Background: Golgi retention mechanism of the core 2 N-acetylglucosaminyltransferase 1 (C2GnT1) is not known.
Results: Golgi phosphoprotein 3 (GOLPH3) controls Golgi localization of C2GnT1 by binding to its cytoplasmic tail.
Conclusion: GOLPH3 determines cell binding properties under dynamic flow by controlling Golgi localization of C2GnT1.
Significance: Cell-cell interactions can be regulated by a protein that controls the Golgi localization of a glycosyltransferase.
Keywords: Carbohydrate Function, Cell Adhesion, Glycoprotein Biosynthesis, Glycosyltransferases, Golgi, C2GnT1, GOLPH3, Golgi Retention Signals, PSGL-1, Cytoplasmic Tail
Abstract
Core 2 N-acetylglucosaminyltransferase 1 (C2GnT1) is a key enzyme participating in the synthesis of core 2-associated sialyl Lewis x (C2-O-sLex), a ligand involved in selectin-mediated leukocyte trafficking and cancer metastasis. To accomplish that, C2GnT1 needs to be localized to the Golgi and this step requires interaction of its cytoplasmic tail (CT) with a protein that has not been identified. Employing C2GnT1 CT as the bait to perform a yeast two-hybrid screen, we have identified Golgi phosphoprotein 3 (GOLPH3) as a principal candidate protein that interacts with C2GnT1 and demonstrated that C2GnT1 binds to GOLPH3 via the LLRRR9 sequence in the CT. Confocal fluorescence microscopic analysis shows substantial Golgi co-localization of C2GnT1 and GOLPH3. Upon GOLPH3 knockdown, C2GnT1 is found mainly in the endoplasmic reticulum and decorated with complex-type N-glycans, indicating that the enzyme has been transported to the Golgi but is not retained. Also, we have found that a recombinant protein consisting of C2GnT1 CT1–16-Leu17–32-Gly33–42-GFP is localized to the Golgi although the same construct with mutated CT (AAAAA9) is not. The data demonstrate that the C2GnT1 CT is necessary and sufficient for Golgi localization of C2GnT1. Furthermore, GOLPH3 knockdown results in reduced synthesis of C2-O-sLex associated with P-selectin glycoprotein ligand-1, reduced cell tethering to and rolling on immobilized P- or E-selectin, and compromised E-selectin-induced activation of spleen tyrosine kinase and cell adhesion to intercellular adhesion molecule-1 under dynamic flow. Our results reveal that GOLPH3 can regulate cell-cell interaction by controlling Golgi retention of C2GnT1.
Introduction
Cell surface carbohydrates play important roles in many biological processes, such as differentiation, immune surveillance, pathogen clearance, and blood group antigen recognition (1–5). Synthesis of these carbohydrates is under a strict control to ensure that they are generated when needed, and vanished when the intended tasks have been completed. Pathological consequences can occur if these carbohydrates are inappropriately produced or maintained (6, 7). One such example is the synthesis of core 2-associated sialyl Lewis x (C2-O-sLex).3 Under resting conditions, circulating leukocytes express a low level of C2-O-sLex. However, following tissue injury or infection, these cells greatly increase the production of C2-O-sLex on their cell surface as a result of up-regulation of the core 2 N-acetylglucosaminyltransferase 1 (C2GnT1) gene (8, 9). The C2-O-sLex is a selectin ligand involved in leukocyte trafficking (10, 11). Binding of C2-O-sLex-containing ligands to selectins initiates a series of signaling events culminating in firm adhesion of activated leukocytes to the inflamed endothelium leading to their extravasation (12). The role of C2GnT1 in this immune-modulating function has been confirmed in a C2GnT1 gene-deficient mouse (13, 14). Deletion of the C2GnT1 gene had its greatest impact on the inflammatory response, resulting in reduction of leukocyte rolling on P-, E-, and L-selectins and migration of these leukocytes to injured sites. On the other hand, elevated expression of the C2GnT1 gene results in altered O-glycans (15, 16), which if persisted would compromise humoral and cellular immunity as found in Wiscott-Aldrich syndrome (17). A similar phenomenon has been confirmed in a C2GnT1 transgenic mouse model (18, 19), where T lymphocytes showed a delayed-type hypersensitivity response, reduced proliferation rates, and weakened interaction with B lymphocytes. In addition, up-regulation of the C2GnT1 gene and C2-O-sLex are frequently observed in metastatic tumors (20, 21). These tumors utilize mechanisms similar to the selectin-mediated leukocyte trafficking to metastasize to distant sites (6). Although regulation of gene expression is a key for controlling the production of C2GnT1, the glycosylation function of this enzyme depends on its localization to the Golgi (22). To date, the Golgi localization mechanism of C2GnT1 is not clear.
Golgi phosphoprotein 3 (GOLPH3/GPP34/GMX33/MIDAS) is a Golgi-associated protein (23) conserved from yeast to human. It was recently recognized as an oncoprotein amplified in various types of human malignancies, including melanoma, breast, non-small cell lung cancer, gliomas, and connective tissue tumors (24–26). GOLPH3 exhibits diverse biological roles. It has been linked to the modulation of mitochondrial mass and lipid synthesis (27), the mammalian target of rapamycin signaling pathway (24), budding of vesicles from the trans-Golgi (28), and recycling transmembrane receptors from endosomes to the trans-Golgi network (24, 29). Depletion of the GOLPH3 yeast homolog Vps74p in yeast cells prevents Golgi localization of mannosyltransferases resulting in the production of hypoglycosylated glycoconjugates. The consensus amino acid sequence (F/L)(L/I/V)XX(R/K) recognized by Vps74p is present in the cytoplasmic tails (CT) of yeast mannosyltransferases (30, 31). This sequence is also found in the C2GnT1 CT of various species (supplemental Fig. S1). This information prompted us to examine whether GOLPH3 could be the protein that determined the Golgi localization of C2GnT1.
In this study, we screened by a yeast two-hybrid method for proteins that interact with C2GnT1 CT and identified GOLPH3 as a principal candidate. We examined the effect of GOLPH3 depletion on Golgi localization of C2GnT1 and its function in the generation of C2-O-sLex on P-selectin glycoprotein ligand-1 (PSGL-1), and its role in cell tethering to and rolling on immobilized selectins, and adhesion of E-selectin-activated cells to immobilized intercellular adhesion molecule-1 (ICAM-1) under physiologically relevant sheer force. We have found that GOLPH3 regulates cell tethering, rolling and adhesion by controlling Golgi localization of C2GnT1.
EXPERIMENTAL PROCEDURES
Materials, Antibodies, and siRNAs
The materials used in this study were obtained from the following suppliers. Primary antibodies (Abs) against GOLPH3 (mouse monoclonal and rabbit polyclonal), GM130, Giantin, mannosidase II (ManII) (all are rabbit polyclonal), and C1GalT1 and St3GalT1 mouse monoclonal antibodies were purchased from Abcam (Cambridge, MA); mouse monoclonal anti-C2GnT1 Ab, Novus Biologicals (Littleton, CO); TGN46 (rabbit polyclonal) and β-actin (mouse monoclonal) Abs, Sigma; mouse C2-O-sLex IgM Ab, R&D Systems (Minneapolis, MN) (32); mouse anti-PSGL-1 (PL1 and PL2) Ab, Santa Cruz Biotechnology (Santa Cruz, CA); rabbit anti-SYK and anti-phospho-Y323-SYK Abs, Cell Signaling Technology (Danvers, MA); donkey anti-mouse and anti-rabbit Dylight 488 and 594 secondary Abs, Jackson ImmunoResearch (West Grove, PA); GOLPH3 siGenome siRNAs and C2GnT1 siGenome siRNAs, Dharmacon (Waltman, MA); and K562 (CCL-243) and KG1a (CCL-246.1) cells, ATCC (Manassas, VA). The lectins used in this study were purchased from E-Y Laboratories (San Mateo, CA) and they include ConA, Canavalia ensiformis agglutinin; PSA, Pisum sativum agglutinin; DSA, Datura stramonium; PNA, Peanut agglutinin; VVA, Vicia villosa agglutinin.
Plasmid Construction and Transient Transfection of KG1a and K562 Cells
The coding region of the human (h) C2GnT1 gene (GenBankTM accession no. NM_001490) was cloned by PCR using the primer set of 5′-ATCTCGAGGCCACCATGCTGAGGACGTTGCTGCGAAGGAG-3′ and 5′-ATGGATCCCCTCCGTGTTTTAATGTCTCCAAAGCTTTGTGTC-3′, and ligated into XhoI and BamHI sites of the pEGFP-N1 vector (Clontech, Mountain View, CA) to generate C2GnT1(1–428)-GFP. cDNA coding the N-terminal region of C2GnT1, which includes CT(1–16), transmembrane domain TMD(17–32), and a part of the stem region(33–70), was cloned into the vector by PCR to generate an insert of C2GnT11–70-GFP. To construct the C2GnT1Δ5–9-GFP, C2GnT1(AARRR9)-GFP, C2GnT1(LLAAA9)-GFP, and C2GnT11–70(AAAAA9)-GFP mutant plasmids, nucleotides of the forward primer were modified to omit amino acids 5–9 or mutate LL6 to AA6, RRR9 to AAA9, or LLRRR9 to AAAAA9. To construct C2GnT11–16-GFP and C2GnT11–16(AAAAA9)-GFP, minigenes encoding C2GnT1 CT1–16, nonspecific TMD Leu17–32 and nonspecific stem Gly33–42, and its mutant, C2GnT11–70(AAAAA9)-GFP, were chemically synthesized (IDT Inc., Coralville, IA). The integrity and orientation of all plasmids were confirmed by restriction digestion and sequencing. The hematopoietic cell lines K562 and KG1a were maintained in Iscove's modified Dulbecco's medium (Hyclone, Waltman, MA) supplemented with 20% fetal bovine serum and 1% penicillin and streptomycin. Cells were maintained at 37 °C, 5% CO2. K562 cells were nucleofected with the Nucleofector system from Amaxa (Lonza Inc., Walkersville, MD). About 106 cells suspended in 0.1 ml of Lonza solution V were mixed with 8 μg of any of the constructs above, before nucleofection using the program T-016. After nucleofection, the cells were immediately transferred to 2 ml of pre-warmed culture medium and cultured until analysis. K562 cells were chosen for this experiment because of its high transfection efficiency and lack of C2GnT1 compared with KG1a cells. For siRNA treatment, KG1a cells (106) suspended in Lonza solution L were mixed with 10 μl of a solution containing 50 pmol of siRNA before nucleofection using the program V-001. After nucleofection, the cells were immediately transferred to 2 ml of pre-warmed culture medium. After 48–72 h, the transfected cells were analyzed by confocal immunofluorescence microscopy.
Cloning, Expression, and Purification of Recombinant GOLPH3 in Escherichia coli
Human RNA isolated from KG1a cells was used for PCR cloning of the cDNA of GOLPH3 (GenBank accession number NM_022130). The forward primer 5′-TAGGATCCATGACCTCGCTGACCCAGCGCAG-3′ and the reverse primer 5′-ATGAATTCTTACTTGGTGAACGCCGCCACCAC-3′ were used. The restriction sites BamHI and EcoRI are underlined. The purified PCR product (897 bp) was cloned into the 3′ end of the GST cDNA flanked by a Precision protease cleavage site in the pGEX-6P-2 vector (GE Healthcare) at these two sites. The protease cleavage site (/) was introduced into a nucleotide sequence, CTGGAAGTTCTGTTCCAGGGG/CCC, that encodes Leu-Glu-Val-Leu-Phe-Gln-Gly/Pro downstream of GST. This pGEX-6P-2 vector carrying the 897-bp insert was transformed into high efficiency competent E. coli DH5 cells (New England Biolabs, Ipswich, MA). The positive clones were confirmed by restriction digestion and sequencing. The pGEX-6P-2 vector and the construct containing the 897-bp GOLPH3 cDNA was then transformed into E. coli strain BL21(DE3) host cells (New England Biolabs). A single clone was cultured in LB medium containing amplicillin (100 μg/ml). The expression of the recombinant protein in the E. coli culture at logarithmic phase (A600 ∼0.6) was induced with 0.5 mm isopropyl β-d-thiogalactoside at 37 °C for 3 h. The cell pellet was dissolved in 10 ml of lysis buffer (10 ml of PBS, 80 μl of 100 mm PMSF, 10 μl of 1 m DTT, and 80 μl of Nonidet P-40) and a tablet of Roche protease inhibitor mixture mini (Indianapolis, IN) and lysed 3 times with a French press (Thermo Scientific). The supernatant (13,000 × g, 40 min at 4 °C) was mixed with 1 ml of glutathione resin (Genscript, Piscataway, NJ) to immobilize the recombinant protein. The PBS-washed beads were suspended in 1.5 ml of PBS followed by addition of 40 μg of GST-tagged Precision protease (GE Healthcare) to cleave the recombinant protein from the resin. The tube was allowed to rock at 4 °C for 16 h. The supernatant was recovered and mixed with 500 μl of fresh glutathione resin and rocked for 2 h at 4 °C to remove the Precision GST-protease. The supernatant that contained GOLPH3 was recovered and analyzed for purity of the recombinant GOLPH3 protein by SDS-10% PAGE followed by Coomassie Blue staining.
Flow Cytometry Analysis
Assessment of cell surface C2-O-sLex and PSGL-1 was performed by flow cytometry using mouse anti-C2-O-sLex (CHO-131) IgM and the F(ab)2 fragment of goat anti-mouse IgM fluorescein isothiocyanate-conjugated secondary Abs and rabbit anti-PSGL-1 and F(ab)2 fragment of goat anti-rabbit phycoerythrin-conjugated secondary Abs. KG1a cells were transfected with either mock (non-targeting) siRNA or GOLPH3 siRNAs and cultured for 48–72 h. The cells (0.5 × 106) were then treated with 1% BSA in PBS for 30 min on ice to block nonspecific sites before exposure to primary Abs. For primary Ab staining, cells were incubated with anti-sLex and anti-PSGL-1 Abs (10 μg/ml in PBS washing buffer containing 1% BSA and 0.09% NaN3) on ice for 30 min and then washed with washing buffer (3 times). Secondary Ab staining was performed with the F(ab)2 fragment of goat anti-mouse IgM FITC or phycoerythrin-conjugated secondary Abs at 1:200 dilution for 30 min on ice and excess Abs were washed with washing buffer. Finally, cells were resuspended in 500 μl of 0.5% paraformaldehyde and analyzed using a FACSCalibur (BD Biosciences) equipped with a 488 argon laser and a 635 diode laser and analyzed using Cell Quest-pro software. The unstained cells, single-stained cells, and cells incubated with only the secondary antibodies were used as controls.
Preparation of E-selectin, P-selectin, and ICAM-1-coated Coverslips
Recombinant human E- and P-selectins/IgG Fc chimeras were purified from a CHO cell line supernatant transfected with P-/E-selectin cDNA (a gift from Dr. Ajit Varki, University of California, San Diego, CA) (33) by immunoaffinity chromatography using protein A-Sepharose (GE Healthcare Life Sciences) (34). The E-selectin, P-selectin, or the ICAM-1 (R&D Systems) dissolved in Dulbecco's phosphate-buffered saline containing 2 mm Ca2+ and 2 mm Mg2+ and used to coat the glass coverslips as described by Ross et al. (35). Twenty μl of E-selectin, P-selectin, or ICAM-1 solution (1 mg/ml) were placed in the flow path (∼160 mm2) on a glass coverslip (GlycoTech, Gaithersburg, MD), followed by incubation at 4 °C for 12 h and then rinsing with 1 ml of Dulbecco's phosphate-buffered saline to remove unbound proteins. Prior to the experiment, slides were incubated with 3% BSA (in Dulbecco's phosphate-buffered saline) for 1 h at room temperature.
Parallel Plate Flow Chamber Assay
Interaction of KG1a cells with E-selectin, P-selectin, or ICAM-1 coated on coverslips was measured by a parallel-plate flow chamber assay as described previously (36) and evaluated according to Krull et al. (37). Briefly, cells were dislodged from the flasks with non-enzymatic cell striper (Cellstripper, Mediatech, Manassas, VA) and washed (3 times) with binding buffer (2 mm HEPES containing 2 mm CaCl2, pH 7.4). A suspension of 2.5 × 105 cells/ml prepared in 4 ml of the binding buffer plus 10 μg/ml of DNase I (MP Biomedicals, Solon, OH) was perfused through the chamber at a constant wall shear stress of 1.0 dyne/cm2 using a Harvard Syringe Pump (Harvard Apparatus, Holliston, MA). The field of observation was chosen randomly and visualized with a phase-contrast video microscope (Olympus 1 × 70 coupled with camera; Q-Imaging, Surrey, BC, Canada) at ×40 magnification and videotaped using Q-pro software. Videotapes were played back at 1/10 speed in Quick-time for analysis. The cells at each of 20 different fields were counted for each experiment (38). Tethering was determined by counting the number of cells that adhered to slides over the first 2–3 s of continuous shear flow to a given field of view and remained tethered for at least 3 s while rolling was defined as five times cell diameters of forward movement, assessed at 1.0 dynes/cm2. Tethering or rolling was expressed as the number of tethered or rolled cells/high-power field during a 10-min observation period (39). E-selectin-activated cells, which were stably adhered to ICAM-1-coated slides for more than 30 s, were recorded as adhesion.
Yeast Two-hybrid Screen
Matchmaker Two-Hybrid System 3 (Clontech) was employed for screening a human universal cDNA library using the human C2GnT1 CT (MLRTLLRRRLFSYPTKY) as the bait. The oligonucleotide coding the wild-type C2GnT1 CT (IDT, Coralville, IA) was hybridized and cloned into pGBKT7 to create a c-Myc-tagged fusion protein with the Gal4(DBD). The library screen was performed under high stringency auxotrophic conditions on minimal medium lacking adenine and amino acids His, Leu, and Trp (SD-Ade/-His/-Leu/-Trp/X-α-Gal), and containing 125 ng/ml of aureobasidin A, where ∼2.5 × 106 clones were screened. Plasmid DNA from positive clones was isolated, propagated in E. coli, and validated by sequencing.
Forward Yeast Two-hybrid Analysis
To investigate the interaction between GOLPH3 and the C2GnT1 CT peptide, oligonucleotides coding the wild-type CT and its AA6 or AAA9 mutants were synthesized (IDT), hybridized, and cloned into pGBKT7 to create plasmids encoding GAL4(DBD)-C2GnT1 (LLRRR9), GAL4(DBD)-C2GnT1(AARRR9), or GAL4(DBD)-C2GnT1(LLAAA9). The plasmids were cotransformed with GAL4(AD)-GOLPH3 by the lithium acetate procedure into yeast strains Y2HGold (Clontech). Co-transformants were selected on plates lacking Leu and Trp at 30 °C. Two to three colonies were picked and suspended in water, equilibrated to the same optical density at 600 nm, and spotted on plates lacking Leu and Trp (+His) as well as plates containing 125 ng/ml of aureobasidin A, and lacking His (-His). The plates were incubated at 30 °C for 3 days before being photographed. The strongly interacting protein pair of Gal4(DBD)-p53 and Gal4(AD)-SV40 large T antigen was used as the positive control, and the non-interacting pair of Gal4(DBD)-Lamin and Gal4(AD)-SV40 large T antigen was used as a negative control.
Peptide Pulldown Assay
The biotinylated N-terminal 20-aa peptide of C2GnT1, which includes the CT and a portion of the transmembrane domain (YFM), was synthesized and tagged with biotin at the C terminus (LifeTein LLC, South Plainfield, NJ). A 21-amino acid control peptide (GHGTGSTGSGSSGTASSEGST) labeled with biotin at the N terminus was obtained from the same company. To perform a GOLPH3 pulldown experiment, 6 μg of C2GnT1 peptide (FW 3000) in dimethyl sulfoxide (4 μm) was mixed with 500 μg of cell lysate (1 μg/μl). After incubation at 4 °C for 1 h, 30 μl of Dynabeads M-280 streptavidin (Invitrogen) was added. Following gentle rotation for 30 min, the beads with immobilized complexes were isolated with a magnet. After washing 5 times with lysis buffer, the proteins captured on Dynabeads were analyzed by SDS-10% PAGE followed by Western blotting with GOLPH3 Abs.
Preparation of Cell Lysates
Cells were washed three times with PBS, then lysed using CHAPS lysis buffer (50 mm Tris, pH 7.4, 110 mm NaCl, 5 mm EDTA, 1% CHAPS (w/v), and 1% (v/v) of mammalian protease inhibitor mixture (Sigma)). Protein concentrations were determined with the Coomassie Plus Protein Assay (Pierce Chemical Co.) using BSA as the standard.
Electrophoresis and Immunoblotting
Proteins were separated on SDS-10% polyacrylamide gels and transferred to PVDF membranes. The membrane was blocked at room temperature for 1 h with PBST (PBS, pH 7.4, 5% nonfat dried milk, 0.1% Tween 20) and incubated overnight at 4 °C in a blocking solution containing primary Abs with appropriate dilutions. After washing with PBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary Abs for 1 h. Immunoreactive bands were detected by Thermo Scientific SuperSignal West Pico Chemo- luminescent Substrate reagents and exposed to BioExpress Blue Basic Autorad chemoluminescence film (Kaysville, UT). Prestained Protein Ladder from Fermentas (Glen Burnie, MD) was used as the molecular weight markers.
Immunoprecipitation, Endoglycosidase H Digestion, and Lectin Blotting
Subconfluent KG1a cells cultured in T-75 flasks were harvested and cellular proteins were extracted in buffer containing 50 mm Tris (pH 7.4), 150 mm NaCl, 5 mm EDTA, 0.5% SDS, 1% Nonidet P-40 (w/w), and 1% (v/v) mammalian protease inhibitor mixture (Sigma). Cell lysates were pre-cleared with non-immune rabbit IgG for 2 h at 4 °C, followed by incubation with protein A/G-agarose (EMD) at 4 °C for 1 h. Precleared protein extracts were immunoprecipitated overnight at 4 °C with the monoclonal Ab against hC2GnT1 (Abcam). Glycosidase digestions were performed following the protocol provided by the supplier. Briefly, the immunoprecipitates were released from protein A/G-agarose (EMD) and antibodies by boiling in 1% SDS, 200 mm dithiothreitol for 10 min. An equal volume of 0.5 m sodium citrate buffer (pH 5.5) was added to the immunoprecipitates followed by 500 units of endoglycosidase H (40) (New England BioLabs) before incubation at 37 °C for 1 h. The proteins in the treated samples were resolved by SDS-10% PAGE, then transferred onto a PVDF membrane. The membrane was incubated overnight at 4 °C in the blocking buffer of 1% BSA (w/v) in TBS containing 0.1% Tween 20. Then, the blots were incubated with 10 μg/ml each of HRP-labeled lectins C. ensiformis agglutinin (ConA) (41, 42) and P. sativum agglutinin (PSA) (43) (E-Y Laboratories, San Mateo, CA) for 2 h at room temperature, in the blocking buffer. Detection of the lectin-bound bands was performed as described above. As a control, parallel cell culture was treated with or without 10 μm sodium orthovanadate, a dynein inhibitor (44) (New England Biolabs, Ipswich, MA) for 1 h at 37 °C and then processed after a 6-h culture.
Confocal Fluorescence Microscopy
Cells were spotted on poly-l-lysine-coated coverslips placed in a 6-well plate, rinsed with PBS, and immediately fixed with 4% paraformaldehyde/PBS for 15 min at room temperature. After being incubated with primary Abs at 37 °C for 1 h, cells were washed three times with PBST and stained with DyLight 488- or DyLight 594-conjugated corresponding secondary Abs. After a final wash in PBST, cells were mounted in ProLong Gold antifade reagent with DAPI (Invitrogen). Stained cells were viewed under a Zeiss 710 Meta Confocal Laser Scanning Microscope performed at the Confocal Microscopy Core Facility of the University of Nebraska Medical Center. For some pictures confocal z-sections covering a depth of 3.5 μm were recorded and combined into a single image, and processed by three-dimensional rendering using Zen2010 and Image J software.
Images of the Golgi stacks were collected at 0.35-μm intervals from the basolateral to apical region using an emission filter of a 505 to 550 nm band pass for green, and a 575 to 615 nm band pass for red. This approach allows us to analyze multiple focal planes and provides a complete picture of the co-distribution of targeted proteins instead of a constrained analysis of one z-section. The overlap of two proteins on each section was analyzed by Mander's coefficient for green (M1) and red (M2) separately. Colocalization of C2GnT1 and GOLPH3 was analyzed in an average of 10 z-sections/cell in 30 different cells from three independent experiments.
Statistical Analysis
All statistical comparisons were conducted using two-sided t test procedure. SigmaPlot software (Systat Software, San Jose, CA) was used for all the statistical analyses and graphing. Unless otherwise indicated in the text, data are expressed as mean ± S.E. Statistical significance was set at p < 0.05.
RESULTS
Yeast Two-hybrid Screen Identifies GOLPH3 as a C2GnT1 Interacting Protein
Golgi glycosyltransferases (GTs) are retained in the Golgi apparatus by interacting with individual GT-specific proteins via their CTs (31, 45–47). To identify the Golgi-retention partner of C2GnT1, a human universal cDNA library in a yeast two-hybrid system was screened using a CT-containing N-terminal peptide of C2GnT1 as the bait. We found no evidence of self-activation of reporter expression in cells transformed with vectors containing either C2GnT1 (1–17 aa) or GOLPH3 cDNA. The C2GnT1 CT interacting proteins identified are listed in supplemental data Table S1. We chose GOLPH3 for further characterization because of the following reasons. First, GOLPH3 is a protein associated with the Golgi (23). Second, GOLPH3 is a human ortholog of yeast Vps74p, which retains yeast mannosyltransferases in the Golgi (30). Third, the CT of C2GnT1 contains a LLRRR9 sequence, which matches the consensus Vps74p recognition sequence (F/L)-(L/I/V)-X-X-(R/K) (31) found in the CTs of yeast mannosyltransferases.
GOLPH3 Interacts with C2GnT1 via the CT
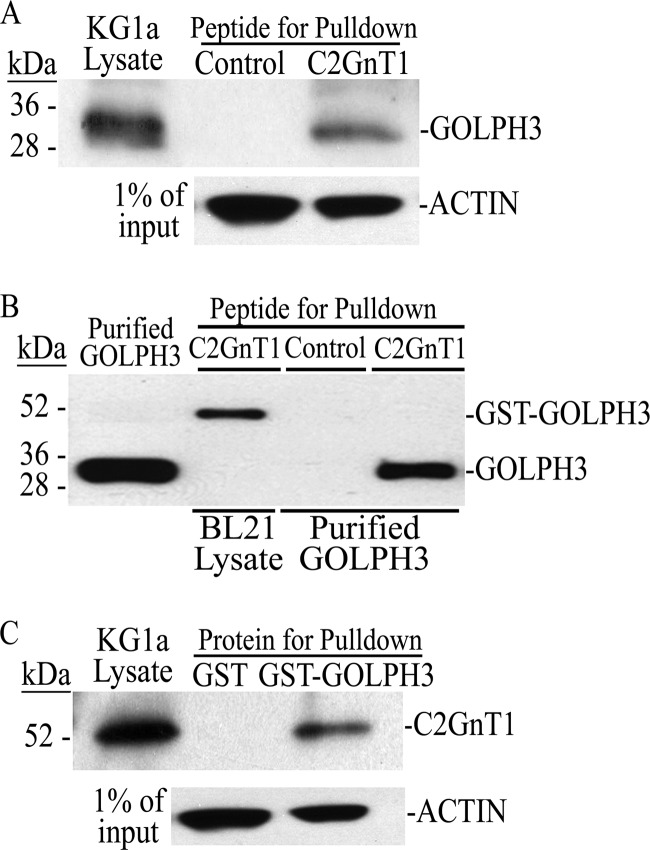
We utilized a biotinylated 21-amino acid peptide that contains the C2GnT1 CT to perform a GOLPH3 pulldown experiment. We found that this C2GnT1 peptide pulled down GOLPH3 from the KG1a cell lysate (Fig. 1A). To determine whether this C2GnT1 peptide directly interacted with GOLPH3, we performed a recombinant GOLPH3 protein pulldown assay using same peptide. As shown in Fig. 1B, this C2GnT1 peptide pulled down purified GOLPH3 as well as GST-GOLPH3 from the lysate of BL21 bacteria that produce GST-GOLPH3. The control peptide did not pull down purified GOLPH3. To further confirm this interaction in mammalian cells, we performed a C2GnT1 pulldown experiment using GST-GOLPH3. As shown in Fig. 1C, GST-GOLPH3 but not GST (negative control), pulled down C2GnT1 from the KG1a cell lysate.
FIGURE 1.
Interaction of GOLPH3 with C2GnT1 CT in vitro. A, GOLPH3 Western blot of the pulldown from KG1a cell lysate with biotinylated control or C2GnT1 1–20-aa peptide. B, GOLPH3 Western blot analysis of the pulldown from the lysate of BL21 bacteria transformed with GST-GOLPH3 cDNA and from purified recombinant GOLPH3 using biotinylated control or C2GnT1(1–20 aa) peptide. C, C2GnT1 Western blot analysis of the pulldown from KG1a cell lysate using GST (control) or GOLPH3-GST immobilized on glutathione-resin.
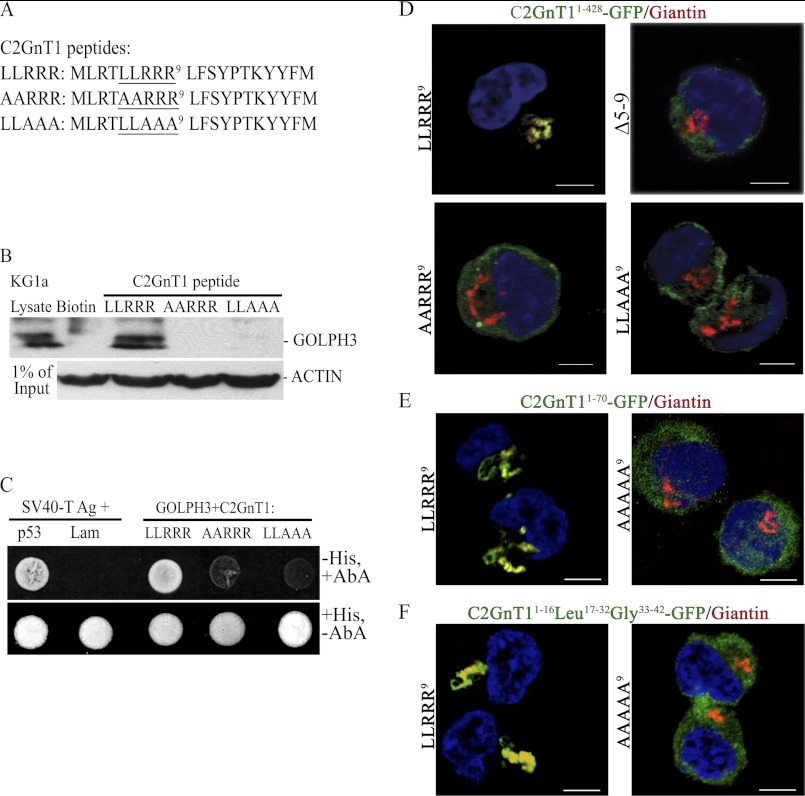
Because the LLRRR9 motif is likely responsible for the interaction of C2GnT1 CT with GOLPH3 based on published results (31), we proceeded to mutate LL6 or RRR9 in the peptide to AA6 or AAA9, respectively (Fig. 2A), and examined the ability of these peptides to interact with GOLPH3 by pulldown and yeast two-hybrid analysis. We found that neither mutated peptide could pull down GOLPH3 from the KG1a cell lysate (Fig. 2B). In addition, neither of these two mutant peptides could bind to GOLPH3, resulting in failure of the yeasts that carried either mutant to grow in media devoid of His but containing aureobasidin A (Fig. 2C). The essential role the interaction of C2GnT1 CT with GOLPH3 plays in Golgi retention of C2GnT1 is shown in Fig. 2D. The C2GnT1 with wild-type CT was localized to the Golgi, but the C2GnT1 with a 5–9-aa deletion, or AA6 or AAA9 mutant was not localized to the Golgi (Fig. 2D). Furthermore, GFP fused with the N-terminal 70 aa of C2GnT1 was localized to the Golgi, but the mutant (AAAAA9) was not (Fig. 2E), indicating that the Golgi localization signal of C2GnT1 resides in this region, especially amino acids 5–9. To address the question of whether the CT itself is not only necessary but also sufficient to localize C2GnT1 to the Golgi, we examined whether a fusion protein consisting of C2GnT1 CT1–16-Leu17–32-Gly33–42-GFP could be localized to the Golgi. As shown in Fig. 2F, this fusion protein was localized to the Golgi, whereas the same protein with mutated CT (AAAAA9) was not.
FIGURE 2.
Identification of the amino acids in the C2GnT1 CT critical for binding to GOLPH3. A, sequences of the biotinylated C2GnT1 peptides used for the experiment; wild type (1–20) and mutants (Ala5,6 and Ala7–9). B, GOLPH3 Western blot analysis of the pulldown from KG1a lysates using biotin (control), biotinylated C2GnT1 1–20-aa peptide, and Ala5,6 and Ala7–9 mutants shown in A. C, forward yeast two-hybrid assay between Gal4(DBD)-C2GnT1(LLRRR9), Gal4(DBD)-C2GnT1 (AARRR9) or Gal4(DBD)-C2GnT1(LLAAA9), and Gal4(AD)-GOLPH3. Yeasts carrying the mutant plasmids did not grow (i.e. failure to interact with GOLPH3) in media lacking His but containing aureobasidin A. SV40 large T antigen and P53 are the positive controls and SV40 large T antigen and Lam are the negative controls. D, confocal fluorescence images of K562 cells transfected with various plasmid constructs showing differential Golgi localization of GFP: GFP is localized to the Golgi with the C2GnT1(1–428)-GFP construct but not the same construct with mutated CT such as deletion of amino acids 5–9, and mutation of AA6 or AAA9. E, GFP is localized to the Golgi using C2GnT11–70-GFP construct, but not the same construct with mutated CT (AAAAA9). E, GFP was localized to the Golgi using C2GnT1CT1–16-Leu17–32-Gly33–42-GFP construct but not the same construct with mutated CT (AAAAA9). Bar = 5 μm.
GOLPH3 Colocalizes with C2GnT1 in the Golgi of KG1a Cells
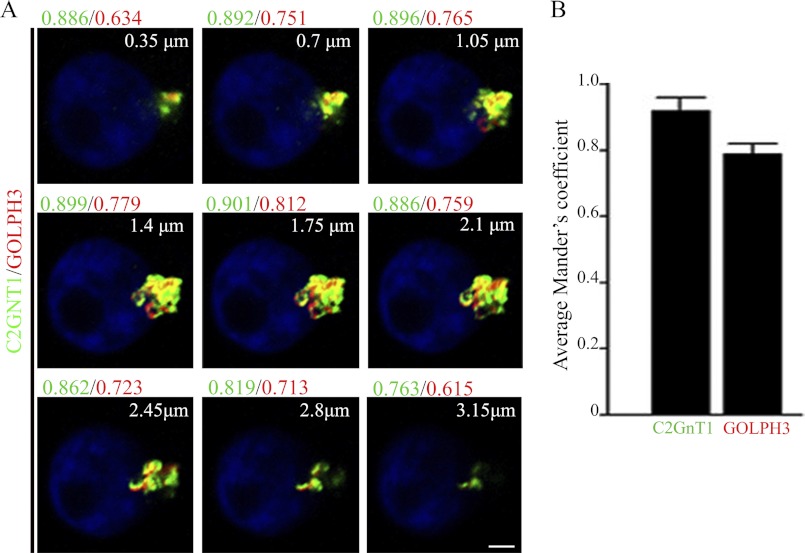
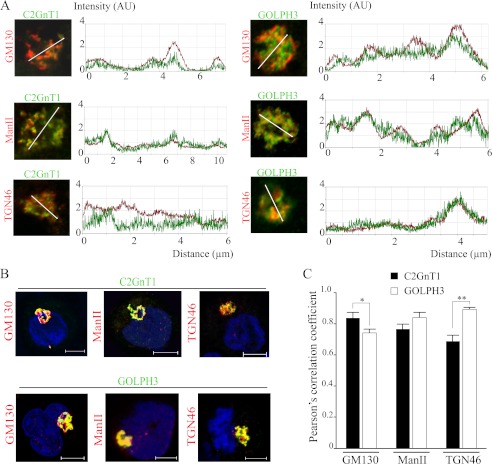
C2GnT1 has been detected in cis/medial Golgi (22), while GOLPH3 was localized to cis and medial as well as trans-Golgi (23). Based on our results, we predicted that C2GnT1 would colocalize with GOLPH3 in the Golgi although the extent of their colocalization remains to be determined. We analyzed the colocalization of C2GnT1 and GOLPH3 across the entire Golgi by confocal fluorescence microscopy (Fig. 3A). We found that the Mander's coefficient, which was an average of all z-sections where Golgi staining of both proteins was present, was 0.92 ± 0.03 for C2GnT1 and 0.79 ± 0.04 for GOLPH3 (Fig. 3B), whereas the degree of the Golgi overlap of these two proteins differed at different z-sections of the Golgi (Fig. 3A). We found that GOLPH3 overlapped more with C2GnT1 in the middle z-sections toward the basal end than other z-sections of the Golgi. In addition, there was a difference in the relative distribution of these two proteins as compared with known markers of different sub-Golgi compartments (Figs. 4, A–C). We found a higher degree of colocalization of a cis-Golgi protein GM130 (48) with C2GnT1 than with GOLPH3 (0.84 ± 0.04 versus 0.74 ± 0.02, respectively, p < 0.01), but colocalization of GOLPH3 and C2GnT1 with a medial Golgi enzyme ManII was not different statistically (0.78 ± 0.03 versus 0.84 ± 0.04, respectively, p > 0.05). Our data also indicate partial co-localization of C2GnT1 with a trans-Golgi marker TGN46, whereas an extensive colocalization of GOLPH3 with TGN46 (0.69 ± 0.03 versus 0.89 ± 0.01, respectively, p < 0.001). Therefore, GOLPH3 exhibited an increasing trend of colocalization with these markers from the cis-Golgi marker (GM130), medial Golgi marker (ManII) to trans-Golgi marker (TGN46). The trend was reversed for C2GnT1 colocalization with these Golgi markers.
FIGURE 3.
Analysis of the overlap of C2GnT1 with GOLPH3 in the Golgi of KG1a cells. A, overlap images of C2GnT1 (green) and GOLPH3 (red) on each Golgi z-section. The Mander's coefficient for green (C2GnT1) and red (GOLPH3) at each z-section is shown. The z-sections were obtained in 0.35-μm increments along the z axis (microscope optical axis) in a basal to apical direction. The Mander's coefficient for each z-section across the entire Golgi obtained from one representative cell is shown. B, average Mander's coefficients (mean ± S.E.) for C2GnT1 and GOLPH3 were analyzed for an average of 10 z-sections/cell in 30 different cells from three independent experiments. Bar = 2 μm.
FIGURE 4.
Colocalization of C2GnT1 and GOLPH3 with markers of various Golgi compartments in KG1a cells. A, the Golgi line scan graphs showing the immunofluorescence intensity of C2GnT1 (left panel) and GOLPH3 (right panel) with cis-Golgi GM130, medial Golgi ManII, and trans-Golgi TGN46 along randomly positioned arrowed lines spanning the Golgi. The intensity profiles shown are representatives of 30 cells from three independent experiments. B, three-dimensional rendering of confocal fluorescence images showing the Golgi colocalization of C2GnT1 (top panel) and GOLPH3 (bottom panel) with GM130, ManII, and TGN46. C, Pearson's correlation coefficients for the colocalization of C2GnT1 or GOLPH3 with Golgi markers; *, p < 0.01; **, p < 0.001. Bar = 5 μm.
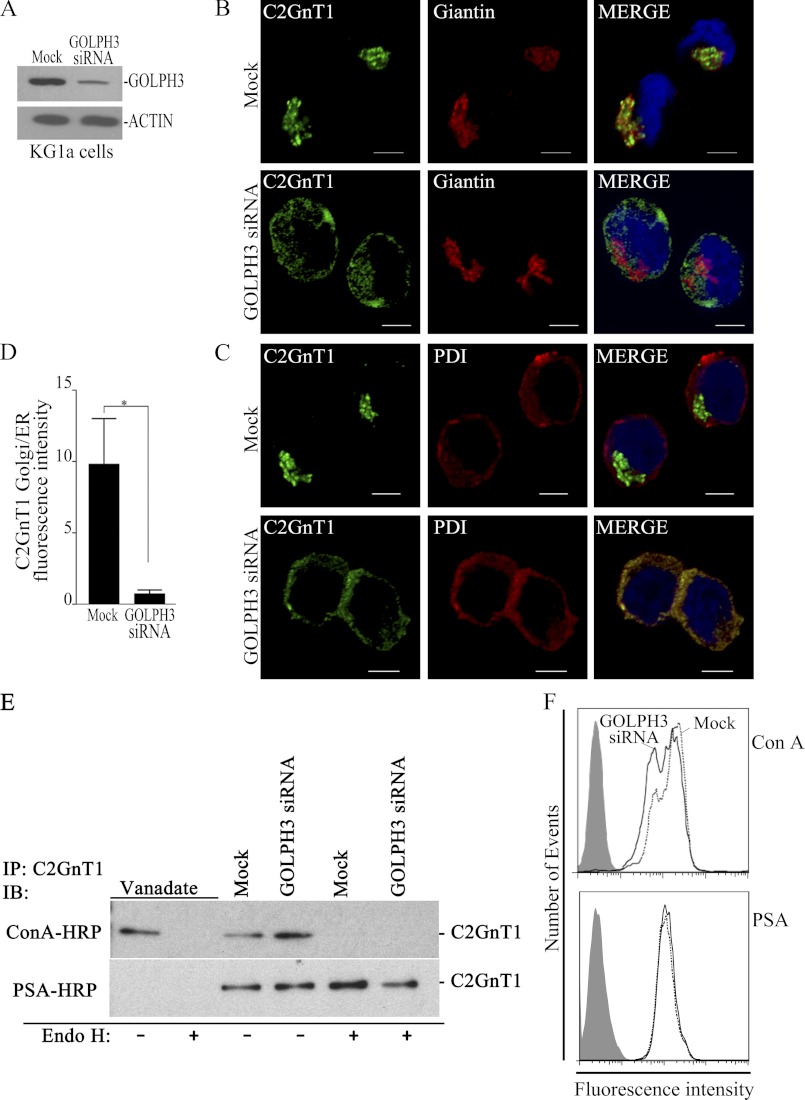
GOLPH3 Retains C2GnT1 in the Golgi of KG1a and K562 Cells
To delineate the functional significance of GOLPH3-mediated Golgi localization of C2GnT1, KG1a cells were treated with siRNAs to knockdown GOLPH3. As depicted in Fig. 5A, GOLPH3 depletion efficacy ranged from 80 to 90%. Knockdown of GOLPH3 mRNA did not have an effect on the morphological integrity of the Golgi as determined by Giantin staining (Fig. 5B) but resulted in localization of C2GnT1 to the ER as revealed by its colocalization with an ER marker, protein-disulfide isomerase (Fig. 5C). A significant decrease of fluorescence intensity of C2GnT1 in the Golgi relative to that in the ER of GOLPH3-depleted versus non-depleted cells was found (Fig. 5D). To discern whether the C2GnT1 detected in the ER of the GOLPH3-depleted cells represents the enzyme that fails to exit the ER or the enzyme that has been transported to the Golgi but fails to be retained at the Golgi, we proceeded to determine whether the enzyme in the GOLPH3-depleted cells was decorated with high mannose or complex-type glycans. As shown in Fig. 5E, the N-glycans on C2GnT1 in the control or GOLPH3 siRNA-treated cells were resistant to endoglycosidase H and detected with PSA lectin, which suggests that C2GnT1 in the GOLPH3-depleted cells was decorated with complex-type N-glycans terminated with GlcNAc. The result indicates that the enzyme has been transported to the Golgi and its high mannose glycans have been processed to the complex-type, but it could not be retained in the Golgi. Fig. 5F shows that there was no apparent change in N-glycans in GOLPH3-depleted cells. Furthermore, depletion of GOLPH3 did not affect Golgi localization of C1GalT1 or ST3GALT1, or cell surface glycans detected with DSA, PNA, Jackalin, or VVA lectins (supplemental Fig. S2), suggesting that the effect of GOLPH3 knockdown is specific to C2GnT1.
FIGURE 5.
GOLPH3 knockdown prevents Golgi localization of C2GnT1 without affecting the Golgi morphology, and analysis of the N-glycan types on C2GnT1 in KG1a cells treated with vanadate, water control, non-targeting or GOLPH3-specific siRNAs. A, GOLPH3 Western blot showing about 80–90% depletion of GOLPH3 in KG1a cells after transfection with GOLPH3 siRNAs as compared with cells transfected with non-targeting siRNAs (Mock). B and C, confocal fluorescence images of KG1a cells showing localization of C2GnT1 to the Golgi (Giantin) but distribution of C2GnT1 to the ER (protein-disulfide isomerase, PDI) after treatment with GOLPH3 siRNAs. D, quantification of the C2GnT1 fluorescence intensity in the ER relative to that of the Golgi in non-targeting or GOLPH3 siRNA-treated cells (n ≥ 25, bar = 5 μm). *, p < 0.01. E, ConA and PSA blots of the KG1a cell lysates with and without endoglycosidase H (Endo H) treatment. These KG1a cells have been treated with vanadate (inhibitor of ER-to-Golgi transport), water, non-targeting siRNAs (Mock), or GOLPH3 siRNAs. F, FACS analysis of KG1a cells after treated with fluorescein-labeled ConA or PSA lectin. Cells transfected with non-targeting siRNA (Mock), dotted line; or GOLPH3 siRNA, solid line. ConA binds high mannose-type N-glycans preferentially, PSA binds bi- and triantennary complex type N-glycans terminated with GlcNAc.
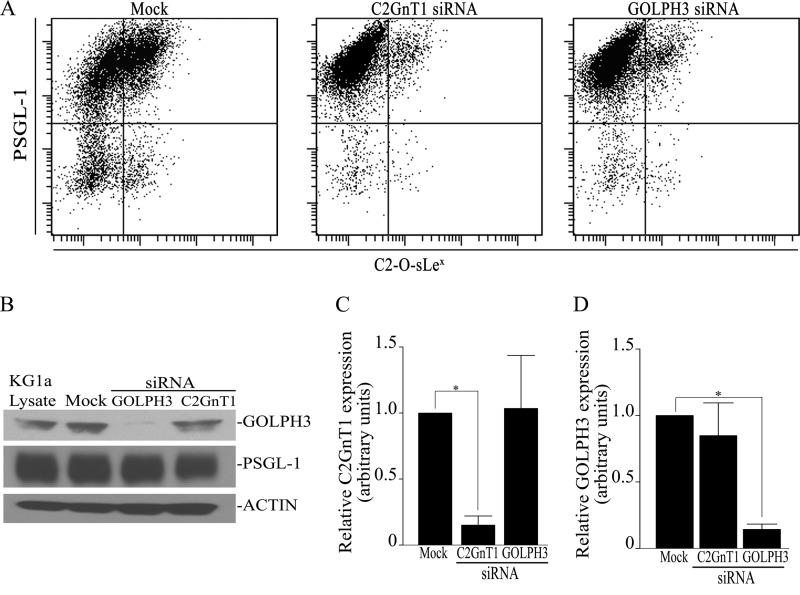
GOLPH3 Controls the Production of C2-O-sLex in KG1a Cells
Because altered Golgi localization of C2GnT1 decreases the synthesis of branched O-glycan (22), we hypothesized that knockdown of GOLPH3 would affect the expression of C2-O-sLex on the surface of KG1a cells. As shown in Fig. 6A, following treatment with GOLPH3 siRNA, there was an 81.9% decrease in PSGL-1-associated C2-O-sLex (40.6 ± 9.7 versus 7.33 ± 1.37%). Similarly, following the treatment with C2GnT1 siRNA, there was a 74.6% decrease in PSGL-1-associated C2-O-sLex (40.6 ± 9.7 versus 10.3 ± 1.10%). The decrease in C2-O-sLex, however, was not due to reduced PSGL-1 protein expression (Fig. 6B). Furthermore, depletion of C2GnT1 had no effect on the mRNA expression level of GOLPH3 and vice versa (Fig. 6, C and D), which suggests that the failure of Golgi targeting of C2GnT1, and not decrease in the PSGL-1 protein level, is responsible for the reduction of C2-O-sLex on PSGL-1. The results clearly show the requirement of Golgi localization of C2GnT1 for the synthesis of C2-O-sLex.
FIGURE 6.
GOLPH3 knockdown decreases PSGL-1-associated C2-O-sLex. A, KG1a cells were transfected with either mock (non-targeting) siRNA, GOLPH3 siRNAs, or C2GnT1 siRNA and cultured for 48–72 h. Cell surface C2-O-sLex and PSGL-1 were analyzed by flow cytometry using mouse anti-C2-O-sLex (CHO-131) IgM and F(ab)2 fragment of goat anti-mouse IgM fluorescein isothiocyanate-conjugated secondary Abs, and rabbit anti-PSGL-1 Abs and F(ab)2 fragment of goat anti-rabbit phycoerythrin-conjugated secondary Abs. GOLPH3 or C2GnT1 siRNA treatment reduced PSGL-1-associated C2-O-sLex from 40.6 to 10.3 and 7.3%, respectively. B, GOLPH3 or C2GnT1 knockdown did not affect the PSGL-1 protein level. C and D, real-time quantitative PCR analysis of C2GnT1 and GOLPH3 in KG1a cells transfected with C2GnT1 or GOLPH3 siRNA, respectively. Knockdown of the mRNA of either gene did not affect the expression level of the other.
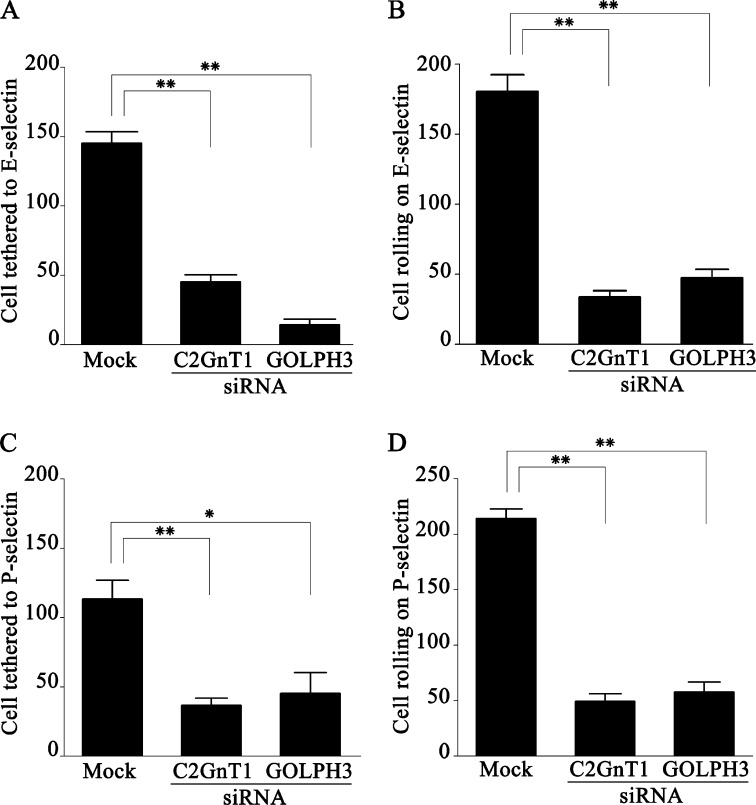
GOLPH3 Controls KG1a Cell Tethering and Rolling on Immobilized Selectins
Because C2-O-sLex is a ligand involved in selectin-mediated leukocyte trafficking (10, 11), we proceeded to examine the effect of knocking down GOLPH3 or C2GnT1 mRNA in KG1a cells on their interaction with immobilized P- or E-selectin in vitro under dynamic flow conditions. We found that knockdown of either C2GnT1 or GOLPH3 in KG1a cells significantly reduced their tethering to and rolling on immobilized E- and P-selectins (Fig. 7). As compared with mock (non-targeting) siRNA-treated cells, the cells treated with siRNA of either C2GnT1 or GOLPH3 exhibited significant reductions in tethering to E-selectin by 68.9 and 90.3% (Fig. 7A) and P-selectin by 67.2 and 60.1% (Fig. 7C), and rolling on E-selectin by 81.1 and 73.8% (Fig. 7B) and P-selectin by 77.1 and 72.9% (Fig. 7D), respectively. The results indicate that GOLPH3 controls the selectin-mediated cell tethering and rolling.
FIGURE 7.
C2GnT1 or GOLPH3 knockdown reduces interaction of KG1a cells with immobilized E- or P- selectin under dynamic flow. C2GnT1 siRNA, GOLPH3 siRNA, or non-targeting siRNA (mock) treated cells (2.5 × 105 cells/ml) were perfused through a parallel-plate flow chamber containing a coverslip coated with E- or P-selectin at a constant wall shear stress of 1.0 dyne/cm2. Tethering or rolling was visualized with a phase-contrast microscope at ×40 magnification and videotaped. A, compared with mock treated cells, C2GnT1 or GOLPH3 siRNA-treated cells that were tethered to immobilized E-selectin were reduced by 68.9 (145 ± 8 versus 45 ± 5 cells, p < 0.001) and 90.3% (145 ± 8 versus 14 ± 4 cells, p < 0.001), respectively. B, the C2GnT1 or GOLPH3 siRNA-treated cells that rolled on immobilized E-selectin were reduced by 81.1 (180 ± 12 versus 34 ± 5 cells, p < 0.001) and 73.8% (180 ± 12 versus 47 ± 6 cells, p < 0.001), respectively. C, C2GnT1 or GOLPH3 siRNA-treated cells that were tethered to immobilized P-selectin were reduced by 67.2 (113 ± 14 versus 37 ± 5 cells, p < 0.001) and 60.1% (113 ± 14 versus 45 ± 15 cells, p < 0.01), respectively. D, C2GnT1 or GOLPH3 siRNA-treated cells that rolled on immobilized P-selectin were reduced by 77.1 (214 ± 9 versus 49 ± 7 cells, p < 0.001) and 72.9% (214 ± 9 versus 58 ± 9 cells, p < 0.001), respectively.
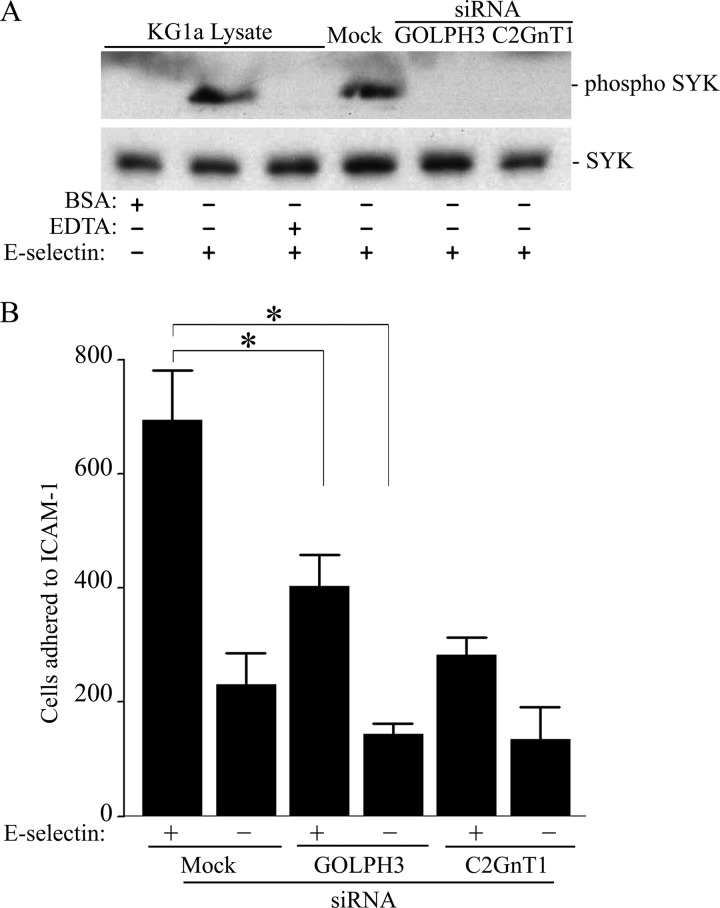
GOLPH3 Controls the Adhesion of E-selectin-activated KG1a Cells to Immobilized ICAM-1
The interaction of P- or E-selectin present on the surface of activated endothelial cells with the selectin ligands on circulating leukocytes leads to phosphorylation of spleen tyrosine kinase (SYK) and activation of β2-integrin (49–51), and then firm adherence to ICAM-1 expressed on activated endothelial cells (52) to facilitate extravasation of these leukocytes. To examine if GOLPH3 was involved in this process, we measured the effect of GOLPH3 knockdown on the activation of SYK and adhesion of E-selectin-treated KG1a cells to immobilized ICAM-1 in a flow chamber. As shown in Fig. 8, the depletion of either GOLPH3 or C2GnT1 prevented the phosphorylation of SYK in KG1a cells activated with E-selectin (Fig. 8A), and consequently, decreased the number of the E-selectin-activated KG1a cells adhered to ICAM-1 under dynamic flow conditions by 41.9 (694 ± 86 versus 403 ± 56 cells; p < 0.05) and 59.2% (694 ± 86 versus 283 ± 30 cells; p < 0.01), respectively (Fig. 8B). The result indicates that GOLPH3 also controls ICAM-1-mediated adhesion of cells activated by E-selectin.
FIGURE 8.
GOLPH3 or C2GnT1 knockdown prevents activation of SYK and reduces the adhesion of E-selectin-treated KG1a cells to immobilized ICAM-1 under dynamic flow. A, EDTA, C2GnT1 siRNA, GOLPH3 siRNA, or non-targeting siRNA (mock)-treated cells were incubated with E-selectin for 20 min. Whole cell lysates were analyzed by Western blotting for phosphorylated SYK. EDTA inhibits phosphorylation of SYK and BSA does not induce SYK phosphorylation. B, C2GnT1 siRNA, GOLPH3 siRNA, or non-targeting siRNA (mock)-treated cells (2.5 × 105 cells/ml) were incubated with E-selectin for 20 min before being perfused through a parallel plate flow chamber containing a coverslip coated with ICAM-1 at a constant wall shear stress of 1.0 dyne/cm2. Compared with mock treated cells, GOLPH3 or C2GnT1-depleted and E-selectin-activated KG1a cells exhibited a decrease in their adherence to immobilized ICAM-1 by 41.9 (694 ± 86 versus 403 ± 56, p < 0.05) and 59.2% (694 ± 86 versus 283 ± 30, p < 0.01), respectively.
DISCUSSION
C2GnT1 is responsible for the synthesis of the mucin core 2 structure on which sLex is decorated (8). The C2-O-sLex formed serves as an important selectin ligand to direct leukocyte trafficking and cancer metastasis (21, 53). The localization of C2GnT1 at its Golgi compartment is required for this enzyme to carry out its glycosylation function (22). As described in this article, we have identified GOLPH3 as the protein that interacts with C2GnT1 via a specific sequence in the CT to localize the enzyme to the Golgi to enable the enzyme to perform its glycosylation function. We show that GOLPH3 colocalizes with C2GnT1 in the Golgi, and controls the formation of C2-O-sLex, cell tethering to and rolling on immobilized selectins, and adhesion of E-selectin-activated cells to ICAM-1. Therefore, GOLPH3 regulates cell tethering, rolling, and adhesion by controlling Golgi localization of C2GnT1.
It has been well recognized that the Golgi retention signals of GTs reside in the N-terminal region consisted of a short CT, a short transmembrane domain, and a stem. Although the stem and transmembrane domain each can contribute to the retention of GTs, they are limited to only few GTs (54, 55). A growing body of evidence in the literature indicates that Golgi localization of a GT is determined primarily by its CT through interaction with an individual GT-specific cytoplasmic protein (31, 45–47). To date, only a few Golgi GTs have their cytoplasmic binding partners identified. The formation of complexes between a GT and a cytosolic protein can have several important biological implications. First, it helps secure GTs to the Golgi because a GT without its tail is no longer retained in the Golgi (Fig. 2D) (56). The binding of yeast Vps74p protein with the CTs of mannosyltransferases is such an example (31). The current study provides another example showing that GOLPH3 retains C2GnT1 at the Golgi by binding to its CT. Second, it stabilizes the Golgi structure. This is represented by the binding of β4GalT to cytoskeletal proteins such as α- and β-tubulins (57). Third, it provides a link of the Golgi to important cellular activities, such as signaling and gene regulation. The Src-suppressed C kinase substrate, a modulator of the cyto-architecture, has been shown to interact with the CT of a galactosyltransferase (58). Furthermore, binding of GA2/GM2/GD2 β3-galactosyltransferase with calsenilin (59) via the CT has linked this GT to modulation of genes involved in Ca2+ regulation (60) and pancreatic prodynorphin expression (61). Adding to this list is our current finding showing regulation of cell binding properties via binding of GOLPH3 to the CT of C2GnT1. Fourth, the binding of a Golgi GT with non-muscle myosin IIA via its CT facilitates the recycling of this Golgi resident protein, a mechanism responsible for Golgi remodeling (62). Therefore, the CT of a GT can interact with cellular proteins to retain the GT at the Golgi and engage in many other cellular functions.
Many functions have been elucidated for GOLPH3 since its identification as a Golgi-associated protein (23). They include regulating the mitochondrial mass by controlling the biosynthesis of cardiolipin, a mitochondria-specific phospholipid (27), promoting cell survival by enhancing growth factor-induced mammalian target of rapamycin signaling pathway in human cancer cells and altering the response to an mammalian target of rapamycin inhibitor in vivo (24, 63), inducing vesicle budding from the Golgi by bridging phosphatidylinositol 4-phosphate in the Golgi and the nonconventional myosin MYO18A linked to the actin cytoskeleton (28, 64), and mediating retrograde transport of transmembrane receptors and other cargo proteins back to the Golgi as it has been reported to interact with Vps29-Vps35-Vps26, the selective subcomplex of retromer (24, 29). The original function of its yeast ortholog Vps74p as the protein that retains mannosyltransferases in the Golgi by interacting with a consensus amino acid sequence, (F/L)(L/I/V)XX(R/K) (31), present in the CTs of these enzymes (30) has never been confirmed in mammalian cells until now. Our current work shows that GOLPH3 binds to a similar sequence in the CT of C2GnT1 to retain the enzyme in the Golgi. The consensus amino acid sequence recognized by GOLPH3 is also present in three other Golgi GTs, including GALNT6 (MRLLRRRH) (NP_009141), ST3GAL3 (MGLLVFVRN) (NP_006270), and B3GNT5 (MRMLVSGRRVKKWQ) (NP_ 114436). The significance of this observation remains to be established.
The reports about the sub-Golgi localization of GOLPH3 in the literature have been inconsistent. GOLPH3 was initially (23) shown to be present at both cis- and trans-Golgi by immunoelectron microscopy and to colocalize with a medial Golgi marker MG160 (65) by confocal fluorescence microscopy, then was assigned as a trans-Golgi-associated protein in subsequent reports (24, 27, 28, 64, 66). To gain a comprehensive insight of the sub-Golgi localization of C2GnT1 and GOLPH3, we carried out a colocalization analysis of these two proteins with well established Golgi subcompartment markers that selectively reside across the entire Golgi apparatus (67). We have found that both GOLPH3 and C2GnT1 are distributed over the entire Golgi with GOLPH3 having a wider distribution than C2GnT1. However, GOLPH3 is the most abundant at the trans-Golgi and the least abundant at the cis-Golgi. The opposite is found for C2GnT1. The wider Golgi distribution of GOLPH3 compared with that of C2GnT1, especially at the trans-Golgi, is consistent with a known function of GOLPH3 at the trans-Golgi, i.e. the interaction of highly enriched phosphatidylinositol 4-phosphate present at the trans-Golgi (28, 64). The preferential distribution of C2GnT1 in the cis-medial Golgi is also consistent with its role in the formation of the core 2 structure in mucin-type glycans. Although in theory all C2GnT1 is expected to colocalize with GOLPH3 because Golgi localization of C2GnT1 requires its binding with GOLPH3, some C2GnT1 was not colocalized with GOLPH3. This is not unexpected because this fraction of C2GnT1 consists of at least two other populations of this enzyme. The first one represents those that bind to non-muscle myosin IIA, which we have shown to colocalize with C2GnT2 at the periphery of the Golgi and be responsible for the recycling of this Golgi enzyme (62). Our preliminary study also shows that C2GnT1 colocalizes with non-muscle myosin IIA at the Golgi,4 suggesting that non-muscle myosin IIA is also involved in the recycling of C2GnT1. The second population represents those that have arrived at the Golgi and their N-glycans are still undergoing processing.
Leukocytes are known to produce C2-O-sLex following activation of the C2GnT1 gene by cytokines (68, 69). These leukocytes are then arrested by the endothelium at the inflamed sites via interactions of P- and E-selectins with the C2-O-sLex present on PSGL-1. Such interactions trigger phosphorylation of SYK (51), which leads to activation of β2-integrin, enhancement of leukocyte adhesion to ICAM-1 (52) (Fig. 9), and their extravasation to the inflamed site (12). The formation of the core 2 branch and subsequent C2-O-sLex depends on not only the amounts of C2GnT1 protein but also its Golgi localization as shown in our study and those of others (22). Because the CT of C2GnT1 interacts directly with GOLPH3, depletion of GOLPH3 prevents the Golgi localization of C2GnT1 and renders the cells unable to generate C2-O-sLex on PSGL-1. Consequently, the binding of PSGL-1-associated C2-O-sLex on leukemic cells to E-selectin, which activates a signaling cascade that culminates in increased adherence of these cells to ICAM-1 present on the surface of endothelial cells, is compromised. Therefore, we conclude that GOLPH3 regulates cell tethering, rolling, and adhesion properties by controlling the Golgi localization of C2GnT1. Similar results were obtained in C2GnT1-depleted cells. These results corroborate with a previous finding in C2GnT1-depleted KM3 human leukemia cells (70).
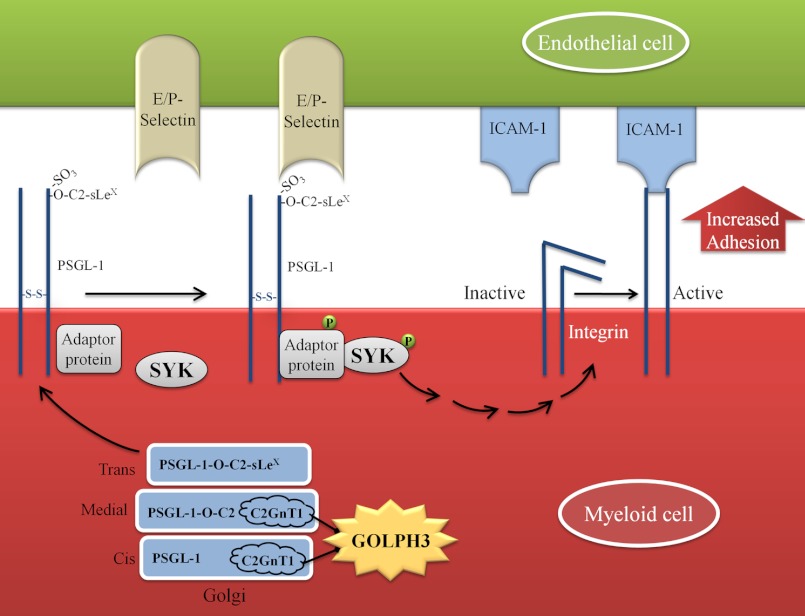
FIGURE 9.
A diagram depicting the function of GOLPH3 in retaining C2GnT1 in the Golgi, and promoting the formation of C2-O-sLex and subsequent interaction with P/E-selectin and ICAM1. GOLPH3 binds to the CT of C2GnT1 to retain this enzyme in the Golgi to synthesize the core 2 structure, which enables the synthesis of selectin ligand (C2-O-sLex) on PSGL-1 to facilitate the interaction of the KG1a cells with P/E-selectin. This interaction results in phosphorylation of SYK, which in turn activates β2-integrin to induce a firm adhesion of the activated cells to the endothelial cells via ICAM-1.
The current study shows that knockdown of GOLPH3, or deletion or mutation of critical amino acids in the CT of C2GnT1, which are recognized by GOLPH3, all results in failure of C2GnT1 to be retained in the Golgi. The detection of complex-type N-glycans terminated with GlcNAc on C2GnT1 in these cells indicates that the C2GnT1 has been transported to the Golgi but was not retained rather than failure of C2GnT1 to exit the ER. The results suggest that processing of N-glycans on the cis/medial Golgi resident protein C2GnT1 takes place before the enzyme is retained in the Golgi by GOLPH3. It is also of interest to note that the C2GnT1 CT is both necessary and sufficient for localizing this enzyme to the Golgi because fusion of this CT to a stretch of hydrophobic amino sequence serving as a transmembrane domain followed by a stretch of glycine serving as the stem and then GFP can localize the GFP to the Golgi.
The results of this study have expanded our understanding of the role of the CT of a GT in localization of this enzyme in the Golgi. Identification of additional CT-binding proteins that are responsible for the localization of other Golgi GTs should further uncover the link of Golgi GTs to other biological processes beyond their recognized enzymatic activities.
Supplementary Material
Acknowledgments
We thank Janice A. Taylor and James R. Talaska of the Confocal Laser Scanning Microscope Core Facility at the University of Nebraska Medical Center for providing assistance in confocal microscopy, Yinbo Zhang for assistance in the production of GOLPH3 in E. coli, Dr. Jaswant Singh for critical review of the manuscript, and the Nebraska Research Initiative and the Eppley Cancer Center for their support of the Core Facilities.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R21HL097238 and 2RO1HL48282, the Office of Research and Development, Medical Research Service, Dept. of Veterans Affairs Grant VA 1I1BX000985, and State of Nebraska Grant LB506 (to P. W. C.).

This article contains supplemental Figs. S1 and S2 and Table S1.
Preliminary experiments in our laboratory show an interaction between non-muscle myosin IIA and C2GnT1 by a pulldown assay and colocalization using confocal fluorescence microscopy.
- C2-O-sLex
- core 2-associated sialyl Lewis x
- C2GnT1
- core 2 N-acetylglucosaminyltransferase 1
- CT
- cytoplasmic tail
- GT
- glycosyltransferase
- GOLPH3
- Golgi phosphoprotein 3
- PSGL-1
- P-selectin glycoprotein ligand-1
- SYK
- spleen tyrosine kinase
- ICAM-1
- intercellular adhesion molecule 1
- ConA
- C. ensiformis agglutinin
- PSA
- P. sativum agglutinin
- DSA
- D. stramonium
- PNA
- P. agglutinin
- VVA
- V. villosa agglutinin
- aa
- amino acid.
REFERENCES
- 1. van den Berg T. K., Döpp E. A., Dijkstra C. D. (2001) Rat macrophages. Membrane glycoproteins in differentiation and function. Immunol. Rev. 184, 45–57 [DOI] [PubMed] [Google Scholar]
- 2. Feizi T. (1980) Structural and biological aspects of blood group I and i antigens on glycolipids and glycoproteins. Rev. Fr. Transfus. Immunohematol. 23, 563–577 [DOI] [PubMed] [Google Scholar]
- 3. Ochs H. D., Wedgwood R. J., Heller S. R., Beatty P. G. (1986) Complement, membrane glycoproteins, and complement receptors. Their role in regulation of the immune response. Clin. Immunol. Immunopathol. 40, 94–104 [DOI] [PubMed] [Google Scholar]
- 4. Chen S., Fukuda M. (2006) Cell type-specific roles of carbohydrates in tumor metastasis. Methods Enzymol. 416, 371–380 [DOI] [PubMed] [Google Scholar]
- 5. Varki A. C. R., Esko J., Freeze H., Stanley P., Bertozzi C., Hart G., Etzler M. (2009) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 6. Cheng P. W., Radhakrishnan P. (2011) Mucin O-glycan branching enzymes. Structure, function, and gene regulation. Adv. Exp. Med. Biol. 705, 465–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beum P. V., Cheng P. W. (2001) Biosynthesis and function of β1,6-branched mucin-type glycans. Adv. Exp. Med. Biol. 491, 279–312 [DOI] [PubMed] [Google Scholar]
- 8. Nakamura M., Kudo T., Narimatsu H., Furukawa Y., Kikuchi J., Asakura S., Yang W., Iwase S., Hatake K., Miura Y. (1998) Single glycosyltransferase, core 2 β1 → 6-N-acetylglucosaminyltransferase, regulates cell surface sialyl-Lex expression level in human pre-B lymphocytic leukemia cell line KM3 treated with phorbol ester. J. Biol. Chem. 273, 26779–26789 [DOI] [PubMed] [Google Scholar]
- 9. Kumar R., Camphausen R. T., Sullivan F. X., Cumming D. A. (1996) Core 2 β1,6-N-acetylglucosaminyltransferase enzyme activity is critical for P-selectin glycoprotein ligand-1 binding to P-selectin. Blood 88, 3872–3879 [PubMed] [Google Scholar]
- 10. McEver R. P., Cummings R. D. (1997) Perspectives series. Cell adhesion in vascular biology. Role of PSGL-1 binding to selectins in leukocyte recruitment. J. Clin. Invest. 100, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li F., Wilkins P. P., Crawley S., Weinstein J., Cummings R. D., McEver R. P. (1996) Post-translational modifications of recombinant P-selectin glycoprotein ligand-1 required for binding to P- and E-selectin. J. Biol. Chem. 271, 3255–3264 [PubMed] [Google Scholar]
- 12. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Getting to the site of inflammation. The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 13. Ellies L. G., Tsuboi S., Petryniak B., Lowe J. B., Fukuda M., Marth J. D. (1998) Core 2 oligosaccharide biosynthesis distinguishes between selectin ligands essential for leukocyte homing and inflammation. Immunity 9, 881–890 [DOI] [PubMed] [Google Scholar]
- 14. Sperandio M., Thatte A., Foy D., Ellies L. G., Marth J. D., Ley K. (2001) Severe impairment of leukocyte rolling in venules of core 2 glucosaminyltransferase-deficient mice. Blood 97, 3812–3819 [DOI] [PubMed] [Google Scholar]
- 15. Piller F., Le Deist F., Weinberg K. I., Parkman R., Fukuda M. (1991) Altered O-glycan synthesis in lymphocytes from patients with Wiskott-Aldrich syndrome. J. Exp. Med. 173, 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Higgins E. A., Siminovitch K. A., Zhuang D. L., Brockhausen I., Dennis J. W. (1991) Aberrant O-linked oligosaccharide biosynthesis in lymphocytes and platelets from patients with the Wiskott-Aldrich syndrome. J. Biol. Chem. 266, 6280–6290 [PubMed] [Google Scholar]
- 17. Park J. K., Rosenstein Y. J., Remold-O'Donnell E., Bierer B. E., Rosen F. S., Burakoff S. J. (1991) Enhancement of T-cell activation by the CD43 molecule whose expression is defective in Wiskott-Aldrich syndrome. Nature 350, 706–709 [DOI] [PubMed] [Google Scholar]
- 18. Tsuboi S., Fukuda M. (1997) Branched O-linked oligosaccharides ectopically expressed in transgenic mice reduce primary T-cell immune responses. EMBO J. 16, 6364–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsuboi S., Fukuda M. (1998) Overexpression of branched O-linked oligosaccharides on T cell surface glycoproteins impairs humoral immune responses in transgenic mice. J. Biol. Chem. 273, 30680–30687 [DOI] [PubMed] [Google Scholar]
- 20. Beum P. V., Singh J., Burdick M., Hollingsworth M. A., Cheng P. W. (1999) Expression of core 2 β-1,6-N-acetylglucosaminyltransferase in a human pancreatic cancer cell line results in altered expression of MUC1 tumor-associated epitopes. J. Biol. Chem. 274, 24641–24648 [DOI] [PubMed] [Google Scholar]
- 21. St. Hill C. A., Baharo-Hassan D., Farooqui M. (January 19, 2011) C2-O-sLeX glycoproteins are E-selectin ligands that regulate invasion of human colon and hepatic carcinoma cells. PLoS One doi: 10.1371/journal.pone.0016281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skrincosky D., Kain R., El-Battari A., Exner M., Kerjaschki D., Fukuda M. (1997) Altered Golgi localization of core 2 β1,6-N-acetylglucosaminyltransferase leads to decreased synthesis of branched O-glycans. J. Biol. Chem. 272, 22695–22702 [DOI] [PubMed] [Google Scholar]
- 23. Bell A. W., Ward M. A., Blackstock W. P., Freeman H. N., Choudhary J. S., Lewis A. P., Chotai D., Fazel A., Gushue J. N., Paiement J., Palcy S., Chevet E., Lafrenière-Roula M., Solari R., Thomas D. Y., Rowley A., Bergeron J. J. (2001) Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 276, 5152–5165 [DOI] [PubMed] [Google Scholar]
- 24. Scott K. L., Kabbarah O., Liang M. C., Ivanova E., Anagnostou V., Wu J., Dhakal S., Wu M., Chen S., Feinberg T., Huang J., Saci A., Widlund H. R., Fisher D. E., Xiao Y., Rimm D. L., Protopopov A., Wong K. K., Chin L. (2009) GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 459, 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kunigou O., Nagao H., Kawabata N., Ishidou Y., Nagano S., Maeda S., Komiya S., Setoguchi T. (2011) Role of GOLPH3 and GOLPH3L in the proliferation of human rhabdomyosarcoma. Oncol. Rep. 26, 1337–1342 [DOI] [PubMed] [Google Scholar]
- 26. Li X. Y., Liu W., Chen S. F., Zhang L. Q., Li X. G., Wang L. X. (2011) Expression of the Golgi phosphoprotein-3 gene in human gliomas. A pilot study. J. Neurooncol. 105, 159–163 [DOI] [PubMed] [Google Scholar]
- 27. Nakashima-Kamimura N., Asoh S., Ishibashi Y., Mukai Y., Shidara Y., Oda H., Munakata K., Goto Y., Ohta S. (2005) MIDAS/GPP34, a nuclear gene product, regulates total mitochondrial mass in response to mitochondrial dysfunction. J. Cell Sci. 118, 5357–5367 [DOI] [PubMed] [Google Scholar]
- 28. Dippold H. C., Ng M. M., Farber-Katz S. E., Lee S. K., Kerr M. L., Peterman M. C., Sim R., Wiharto P. A., Galbraith K. A., Madhavarapu S., Fuchs G. J., Meerloo T., Farquhar M. G., Zhou H., Field S. J. (2009) GOLPH3 bridges phosphatidylinositol 4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell 139, 337–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bugarcic A., Zhe Y., Kerr M. C., Griffin J., Collins B. M., Teasdale R. D. (2011) Vps26A and Vps26B subunits define distinct retromer complexes. Traffic 12, 1759–1773 [DOI] [PubMed] [Google Scholar]
- 30. Schmitz K. R., Liu J., Li S., Setty T. G., Wood C. S., Burd C. G., Ferguson K. M. (2008) Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell 14, 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tu L., Tai W. C., Chen L., Banfield D. K. (2008) Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science 321, 404–407 [DOI] [PubMed] [Google Scholar]
- 32. Walcheck B., Leppanen A., Cummings R. D., Knibbs R. N., Stoolman L. M., Alexander S. R., Mattila P. E., McEver R. P. (2002) The monoclonal antibody CHO-131 binds to a core 2 O-glycan terminated with sialyl-Lewis x, which is a functional glycan ligand for P-selectin. Blood 99, 4063–4069 [DOI] [PubMed] [Google Scholar]
- 33. Borsig L., Wong R., Hynes R. O., Varki N. M., Varki A. (2002) Synergistic effects of L- and P-selectin in facilitating tumor metastasis can involve non-mucin ligands and implicate leukocytes as enhancers of metastasis. Proc. Natl. Acad. Sci. U.S.A. 99, 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moll T., Vestweber D. (1999) Construction and purification of adhesion molecule immunoglobulin chimeric proteins. Methods Mol. Biol. 96, 77–84 [DOI] [PubMed] [Google Scholar]
- 35. Ross J. M., McIntire L. V., Moake J. L., Kuo H. J., Qian R. Q., Glanville R. W., Schwartz E., Rand J. H. (1998) Fibrillin containing elastic microfibrils support platelet adhesion under dynamic shear conditions. Thromb. Haemost. 79, 155–161 [PubMed] [Google Scholar]
- 36. Wiese G., Barthel S. R., Dimitroff C. J. (February 11, 2009) Analysis of physiologic E-selectin-mediated leukocyte rolling on microvascular endothelium. J. Vis. Exp., 10.3791/1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krüll M., Klucken A. C., Wuppermann F. N., Fuhrmann O., Magerl C., Seybold J., Hippenstiel S., Hegemann J. H., Jantos C. A., Suttorp N. (1999) Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J. Immunol. 162, 4834–4841 [PubMed] [Google Scholar]
- 38. Lawrence M. B., Springer T. A. (1991) Leukocytes roll on a selectin at physiologic flow rates. Distinction from and prerequisite for adhesion through integrins. Cell 65, 859–873 [DOI] [PubMed] [Google Scholar]
- 39. Sriramarao P., Norton C. R., Borgstrom P., DiScipio R. G., Wolitzky B. A., Broide D. H. (1996) E-selectin preferentially supports neutrophil but not eosinophil rolling under conditions of flow in vitro and in vivo. J. Immunol. 157, 4672–4680 [PubMed] [Google Scholar]
- 40. Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr. (1989) Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 41. Ohyama Y., Kasai K., Nomoto H., Inoue Y. (1985) Frontal affinity chromatography of ovalbumin glycoasparagines on a concanavalin A-Sepharose column. A quantitative study of the binding specificity of the lectin. J. Biol. Chem. 260, 6882–6887 [PubMed] [Google Scholar]
- 42. Mega T., Oku H., Hase S. (1992) Characterization of carbohydrate-binding specificity of concanavalin A by competitive binding of pyridylamino sugar chains. J. Biochem. 111, 396–400 [DOI] [PubMed] [Google Scholar]
- 43. Tateno H., Nakamura-Tsuruta S., Hirabayashi J. (2009) Comparative analysis of core-fucose-binding lectins from Lens culinaris and Pisum sativum using frontal affinity chromatography. Glycobiology 19, 527–536 [DOI] [PubMed] [Google Scholar]
- 44. Wang X. M., Huang T. H., Xie Q. D., Zhang Q. J., Ruan Y. (2004) Effect of dynein inhibitor on mouse oocyte in vitro maturation and its cyclin B1 mRNA level. Biomed. Environ. Sci. 17, 341–349 [PubMed] [Google Scholar]
- 45. Osman N., McKenzie I. F., Mouhtouris E., Sandrin M. S. (1996) Switching amino-terminal cytoplasmic domains of α-1,2-fucosyltransferase and α1,3-galactosyltransferase alters the expression of H substance and Galα1,3Gal. J. Biol. Chem. 271, 33105–33109 [DOI] [PubMed] [Google Scholar]
- 46. Uliana A. S., Giraudo C. G., Maccioni H. J. (2006) Cytoplasmic tails of SialT2 and GalNAcT impose their respective proximal and distal Golgi localization. Traffic 7, 604–612 [DOI] [PubMed] [Google Scholar]
- 47. Okamoto M., Yoko-o T., Miyakawa T., Jigami Y. (2008) The cytoplasmic region of α1,6-mannosyltransferase Mnn9p is crucial for retrograde transport from the Golgi apparatus to the endoplasmic reticulum in Saccharomyces cerevisiae. Eukaryot. Cell 7, 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. (1995) Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Urzainqui A., Serrador J. M., Viedma F., Yáñez-Mó M., Rodríguez A., Corbí A. L., Alonso-Lebrero J. L., Luque A., Deckert M., Vázquez J., Sánchez-Madrid F. (2002) ITAM-based interaction of ERM proteins with Syk mediates signaling by the leukocyte adhesion receptor PSGL-1. Immunity 17, 401–412 [DOI] [PubMed] [Google Scholar]
- 50. Yago T., Shao B., Miner J. J., Yao L., Klopocki A. G., Maeda K., Coggeshall K. M., McEver R. P. (2010) E-selectin engages PSGL-1 and CD44 through a common signaling pathway to induce integrin αLβ2-mediated slow leukocyte rolling. Blood 116, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zarbock A., Lowell C. A., Ley K. (2007) Spleen tyrosine kinase Syk is necessary for E-selectin-induced αLβ2 integrin-mediated rolling on intercellular adhesion molecule-1. Immunity 26, 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gopalan P. K., Smith C. W., Lu H., Berg E. L., McIntire L. V., Simon S. I. (1997) Neutrophil CD18-dependent arrest on intercellular adhesion molecule 1 (ICAM-1) in shear flow can be activated through L-selectin. J. Immunol. 158, 367–375 [PubMed] [Google Scholar]
- 53. Carlow D. A., Gossens K., Naus S., Veerman K. M., Seo W., Ziltener H. J. (2009) PSGL-1 function in immunity and steady state homeostasis. Immunol. Rev. 230, 75–96 [DOI] [PubMed] [Google Scholar]
- 54. Tu L., Banfield D. K. (2010) Localization of Golgi-resident glycosyltransferases. Cell. Mol. Life Sci. 67, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Colley K. J. (1997) Golgi localization of glycosyltransferases. More questions than answers. Glycobiology 7, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zerfaoui M., Fukuda M., Langlet C., Mathieu S., Suzuki M., Lombardo D., El-Battari A. (2002) The cytosolic and transmembrane domains of the β1,6 N-acetylglucosaminyltransferase (C2GnT) function as a cis to medial/Golgi-targeting determinant. Glycobiology 12, 15–24 [DOI] [PubMed] [Google Scholar]
- 57. Yamaguchi N., Fukuda M. N. (1995) Golgi retention mechanism of β1,4-galactosyltransferase. Membrane-spanning domain-dependent homodimerization and association with α- and β-tubulins. J. Biol. Chem. 270, 12170–12176 [DOI] [PubMed] [Google Scholar]
- 58. Wassler M. J., Foote C. I., Gelman I. H., Shur B. D. (2001) Functional interaction between the SSeCKS scaffolding protein and the cytoplasmic domain of β1,4-galactosyltransferase. J. Cell Sci. 114, 2291–2300 [DOI] [PubMed] [Google Scholar]
- 59. Quintero C. A., Valdez-Taubas J., Ferrari M. L., Haedo S. D., Maccioni H. J. (2008) Calsenilin and CALP interact with the cytoplasmic tail of UDP-Gal:GA2/GM2/GD2 β1,3-galactosyltransferase. Biochem. J. 412, 19–26 [DOI] [PubMed] [Google Scholar]
- 60. Sours-Brothers S., Ma R., Koulen P. (2009) Ca2+-sensitive transcriptional regulation. Direct DNA interaction by DREAM. Front. Biosci. 14, 1851–1856 [DOI] [PubMed] [Google Scholar]
- 61. Jacobson D. A., Cho J., Landa L. R., Jr., Tamarina N. A., Roe M. W., Buxbaum J. D., Philipson L. H. (2006) Downstream regulatory element antagonistic modulator regulates islet prodynorphin expression. Am. J. Physiol. Endocrinol. Metab. 291, E587–595 [DOI] [PubMed] [Google Scholar]
- 62. Petrosyan A., Ali M. F., Verma S. K., Cheng H., Cheng P. W. (2012) Non-muscle myosin IIA transports a Golgi glycosyltransferase to the endoplasmic reticulum by binding to its cytoplasmic tail. Int. J. Biochem. Cell Biol. 44, 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scott K. L., Chin L. (2010) Signaling from the Golgi. Mechanisms and models for Golgi phosphoprotein 3-mediated oncogenesis. Clin. Cancer Res. 16, 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wood C. S., Schmitz K. R., Bessman N. J., Setty T. G., Ferguson K. M., Burd C. G. (2009) PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gonatas J. O., Mezitis S. G., Stieber A., Fleischer B., Gonatas N. K. (1989) MG-160. A novel sialoglycoprotein of the medial cisternae of the Golgi apparatus (published eeratum appears in J. Biol. Chem. 1989 Mar 5;264(7):4264). J. Biol. Chem. 264, 646–653 [PubMed] [Google Scholar]
- 66. Graham T. R., Burd C. G. (2011) Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 21, 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dejgaard S. Y., Murshid A., Dee K. M., Presley J. F. (2007) Confocal microscopy-based linescan methodologies for intra-Golgi localization of proteins. J. Histochem. Cytochem. 55, 709–719 [DOI] [PubMed] [Google Scholar]
- 68. Tarr J. M., Ding N., Kaul K., Antonell A., Pérez-Jurado L. A., Chibber R. (2012) Cellular crosstalk between TNF-α, NADPH oxidase, PKCβ2, and C2GNT in human leukocytes. Cell. Signal. 24, 873–878 [DOI] [PubMed] [Google Scholar]
- 69. Radhakrishnan P., Chachadi V., Lin M. F., Singh R., Kannagi R., Cheng P. W. (2011) TNFα enhances the motility and invasiveness of prostatic cancer cells by stimulating the expression of selective glycosyl- and sulfotransferase genes involved in the synthesis of selectin ligands. Biochem. Biophys. Res. Commun. 409, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kikuchi J., Ozaki H., Nonomura C., Shinohara H., Iguchi S., Nojiri H., Hamada H., Kiuchi A., Nakamura M. (2005) Transfection of antisense core 2 β1,6-N-acetylglucosaminyltransferase-1 cDNA suppresses selectin ligand expression and tissue infiltration of B-cell precursor leukemia cells. Leukemia 19, 1934–1940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.