Background: 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates intracellular signaling through uncharacterized pathways similar to those engaged by bacterial pathogens.
Results: Mitochondrial targeting agents and absence of STING impair the response to DMXAA in mouse macrophages.
Conclusion: Mitochondrial membrane potential is required for optimal response to DMXAA.
Significance: This study illustrates that mitochondrial physiology is pivotal in the host response to DMXAA and possibly bacterial pathogens.
Keywords: Antioxidants, Cytokines/Interferon, Inflammation, Innate Immunity, Macrophages, Mitochondria, Pattern Recognition Receptor, Reactive Oxygen Species (ROS)
Abstract
The chemotherapeutic agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA) is a potent inducer of type I IFNs and other cytokines. This ability is essential for its chemotherapeutic benefit in a mouse cancer model and suggests that it might also be useful as an antiviral agent. However, the mechanism underlying DMXAA-induced type I IFNs, including the host proteins involved, remains unclear. Recently, it was reported that the antioxidant N-acetylcysteine (NAC) decreased DMXAA-induced TNF-α and IL-6, suggesting that oxidative stress may play a role. The goal of this study was to identify host proteins involved in DMXAA-dependent signaling and determine how antioxidants modulate this response. We found that expression of IFN-β in response to DMXAA in mouse macrophages requires the mitochondrial and endoplasmic reticulum resident protein STING. Addition of the antioxidant diphenylene iodonium (DPI) diminished DMXAA-induced IFN-β, but this decrease was independent of both the NADPH oxidase, Nox2, and de novo generation of reactive oxygen species. Additionally, IFN-β up-regulation by DMXAA was inhibited by agents that target the mitochondrial electron transport chain and, conversely, loss of mitochondrial membrane potential correlated with diminished innate immune signaling in response to DMXAA. Up-regulation of Ifnb1 gene expression mediated by cyclic dinucleotides was also impaired by DPI, whereas up-regulation of Ifnb1 mRNA due to cytosolic double-stranded DNA was not. Although both stimuli signal through STING, cyclic dinucleotides interact directly with STING, suggesting that recognition of DMXAA by STING may also be mediated by direct interaction.
Introduction
5,6-Dimethylxanthenone-4-acetic acid (DMXAA)2 is a chemotherapeutic compound classified as a vascular disrupting agent for its ability to induce apoptosis in endothelial cells (1–3), disrupting the blood vessels that are essential for supplying the requisite nutrients to the growing tumor. In addition to this rapid apoptotic event, it has been shown that DMXAA is also a potent inducer of cytokines, like TNF-α and IFN-β (4–6), that are also typically induced during infection with bacteria and viruses. Expression of this latter cytokine by DMXAA causes IFN-β-dependent growth inhibition of Lewis lung carcinoma cells injected subcutaneously into mice (7). Despite showing promise in initial phase II trials for treatment of non-small cell lung cancer in combination with the current chemotherapeutic regimen of carboplatin and paclitaxel (8, 9), DMXAA did not show efficacy in the most recent phase III clinical trial (10). However, induction of IFN-β also allows DMXAA to act as an antiviral agent during in vitro infection of multiple cell types with a variety of influenza strains (including a Tamiflu-resistant strain) and during in vivo infection of mice with mouse-adapted influenza A virus (11, 12). Combined with its use as a tool to study signaling pathways activated by intracellular pathogens, understanding the mechanism of DMXAA-mediated up-regulation of IFN-β would benefit multiple fields of study.
DMXAA treatment of primary macrophages leads to up-regulated expression of IFN-β (4), suggesting that the antitumor and antiviral effects of DMXAA may be mediated, in part, by macrophage-derived IFN-β. IFN-β is expressed at very low levels basally in macrophages, but can be induced by ligation of host pattern recognition receptors (PRRs) by “pathogen-associated molecular patterns” present during viral or bacterial infection. PRRs can be further subdivided into transmembrane and cytosolic members. Examples of transmembrane PRRs include Toll-like receptor 4 (TLR4)-mediated recognition of bacterial lipopolysaccharide (13), recognition of lipoproteins by TLR2 (14), and of flagellin by TLR5 (15). TLR3-mediated recognition of double-stranded viral RNA occurs endosomally (16), and similarly, double-stranded viral RNA can also be recognized by cytosolic pattern receptors such as the RNA helicases, retinoic acid inducible gene-I, and melanoma differentiation-associated protein 5 (17, 18). The signal transduction pathways for these two PRRs utilize the mitochondrial adapter protein mitochondrial antiviral signaling protein (MAVS) (19–22). In addition to cytosolic dsRNA, up-regulation of IFN-β can also occur in response to cytosolic delivery of either dsDNA or the bacterial cyclic dinucleotides, cyclic di-AMP and cyclic di-GMP, in a manner requiring the mitochondrial and endoplasmic reticulum resident protein STING (23–26). STING is also necessary for IFN-β during infection with the intracellular pathogens herpes simplex virus 1 (27), Listeria monocytogenes (26), Legionella pneumophila (28), Brucella abortus (29), Francisella tularensis (30), and Chlamydia muridarum (31), suggesting its involvement in numerous cytosolic sensing pathways. However, it has been shown that IFN-β expression in macrophages treated with DMXAA occurs independently of TLRs and RNA helicases (4), but rather, is dependent upon the downstream signaling components IFN regulatory factor 3 (IRF3) and TANK-binding kinase-1 (TBK-1) that are common to multiple PRR pathways. To date, a host PRR has not yet been identified that can recognize DMXAA and trigger induction of IFN-β.
Although signal transduction pathways are initiated by PRRs, there are other cellular elements that can modulate the ensuing signal, such as reactive oxygen species (ROS). This was highlighted in a seminal paper by Ehrt and colleagues (32) who demonstrated a pervasive role for ROS in macrophage transcriptional activation in response to such diverse stimuli as IFN-γ and Mycobacterium tuberculosis. Mechanistically, this is frequently manifested by covalent modification of cysteine residues of proteins including reversible oxidation, formation of disulfide linkages, or S-glutathionylation. Paradoxically, ROS has been shown to activate signaling pathways like MAPK and EGF by inactivation of cellular phosphatases (33–35), whereas inactivating downstream transcription factors such as NF-κB and AP-1 (36, 37). This illustrates the intricate and complex role ROS plays in cell signaling. The role for ROS in antiviral signaling pathways has been an emerging field of study, showing regulation at multiple levels by various cellular sources. Professional phagocytes can deliberately produce ROS using a multiprotein complex featuring NADPH oxidase 2 (Nox2). Nox2-dependent ROS is crucial for antiviral signaling by maintaining basal levels of viral RNA helicase signaling adapter MAVS (38). MAVS and translocases of outer membrane 70 are two proteins located in the mitochondria that are important for activation of antiviral innate immune pathways (19–22, 39). In addition to the inducible generation of ROS by Nox2, the mitochondria can also be a source of basal ROS in the cell during the process of cellular respiration. It has been shown that optimal MAVS signaling requires intact mitochondrial membrane potential (40), whereas fragmenting the mitochondria also dampens MAVS signaling (41), showing the prominent role mitochondrial physiology can play in innate antiviral signaling. Another potential site for ROS generation is the peroxisome and this organelle has also been shown to participate in MAVS signaling (42). With respect to DMXAA signaling, much less has been characterized. N-Acetylcysteine (NAC), the precursor of the cellular antioxidant glutathione, can be added to analyze the affects of oxidative stress. Previously, it has been shown that NAC treatment of the macrophage cell line RAW 264.7 decreases the expression of numerous cytokine genes such as Il6 and Tnf in response to DMXAA (43). This data suggests that antioxidants might also influence the expression of IFN-β in response to DMXAA. The goal of the present study is to elucidate the mechanism of DMXAA-dependent signaling pathways by further characterizing the host proteins involved in this process and by examining how antioxidants modulate IFN-β up-regulation in response to DMXAA.
EXPERIMENTAL PROCEDURES
Reagents
The following reagents were purchased from Sigma, unless otherwise noted: DMXAA (catalog number D5817), rotenone (number R8875), antimycin A (number A8674), 2-thenoyltrifluoroacetone (TTFA) (number T9888), oligomycin (number 9996S, Cell Signaling, Danvers, MA), methylthiazolyldiphenyl tetrazolium bromide (MTT) (number 2128), carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (number C2759) and nitro blue tetrazolium (NBT) (number N6876). Diphenyleneiodonium chloride (DPI) (number BML-CN240) and the mitochondrial membrane potential sensitive dye JC-10 (number ENZ-52305) were purchased from Enzo Life Sciences (Farmingdale, NY). The intracellular ROS detecting fluorescent dye 2′,7′-dichlorodihydrofluorescein diacetate (H2-DCFDA) (number D399) and the mitochondrial membrane staining dye MitoTracker Green used to measure the total amount cellular mitochondria (number M7514) were purchased from Invitrogen. Phorbol 12-myristate 13-acetate (number 4174S), which was used to induce macrophage ROS, was purchased from Cell Signaling Technology. The ligands used to examine activation of STING-dependent pathways were the synthetic dsDNA species poly(dA:dT) complexed to the transfection reagent LyoVec (number tlrl-patc, Invivogen, San Diego, CA) to facilitate cytosolic delivery and cyclic di-GMP (number ED0001, Kerafast, Winston-Salem, NC). The latter compound was transfected into the cell using SuperFect transfection reagent (number 301305, Qiagen, Valencia, CA).
Primary Macrophage Isolation and in Vitro Tissue Culture
All experiments using animals were carried out with institutional approval from the IACUC at University of Maryland, Baltimore, MD. Macrophages were harvested as previously described (44) with minor modifications. Briefly, C57BL/6J mice (stock number 000664, Jackson Laboratories, Bar Harbor, ME) or Nox2 knock-out mice (45) (stock number 002365, C57BL/6 background, Jackson Laboratories) were injected intraperitoneally with 3 ml of 3% thioglycollate (Remel Products, Lenexa, KS). After 4 days, the peritoneal exudates of the mice were obtained by lavage with macrophage medium (RPMI media supplemented with 10% FBS, 10 mm HEPES buffer, 1% nonessential amino acids, 2 mm l-glutamine, 1 mm sodium pyruvate, and 1% penicillin/streptomycin). Macrophages isolated from multiple mice were pooled and then subsequently plated at 1 × 106 macrophages per well in 1 ml of macrophage media in a 24-well tissue culture dish. Nonadherent cells were removed by aspiration when the media was changed after a 2-h incubation at 37 °C and 5% CO2. All experiments were performed on the day after isolation or the day after thawing of macrophages frozen immediately after isolation. Bone marrow-derived macrophages from STING KO (27) and littermate controls were isolated and cultured as previously described (46). Macrophages were treated with 100 μg/ml of DMXAA, which was previously demonstrated to be the optimal dose for examining up-regulation of IFN-β (4). Two hundred hemagglutinin units of Sendai virus (Cantrell Strain, Charles River Laboratories, Wilmington, MA) and transfection of either dsDNA or cyclic di-GMP were also used to stimulate IFN-β expression. Where indicated, cells were pretreated for 1 h (oligomycin, CCCP, DPI, TTFA, and antimycin A) or 6 h (rotenone) prior to treatment with DMXAA. The media was not changed at any point after the initial prestimulation.
Expression Analysis by Quantitative Real-time PCR (qRT-PCR) and ELISA
For expression analysis, 0.5 ml of the TriPure isolation reagent (catalog number 11667157001, Roche Applied Science) was added directly to cell monolayers in 24-well plates immediately after the supernatants were removed. The harvested lysates were frozen at −80 °C and stored until analysis was performed. RNA was isolated according to the manufacturer's protocol. To remove any residual DNA before the reverse transcription reaction, the sample RNA was treated with RNase-free DNase (M6101, Promega, Madison, WI) at 37 °C for 30 min followed by 10 min at 70 °C to inactivate the enzyme. The reverse transcription reaction using the DNase-treated RNA samples was performed with the Superscript III first strand synthesis system for real time PCR (catalog number 18080-051, Invitrogen) according to the manufacturer's supplied instructions. Quantitative PCR took place in the 7900HT Fast Real-time PCR system (Applied Biosystems, Carlsbad CA) using the 2× Power SYBR Green mix PCR master mix (catalog number 4367660, Invitrogen), and the following primer sequences: hypoxanthine-guanine phosphoribosyltransferase sense, 5′-GCTGACCTGCTGGATTACATTAA-3′, hypoxanthine-guanine phosphoribosyltransferase antisense, 5′-TGATCATTACAGTAGCTCTTCAGTCT-3′; Ifnb1 sense, 5′-CACTTGAAGAGCTATTACTGGAGGG-3′, Ifnb1 antisense, 5′-CTCGGACCACCATCCAGG-3′; Tnf sense, 5′-GACCCTCACACTCAGATCATCTTCT-3′, Tnf antisense, 5′-CCACTTGGTGGTTTGCTACGA-3′. Nox1 sense, 5′-AATGCCCAGGATCGAGGT-3′; Nox1 antisense, 5′-GATGGAAGCAAAGGGAGTGA-3′, Nox2 sense, 5′-ACCTTACTGGCTGGGATGAA-3′, Nox2 antisense, 5′-TGCAATGGTCTTGAACTCGT-3′; Nox3 sense, 5′-TCCACATTGTGGCACTTGTT-3′, Nox3 antisense, 5′-AAAGGTGCGGACTGGATTGAG-3′; Nox4 sense, 5′-GATCACAGAAGGTCCCTAGCAG-3′, Nox4 antisense, 5′-GTTGAGGGCATTCACCAAGT-3′; Duox1 sense, 5′-AGAGAAGTTTCAACGCAGCCG-3′, Duox1 antisense, 5′-TGTAAAACACCGCCACACAGC-3′; Duox2 sense, 5′-TGCCTGTCGAGTCTCGTTCAT-3′, Duox2 antisense, 5′-AATGTGGCGCCGGTAGTTT-3′. The total amount of mRNA was calculated using the comparative Ct method and expressed as fold-induction compared with untreated cells (47). Measurement of IFN-β protein by ELISA was assayed as previously described (4).
Mitochondrial Isolation and Western Blotting
Intact mitochondria were isolated from 2.0 × 107 primary peritoneal macrophages using the Mitochondria Isolation Kit for Cultured Cells (number 89874, Pierce Biotechnology) according to the manufacturer's instructions. The mitochondria were lysed and the mitochondrial and cytosolic fractions were separated by SDS-PAGE and transferred to a nitrocellulose membrane for Western analysis. The membrane was probed with primary antibodies directed against STING (number 3337, Cell Signaling Technology) and the endoplasmic reticulum chaperone protein calreticulin (number 2981, Cell Signaling Technology). The bands were visualized using the peroxidase-AffiniPure goat anti-rabbit IgG (H+L) secondary antibody (number 111-035-003, Jackson ImmunoResearch Laboratories) for both.
Mitochondrial Dehydrogenase Assay
A colorimetric assay to measure mitochondrial dehydrogenase activity was performed as previously described to monitor the activity of mitochondrial succinate dehydrogenase by the MTT assay (48). Briefly, after incubation for 1 h with 1 mg/ml of MTT solution dissolved in phosphate-buffered saline (number 21-040-CV, Mediatech, Inc., Herndon, VA), the water-insoluble formazan products were dissolved with isopropyl alcohol, yielding a purple solution whose absorbance was quantified at 595 nm on a Biotek microplate reader.
Measurement of Cellular ATP and Staining for Mitochondrial Membrane and Mitochondrial Membrane Potential
Levels of cellular ATP were quantified using the ATP-GLOTM Bioluminometric Assay kit (number 30020-1, Biotium, Inc., Hayward, Ca) according to the manufacturer's instructions. Staining for total membrane potential was accomplished using MitoTracker Green, which stains the mitochondrial membrane independently of its membrane potential. Stock MitoTracker Green was dissolved in ultrapure DMSO at a concentration of 1 mm. MitoTracker Green was further diluted in Hanks' balanced salt solution to a working concentration of 100 nm. After treatment of cells with CCCP for 2 h to dissipate the mitochondrial membrane potential, media was withdrawn and replaced with the MitoTracker Green staining solution for an additional 30-min incubation at 37 °C, before being washed once with buffer and read on a microplate reader with an excitation filter of 485 nm and emission filter of 516 nm. The JC-10 dye exhibits two staining spectra. In normally respiring cells, the dye forms aggregates in the mitochondrial membrane, exhibiting orange fluorescence. When membrane potential is lost, monomeric JC10 forms in the cytosol, exhibiting green fluorescence. Stock JC-10 was prepared by dissolving JC-10 at a concentration of 5 mg/ml in ultrapure dimethyl sulfoxide. Staining of the cells was performed by diluting the stock solution to 5 mg/ml in macrophage culture medium for 10 min at 37 °C. Cells were washed twice with PBS, and resuspended in fresh culture medium and read immediately on a microplate reader with an excitation filter of 485 nm and emission filter of 527 nm.
Microplate Assays for Detection of Intracellular ROS Generation
Intracellular ROS generation was determined using fluorescent and colorimetric assays. The fluorescent assay utilized the fluorescein derivative H2-DCFDA. Solutions of H2-DCFDA were made fresh immediately prior to use by dissolving H2-DCFDA in 10 mm dimethyl sulfoxide. This stock solution was diluted to a working concentration of 10 μm in Hanks' balanced salt solution (number 21-022-CV, Mediatech, Inc., Herndon, VA) and added to 1.50 × 105 cells cultured in a 96-black well tissue culture plate for an incubation of 30 min after the initial stimulation with NAC, DPI, rotenone, or DMXAA was completed. After the cells were washed with pre-warmed Hanks' balanced salt solution, the cells were cultured in macrophage media for an additional 30 min for intracellular host esterases to activate the fluorescent dye. Fluorescence was measured on a Fluoroskan Ascent FL microplate reader (ThermoFisher Scientific) with an excitation filter of 485 nm and emission filter of 527 nm. The colorimetric assay for ROS detection is a variation of the NBT reduction assay (49), which is used to diagnose chronic granulomatous disease in humans (50). The NBT reagent was dissolved in macrophage media lacking phenol red immediately prior to use at a concentration of 2.0 mg/ml and dispensed into wells of a 96-well tissue culture plate containing 1.5 × 105 cells in an equal volume macrophage media for a final NBT concentration of 1.0 mg/ml. Unlike the fluorescent assay, the macrophages were stimulated with DMXAA or phorbol 12-myristate 13-acetate (PMA) concurrently with NBT. Monolayers were monitored kinetically for water-insoluble formazan formation by reading the absorbance at 595 nm using a Biotek plate reader.
siRNA-mediated Knockdown of STING
Knockdown of STING was performed in the macrophage cell line RAW 264.7 using the Amaxa Nucleofector device (Lonza Inc., Cologne, Germany) according to the manufacturer's supplied instructions. Briefly, 2 × 106 cells were electroporated with the Amaxa Cell Line Nucleofector Kit V (Lonza Inc., number VCA-1003) using program D-032 on the electroporator device and then plated in a 6-well dish containing fresh media. Cells were treated with either 250 pmol of a smartpool targeting STING (aka TMEM173) (Thermo Scientific, Dharmacon RNAi Technologies, catalog number L-055528-00-0005) or an siRNA duplex targeting the housekeeping gene GAPDH (Thermo Scientific, catalog number D-001830-02-05). Cells were allowed to rest for 48 h before being stimulated with DMXAA.
Statistical Analysis
Experiments were set up and performed in duplicate for RNA analysis. For microplate experiments, experiments were set up in at least triplicate. Values obtained from these experiments were analyzed for significance using either a one-way analysis of variance or a two-tailed Student's t test depending on the number of experimental groups using GraphPad Prism 4 (GraphPad Software Inc., La Jolla, CA). Significance was determined to be p < 0.05. For the one-way analysis of variance, if the data met this standard, a Tukey's post hoc test was used to test for significance between experimental groups in the data set. The resulting p values between groups are listed in the figures or figure legends for the individual experiments.
RESULTS
The Antioxidant DPI Diminishes DMXAA-mediated Signaling in Primary Macrophages
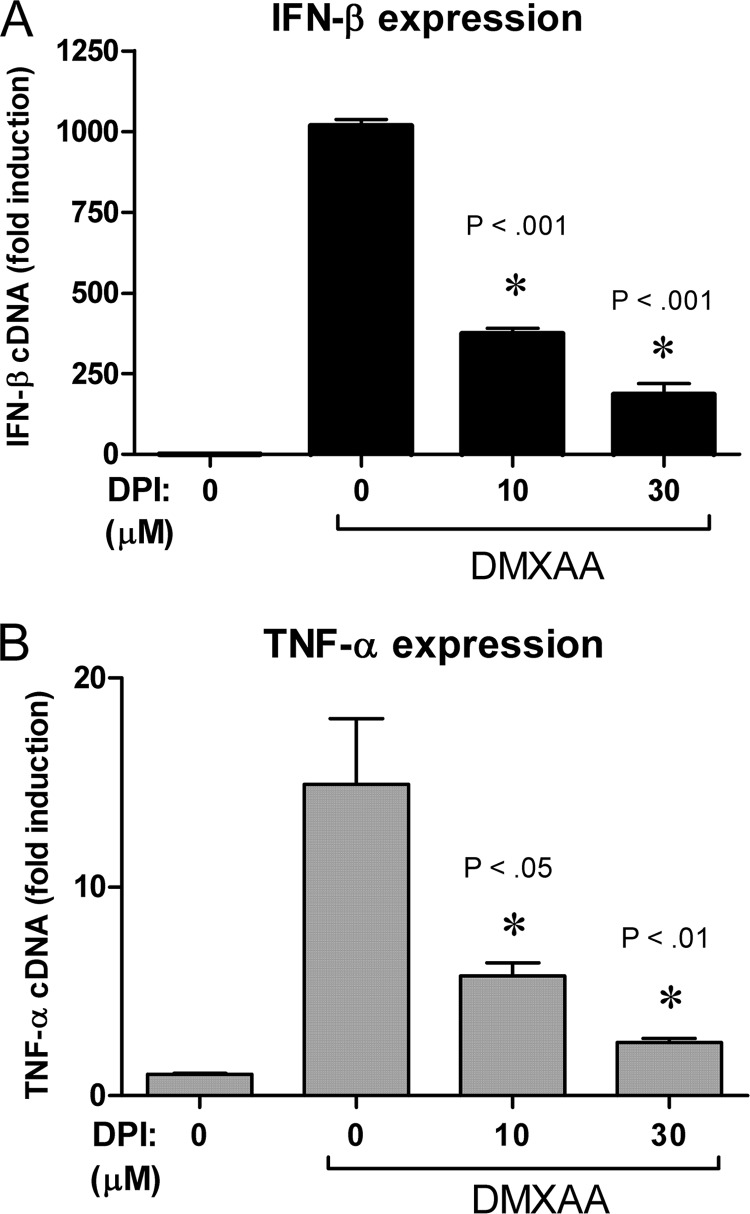
It has been shown previously that the antioxidant NAC inhibits DMXAA-mediated expression of cytokine genes such as Tnf in the macrophage cell line RAW 264.7 (43), suggesting that ROS could be regulating signaling in this pathway. Potential sources of cellular ROS in professional phagocytic cells are NADPH oxidases. Because DPI acts by inhibiting NADPH oxidases, it was hypothesized that treatment of DMXAA-treated macrophages with DPI (51) might also diminish DMXAA-dependent signaling. In agreement with this prediction, addition of DPI inhibited DMXAA-mediated expression of both Ifnb1 (Fig. 1A) and Tnf (Fig. 1B) mRNA in a dose-dependent manner.
FIGURE 1.
DMXAA mediated signaling is inhibited following exposure to the antioxidant DPI. Peritoneal macrophages were stimulated with 100 μg/ml of DMXAA for 2 h following pretreatment with the indicated dose of DPI. Expression of Ifnb1 mRNA (A) or Tnf mRNA (B) was quantified by qRT-PCR. Data represents mean ± S.D. for experiments performed in duplicate. An asterisk denotes the significance compared with the DMXAA-treated control at the indicated p value. A representative example of at least 3 independent experiments is shown.
DPI-mediated Inhibition of DMXAA Signaling Is Independent of Nox2 and de Novo Generation of Cellular ROS
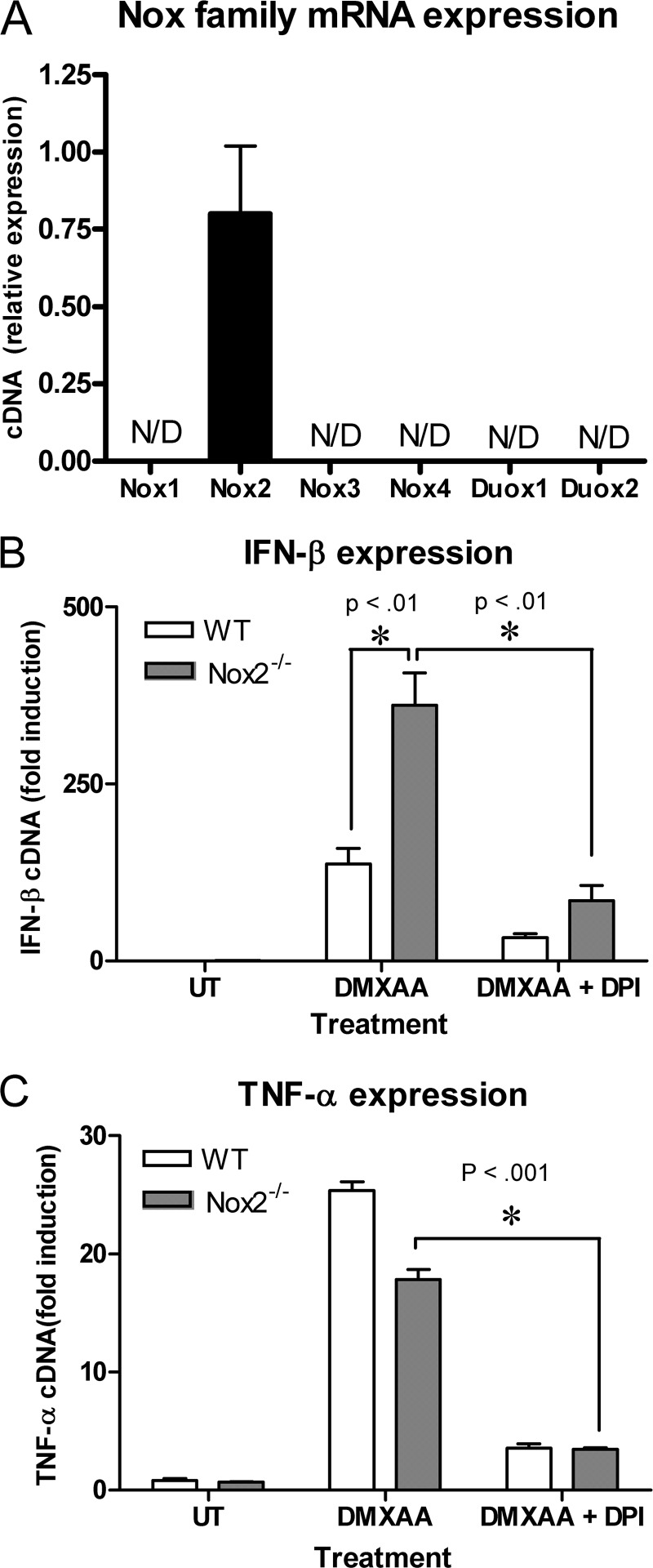
The finding that DPI inhibited cytokine up-regulation in response to DMXAA suggested the involvement of a NADPH oxidase in this pathway. To explore this possibility, we initially examined expression of various candidate macrophage NADPH oxidases by RT-PCR. As measured by qRT-PCR, primary macrophages expressed mRNA for only the professional phagocyte NADPH oxidase, Nox2 (Fig. 2A). Therefore, to test the involvement of Nox2 in DMXAA-mediated signaling, macrophages isolated from wild-type (WT) and Nox2−/− mice were stimulated with DMXAA. DMXAA-induced Ifnb1 mRNA was not inhibited in the Nox2−/− macrophages compared with WT controls, and in fact, was significantly enhanced (Fig. 2B). This data clearly indicates that Nox2 regulates signaling induced by DMXAA. DPI inhibited DMXAA-dependent IFN-β expression in both WT and Nox2−/− macrophages (Fig. 2B), further confirming that the target for DPI is independent of Nox2. Similarly, DPI also decreased DMXAA-induced TNF-α in Nox2−/− macrophages (Fig. 2C).
FIGURE 2.
DPI inhibits DMXAA-mediated cytokine production independent of Nox2. Resting peritoneal macrophages were examined for expression of the indicated NADPH oxidase family members by qRT-PCR. N/D indicates that no PCR product was detected after 40 PCR cycles. Peritoneal macrophages from wild type (WT) and Nox2 KO (Nox2−/−) mice were stimulated for 2 h with either DMXAA alone (100 μg/ml) or DMXAA following pretreatment with 30 μm DPI. Ifnb1 (B) and Tnf mRNA (C) expressions were quantified by qRT-PCR. Data represents mean ± S.D. for experiments performed in duplicate. An asterisk denotes a significant difference at the indicated p value. All panels show a representative example of 3 independent experiments.
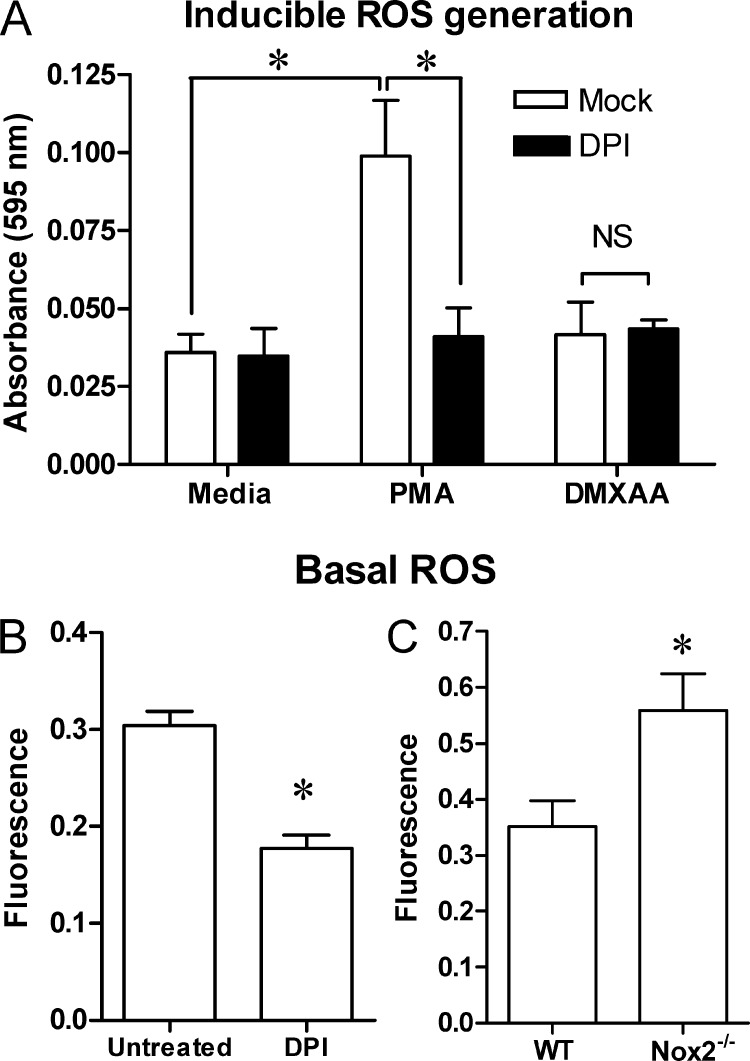
In addition to NADPH oxidases, DPI can also inhibit flavin-containing proteins of the mitochondrial electron transport chain (52, 53), albeit with lesser potency (51), indicating that DMXAA might trigger ROS production from the mitochondria, independent of Nox2 involvement. PMA has been reported to stimulate the activity of host NADPH oxidases to produce ROS in a Nox2-dependent fashion (54, 55). However, stimulation of macrophages with PMA, but not DMXAA, induced ROS production that was inhibited by DPI (Fig. 3A). DPI (Fig. 3B) and NAC (data not shown) both inhibited basal ROS levels in a fluorescence-based assay, similar to what was previously seen upon NAC treatment of RAW 264.7 cells (43). The fact that Nox2−/− macrophages actually exhibited increased basal ROS levels when compared with WT cells (Fig. 3C) suggests that the inhibitory effect of DPI on basal ROS levels is independent of Nox2 and indicates that increased basal ROS levels may explain why Nox2−/− macrophages expressed increased Ifnb1 in response to DMXAA (Fig. 2B).
FIGURE 3.
The antioxidant DPI impairs basal cellular ROS levels but not DMXAA-induced ROS generation. A, peritoneal macrophages were treated for 2 h with PMA (10 μm) or DMXAA (100 μg/ml) with or without concurrent treatment with DPI (30 μm) in media containing NBT (1 mg/ml) to measure inducible ROS generation. The formation of ROS-dependent formazan crystals was measured by absorbance readings taken at 595 nm. B, macrophages were treated with DPI (30 μm) for 2 h without any DMXAA present. C, separately, macrophages from WT or Nox2 KO mice were cultured without any subsequent treatment. Basal ROS levels were measured in B and C using the fluorescent dye H2-DCFDA as described under “Experimental Procedures.” Data represents the mean ± S.D. for experiments performed in replicates of 8 (A and B) or 6 (C). An asterisk denotes a statistically significant difference (p < 0.001). NS denotes not significant. A representative example of 3 independent experiments is shown.
DPI Inhibits Cyclic Dinucleotide-induced Activation of STING Signaling, but Does Not Broadly Inhibit All Innate Signaling Pathways
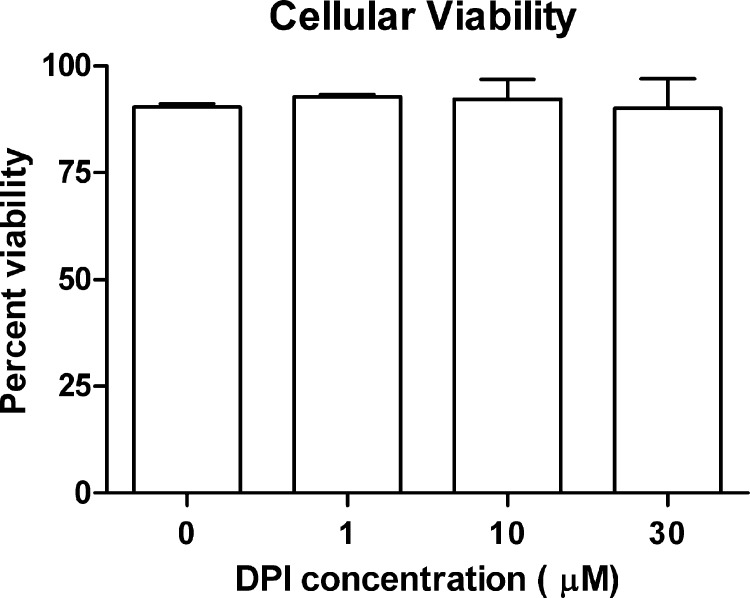
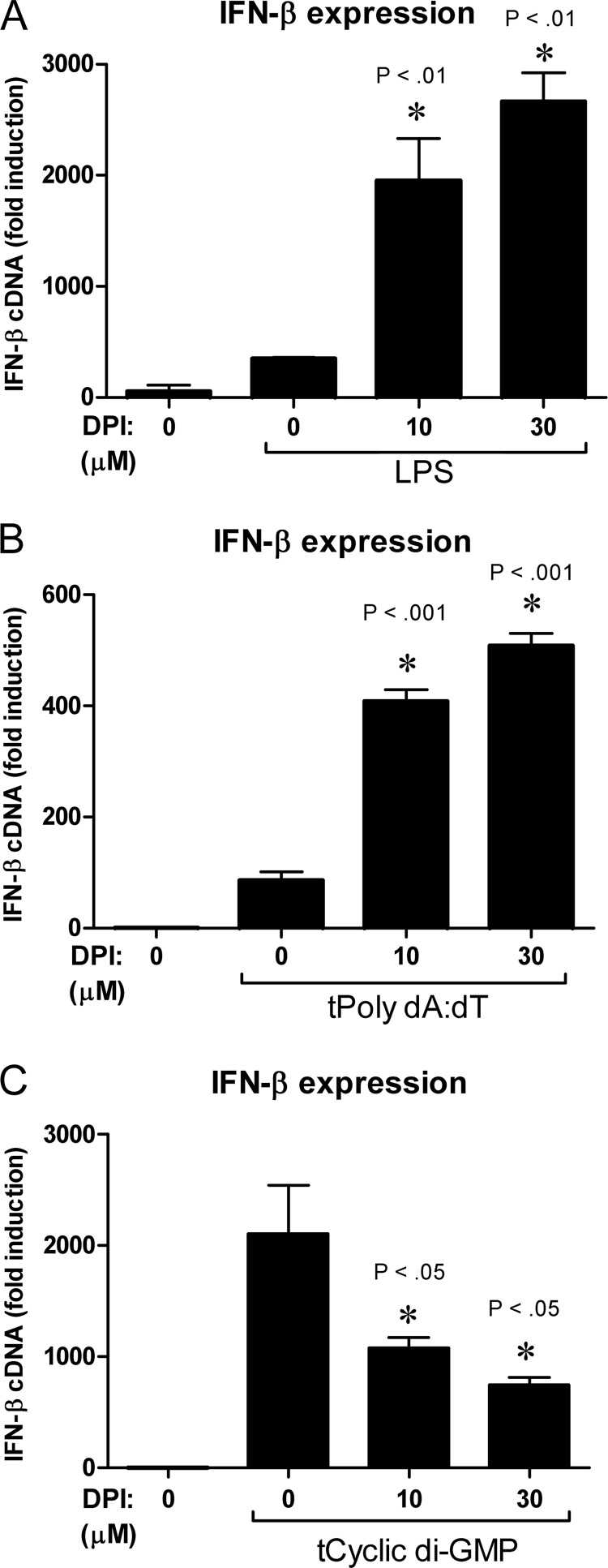
One possible explanation for a lack of DMXAA-mediated signaling upon treatment with DPI is that DPI exerts a cytotoxic effect on the cell, impairing signaling pathways globally. However, addition of DPI did not alter the cellular viability during the time frame of the experiments (Fig. 4). In support of this data, DPI did not inhibit signaling globally because DPI actually enhanced Ifnb1 mRNA up-regulation by the prototypical TLR4 agonist, LPS (Fig. 5A). However, because DMXAA is thought to activate non-TLR intracellular signaling pathways (4), the effect of DPI on intracellular signaling downstream of STING was next examined. STING mediates signaling in response to both dsDNA and cyclic dinucleotides independent of TLRs (23, 24, 27, 56, 57). In response to synthetic dsDNA transfected into the cytosol (tPoly(dA:dT)), DPI actually promoted STING-mediated up-regulation of Ifnb1 (Fig. 5B). In contrast, DPI dose-dependently decreased the up-regulation of Ifnb1 in response to transfected cyclic di-GMP (Fig. 5C).
FIGURE 4.
DPI does not decrease cell viability. Peritoneal macrophages were stimulated with the indicated doses of DPI for 3 h. At the end of this incubation cellular viability was calculated by measuring of trypan blue exclusion. The data represents the mean ± S.D. for experiments performed in duplicate. A representative of 3 independent experiments is shown for each panel.
FIGURE 5.
DPI has contrasting effects on IFN-β expression in response to LPS, dsDNA, and cyclic di-GMP. Ifnb1 expression was quantified by qRT-PCR in macrophages that were stimulated with either 100 ng/ml of lipopolysaccharide for 2 h (A), 2 μg/ml of transfected dsDNA for 4 h (B), or 10 μg/ml of transfected cyclic di-GMP for 6 h (C). Data represents mean ± S.D. for experiments performed in duplicate. An asterisk denotes a statistically significant difference compared with LPS, poly(dA:dT), or cyclic di-GMP treated control at the indicated p value. A representative of 3 independent experiments is shown for all panels.
DMXAA Signaling Requires the Mitochondrial/ER Protein STING
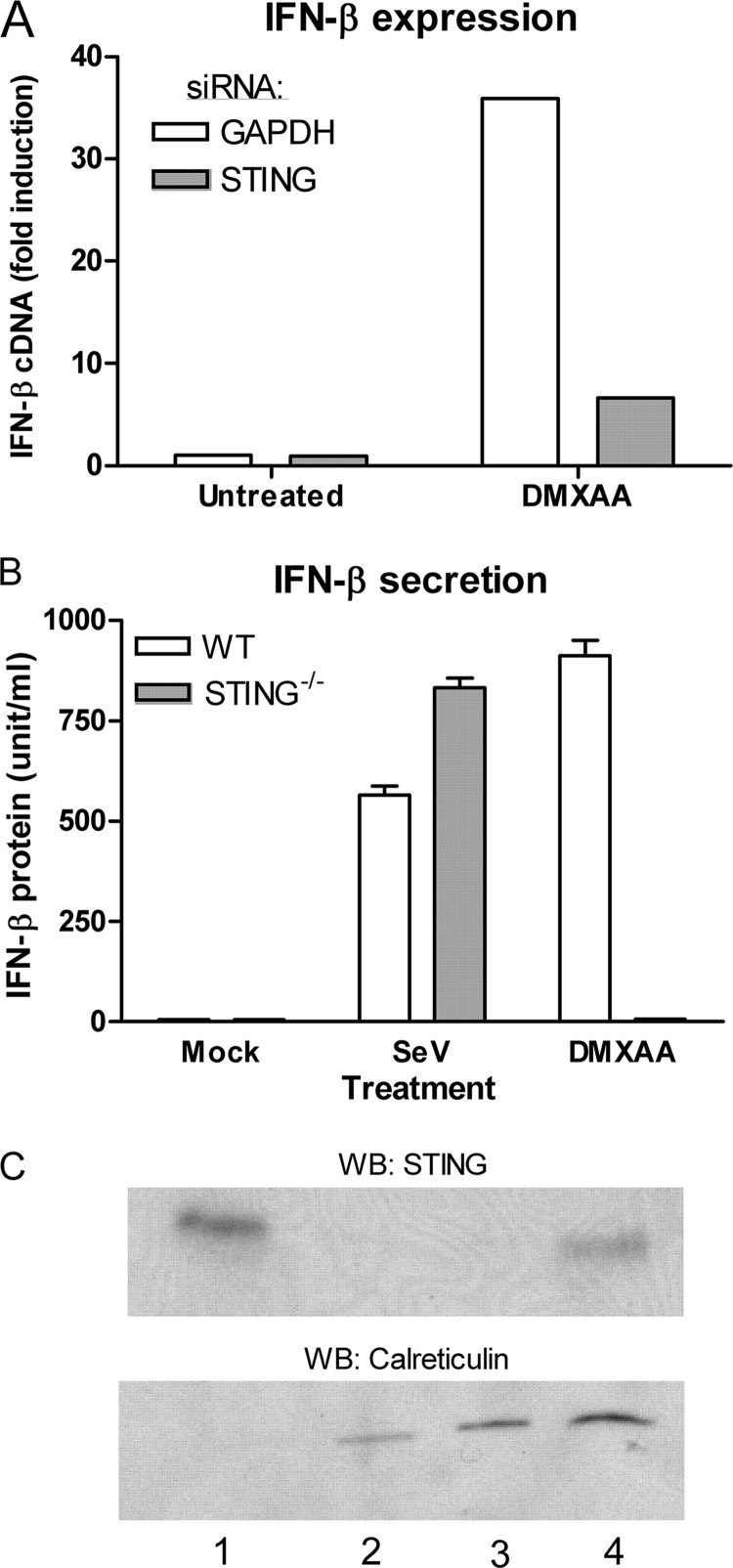
Because cyclic dinucleotides bind to STING directly to stimulate signaling (58), whereas dsDNA involves recognition by accessory PRRs upstream of STING (59–61), we hypothesized that DMXAA-induced signaling might also be mediated by STING. To test whether STING was necessary for DMXAA-mediated signaling, STING expression was initially knocked down in the RAW 264.7 macrophage cell line. STING knockdown led to a decreased transcriptional up-regulation of Ifnb1 in response to DMXAA when compared with cells treated with a control siRNA duplex targeting Gapdh (Fig. 6A). As siRNA can sometimes have off-target effects, macrophages from STING−/− mice (27) were used to corroborate this finding. Cells lacking STING demonstrated significantly less IFN-β secretion by macrophages stimulated by DMXAA than STING-sufficient cells (Fig. 6B). STING can be detected by Western blot from mitochondria isolated from primary mouse macrophages (Fig. 6C).
FIGURE 6.
The mitochondrial protein STING mediates DMXAA signaling. A, RAW264.7 macrophages were electroporated with siRNA targeting either GAPDH or STING and treated with DMXAA (100 μg/ml) or solvent (mock) for 1 h. Ifnb1 mRNA was quantified by qRT-PCR. A representative of 3 independent experiments is shown. B, bone marrow-derived macrophages were cultured from STING KO (STING−/−) mice or STING sufficient littermate controls (WT) and subsequently treated with DMXAA (100 μg/ml) or infected with 200 hemagglutinin units of Sendai virus (SeV). Supernatants were collected after 12 h and the amount of IFN-β was measured by ELISA. C, intact mitochondria were isolated from primary peritoneal macrophages and the cell fractions were analyzed by Western blot to detect either STING or the endoplasmic reticulum chaperone calreticulin. Lane 1, mitochondrial fraction; lane 2, cytosolic fraction; lane 3, total cell lysate ∼1 × 105 cells; lane 4, total cell lysate ∼1 × 106 cells.
Mitochondrial Oxidative Phosphorylation Correlates with Optimal DMXAA-mediated Signaling
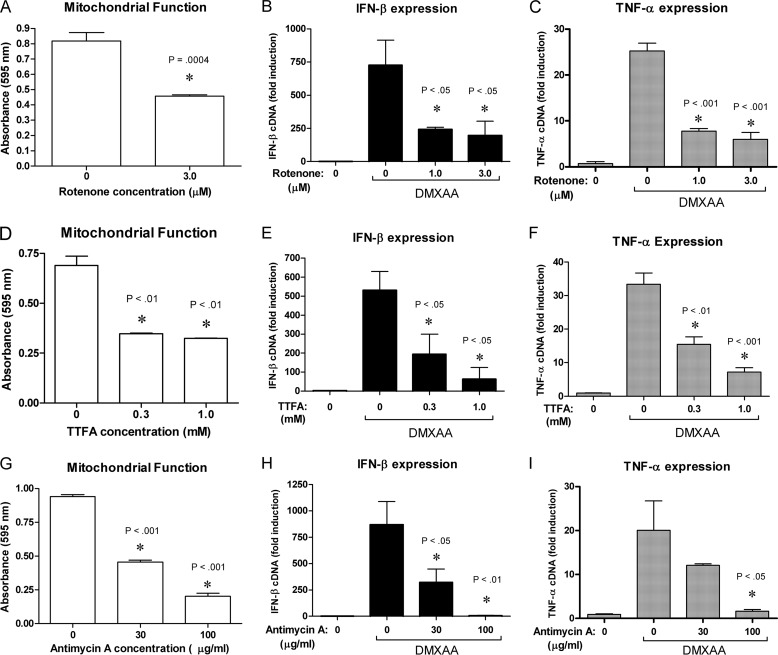
As the mitochondria is thought to be a primary source of basal ROS via incomplete oxidation of molecular oxygen in the electron transport chain, we hypothesized that DPI suppresses endogenous ROS levels by impairing mitochondrial oxidative phosphorylation. If reagents that more specifically target the mitochondria also inhibit DMXAA signaling, this would lend credence to the theory that DPI exerts its effect on signaling by impacting mitochondrial physiology. The electron transport chain involves pumping of protons to form ATP by oxidative phosphorylation, but can also lead to formation of ROS by incomplete reduction of molecular oxygen. Therefore, to address this possibility, rotenone, an inhibitor of complex I of the electron transport chain (62), was added to macrophages prior to addition of DMXAA. The primary molecular target of each of the pharmacological agents used is listed in Table 1. Rotenone significantly inhibited mitochondrial dehydrogenase activity as measured by the MTT assay (Fig. 7A) and also inhibited the induction of Ifnb1 (Fig. 7B) and Tnf (Fig. 7C) mRNA in response to DMXAA. TTFA and antimycin A inhibit complexes II and III of the electron transport chain, respectively (63, 64). Similar to rotenone, both TTFA (Fig. 7, D–F) and antimycin A (Fig. 7, G–I) dose dependently inhibited the cellular mitochondrial dehydrogenase activity and the expression of Ifnb1 and Tnf mRNA in response to DMXAA.
TABLE 1.
Pharmacologic agents used in this study and their putative targets
| Compound | Pharmacologic target(s) | Ref. no. | Figs. |
|---|---|---|---|
| DPI | 1) NADPH oxidases | 1) 52 | 1 and 2 |
| 2) Mitochondrial complex I | 2) 71 | ||
| Rotenone | Mitochondrial complex I | 62 | 7 |
| TTFA | Mitochondrial complex II | 63 | 7 |
| Antimycin A | Mitochondrial complex III | 64 | 7 |
| Oligomycin | Mitochondrial F1F0-ATPase | 68 | 8 |
| CCCP | Mitochondrial membrane potential | 69 | 9 |
FIGURE 7.
Inhibitors of the electron transport chain impair DMXAA signaling. A, D, and G, mitochondrial dehydrogenase activity was assessed using the MTT assay on peritoneal macrophages treated with the indicated doses of rotenone (A), TTFA (D), or antimycin A (G) or solvent control. Data represents the mean optical density ± S.D. for experiments performed in duplicate except for A, which was performed in triplicate. Ifnb1, and Tnf mRNA were analyzed by qRT-PCR in macrophages pretreated with increasing doses of rotenone (B and C), TTFA (E and F), or antimycin A (H and I) prior to a 2-h exposure to DMXAA, as indicated by the bar. Data represents the mean ± S.D. for experiments performed in duplicate. An asterisk denotes a statistically significant difference compared with untreated control (A, D, and G), or compared with DMXAA treated control (B, C, E, F, H, and I) at the indicated p value. A representative of 3 independent experiments is shown for each panel.
DMXAA-mediated Signaling Is Independent of ATP Levels, but Requires an Intact Mitochondrial Membrane Potential
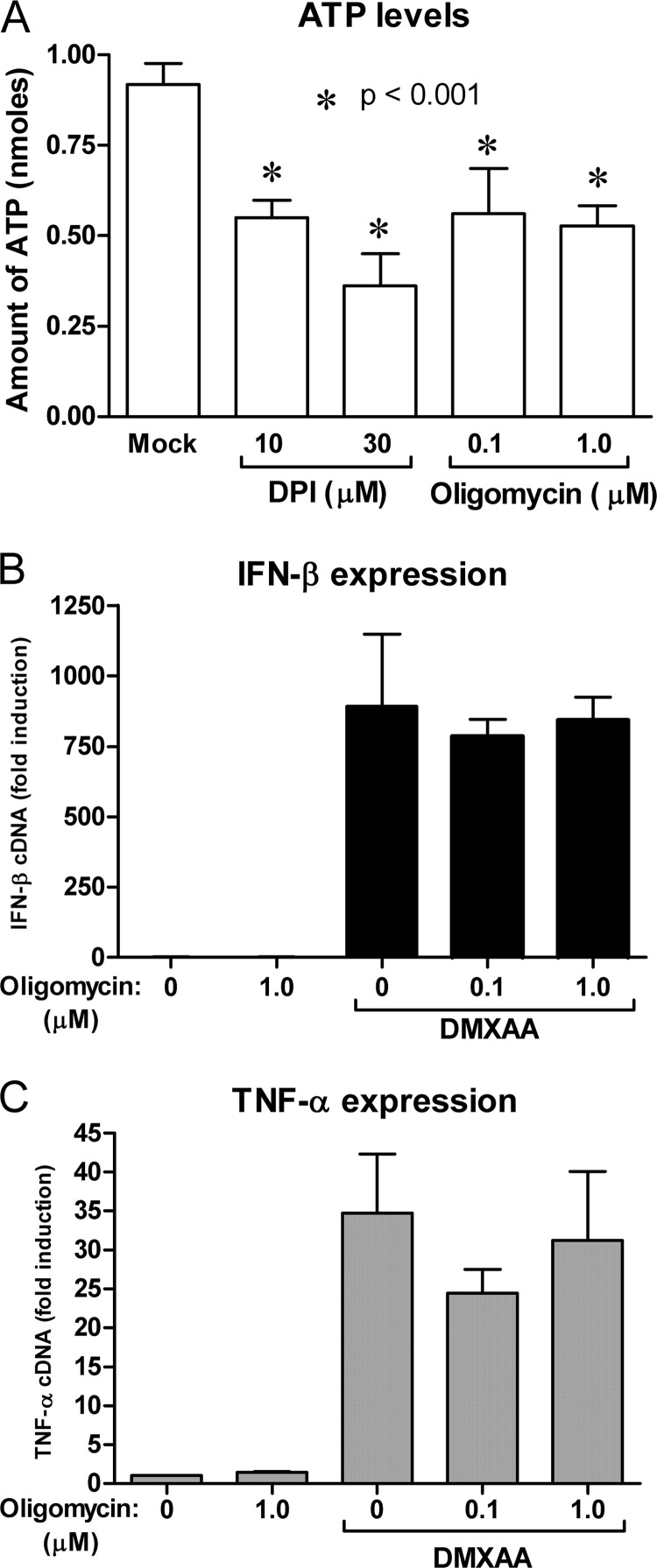
In addition to targeting the electron transport chain, antimycin A, TTFA, and rotenone have all been shown to reduce ATP generation (65) or mitochondrial membrane potential (66, 67) within the cell. DPI treatment also leads to a decrease in cellular ATP with a dose-response relationship that is similar to its repression of DMXAA-mediated Ifnb1 mRNA induction (Fig. 8A). A specific inhibitor of mitochondrial ATP synthase, oligomycin (68), also decreased the level of cellular ATP (Fig. 8A). Oligomycin should not affect the pumping of protons by the individual electron transport chain complexes. In contrast to DPI treatment, ATP synthase inhibition did not alter the expression of Ifnb1 or Tnf mRNA in response to DMXAA (Fig. 8, B and C).
FIGURE 8.
DPI inhibits ATP levels within the cell, but DMXAA signaling occurs independently of ATP synthesis. Peritoneal macrophages were treated with the DPI mitochondrial F1F0-ATPase inhibitor oligomycin at the indicated doses. ATP levels (A) were measured by luminescent assay after 2 h of incubation. Data represents the mean ± S.D. for experiments performed in triplicate. An asterisk denotes a statistically significant difference compared with solvent treated control at the indicated p value. Macrophages were stimulated with DXMAA for 2 h following a 1-h pretreatment with the indicated doses of oligomycin. Expression of Ifnb1 (B) and Tnf (C) were quantified by qRT-PCR. Data represents the mean ± S.D. for experiments performed in duplicate. For each panel a representative of 3 independent experiments is shown.
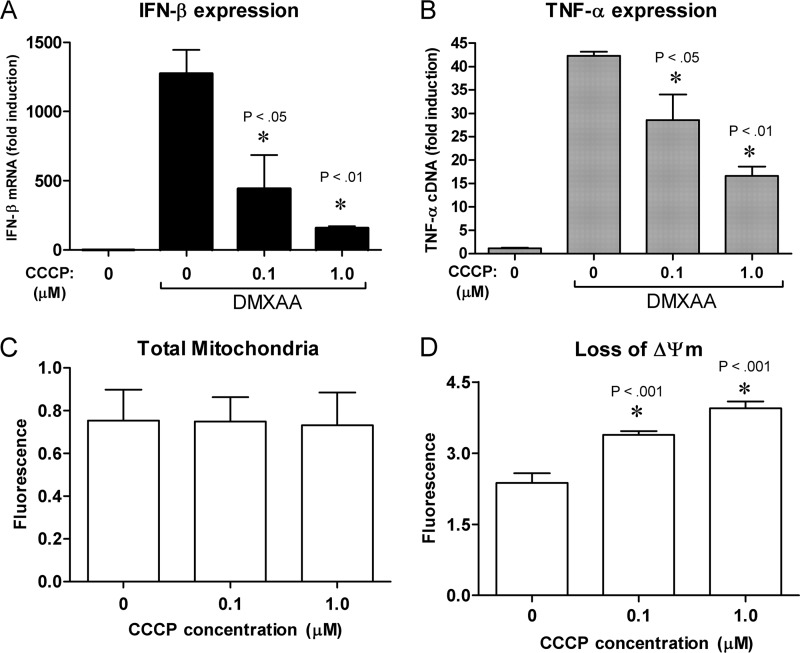
Based on this data, we hypothesize that mitochondrial membrane potential, and not ATP synthesis, may be required for signaling by DMXAA. To test this hypothesis, cells were treated with the proton ionophore CCCP that acts by dissipating the membrane potential by transporting protons into the matrix of the mitochondria (69). CCCP pretreatment of cells decreased DMXAA-induced expression of Ifnb1 and Tnf mRNA in a dose-dependent manner (Fig. 9, A and B). Total mitochondrial membrane was unchanged as measured by staining with MitoTracker Green (Fig. 9C), whereas loss of mitochondrial membrane potential (ΔΨm) increased (Fig. 9D) with a similar dose dependence as Ifnb1 mRNA expression. These data indicate that DMXAA signaling requires an intact mitochondrial membrane potential.
FIGURE 9.
DMXAA-dependent Ifnb1 expression requires an intact mitochondrial membrane potential. Peritoneal macrophages were pretreated for 1 h with the indicated doses of the proton ionophore CCCP or mock solvent. Ifnb1 (A) and Tnf (B) expression were quantified by qRT-PCR after a 2-h incubation with DMXAA. Data represents mean ± S.D. for experiments performed in duplicate. Total mitochondria (C) was quantified by MitoTracker Green staining after 3 h after treatment of the indicated doses of CCCP. Data represents mean ± S.D. for experiments performed in replicates of 5. Loss of mitochondrial membrane potential (ΔΨm) (D) was quantified by measuring the amount of monomeric JC-10 dye after 2 h of treatment with the indicated doses of CCCP. Data represents mean ± S.D. for experiments performed in replicates of 8. An asterisk denotes a statistically significant difference compared with DMXAA treated control (A and B) or solvent treated control (D) at the indicated p value. For each panel a representative of 3 independent experiments is shown.
DISCUSSION
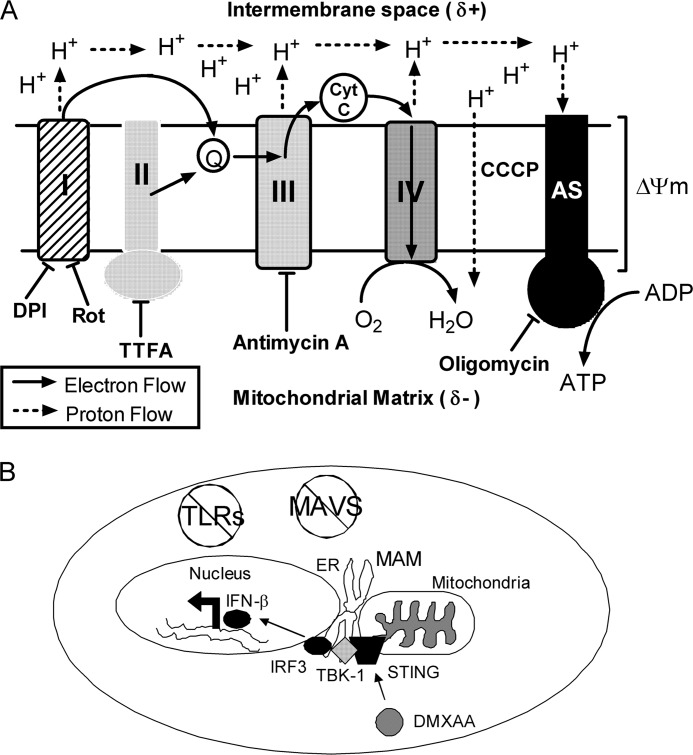
Because the mechanisms governing IFN-β induction in response to DMXAA remain largely uncharacterized, this study attempted to clarify how DMXAA mediates cytokine up-regulation in primary mouse macrophages. To address this issue, much of this study involves the use of pharmacologic agents. The molecular targets of those used in this study are listed in Table 1 and diagrammed in Fig. 10A. It is important to note that any time pharmacologic agents play a prominent role in an experiment, off-target effects must be taken into account. In the RAW 264.7 macrophage cell line, it has been demonstrated that addition of the antioxidant NAC inhibited production of many cytokines such as TNF-α, IL-6, and IL-1β in response to stimulation with DMXAA (43). This indicates that ROS could potentially be modulating the signaling pathway mediated by DMXAA. Inhibition of DMXAA-induced Ifnb1 and Tnf by DPI (Fig. 1) supports this notion. Interestingly, when used at high doses, NAC can also lead to impaired mitochondrial function, including loss of mitochondrial membrane potential due to inhibition of cation transport within the organelle (70). Although DPI is classically used as a potent NADPH oxidase inhibitor (51), at higher concentrations, DPI can likewise target the flavin-containing enzymes in the mitochondrial electron transport chain (52, 53). As a result, DPI has been shown to inhibit both mitochondrial membrane potential (71) and ATP generation within the cell (72). In this study, the effect of DPI on cytokine up-regulation in response to DMXAA was demonstrated to be independent of the NADPH oxidase, Nox2, suggesting that it may be exerting its affect through a mitochondrial mechanism (Fig. 3). In support of this possibility, DMXAA-dependent signaling has been determined previously to be sensitive to the presence of salicylic acid (4) that has also been linked to inhibition of the electron transport chain at complex III (73).
FIGURE 10.
Multiple pharmacologic inhibitors affect mitochondrial physiology. A, the electron transport chain is pictured, showing the flow of electrons and protons through the respective enzyme complexes during respiration. Also pictured are inhibitors that effect mitochondrial physiology and lead to decreased cytokine induction following DMXAA treatment. The resulting hypothesis is that ΔΨm must be intact for optimal signaling to occur in response to DMXAA. Abbreviations: Q, coenzyme Q; Cyt C, cytochrome c; AS, ATP synthase. B, STING is the missing TLR- and RLR-independent element connecting DMXAA to activation of IRF3, leading to induction of IFN-β.
To strengthen the link between the mitochondria and innate immune signaling in response to DMXAA, macrophages were treated with inhibitors that individually target three of the electron transport chain enzyme complexes (I, II, and III). Similar to DPI and NAC, these inhibitors also diminished DMXAA-dependent cytokine up-regulation (Fig. 7). The most likely conclusion to be drawn from these cumulative studies is that mitochondrial physiology is central for the innate immune response to DMXAA. The crucial issue to address is the mechanism for how targeting the mitochondrial electron transport chain alters the response to DMXAA. The three most likely possibilities are as follows. 1) The inhibitors cause cytotoxicity and a global decrease in innate signaling pathways; 2) de novo ATP levels influence the signaling through this pathway; or 3) mitochondrial membrane potential is required for signaling. Although prolonged DPI exposure might lead to signs of apoptosis, over the relatively short time frame of these experiments, there was no cytotoxicity associated with the treatment and no global impairment of other innate pathways like TLR4. DPI was shown to diminish the level of cellular ATP, but oligomycin does this to a similar extent without affecting cytokine levels, implying that this is an effect of DPI treatment and not a specific cause of cytokine repression. Oligomycin does not impair the proton pumping and, thus, the mitochondrial membrane potential. Therefore, the fact that the proton ionophore CCCP represses cytokine up-regulation at the same dose as it alters the mitochondrial membrane potential suggests a causal relationship. Interestingly, it has been reported that MAVS-dependent signaling was also found to be regulated by the mitochondrial membrane potential (40) in a manner very similar to the findings in this study. This effect was hypothesized to work at the level of MAVS, without directly affecting downstream signaling elements like TBK1 and IRF3 (40).
The Ifnb1 promoter contains positive regulatory elements for transcription factors NF-κB, AP-1, and IRF family members (74–76). Although capable of activating MAPK- and NF-κB-dependent pathways (77, 78), it was previously shown in primary mouse macrophages that DMXAA triggers potent IRF3 activation that requires TBK-1, but is independent of all known TLRs and MAVS-dependent RNA helicases (4). On the surface, this finding is reminiscent of how IFN-β is regulated in response to cytosolic dsDNA (23, 24) or bacterial cyclic dinucleotides (26) and the intracellular pathogens C. muridarum (31), L. monocytogenes (26), F. tularensis (30), B. abortus (29), and L. pneumophila (28). One commonality among those diverse stimuli is the involvement of the host protein STING in the expression of IFN-β. Additionally, our finding that DPI inhibited the up-regulation of Ifnb1 in response to cyclic dinucleotides (Fig. 5C) supports the hypothesis that STING might also be a crucial protein in the DMXAA-mediated signaling pathway. In agreement with this prediction, cells lacking STING, either due to siRNA-mediated knockdown or due to genetic ablation, were impaired for up-regulation of Ifnb1 mRNA or protein by DMXAA treatment (Fig. 6, A and B). Overall, this study demonstrates that STING is a necessary component in the pathway that DMXAA uses to activate the downstream components IRF3 and TBK-1, and to up-regulate Ifnb1 (Fig. 10B). Direct binding of STING by bacterial cyclic dinucleotides is necessary and sufficient for IFN-β expression (58). However, dsDNA does not compete for binding with cyclic dinucleotides to STING, and different amino acids seem to be important for rescuing null mutants in response to the two different stimuli, illustrating that the two pathways are dissociable (58). In the case of dsDNA sensing, STING seems to require accessory DNA-binding PRRs such as DAI (59), DDX41 (61), and IFI16 (60). Because DMXAA signaling appears to mimic cyclic dinucleotide signaling, we speculate that DMXAA activates STING-dependent pathways through direct binding. This would potentially explain why DMXAA signals in a different manner than dsDNA (Fig. 5B), whose recognition by STING is mediated by other dsDNA sensors that, in turn, engage STING. We further theorize that mitochondrial membrane potential mediates the binding between DMXAA and STING in a redox-dependent fashion whether in the mitochondria or in the neighboring endoplasmic reticulum fractions associated with the mitochondria (Fig. 10B). In support of this hypothesis, we were able to detect STING in mitochondria isolated from primary mouse macrophages (Fig. 6C). The cellular localization of STING has not been completely characterized, particularly in primary cells. Although, we cannot definitively rule out that a small, undetectable amount of endoplasmic reticulum was contaminating the mitochondrial fraction and contributing to the total amount of STING protein, the evidence clearly suggests that STING resides in the mitochondria. Trafficking studies have been performed looking at STING localization in response to dsDNA (79), but it is unclear whether these results would translate to the distinct direct binding pathways triggered by cyclic dinucleotides or, by extension, DMXAA.
Alternatively, other possibilities could explain the mitochondrial membrane dependence for maximal DMXAA signaling. For instance, proteins that gain or lose association with the mitochondria when membrane potential is lost, such as the pro-apototic Bcl-2 family members Bax and Bak (80) or apoptosis inducing factor (81), might also play a role in regulating innate signaling. Dissociation of these proteins could influence neighboring organelles like the endoplasmic reticulum in addition to the mitochondria. Additionally, it is intriguing that apoptosis inducing factor is a mitochondrial inner membrane flavoprotein with NADH oxidase activity (82) and could be a direct target of DPI. Studies are ongoing to determine these potential contributions to the innate signaling pathways activated by DMXAA.
A separate future avenue of study is how mitochondrial physiology may possibly affect how the infected cell responds to intracellular bacterial or viral pathogens. DPI has been shown to alter the activation of IRF3 during infection with respiratory syncytial virus, suggesting a potential involvement in that infection model (83). Theoretically, a pathogen targeting the mitochondria might also be able to suppress the innate immune response generated by its presence. In conclusion, the findings from this study have demonstrated that STING mediates the signaling pathway activated by DMXAA and antioxidants impair this response independently of de novo ROS generation.
Acknowledgment
We thank Dr. Alan Cross for critical review of the manuscript prior to submission.
This work was supported, in whole or in part, by National Institutes of Health Grants AI-18797 (to S. N. V.), AI-070823 (to M. S. W.), and AI-067497 (to K. A. F.) and a Ruth L. Kirchstein National Research Service Award for Individual Postdoctoral Fellows (to D. P.).
- DMXAA
- dimethylxanthenone-4-acetic acid
- CCCP
- carbonyl cyanide 3-chlorophenylhydrazone
- DPI
- diphenylene iodonium
- IRF3
- interferon regulatory factor 3
- MAVS
- mitochondrial antiviral signaling protein
- NAC
- N-acetylcysteine
- NBT
- nitro blue tetrazolium
- Nox2
- NADPH oxidase 2
- PMA
- phorbol 12-myristate 13-acetate
- PRR
- pattern recognition receptors
- ROS
- reactive oxygen species
- STING
- stimulator of interferon gene
- TBK-1
- TANK-binding kinase-1
- TLR
- Toll-like receptor
- TTFA
- 2-thenoyltrifluoroacetone
- dsDNA
- double-stranded DNA
- MTT
- methylthiazolyldiphenyl tetrazolium bromide
- H2-DCFDA
- 2′,7′-dichlorodihydrofluorescein diacetate
- qRT
- quantitative real-time.
REFERENCES
- 1. Baguley B. C. (2003) Antivascular therapy of cancer. DMXAA. Lancet Oncol. 4, 141–148 [DOI] [PubMed] [Google Scholar]
- 2. Ching L. M., Cao Z., Kieda C., Zwain S., Jameson M. B., Baguley B. C. (2002) Induction of endothelial cell apoptosis by the antivascular agent 5,6-dimethylxanthenone-4-acetic acid. Br. J. Cancer 86, 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ching L. M., Zwain S., Baguley B. C. (2004) Relationship between tumour endothelial cell apoptosis and tumour blood flow shutdown following treatment with the antivascular agent DMXAA in mice. Br. J. Cancer 90, 906–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roberts Z. J., Goutagny N., Perera P. Y., Kato H., Kumar H., Kawai T., Akira S., Savan R., van Echo D., Fitzgerald K. A., Young H. A., Ching L. M., Vogel S. N. (2007) The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J. Exp. Med. 204, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perera P. Y., Barber S. A., Ching L. M., Vogel S. N. (1994) Activation of LPS-inducible genes by the antitumor agent 5,6-dimethylxanthenone-4-acetic acid in primary murine macrophages. Dissection of signaling pathways leading to gene induction and tyrosine phosphorylation. J. Immunol. 153, 4684–4693 [PubMed] [Google Scholar]
- 6. Jassar A. S., Suzuki E., Kapoor V., Sun J., Silverberg M. B., Cheung L., Burdick M. D., Strieter R. M., Ching L. M., Kaiser L. R., Albelda S. M. (2005) Activation of tumor-associated macrophages by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid induces an effective CD8+ T-cell-mediated antitumor immune response in murine models of lung cancer and mesothelioma. Cancer Res. 65, 11752–11761 [DOI] [PubMed] [Google Scholar]
- 7. Roberts Z. J., Ching L. M., Vogel S. N. (2008) IFN-beta-dependent inhibition of tumor growth by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA). J. Interferon Cytokine Res. 28, 133–139 [DOI] [PubMed] [Google Scholar]
- 8. McKeage M. J., Reck M., Jameson M. B., Rosenthal M. A., Gibbs D., Mainwaring P. N., Freitag L., Sullivan R., Von Pawel J. (2009) Phase II study of ASA404 (vadimezan, 5,6-dimethylxanthenone-4-acetic acid/DMXAA) 1800 mg/m(2) combined with carboplatin and paclitaxel in previously untreated advanced non-small cell lung cancer. Lung Cancer 65, 192–197 [DOI] [PubMed] [Google Scholar]
- 9. McKeage M. J., Von Pawel J., Reck M., Jameson M. B., Rosenthal M. A., Sullivan R., Gibbs D., Mainwaring P. N., Serke M., Lafitte J. J., Chouaid C., Freitag L., Quoix E. (2008) Randomised phase II study of ASA404 combined with carboplatin and paclitaxel in previously untreated advanced non-small cell lung cancer. Br. J. Cancer 99, 2006–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lara P. N., Jr., Douillard J. Y., Nakagawa K., von Pawel J., McKeage M. J., Albert I., Losonczy G., Reck M., Heo D. S., Fan X., Fandi A., Scagliotti G. (2011) Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small cell lung cancer. J. Clin. Oncol. 29, 2965–2971 [DOI] [PubMed] [Google Scholar]
- 11. Shirey K. A., Nhu Q. M., Yim K. C., Roberts Z. J., Teijaro J. R., Farber D. L., Blanco J. C., Vogel S. N. (2011) The anti-tumor agent, 5,6-dimethylxanthenone-4-acetic acid (DMXAA), induces IFN-β-mediated antiviral activity in vitro and in vivo. J. Leukocyte Biol. 89, 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng G., Wang L. C., Fridlender Z. G., Cheng G. S., Chen B., Mangalmurti N. S., Saloura V., Yu Z., Kapoor V., Mozdzanowska K., Moon E., Sun J., Kreindler J. L., Cohen N. A., Caton A. J., Erikson J., Albelda S. M. (2011) Pharmacologic activation of the innate immune system to prevent respiratory viral infections. Am. J. Respir. Cell Mol. Biol. 45, 480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. (1999) Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide. Evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752 [PubMed] [Google Scholar]
- 14. Lien E., Sellati T. J., Yoshimura A., Flo T. H., Rawadi G., Finberg R. W., Carroll J. D., Espevik T., Ingalls R. R., Radolf J. D., Golenbock D. T. (1999) Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 274, 33419–33425 [DOI] [PubMed] [Google Scholar]
- 15. Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 16. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 17. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 18. Andrejeva J., Childs K. S., Young D. F., Carlos T. S., Stock N., Goodbourn S., Randall R. E. (2004) The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc. Natl. Acad. Sci. U.S.A. 101, 17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 20. Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 21. Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 22. Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 23. Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H. B. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 [DOI] [PubMed] [Google Scholar]
- 24. Ishikawa H., Barber G. N. (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin L., Hill K. K., Filak H., Mogan J., Knowles H., Zhang B., Perraud A. L., Cambier J. C., Lenz L. L. (2011) MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic di-AMP and cyclic di-GMP. J. Immunol. 187, 2595–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sauer J. D., Sotelo-Troha K., von Moltke J., Monroe K. M., Rae C. S., Brubaker S. W., Hyodo M., Hayakawa Y., Woodward J. J., Portnoy D. A., Vance R. E. (2011) The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishikawa H., Ma Z., Barber G. N. (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lippmann J., Muller H., Naujoks J., Tabeling C., Shin S., Witzenrath M., Hellwig K., Kirschning C. J., Taylor G. A., Barchet W., Bauer S., Suttorp N., Roy C. R., Opitz B. (2012) Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell Microbiol. 13, 1668–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Almeida L. A., Carvalho N. B., Oliveira F. S., Lacerda T. L., Vasconcelos A. C., Nogueira L., Bafica A., Silva A. M., Oliveira S. C. (2011) MyD88 and STING signaling pathways are required for IRF3-mediated IFN-β induction in response to Brucella abortus infection. PLoS One 6, e23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones J. W., Kayagaki N., Broz P., Henry T., Newton K., O'Rourke K., Chan S., Dong J., Qu Y., Roose-Girma M., Dixit V. M., Monack D. M. (2010) Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. U.S.A. 107, 9771–9776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prantner D., Darville T., Nagarajan U. M. (2010) Stimulator of IFN gene is critical for induction of IFN-β during Chlamydia muridarum infection. J. Immunol. 184, 2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ehrt S., Schnappinger D., Bekiranov S., Drenkow J., Shi S., Gingeras T. R., Gaasterland T., Schoolnik G., Nathan C. (2001) Reprogramming of the macrophage transcriptome in response to interferon-γ and Mycobacterium tuberculosis. Signaling roles of nitric-oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194, 1123–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. (2003) Redox regulation of protein-tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423, 769–773 [DOI] [PubMed] [Google Scholar]
- 34. Lee S. R., Kwon K. S., Kim S. R., Rhee S. G. (1998) Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 273, 15366–15372 [DOI] [PubMed] [Google Scholar]
- 35. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Reactive oxygen species promote TNFα-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 36. Abate C., Patel L., Rauscher F. J., 3rd, Curran T. (1990) Redox regulation of fos and jun DNA-binding activity in vitro. Science 249, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 37. Toledano M. B., Leonard W. J. (1991) Modulation of transcription factor NF-κB binding activity by oxidation-reduction in vitro. Proc. Natl. Acad. Sci. U.S.A. 88, 4328–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soucy-Faulkner A., Mukawera E., Fink K., Martel A., Jouan L., Nzengue Y., Lamarre D., Vande Velde C., Grandvaux N. (2010) Requirement of NOX2 and reactive oxygen species for efficient RIG-I-mediated antiviral response through regulation of MAVS expression. PLoS Pathog. 6, e1000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu X. Y., Wei B., Shi H. X., Shan Y. F., Wang C. (2010) Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 20, 994–1011 [DOI] [PubMed] [Google Scholar]
- 40. Koshiba T., Yasukawa K., Yanagi Y., Kawabata S. (2011) Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci. Signal 4, ra7. [DOI] [PubMed] [Google Scholar]
- 41. Castanier C., Garcin D., Vazquez A., Arnoult D. (2010) Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 11, 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dixit E., Boulant S., Zhang Y., Lee A. S., Odendall C., Shum B., Hacohen N., Chen Z. J., Whelan S. P., Fransen M., Nibert M. L., Superti-Furga G., Kagan J. C. (2010) Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brauer R., Wang L. C., Woon S. T., Bridewell D. J., Henare K., Malinger D., Palmer B. D., Vogel S. N., Kieda C., Tijono S. M., Ching L. M. (2010) Labeling of oxidizable proteins with a photoactivatable analog of the antitumor agent DMXAA. Evidence for redox signaling in its mode of action. Neoplasia 12, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prantner D., Nagarajan U. M. (2009) Role for the chlamydial type III secretion apparatus in host cytokine expression. Infect. Immun. 77, 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pollock J. D., Williams D. A., Gifford M. A., Li L. L., Du X., Fisherman J., Orkin S. H., Doerschuk C. M., Dinauer M. C. (1995) Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9, 202–209 [DOI] [PubMed] [Google Scholar]
- 46. Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. (2003) LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the Toll adapters TRAM and TRIF. J. Exp. Med. 198, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 48. Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival. Application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 49. Pick E., Charon J., Mizel D. (1981) A rapid densitometric microassay for nitro blue tetrazolium reduction and application of the microassay to macrophages. J. Reticuloendothel. Soc. 30, 581–593 [PubMed] [Google Scholar]
- 50. Nathan D. G., Baehner R. L., Weaver D. K. (1969) Failure of nitro blue tetrazolium reduction in the phagocytic vacuoles of leukocytes in chronic granulomatous disease. J. Clin. Invest. 48, 1895–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hancock J. T., Jones O. T. (1987) The inhibition by diphenyleneiodonium and its analogues of superoxide generation by macrophages. Biochem. J. 242, 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ragan C. I., Bloxham D. P. (1977) Specific labelling of a constituent polypeptide of bovine heart mitochondrial reduced nicotinamide-adenine dinucleotide-ubiquinone reductase by the inhibitor diphenyleneiodonium. Biochem. J. 163, 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Holland P. C., Sherratt H. S. (1972) Biochemical effects of the hypoglycaemic compound diphenyleneiodonnium. Catalysis of anion-hydroxyl ion exchange across the inner membrane of rat liver mitochondria and effects on oxygen uptake. Biochem. J. 129, 39–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Porter C. D., Parkar M. H., Levinsky R. J., Collins M. K., Kinnon C. (1993) X-linked chronic granulomatous disease. Correction of NADPH oxidase defect by retrovirus-mediated expression of gp91-phox. Blood 82, 2196–2202 [PubMed] [Google Scholar]
- 55. McPhail L. C., DeChatelet L. R., Shirley P. S. (1976) Further characterization of NADPH oxidase activity of human polymorphonuclear leukocytes. J. Clin. Invest. 58, 774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McWhirter S. M., Barbalat R., Monroe K. M., Fontana M. F., Hyodo M., Joncker N. T., Ishii K. J., Akira S., Colonna M., Chen Z. J., Fitzgerald K. A., Hayakawa Y., Vance R. E. (2009) A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic di-GMP. J. Exp. Med. 206, 1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takaoka A., Wang Z., Choi M. K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 [DOI] [PubMed] [Google Scholar]
- 60. Unterholzner L., Keating S. E., Baran M., Horan K. A., Jensen S. B., Sharma S., Sirois C. M., Jin T., Latz E., Xiao T. S., Fitzgerald K. A., Paludan S. R., Bowie A. G. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y. J. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gutman M., Singer T. P., Beinert H., Casida J. E. (1970) Reaction sites of rotenone, piericidin A, and amytal in relation to the nonheme iron components of NADH dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 65, 763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Baginsky M. L., Hatefi Y. (1969) Reconstitution of succinate-coenzyme Q reductase (complex II) and succinate oxidase activities by a highly purified, reactivated succinate dehydrogenase. J. Biol. Chem. 244, 5313–5319 [PubMed] [Google Scholar]
- 64. Ragan C. I., Heron C. (1978) The interaction between mitochondrial NADH-ubiquinone oxidoreductase and ubiquinol-cytochrome c oxidoreductase. Evidence for stoichiometric association. Biochem. J. 174, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J. A., Robinson J. P. (2003) Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278, 8516–8525 [DOI] [PubMed] [Google Scholar]
- 66. Zhou R., Yazdi A. S., Menu P., Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 67. Byun H. O., Kim H. Y., Lim J. J., Seo Y. H., Yoon G. (2008) Mitochondrial dysfunction by complex II inhibition delays overall cell cycle progression via reactive oxygen species production. J. Cell. Biochem. 104, 1747–1759 [DOI] [PubMed] [Google Scholar]
- 68. Chappell J. B., Crofts A. R. (1965) The effect of atractylate and oligomycin on the behaviour of mitochondria towards adenine nucleotides. Biochem. J. 95, 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kessler R. J., Tyson C. A., Green D. E. (1976) Mechanism of uncoupling in mitochondria. Uncouplers as ionophores for cycling cations and protons. Proc. Natl. Acad. Sci. U.S.A. 73, 3141–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Löhrke B., Xu J., Weitzel J. M., Krüger B., Goldammer T., Viergutz T. (2010) N-Acetylcysteine impairs survival of luteal cells through mitochondrial dysfunction. Cytometry A 77, 310–320 [DOI] [PubMed] [Google Scholar]
- 71. Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J. A., Robinson J. P. (2003) DPI induces mitochondrial superoxide-mediated apoptosis. Free Radic. Biol. Med. 34, 465–477 [DOI] [PubMed] [Google Scholar]
- 72. Hutchinson D. S., Csikasz R. I., Yamamoto D. L., Shabalina I. G., Wikström P., Wilcke M., Bengtsson T. (2007) Diphenylene iodonium stimulates glucose uptake in skeletal muscle cells through mitochondrial complex I inhibition and activation of AMP-activated protein kinase. Cell. Signal. 19, 1610–1620 [DOI] [PubMed] [Google Scholar]
- 73. de Souza W. R., Vessecchi R., Dorta D. J., Uyemura S. A., Curti C., Vargas-Rechia C. G. (2011) Characterization of Rubus fruticosus mitochondria and salicylic acid inhibition of reactive oxygen species generation at complex III/Q cycle. Potential implications for hypersensitive response in plants. J. Bioenerg. Biomembr. 43, 237–246 [DOI] [PubMed] [Google Scholar]
- 74. Lenardo M. J., Fan C. M., Maniatis T., Baltimore D. (1989) The involvement of NF-κB in β-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 57, 287–294 [DOI] [PubMed] [Google Scholar]
- 75. Du W., Maniatis T. (1992) An ATF/CREB binding site is required for virus induction of the human interferon β gene (corrected). Proc. Natl. Acad. Sci. U.S.A. 89, 2150–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wathelet M. G., Lin C. H., Parekh B. S., Ronco L. V., Howley P. M., Maniatis T. (1998) Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1, 507–518 [DOI] [PubMed] [Google Scholar]
- 77. Cheng G., Sun J., Fridlender Z. G., Wang L. C., Ching L. M., Albelda S. M. (2010) Activation of the nucleotide oligomerization domain signaling pathway by the non-bacterially derived xanthone drug 5′,6-dimethylxanthenone-4-acetic acid (Vadimezan). J. Biol. Chem. 285, 10553–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sun J., Wang L. C., Fridlender Z. G., Kapoor V., Cheng G., Ching L. M., Albelda S. M. (2011) Activation of mitogen-activated protein kinases by 5,6-dimethylxanthenone-4-acetic acid (DMXAA) plays an important role in macrophage stimulation. Biochem. Pharmacol. 82, 1175–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H., Matsunaga K., Kageyama S., Omori H., Noda T., Yamamoto N., Kawai T., Ishii K., Takeuchi O., Yoshimori T., Akira S. (2009) Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 106, 20842–20846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Proapoptotic BAX and BAK. A requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Susin S. A., Lorenzo H. K., Zamzami N., Marzo I., Snow B. E., Brothers G. M., Mangion J., Jacotot E., Costantini P., Loeffler M., Larochette N., Goodlett D. R., Aebersold R., Siderovski D. P., Penninger J. M., Kroemer G. (1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 441–446 [DOI] [PubMed] [Google Scholar]
- 82. Miramar M. D., Costantini P., Ravagnan L., Saraiva L. M., Haouzi D., Brothers G., Penninger J. M., Peleato M. L., Kroemer G., Susin S. A. (2001) NADH oxidase activity of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 276, 16391–16398 [DOI] [PubMed] [Google Scholar]
- 83. Indukuri H., Castro S. M., Liao S. M., Feeney L. A., Dorsch M., Coyle A. J., Garofalo R. P., Brasier A. R., Casola A. (2006) IKKϵ regulates viral-induced interferon regulatory factor-3 activation via a redox-sensitive pathway. Virology 353, 155–165 [DOI] [PubMed] [Google Scholar]