemental Table S1A) (16).
FIGURE 2.
Proteomics results. A, number of proteins identified in total form and assembled form in YtA and YtB. B, BN gel showing the six fractions cut for proteomic analysis. C, functional categories of identified proteins.
To provide independent experimental evidence validating quantitative ratios based on MS data, Western blot analysis was conducted on representative mitochondrial proteins, including ATP synthase α subunit (ATP1) (Osm1g00580.1), ORFB (Osm1g00170.1), NADH dehydrogenase subunit 9 (NAD9) (Osm1g00590.1), cytochrome c (CYTC) (Os05g34770.1), prohibitin 2 (PHB2) (Os07g15880.1), and T-complex protein (HSP60) (Os10g32550.1). Equal amounts of mitochondrial proteins from YtA and YtB were loaded and resolved by Tricine/SDS-PAGE. Coomassie Blue staining patterns indicated that there were no visible differences between the total proteins of YtA and YtB (supplemental Fig. S2). Comparison of the relative Western blot densities obtained with the ratios generated by LC-MS/MS analysis demonstrated that mass spectrometry quantitation data can be validated by Western blot analysis (supplemental Fig. S2).
In the mitochondrial complexes group, starting from 1 mg of isolated mitochondria, protein complexes were extracted and separated on BN gels. Protein bands above 100 kDa were divided into six fractions, and labeled as “BN1” to “BN6”, respectively (Fig. 2B). In-gel tryptic digestion was performed on each fraction, and peptides were labeled with either the light or heavy dimethyl group and mixed for LC-MS/MS analysis. In all the BN fractions combined, we identified 191 proteins with 138 proteins confirmed as mitochondrial proteins (Fig. 2A; a list of these proteins is shown in supplemental Table S2A, with duplicated data alignment shown in supplemental Fig. S1). The false-positive rates of peptide spectrum matches determined by data search against a decoy database were 0.71 and 0.72%, respectively, for the two mitochondrial protein complex analysis data sets. Among the mitochondrial proteins in the two groups, 122 proteins were in common (Fig. 2A).
To establish a structural linkage between the protein subunits and their relevant complexes, a heat map was generated showing the number of identified peptides from each protein and their corresponding BN gel location (supplemental Fig. S3). Proteins of mitochondrial complex I and complex III2 were concentrated in two fractions, BN1 and BN3. Therefore, we concluded that the relative protein ratio in a BN fraction should correlate to the ratio of their corresponding complexes. Mitochondrial complex V was identified primarily in fractions BN2, BN3, and BN4. This distribution pattern is consistent with previous reports that there are three forms for this complex (19, 25).
The function of all the identified proteins was assigned to 12 functional categories based on functional assignments previously established (Fig. 2C) by Huang et al. (16). From the number of proteins identified in each category, as expected, mitochondrial complex associated proteins or membrane proteins (“oxidative phosphorylation complexes” and “carriers and transporters”) were well represented in data from BN analysis. Although some proteins normally not considered as assembled complex members were also identified from BN-PAGE fractions, their distributions were not BN fraction-specific, indicating these proteins may be loosely attached to mitochondrial complexes.
Mitochondrial Protein Assembly Analysis
A biological process is usually not performed by a single protein. Multiple proteins, either as an assembled complex or working together, are often needed to perform biological functions. A good example is the mitochondrial oxidative phosphorylation system that consists of a series of protein complexes and super complexes (26). Therefore, in addition to the total mitochondrial proteome analysis and comparison, we also examined how the mitochondrial protein complexes were formed and whether there were any differences between the YtA and the YtB samples.
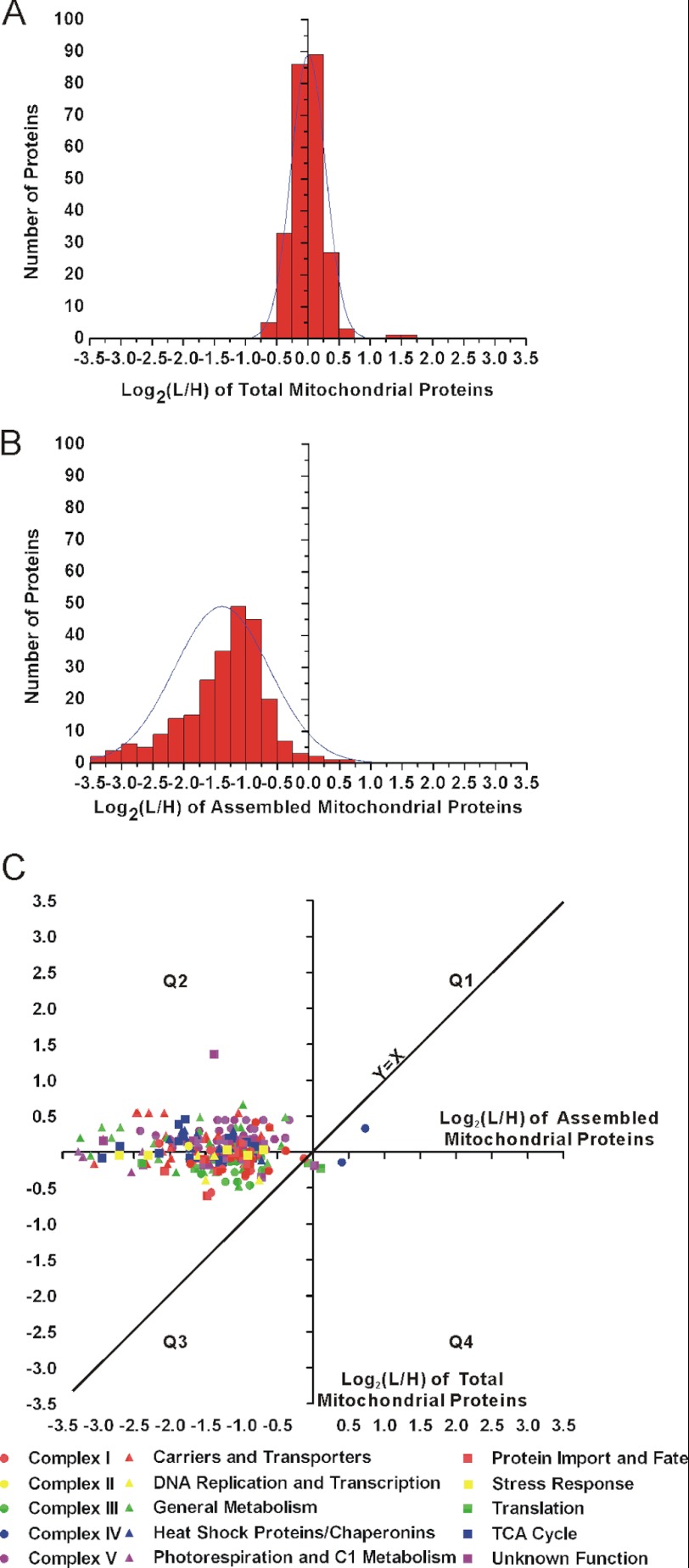
The stable isotope labeling technique we employed provided relative quantitative information. The light/heavy (L/H) ratio from each peptide can provide information about protein levels. We analyzed the L/H ratio (or YtA/YtB ratio) for both the total and assembled form of mitochondrial proteins. The number of identified proteins that fall into each 0.25-interval of log2(L/H) was displayed in Fig. 3, A and B. In the group of total mitochondrial proteins, proteins exhibiting positive log2(L/H) (L,YtA; H, YtB) was almost the same number as those having negative values (Fig. 3A). Most were localized between −1.0 and 1.0, indicating that the sterile line (YtA) contained a mitochondrial proteome similar to the fertile line, which was confirmed by the total mitochondria SDS-PAGE assay (supplemental Fig. S2). However, the proteins in assembled form showed a strong deviation in the negative direction, most of which fell into the section between −3.5 and 0 (Fig. 3B). This difference suggested that the total amount of mitochondrial proteins were nearly the same between the sterile line (YtA) and the fertile line (YtB) but that much less functional mitochondrial complexes were assembled in YtA.
FIGURE 3.
Comparison of the ratio correlation between total and assembled mitochondrial proteins. A, protein L/H ratio (or YtA/YtB ratio) distribution for the total mitochondrial proteins. y axis shows the number of proteins identified that fall into each 0.25-interval of log2(L/H). B, protein L/H ratio (or YtA/YtB ratio) distribution for the assembled form of mitochondrial proteins. y axis shows the number of proteins identified that fall into each 0.25-interval of log2(L/H). C, scatter plot based on the log2(L/H) (or YtA/YtB) value of 122 proteins shared by both total mitochondrial proteins (y axis) and assembled mitochondrial proteins (x axis). Each spot represents a protein, and its assigned functional category was indicated. Four quadrants (Q1–Q4) were assigned as indicated.
To identify proteins which were selectively reduced in mitochondrial assembled protein complexes of YtA, we created a scatter plot based on the log2(L/H) (or YtA/YtB) values of 122 proteins shared by both total mitochondrial proteins (y axis) and assembled mitochondrial proteins (x axis; Fig. 3C). In this plot, if a protein is found along the diagonal line (y = x line), it would suggest that the protein has the same relative ratio distribution in the total mitochondrial protein and in the assembled mitochondrial complexes. Instead, we found 57% of these data points fell into quadrant 2 (Q2), which has a positive log2(L/H) in total mitochondrial proteins but a negative log2(L/H) in assembled complexes. This indicated that these proteins were increased in total mitochondrial form in the sterile line (YtA) but decreased in the assembled form. For example, for the complex V, 11 subunits were identified in both experiments but exhibited a lower ratio as assembled mitochondrial proteins in different BN fractions (Fig. 3C).
Statistic analysis of the YtA/YtB ratio indicated that ∼12% of mitochondria total protein were considered as significantly different from a ratio of 1, but ∼63% of mitochondria assembled protein were considered as significantly different from ratio of 1 (supplemental Tables S1 and S2). This provided additional evidence suggesting that although the total amount of mitochondrial proteins was similar between the sterile line (YtA) and the fertile line (YtB), assembled mitochondrial protein complexes were reduced in YtA.
Discovery of ORFH79 Complexes
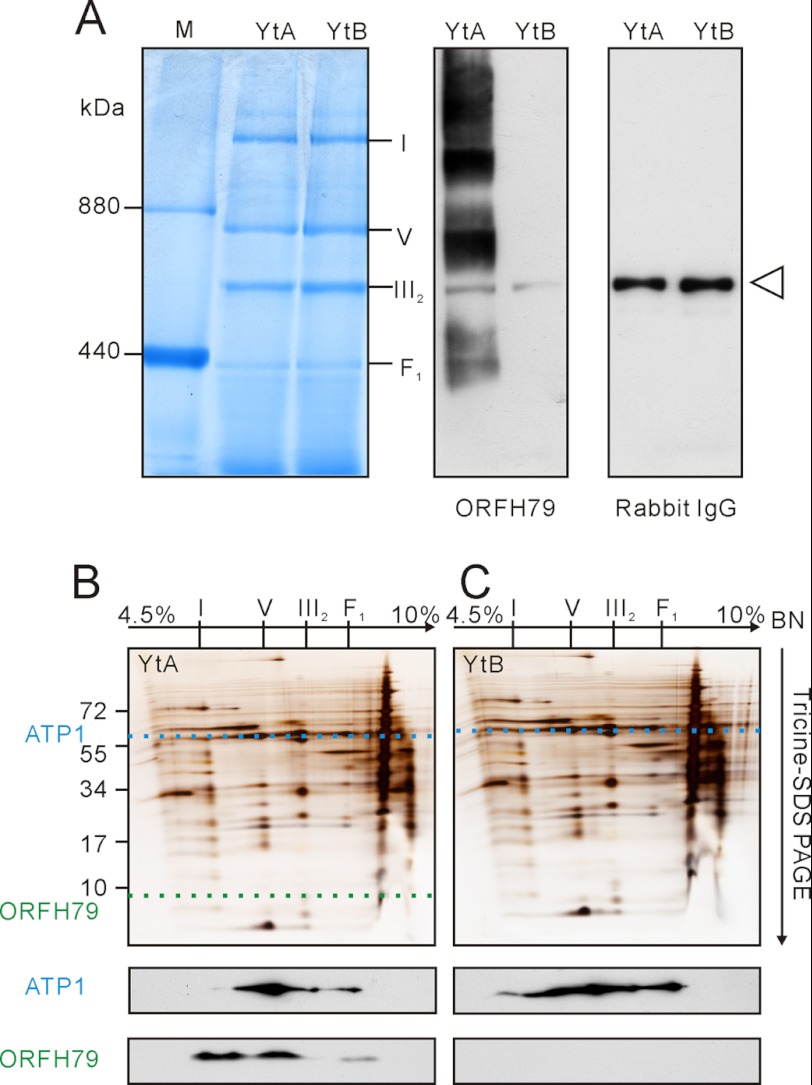
Previous studies have clearly linked the CMS protein ORFH79 to YtA, but we failed to identify the ORFH79 protein in our proteomics analyses described above. Because ORFH79 was localized to the mitochondrial membrane (13), it was possible that ORFH79 was in low abundance and below the limits of MS detection. Therefore, in searching for the ORFH79 protein, we performed Western blotting, which is normally more sensitive (Fig. 4, A–C). Based upon the Western blot data, the ORFH79-containing complexes were estimated to be ∼350, 600, and 900 kDa and 1.1 MDa (Fig. 4A). Use of Western blotting and two-dimensional BN-SDS-PAGE showed ORFH79 protein matches in the vertical location (a potential indication of the same complex) spanning from the BN1 band to the BN4 band.
FIGURE 4.
Comparison of ORFH79 and ATP1 from YtA and YtB in one- and two-dimensional BN-SDS gels. A, mitochondrial complexes resolved on one-dimensional BN gel were transferred to a PVDF membrane and immunoblotted with anti-ORFH79 serum directly. The unspecific bands detected with antibodies in both YtA and YtB were indicated with an open triangle. The nonspecificity was validated with normal rabbit IgG. B and C, BN strips of YtA (B) and YtB (C) mitochondria were denatured and run on Tricine-SDS-PAGE. The two-dimensional gels were silver-stained or transferred to PVDF membranes and immunoblotted with anti-ORFH79 serum and ATP1, respectively. The positions of molecular mass markers are indicated on the left. Corresponding complexes are indicated on the top.
Based on our proteomics results, many well known protein complexes were identified from BN1 to BN4 fractions, including the prohibitin (PHB) complex, complex I, the heat shock protein (HSP) complex, complex V, complex III2, and the F1 part of complex V (7, 19, 25). It was possible that ORFH79 was co-localized with these complexes. ATP1 protein, an F1 sector component of complex V, is often used as a marker for complex V (27). We unexpectedly found by Western blotting that there was a correlation of ATP1 with that of ORFH79 on the BN gel (Fig. 4, B and C). Western blot analysis after two-dimensional BN-SDS also showed minor differences between YtA and YtB on ATP1 staining (Fig. 4, B and C), suggesting possible structural differences of complex V between YtA and YtB.
As an alternative strategy, anti-ORFH79 serum was used to immunoprecipitate mitochondrial membrane proteins, and Western blotting was used to detect PHB2, HSP60, NAD9, ATP1, and CYTC (members of the PHB complex, the HSP complex, complex I, complex V, and complex III2, respectively). As expected, ORFH79 protein was immunoprecipitated and detected in YtA but not in YtB. Unfortunately, none of the other proteins tested were found to be YtA-specific, with a number of proteins detected in the pre-immune serum IP lane (supplemental Fig. S5). Therefore, no ORFH79-specific protein partner information could be derived from these data.
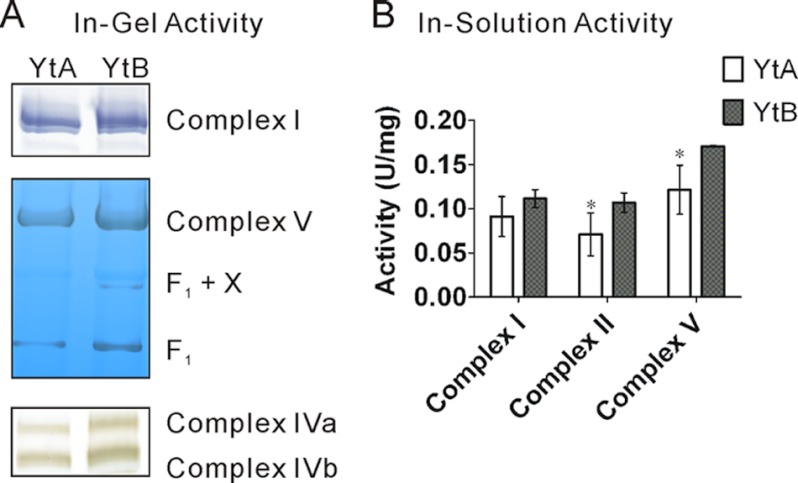
Enzyme Activities of Several Mitochondrial Respiratory Complexes
Enzyme activities of several mitochondrial respiratory complexes were compared in-gel and in-solution between the fertile and sterile lines. The activity of oxidative phosphorylation complexes, particularly complex V, was reduced in the sterile line (Fig. 5). This observation suggested that reduction in enzyme activity was accompanied by a defect in complex assembly in the sterile line (YtA), which provided a correlation between enzyme activity and the quantitative proteomic analyses.
FIGURE 5.
Respiratory complex activity assays. A, in-gel enzyme activities of complex I, V, and IV. B, in-solution enzyme activities of complex I, II, and V. Each bar represents the mean ± S.D. (n = 3). The asterisk indicates that the difference between YtA and YtB is significant (p < 0.05).
Additional assays were performed with mitochondria purified from transgenic callus carrying the orfH79 gene. Silver staining indicated little or no difference between total proteins of the orfH79 transgenic line (Trans79) and the blank vector transgenic line (TransBL). Western blotting showed equal amounts of ATP1 in both transgenic lines. However, the in-gel enzyme activity assay showed a reduced activity of complex V in Trans79 (supplemental Fig. S4). Taken together, the results suggested that the assembly defect is directly linked to the presence of ORFH79.
Altered Mitochondrial Morphology in the Sterile Line
It has been reported that mitochondrial ultrastructure is closely related to its function (28). The results of this study indicated that mitochondria of the sterile line (YtA) contained reduced functional protein complexes, while gaining an additional ORFH79-containing complex. Transmission electron microscopy was used to compare mitochondrial ultrastructure between YtA and YtB in etiolated seedlings at 10 days (Fig. 6). No significant differences in the number of mitochondria per cell were observed. However, most of the mitochondria in YtA displayed a spherical morphology with reduced cristae, whereas in the control YtB, sausage-shaped mitochondria were observed with normal cristae. The morphology of etioplasts in YtA and YtB were similar (Fig. 6). These results suggested that complex assembly defects in the sterile line may have influenced its morphology.
FIGURE 6.
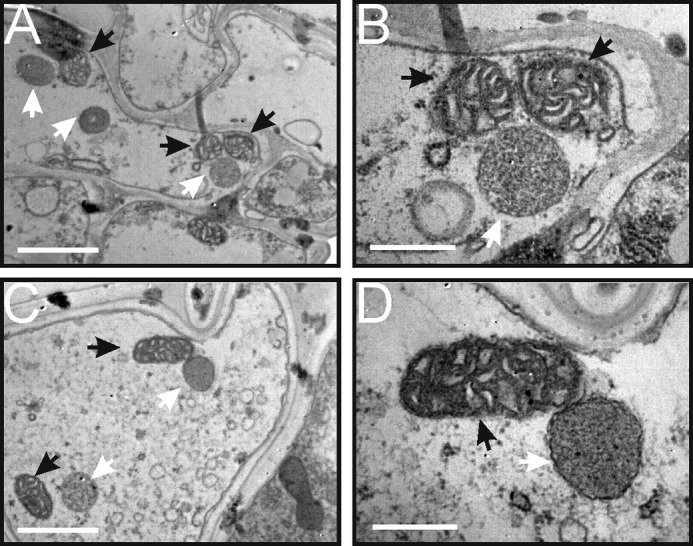
Mitochondrial ultrastructure in YtA and YtB. Transmission electron micrographs of mesophyll cells from YtA (A and B) and YtB (C and D). Mitochondria are indicated with black arrows and etioplasts with white arrows. Bars, 1000 nm in A and C and 400 nm in B and D.
Abnormal Jasmonic Acid Biosynthesis in the Sterile Line
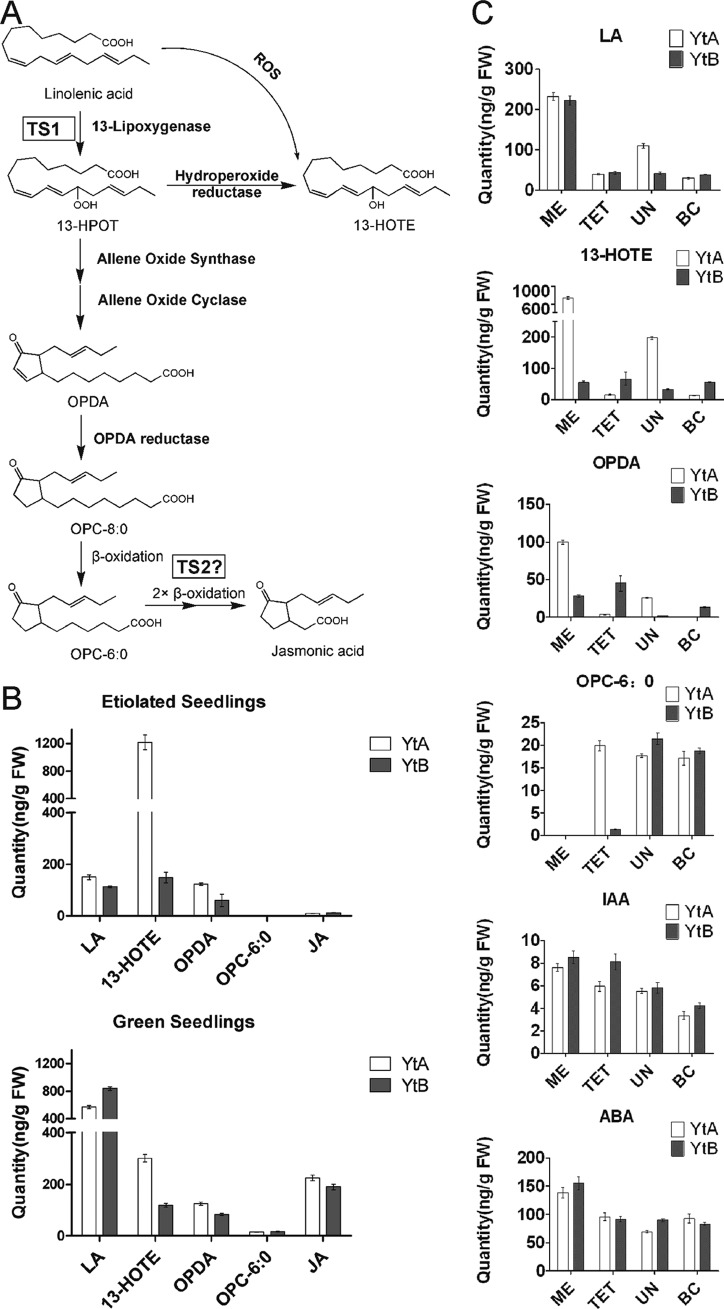
From total mitochondrial proteomic analyses, only two proteins were identified with a greater than 2-fold increase in the sterile line YtA (supplemental Table S1A). These include a protein of unknown function (Os05g51540.1, 2.5-fold), and sex determination protein TASSELSEED-2 (Os07g46920.1, 3.2-fold) (16, 21). Mass spectra for the identification and quantitation of sex determination protein TASSELSEED-2 is shown in supplemental Fig. S6. A homolog of this sex determination protein TASSELSEED-2 in maize, TASSELSEED2 (TS2), was reported to be involved in floral organ determination (29) and may participate in JA biosynthesis (30, 31).
We hypothesized that the up-regulation of the TS2 homolog in YtA may influence the JA biosynthesis in this sterile line (Fig. 7A). Therefore, the abundances of major components in the JA pathway were compared between the etiolated seedlings of YtA and YtB. Although the JA abundance was too low to compare, we observed significant up-regulation of several of its precursors such as hydroxyoctadecatrienoic acid and cis-(+)-12-oxophytodienoic acid in the sterile line YtA compared with YtB (Fig. 7B). In green seedlings, there were also various alterations in JA pathway intermediates between the YtA and YtB lines (Fig. 7B).
FIGURE 7.
Quantitation of JA-related intermediates. A, biosynthesis of JA through the octadecanoid pathway (adapted from Refs. 30, 36–38). B, absolute quantitation of JA-related intermediates from etiolated seedlings and green seedlings. C, absolute quantitation of JA-related intermediates, indole acetic acid (IAA), and abscisic acid (ABA) from developing tassels in different stages. ROS, reactive oxygen species; ME, meiosis stage; TET, tetrad stage; UN, uninucleate stage; BC, bicellular pollen stage. The data are shown as mean ± S.D. (n = 3). 13-HPOT, hydroperoxy octadecatrienoic acid; 13-HOTE, hydroxy octadecatrienoic acid; OPDA, cis-(+)-12-oxophytodienoic acid; LA, linolenic acid.
To investigate whether these JA precursor differences are also present in reproductive organs, tassels from meiosis in the bicellular pollen stage were collected, and the JA-related components were analyzed (Fig. 7C). Indole acetic acid and abscisic acid, two plant hormones not related to JA pathways, were included as controls (Fig. 7C). In the indole acetic acid and abscisic acid samples, only minor differences were observed between the YtA and YtB lines in the four stages examined. In contrast, significant differences among all the JA precursors in all the stages except the bicellular pollen stage were detected. In the meiosis state, excess of hydroxyoctadecatrienoic acid and cis-(+)-12-oxophytodienoic acid accumulation were found in the YtA line compared with the YtB line. In the tetrad stage, OPC-6:0, the last precursor leading to JA, was elevated in YtA. In the uninucleate stage, most of the JA precursors except OPC-6:0 were nearly undetectable in YtB but not in YtA. In the final bicellular pollen stage, all the JA precursors were at similar levels in both lines. Taken together, the results clearly indicated that the JA biosynthetic pathway is differentially regulated in the sterile line.
Additional Altered Mitochondrial Proteins
Electron transfer flavoprotein subunit β (Os04g10400.1) and superoxide dismutase (Os05g25850.1), two proteins that involved in reactive oxygen species related function, are also found as being up-regulated by 1.3- and 1.45-fold, respectively. The increase of superoxide dismutase protein level is consistent with the transcript level increase reported at the initial stage (meiosis stage) of pollen abortion (17).
DISCUSSION
How CMS-associated proteins cause male sterility in plants is an important but difficult mechanism to characterize, mainly due to the technical problems of obtaining sufficient quantities of viable reproductive tissue in a short time (3, 32). Studies using mitochondria isolated from vegetative tissues that constitutively express CMS-associated proteins are therefore an excellent system to study the mechanism of aborted pollen development (7, 32, 33). In this study, a quantitative proteomics approach was used to compare intact mitochondrial proteins and mitochondrial protein complexes between the HL-type CMS rice YtA and its isogenic fertile line YtB. A major finding of this study is that, compared with YtB, YtA shows decreases in the ability to assemble mitochondrial complexes.
The finding that the ORFH79 protein can be a member of protein complexes ranging from ∼400 kDa to ∼1.2 MDa was unexpected. Another CMS protein, ORF138 in rapeseed, was found as part of a 750-kDa complex but not associated with a wide range of complexes (7). A previous study showed that the membrane potential is decreased in YtA (13). Whether this membrane potential decrease is related to ORFH79 remains unclear. When mitochondrial lysates of YtA were analyzed by SDS-PAGE in nonreducing conditions, no evidence of oligomerization could be found (data not shown). It is therefore unlikely that the ORFH79 protein acts as an uncoupling protein similar to ORF138 in rapeseed and T-URF13 in T-maize (5, 7).
Several independent reports have suggested a linkage between ORFH79 and complex V. First, a phenotype of programmed cell death in microspores was observed in YtA (12). It is possible that ORFH79 acts in a similar manner as ORF522, which is thought to compete with normal subunits of complex V and result in programmed cell death in tapetal cells in PET1-CMS sunflower (8, 34). Second, the amount and activities of each form of complex V were reduced in the sterile line (Figs. 3 and 5). Third, our BN-PAGE and two-dimensional BN-SDS-PAGE analyses showed that ORFH79-containing complexes co-migrated with a different ATP1 protein, which is a marker for the F1 sector of complex V (Fig. 4). Fourth, complex V is one of the factors that could affect different mitochondrial morphologies, such as onion-shaped and reduced cristae (35). YtA mitochondria are mostly spherical shaped and hold reduced cristae, which implies that ORFH79 might change the ultrastructure of mitochondria by interfering with complex V. These results are suggestive, but further biochemical and structural studies are needed fully resolve whether ORFH79 proteins are involved in the assembly of complex V.
The up-regulation of one short chain alcohol dehydrogenase, which was annotated as sex determination protein TASSELSEED-2, was another important contribution in understanding the mechanism of HL-CMS. Furthermore, the JA biosynthetic pathway has been shown to be altered in YtA. JA biosynthesis is critical for pollen development and anther dehiscence, so mutation of several enzymes involved in this pathway could lead to male sterility (36).
In summary, our proteomic analyses of HL-type CMS rice mitochondria has provided important new information regarding the biochemical mechanisms underlying the CMS phenotype. From ORFH79 to alterations in the mitochondrial protein complex and to changes in JA production, it is likely that HL-type CMS is the consequence of multiple alterations in biological processes. Further study is needed to connect these processes together, to determine whether there is a single underlying genetic cause, or multiple genetic defects involved in this disorder.
Supplementary Material
Acknowledgments
We thank Dr. S. Huang and Prof. A. H. Millar for the technical help with mitochondrial isolation and BN-PAGE. We thank Dr. T. Elthon for the ATP1 antibody and Prof. J. M. Grienenberger for the NAD9 antibody. We thank Xiu-Wen Wang and Yu-Qing He for the technical help with transmission EM. We thank Ming-Liang Wan, Hai-Ning Liu, Hao-Xin Li, and Dan Li for critical reading of the manuscript.
This work was supported in part by the National Basic Research Program of China Grants 2007CB109005 and 2013CB911102, National Natural Science Foundation of China Grants 31071391, 91019013, 91017013, 31070327, and 30921001, the National High-Tech Research and Development Project Grant 2009AA101101, and the 111 Project of China Grant B06018.

This article contains supplemental Figs. S1–S6 and Tables S1 and S2.
- CMS
- cytoplasmic male sterility
- BN-PAGE
- blue native-PAGE
- PHB
- prohibitin
- HL
- Honglian
- DDM
- dodecyl maltoside
- JA
- jasmonic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- YtA
- Yuetai A
- YtB
- Yuetai B
- L/H
- light/heavy
- SCX
- strong cation exchange.
REFERENCES
- 1. Havey M. J. (2004) In Molecular Biology and Biotechnology of Plant Organelles (Daniell H., Chase C., eds.) pp. 623–634, Springer, Dordrecht, The Netherlands [Google Scholar]
- 2. Wang Z., Zou Y., Li X., Zhang Q., Chen L., Wu H., Su D., Chen Y., Guo J., Luo D., Long Y., Zhong Y., Liu Y. G. (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18, 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanson M. R., Bentolila S. (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16, S154–S169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Budar F., Berthomé R. (2007) In Plant Mitochondria (Logan D. C., ed.) pp. 278–307, John Wiley & Sons, Inc. New York [Google Scholar]
- 5. Levings C. S., 3rd, Siedow J. N. (1992) Molecular basis of disease susceptibility in the Texas cytoplasm of maize. Plant Mol. Biol. 19, 135–147 [DOI] [PubMed] [Google Scholar]
- 6. Rhoads D. M., Levings C. S., 3rd, Siedow J. N. (1995) URF13, a ligand-gated, pore-forming receptor for T-toxin in the inner membrane of CMS-T mitochondria. J. Bioenerg. Biomembr. 27, 437–445 [DOI] [PubMed] [Google Scholar]
- 7. Duroc Y., Hiard S., Vrielynck N., Ragu S., Budar F. (2009) The Ogura sterility-inducing protein forms a large complex without interfering with the oxidative phosphorylation components in rapeseed mitochondria. Plant Mol. Biol. 70, 123–137 [DOI] [PubMed] [Google Scholar]
- 8. Sabar M., Gagliardi D., Balk J., Leaver C. J. (2003) ORFB is a subunit of F1F(O)-ATP synthase. Insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep. 4, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li S., Yang D., Zhu Y. (2007) Characterization and use of male sterility in hybrid rice breeding. J. Integrative Plant Biol. 49, 791–804 [Google Scholar]
- 10. Yi P., Wang L., Sun Q., Zhu Y. (2002) Discovery of mitochondrial chimeric-gene associated with cytoplasmic male sterility of HL-rice. Chin. Sci. Bull. 47, 744–747 [Google Scholar]
- 11. Li X. M., Zhen Y. L., Zhang F. D., Zhu Y. G. (2000) RFLP analysis for mitochondria genome of CMS rice Honglian Type, Hereditas (Beijing) 22, 201–204 [Google Scholar]
- 12. Li S., Wan C., Kong J., Zhang Z., Li Y., Zhu Y. (2004) Programmed cell death during microsporogenesis in a Honglian CMS line of rice is correlated with oxidative stress in mitochondria. Funct. Plant Biol. 31, 369–376 [DOI] [PubMed] [Google Scholar]
- 13. Peng X., Wang K., Hu C., Zhu Y., Wang T., Yang J., Tong J., Li S., Zhu Y. (2010) The mitochondrial gene orfH79 plays a critical role in impairing both male gametophyte development and root growth in CMS-Honglian rice. BMC Plant Biol. 10, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mackenzie S., McIntosh L. (1999) Higher plant mitochondria. Plant Cell 11, 571–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mihr C., Baumgartner M., Dieterich J. H., Schmitz U. K., Braun H. P. (2001) Proteomic approach for investigation of cytoplasmic male sterility (CMS) in Brassica. J. Plant Physiol. 158, 787–794 [Google Scholar]
- 16. Huang S., Taylor N. L., Narsai R., Eubel H., Whelan J., Millar A. H. (2009) Experimental analysis of the rice mitochondrial proteome, its biogenesis, and heterogeneity. Plant Physiol. 149, 719–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wan C., Li S., Wen L., Kong J., Wang K., Zhu Y. (2007) Damage of oxidative stress on mitochondria during microspores development in Honglian CMS line of rice. Plant Cell Rep. 26, 373–382 [DOI] [PubMed] [Google Scholar]
- 18. Heazlewood J. L., Howell K. A., Whelan J., Millar A. H. (2003) Towardan analysis of the rice mitochondrial proteome. Plant Physiol. 132, 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jänsch L., Kruft V., Schmitz U. K., Braun H. P. (1996) New insights into the composition, molecular mass, and stoichiometry of the protein complexes of plant mitochondria. Plant J. 9, 357–368 [DOI] [PubMed] [Google Scholar]
- 20. Chen X., Wu D., Zhao Y., Wong B. H., Guo L. (2011) Increasing phosphoproteome coverage and identification of phosphorylation motifs through combination of different HPLC fractionation methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 879, 25–34 [DOI] [PubMed] [Google Scholar]
- 21. Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., Thibaud-Nissen F., Malek R. L., Lee Y., Zheng L., Orvis J., Haas B., Wortman J., Buell C. R. (2007) The TIGR rice genome annotation resource. Improvements and new features. Nucleic Acids Res. 35, D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sabar M., Balk J., Leaver C. J. (2005) Histochemical staining and quantification of plant mitochondrial respiratory chain complexes using blue-native polyacrylamide gel electrophoresis. Plant J. 44, 893–901 [DOI] [PubMed] [Google Scholar]
- 23. Sosso D., Mbelo S., Vernoud V., Gendrot G., Dedieu A., Chambrier P., Dauzat M., Heurtevin L., Guyon V., Takenaka M., Rogowsky P. M. (2012) PPR2263, a DYW subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24, 676–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen M. L., Huang Y. Q., Liu J. Q., Yuan B. F., Feng Y. Q. (2011) Highly sensitive profiling assay of acidic plant hormones using a novel mass probe by capillary electrophoresis-time of flight-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 879, 938–944 [DOI] [PubMed] [Google Scholar]
- 25. Eubel H., Jänsch L., Braun H. P. (2003) New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 133, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lenaz G., Genova M. L. (2009) Structural and functional organization of the mitochondrial respiratory chain. A dynamic super-assembly. Int. J. Biochem. Cell Biol. 41, 1750–1772 [DOI] [PubMed] [Google Scholar]
- 27. Wittig I., Schägger H. (2008) Structural organization of mitochondrial ATP synthase. Biochim. Biophys. Acta 1777, 592–598 [DOI] [PubMed] [Google Scholar]
- 28. Zick M., Rabl R., Reichert A. S. (2009) Cristae formation-linking ultrastructure and function of mitochondria. Biochim. Biophys. Acta 1793, 5–19 [DOI] [PubMed] [Google Scholar]
- 29. DeLong A., Calderon-Urrea A., Dellaporta S. L. (1993) Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74, 757–768 [DOI] [PubMed] [Google Scholar]
- 30. Acosta I. F., Laparra H., Romero S. P., Schmelz E., Hamberg M., Mottinger J. P., Moreno M. A., Dellaporta S. L. (2009) tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323, 262–265 [DOI] [PubMed] [Google Scholar]
- 31. Wu X., Knapp S., Stamp A., Stammers D. K., Jörnvall H., Dellaporta S. L., Oppermann U. (2007) Biochemical characterization of TASSELSEED 2, an essential plant short-chain dehydrogenase/reductase with broad spectrum activities. FEBS J 274, 1172–1182 [DOI] [PubMed] [Google Scholar]
- 32. Dewey R. E., Timothy D. H., Levings C. S. (1987) A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc. Natl. Acad. Sci. U.S.A. 84, 5374–5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wise R. P., Fliss A. E., Pring D. R., Gengenbach B. G. (1987) Urf13-T of T-cytoplasm maize mitochondria encodes a 13 kd polypeptide. Plant Mol. Biol. 9, 121–126 [DOI] [PubMed] [Google Scholar]
- 34. Balk J., Leaver C. J. (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mannella C. A. (2006) Structure and dynamics of the mitochondrial inner membrane cristae. Biochim. Biophys. Acta 1763, 542–548 [DOI] [PubMed] [Google Scholar]
- 36. Wasternack C. (2007) Jasmonates. An update on biosynthesis, signal transduction, and action in plant stress response, growth, and development. Ann. Bot. 100, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Møller I. M., Jensen P. E., Hansson A. (2007) Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58, 459–481 [DOI] [PubMed] [Google Scholar]
- 38. Schaller F. (2001) Enzymes of the biosynthesis of octadecanoid-derived signaling molecules. J. Exp. Bot. 52, 11–23 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.