Background: In addition to LPS, MDP in the bacterial cell wall induces a tolerant state in host cells.
Results: Upon MDP stimulation, the MDP receptor Nod2 is rapidly ubiquitinated and degraded.
Conclusion: The rapid degradation of Nod2 confers MDP tolerance.
Significance: This serves as a defense mechanism to prevent septic shock.
Keywords: Cytokine, Endotoxin, Inflammation, Nod-like Receptors (NLR), Ubiquitination, MDP, Nod2, Bacterial Cell Wall, Endotoxin Tolerance, Septic Shock
Abstract
The innate immune system serves as the first line of defense by detecting microbes and initiating inflammatory responses. Although both Toll-like receptor (TLR) and nucleotide binding domain and leucine-rich repeat (NLR) proteins are important for this process, their excessive activation is hazardous to hosts; thus, tight regulation is required. Endotoxin tolerance is refractory to repeated lipopolysaccharide (LPS) stimulation and serves as a host defense mechanism against septic shock caused by an excessive TLR4 response during Gram-negative bacterial infection. Gram-positive bacteria as well as their cell wall components also induce shock. However, the mechanism underlying tolerance is not understood. Here, we show that activation of Nod2 by its ligand, muramyl dipeptide (MDP) in the bacterial cell wall, induces rapid degradation of Nod2, which confers MDP tolerance in vitro and in vivo. Nod2 is constitutively associated with a chaperone protein, Hsp90, which is required for Nod2 stability and protects Nod2 from degradation. Upon MDP stimulation, Hsp90 rapidly dissociates from Nod2, which subsequently undergoes ubiquitination and proteasomal degradation. The SOCS-3 protein induced by Nod2 activation further facilitates this degradation process. Therefore, Nod2 protein stability is a key factor in determining responsiveness to MDP stimulation. This indicates that TLRs and NLRs induce a tolerant state through distinct molecular mechanisms that protect the host from septic shock.
Introduction
As the first line of host defense, the innate immune system detects and responds to microbes through innate immune receptors, such as Toll-like receptors (TLRs)3 and nucleotide binding domain and leucine-rich repeat (NBD-LRR or NLR) protein families (1). Stimulation of TLRs and NLRs with their ligands, such as lipopolysaccharide (LPS) for TLR4 and muramyl dipeptide (MDP) for Nod2, activates downstream signaling cascades including NF-κB and mitogen-activated protein kinases. This results in the secretion of inflammatory cytokines, such as TNF-α, interleukin-6 (IL-6), and IL-8 (1–3). Although the production of inflammatory cytokines serves as an important mechanism in protecting the host from bacterial pathogens, excessive production of these cytokines without proper regulation is detrimental to the host and may lead to microcirculatory dysfunction, tissue damage, shock, or even death (4, 5). Therefore, innate immune responses against bacterial infection must be tightly regulated by a negative feedback mechanism.
Endotoxin tolerance (or LPS tolerance) is one such protective mechanism during Gram-negative bacterial infection. Prior exposure to LPS induces refractoriness to further stimulation with LPS in vitro and in vivo (6–8), and the mechanism of endotoxin tolerance has been investigated extensively at the molecular and cellular levels using cultured macrophages, animals, and humans (7, 9). Tolerogenic characteristics of endotoxin tolerance include the down-regulation of inflammatory mediators (such as TNF-α, IL-1β, or CXCL10) (8, 10, 11), the up-regulation of anti-inflammatory cytokines (such as IL-10 and TGF-β) (12–14), and impaired antigen presentation (15–17). Endotoxin tolerance is caused by an increase in the expression levels of negative regulators, IRAK-M, ST2, and A20, for example (18–21), and a decrease in TLR4 surface expression (22). Recent studies reported that altered accessibility to gene loci by chromatin modification and microRNA (e.g. miR146, miR155, and miR125b)-mediated regulation of target genes are also possible negative regulatory mechanisms of inflammation at the transcriptional and post-transcriptional levels, respectively (23–26).
In addition to Gram-negative bacteria, Gram-positive bacteria, which lack LPS, also cause septic shock via inflammatory toxicity of their exotoxins and cell wall components (27). Nod2, a cytoplasmic NLR, senses the component of bacterial cell wall peptidoglycan called MDP, which consists of N-acetylmuramyl-l-Ala-d-Gln (28). The N-terminal part of Nod2 contains two caspase recruitment domains (CARDs) responsible for interacting with a downstream effector, a centrally located NBD required for nucleotide binding and self-oligomerization, and a C-terminal portion containing LRRs involved in ligand recognition (29). Upon recognition of MDP, Nod2 activates downstream signaling cascades through the Rip2 kinase (29, 30). The Nod2 pathway is critical for bacterial sensing and host defense (31, 32) as well as for the maintenance of intestinal homeostasis (33, 34). Moreover, genetic studies support the importance of Nod2 in inflammatory diseases; its loss of function mutation is strongly associated with Crohn disease (35, 36), and its gain of function mutation is associated with Blau syndrome and early onset sarcoidosis (37–39).
It is known that, similar to LPS, exposure to MDP causes an impaired response to subsequent MDP administration (MDP tolerance) (40, 41). However, the mechanism behind MDP tolerance is not known. Here, we show that MDP tolerance is caused by the rapid proteasomal degradation of Nod2 in a ligand-specific manner. The rapid dissociation of heat shock protein (Hsp) 90 from Nod2 and recruitment of SOCS-3 are involved in the ubiquitination and degradation of Nod2.
EXPERIMENTAL PROCEDURES
Cell Culture and Mouse Strains
RAW264.7 cells and HEK293T cells were maintained in DMEM (Invitrogen) containing 10% (v/v) heat-inactivated FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin at 37 °C under 5% CO2. Bone marrow-derived macrophages were prepared as described (42). Briefly, mouse bone marrow was obtained by flushing the tibia and femur of a C57BL/6 mouse (Taconic) with DMEM supplemented with 10% heat-inactivated FBS (Invitrogen). Bone marrow cells were cultured in 10 ml of DMEM supplemented with 10% FBS, glutamine (both from Invitrogen), and 30% L929 cell supernatant containing macrophage colony-stimulating factor at an initial density of 1 × 106 cells/ml in 100-mm Petri dishes (BD Biosciences) at 37 °C in humidified 5% CO2 for 6 days. Cells were harvested with cold PBS (Invitrogen), washed, resuspended in DMEM supplemented with 10% FBS, and used at a density of 2 × 105 cells/ml. SW480 human epithelial cells were maintained in Leibovitz's L-15 medium (ATCC, Manassas, VA) containing 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin at 37 °C under 5% CO2. C57BL/6 mice were purchased from Taconic Farms. Nod2-deficient mice (kindly provided by Richard Flavell, Yale University) were backcrossed to C57BL/6 for 12 generations, rederived into specific pathogen-free conditions, and maintained in isolated barrier units thereafter at Taconic Farms.
Reagents
MDP (N-acetylmuramyl-l-alanyl-d-isoglutamine) was purchased from Sigma-Aldrich and resuspended in endotoxin-free water. LPS was from Enzo Life Sciences (Plymouth Meeting, PA). Phosphorothioate-modified CpG oligonucleotide DNA (TCCATGACGTTCCTGACGTT) was synthesized at Integrated DNA Technologies. Pam3CSK4, flagellin, and MDP-dd (N-acetylmuramyl-d-alanyl-d-isoglutamine) were from InvivoGen (San Diego, CA). 17-Allylamino-17-demethoxygeldanamycin (17AAG), radicicol, cycloheximide, thioglycollate, and mouse monoclonal anti-FLAG antibody were purchased from Sigma-Aldrich. The proteasome inhibitor MG-132 was obtained from Calbiochem. Rabbit polyclonal anti-IκBα, anti-p65, mouse monoclonal anti-Hsp90, anti-ubiquitin (Ub), goat polyclonal anti-SOCS-3, and anti-actin antibodies and siRNAs targeting Hsp90 or SOCS-3 gene were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-p-ERK and anti-p-SAPK/JNK antibodies were from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal anti-inducible nitric-oxide synthase (iNOS) was from Abcam (Cambridge, MA). Mouse monoclonal anti-hemagglutinin (HA) antibody was from Covance (Princeton, NJ). Rabbit polyclonal anti-Rip2 antibody was obtained from Enzo Life Sciences. Recombinant mouse interferon-γ (IFN-γ) and rat polyclonal anti-Nod2 antibody were from eBioscience (San Diego, CA). Goat anti-rabbit/mouse/goat secondary antibodies conjugated with horseradish peroxidase were from Santa Cruz Biotechnology. pCMV-FLAG-SOCS-3 was purchased from Addgene (Cambridge, MA). The following expression vectors for the Nod2 deletion mutants were kindly provided by Dr. Naohiro Inohara (University of Michigan): pcDNA3-Fpk3-Myc Nod2 mutants (129–1040 (ΔCARD1), Δ125–214 (ΔCARD2), 1–125 (CARD1), 1–744 (ΔLRR), 265–1040 (ΔCARDs), 126–301 (CARD2), and 265–744 (NBD)) and pcDNA3-HA Nod2 mutants (1–301 (CARDs) and 744–1040 (LRR)).
Bacterial Strain
Bacillus subtilis were grown in LB at 37 °C. Bacterial growth was monitored by absorbance at 600 nm. The bacterial pellets were resuspended in PBS and heat-inactivated at 70 °C for 20 min.
Determination of Cytokine Secretion
Cytokine levels in culture supernatants were determined using an ELISA kit (R&D Systems) according to the manufacturer's instructions.
Immunoblot Analysis and Immunoprecipitation
For the immunoblot analysis, 30 μg of protein were resolved by 4–12% gradient SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk, PBS, and 0.1% Tween 20 for 1 h before incubation overnight at 4 °C with primary antibodies in 5% skim milk, PBS, and 0.1% Tween 20. The membranes were then washed three times in 1× PBS and 0.1% Tween 20 and incubated with horseradish peroxidase-conjugated secondary antibodies in 5% skim milk, PBS, and 0.1% Tween 20 for 1 h. After successive washes, the membranes were developed using a SuperSignal West Pico Chemiluminescent kit (Thermo Scientific). Immunoprecipitations with anti-Nod2, anti-Hsp90, and anti-FLAG antibodies were performed on RAW264.7 cells or HEK293T cells. After rotating samples at 4 °C overnight, Protein A/G UltraLink Resin (Thermo Scientific, Rockford, IL) was added to each tube and rotated at 4 °C for 3 h. The beads were washed three times sequentially in cell lysis buffer and washing buffer (20 mm Tris-HCl (pH 7.4) and 0.1% Nonidet P-40), and samples were boiled for 10 min in 20 μl of loading buffer and subjected to SDS-PAGE and immunoblot analysis.
Immunofluorescent Staining for p65
The cells grown in 35-mm dishes were fixed in methanol. The cells were incubated with rabbit polyclonal anti-p65 antibody diluted 1:100 in 3% BSA for 24 h. The cells were incubated with rhodamine isothiocyanate-conjugated goat anti-rabbit immunoglobulin G antibody diluted 1:100 in 3% BSA for 30 min. After mounting with 50% glycerol, the slides were analyzed with a fluorescence microscope (Nikon Eclipse TE300).
Real Time Quantitative PCR
RNA was isolated using TRIzol reagent (Invitrogen) and ethanol-precipitated. cDNA synthesis was performed using the qScript Flex cDNA synthesis kit (Quanta Biosciences) according to the manufacturer's instructions. RNA expression was quantified on the 7300 Real-Time PCR System (Applied Biosystems) using the PerfeCTa SYBR Green SuperMix with ROX (Quanta Biosciences). Primer pairs used in the quantitative PCR analysis were as follows: Nod2 forward, 5′-TGACTGTGGCTAATGTCCTTTGTG-3′; Nod2 reverse, 5′-TTCTATCGCCTTCTTGACGAGTTC-3′; β-actin forward, 5′-GCTGTGCTGTCCCTGTATGCCTCT-3′; and β-actin reverse, 5′-CTTCTCAGCTGTGGTGGTGAAGC-3′.
Luciferase Assay
Cells cultured in 6-well plates were transfected with the NF-κB reporter plasmid (pBVI-Luc) and pRLNull plasmids according to the manufacturer's specifications. Luciferase activity was determined using a Dual-Luciferase assay kit (Promega, Madison, WI).
Statistical Analysis
Data were subjected to Student's t test for analysis of statistical significance, and a p value of <0.05 was considered to be significant.
RESULTS
MDP Pretreatment Induces Refractoriness to Subsequent MDP Challenge
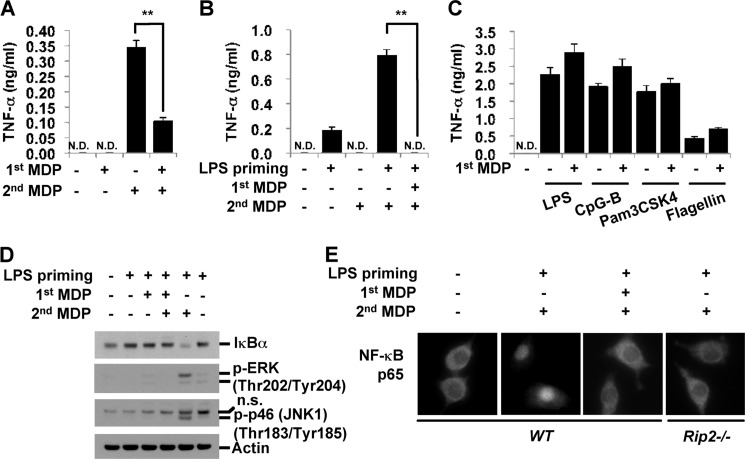
To determine whether MDP pretreatment induces resistance to subsequent stimulation with MDP, we treated RAW264.7 mouse macrophage cells with MDP for 4 h. Cells were washed and restimulated with MDP for 24 h. Consistent with a previous report (43), MDP pretreatment induced a tolerant state in RAW macrophages, resulting in the reduced release of TNF-α upon subsequent MDP stimulation (Fig. 1A). Primary macrophages were also examined for MDP tolerance. Because the level of Nod2 expression is relatively low in bone marrow-derived macrophages (BMDM), cells were primed with a low dose of LPS to increase Nod2 expression levels. Similar to RAW cells, MDP pretreatment for 4 h induced MDP tolerance in BMDM, which produced undetectable levels of TNF-α upon subsequent MDP stimulation (Fig. 1B). Unlike LPS tolerance, which can be induced in 6–24 h depending on the cell type and stimulation conditions, MDP tolerance is induced rapidly within 4 h after MDP treatment in both RAW cells and BMDM. This MDP-induced rapid tolerance is specific to MDP responses because MDP-pretreated RAW264.7 were competent to produce TNF-α upon subsequent stimulation with TLR ligands including Pam3CSK4 (a ligand for TLR1/2), LPS (for TLR4), flagellin (for TLR5), and CpG oligo DNA (for TLR9) (Fig. 1C). Nod2 activation induces proinflammatory cytokine production via the activation of mitogen-activated protein kinases (MAPKs) and phosphorylation-mediated degradation of IκBα that allows nuclear translocation of the p65 NF-κB subunit (1). To investigate whether a decrease in proinflammatory cytokine production in MDP-tolerant cells is due to the inhibition of these signaling mediators, we examined the expression level of IκBα and the level of active phosphorylated extracellular signal-regulated kinase (p-ERK) and c-Jun N-terminal kinase (p-JNK). As shown in Fig. 1D, MDP pretreatment in LPS-primed cells completely blocked IκBα degradation and MAPK activation. MDP pretreatment also inhibited p65 nuclear translocation upon a secondary MDP stimulation (Fig. 1E). Taken together, the pretreatment of macrophages with MDP induced a state of rapid tolerance to subsequent MDP stimulation that is mediated by the inhibition of upstream events in the IκBα and MAPK signaling cascades.
FIGURE 1.
Pretreatment with MDP inhibits proinflammatory responses to subsequent MDP treatment. A and C, RAW264.7 cells were pretreated with MDP (100 μg/ml) for 4 h, washed, and restimulated with MDP (100 μg/ml), LPS (10 ng/ml), CpG-B (1 μm), Pam3CSK4 (10 μg/ml), or flagellin (3 μg/ml) for 24 h. B, BMDM were stimulated with LPS (0.5 ng/ml) for 6 h and then incubated with MDP (100 μg/ml) for 4 h in the absence of LPS. After several washes, the cells were restimulated with MDP (100 μg/ml) for 24 h. Supernatants were subjected to ELISA for TNF-α. D and E, wild-type BMDM or Rip2-deficient BMDM were pretreated with LPS (0.1 ng/ml) for 6 h and stimulated with MDP for 4 h in the absence of LPS. The cells were washed and restimulated with MDP for 30 min. D, cell extracts were subjected to Western blot analysis for IκBα, p-ERK, p-JNK, and actin. E, cells were fixed and permeabilized for 5 min. Immunofluorescent staining for p65 was performed using anti-p65 Ab followed by Alexa Fluor 594 detection antibody. Cells were analyzed under a fluorescence microscope. Error bars represent the mean ± S.D. of triplicates. **, p < 0.005. Data are representative of three independent experiments with similar results. n.s., nonspecific; N.D., not detected.
MDP Treatment Rapidly Reduces Expression Levels of Nod2
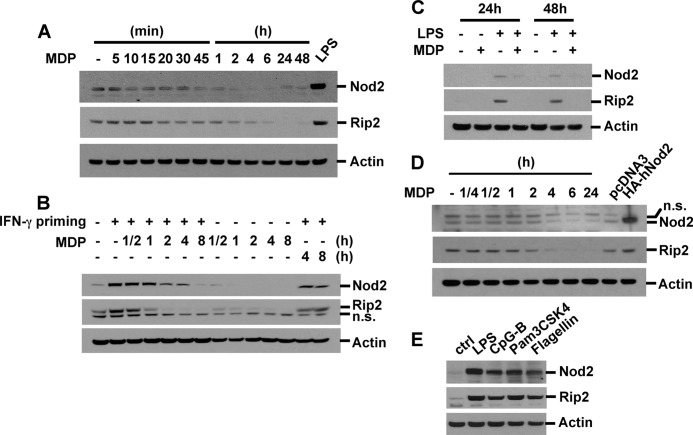
Although it has been reported that full LPS tolerance in macrophages induced by LPS stimulation takes 6–24 h (44–46), 4-h incubation with MDP was sufficient to induce MDP self-tolerance (Fig. 1, A and B), indicating that the tolerance mechanism of MDP is different from that induced by a TLR ligand. One possible explanation is the rapid alteration in the expression of molecules upstream of IκBα and MAPKs after MDP stimulation. To evaluate this, we treated RAW264.7 cells, BMDM, and SW480 cells (a human colon epithelial cell line) with MDP for various periods from 5 min up to 48 h and examined the expressions of Nod2 and Rip2. MDP stimulation induced a rapid decrease in basal levels of Nod2 and Rip2 in RAW264.7 cells (Fig. 2A) and SW480 cells (Fig. 2D) in a time-dependent manner. To test the effect of MDP in inducing Nod2 and Rip2 expression, we primed cells with IFN-γ (100 units/ml) or a low dose of LPS for 6 h and restimulated the cells with MDP for the indicated periods. MDP stimulation rapidly decreased Nod2 and Rip2 expression levels in IFN-γ- or LPS-primed macrophages (Fig. 2B and supplemental Fig. S1). Co-stimulation of BMDM with LPS and MDP also resulted in Nod2 and Rip2 down-regulation (Fig. 2C). On the other hand, stimulation with ligands for TLR2, -4, -5, or -9 instead up-regulated the expression of Nod2 and Rip2 in a dose- and time-dependent manner (Fig. 2E and data not shown).
FIGURE 2.
MDP treatment induces degradation of Nod2 and Rip2 via the proteasomal pathway. A, RAW264.7 cells were treated with MDP (100 μg/ml) for the indicated times or with LPS (10 ng/ml) for 6 h. B, RAW264.7 cells were pretreated with IFN-γ (100 units/ml) for 6 h and then stimulated with MDP for the indicated times. C, BMDM were co-treated with MDP (100 μg/ml) in the presence or absence of LPS (0.5 ng/ml) for the indicated times. D, SW480 cells were treated with MDP for the indicated times. Cells were transfected with pcDNA3 or HA-human Nod2 plasmid vectors. E, RAW264.7 cells were stimulated with LPS (10 ng/ml), CpG-B (1 μm), Pam3CSK4 (10 μg/ml), or flagellin (1 μg/ml) for 6 h. Cell extracts were subjected to Western blot analysis for Nod2, Rip2, and actin. n.s., nonspecific; ctrl, control.
MDP Treatment Induces Proteasomal Degradation of Nod2 and Rip2
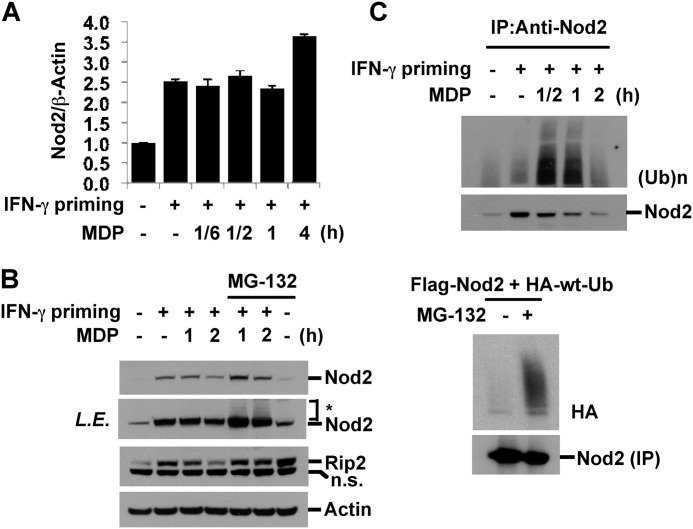
To investigate whether down-regulation of Nod2 and Rip2 is due to reduced levels of transcription, IFN-γ-primed RAW264.7 cells were treated with MDP for the indicated periods, and the levels of Nod2 mRNA were analyzed by quantitative real time PCR. Upon IFN-γ stimulation, transcripts of Nod2 were induced, and this induction was not significantly altered by subsequent MDP stimulation (Fig. 3A). Moreover, long term incubation with MDP (24 h) slightly increased the amounts of Nod2 transcript, indicating that transcriptional regulation is not a cause of MDP-mediated Nod2 down-regulation (data not shown). Many proteins with a short half-life experience phosphorylation, polyubiquitination, and targeted degradation through the proteasome. To test whether Nod2 down-regulation is due to enhanced protein degradation, a 20 S proteasome inhibitor, MG-132, was added to the culture media of IFN-γ-primed MDP-stimulated cells. MG-132 increased the basal levels of Nod2 and Rip2 and suppressed MDP-induced degradation of Nod2 and Rip2 (Fig. 3B). Interestingly, MG-132 treatment induced a slowly migrating Nod2 on SDS-PAGE, indicating a possible ubiquitination of Nod2. Therefore, we next investigated whether Nod2 undergoes polyubiquitination after MDP treatment. Cell extracts from IFN-γ-primed, MDP-stimulated RAW264.7 cells were immunoprecipitated with anti-Nod2 antibody, and the level of polyubiquitination was examined by Western blotting for Ub. MDP stimulation rapidly induced Nod2 polyubiquitination, which preceded its degradation (Fig. 3C, upper). We confirmed this polyubiquitination of Nod2 by the co-transfection of FLAG-tagged Nod2 and HA-tagged wild type (WT) Ub in HEK293T cells and the immunoprecipitation of Nod2 with anti-FLAG antibody. In the presence of MG-132, ubiquitinated Nod2 was observed (Fig. 3C, lower). Taken together, these results indicated that MDP stimulation causes rapid polyubiquitination of Nod2, which subsequently undergoes proteasome-mediated degradation.
FIGURE 3.
MDP treatment induces proteasomal degradation of Nod2 and Rip2. A and C, upper, RAW264.7 cells were pretreated with IFN-γ for 6 h and then stimulated with MDP for the indicated times. A, the expression of Nod2 was examined by quantitative real time PCR. Data were normalized to the expression of the β-actin gene. Data represent the mean ± S.D. of triplicates. B, cells were pretreated with IFN-γ for 6 h and then stimulated with MDP for the indicated times in the presence or absence of MG-132 (10 μm). Cell extracts were subjected to Western blot analysis for Nod2, Rip2, and actin. *: slowly migrating Nod2. C, upper, cell extracts were immunoprecipitated (IP) with anti-Nod2 antibody. The level of ubiquitination on Nod2 was analyzed. C, lower, HEK293T cells were transfected with FLAG-Nod2 and HA-WT-Ub plasmid vectors. Forty-eight hours after transfection, cells were treated with MG-132 (10 μm) for 2 h. Cell extracts were immunoprecipitated with anti-Nod2 antibody. The level of Nod2 ubiquitination was analyzed. Error bars represent the mean ± S.D. of triplicates. Data are representative of three independent experiments with similar results. L.E., long exposure; n.s., nonspecific.
Degradation of Nod2 Is Specific to MDP Stimulation
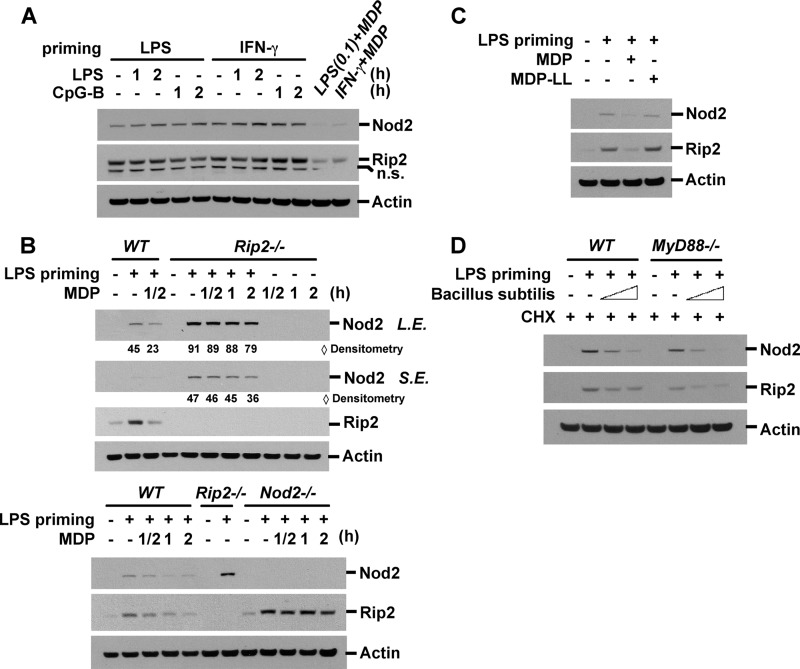
We next determined whether degradation of Nod2 and Rip2 is ligand-specific. Stimulation with LPS (ligand for TLR4) and CpG-B (TLR9) did not induce down-regulation of Nod2 and Rip2 in RAW macrophage cells primed with LPS, CpG, or IFN-γ (Fig. 4A and supplemental Fig. S2). In Nod1-deficient BMDM, MDP-induced Nod2 and Rip2 degradation was similar to that in wild-type BMDM (data not shown). On the other hand, Rip2 degradation was completely blocked in Nod2-deficient macrophages (Fig. 4B, lower), confirming that Nod2 is critical for the recognition of MDP and that Nod2 activation is required for the down-regulation of Nod2 and Rip2. Interestingly, LPS-induced Nod2 expression was slightly more elevated in Rip2-deficient BMDM than in wild-type BMDM, possibly due to an unknown compensatory mechanism for Nod2 expression (Fig. 4B, upper). In Rip2-deficient BMDM, Nod2 degradation by MDP still occurred but was delayed compared with wild-type BMDM, indicating that Nod2 down-regulation is regulated by both Rip2-dependent and -independent mechanisms (Fig. 4B). To evaluate the specificity of MDP for Nod2 and Rip2 degradation, we used MDP-ll, an inactive chiral isomer of MDP. MDP-ll could not decrease the levels of Nod2 and Rip2 expression, indicating that only biologically active MDP can induce Nod2 down-regulation (Fig. 4C). MDP exists in the peptidoglycan of both Gram-negative and Gram-positive bacteria cell walls, which suggests that bacteria might induce MDP tolerance. To test the effect of bacteria on Nod2 and Rip2 expression, we challenged LPS-primed BMDM with heat-killed B. subtilis, which has MDP in its bacterial cell wall and is thereby capable of activating Nod2 (47). To exclude the possibility of transcriptional/translational regulation mediated via TLR ligands in the bacteria, we pretreated the cells with cycloheximide after LPS priming. LPS priming increased the expression of Nod2 and Rip2 in MyD88-dependent and MyD88-independent manners (Fig. 4D). B. subtilis induced Nod2 and Rip2 degradation in wild-type and MyD88-deficient BMDM in a dose-dependent manner (Fig. 4D). These data suggest that degradation of Nod2 is specific to active MDP stimulation and that the bacterial cell wall can induce a similar effect.
FIGURE 4.
MDP-induced degradation of Nod2 and Rip2 is specific to MDP stimulation. A, RAW264.7 cells were pretreated with LPS (0.1 ng/ml) or IFN-γ (100 units/ml) for 6 h and then stimulated with LPS, CpG-B, or MDP for the indicated times. B, wild-type, Nod2-deficient, or Rip2-deficient BMDM were pretreated with LPS (0.2 ng/ml) for 6 h and then stimulated with MDP (100 μg/ml) for the indicated times. C, RAW264.7 cells were pretreated with LPS (1 ng/ml) for 6 h and then incubated with MDP (MDP-ld) or biologically inactive MDP-ll (100 μg/ml) for 3 h. D, wild-type or MyD88-deficient BMDM were stimulated with LPS for 6 h and then infected with heat-killed B. subtilis (5 × 106 or 20 × 106 cells/ml) in the presence of cycloheximide (CHX; 20 μg/ml) for 4 h. Cell extracts were subjected to Western blot analysis for Nod2, Rip2, and actin. Data are representative of three independent experiments with similar results. S.E., short exposure; L.E., long exposure; n.s., nonspecific.
MDP Treatment Induces Rapid Dissociation of the Nod2·Hsp90 Complex
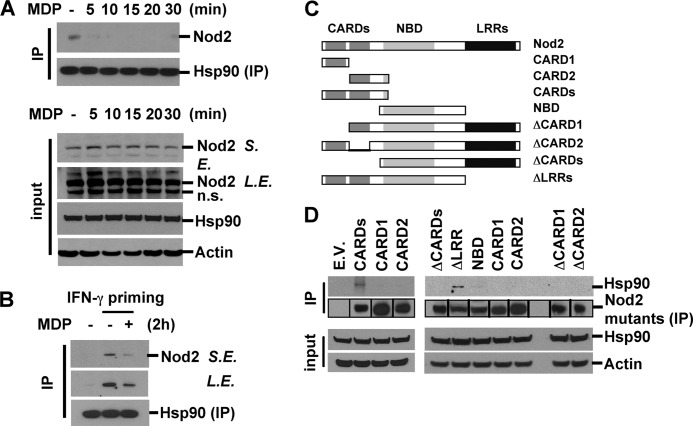
NLRs form a stable complex with several proteins (48). To identify the factors that might be involved in maintaining the stability of Nod2, we transfected HEK293T cells with a FLAG-tagged Nod2 expression vector. Cell lysates were immunoprecipitated with anti-FLAG antibody and subjected to SDS-PAGE followed by silver staining. Among the multiple bands indicating Nod2-associated proteins, we found a band at ∼90 kDa (supplemental Fig. S3). This prompted us to consider that a chaperone protein, Hsp90, might be a component of the Nod2 complex. RAW264.7 cells were stimulated with MDP for various periods, and the interaction between Nod2 and Hsp90 was evaluated by immunoprecipitation with anti-Hsp90 antibody following Western blotting with anti-Nod2 antibody. In unstimulated cells, endogenous Nod2 physically interacted with Hsp90. Notably, MDP stimulation caused the rapid dissociation of Nod2 from Hsp90 within 5 min (Fig. 5A, upper). Similar to unprimed macrophages, in IFN-γ-primed macrophages, Hsp90 was associated with Nod2, and MDP stimulation induced dissociation of Nod2 from Hsp90 (Fig. 5B). To investigate which domain(s) of Nod2 interacts with Hsp90, we co-transfected HEK293T cells with plasmids expressing various truncated forms of Nod2 (Fig. 5C). Nod2 was immunoprecipitated, and Nod2-interacting Hsp90 was detected by Western blotting. The Nod2 deletion mutant containing two CARDs interacted with Hsp90, although the deletion mutant that lacked either one of the CARDs did not, indicating that a double CARD motif is necessary and sufficient for association with Hsp90 (Fig. 5D). We did not observe Rip2/Hsp90 interaction in Myc-tagged Rip2-overexpressing HEK293T cells (data not shown). Taken together, these results indicate that Nod2 associates with Hsp90 via CARDs at a resting state. The activation of Nod2 by MDP stimulation leads to the rapid dissociation of Hsp90.
FIGURE 5.
MDP treatment induces rapid dissociation of the Nod2·Hsp90 complex. A, RAW264.7 cells were treated with MDP for the indicated times. B, cells were pretreated with IFN-γ for 6 h and then stimulated with MDP for the indicated times. Cell lysates were immunoprecipitated (IP) with anti-Hsp90 antibody, and associated Nod2 was analyzed by Western blotting. C, the expression vectors for Nod2 deletion mutants used in the study. D, HEK293T cells were transfected with the indicated plasmids. Forty-eight hours after transfection, total cellular proteins were extracted. Mutant Nod2 proteins in the lysates were immunoprecipitated, and associated endogenous Hsp90 protein was analyzed by Western blotting. Data are representative of three independent experiments with similar results. S.E., short exposure; L.E., long exposure; n.s., nonspecific; E.V., empty vector.
Hsp90 Is Required for the Maintenance of Endogenous and Inducible Levels of Nod2 and for Responses to MDP Stimulation
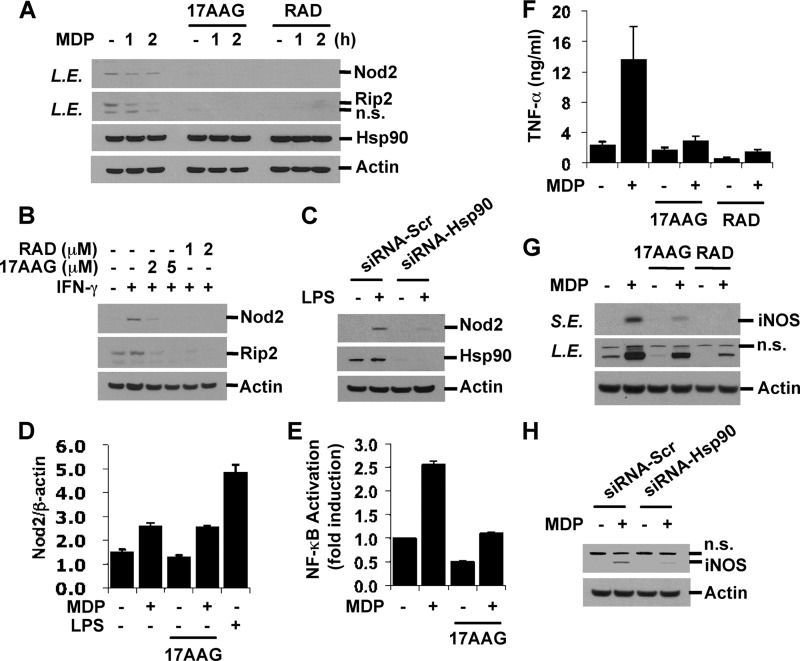
Hsp90 is one of the most abundant proteins in the cytoplasm and serves as a chaperone protein. Its main functions include the regulation of protein activity, prevention of aggregation of refolded peptides, and maintenance of protein stability (49) To evaluate the effect of Hsp90 dissociation from Nod2, we treated cells with Hsp90 inhibitors, 17AAG and radicicol (RAD), to disrupt the interaction between Hsp90 and its bound protein. Basal and IFN-γ-inducible levels of Nod2 and Rip2 were determined by Western blotting. As shown in Fig. 6, A and B, 17AAG and radicicol significantly reduced Nod2 and Rip2 expression levels. Furthermore, knockdown of Hsp90 by specific siRNAs but not by nonspecific scrambled siRNAs significantly reduced Nod2 expression in LPS-primed RAW264.7 cells (Fig. 6C). The concentrations of 17AAG (∼2 μm) and RAD (∼1 μm) used in the experiments did not alter cell morphology and did not induce apoptotic cell death as analyzed by Western blotting for active caspase-3, suggesting that this decrease in Nod2 expression was not due to cell toxicity (supplemental Fig. S4). 17AAG- and radicicol-mediated reduced expression was specific to Nod2 and Rip2 as the expression of other Nod2 and TLR signaling molecules, such as ERK, p38, TRAF6, and MyD88, was not altered by 17AAG or radicicol treatment (supplemental Fig. S4). 17AAG treatment did not inhibit an increase in Nod2 transcripts by MDP stimulation, indicating that the transcriptional machinery was not inhibited by 17AAG (Fig. 6D). We next tested the effect of Hsp90 inhibition on MDP-induced NF-κB activity and NF-κB target gene expression, such as that of TNF-α and iNOS. NF-κB activity was examined by measuring luciferase activity in HEK293T cells transiently co-transfected with an NF-κB reporter plasmid. MDP stimulation induced increases in NF-κB activity, and pretreatment with 17AAG abrogated this induction (Fig. 6E). The effect of Hsp90 inhibitors on MDP-induced NF-κB is not cell type-specific because 17AAG and RAD suppressed NF-κB activation upon stimulation with MDP in intestinal epithelial cell lines including HCT116, LS174T, and SW480 (supplemental Fig. S5). Moreover, treatment with 17AAG or RAD inhibited TNF-α release and the induction of iNOS expression by MDP stimulation in RAW macrophages (Fig. 6, F and G). The requirement of Hsp90 for Nod2-mediated iNOS induction was further confirmed by the knockdown of Hsp90 (Fig. 6H). Taken together, these results suggest that the interaction of Nod2 and Hsp90 is critical for the maintenance of Nod2 and Rip2 stability. Dissociation of Hsp90 leads to decreased expressions of Nod2 and Rip2, resulting in a poor response to MDP.
FIGURE 6.
Hsp90 inhibition decreases endogenous and inducible levels of Nod2 and Rip2 and blocks MDP response. A and B, RAW264.7 cells were pretreated with 17AAG (2 or 5 μm) or RAD (1 or 2 μm) for 8 h and then stimulated with MDP (100 μg/ml) for the indicated times or IFN-γ (100 units/ml) for 6 h in the presence of Hsp90 inhibitors. C, RAW264.7 cells were transfected with siRNAs targeting Hsp90 or control siRNAs. Forty-eight hours after transfection, cells were stimulated with LPS (0.1 ng/ml) for 6 h. Cell extracts were subjected to Western blot analysis for Nod2, Rip2, Hsp90, and actin. D, RAW264.7 cells were pretreated with 17AAG (2 μm) and then stimulated with MDP for 5 h in the presence or absence of 17AAG. LPS (10 ng/ml; 5 h) was used as a positive control. E, HEK293T cells were co-transfected with pBVI-Luc reporter and pRLNull plasmids. Twenty-four hours after transfection, cells were co-treated with MDP and 17AAG (2 μm) for 20 h. NF-κB activity was measured by Dual-Luciferase assay. F and G, RAW264.7 cells were pretreated with 17AAG (2 μm) or RAD (1 μm) and then stimulated with MDP for 24 h. TNF-α in the media was measured by ELISA. Cell extracts were subjected to Western blot analysis for iNOS and actin. H, RAW264.7 cells were transfected as in C. Cells were stimulated with MDP for 24 h. The iNOS level was detected by Western blotting. Error bars represent the mean ± S.D. of triplicates. Data are representative of three independent experiments with similar results. L.E., long exposure; S.E., short exposure; n.s., nonspecific; Scr, scrambled.
MDP-induced SOCS-3 Accelerates Nod2 Degradation
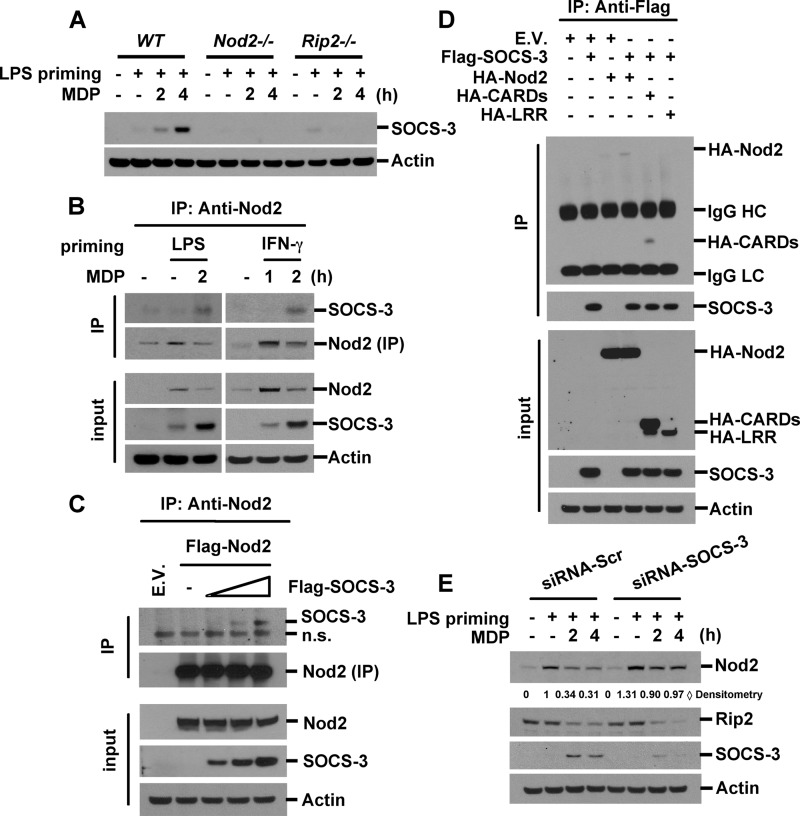
The clarification of Nod2 degradation in both Rip2-dependent and -independent mechanisms (Fig. 4B) led us to search for the MDP-inducible molecule that contributes to the degradation of Nod2. SOCS molecules are well known negative regulators of cytokine signaling. SOCS-1 has been reported to inhibit TLR2 and TLR4 signaling by inducing the degradation of signaling molecules (50). We investigated whether MDP stimulation could up-regulate SOCS proteins. We found that SOCS-3 was highly induced upon MDP stimulation in LPS-primed BMDM (Fig. 7A). MDP treatment alone also slightly increased SOCS-3 expression levels (data not shown). Moreover, we found that MDP-induced SOCS-3 was totally dependent on Nod2 and Rip2 as MDP stimulation failed to induce SOCS-3 in Nod2- or Rip2-deficient macrophages (Fig. 7A). SOCS proteins are composed of an N-terminal domain, a central SH2 domain, and a C-terminal SOCS box. The SOCS box acts as a linker between the substrate and E3 ligase components (51, 52). SOCSs bind to target proteins via an SH2 domain and regulate the turnover of target proteins by mediating polyubiquitination and subsequent degradation (52). Therefore, we evaluated whether SOCS-3 interacts with Nod2. After MDP treatment in LPS- or IFN-γ-primed RAW264.7 cells, total cell lysates were immunoprecipitated with anti-Nod2 antibody, and Nod2-associated SOCS-3 was detected by Western blotting. The levels of SOCS-3 bound to Nod2 increased after MDP stimulation (Fig. 7B). We confirmed the interaction between Nod2 and SOCS-3 by transient transfection with expression vectors for Nod2 and SOCS-3 in HEK293T cells (Fig. 7C). Transfection of various deletion mutants of Nod2 revealed that SOCS-3 interacted with the CARD domains of Nod2 (Fig. 7D). On the other hand, we did not observe Rip2/SOCS-3 interaction by immunoprecipitation assay (data not shown). To test the role of SOCS-3 in MDP-induced Nod2 degradation, we induced silencing of SOCS-3 by transfection with siRNAs targeting SOCS-3. Knockdown of SOCS-3 partially blocked MDP-induced Nod2 degradation (Fig. 7E). Taken together, these results indicate that SOCS-3 is induced by MDP stimulation and then subsequently facilitates the degradation of Nod2.
FIGURE 7.
Knockdown of SOCS-3 suppresses MDP-mediated Nod2 degradation. A, wild-type, Nod2-deficient, or Rip2-deficient BMDM were pretreated with LPS (0.2 ng/ml) for 6 h and then stimulated with MDP (100 μg/ml) for the indicated times. Cell extracts were subjected to Western blot analysis for SOCS-3 and actin. B, RAW264.7 cells were pretreated with LPS or IFN-γ for 6 h and then incubated with MDP for the indicated times. Nod2 was immunoprecipitated (IP) with anti-Nod2 antibody. Nod2-associated SOCS-3 was detected by Western blotting. C, HEK293T cells were transfected with FLAG-Nod2 and various concentrations of FLAG-SOCS-3 plasmids. Forty-eight hours after transfection, total cellular proteins were extracted. Cell extracts were immunoprecipitated with anti-Nod2 antibody. Co-precipitated proteins were analyzed by Western blotting with anti-SOCS-3 antibody. D, HEK293T cells were transfected with the expression vectors for FLAG-SOCS-3 and HA-Nod2 deletion mutants. SOCS-3 was immunoprecipitated with anti-FLAG antibody. SOCS-3-associated Nod2 deletion mutants were detected by Western blotting using anti-HA antibody. E, RAW264.7 cells were transfected with siRNAs for SOCS-3 and control siRNAs. Forty-eight hours after transfection, cells were treated with LPS (0.5 ng/ml) for 6 h and then stimulated with MDP for the indicated times. Cell extracts were subjected to Western blot analysis for Nod2, Rip2, SOCS-3, and actin. Data are representative of three independent experiments with similar results. n.s., nonspecific; E.V., empty vector; Scr, scrambled.
Reduced Expression of Nod2 Mediates MDP Tolerance in Vivo
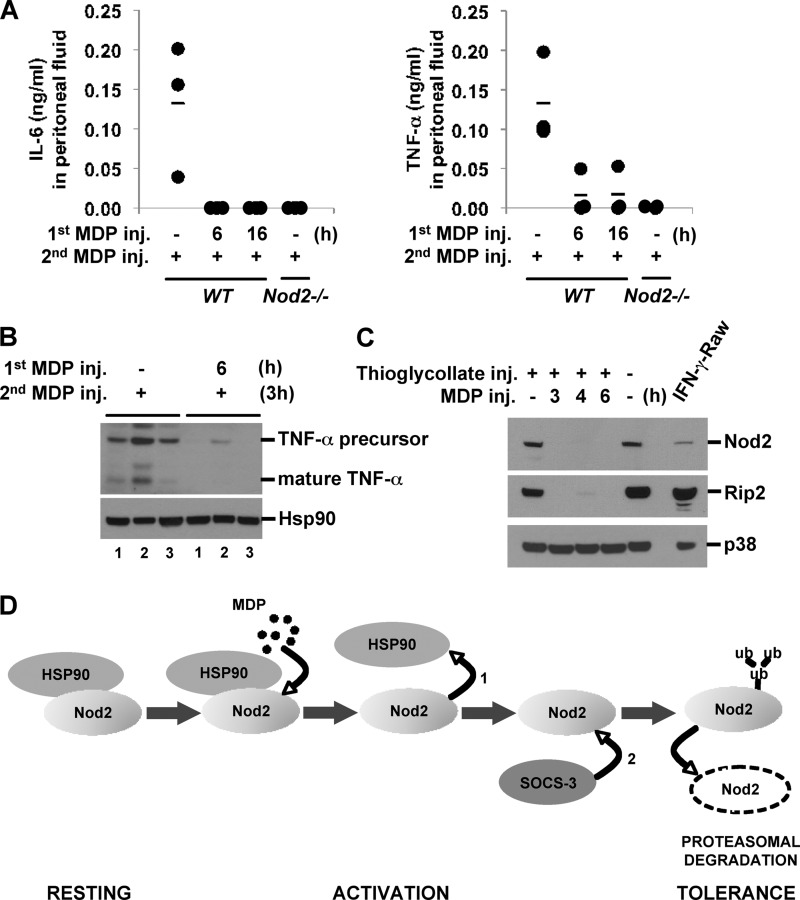
To test whether in vivo MDP tolerance is also caused by the rapid degradation of Nod2, we injected MDP intraperitoneally into mice, which were subsequently challenged with a second MDP injection after 6 or 16 h. Peritoneal fluid and peritoneal macrophages were analyzed for proinflammatory cytokine levels and Nod2 and Rip2 expression 3 h after the second MDP injection. MDP induced the release of IL-6 and TNF-α in peritoneal fluid (Fig. 8A). As a negative control, we used Nod2-deficient mice. Similar to in vitro MDP tolerance, the pretreatment of mice with MDP suppressed the release of IL-6 and TNF-α (Fig. 8A). In contrast, MDP pretreatment did not change the responsiveness to successive LPS injection, indicating that Nod2 activation does not induce cross-tolerance in vivo under the same conditions (supplemental Fig. S6). Intracellular TNF-α levels in peritoneal macrophages peaked at 3 h after MDP injection and gradually decreased thereafter (data not shown). Preinjection with MDP significantly reduced the expression of both precursor and mature forms of TNF-α induced upon the second MDP injection in peritoneal macrophages (Fig. 8B). Strikingly, MDP administration effectively decreased the expression levels of Nod2 and Rip2 in vivo (Fig. 8C). Taken together, these data suggest that the rapid reduction in Nod2 and Rip2 expression is induced by in vivo MDP stimulation and is associated with MDP tolerance.
FIGURE 8.
MDP tolerance is mediated by Nod2 and Rip2 down-regulation in vivo. Wild-type or Nod2-deficient mice were injected with 2 ml of autoclaved 4% thioglycollate intraperitoneally. Five days after thioglycollate treatment, MDP (35 mg/kg) was administered to the mice intraperitoneally for the indicated times, and then the mice were reinjected with MDP. Three hours after the second MDP injection, peritoneal fluid and macrophages were collected (n = 3 per group). A, IL-6 and TNF-α in peritoneal fluid were measured by ELISA. Mean value is indicated by a bar. B and C, cell extracts were subjected to Western blot analysis for TNF-α, Nod2, Rip2, Hsp90, and p38. RAW264.7 cells treated with IFN-γ (100 units/ml) were used as a positive control. D, model of MDP tolerance mediated by rapid Nod2 degradation. MDP stimulation causes rapid dissociation of the Nod2·Hsp90 complex (1). MDP-induced SOCS-3 binds to Nod2 (2). Nod2 undergoes polyubiquitination and proteasome-mediated degradation, resulting in reduced responsiveness to subsequent MDP. inj., injection.
DISCUSSION
Endotoxin tolerance is regarded as an important protection mechanism against septic shock during Gram-negative bacterial infection to limit tissue damage by an overactive immune response (53–55). The molecular mechanism behind endotoxin tolerance has been intensively investigated in the last decade, and it became apparent that not a single but rather multiple molecular mechanisms induce endotoxin tolerance at various stages (7, 9). In comparison with endotoxin tolerance, very little is known about the mechanism of MDP tolerance. In this study, we showed that rapid proteasomal degradation of Nod2 is a cause of MDP tolerance. We confirmed MDP-induced Nod2 degradation in a macrophage cell line, primary macrophages, and intestinal epithelial cell lines as well in in vivo experiments where mice were peritoneally injected with MDP. Nod2 is constitutively associated with the chaperone protein Hsp90, which protects Nod2 from degradation. Upon MDP stimulation, Hsp90 rapidly dissociates in 5 min, and then Nod2 undergoes ubiquitination and subsequent proteasomal degradation. We found that degradation occurs in both a Rip2-dependent and -independent manner (Fig. 4B). Rip2 does not phosphorylate Nod2 (supplemental Fig. S7, Supplemental Experimental Procedures). Instead, its downstream activation via Rip2 autophosphorylation appears to be a part of the mechanism behind Nod2 degradation. Indeed, up-regulated SOCS-3 upon MDP stimulation further enhanced Nod2 degradation, and thus SOCS-3 is a negative regulator of the Nod2 pathway (Fig. 7, A and E). Therefore, we propose the following model for Nod2 activation and MDP tolerance: At a resting stage, Nod2 is constitutively associated with Hsp90 (Fig. 8D, resting stage). Upon MDP stimulation, Nod2 changes its conformation, which enables NTP hydrolysis and further conformational changes. This results in a reduced affinity to Hsp90, which rapidly dissociates from Nod2 (activation stage). Activated Nod2 can signal downstream factors by generating a protein complex with Rip2 kinase, which activates cascades of NF-κB and MAPK, leading to the transcriptional activation of immune response genes. One transcriptional target is SOCS-3, which promotes Nod2 degradation. In the absence of Hsp90, Nod2 becomes ubiquitinated and undergoes proteasomal degradation. SOCS-3 associates with Nod2 but not Rip2 (data not shown) and facilitates this process, presumably by recruiting the ubiquitin machinery to Nod2 via its SOCS box domain. SOCS-3 itself does not have the ability to induce Nod2 degradation because IFN-γ priming up-regulates both SOCS-3 and Nod2 proteins (data not shown). However, once Nod2 is activated by MDP, SOCS-3 facilitates proteasomal degradation of Nod2. The Nod2·Hsp90 complex and Nod2·SOCS-3 complex are two sequential complexes as they exist at the resting stage and after the activation stage, respectively. Reduced Nod2 expression results in impaired responses to subsequent MDP stimulation, and in this manner, cells become tolerant (Fig. 8D, tolerance stage).
Hsps are a group of proteins that function as molecular chaperones for other proteins. It is interesting that Hsp90 is evolutionarily important for NLR protein function. Another NLR protein, NLRP3, also associates with Hsp90 (56). Moreover, structurally related plant R (resistance) proteins have been reported to make a functional complex together with Hsp90 and co-chaperone-like, ubiquitin ligase-associated protein SGT1 (57–59). Hsp90 inactivation induces dissociation of its client proteins from the Hsp90 complex, and the dissociated proteins are targeted for ubiquitination and proteasome-mediated degradation in a manner similar to what is shown in this study for Nod2 (Fig. 6). Therefore, Hsp90 is a key molecule in stabilizing the NLR protein until it is required for innate immune responses by microbial components.
There is significant cross-talk between the TLR and NLR systems. This is probably because microbes are composed of multiple ligands that are detectable by the innate systems. Although LPS priming induces Nod2 expression, TLR stimulation augments NLR responses (60) either by the up-regulation of NLRs, such as Nod2, or by inducing pro-IL-1β for IL-1β maturation by inflammasome NLRs (61). Nod2 stimulation also enhances TLR responses. A short incubation with MDP enhances the cellular response to subsequent TLR ligands (Fig. 1C). In vivo, it is well known that MDP pretreatment for a short period greatly enhances the toxicity of subsequent LPS injections (62–64). However, depending on cell types and conditions, a longer incubation with the Nod2 ligand will induce refractoriness to subsequent stimulation with TLR ligands (cross-tolerance) in vitro and in vivo (43, 65, 66). It has been proposed that cross-tolerance is mediated by reduced IRAK-1 activity (43); secretory mediators, such as IL-1β, IL-10, TGF-β, and IL-1Ra (67); or up-regulation of inhibitory molecules of TLR signaling, such as regulatory factor 4 (IRF4) or IRAK-M (43, 65). It is unlikely that these inhibitory molecules are involved in MDP self-tolerance because these molecules regulate only TLR signaling; for example, IRAK-M targets IRAK-1 in TLR signaling. Also, we found that MDP could induce Nod2 activation normally even in LPS-stimulated macrophages in which IRAK-1 is absent due to proteasomal degradation after activation (data not shown).
The difference between endotoxin tolerance and MDP tolerance in terms of required time after stimulation is intriguing. Endotoxin tolerance in culture cells requires 6–24 h depending on the report, whereas MDP tolerance is induced in as short as 4 h (43) (Fig. 1, A and B). Although we can only speculate on the reason behind this, the mode of ligand generation might be involved. LPS can be shed from Gram-negative bacteria and thus acts as a signal to innate immune cells even if the bacteria themselves are not present. It has been reported that enzymatic cleavage of the bacterial cell wall in the phagolysosome generates MDP (68, 69). Therefore, perhaps compared with LPS-stimulated cells, MDP-sensing cells are likely to have bacteria already in the cell, and thus refractoriness is needed to be rapidly induced. It is feasible that proteasomal degradation was selected for rapid induction of refractoriness compared with LPS tolerance, which mostly relies on the induction of negative regulators.
In conclusion, we identified that Nod2 protein degradation is a key mechanism behind MDP tolerance. To detect microbes, which have multiple ligands, the innate immune system has evolved to possess both TLR and NLR systems. Our study showed that although both systems utilize a tolerant state to protect the host from septic shock, they have distinct molecular mechanisms.
Supplementary Material
Acknowledgments
We thank Torsten B. Meissner for helpful discussions, Jonathan Kagan for critical reading, and Andrea Dearth for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK074738 (to K. S. K.). This work was also supported by a grant from the Crohn's and Colitis Foundation of America (to K. S. K.).

This article contains supplemental Experimental Procedures and Figs. S1–S7.
- TLR
- Toll-like receptor
- NLR
- nucleotide binding domain (NBD) and leucine-rich repeat (LRR)
- Nod2
- nucleotide-binding oligomerization domain-containing 2
- MDP
- muramyl dipeptide (N-acetylmuramyl-l-Ala-d-Gln (MDP-ld))
- IRAK
- interleukin-1 receptor-associated kinase
- CARD
- caspase recruitment domain
- Ub
- ubiquitin
- iNOS
- inducible nitric-oxide synthase
- BMDM
- bone marrow-derived macrophages
- MDP-dd
- N-acetylmuramyl-d-alanyl-d-isoglutamine
- MDP-ll
- N-acetylmuramyl-l-alanyl-l-isoglutamine
- 17AAG
- 17-allylamino-17-demethoxygeldanamycin
- RAD
- radicicol
- SOCS
- suppressor of cytokine signaling
- SH2
- Src homology 2.
REFERENCES
- 1. Kawai T., Akira S. (2009) The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21, 317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akira S., Takeda K., Kaisho T. (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 [DOI] [PubMed] [Google Scholar]
- 3. Akira S., Takeda K. (2004) Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 4. Beutler B., Milsark I. W., Cerami A. C. (1985) Passive immunization against cachectin/tumor necrosis factor protects mice from lethal effect of endotoxin. Science 229, 869–871 [DOI] [PubMed] [Google Scholar]
- 5. Danner R. L., Elin R. J., Hosseini J. M., Wesley R. A., Reilly J. M., Parillo J. E. (1991) Endotoxemia in human septic shock. Chest 99, 169–175 [DOI] [PubMed] [Google Scholar]
- 6. West M. A., Heagy W. (2002) Endotoxin tolerance: a review. Crit. Care Med. 30, (suppl.) S64–S73 [PubMed] [Google Scholar]
- 7. Fan H., Cook J. A. (2004) Molecular mechanisms of endotoxin tolerance. J. Endotoxin Res. 10, 71–84 [DOI] [PubMed] [Google Scholar]
- 8. Draisma A., Pickkers P., Bouw M. P., van der Hoeven J. G. (2009) Development of endotoxin tolerance in humans in vivo. Crit. Care Med. 37, 1261–1267 [DOI] [PubMed] [Google Scholar]
- 9. Biswas S. K., Lopez-Collazo E. (2009) Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 [DOI] [PubMed] [Google Scholar]
- 10. Foster S. L., Hargreaves D. C., Medzhitov R. (2007) Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 [DOI] [PubMed] [Google Scholar]
- 11. Mages J., Dietrich H., Lang R. (2007) A genome-wide analysis of LPS tolerance in macrophages. Immunobiology 212, 723–737 [DOI] [PubMed] [Google Scholar]
- 12. Shimauchi H., Ogawa T., Okuda K., Kusumoto Y., Okada H. (1999) Autoregulatory effect of interleukin-10 on proinflammatory cytokine production by Porphyromonas gingivalis lipopolysaccharide-tolerant human monocytes. Infect. Immun. 67, 2153–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frankenberger M., Pechumer H., Ziegler-Heitbrock H. W. (1995) Interleukin-10 is upregulated in LPS tolerance. J. Inflamm. 45, 56–63 [PubMed] [Google Scholar]
- 14. Randow F., Syrbe U., Meisel C., Krausch D., Zuckermann H., Platzer C., Volk H. D. (1995) Mechanism of endotoxin desensitization: involvement of interleukin 10 and transforming growth factor β. J. Exp. Med. 181, 1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. del Fresno C., García-Rio F., Gómez-Piña V., Soares-Schanoski A., Fernández-Ruíz I., Jurado T., Kajiji T., Shu C., Marín E., Gutierrez del Arroyo A., Prados C., Arnalich F., Fuentes-Prior P., Biswas S. K., López-Collazo E. (2009) Potent phagocytic activity with impaired antigen presentation identifying lipopolysaccharide-tolerant human monocytes: demonstration in isolated monocytes from cystic fibrosis patients. J. Immunol. 182, 6494–6507 [DOI] [PubMed] [Google Scholar]
- 16. Monneret G., Finck M. E., Venet F., Debard A. L., Bohé J., Bienvenu J., Lepape A. (2004) The anti-inflammatory response dominates after septic shock: association of low monocyte HLA-DR expression and high interleukin-10 concentration. Immunol. Lett. 95, 193–198 [DOI] [PubMed] [Google Scholar]
- 17. Wolk K., Döcke W. D., von Baehr V., Volk H. D., Sabat R. (2000) Impaired antigen presentation by human monocytes during endotoxin tolerance. Blood 96, 218–223 [PubMed] [Google Scholar]
- 18. Escoll P., del Fresno C., García L., Vallés G., Lendínez M. J., Arnalich F., López-Collazo E. (2003) Rapid up-regulation of IRAK-M expression following a second endotoxin challenge in human monocytes and in monocytes isolated from septic patients. Biochem. Biophys. Res. Commun. 311, 465–472 [DOI] [PubMed] [Google Scholar]
- 19. López-Collazo E., Fuentes-Prior P., Arnalich F., del Fresno C. (2006) Pathophysiology of interleukin-1 receptor-associated kinase-M: implications in refractory state. Curr. Opin. Infect. Dis. 19, 237–244 [DOI] [PubMed] [Google Scholar]
- 20. Brint E. K., Xu D., Liu H., Dunne A., McKenzie A. N., O'Neill L. A., Liew F. Y. (2004) ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nat. Immunol. 5, 373–379 [DOI] [PubMed] [Google Scholar]
- 21. Wang J., Ouyang Y., Guner Y., Ford H. R., Grishin A. V. (2009) Ubiquitin-editing enzyme A20 promotes tolerance to lipopolysaccharide in enterocytes. J. Immunol. 183, 1384–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nomura F., Akashi S., Sakao Y., Sato S., Kawai T., Matsumoto M., Nakanishi K., Kimoto M., Miyake K., Takeda K., Akira S. (2000) Cutting edge: endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface toll-like receptor 4 expression. J. Immunol. 164, 3476–3479 [DOI] [PubMed] [Google Scholar]
- 23. Baltimore D., Boldin M. P., O'Connell R. M., Rao D. S., Taganov K. D. (2008) MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9, 839–845 [DOI] [PubMed] [Google Scholar]
- 24. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- 27. Bone R. C. (1994) Gram-positive organisms and sepsis. Arch. Intern. Med. 154, 26–34 [PubMed] [Google Scholar]
- 28. Ogawa C., Liu Y. J., Kobayashi K. S. (2011) Muramyl dipeptide and its derivatives: peptide adjuvant in immunological disorders and cancer therapy. Curr. Bioact. Compd. 7, 180–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 30. Kobayashi K., Inohara N., Hernandez L. D., Galán J. E., Núñez G., Janeway C. A., Medzhitov R., Flavell R. A. (2002) RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199 [DOI] [PubMed] [Google Scholar]
- 31. Wilmanski J. M., Petnicki-Ocwieja T., Kobayashi K. S. (2008) NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J. Leukoc. Biol. 83, 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Franchi L., Warner N., Viani K., Nuñez G. (2009) Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 227, 106–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petnicki-Ocwieja T., Hrncir T., Liu Y. J., Biswas A., Hudcovic T., Tlaskalova-Hogenova H., Kobayashi K. S. (2009) Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. U.S.A. 106, 15813–15818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Biswas A., Liu Y. J., Hao L., Mizoguchi A., Salzman N. H., Bevins C. L., Kobayashi K. S. (2010) Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc. Natl. Acad. Sci. U.S.A. 107, 14739–14744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 36. Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 37. Miceli-Richard C., Lesage S., Rybojad M., Prieur A. M., Manouvrier-Hanu S., Häfner R., Chamaillard M., Zouali H., Thomas G., Hugot J. P. (2001) CARD15 mutations in Blau syndrome. Nat. Genet. 29, 19–20 [DOI] [PubMed] [Google Scholar]
- 38. Rosé C. D., Doyle T. M., McIlvain-Simpson G., Coffman J. E., Rosenbaum J. T., Davey M. P., Martin T. M. (2005) Blau syndrome mutation of CARD15/NOD2 in sporadic early onset granulomatous arthritis. J. Rheumatol. 32, 373–375 [PubMed] [Google Scholar]
- 39. Kanazawa N., Okafuji I., Kambe N., Nishikomori R., Nakata-Hizume M., Nagai S., Fuji A., Yuasa T., Manki A., Sakurai Y., Nakajima M., Kobayashi H., Fujiwara I., Tsutsumi H., Utani A., Nishigori C., Heike T., Nakahata T., Miyachi Y. (2005) Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-κB activation: common genetic etiology with Blau syndrome. Blood 105, 1195–1197 [DOI] [PubMed] [Google Scholar]
- 40. Kim Y. G., Park J. H., Shaw M. H., Franchi L., Inohara N., Núñez G. (2008) The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity 28, 246–257 [DOI] [PubMed] [Google Scholar]
- 41. Ferreira M. E., Coelho M. M., Pelá I. R. (2001) Role of the hepatic function in the development of the pyrogenic tolerance to muramyl dipeptide. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R162–R169 [DOI] [PubMed] [Google Scholar]
- 42. Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 43. Hedl M., Li J., Cho J. H., Abraham C. (2007) Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc. Natl. Acad. Sci. U.S.A. 104, 19440–19445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A., Jr., Medzhitov R., Flavell R. A. (2002) IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 45. Virca G. D., Kim S. Y., Glaser K. B., Ulevitch R. J. (1989) Lipopolysaccharide induces hyporesponsiveness to its own action in RAW 264.7 cells. J. Biol. Chem. 264, 21951–21956 [PubMed] [Google Scholar]
- 46. Haas J. G., Baeuerle P. A., Riethmüller G., Ziegler-Heitbrock H. W. (1990) Molecular mechanisms in down-regulation of tumor necrosis factor expression. Proc. Natl. Acad. Sci. U.S.A. 87, 9563–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hasegawa M., Yang K., Hashimoto M., Park J. H., Kim Y. G., Fujimoto Y., Nuñez G., Fukase K., Inohara N. (2006) Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J. Biol. Chem. 281, 29054–29063 [DOI] [PubMed] [Google Scholar]
- 48. Kadota Y., Shirasu K., Guerois R. (2010) NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 35, 199–207 [DOI] [PubMed] [Google Scholar]
- 49. Whitesell L., Lindquist S. L. (2005) HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 50. Mansell A., Smith R., Doyle S. L., Gray P., Fenner J. E., Crack P. J., Nicholson S. E., Hilton D. J., O'Neill L. A., Hertzog P. J. (2006) Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat. Immunol. 7, 148–155 [DOI] [PubMed] [Google Scholar]
- 51. Fujimoto M., Naka T. (2003) Regulation of cytokine signaling by SOCS family molecules. Trends Immunol. 24, 659–666 [DOI] [PubMed] [Google Scholar]
- 52. Ilangumaran S., Ramanathan S., Rottapel R. (2004) Regulation of the immune system by SOCS family adaptor proteins. Semin. Immunol. 16, 351–365 [DOI] [PubMed] [Google Scholar]
- 53. Henricson B. E., Benjamin W. R., Vogel S. N. (1990) Differential cytokine induction by doses of lipopolysaccharide and monophosphoryl lipid A that result in equivalent early endotoxin tolerance. Infect. Immun. 58, 2429–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gustafson G. L., Rhodes M. J., Hegel T. (1995) Monophosphoryl lipid A as a prophylactic for sepsis and septic shock. Prog. Clin. Biol. Res. 392, 567–579 [PubMed] [Google Scholar]
- 55. Salkowski C. A., Detore G., Franks A., Falk M. C., Vogel S. N. (1998) Pulmonary and hepatic gene expression following cecal ligation and puncture: monophosphoryl lipid A prophylaxis attenuates sepsis-induced cytokine and chemokine expression and neutrophil infiltration. Infect. Immun. 66, 3569–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mayor A., Martinon F., De Smedt T., Pétrilli V., Tschopp J. (2007) A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 8, 497–503 [DOI] [PubMed] [Google Scholar]
- 57. Schulze-Lefert P. (2004) Plant immunity: the origami of receptor activation. Curr. Biol. 14, R22–R24 [PubMed] [Google Scholar]
- 58. Sangster T. A., Queitsch C. (2005) The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 8, 86–92 [DOI] [PubMed] [Google Scholar]
- 59. Shirasu K., Schulze-Lefert P. (2003) Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci. 8, 252–258 [DOI] [PubMed] [Google Scholar]
- 60. Tsai W. H., Huang D. Y., Yu Y. H., Chen C. Y., Lin W. W. (2011) Dual roles of NOD2 in TLR4-mediated signal transduction and -induced inflammatory gene expression in macrophages. Cell. Microbiol. 13, 717–730 [DOI] [PubMed] [Google Scholar]
- 61. Davis B. K., Wen H., Ting J. P. (2011) The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Takada H., Galanos C. (1987) Enhancement of endotoxin lethality and generation of anaphylactoid reactions by lipopolysaccharides in muramyl-dipeptide-treated mice. Infect. Immun. 55, 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Parant M. A., Pouillart P., Le Contel C., Parant F. J., Chedid L. A., Bahr G. M. (1995) Selective modulation of lipopolysaccharide-induced death and cytokine production by various muramyl peptides. Infect. Immun. 63, 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takada H., Yokoyama S., Yang S. (2002) Enhancement of endotoxin activity by muramyldipeptide. J. Endotoxin Res. 8, 337–342 [DOI] [PubMed] [Google Scholar]
- 65. Watanabe T., Asano N., Murray P. J., Ozato K., Tailor P., Fuss I. J., Kitani A., Strober W. (2008) Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J. Clin. Investig. 118, 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meshcheryakova E., Guryanova S., Makarov E., Alekseeva L., Andronova T., Ivanov V. (2001) Prevention of experimental septic shock by pretreatment of mice with muramyl peptides. Int. Immunopharmacol. 1, 1857–1865 [DOI] [PubMed] [Google Scholar]
- 67. Hedl M., Abraham C. (2011) Secretory mediators regulate Nod2-induced tolerance in human macrophages. Gastroenterology 140, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gallis H. A., Miller S. E., Wheat R. W. (1976) Degradation of 14C-labeled streptococcal cell walls by egg white lysozyme and lysosomal enzymes. Infect. Immun. 13, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Herskovits A. A., Auerbuch V., Portnoy D. A. (2007) Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 3, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.