Background: ChChd3 is an inner mitochondrial membrane protein involved in maintaining crista biogenesis.
Results: Myristoylation and the CHCH domain are essential for the import; ChChd3 binds to Mia40 for oxidative folding and structural stability of the CHCH domain.
Conclusion: Myristoylation represents a novel targeting mechanism for the intermembrane space CHCH domain-containing proteins.
Significance: This study identifies ChChd3 as a Mia40 substrate and advances our understanding of ChChd3.
Keywords: Disulfide, Imaging, Mitochondria, Mitochondrial Metabolism, Protein Myristoylation, CHCH Domain, Disulfide Bonds, Disulfide Relay, Mia40, Protein Import
Abstract
Coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) is a mitochondrial inner membrane (IM) protein facing toward the intermembrane space (IMS). In the IMS, ChChd3 complexes with multiple proteins at the crista junctions and contact sites and plays a key role in maintaining crista integrity. ChChd3 is myristoylated at the N terminus and has a CHCH domain with twin CX9C motifs at its C terminus. The CHCH domain proteins are traditionally imported and trapped in the IMS by using a disulfide relay system mediated by Mia40 and Erv1. In this study, we systematically analyzed the role of the myristoylation and the CHCH domain in the import and mitochondrial localization of ChChd3. Based on our results, we predict that myristoylation promotes binding of ChChd3 to the outer membrane and that the CHCH domain translocates the protein across the outer membrane. By analysis of the CHCH domain cysteine mutants, we further show that they have distinct roles in binding to Mia40 in the IMS and proper folding of the protein. The transient disulfide-bonded intermediate with Mia40 is formed preferentially between the second cysteine in helix 1, Cys193, and the active site cysteine in Mia40, Cys55. Although each of the four cysteines is essential for folding of the protein and binding to mitofilin and Sam50, they are not involved in import. Together our results indicate that both the myristoylation and the CHCH domain are essential for the import and mitochondrial localization of ChChd3. Once imported, ChChd3 binds to Mia40 for further folding and assembly into macromolecular complexes.
Introduction
Coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3),2 initially identified as a substrate for cAMP-dependent protein kinase (PKA), is a ubiquitous protein in the mitochondria and plays a prominent role in maintaining cristae integrity and mitochondrial function (1, 2). In mitochondria, ChChd3 is predominantly localized to the inner membrane (IM), facing toward the intermembrane space (IMS), and is part of the large protein complex now called as MINOS (mitochondrial inner membrane organizing system) (3), or MIB (mitochondrial intermembrane space bridging) (4). Recent studies have suggested that the MINOS/MIB complex is localized at the crista junctions and acts as a scaffold for crista junction formation (3, 4). All the components of the MINOS/MIB: mitofilin/Fcj1, MINOS1, ChChd3/MINOS3, and Sam50, have been shown to affect the IM crista morphology (2–7). The majority of the proteins in this complex are localized on the IM, facing towards the IMS (2, 3, 6). The subunits of the SAM complex Sam50, metaxin 1 and metaxin 2, however, are located on the outer membrane (OM) (8) and have been shown to co-purify with the IM proteins mitofilin and ChChd3 (2, 9). siRNA depletion of ChChd3 resulted in significant reduction in both mitofilin and Sam50 protein levels accompanied by aberrant crista and crista junctions (2). These findings led us to propose the model that ChChd3 is a scaffolding protein at crista junction complexes, as well as at the contact sites, and support the role of these proteins in crista junction and contact site formation (2). This hypothesis was further supported in a recent study by Harner et al. (10) who isolated contact site complexes from S. cerevisiae mitochondria and showed that these complexes are preferentially located close to the crista junctions and are essential for crista junction formation. Although ChChd3 is not found in yeast, and so far the homologs for this are not identified, it has been suggested that Mcs19 (mitochondrial contact site 19), isolated as a novel protein in the mitochondrial contact site complex (MICOS) complex, may play a similar role based on the myristoylation motif and the C-terminal CHCH domain. In addition to mitofilin and Sam50, ChChd3 has also been found to associate with optic atrophy 1 (OPA1) (2), a key player in regulating mitochondrial fusion (11), and sphingosine kinase interacting protein 1 (SKIP) (12), a recently characterized A kinase anchoring protein (AKAP) (12, 13). SKIP was shown to localize to mitochondria, and this interaction was shown to be essential for phosphorylation of ChChd3 by PKA (12).
All the above mentioned studies clearly establish the role of ChChd3 as a major scaffold in the IMS and a key protein for maintaining structure and function of mitochondria. Having established this, a subsequent goal was to determine the requirements for localization to this compartment. ChChd3 is myristoylated at the amino terminus and has a carboxyl-terminal CHCH domain (2). The CHCH domain proteins are commonly seen in the IMS of mitochondria (14). Each helix in the CHCH domain contains two of the four conserved cysteines arranged in a CX9C or CX3C manner. In the functionally mature protein, the cysteines are oxidized, forming two intramolecular disulfide bonds as evidenced by the known structures of Cox17 and the Tim9-Tim10 complex (15, 16).
The disulfide bonds in the CHCH domain are crucial for proper folding, structural stabilization, and subsequent trapping of the protein in the IMS. The IMS proteins Mia40 (mitochondrial intermembrane assembly protein 40) along with the sulfhydryl oxidase Erv1 (essential for respiration and vegetative growth) in S. cerevisiae or the human homolog ALR (augmenter of liver regeneration) have been characterized to be the key components for this mechanism (17). Specifically, the oxidoreductase Mia40 is responsible for introducing disulfide bonds in the CHCH domain proteins. Mia40 possess a C-terminal core domain of about 60 amino acids with 6 conserved cysteines in a CPC-CX9C-CX9C fold (18). The CPC motif participates in the formation of mixed disulfide-bonded intermediate with the substrate proteins in the IMS.
Although studies from the past decade have established the essential role of Mia40-Erv1 in import and trapping of CHCH domain proteins into the IMS of mitochondria in yeast, these studies are primarily focused on Cox17 and small Tim proteins. Furthermore, Mia40 substrates are assumed to be small molecular mass proteins (<20 kDa) such as the well studied Cox17 (8 kDa) and Tim9 and Tim10 (10 kDa). ChChd3 (27 kDa) is the largest member of this family and includes a large DUF domain that precedes the CHCH motif. Additionally, ChChd3 is myristoylated at the N terminus. N-Myristoylation is a co-translational modification that has been known to play a prominent role in protein-protein interactions and anchoring proteins to the membranes (19). In addition to this, myristoylation has also been involved in targeting proteins to subcellular organelles including mitochondria (20, 21). Hence, in this study, we analyzed the role of myristoylation and the CHCH domain in targeting and import of ChChd3 into the mitochondria. Our results show that both the myristoylation and the CHCH domain are essential for the mitochondrial localization of ChChd3. We further show that ChChd3 forms disulfide-bonded intermediates with Mia40 where the inner cysteine from the helix 1, Cys193, binds to the active site cysteine in Mia40, Cys55. We also show that the cysteines in the IMS are not required for import. They are essential for proper folding and forming the protein complexes in the IMS. We predict that when the unique oxidizing environment of the IMS is disrupted, the trapped disulfide-bonded proteins will be reduced, and thus released from the mitochondria. This is likely to be an important step in apoptosis that allows for many IMS proteins such as AIF and cytochrome c to be released.
EXPERIMENTAL PROCEDURES
Antibodies and Plasmids
The following monoclonal (mAb) and polyclonal antibodies (pAb) were used in this study: mouse mAb FLAG and FLAG-agarose (Sigma), mouse mAb HA and HA agarose (Sigma), rabbit pAb Mia40 (Santa Cruz Biotechnology), rabbit pAb Lamp1 (Abcam), rabbit pAb Cox-IV (Abcam), and rabbit pAb ChChd3 (2). The cDNA clones for Tim10 and subunit 9 dihydrofolate reductase (su9-DHFR) are kind gifts from Prof. Carla Koehler, at the University of California, Los Angeles. The mouse cDNA clone for ChChd3 (BC021941) in pCMV-SP6 vector was procured from Life Technologies (catalog number, 5124504) and subsequently subcloned into a modified C-terminally FLAG-tagged pCMV-SP1 vector by using EcoRI and SalI restriction sites. The human cDNA clone for Mia40 was purchased from OriGene (catalog number, SC316333; Reference Sequence, NM_001098502) and was PCR-amplified and subcloned into the N-terminally HA-tagged phCMV2-XiClone vector (Genlantis) by following the manufacturer's recommendations. Point mutations were introduced by QuikChange site-directed mutagenesis (Stratagene). ΔCT and ΔNT mutants were made by PCR amplification and subcloning using standard protocols. All constructs were sequence verified (Eton Biosciences).
Immunostaining and Confocal Microscopy
For confocal microscopy experiments, HeLa cells maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 2 mm GlutaMAX were grown to 50–60% confluency on glass coverslips and transfected using FuGENE 6 (Roche Applied Science) transfection reagent by following the manufacturer's protocol. Approximately 20 h after transfection, cells were fixed with 4% paraformaldehyde. For immunostaining, cells were permeabilized with 0.3% Triton X-100 and blocked with 1% normal donkey serum, 0.5% BSA, and 50 mm glycine. Cells were stained with primary antibodies overnight at 4 °C and secondary antibodies for 1 h at room temperature. Coverslips were mounted on glass slides using ProLong Gold antifade reagent (Invitrogen). Images were acquired using FluoView 1000 confocal laser scanning microscope on a 60× objective lens with an NA of 1.2 (Olympus). Stacks of 10–15 slices were acquired (0.4 μm) using sequential scanning method to prevent bleed through between the channels. The images were processed on a customized ImageJ macro by maximum intensity projection method.
Immunoprecipitation
For immunoprecipitation of transiently expressed proteins, HEK 293 cells maintained in DMEM supplemented with 10% FBS and 2 mm GlutaMAX were grown to 80% confluency and transfected with Lipofectamine 2000 (Life Technologies) by following the manufacturer's protocols. Approximately 20 h after transfection, cells were washed with DPBS containing 1 mm N-ethylmaleimide and harvested in the buffer (50 mm Tris-HCl, 150 mm NaCl, and 1% Triton X-100, pH 7.4) containing 10 mm N-ethylmaleimide and protease inhibitor cocktail (Calbiochem). The cell lysates were incubated with anti-FLAG or HA agarose resin (Sigma) for 2 h at 4 °C. The resin was washed with wash buffer (50 mm Tris-HCl, 150 mm NaCl, and 1 mm N-ethylmaleimide, 4×) and eluted with Laemmli sample buffer. Samples were analyzed on SDS-PAGE followed by immunoblot. Wherever indicated, samples were reduced with 100 mm DTT and heated at 70 °C for 10 min before loading on SDS-PAGE.
Protein Import into Mitochondria
Mitochondria from mouse liver were isolated, and Histodenz purified as described (2). The precursor proteins were synthesized in a cell-free system from rabbit reticulocyte lysates using the TnT SP6 quick coupled transcription and translation mix (Promega) in the presence of [35S]methionine. Radiolabeled proteins were imported into freshly isolated and purified mouse liver mitochondria as described (22). In brief, radiolabeled proteins were incubated with freshly isolated mitochondria in import buffer (0.6 m sorbitol, 150 mm KCl, 10 mm MgCl2, 2.5 mm EDTA, 2 mm ATP, 2 mm NADH, 1 mg/ml fatty acid-free BSA, and 20 mm Hepes-KOH, pH 7.4) at 30 °C. To analyze the Mia40-ChChd3 intermediate, towards the end of the reactions, samples were centrifuged at 10,000 × g for 10 min, and the mitochondrial pellet was resuspended in Laemmli sample buffer and analyzed on SDS-PAGE followed by autoradiography. To remove nonimported precursor proteins, samples were digested with 10 μg/ml trypsin for 30 min on ice followed by 20 μg/ml soybean trypsin inhibitor. The IMS-targeted protein, Tim10, and the matrix-targeted protein, su9-DHFR, were used as model substrates to test the import competency of the mitochondria and the efficiency of trypsin digestion in the reaction. Mitochondria from trypsin-digested and nondigested samples were isolated by centrifugation at 10,000 × g for 5 min, resuspended in Laemmli sample buffer with 100 mm DTT, and analyzed by SDS-PAGE followed by autoradiography.
RESULTS
Myristoylation and the CHCH Domain Are Essential for Mitochondrial Localization of ChChd3
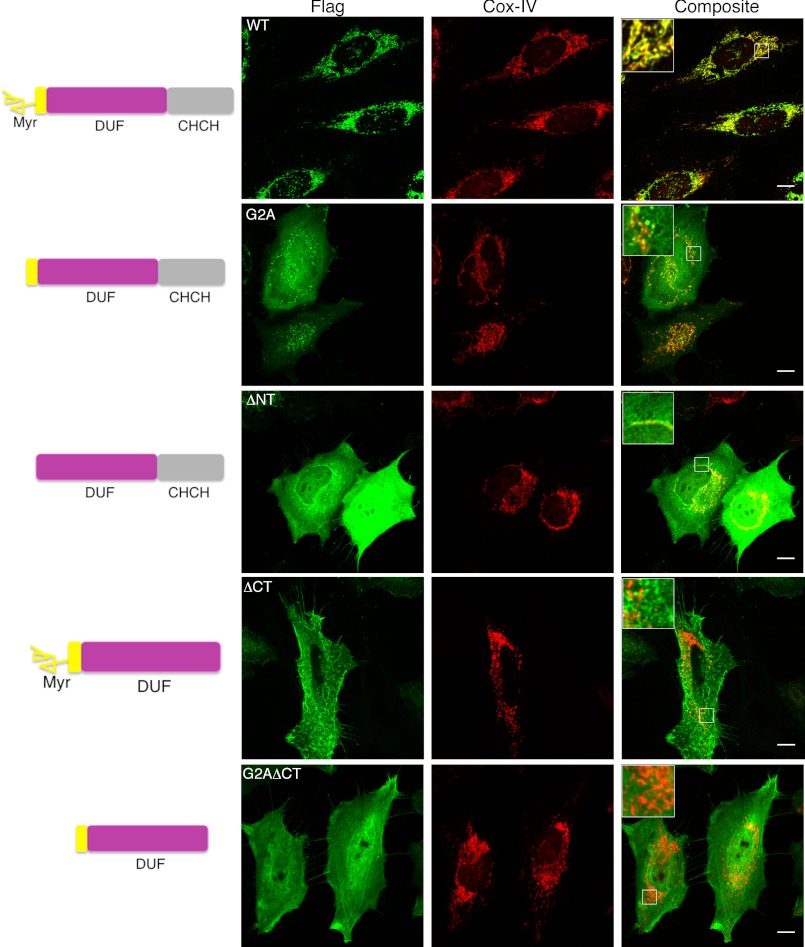
Although carboxyl-terminally FLAG-tagged ChChd3 is exclusively localized to mitochondria, fusion of GFP to either the amino or the carboxyl terminus interfered with the targeting of protein to mitochondria (data not shown). This suggested that both these segments might be essential for mitochondrial targeting and/or localization. We previously showed that ChChd3 is myristoylated and phosphorylated at the amino-terminal motif (1, 2). Our previous studies also indicated that the myristoylation is essential for binding of the protein to Sam50 on the OM of mitochondria. Various studies have shown that myristoylation enhances membrane binding and thus can play a role in localization and biological function of proteins (20). Additionally, phosphorylation at the myristoylation motif has been shown to participate in a “switch” mechanism and cycle the myristoylated protein between membrane and cytosol (23). Thus, we speculated that myristoylation and phosphorylation at the amino-terminal motif could potentially regulate the localization and/or targeting of ChChd3 to the IMS. To test this, we introduced mutations in the myristoylation motif of C-terminally FLAG-tagged ChChd3 and analyzed the localization of the mutant proteins by indirect immunofluorescence. Although WT ChChd3 is exclusively localized to mitochondria, the glycine to alanine mutant protein (G2A), which blocks myristoylation, and the N-terminal truncated protein (ΔNT), lacking the myristoylation motif, showed only partial mitochondrial localization (Fig. 1). These mutant proteins diffused throughout the cell, including the nucleus, indicating that the myristoylation in ChChd3 plays a significant role in the mitochondrial targeting and/or retention inside the mitochondria. The localization of the phosphorylation site mutated proteins T11A and T11E was similar to that of WT protein, indicating that the phosphorylation does not regulate the mitochondrial targeting of ChChd3 (data not shown).
FIGURE 1.
The N-terminal myristoylation (Myr) and the C-terminal CHCH domain are essential for targeting and localization of ChChd3 to mitochondria. Representative confocal microscopic images of HeLa cells transiently expressing FLAG-tagged ChChd3 mutants are shown. ChChd3 localization was analyzed by indirect immunofluorescence by staining against FLAG tag. Scale bar, 10 μm.
Next we asked whether myristoylation itself is sufficient or the CHCH domain is also necessary for the mitochondrial localization of ChChd3. To test this, we generated a FLAG-tagged mutant lacking the CHCH domain (ΔCT) and analyzed its cellular localization. Deletion of the CHCH domain completely excluded the protein from mitochondria. The ΔCT appeared to be primarily localized to membranes. Further mutating the myristoylation site (G2AΔCT) disrupted the membrane targeting and resulted in a diffused pattern (Fig. 1). These results support that both the myristoylation and the CHCH domain are essential for the mitochondrial localization of ChChd3.
Is ChChd3 a Substrate for Mia40?
Because Mia40 plays a crucial role in the import and folding of CX3C and CX9C motif proteins, and we found that CHCH domain is essential for mitochondrial localization of ChChd3, the next goal was to analyze whether ChChd3 is a substrate for Mia40.
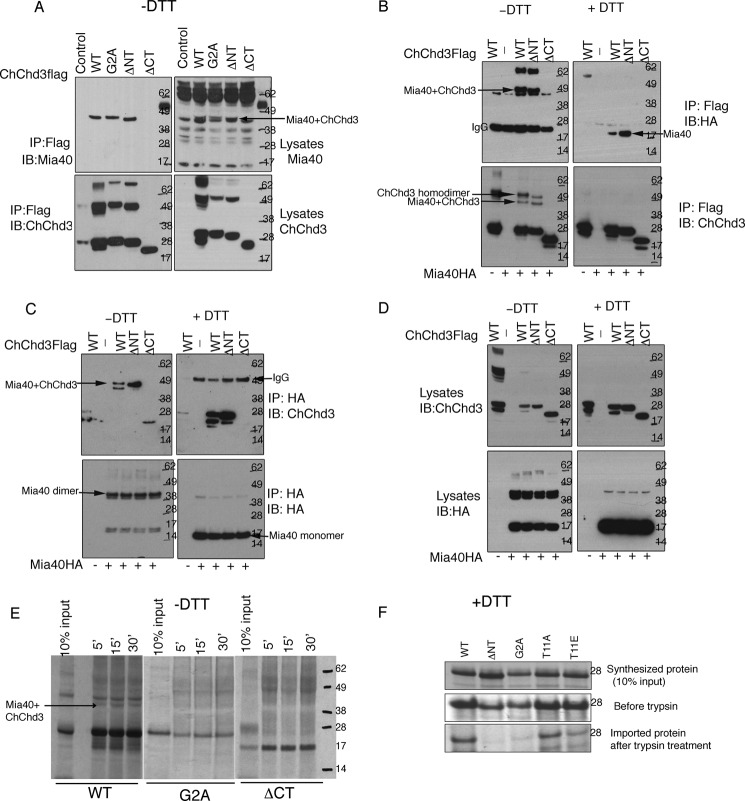
We sought to characterize the disulfide-bonded intermediate of ChChd3 and Mia40 in total cell lysates and immunoprecipitated samples from HEK 293 cells under nonreducing and reducing conditions. Because our results indicated that myristoylation is essential for mitochondrial localization, we addressed how this modification affects the formation of Mia40-ChChd3 intermediate and the import of ChChd3. FLAG-tagged proteins were expressed in HEK 293 cells, harvested in the presence of 10 mm N-ethylmaleimide to prevent oxidation in sample handling, and immunoprecipitated with FLAG resin. Analysis of the FLAG immunoprecipitated samples on a nonreducing SDS-PAGE revealed that both the WT and the nonmyristoylated form of ChChd3 mutant proteins, G2A and ΔNT, form disulfide-bonded products with Mia40 at ∼45 kDa, corresponding to the total molecular mass of ChChd3 (∼28 kDa) and Mia40 (∼17 kDa) (Fig. 2A). Although HEK 293 cells express extremely low levels of Mia40 that were hard to detect in lysates, ChChd3 immunoprecipitated complexes from HEK 293 cells clearly revealed the presence of the Mia40-ChChd3 intermediate using Mia40 antibody under nonreducing conditions from both the immunoprecipitated samples and total cell lysates (Fig. 2A, top panel). However, when these samples were reduced with 100 mm DTT, the Mia40 antibody failed to detect any protein. This is mainly due to the specificity of Mia40 antibody toward the oxidized protein. We verified this by using the Escherichia coli purified His-tagged Mia40 where the Mia40 antibody preferentially recognized the nonreduced protein when compared with that of the reduced form (data not shown). To further verify the Mia40-ChChd3 disulfide-bonded intermediates, we co-expressed HA-tagged Mia40 with FLAG-tagged ChChd3 in HEK 293 cells and immunoprecipitated the proteins using HA or FLAG resin (Fig. 2, B–D). Under nonreducing conditions, both the FLAG and the HA immunoprecipitated samples showed the disulfide-bonded intermediate at approximately the same molecular mass (∼45 kDa, Fig. 2, B and C, top left panels), which is similar to that of the endogenous Mia40-ChChd3 FLAG intermediate (Fig. 2A). Reducing these samples with DTT revealed the presence of monomeric proteins with both with the HA and the ChChd3 antibodies (Fig. 2, B and C, top right panel). As expected, the CHCH domain deletion mutant, ΔCT, did not show any binding to Mia40 (Fig. 2, A–C).
FIGURE 2.
ChChd3 binds to Mia40 in HEK 293 cells and in isolated mouse liver mitochondria. A–D, characterization of ChChd3-Mia40 disulfide-bonded product in HEK 293 cells. A, FLAG-tagged ChChd3 mutants were expressed in HEK 293 cells and immunoprecipitated (IP) with FLAG resin. Eluted samples and 10% input from total cell lysates were analyzed on SDS-PAGE under nonreduced conditions followed by immunoblotting (IB) against Mia40 and ChChd3 antibodies. B–D, HEK 293 cells co-expressing FLAG-tagged ChChd3 and HA-tagged Mia40 were subjected to immunoprecipitation on FLAG (B) or HA resin (C). Immunoisolated samples were analyzed on SDS-PAGE under nonreduced (left panel, −DTT) as well as reduced conditions (right panel, 100 mm DTT (+DTT)) followed by immunoblotting against HA and ChChd3 antibodies. 10% input from the total cell lysates is shown in D. E and F, in vitro import of ChChd3 mutants into isolated mouse liver mitochondria. 35S-radiolabeled proteins were incubated with freshly isolated mitochondria at the indicated time points and analyzed by SDS-PAGE and autoradiography under nonreduced conditions to analyze Mia40-ChChd3 disulfide-bonded intermediate product (E), and nonimported precursor proteins were removed by digesting with trypsin (F).
Myristoylation and the CHCH Domain Are Essential for the Import of ChChd3 into Mitochondria
Although the nonmyristoylated ChChd3 mutant proteins G2A and ΔNT are only partially localized to mitochondria, in HEK 293 cells, these mutants bind Mia40 with similar efficiency to that of WT. There are two likely explanations. These mutants can pass through the translocase of outer membrane (TOM) complex and interact with Mia40, but are not retained inside the mitochondria, as they cannot attain a stable conformation by binding to mitochondrial membranes or binding to other proteins. Alternatively, as both ChChd3 and Mia40 are nuclear-encoded proteins, the oxidative environment of the endoplasmic reticulum lumen or even cytosol can facilitate the binding and can engage the protein outside the mitochondria. To verify this, we imported radiolabeled WT and G2A mutant proteins into isolated mouse liver mitochondria. Analysis by SDS-PAGE under nonreducing conditions revealed that WT protein formed high molecular mass product inside the mitochondria as fast as 5 min and was stable up to 30 min (Fig. 2E). Under similar conditions, we failed to detect this product with G2A and ΔCT mutants. When the nonimported precursor proteins or the proteins that bound to the OM were removed by digesting with trypsin and analyzed under reducing conditions, the nonmyristoylated mutant proteins were completely absent in the mitochondria (Fig. 2F). Under similar conditions, we were able to detect WT protein and the phosphorylation site mutants T11A and T11E. This suggested that myristoylation is essential for import of ChChd3 into mitochondria and that the binding to Mia40 with nonmyristoylated mutants in the immunoprecipitated samples is likely outside the mitochondria. Once imported into the mitochondrial IMS, myristoylation may confer further stability to ChChd3 through protein-protein interactions.
Mia40 Binds Cys193 from Helix 1 of ChChd3
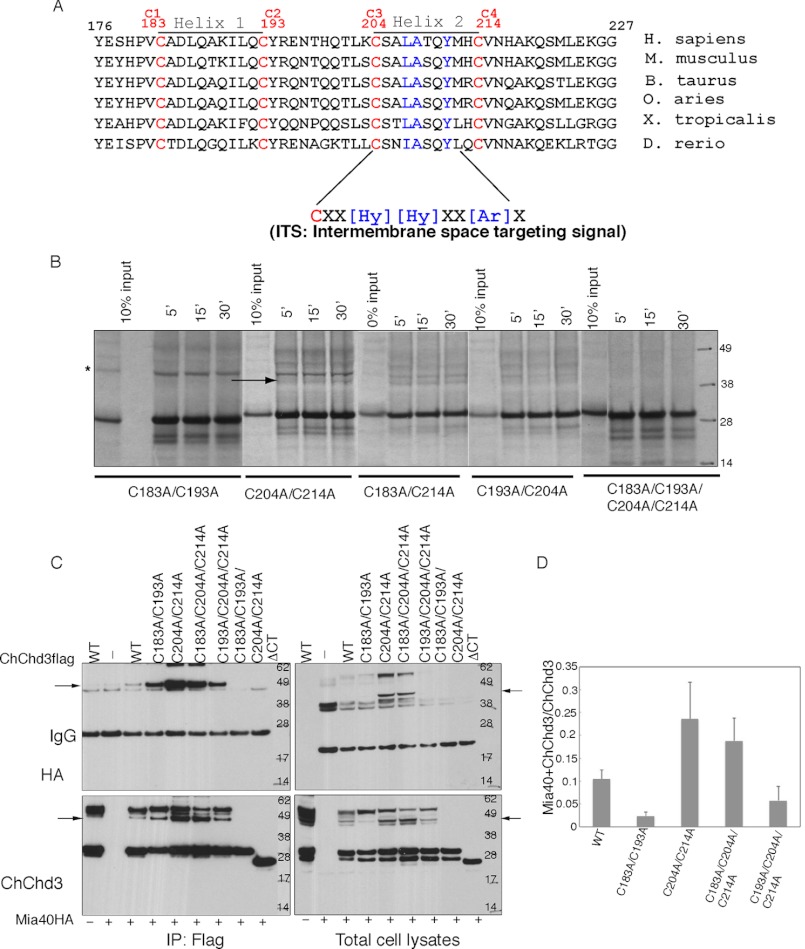
Mia40 recognizes substrates in a cysteine-dependent manner. In yeast Cox17, the third cysteine that pairs with the second cysteine interacts with Mia40 (24), whereas in Tim9 and Tim10, the N-terminal cysteine is known to bind Mia40 (25), clearly indicating the differential binding of cysteines in different proteins. The solution structure of the human Mia40 (24) and the crystal structure of the yeast Mia40 (26) revealed the presence of three disulfide bonds: C1-C2 (CPC), C3-C6, and C4-C6 (CX9C motif), and a shallow hydrophobic cleft adjacent to the CPC motif. The hydrophobic cleft was proposed to bind substrates. Recent studies have suggested that the hydrophobic residues upstream of the docking cysteine from the substrate proteins direct them toward the Mia40 hydrophobic binding pocket and position the specific cysteine for binding. The mitochondrial intermembrane space sorting (MISS)/intermembrane space targeting sequence (ITS) sequence predicted based on the studies from Tim9 and Tim10 and Cox17 sequence include a 9-amino acid region with the hydrophobic amino acids at the upstream of the docking cysteine (−4 position) for MISS (27) and the hydrophobic and aromatic amino acids at the upstream or downstream of the docking cysteine for ITS (28). Analysis of the CHCH domain sequence in ChChd3 identified a well conserved ITS in helix 2 at the upstream of Cys204 (Fig. 3A). The +3 and +4 positions of Cys204 in helix 2 are occupied by hydrophobic amino acids (Leu207 and Ala208), and at the +7 position, there is a tyrosine (Tyr211). Based on the ITS predictions, Cys204 has a very strong docking site with the active site cysteine in Mia40. To test the specific cysteine of ChChd3 that binds Mia40, we generated several FLAG-tagged ChChd3 mutants and tested for their ability to interact with Mia40 in isolated mouse liver mitochondria and HEK 293 cells. [35S]-radiolabeled mutant proteins synthesized in rabbit reticulocyte lysate system were imported into mouse liver mitochondria and analyzed for ChChd3-Mia40 disulfide-bonded intermediates under nonreducing conditions. Surprisingly, the Mia40-ChChd3 intermediate was clearly identified when both the cysteines in helix 2 were replaced with alanine (C204A/C214A), and it was nearly absent when the helix 1 cysteines were replaced (C183A/C193A) (Fig. 3B). This ruled out the involvement of helix 2 cysteines in binding to Mia40. We next generated the double cysteine mutant C183A/C214A where the outer cysteine pair is mutated, and C193A/C204A where the inner cysteine pair is mutated. As shown in Fig. 3B, we were able to detect the C183A/C214A-Mia40 intermediate inside the mitochondria, whereas no adduct with Mia40 was observed with the C193A/C204A variant. Because the intermediate is formed with the double mutant C204A/C214A, and did not form in C193A/C204A, it is presumed that Mia40 binds through the inner cysteine, Cys193.
FIGURE 3.
ChChd3 binds with Mia40 primarily through Cys193. A, sequence alignment of CHCH domain of ChChd3 shows the presence of ITS upstream of the Cys204. H. sapiens, Homo sapiens; M. musculus, Mus musculus; B. taurus, Bos taurus; O. aries, Ovis aries; X. tropicalis, Xenopus tropicalis; D. rerio, Danio rerio. B, analysis of the CX9C motif cysteine-Mia40 intermediate in isolated mouse liver mitochondria. Radiolabeled precursor proteins were imported into freshly isolated mouse liver mitochondria at the indicated time points and analyzed under nonreducing conditions. C, analysis of the CX9C motif cysteine-Mia40 intermediate in HEK 293 cells. HEK 293 cells co-expressing FLAG-tagged ChChd3 mutants and HA-tagged Mia40 were subjected to immunoprecipitation (IP) on FLAG resin. Immunoisolated samples and the total cell lysates were analyzed on nonreducing SDS-PAGE followed by immunoblotting against HA and ChChd3 antibodies. 10% input from the total cell lysates is shown. D, Mia40-ChChd3 disulfide intermediate was quantified from the total cell lysates as the ratio of dimer to monomer. Error bars represent S.D. from three independent experiments. Arrowhead indicates ChChd3-Mia40 disulfide-bonded dimer. * corresponds to the background labeling of the 42-kDa protein from [35S]methionine in rabbit reticulocyte lysate system.
Analysis of the cell lysates and immunoisolated complexes from HEK 293 cells provided additional evidence that Mia40 covalently binds ChChd3 through Cys193 (Fig. 3, C and D). Helix 1 variants did not show a significant difference in binding to Mia40 when compared with that of WT protein. However, helix 2 mutants consistently showed increased binding when compared with that of WT or the helix 1 mutants (Fig. 3C, left) Similar increase was observed in the intermediate from the total cell lysates (Fig. 3C, right). This suggested that helix 2 cysteines Cys204 and Cys214 are dispensable for binding to Mia40 in binding to ChChd3. In C204A and C204A/C214A, the intermediate is likely be trapped as these cysteines are not available for intramolecular disulfide bonding and subsequent folding of the protein. Further analysis with the triple cysteine mutants C183A/C204A/C214A and C193A/C204A/C214A showed that Mia40 binds ChChd3 through Cys193. Although C183A/C204A/C214A can bind Mia40 with similar efficiency as that of the helix 2 cysteine mutants, C193A/C204A/C214A showed much weaker binding. As expected, the tetracysteine mutant, C183A/C193A/C204A/C214A, and ΔCT did not show any binding to Mia40, indicating that the interaction of Mia40 with ChChd3 is only through the disulfide bonds (Fig. 3C). Together these results suggest that Mia40 preferentially binds ChChd3 through Cys193 in helix 1 that pairs with Cys204 in helix 2.
Hydrophobic Residues in Helix 1 Are Essential for Docking Cys193 onto Mia40
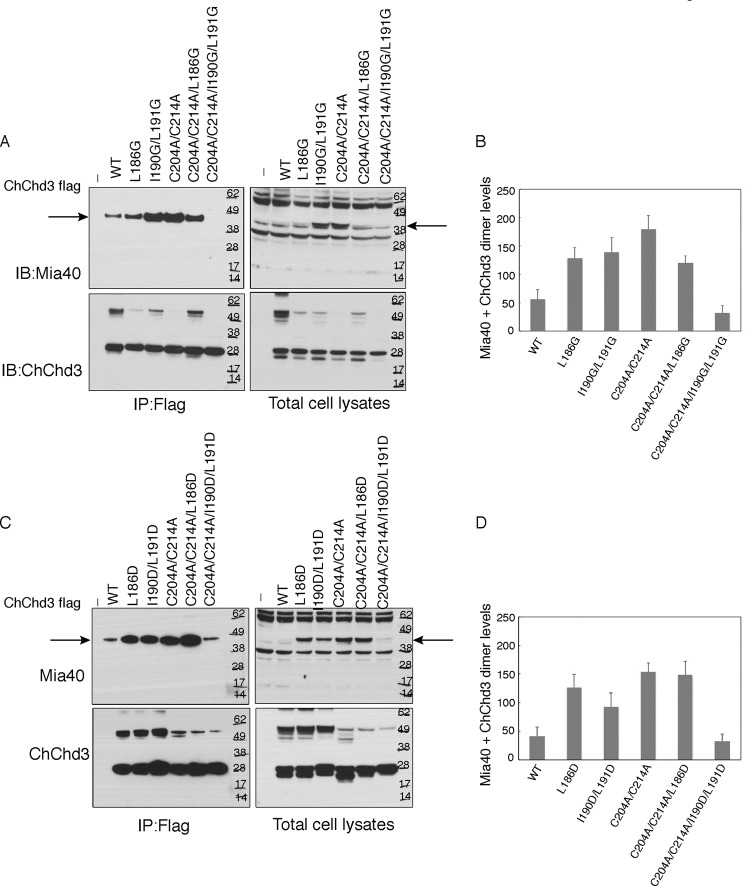
Analysis of the helix 1 in ChChd3 identified the hydrophobic amino acids, Leu191, Ile190, and Leu186, at the −2, −3, and −7 positions of Cys193. We analyzed the role of these residues in binding to Mia40. We mutated these residues to glycine (L191G, I190G/I191G) and aspartic acid (L191D, I191D/I191D) and analyzed the binding of the mutant proteins to Mia40 in HEK 293 cells. Because the helix 2 cysteine mutants bind strongly to Mia40, we also introduced the same mutations in C204A/C214A background (L186G/C204A/C214A, I190G/I191G/C204A/C214A, L186D/C204A/C214A, I190D/I191D/C204A/C214A). When L191G, I190G/I191G, L191D, and I191D/I191D in WT background were analyzed for binding to Mia40, surprisingly, they showed increased binding to Mia40 (Fig. 4). When the same mutations were introduced in C204A/C214A, the binding to Mia40 was reduced. L186G and L186D mutations did not significantly affect the binding. However, I190G/L191G and I190D/L191D variants in C204A/C214A background showed a significant decrease in binding to Mia40 when compared with that of C204A/C214A. It is likely that in WT, when the hydrophobic amino acids are mutated in helix 1, helix 2 cysteines can slide onto the Mia40 substrate binding pocket and can bind through Cys204.
FIGURE 4.
Hydrophobic amino acids Leu190 and Ile191 in helix 1 position Cys193 toward Mia40 hydrophobic binding pocket. A and C, HEK 293 cells expressing FLAG-tagged ChChd3 mutants were subjected to immunoprecipitation (IP) on FLAG resin. Immunoisolated samples and the cell lysates were analyzed on a nonreducing SDS-PAGE and immunoblotted (IB) against Mia40 and ChChd3 antibodies. 10% input from the total cell lysates is shown. B and D, quantification of Mia40-ChChd3 dimer levels normalized to ChChd3 protein levels. Error bars represent S.D. from three different experiments. Arrowhead indicates ChChd3-Mia40 disulfide-bonded dimer.
ChChd3 Interacts with the Cys55 of the Mia40 CPC Motif
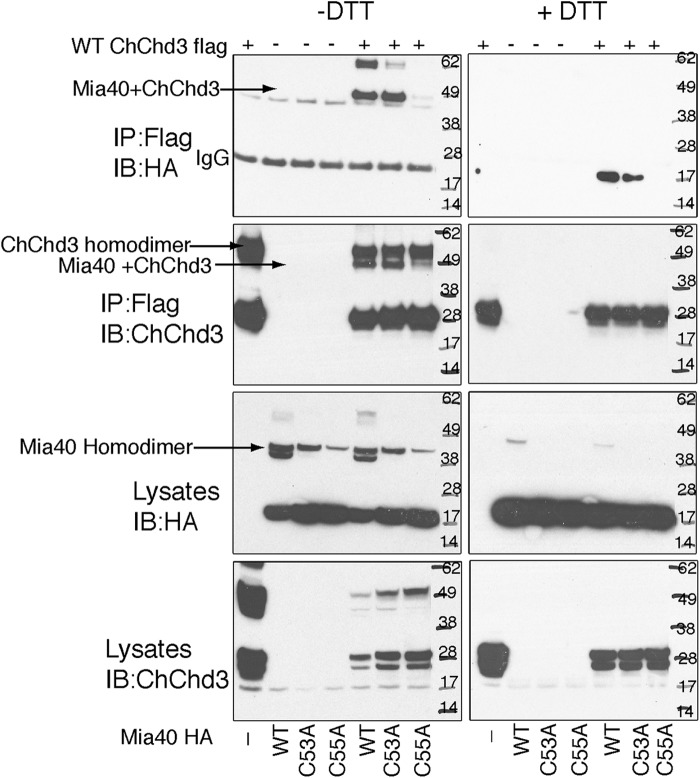
Mia40 possess a well conserved CPC-CX9C-CX9C motif. Although the cysteines in CX9C-CX9C motif are involved in structural stabilization, the CPC cysteines are involved in oxidation of the substrate proteins in the IMS. Furthermore, the second cysteine in the CPC motif, the Cys55, has been shown to be indispensable in binding to substrates. To address the specificity of the ChChd3-Mia40 disulfide-bonded intermediate and to analyze the specific cysteine of Mia40 that interacts with the Cys193 of ChChd3, we mutated the CPC cysteines to alanine (C53A and C55A) and analyzed their binding to ChChd3 in HEK 293 cells. When the HA-tagged Mia40 mutants were co-expressed with FLAG-tagged ChChd3, both the immunoprecipitated samples and the total cell lysates showed that mutating Cys55 completely abolished formation of the Mia40-ChChd3 intermediate (Fig. 5). The C53A variant on the other hand showed no effect in forming the disulfide-bonded adduct, suggesting that Mia40 binds ChChd3 specifically through Cys55.
FIGURE 5.
Mia40 interacts with ChChd3 through its active site cysteine, Cys55. HEK 293 cells co-expressing FLAG-tagged ChChd3 and HA-tagged Mia40 mutants were subjected to immunoprecipitation (IP) on FLAG resin. Immunoisolated samples were analyzed on SDS-PAGE under nonreduced (left panel, −DTT) as well as reduced conditions (right panel, 100 mm DTT (+DTT)) followed by immunoblotting (IB) against HA and ChChd3 antibodies. 10% input from the total cell lysates is shown.
Requirement of CHCH Domain Cysteines for Binding to Mitofilin and Sam50
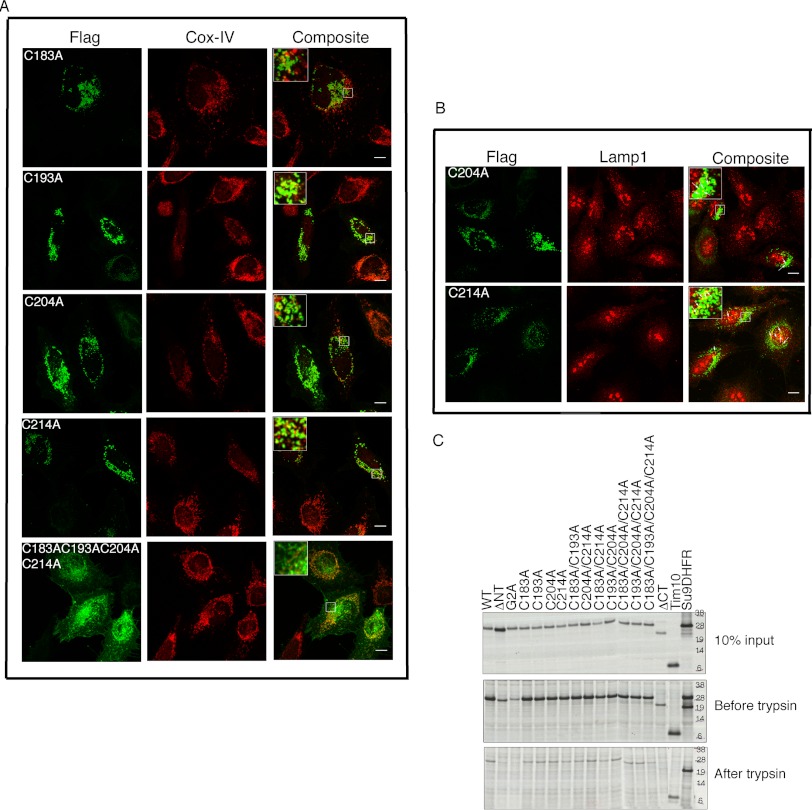
After our initial report (2), several other studies (3, 10, 12, 29) have now established that ChChd3 exists in a large protein complex together with the IM protein mitofilin and the OM protein Sam50 at crista junction and/or contact sites. We also showed that the CHCH domain is essential for binding to mitofilin, whereas it is dispensable for association with Sam50. As the cysteine mutants are imported into the IMS and are able to preferentially bind Mia40, we next asked whether these mutants have similar specificity in complex formation with mitofilin and Sam50. For this purpose, we immunoprecipitated the transiently expressed cysteine mutants from HEK 293 cells and analyzed the binding toward endogenous mitofilin and Sam50. None of the cysteine mutants showed efficient binding to either mitofilin or Sam50 (Fig. 6). Although binding of Sam50 to ChChd3 was not affected when the CHCH domain was completely deleted, mutating the cysteines in the CHCH domain resulted in much reduced binding, indicating that cysteine mutants may be misfolded and/or aggregated. To confirm this, we analyzed the cellular localization of the cysteine mutants by indirect immunofluorescence. Confirming our hypothesis, when expressed in HeLa cells, the cysteine mutants, although localized to mitochondria and partially co-localized with mitochondrial IM protein Cox-IV, were predominantly aggregated and resulted in mitochondrial fragmentation (Fig. 7A). When all four cysteines in the CHCH domain were mutated, the mutant protein (C183A/C193A/C204A/C214A) mainly localized to membrane ruffles similar to that of ΔCT, with a partial co-localization with Cox-IV. This suggested that the aggregation in the cysteine mutants might be due to intermolecular disulfide bonding. Further, when the cysteine mutants were co-stained with the lysosomal marker protein Lamp1, a partial overlap was noticed, suggesting that the protein aggregates may be targeted to lysosomes for final degradation (Fig. 7B). These results suggest that although ChChd3 can be imported into the IMS, as evidenced by the disulfide-bonded intermediates with Mia40, it requires all the cysteines for intramolecular disulfide bonding, structural integrity, and assembly into macromolecular complexes.
FIGURE 6.
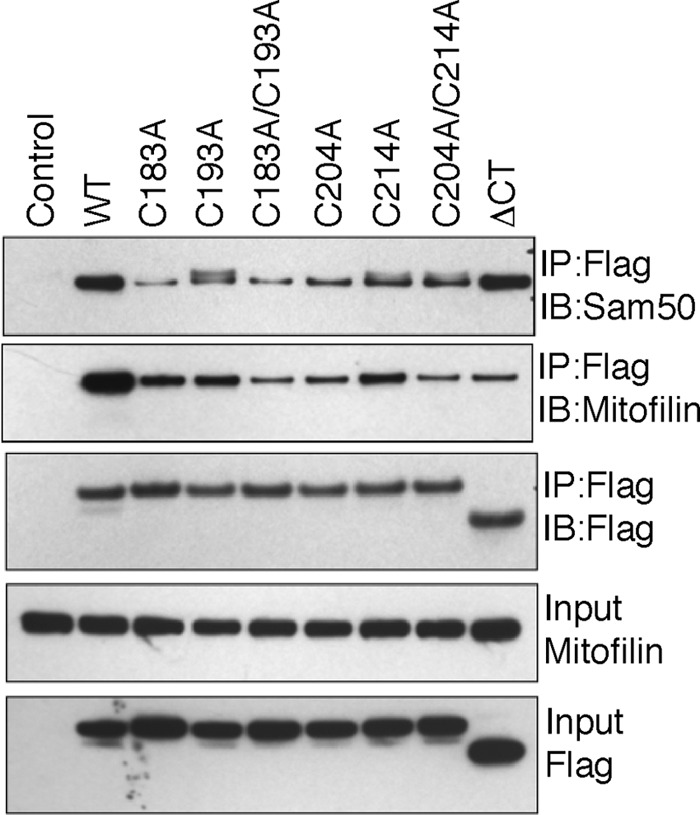
The CHCH domain cysteines are essential for binding to mitofilin and Sam50. HEK 293 cells expressing FLAG-tagged ChChd3 mutants were subjected to immunoprecipitation (IP) on FLAG resin. Immunoisolated samples were analyzed on SDS-PAGE followed by immunoblotting (IB) against the indicated antibodies. 10% input from total cell lysates is shown. Sam50 levels are not shown in the input as the antibody used for Sam50 failed to detect endogenous protein in total cell lysates from HEK 293 cells.
FIGURE 7.
The CHCH domain cysteines are essential for intramolecular disulfide bonding and structural stability of ChChd3 but not for import into the IMS of mitochondria. A and B, cellular localization of ChChd3 cysteine mutants. A, representative confocal microscopic images of HeLa cells transiently expressing FLAG-tagged ChChd3 cysteine mutants showing protein aggregation and partial co-localization with mitochondrial marker protein COX-IV. B, cysteine mutants are also partially localized to lysosomes shown by co-localization with the lysosomal marker protein Lamp1 (red). Localization of the mutant proteins was analyzed by staining against FLAG antibody (green). Scale bar, 10 μm. C, in vitro import of ChChd3 mutants into isolated mouse liver mitochondria. 35S-radiolabeled proteins were incubated with freshly isolated mitochondria. Nonimported precursor proteins were removed by trypsin and analyzed on SDS-PAGE followed by autoradiography under reduced conditions. The inner membrane-targeted protein, Tim10, and the matrix-targeted protein, su9-DHFR, were used as control for the import reaction.
Cysteines Are Not Essential for the Import of ChChd3 into the Mitochondrial IMS
As mutating any of the cysteines in the CHCH domain resulted in protein aggregates, we next analyzed whether these aggregates are trapped in the IMS or on the OM. [35S]-radiolabeled mutant proteins were imported into freshly isolated mouse liver mitochondria, and surface-labeled proteins were digested by treatment of trypsin and analyzed under reduced conditions. All the cysteine mutants tested were imported into the mitochondria and protected from protease treatment with similar efficiency as that of the WT protein (Fig. 7C). From Fig. 7C, it is evident that the myristoylation and the CHCH domain are essential for the import of ChChd3 into mitochondria as ΔNT, G2A, and ΔCT were almost completely digested upon trypsin treatment. It was interesting to note that the tetracysteine mutant is imported into the IMS and protected from trypsin digestion, whereas ΔCT was almost completely digested. This suggested that the CHCH domain but not the cysteines act as the targeting signal to the IMS import. Once in the IMS, cysteines are essential for binding to Mia40 and further folding and retention of the protein. We often noticed that the levels of ΔNT and G2A were low even before treatment with the protease, indicating that the myristoylation promotes binding of ChChd3 to the mitochondria. The CHCH domain can act as a secondary targeting sequence and translocate the protein across the membrane. Once imported in the IMS, Mia40 binds ChChd3 and catalyzes the formation of intramolecular disulfide bonds.
DISCUSSION
Previous studies defined ChChd3 as an essential protein in the IM that faces toward the IMS and serves as a scaffold for crista junction and contact site protein complexes by associating with several key mitochondrial proteins including mitofilin, Sam50, OPA1, and SKIP (2, 12). Originally identified as a PKA substrate, it is now recognized to be an important protein for crista biogenesis (1, 2). ChChd3 is myristoylated at the amino terminus, and the carboxyl terminus has a CHCH domain (2). The CHCH domain proteins are commonly seen in the IMS of mitochondria (14). They typically lack a mitochondrial targeting sequence at the amino terminus and are imported into the IMS by using the cysteines in the CHCH motif. In addition, myristoylation, often a co-translation modification, has been shown to be involved in targeting proteins to subcellular organelles including mitochondria (20, 21). In this study, we analyzed the role of these two motifs for targeting and import of ChChd3 into the mitochondria. Our results show that both the myristoylation and the CHCH domain are essential for the import into IMS and mitochondrial localization of ChChd3. We further show that Mia40 in the IMS forms disulfide-bonded intermediate with ChChd3 and the cysteines in the CHCH domain are essential for structural stabilization.
From the confocal microscopic analysis and the in vitro import of the mutant proteins into isolated mitochondria, it is evident that for mitochondrial localization of ChChd3, both the CHCH domain and myristoylation are essential. Deleting the CHCH domain or mutating all of the cysteines in the CHCH domain fails to localize the protein to mitochondria, and the resulting protein is targeted to membrane ruffles. Because myristoylated proteins are typically associated with membranes, it is likely that the CHCH domain serves as the mitochondrial membrane-targeting signal, and myristoylation further enhances the binding to the membranes. It is interesting to note that unlike the other CHCH domain proteins, in ChChd3, the CHCH domain alone is not sufficient to localize the protein to mitochondria.
Recent studies have suggested that typical substrates of the disulfide relay system are targeted to the IMS by specific import signal sequences that form an amphipathic helix with crucial hydrophobic residues (27, 28). However, based on our results, there appears to be considerable flexibility in the requirements for docking of ChChd3 to the active site of Mia40. In ChChd3, although Cys204 in helix 2 fulfills ITS requirements (Fig. 3A), mutating this cysteine showed increased binding to Mia40 when compared with that of WT (Fig. 3, C and D), suggesting that Cys204 is not involved in binding. Further mutations of other cysteines proved that Cys193 forms intermolecular disulfide bond with Mia40.
The sequence around Cys193 in helix 1 is not identical to either ITS or MISS predictions. However, there are hydrophobic amino acids at the −2, −3, and −7 positions relative to Cys193. Hence, it is likely that the hydrophobic amino acids around the cysteine are sufficient for directing specific proteins to the substrate binding pocket of Mia40. In ChChd3, hydrophobic amino acids in helix 1, Ile190 and Leu191, likely position the Cys193 to the Mia40 Cys55. In the absence of these residues, the hydrophobic residues in helix 2 may also position the cysteines in helix 2 to Mia40. This is apparent because when Ile190 and Leu191 were mutated in the WT protein, the mutant proteins showed stronger binding to Mia40 (Fig. 4). In contrast, when the same hydrophobic residues were mutated along with helix 2 cysteines, Mia40 showed reduced binding, although helix 1 cysteines were still available. It is likely that DUF domain plays a role in directing the helix 1 cysteine, Cys193, toward Mia40. It is important to note that the MISS and ITS sequence were analyzed based on the small molecular mass proteins Cox17 and Tim9 and Tim10 that do not have any other functional domains. Similar findings were reported by Kloppel et al. (30) for copper chaperone of super oxide dismutase 1 (Ccs1), which has been shown to import into the IMS by using the disulfide relay pathway. Ccs1 does not have the conventional CX9C/CX3C motifs or does not possess the ITS/MISS sequence. However, it contains hydrophobic amino acids at the upstream of the docking cysteine similar to that of ChChd3.
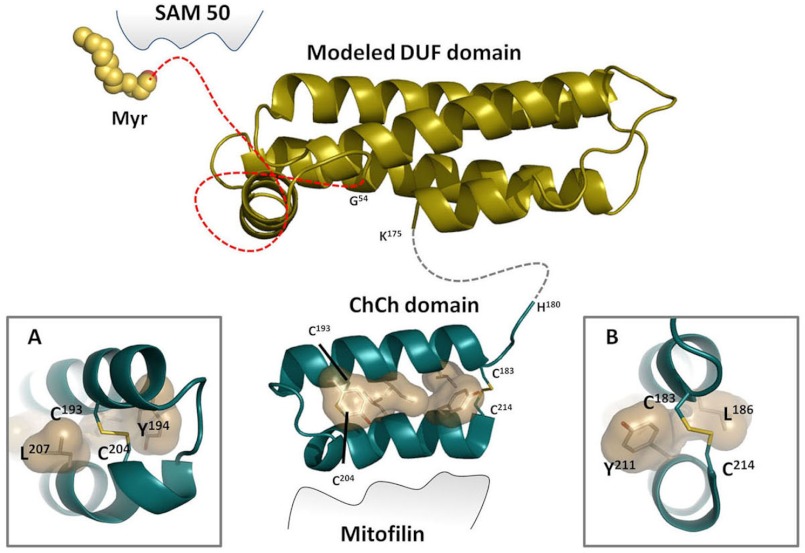
Based on our findings and on other recent descriptions of the MIB complex, we predict that these three proteins, ChChd3, mitofilin, and Sam50, are essential components of the bridging complex with the N terminus of ChChd3 docking onto Sam50, which is embedded in the OM, and the disulfide-bonded CHCH domain binding to mitofilin, which is anchored to the IM (Fig. 8). Although the structure of the DUF domain in ChChd3 is unknown, it is predicted to be helical. When we searched the Protein Data Bank (PDB) with a threading program, PHYRE2 (31), for proteins having a similar fold, we found no native protein hits. However, we did find an engineered helical protein that was designed to be a receptor for IL-4 (32). The program predicted a 93% certainty for this fold. We thus modeled the DUF domain based on this structure and modeled the CHCH domain based on the structure of Cox17 (Fig. 8). Modeling of the CHCH domain also shows that the hydrophobic residues are likely to play an important role in stabilizing the helix in addition to their potential role in docking to Mia40. Based on this model, we predict that the DUF domain will serve as a major scaffold for many other proteins that ChChd3 is known to bind to.
FIGURE 8.
Model of the ChChd3 depicting the myristoylation (Myr)motif, DUF domain, and CHCH domain. The DUF domain is modeled based on the engineered protein that was designed for IL-4 receptor. The CHCH domain is modeled based on the Cox17 structure. A and B, the insets show the hydrophobic residues in the CHCH domain helices that contribute to recognition by Mia40.
Elucidating the mechanism that allows ChChd3 to be retained in the IMS emphasizes the unique oxidizing environment of the IMS, which is reminiscent of the lumen of the endoplasmic reticulum. The reducing environment of the rest of the cell prevents disulfide bonds from forming readily, yet in the IMS, disulfide bonds are specifically formed, and this mechanism is used to retain proteins in this space. All CHCH domain proteins presumably are targeted selectively to this compartment. Furthermore, these proteins should be released when the oxidizing environment is destroyed as occurs in apoptosis.
Acknowledgments
We thank Dr. Alexander Kornev for providing Fig. 8, Susanna Petrosyan for helping with the mitochondrial preparations, and Dr. Hiroyuki Hakozaki for assistance with the confocal imaging.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 DK54441 (to S. S. T.).
- ChChd3
- coiled-coil helix coiled-coil helix domain-containing protein 3
- OM
- outer membrane
- IM
- inner membrane
- IMS
- intermembrane space
- Mia40
- mitochondrial intermembrane space import and assembly 40
- SAM
- sorting and assembly machinery
- SKIP
- sphingosine kinase interacting protein
- Tim
- translocase of inner membrane
- MISS
- mitochondrial intermembrane space sorting
- ITS
- intermembrane space targeting sequence
- MINOS
- mitochondrial inner membrane organizing system
- MIB
- mitochondrial intermembrane space bridging
- DUF
- domain of unknown function;su9-DHFR, subunit 9 dihydrofolate reductase.
REFERENCES
- 1. Schauble S., King C. C., Darshi M., Koller A., Shah K., Taylor S. S. (2007) Identification of ChChd3 as a novel substrate of the cAMP-dependent protein kinase (PKA) using an analog-sensitive catalytic subunit. J. Biol. Chem. 282, 14952–14959 [DOI] [PubMed] [Google Scholar]
- 2. Darshi M., Mendiola V. L., Mackey M. R., Murphy A. N., Koller A., Perkins G. A., Ellisman M. H., Taylor S. S. (2011) ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 286, 2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alkhaja A. K., Jans D. C., Nikolov M., Vukotic M., Lytovchenko O., Ludewig F., Schliebs W., Riedel D., Urlaub H., Jakobs S., Deckers M. (2012) MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell 23, 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ott C., Ross K., Straub S., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M. J., Krohne G., Rudel T., Kozjak-Pavlovic V. (2012) Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell Biol. 32, 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W., Cyrklaff M., Neupert W., Reichert A. S. (2009) Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185, 1047–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. John G. B., Shang Y., Li L., Renken C., Mannella C. A., Selker J. M., Rangell L., Bennett M. J., Zha J. (2005) The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Head B. P., Zulaika M., Ryazantsev S., van der Bliek A. M. (2011) A novel mitochondrial outer membrane protein, MOMA-1, that affects cristae morphology in Caenorhabditis elegans. Mol. Biol. Cell 22, 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozjak-Pavlovic V., Ross K., Benlasfer N., Kimmig S., Karlas A., Rudel T. (2007) Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 8, 576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xie J., Marusich M. F., Souda P., Whitelegge J., Capaldi R. A. (2007) The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6, and DnaJC11. FEBS Lett. 581, 3545–3549 [DOI] [PubMed] [Google Scholar]
- 10. Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., Neupert W. (2011) The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30, 4356–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song Z., Chen H., Fiket M., Alexander C., Chan D. C. (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Means C. K., Lygren B., Langeberg L. K., Jain A., Dixon R. E., Vega A. L., Gold M. G., Petrosyan S., Taylor S. S., Murphy A. N., Ha T., Santana L. F., Tasken K., Scott J. D. (2011) An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, E1227–E1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kovanich D., van der Heyden M. A., Aye T. T., van Veen T. A., Heck A. J., Scholten A. (2010) Sphingosine kinase interacting protein is an A-kinase anchoring protein specific for type I cAMP-dependent protein kinase. Chembiochem. 11, 963–971 [DOI] [PubMed] [Google Scholar]
- 14. Banci L., Bertini I., Ciofi-Baffoni S., Tokatlidis K. (2009) The coiled coil-helix-coiled coil-helix proteins may be redox proteins. FEBS Lett. 583, 1699–1702 [DOI] [PubMed] [Google Scholar]
- 15. Webb C. T., Gorman M. A., Lazarou M., Ryan M. T., Gulbis J. M. (2006) Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed α-propeller. Mol. Cell 21, 123–133 [DOI] [PubMed] [Google Scholar]
- 16. Banci L., Bertini I., Ciofi-Baffoni S., Janicka A., Martinelli M., Kozlowski H., Palumaa P. (2008). A structural-dynamical characterization of human Cox17. J. Biol. Chem. 283, 7912–7920 [DOI] [PubMed] [Google Scholar]
- 17. Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 18. Hofmann S., Rothbauer U., Mühlenbein N., Baiker K., Hell K., Bauer M. F. (2005) Functional and mutational characterization of human MIA40 acting during import into the mitochondrial intermembrane space. J. Mol. Biol. 353, 517–528 [DOI] [PubMed] [Google Scholar]
- 19. Resh M. D. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 20. Zha J., Weiler S., Oh K. J., Wei M. C., Korsmeyer S. J. (2000) Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 21. Colombo S., Longhi R., Alcaro S., Ortuso F., Sprocati T., Flora A., Borgese N. (2005) N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J. Cell Biol. 168, 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stuart R. A., Koehler C. M. (2007). In vitro analysis of yeast mitochondrial protein import. Curr. Protoc. Cell Biol. Chapter 11, Unit 11.19 [DOI] [PubMed] [Google Scholar]
- 23. Thelen M., Rosen A., Nairn A. C., Aderem A. (1991) Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature 351, 320–322 [DOI] [PubMed] [Google Scholar]
- 24. Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N., Tokatlidis K. (2009) MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16, 198–206 [DOI] [PubMed] [Google Scholar]
- 25. Milenkovic D., Gabriel K., Guiard B., Schulze-Specking A., Pfanner N., Chacinska A. (2007) Biogenesis of the essential Tim9-Tim10 chaperone complex of mitochondria: site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J. Biol. Chem. 282, 22472–22480 [DOI] [PubMed] [Google Scholar]
- 26. Kawano S., Yamano K., Naoé M., Momose T., Terao K., Nishikawa S., Watanabe N., Endo T. (2009) Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc. Natl. Acad. Sci. U.S.A. 106, 14403–14407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milenkovic D., Ramming T., Müller J. M., Wenz L. S., Gebert N., Schulze-Specking A., Stojanovski D., Rospert S., Chacinska A. (2009) Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell 20, 2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., Banci L., Bertini I., Tokatlidis K. (2009) A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. An J., Shi J., He Q., Lui K., Liu Y., Huang Y., Sheikh M. S. (2012) CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J. Biol. Chem. 287, 7411–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klöppel C., Suzuki Y., Kojer K., Petrungaro C., Longen S., Fiedler S., Keller S., Riemer J. (2011) Mia40-dependent oxidation of cysteines in domain I of Ccs1 controls its distribution between mitochondria and the cytosol. Mol. Biol. Cell 22, 3749–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 32. Laporte S. L., Forsyth C. M., Cunningham B. C., Miercke L. J., Akhavan D., Stroud R. M. (2005) De novo design of an IL-4 antagonist and its structure at 1.9 Ä. Proc. Natl. Acad. Sci. U.S.A. 102, 1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]