Background: GABAB receptors in the brain assemble with auxiliary KCTD8, 12, 12b, and 16 subunits that influence the receptor response in distinct ways.
Results: Distinct KCTD domains exert opposing effects on desensitization of the receptor response.
Conclusion: KCTD12 and -12b acquired desensitizing properties by disposing of a C-terminal inhibitory domain.
Significance: This study defines the domains and motifs in KCTD proteins that generate functionally distinct GABAB receptor subtypes.
Keywords: G Protein-coupled Receptors (GPCR), GABA Receptors, Molecular Pharmacology, Neurotransmitter Receptors, Receptor Desensitization, GABAB
Abstract
GABAB receptors assemble from principle and auxiliary subunits. The principle subunits GABAB1 and GABAB2 form functional heteromeric GABAB(1,2) receptors that associate with homotetramers of auxiliary KCTD8, -12, -12b, or -16 (named after their K+ channel tetramerization domain) subunits. These auxiliary subunits constitute receptor subtypes with distinct functional properties. KCTD12 and -12b generate desensitizing receptor responses while KCTD8 and -16 generate largely non-desensitizing receptor responses. The structural elements of the KCTDs underlying these differences in desensitization are unknown. KCTDs are modular proteins comprising a T1 tetramerization domain, which binds to GABAB2, and a H1 homology domain. KCTD8 and -16 contain an additional C-terminal H2 homology domain that is not sequence-related to the H1 domains. No functions are known for the H1 and H2 domains. Here we addressed which domains and sequence motifs in KCTD proteins regulate desensitization of the receptor response. We found that the H1 domains in KCTD12 and -12b mediate desensitization through a particular sequence motif, T/NFLEQ, which is not present in the H1 domains of KCTD8 and -16. In addition, the H2 domains in KCTD8 and -16 inhibit desensitization when expressed C-terminal to the H1 domains but not when expressed as a separate protein in trans. Intriguingly, the inhibitory effect of the H2 domain is sequence-independent, suggesting that the H2 domain sterically hinders desensitization by the H1 domain. Evolutionary analysis supports that KCTD12 and -12b evolved desensitizing properties by liberating their H1 domains from antagonistic H2 domains and acquisition of the T/NFLEQ motif.
Introduction
GABAB receptors are the G-protein-coupled receptors (GPCRs)5 for GABA, the main inhibitory neurotransmitter in the central nervous system. They are widely distributed throughout the brain and have been implicated in a variety of disorders including cognitive impairments, addiction, anxiety, depression, and epilepsy (1, 2). GABAB receptors activate Gαi/o-type G-proteins that inhibit adenylyl cyclase and efficiently gate ion channels (3–5). Native GABAB receptors are known to comprise principal and auxiliary subunits that influence receptor properties in distinct ways (5, 6). The principal subunits form two core receptors, GABAB(1a,2) and GABAB(1b,2), that bind all GABAB ligands, couple to G-proteins and regulate classical GABAB receptor effectors, including G-protein coupled inwardly rectifying K+ channels (GIRK channels, also known as Kir3 channels) and voltage-gated Ca2+ channels. The auxiliary subunits KCTD8, -12, -12b, and -16 are cytosolic proteins that modulate agonist potency and kinetic properties of the receptor response in distinct ways (6). In particular, KCTD12 or -12b produce fast desensitizing GABAB receptor-mediated Kir3 currents characterized by time constants of seconds while KCTD8 and -16 produce currents with little desensitization. The molecular determinants in the KCTD proteins and the mechanism underlying these kinetic differences in GABAB responses are unknown. Importantly, fast desensitization of GABAB receptor-mediated K+ currents is also observed with neurons (7, 8) expressing KCTD12 (9). However, it has not been specifically addressed whether this fast desensitization is due to the presence of KCTD12 in the receptor. In addition, desensitization of GABAB responses was shown to be regulated by GRK4 (10), RGS proteins (11–14) or phosphorylation of the GABAB2 subunit (15, 16).
KCTD8, -12, -12b, and -16 constitute a subfamily of the KCTD family of proteins. All KCTD proteins contain a N-terminal bric-a-brac, tramtrak, and broad complex (BTB) domain that is most similar in sequence to the T1 tetramerization domain of voltage-gated K+ channels (17, 18). In voltage-gated K+ channels, T1 domains are responsible for assembly of the four subunits around a central channel pore (19). Likewise, the T1 domains of auxiliary GABAB receptor subunits assemble into a homotetramer that tightly binds to the C-terminal intracellular domain of GABAB2 (6). All auxiliary GABAB receptor subunits contain a second conserved domain, designated the H1 homology domain, which is separated from the T1 domain by a non-conserved linker region. KCTD8 and -16 comprise an additional C-terminal conserved H2 homology domain. The H1 and H2 domains exhibit no sequence similarities to each other or to other proteins, thus giving no hints regarding their functions (5). Here we studied the influence of the H1 and H2 domains on desensitization of the receptor response. We found that H1 and H2 domains have opposite effects on the desensitization kinetics of the receptor response. The evolutionary analysis of protein sequences suggests that KCTD12 and -12b acquired desensitizing properties by disposing of their inhibitory H2 domains and selecting the T/NFLEQ motif in their H1 domains.
EXPERIMENTAL PROCEDURES
Generation of Expression Plasmids
All KCTD cDNAs were cloned in-frame with the N-terminal tag of three c-Myc epitopes (MEQKLISEEDLGEQKLISEEDLLEQKLISEEDLAAEF) into the cytomegalovirus-based expression vector pCI (Promega). Mutant constructs were generated using overlap extension PCR (20). To generate 12T1–16H1H2, residues Arg207 to Glu327 in KCTD12 were replaced by residues Arg161 to Leu427 of KCTD16. To generate 16T1–12H1, residues Arg161 to Leu427 in KCTD16 were replaced by residues Arg207 to Glu327 of KCTD12. To generate 12–16H2, residues Pro280 to Leu427 of KCTD16 were added in-frame at the C terminus of KCTD12. To generate 16H2, residues Pro280 to Leu427 of KCTD16 were cloned in-frame with the N-terminal c-Myc epitopes. To generate 16ΔH2, a stop codon was inserted after residue Glu279 in KCTD16. To generate 12–16H2Δ60 and 12–16H2Δ113, a stop codon was inserted in 12–16H2 after residues Thr367 and Cys314, respectively. To generate 12-Luc and 12-Venus, the cDNA of the Renilla reniformis Luciferase (Luc) or the GFP variant Venus from Aequorea victoria (Venus) was added in-frame via a flexible peptide linker (GGGSGGGGS) to the C terminus of KCTD12. To generate Luc-12 and Venus-12, the cDNA of Luc and Venus, respectively, was added in-frame via a GGGSGGGGS peptide linker to the N terminus of KCTD12. N-terminally tagged KCTD12 constructs did not contain the three c-Myc epitope tag. To generate 16T1–16/12H1G, residues Gly244 to Glu279 in 16ΔH2 were exchanged by residues Gly290 to Glu327 of KCTD12. To generate 16T1–16/12H1N residues Lys231 to Glu279 in KCTD16ΔH2 were exchanged by residues Asn277 to Glu327 of KCTD12. To generate 8ΔH2, a stop codon was inserted after residue Pro325 in KCTD8. To generate 8ΔH2F, Tyr278 in the H1 domain of 8ΔH2 was mutated to Phe. To generate 16ΔH2F, His232 in the H1 domain of 16ΔH2 was mutated to Phe. To generate 16ΔH2NFQ, Lys231, His232, and Arg235 in the H1 domain of 16ΔH2 were mutated to Asn, Phe, and Gln, respectively. To generate 12H, Phe278 in the H1 domain of KCTD12 was mutated to His. To generate 12KHR, Asn277, Phe278, and Gln281 in the H1 domain of KCTD12 were mutated to Lys, His, and Arg, respectively.
Cell Culture
CHO-K1 cells stably expressing human GABAB1b and rat GABAB2 were maintained in Dulbecco's modified Eagle's medium (DMEM) with 500 μm l-glutamine, 40 μg/ml l-proline, 0.5 mg/ml G418, 0.25 mg/ml zeocine, and 10% FCS in a humidified atmosphere of 5% CO2 at 37 °C (21). Cells were transfected in 24-well plates at 80–90% confluency using 3 μl of Lipofectamine 2000 (Invitrogen) and 1.2 μg of Kir3.1/3.2 concatamer in pcDNA3.1 (22), 2-μg KCTD constructs in pCI and 0.3 μg of pEGFP-N1 (Clontech) to visualize transfected cells. 6 h after transfection the cells were plated onto plastic coverslips (Thermanox, Nalge Nunc International) at a dilution of 1:5 in 35 mm dishes and used for electrophysiological recordings 24–48 h later.
HEK293T cells (ATCC CRL-11268) were cultured in DMEM supplemented with 10% FCS and 500 μm l-glutamine in a humidified atmosphere of 5% CO2 at 37 °C. Cells were transfected at 80–90% confluency using Lipofectamine 2000. For transfection in 6-cm dishes, 12 μl of Lipofectamine and 1.5 μg of plasmid DNA were used. Cells were harvested after 48 h for co-immunoprecipitation and Western blot analysis.
Co-immunoprecipitation and Western Blot Analysis
HEK293T cells were harvested, washed in PBS, and subsequently lysed in a Nonidet P-40 buffer (100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 20 mm Tris/HCl, pH 7.4) supplemented with complete EDTA-free protease inhibitor mixture (Roche). After rotation for 10 min at 4 °C, the lysates were cleared by centrifugation at 16,000 × g for 10 min at 4 °C. Lysates were then directly used for Western blot analysis or precleared for 1 h using 30 μl of a 1:1 mixture of protein-A- and protein-G-agarose (GE Healthcare) to be used in co-immunoprecipitation experiments. Precleared lysates were immunoprecipitated with anti-GB2 antibody (Millipore, AB5394, 1 μg, 3 h of incubation at 4 °C) and protein-A- and protein-G-Sepharose (10 μl, 1 h of incubation). Lysates and immunoprecipitates were resolved using standard SDS-PAGE, and probed with the primary antibodies mouse anti-Myc (F1804, Sigma, 1:1000), mouse anti-R. reniformis Luciferase (MAB4410, Millipore, 1:2000) or rabbit anti-GFP (A11122, Invitrogen, 1:1000) and peroxidase-coupled secondary antibodies (NA931V and NA9340V, Amersham Biosciences, 1:10000). The antibody incubation was in 5% nonfat dry milk in PBS containing 0.1% Tween-20. The chemiluminescence detection kit (Pierce) was used for visualization.
Electrophysiology
EGFP-expressing CHO cells were identified via epifluorescence using a FITC filter set and patched under oblique illumination optics (BX51WI; Olympus). Kir3 currents were recorded at 30–32 °C in artificial cerebrospinal fluid containing (in mm): 119 NaCl, 2.7 KCl, 1.3 MgCl2, 2.5 CaCl2, 1 NaH2PO4, 26,2 NaHCO3, 11 glucose, pH 7.3, equilibrated with 95% O2/5% CO2. Patch pipettes were pulled from borosilicate glass capillaries (resistance of 3–5 MΩ) and filled with a solution containing (in mm): 140 K-gluconate, 4 NaCl, 5 HEPES, 2 MgCl2, 1.1 EGTA, 2 Na2-ATP, 5 phosphocreatine, 0.6 Na3-GTP, at pH 7.25 (adjusted with KOH). GABAB responses were evoked at −50 mV by fast application of 100 μm GABA (Sigma) for 40 s with a pressure pipette (standard patch pipette; 3 PSI, Picospritzer III, Intracell).
Data were acquired with a MultiClamp 700B (Molecular Devices), low-pass filtered at 2 kHz and digitized at 10 kHz using a Digidata 1322A interface (Molecular Devices) driven by pClamp 10.2 software. Whole-cell currents were analyzed using Clampfit 10.2 software (Molecular Devices). All values are expressed as mean ± S.D. Data were analyzed by one-way ANOVA followed by Dunnett's test for pairwise comparison with the control group (Igor Pro software). p values < 0.05 were considered as statistically significant.
Protein Alignments and Phylogenetic Trees
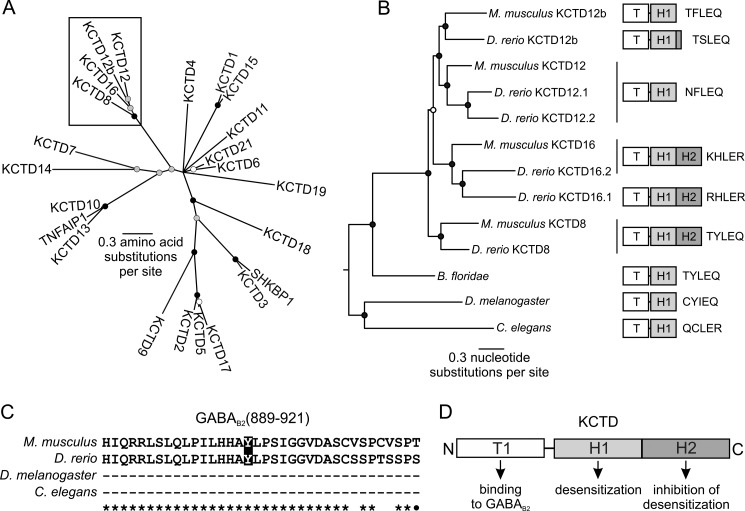
For the tree of all KCTD proteins, all annotated human KCTD protein sequences were used to query the NCBI nr protein database with blastp and all sequences with an e-value less than 1e-5 were retrieved and aligned using MUSCLE (23). Deeply diverging sequences that could not be reliably aligned were discarded (this set included all KCTD20 sequences). The sequences were re-aligned, and used as input into the tree-building program MrBayes 3.0 (24). The tree shown in Fig. 5A has been pruned to retain only the KCTDs from Mus musculus.
FIGURE 5.
KCTD8, -12, -12b, and -16 recently diversified from a single common ancestor. A, phylogenetic tree based on an amino acid alignment of the T1 domains of all KCTD proteins. Most KCTD proteins diverged deeply in time; however, a few groups have diverged more recently, including the group formed by KCTD8, -12, -12b, and -16 (box). TNFAIP1 and SHKBP1 are grouped with the KCTD proteins due to T1 domain-related sequences B, phylogenetic tree based on a nucleotide alignment of KCTD8, -12, -12b, and -16. For clarity, this tree is midpoint rooted. KCTD8, -12, -12b, and -16 evolved through the addition of the H2 domain after the origin of chordates and rapidly diversified into the KCTD8, -12, and -16 lineages, with the H2 domain being lost in KCTD12 and -12b. Of note, D. rerio, but not M. musculus retained part of the H2 domain in KCTD12b at the genomic level. In both A and B, the circles at each node indicate the posterior probabilities (the probability that the descendent proteins are more closely related to each other than to other proteins in the tree): black, greater than 95%; gray, between 75 and 95%; white, between 50 and 75%. Two copies of KCTD12 and 16 are present in D. rerio. These duplicate copies are also present in the genomes of other fish, due to an ancient genome duplication (not shown). C, sequence alignment of the GABAB2 C-terminal domains of M. musculus, D. rerio, D. melanogaster, and C. elegans. Identical and similar amino acids are indicated by stars and dots, respectively. Amino acid Tyr902, which is critical for KCTD binding to GABAB2 is only found in vertebrates and highlighted in black. D, functional model of vertebrate KCTD proteins. The T1 tetramerization domain binds to GABAB2, the H1 domain promotes the desensitization of GABAB responses and the H2 domain prevents desensitization when expressed in cis with a desensitizing H1 domain. TNFAIP1, Tumor necrosis factor α-induced protein 1; SHKBP1, SH3 domain-containing kinase-binding protein 1.
For the tree of KCTD8, -12, -12b, and -16, the H1 domain from human KCTD16 was used to query the NCBI nr protein database with blastp. The nucleotide sequences from all hits with an e-value less than 1e-10 were retrieved and aligned using MUSCLE. GBlocks (25) was used to retain only well-aligned blocks. This alignment was used as input into MrBayes. The tree shown in Fig. 5B has been pruned to retain only the KCTDs of M. musculus, Danio rerio, Branchiostoma floridae, Drosophila melanogaster, and Caenorhabditis elegans.
For the alignment of GABAB2 C termini, the human GABAB2 was used to query the NCBI nr protein database with blastp, and sequences from several species were aligned using MUSCLE. The C termini of non-vertebrates do not align with vertebrates and are not shown in the alignment.
The alignment of vertebrate KCTD H2 domain sequences was performed on the most recent ENSEMBL draft genomes of the species indicated. These sequences were blasted with the KCTD12b protein sequence from Oryzias latipes using tblastn. All hits which had H2-like sequences within the same contig were retained. All potential H2-like open reading frames were separated by 9.4 (Tetraodon nigroviridis) to 103 kb (Ornithorhyncus anatinus) from the open reading frame containing the T1 and H1 domains. The retained sequences grouped with KCTD12b and not KCTD8 or -16, suggesting that they are indeed KCTD12b sequences.
RESULTS
Distinct KCTD Protein Domains Influence Desensitization of the GABAB Receptor Response
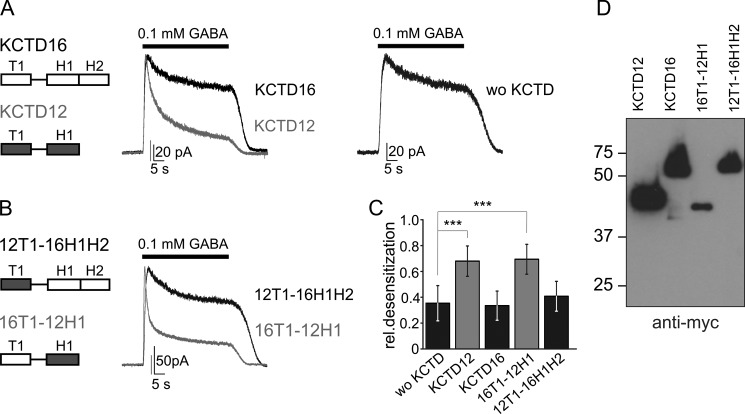
KCTD subunits are built from T1, H1, and H2 domains, whereby only KCTD8 and 16 contain a H2 domain. The domain organization of KCTD12 and KCTD16 is illustrated in Fig. 1A. Previous work indicated that the lack of the H2 domain in KCTD12 and -12b correlates with strong desensitization of the receptor response (6). We therefore hypothesized that H1 domains facilitate and H2 domains inhibit desensitization. We tested this hypothesis using patch-clamp electrophysiology and CHO cells expressing GABAB(1b,2) receptors and effector Kir3 channels in the presence and absence of wild-type and mutant KCTD proteins. In the absence of KCTD proteins, activation of GABAB receptors for 40 s by GABA (0.1 mm) elicited Kir3 currents that slightly decreased in amplitude over time (Fig. 1A). The relative desensitization of GABA-activated Kir3 currents was calculated as the reduction in amplitude measured at the end of the GABA application normalized to the peak amplitude (Fig. 1C). In agreement with reported results (6), co-expression of KCTD12 significantly increased the desensitization of GABA-activated Kir3 currents, while KCTD16 had no significant effect on desensitization (Fig. 1, A and C; p < 0.001, compared with cells without KCTD). To identify the KCTD12 domain(s) responsible for desensitization we generated two chimeric proteins, 16T1–12H1 and 12T1–16H1H2, in which the H1 domain of KCTD12 and the H1/H2 domains of KCTD16 are swapped. The desensitization of GABA-activated Kir3 currents was significantly increased in cells expressing the chimeric protein 16T1–12H1 but not in cells expressing 12T1–16H1H2 (Fig. 1, B and C). Of note, both chimeric proteins contained the linker region of KCTD16. Western blot analysis confirmed that the chimeric KCTD proteins were expressed (Fig. 1D). In summary, these data show that the desensitizing properties of KCTD12 segregate with its H1 domain.
FIGURE 1.
H1 and H2 domains have opposite effects on KCTD12-mediated desensitization of the GABAB response. A and B, representative traces of GABAB-activated Kir3 currents recorded at −50 mV from CHO cells co-expressing GABAB(1b,2), Kir3 channels and KCTD proteins. In the presence of KCTD16 or 12T1–16H1H2 Kir3 currents exhibit modest desensitization in response to continuous GABA (0.1 mm for 40 s) application, comparable to the current desensitization observed without KCTD proteins (wo KCTD). In the presence of KCTD12 or 16T1–12H1, GABAB-activated Kir3 currents exhibit significantly increased desensitization. The scheme depicts the T1 tetramerization domains and H1 and H2 homology domains of KCTD12 (gray) and KCTD16 (white). C, bar graph summarizing the desensitization of Kir3 currents in the absence and presence of KCTD proteins. Data are expressed as mean ± S.D.; ***, p < 0.001 compared with cells without KCTD proteins (Dunnett's multiple comparison test). D, Western blot analysis of wild-type and chimeric KCTD proteins using anti-Myc antibodies. The molecular mass is indicated on the left (in kDa).
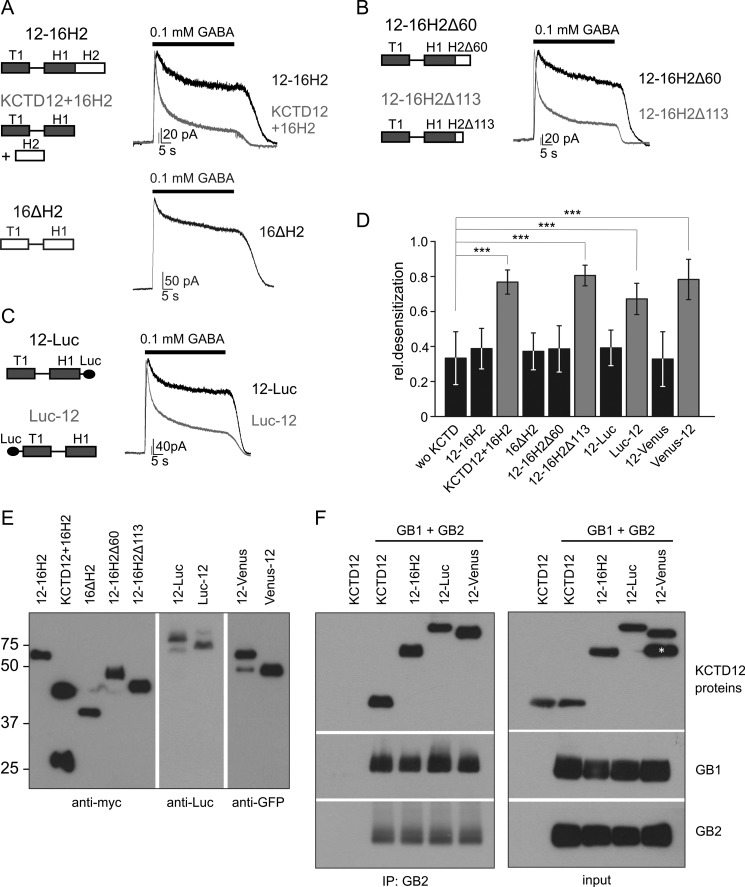
We next tested whether the H2 domain exerts an inhibitory influence on desensitization. We generated the mutant 12–16H2 with the H2 domain of KCTD16 attached to the C terminus of KCTD12. 12–16H2 lacks desensitizing properties, in line with a dominant inhibitory effect of the H2 domain on KCTD12-mediated desensitization (Fig. 2, A and D). However, removal of the H2 domain from KCTD16 in the mutant 16ΔH2 does not produce a desensitizing KCTD protein (Fig. 2, A and D). Therefore, the H1 domain of KCTD16 is not sufficient for desensitization and differs in its functional properties from the H1 domain of KCTD12.
FIGURE 2.
The H2 domain inhibits desensitization by the KCTD proteins. A-C, representative traces of GABAB-activated Kir3 currents recorded at −50 mV from CHO cells co-expressing GABAB(1b,2), Kir3 channels, and KCTD proteins. In the presence of 12–16H2, a chimeric protein consisting of KCTD12 and the H2 domain of KCTD16, Kir3 currents exhibit modest desensitization. In contrast, when the H2 domain of KCTD16 was co-transfected with KCTD12 (KCTD12 + 16H2), currents show significantly increased desensitization. This indicates that the H2 domain exerts an inhibitory influence on desensitization in cis but not in trans. However, KCTD16 lacking its H2 domain (16ΔH2) does not induce current desensitization (A). Deletion of 113 amino acids (12–16H2Δ113) but not of 60 amino acids (12–16H2Δ60) from the C terminus of the H2 domain in 12–16H2 restored the ability of the KCTD protein to induce current desensitization (B). Tagging KCTD12 with Luciferase at the C terminus (12-Luc) but not at the N terminus (Luc-12) eliminated its ability to induce desensitization (C). D, bar graph summarizing the desensitization of Kir3 currents in the absence and presence of KCTD proteins; wo KCTD, without KCTD. Data are expressed as mean ± S.D.; ***, p < 0.001 compared with cells without KCTD (Dunnett's multiple comparison test). E, Western blot analysis of chimeric, truncated, or tagged KCTD proteins using anti-myc, anti-Luc or anti-GFP antibodies. The molecular mass is indicated on the left (in kDa). F, co-immunoprecipitation of C-terminally extended KCTD12 proteins with GABAB2. The indicated Myc-tagged KCTD proteins were co-expressed with GABAB1b and GABAB2 (GB1 + GB2). Immunoprecipitation was performed with antibodies against GB2 and immunoprecipitates were analyzed by Western blot with antibodies against GB1, GB2, and the Myc epitope. The asterisk likely indicates a truncated fragment of the 12-Venus protein that is not co-immunoprecipitated with GB2. Luc, Luciferase; Venus, Venus-GFP variant.
We next addressed whether the H2 domain of KCTD16 not only prevents KCTD12-mediated desensitization in cis but also in trans. When the H2 domain of KCTD16 is co-expressed with KCTD12 as an independent 16H2 protein GABAB-activated Kir3 currents still desensitize (KCTD12 + 16H2; Fig. 2, A and D; p < 0.001, compared with cells without KCTD). Expression of the 16H2 protein was confirmed by Western blot analysis (Fig. 2E). Therefore, the H2 domain only prevents KCTD12-mediated desensitization in cis but not in trans. In order to map the minimal size of the H2 domain preventing desensitization in cis, we generated C-terminal truncations of the chimeric 12–16H2 protein. Truncation of 60 of the 148 amino acid residues of the H2 domain in the 12–16H2Δ60 protein is insufficient to restore desensitization (Fig. 2, B and D). However, truncation of 113 amino acid residues in the 12–16H2Δ113 protein fully restored desensitization (Fig. 2, B and D). This shows that the size of the H2 domain is critical for inhibition of KCTD12-mediated desensitization. We tested whether adding KCTD-unrelated protein domains to the C terminus of KCTD12 also prevents desensitization. Adding Luciferase (Luc) or the GFP variant Venus to the C terminus of KCTD12 in the 12-Luc and 12-Venus proteins completely prevented desensitization, suggestive of a sequence-unrelated steric hindrance of desensitization (Fig. 2, C and D). The 12–16H2, 12-Luc and 12-Venus proteins co-immunoprecipitate with GABAB2 (Fig. 2F). This demonstrates that the lack of desensitization of these proteins is not due to a lack of association with the receptor. Adding the Luc or Venus domains to the N terminus of KCTD12 in the Luc-12 and Venus-12 proteins did not prevent desensitization, showing that only protein domains C-terminal of the H1 domain are inhibitory (Fig. 2, C and D).
The Desensitization Motif of H1 Domains
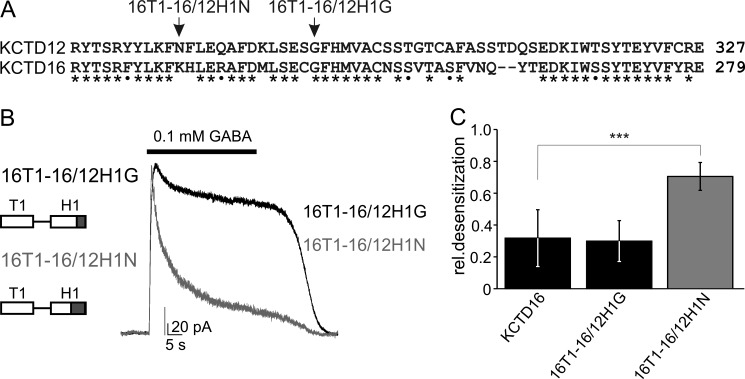
We next determined the amino acid residues in the H1 domain of KCTD12 that differ from KCTD16 and mediate the desensitization. Sequence alignment revealed significant differences between KCTD12 and KCTD16 within the C-terminal half of their H1 domains (Fig. 3A). We therefore tested whether desensitizing properties can be transferred from KCTD12 to KCTD16 by replacing the C-terminal half of the H1 domain of KCTD16 with the corresponding sequence of KCTD12. We generated the two chimeric proteins 16T1–16/12H1G and 16T1–16/12H1N in which the 36 and 49 C-terminal residues, respectively, in the H1 domain of KCTD16 were replaced with those of KCTD12. In addition, we omitted in these chimeric proteins the H2 domain of KCTD16 to avoid its inhibitory effect on desensitization. The desensitization of GABAB receptor-activated Kir3 currents was small in CHO cell expressing 16T1–16/12H1G protein or the KCTD16. In contrast, cells expressing the 16T1–16/12H1N protein exhibited significantly more desensitization (Fig. 3, B and C; p < 0.001, compared with KCTD16). This result shows that the ability to desensitize GABAB-activated Kir3 currents can be transferred from the KCTD12 to KCTD16 H1 domain by exchanging the 49 C-terminal residues of the H1 domains. In addition, the result points at the 13 amino acid residues between Asn277 and Ser289 in the H1 domain of KCTD12 as being critical for desensitization. Sequence alignment of these 13 amino acid residues in KCTD8, 12, 12b, and 16 reveals that only Tyr278 in KCTD8 is not identical or highly conserved with either KCTD12 or KCTD12b, which harbor an Phe residue at this position (Fig. 4A). We therefore addressed whether substitution of Tyr278 in KCTD8 with Phe renders the H1 domain in KCTD8 desensitizing. We first established that deletion of the H2 domain of KCTD8 in the 8ΔH2 protein was insufficient to convert KCTD8 into a desensitizing subunit (Fig. 4, B and D), similar as already observed with KCTD16 (Fig. 2). Strikingly, substitution of Tyr278 with Phe in the 8ΔH2F protein generated receptor responses with increased desensitization (Fig. 4, B and D; p < 0.05, compared with cells without KCTD). KCTD16 exhibits additional sequence divergence with KCTD12 and KCTD12b in the 13 amino acid residues under scrutiny (Fig. 4A). Accordingly, single amino acid substitution of His232, the residue homologous to Tyr278 in KCTD8, with Phe in the 16ΔH2F protein was insufficient to render the H1 domain of KCTD16 desensitizing (Fig. 4, C and D). However, additional substitution of two neighboring non-conserved residues (Asn for Lys231 and Gln for Arg235) in 16ΔH2NFQ also rendered the H1 domain of KCTD16 desensitizing (Fig. 4, C and D; p < 0.01, compared with cells without KCTD). Finally, the converse substitution of these three residues in KCTD12 with the ones of KCTD16, but not the single amino acid substitution of Phe278 by His, rendered the H1 domain of KCTD12 non-desensitizing (12KHR and 12H; Fig. 4, E and F; p < 0.001, compared with KCTD12). These experiments identify the motif T/NFLEQ in the H1 domain as a critical sequence element for KCTD-mediated desensitization.
FIGURE 3.
Swapping experiments identify a region in the H1 domain that is critical for desensitization. A, sequence alignment of the C-terminal half of the H1 domains of KCTD12 and KCTD16. Identical and similar amino acids are marked with stars and dots, respectively. Arrows indicate the KCTD16/KCTD12 boundaries in the chimeric proteins 16T1–16/12H1N and 16T1–16/12H1G, which both lack the H2 domain of KCTD16. B, representative traces of GABAB-activated Kir3 currents recorded at −50 mV from CHO cells expressing GABAB(1b,2), Kir3 channels, and KCTD proteins. Kir3 currents exhibit strong desensitization in the presence of 16T1–16/12H1N but not 16T1–16/12H1G. C, bar graph summarizing the desensitization of Kir3 currents in the presence of KCTD proteins. Data are expressed as mean ± S.D.; ***, p < 0.001 compared with cells transfected with KCTD16 (Dunnett's multiple comparison test).
FIGURE 4.
Identification of a desensitization motif in the H1 domains of KCTD12 and -12b. A, sequence alignment of residues Asn277-Ser289 in the KCTD12 H1 domain with the corresponding sequences in KCTD8, -16, and -12b. Stars and dots indicate identical and similar amino acids, respectively. B and C, representative traces of GABAB-activated Kir3 currents recorded at −50 mV from CHO cells expressing GABAB(1b,2), Kir3 channels and KCTD proteins. Substitution of Tyr278 by Phe in KCTD8 lacking its H2 domain (8ΔH2 and 8ΔH2F; B) induces Kir3-current desensitization. Substitution of Lys231, His232, and Arg235 by NFQ in KCTD16 lacking its H2 domain (16ΔH2NFQ) induces current desensitization, while substitution of His232 by Phe alone (16ΔH2F) is insufficient for this (C). D, bar graph summarizing the desensitization of Kir3 currents in the presence of KCTD proteins. Data are expressed as mean ± S.D.; *, p < 0.05; **, p < 0.01 compared with cells without KCTD proteins (Dunnett's multiple comparison test). E, substitution of Asn277, Phe278, and Gln281 by KHR in KCTD12 (12KHR) eliminates Kir3 current desensitization, while substitution of Phe278 by H alone (12H) is insufficient for this. F, bar graph summarizing the desensitization of Kir3 currents in the presence of KCTD proteins. Data are expressed as mean ± S.D.; ***, p < 0.001 compared with cells transfected with KCTD12 (Dunnett's multiple comparison test).
Molecular Evolution of the KCTD Subunits
Our experiments show that the H1 domain is the functional unit responsible for desensitization of the receptor response. The H1 domains of KCTD8 and 16 lack desensitizing properties due to one or three amino acid substitutions, respectively, in the T/NFLEQ motif. The H2 domains in KCTD8 and -16 have antagonistic effects and inhibit desensitization by the H1 domains. To understand how the KCTD proteins acquired these regulatory domains we investigated their evolutionary history.
Analysis of the human and zebrafish KCTD proteins revealed that they are distinct from voltage-gated K+ channels, due to differences in their T1 domains and the absence of transmembrane domains (17, 26, 27). Our phylogenetic analysis based on the amino acid alignment of the T1 domains of all annotated human KCTD proteins and their orthologues revealed that they diverged deeply in time, preceding the split of animals from plants. However, some KCTD proteins, including the subfamily formed by KCTD8, -12, -12b, and -16, diverged more recently (Fig. 5A). An ancestral KCTD protein with T1 and H1 domains having homology to this subfamily of KCTD proteins is found in nematodes, insects as well as invertebrate chordates (Fig. 5B; C. elegans, D. melanogaster, and B. floridae, a lancelet). However, the C-terminal GABAB2 domain mediating the interaction with the T1 domain (6, 28) is absent in invertebrate GABAB2 (Fig. 5C). Thus, it appears that the ancestral KCTD protein is not part of the invertebrate GABAB receptor complex.
Our phylogenetic analysis shows that soon after the emergence of vertebrates a number of events, occurring almost simultaneously in evolutionary terms, changed the structure of GABAB receptors. The C-terminal domain was added to the GABAB2 subunit, thus enabling interaction between the ancestral KCTD protein and GABAB receptors. Of note, the Tyr902 residue in GABAB2 that is critical for binding to the KCTD proteins (6) is conserved in all vertebrates. In addition, the H2 domain was added to the ancestral KCTD protein. The ancestral KCTD protein then diversified into the KCTD8, 12, and 16 lineages (Fig. 5B). A suite of amino acid changes occurred in the H1 domain of two of these lineages: in the KCTD16 lineage, the TYLEQ motif changed to K/RHLER; in the KCTD12 lineage, the TYLEQ motif changed to NFLEQ (KCTD12) or TF/SLEQ (KCTD12b). In the KCTD8 lineage, the ancestral TYLEQ motif was kept. The final event in the evolution of this KCTD subfamily was a split of KCTD12 and -12b and the removal of the H2 domain in both sub-lineages. With the exception of placental mammals, most vertebrates retained a small part of the H2 domain as an open reading frame in their KCTD12b genes (Fig. 6). This demonstrates that the H2 domain was initially present in the KCTD12 linage. It remains to be addressed whether the residual H2 domain sequences in the KCTD12b genes of vertebrates are transcribed and translated. In this respect, it is interesting to note that all H2 domain sequences analyzed, including those of KCTD8 and -16, are encoded by a separate exon downstream of the exon encoding the T1 and H1 domains. Therefore it is possible that multiple KCTD8, -16, or -12b variants are generated by alternative splicing. In conclusion, our phylogenetic analysis shows that desensitizing KCTD12 and -12b proteins evolved from non-desensitizing KCTD proteins by disposal of the inhibitory H2 domains and acquisition of the T/NFLEQ motif in their desensitizing H1 domains.
FIGURE 6.
Alignment of vertebrate KCTD H2 domain amino acid sequences. The C-terminal part of the H2 domain is retained in KCTD12b in most vertebrates, excluding placental mammals (e.g. M. musculus). Residues are numbered according to the H2 domain of D. rerio KCTD8. Conserved residues are highlighted: dark gray, present in more than 80% of the sequences; light gray, present in more than 50% of the sequences. The letters X in KCTD12b of Gadua morhua and Takifugu rubripes result from unspecified nucleotides in the genomic sequences. M. musculus, mouse; D. rerio, zebrafish; G. morhua, cod; Oreochromis niloticus, tilapia; T. rubripes, tiger pufferfish; T. nigroviridis, green-spotted pufferfish; O. latipes, medaka; Xenopus tropicalis, clawed frog; Pelodiscus sinensis, soft-shelled turtle; Gallus gallus, chicken; Taeniopygia guttata, zebra finch; O. anatinus, duck-billed platypus; Sarcophilus harrisii, tasmanian devil.
DISCUSSION
Association of auxiliary KCTD8, -12, -12b, and -16 subunits with principal GABAB receptor subunits was recently shown to generate molecularly and functionally distinct receptor subtypes (5, 6, 9, 28). The four KCTD proteins are built from obligatory T1 and H1 domains and optional H2 domains. The T1 domains bind as tetramers to the GABAB2 subunit and thus are crucial for formation of the receptor complex. No functional roles have been assigned to the H1 and H2 domains of the KCTD subunits yet. In this study we show that the H1 and H2 domains have opposite effects on fast desensitization of the receptor response. H1 domains containing the T/NFLEQ motif mediate the desensitization while the H2 domains antagonize this desensitization. The antagonistic effect of the H2 domain is only observed when the domain is expressed in cis with the H1 domain but not when the H2 domain is expressed as a separate protein in trans together with KCTD12. This suggests that the H2 domain does not inhibit desensitization through the binding to a specific site. More likely, the H2 domain acts by sterically hindering the binding of the H1 domain to a downstream effector responsible for fast desensitization such as, for example, proteins involved in G-protein signaling. In agreement with a steric hindrance of desensitization by the H2 domain KCTD-unrelated protein domains can substitute for the H2 domain and prevent KCTD12-mediated desensitization when tethered to the C terminus of KCTD12. It is unclear whether desensitizing and non-desensitizing KCTD proteins can simultaneously bind to the same receptor complex (6). If this is the case, our results suggest that the H2 domains of KCTD8 or -16 will be unable to inhibit desensitization by KCTD12 or -12b in trans. From an evolutionary perspective, it appears that the H2 domain was first acquired in an ancestral KCTD protein with non-desensitizing properties and subsequently lost in KCTD12 and -12b. It is therefore unlikely that the prime function of the H2 domain in KCTD8 and -16 is to antagonize desensitization.
KCTD12 and -12b evolved the T/NFLEQ motif within their H1 domain, which is necessary for KCTD-mediated desensitization of the receptor response. One to three amino acid substitutions within this motif can turn a desensitizing into a non-desensitizing H1 domain and vice versa. Secondary structure analysis predicts that the motif is part of a helix with amphipathic characteristics. It appears that an excess of positively charged amino acids within this helix, such as Arg, His, and Lys in KCTD16, is not permissive for desensitization. It is possible that these positively charged amino acids interact with negatively charged phospholipids of the plasma membrane and reduce the mobility of the helix. In addition, it appears that specific residues at the interface between the hydrophilic and the hydrophobic side of the helix are crucial for desensitization. Thus, substitution of Tyr278 with Phe was sufficient to render the H1 domain of KCTD8 desensitizing. Interestingly, amphipathic helices are widely found in proteins participating in membrane-associated biological processes. In particular, amphipathic helices within GPCRs, G-protein subunits or naturally occurring peptides were shown to regulate the G-protein activation-deactivation cycle (29–34). It is thus possible that the amphipathic helix in the H1 domain of KCTD12 and -12b directly regulates the G-protein that binds in its proximity to GABAB2 (35–37). However, no binding partners for the H1 domain have yet been identified.
Our evolutionary analysis shows that receptor subtypes owing to auxiliary KCTD subunits emerged with the appearance of vertebrates. In this respect it is interesting to note that GABAB1 subunit isoforms regulating axonal versus dendritic distribution of GABAB receptors (38) also first evolved in vertebrates. This suggests that it became essential to control GABAB receptor desensitization with the emergence of localized signaling. KCTD12 and -16 proteins appear to be present in pre- and postsynaptic GABAB receptors (6) albeit to differing degrees (9). Biochemical data support that certain GABAB receptors in the brain contain KCTD12 and others KCTD16 (6). However, whether association with specific KCTDs is responsible for the differences in desensitization between pre- and postsynaptic GABAB receptors remains to be addressed (39–41). In conclusion, whereas the heteromeric nature and the activation mechanism of the GABAB core receptor are conserved in evolution (42–46), only the vertebrate GABAB receptors recruit functionally distinct auxiliary KCTD subunits and generate receptor subtypes.
Acknowledgments
We thank Audrée Pinard and Klara Ivankova for critical reading of the manuscript.
This work was supported by grants from the Swiss National Science Foundation (3100A0-117816), the National Center for Competence in Research (NCCR) “Synapsy, Synaptic Bases of Mental Health Disease” (to B. B.), the European Community's Seventh Framework Programme (FP7/2007-2013) under Grant Agreement 201714 (to B. B.), an Ambizione fellowship from the Swiss National Science Foundation (to O. K. S.), and a Marie Heim-Vögtlin fellowship from the Swiss National Science Foundation (to V. J.).
- GPCR
- G-protein-coupled receptor
- KCTD
- K+ channel tetramerization domain
- GIRK
- G-protein activated inwardly rectifying K+
- Kir
- K+ inwardly rectifying
- GRK
- G-protein-coupled receptor kinase
- RGS
- regulator of G-protein signaling
- BTB
- bric-a-brac, tramtrak, and broad complex
- Luc
- Luciferase.
REFERENCES
- 1. Bettler B., Kaupmann K., Mosbacher J., Gassmann M. (2004) Molecular structure and physiological functions of GABAB receptors. Physiol. Rev. 84, 835–867 [DOI] [PubMed] [Google Scholar]
- 2. Bowery N. G., Bettler B., Froestl W., Gallagher J. P., Marshall F., Raiteri M., Bonner T. I., Enna S. J. (2002) International Union of Pharmacology. XXXIII. Mammalian γ-aminobutyric acidB receptors: structure and function. Pharmacol. Rev. 54, 247–264 [DOI] [PubMed] [Google Scholar]
- 3. Chalifoux J. R., Carter A. G. (2011) GABAB receptor modulation of synaptic function. Curr. Opin. Neurobiol. 21, 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Couve A., Moss S. J., Pangalos M. N. (2000) GABAB receptors: a new paradigm in G protein signaling. Mol. Cell. Neurosci. 16, 296–312 [DOI] [PubMed] [Google Scholar]
- 5. Gassmann M., Bettler B. (2012) Regulation of neuronal GABAB receptor functions by subunit composition. Nat. Rev. Neurosci. 13, 380–394 [DOI] [PubMed] [Google Scholar]
- 6. Schwenk J., Metz M., Zolles G., Turecek R., Fritzius T., Bildl W., Tarusawa E., Kulik A., Unger A., Ivankova K., Seddik R., Tiao J. Y., Rajalu M., Trojanova J., Rohde V., Gassmann M., Schulte U., Fakler B., Bettler B. (2010) Native GABAB receptors are heteromultimers with a family of auxiliary subunits. Nature 465, 231–235 [DOI] [PubMed] [Google Scholar]
- 7. Sickmann T., Alzheimer C. (2003) Short-term desensitization of G-protein-activated, inwardly rectifying K+ (GIRK) currents in pyramidal neurons of rat neocortex. J. Neurophysiol. 90, 2494–2503 [DOI] [PubMed] [Google Scholar]
- 8. Sodickson D. L., Bean B. P. (1996) GABAB receptor-activated inwardly rectifying potassium current in dissociated hippocampal CA3 neurons. J. Neurosci. 16, 6374–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Metz M., Gassmann M., Fakler B., Schaeren-Wiemers N., Bettler B. (2011) Distribution of the auxiliary GABAB receptor subunits KCTD8, 12, 12b, and 16 in the mouse brain. J. Comp. Neurol. 519, 1435–1454 [DOI] [PubMed] [Google Scholar]
- 10. Perroy J., Adam L., Qanbar R., Chénier S., Bouvier M. (2003) Phosphorylation-independent desensitization of GABAB receptor by GRK4. EMBO J. 22, 3816–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Labouèbe G., Lomazzi M., Cruz H. G., Creton C., Luján R., Li M., Yanagawa Y., Obata K., Watanabe M., Wickman K., Boyer S. B., Slesinger P. A., Lüscher C. (2007) RGS2 modulates coupling between GABAB receptors and GIRK channels in dopamine neurons of the ventral tegmental area. Nat. Neurosci. 10, 1559–1568 [DOI] [PubMed] [Google Scholar]
- 12. Maity B., Stewart A., Yang J., Loo L., Sheff D., Shepherd A. J., Mohapatra D. P., Fisher R. A. (2012) Regulator of G protein signaling 6 (RGS6) protein ensures coordination of motor movement by modulating GABAB receptor signaling. J. Biol. Chem. 287, 4972–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mutneja M., Berton F., Suen K. F., Lüscher C., Slesinger P. A. (2005) Endogenous RGS proteins enhance acute desensitization of GABAB receptor-activated GIRK currents in HEK-293T cells. Pflugers Arch. 450, 61–73 [DOI] [PubMed] [Google Scholar]
- 14. Xie K., Allen K. L., Kourrich S., Colón-Saez J., Thomas M. J., Wickman K., Martemyanov K. A. (2010) Gβ5 recruits R7 RGS proteins to GIRK channels to regulate the timing of neuronal inhibitory signaling. Nat. Neurosci. 13, 661–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Couve A., Thomas P., Calver A. R., Hirst W. D., Pangalos M. N., Walsh F. S., Smart T. G., Moss S. J. (2002) Cyclic AMP-dependent protein kinase phosphorylation facilitates GABAB receptor-effector coupling. Nat. Neurosci. 5, 415–424 [DOI] [PubMed] [Google Scholar]
- 16. Fairfax B. P., Pitcher J. A., Scott M. G., Calver A. R., Pangalos M. N., Moss S. J., Couve A. (2004) Phosphorylation and chronic agonist treatment atypically modulate GABAB receptor cell surface stability. J. Biol. Chem. 279, 12565–12573 [DOI] [PubMed] [Google Scholar]
- 17. Bayón Y., Trinidad A. G., de la Puerta M. L., Del Carmen Rodríguez M., Bogetz J., Rojas A., De Pereda J. M., Rahmouni S., Williams S., Matsuzawa S., Reed J. C., Crespo M. S., Mustelin T., Alonso A. (2008) KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 275, 3900–3910 [DOI] [PubMed] [Google Scholar]
- 18. Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Privé G. G. (2005) Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bixby K. A., Nanao M. H., Shen N. V., Kreusch A., Bellamy H., Pfaffinger P. J., Choe S. (1999) Zn2+-binding and molecular determinants of tetramerization in voltage-gated K+ channels. Nat. Struct. Biol. 6, 38–43 [DOI] [PubMed] [Google Scholar]
- 20. Horton R. M., Cai Z. L., Ho S. N., Pease L. R. (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8, 528–535 [PubMed] [Google Scholar]
- 21. Urwyler S., Mosbacher J., Lingenhoehl K., Heid J., Hofstetter K., Froestl W., Bettler B., Kaupmann K. (2001) Positive allosteric modulation of native and recombinant γ-aminobutyric acidB receptors by 2,6-Di-tert-butyl-4-(3-hydroxy-2,2-dimethyl-propyl)-phenol (CGP7930) and its aldehyde analog CGP13501. Mol. Pharmacol. 60, 963–971 [PubMed] [Google Scholar]
- 22. Wischmeyer E., Döring F., Spauschus A., Thomzig A., Veh R., Karschin A. (1997) Subunit interactions in the assembly of neuronal Kir3.0 inwardly rectifying K+ channels. Mol. Cell. Neurosci. 9, 194–206 [DOI] [PubMed] [Google Scholar]
- 23. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ronquist F., Huelsenbeck J. P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 25. Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- 26. Gamse J. T., Kuan Y. S., Macurak M., Brösamle C., Thisse B., Thisse C., Halpern M. E. (2005) Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development 132, 4869–4881 [DOI] [PubMed] [Google Scholar]
- 27. Taylor R. W., Qi J. Y., Talaga A. K., Ma T. P., Pan L., Bartholomew C. R., Klionsky D. J., Moens C. B., Gamse J. T. (2011) Asymmetric inhibition of Ulk2 causes left-right differences in habenular neuropil formation. J. Neurosci. 31, 9869–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bartoi T., Rigbolt K. T., Du D., Köhr G., Blagoev B., Kornau H. C. (2010) GABAB receptor constituents revealed by tandem affinity purification from transgenic mice. J. Biol. Chem. 285, 20625–20633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamm H. E. (2001) How activated receptors couple to G proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 4819–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnston C. A., Willard F. S., Jezyk M. R., Fredericks Z., Bodor E. T., Jones M. B., Blaesius R., Watts V. J., Harden T. K., Sondek J., Ramer J. K., Siderovski D. P. (2005) Structure of Gαi1 bound to a GDP-selective peptide provides insight into guanine nucleotide exchange. Structure 13, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kisselev O. G., Downs M. A. (2003) Rhodopsin controls a conformational switch on the transducin γ subunit. Structure 11, 367–373 [DOI] [PubMed] [Google Scholar]
- 32. Kisselev O. G., Kao J., Ponder J. W., Fann Y. C., Gautam N., Marshall G. R. (1998) Light-activated rhodopsin induces structural binding motif in G protein α subunit. Proc. Natl. Acad. Sci. U.S.A. 95, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kusunoki H., Wakamatsu K., Sato K., Miyazawa T., Kohno T. (1998) G protein-bound conformation of mastoparan-X: heteronuclear multidimensional transferred nuclear overhauser effect analysis of peptide uniformly enriched with 13C and 15N. Biochemistry 37, 4782–4790 [DOI] [PubMed] [Google Scholar]
- 34. Okuno T., Ago H., Terawaki K., Miyano M., Shimizu T., Yokomizo T. (2003) Helix 8 of the leukotriene B4 receptor is required for the conformational change to the low affinity state after G-protein activation. J. Biol. Chem. 278, 41500–41509 [DOI] [PubMed] [Google Scholar]
- 35. Duthey B., Caudron S., Perroy J., Bettler B., Fagni L., Pin J. P., Prézeau L. (2002) A single subunit (GB2) is required for G-protein activation by the heterodimeric GABAB receptor. J. Biol. Chem. 277, 3236–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galvez T., Duthey B., Kniazeff J., Blahos J., Rovelli G., Bettler B., Prézeau L., Pin J. P. (2001) Allosteric interactions between GB1 and GB2 subunits are required for optimal GABAB receptor function. EMBO J. 20, 2152–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robbins M. J., Calver A. R., Filippov A. K., Hirst W. D., Russell R. B., Wood M. D., Nasir S., Couve A., Brown D. A., Moss S. J., Pangalos M. N. (2001) GABAB2 is essential for G-protein coupling of the GABAB receptor heterodimer. J. Neurosci. 21, 8043–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Biermann B., Ivankova-Susankova K., Bradaia A., Abdel Aziz S., Besseyrias V., Kapfhammer J. P., Missler M., Gassmann M., Bettler B. (2010) The Sushi domains of GABAB receptors function as axonal targeting signals. J. Neurosci. 30, 1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cruz H. G., Ivanova T., Lunn M. L., Stoffel M., Slesinger P. A., Lüscher C. (2004) Bi-directional effects of GABAB receptor agonists on the mesolimbic dopamine system. Nat. Neurosci. 7, 153–159 [DOI] [PubMed] [Google Scholar]
- 40. Pennock R. L., Dicken M. S., Hentges S. T. (2012) Multiple inhibitory G-protein-coupled receptors resist acute desensitization in the presynaptic but not postsynaptic compartments of neurons. J. Neurosci. 32, 10192–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wetherington J. P., Lambert N. A. (2002) GABAB receptor activation desensitizes postsynaptic GABAB and A1 adenosine responses in rat hippocampal neurones. J. Physiol. 544, 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dittman J. S., Kaplan J. M. (2008) Behavioral impact of neurotransmitter-activated G-protein-coupled receptors: muscarinic and GABAB receptors regulate Caenorhabditis elegans locomotion. J. Neurosci. 28, 7104–7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mezler M., Müller T., Raming K. (2001) Cloning and functional expression of GABAB receptors from Drosophila. Eur. J. Neurosci. 13, 477–486 [DOI] [PubMed] [Google Scholar]
- 44. Schultheis C., Brauner M., Liewald J. F., Gottschalk A. (2011) Optogenetic analysis of GABAB receptor signaling in Caenorhabditis elegans motor neurons. J. Neurophysiol. 106, 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vashlishan A. B., Madison J. M., Dybbs M., Bai J., Sieburth D., Ch'ng Q., Tavazoie M., Kaplan J. M. (2008) An RNAi screen identifies genes that regulate GABA synapses. Neuron 58, 346–361 [DOI] [PubMed] [Google Scholar]
- 46. Wilson R. I., Laurent G. (2005) Role of GABAergic inhibition in shaping odor-evoked spatiotemporal patterns in the Drosophila antennal lobe. J. Neurosci. 25, 9069–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]