Background: ELMOD family proteins function either as Rac guanine nucleotide exchange factors or Arf GTPase-activating proteins.
Results: The ELMOD family spans eukaryotic diversity and contains a putative catalytic arginine, essential for Arf GAP function.
Conclusion: The ELMOD family is ancient, and GAP activity lies within the ELMO domain.
Significance: This study establishes a function of the ELMO domain as a GTPase activating domain.
Keywords: ARF, Golgi, GTPase, Lipid Droplets, Phylogenetics, Arl2, Arl3, ELMO, GTPase-activating Protein (GAP)
Abstract
The human family of ELMO domain-containing proteins (ELMODs) consists of six members and is defined by the presence of the ELMO domain. Within this family are two subclassifications of proteins, based on primary sequence conservation, protein size, and domain architecture, deemed ELMOD and ELMO. In this study, we used homology searching and phylogenetics to identify ELMOD family homologs in genomes from across eukaryotic diversity. This demonstrated not only that the protein family is ancient but also that ELMOs are potentially restricted to the supergroup Opisthokonta (Metazoa and Fungi), whereas proteins with the ELMOD organization are found in diverse eukaryotes and thus were likely the form present in the last eukaryotic common ancestor. The segregation of the ELMO clade from the larger ELMOD group is consistent with their contrasting functions as unconventional Rac1 guanine nucleotide exchange factors and the Arf family GTPase-activating proteins, respectively. We used unbiased, phylogenetic sorting and sequence alignments to identify the most highly conserved residues within the ELMO domain to identify a putative GAP domain within the ELMODs. Three independent but complementary assays were used to provide an initial characterization of this domain. We identified a highly conserved arginine residue critical for both the biochemical and cellular GAP activity of ELMODs. We also provide initial evidence of the function of human ELMOD1 as an Arf family GAP at the Golgi. These findings provide the basis for the future study of the ELMOD family of proteins and a new avenue for the study of Arf family GTPases.
Introduction
The human family of ELMO proteins consists of six members defined by the presence of the ELMO domain. These six proteins have been further divided into two subgroups, ELMOs and ELMODs,3 based on protein size and domain architecture (Fig. 1). ELMOs are approximately twice as large as ELMODs and contain multiple domains, including a domain of unknown function 3361, predicted to contain Armadillo repeats, and a PH domain (1, 2). The ELMODs consist of little more than the ELMO domain itself and are typically ∼300 residues in length. The only sequence homology between the ELMOs and ELMODs lies within the ELMO domain, and thus, this domain is the only common feature linking these two groups of proteins (3). However, no biochemical or cellular function or activity has been linked to the ELMO domain, and it remains a domain of unknown function.
FIGURE 1.
Six human ELMO domain-containing family members are equally divided into three ELMO and three ELMOD proteins. Each member of the family contains the ELMO domain (solid black). The three ELMOs also contain the domain of unknown function (DUF) 3361 (gray shading) and a pleckstrin homology (PH) domain (hatched). The presence of DUF3361 and PH domains in ELMO proteins is highly conserved in opisthokonts.
ELMO1 regulates actin rearrangement and is essential for phagocytosis and cell migration in Caenorhabditis elegans (4–7). ELMO1 is also essential for the clearance of apoptotic germ cells and spermatogenesis (8, 9) and has been implicated in the regulation of fibronectin expression levels (10, 11). When bound to Dock180, the heterodimer functions as an unconventional guanine nucleotide exchange factor (GEF) for Rac1, a key regulator of actin dynamics, despite the absence of the Dbl homology domain (a characteristic of other Rac GEFs) on either ELMO or Dock180 (1). Instead, the ELMO1-Dock180 complex binds nucleotide-free Rac1 through the Docker domain of Dock180 to facilitate GTP loading (1, 2). The Rac GEF activity is found within the Dock180 protein, although it is stabilized or increased by its binding to ELMO (1, 5, 12, 13). ELMO2 has also been shown to bind Dock180 and regulate the same pathways as ELMO1, but no functional information is currently available for ELMO3 (5). The regions of ELMO1 required to mediate both Rac1 GEF activity and changes to the actin cytoskeleton have been mapped to areas outside of the ELMO domain (5, 7, 12, 14). Thus, the function of the ELMO domain in these or other processes remains unknown.
ELMOD2 was purified based on its GTPase-activating protein (GAP) activity for Arl2, a member of the Arf family of regulatory GTPases (3). The Arf family consists of at least 30 members in mammals, including 6 Arf, 22 Arl, and 2 Sar proteins (15). Arf and Sar regulate membrane traffic at virtually every step of the endocytic and secretory pathways to initiate carrier formation by coordinate recruitment of soluble adaptors and coat machinery to the surface of the membrane as well as localized changes to phospholipid metabolism (16, 17). Arls are largely functionally distinct from Arf and Sar and are involved in a diverse array of cellular functions, including energy metabolism, cytoskeleton dynamics (18), cytokinesis, lipid droplet formation, cilia functions (19, 20), recruitment of Golgins to the Golgi (21, 22), and other aspects of membrane traffic (23). ELMOD2 represents the first GAP identified for any of the 22 mammalian Arl proteins, and tests of the other ELMO family members revealed that only ELMOD1 shared this Arl2 GAP activity (3). To date, only one other GAP for any mammalian Arl protein has been identified, retinitis pigmentosa protein 2 (RP2), which has GAP activity for Arl3 (24), although it shares no sequence homology with ELMOD1 or ELMOD2.
Tests of the specificity of ELMOD2 for a limited number of other Arf family members revealed that ELMOD2 also had GAP activity for each of the Arf family members tested, including Arl3, Arf1, and Arf6 (3). Such specificity for both Arfs and Arls is unprecedented, as none of the 31 known human Arf GAPs have been shown to exhibit GAP activity for any Arl proteins. The Arf GAP Gcs1 in Saccharomyces cerevisiae was shown to possess GAP activity for both Arfs and Arl1, a key regulator of membrane traffic at the Golgi, but this was not completely unexpected given their close sequence conservation and functional relatedness (25). ELMOD2 lacks the canonical Arf GAP domain found in every other known Arf GAP, consisting of a zinc finger motif of four cysteine residues with specific spacing culminating in a highly conserved arginine residue that functions as a “catalytic arginine” residue (CX2CX16–18CX2CX4R) (26, 27). The catalytic arginine is the only residue that the GAP contributes to the site of GTP hydrolysis, and even subtle mutations of this arginine are usually sufficient to reduce GAP activity by several orders of magnitude (28). Thus, the catalytic arginine is critical for the biochemical GAP activity of the Arf GAPs. This catalytic arginine mechanism extends well beyond the Arf family and is a very common, but not universal, feature of GAPs acting on other small regulatory GTPases, including Rho and Ras family members, despite the highly divergent sequences and structures of the different GAPs (28).
Considering that ELMODs consist of little more than the ELMO domain itself, it is likely that the GAP activity of these proteins resides within the ELMO domain, but Arl2 GAP activity was not detected for any of the ELMOs. Thus, the defining domain of the family may have distinct cellular and biochemical functions between ELMOs and ELMODs and potentially even within the subgroups.
ELMO family members have only been described in metazoans (primarily mammals and C. elegans) and in two studies with the amoeba, Dictyostelium discoideum (29, 30), suggesting some evolutionary conservation. One study examined the phylogenetics of the ELMO family but only examined five species sampling fungi, metazoa, and D. discoideum (31). The diversity of eukaryotes extends well beyond this range, encompassing parasites, algae, and plants in six large taxonomic divisions or “supergroups” of eukaryotes (32). Molecular evolutionary studies of proteins involved in membrane traffic have demonstrated that, with some important exceptions (33), much of the protein machinery implicated in vesicular transport is conserved across this span implying generality of cell biological models and a surprisingly complex endomembrane system in the LECA (34, 35).
In this study, we used homology searching and phylogenetic analysis of ELMO domains to discover that ELMODs are an ancient family and that the ELMOs emerged as a distinct clade only in the Opisthokonta and are thus predicted to have acquired a distinct set of cellular functions. We also identify the GAP domain of the ELMODs, perform initial characterization of the most highly conserved arginine in ELMODs throughout eukaryotic evolution, and test it for potential function as a catalytic arginine. Results of this study also offer initial evidence for the function of ELMOD1 as an Arf GAP in cells.
EXPERIMENTAL PROCEDURES
Identification of ELMO Family Members
Candidate ELMO family members were identified by protein Basic Local Alignment Search Tool (BLASTp) of each of the six full-length human ELMO family members against predicted proteomes spanning as much eukaryotic diversity as possible. Based on the present understanding of eukaryotic relationships (36, 37), our sampling represents the full span of eukaryotic diversity. Candidate protein sequences were identified by initial BLASTp, with an expected value of ≤0.05 for any single human ELMO family member. Candidate protein sequences were verified by reciprocal BLASTp, with expected values of ≤0.05. The full list of species searched, along with all putative ELMO and ELMOD candidate homologs and their relevant accession number, is found in supplemental Table S1.
Phylogenetic Analysis
The ELMO domain from each protein sequence was identified and isolated by BLAST against the Conserved Domain Database. Protein sequences without ELMO domains recognized by the Conserved Domain Database were excluded from the analysis (e.g. 4 of 75 proteins). Isolated ELMO domains were aligned using MUSCLE version 3.6 (38) and manually adjusted to only include regions of unambiguous homology. An initial set of 71 sequences from 24 species spanning eukaryotic diversity consisting of 123 homologous positions was used to analyze the distribution of ELMOs and ELMODs among eukaryotes. A second analysis was then performed using a set of 20 genes identified as ELMOs from 11 species of metazoa consisting of 709 homologous positions, and it was used to resolve the order of emergence of metazoan ELMOs. For each analysis, ProtTest (version 2.4 (39)) was used to determine the optimal model for sequence evolution. Three phylogenetic algorithms were used to analyze each dataset as follows: PhyML (version 2.44 (40)), RaxML (version 2.2.3 (41)), and MrBayes (version 3.2.1 (42)). The latter was used to determine tree topology and posterior probability values. For PhyML and RaxML, 100 pseudoreplicates were analyzed. For MrBayes, 1 × 106 generations were used, and burnin values were assessed by removing trees before a graphically determined plateau. Convergence was determined by ensuring a splits frequency less than 0.1 in all cases.
Antibodies, Cells, and Reagents
All chemicals used were purchased from commercial sources. Rabbit polyclonal antibodies to human ELMOD1 were generated by immunization of rabbits with a purified recombinant fusion protein of human ELMOD1 tagged at the C terminus with maltose-binding protein, ELMOD1-MBP. Immunizations and sera collections were performed by Strategic Biosolutions. Other antibodies used were mouse monoclonals GM130 (BD Biosciences), β-tubulin (Sigma), SC35 (BD Biosciences), and rabbit polyclonals raised against Arf1 (43) or ATGL (Cell Signaling Technologies).
Cloning and Plasmids
The plasmids directing the expression of ELMOD1-myc/His and ELMOD2-myc/His were previously described (3), and plasmids for the expression of ELMOD1 and ELMOD2 mutants were derived from these plasmids using the QuikChange site-directed mutagenesis kit (Stratagene). All mutations were confirmed by DNA sequencing to ensure the desired mutation was incorporated and that others were not. Plasmids for the bacterial expression of trigger factor fusions of ELMOD1 and ELMOD2 were constructed using the pColdTM TF vector (Takara Bio Inc.). The Tet-On inducible expression plasmid for ELMOD1-HA for lentiviral infection was constructed from ELMOD1-myc/His using the tCMV/GFP/Ubi/rtTA2SM2 expression plasmid (Emory Viral Vector Core), and the expression plasmid for ELMOD1(R174K)-HA was derived from this plasmid. Lentiviruses were made by the Emory Viral Vector Core. The plasmid used for the bacterial expression of GST-GGA3 was a kind gift from James Casanova (University of Virginia) (44).
Preparation of Recombinant Proteins
Purified recombinant Arl2 was prepared as described previously (45). Expression plasmid for GST-GGA3 was transformed into Escherichia coli BL21(DE3), and a single colony was picked into LB medium. Cultures were grown at 37 °C until A600 = 0.6 when they were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3.5 h. Cells were then harvested and lysed by multiple passes through a French press. GST-GGA3-Sepharose beads were made by incubating lysate with glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at 4 °C with rocking. GST-GGA3 conjugated beads were spun, washed, and stored at −80 °C for no more than 30 days. Plasmids directing expression of trigger factor fusions of ELMOD1 and ELMOD2 were transformed into E. coli BL21(DE3), and a single colony was picked into LB medium. Cultures were grown at 37 °C until A600 = 0.4 when they were cooled to 15 °C for 30 min. Protein expression was induced with 0.5 mm 1-thio-β-d-galactopyranoside at 15 °C for 16 h. Cells were then harvested and lysed by multiple passes through a French press, and protein was purified using a nickel-Sepharose column.
Cell Culture and Immunofluorescence
HeLa and NRK cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (Invitrogen). Cells were transfected with Lipofectamine 2000 according to the manufacturer's directions (Invitrogen) for exogenous expression. NRK cells were transfected with Dharmafect 2 and Dharmacon OnTarget Plus Smartpool directed against rat ELMOD1 according to the manufacturer's directions (Dharmacon). HeLa cell lines capable of the inducible expression of ELMOD1-HA and ELMOD1(R174K)-HA were generated by infecting cells with ELMOD1-HA or ELMOD1(R174K)-HA lentivirus. Protein expression was induced with 2 μg/ml doxycycline. Where indicated, HeLa cells were treated with 10 μm brefeldin A (BFA) for 10 min at 37 °C. For immunofluorescence, cells were fixed using 2% paraformaldehyde at room temperature for 15 min, washed four times in PBS, and then permeabilized with 0.05% saponin for 30 min at room temperature in blocking buffer containing 1% BSA in PBS. Cells were incubated in primary antibody overnight in blocking buffer, washed four times with a solution of 0.05% saponin in PBS (wash buffer), and labeled with Alexa Fluor 488 and Alexa Fluor 594 secondary antibodies (Invitrogen) at room temperature for 1 h in blocking buffer. Cells were then washed twice and labeled with 1 μm Hoechst nuclear stain for 5 min in wash buffer. Cells were washed two more times with wash buffer and then once with PBS before being mounted with Mowiol (46).
GAP Assays
HeLa cell lysates were prepared from cells overexpressing ELMOD1-myc/His or ELMOD2-myc/His or their respective mutants and from mock-transfected cells by incubating cells in 25 mm HEPES, pH 7.4, 100 mm NaCl, 1% CHAPS on ice for 15 min followed by centrifugation at 14,000 × g. The assay was performed as described previously (3) using recombinant Arl2 as a substrate, and HeLa lysate or purified recombinant trigger factor fusion proteins as a source of GAP. Briefly, recombinant Arl2 was loaded with [γ-32P]GTP at 30 °C for 15 min in 25 mm HEPES, pH 7.4, 100 mm NaCl, 2.5 mm MgCl2, 1 mm dithiothreitol, ∼2 μm Arl2, and ∼0.3 μCi/μl [γ-32P]GTP (6000 Ci/mmol; PerkinElmer Life Sciences). HeLa lysate (25 μl, 20 μg) or trigger factor fusion protein (25 μl, 20 μg of TF-ELMOD1 or 2 μg of TF-ELMOD2) was then mixed with 20 μl of reaction buffer (62.5 mm HEPES, pH 7.4, 6.25 mm MgCl2, 2.5 mm dithiothreitol, 4.2 mm ATP, and 2.5 mm GTP) on ice. The reaction was started by adding 5 μl of pre-loaded Arl2-[γ-32P]GTP to the HeLa lysate in the reaction mixture, briefly vortexing, and placing in a 30 °C water bath. The reaction was carried out for 4 min, at which time it was stopped by the addition of 750 μl of a suspension of ice-cold activated charcoal (5% in 50 mm NaH2PO4). The charcoal was pelleted, and the amount of 32Pi in 400 μl of supernatant was determined by scintillation counting. In a parallel set of triplicate tubes, an equal amount of [γ-32P]GTP to that used in the assay was incubated in the absence of Arl2 or cell lysates and treated with charcoal as described. The resulting free 32Pi was quantified and subtracted from each value in the GAP assay as it reflects the level of GTP hydrolyzed prior to use in the assay.
GST-GGA3 Pulldown Assay for Activated Arf
ELMOD1-HA or ELMOD1(R174K)-HA was expressed in HeLa cells infected with lentivirus by induction with doxycycline. Levels of activated Arf in cell lysates were determined using a GST fusion of the Arf binding domain of the adaptor protein GGA3 as described previously (47). The levels of activated Arf in the pulldowns were determined by Western blotting with a polyclonal antibody against Arf1 (43). The level of ELMOD1 expression was determined by loading 4% of cleared starting lysate and was also immunoblotted using our polyclonal ELMOD1 antibody. Cleared lysate was also immunoblotted for Arf1 and with a monoclonal antibody against β-tubulin as a loading control.
NRK Cell Fractionation and Lipid Droplet Purification
For cell fractionation, NRK cells were lysed via glass/glass Dounce homogenizer (40 strokes) in 10 mm Tris/MOPS, pH 7.4, 1 mm EGTA/Tris, 200 mm sucrose, protease inhibitor mixture (Sigma). Cell homogenates were spun at 600 × g for 10 min at 4 °C, and supernatants were collected as post-nuclear supernatants. Post-nuclear supernatant was spun at 11,000 × g for 10 min at 4 °C, and the supernatant and pellet were collected as S11 and P11, respectively. The S11 was then spun at 100,000 × g for 1 h at 4 °C, and the supernatant and pellet were collected as S100 and P100. The protocol for lipid droplet purification was adapted from Ref. 48; HeLa cells were lysed via Potter-Elvehjem homogenizer with 8 strokes by hand in buffer (20 mm Tris-HCl, pH 7.4, 1 mm EDTA, and protease inhibitor mixture (Sigma)). Homogenate was spun at 1,000 × g for 10 min at 4 °C. The post-nuclear supernatant was collected, and a solution of 60% sucrose in the same buffer was added to bring the final concentration of sucrose to 20%, which was then overlaid with 5 ml of 5% sucrose and 6 ml of 0% sucrose all in the same buffer. The gradient was spun at 28,000 × g for 30 min at 4 °C with no braking, and 1-ml fractions were collected. Fractions with lipid droplets were determined by immunoblot of fractions using an antibody against the lipid droplet marker ATGL.
ELMOD1 Knockdown
HeLa or NRK cells were transfected with each of six pSUPER-based (49) constructs targeting the coding sequence of human ELMOD1 or ON-TARGET plus SMART pools targeting human and rat ELMOD1 (Dharmacon L-013812-01 and L-0-101035-02) according to the manufacturer's directions. Cell lysates were prepared after 24, 48, or 72 h by incubating cells in 25 mm HEPES, pH 7.4, 100 mm NaCl, 1% CHAPS on ice for 15 min followed by centrifugation at 14,000 × g. The level of endogenous ELMOD1 expression was determined by immunoblot using our polyclonal ELMOD1 antibody.
RESULTS
Phylogenetic Analysis of the ELMO Family of Proteins
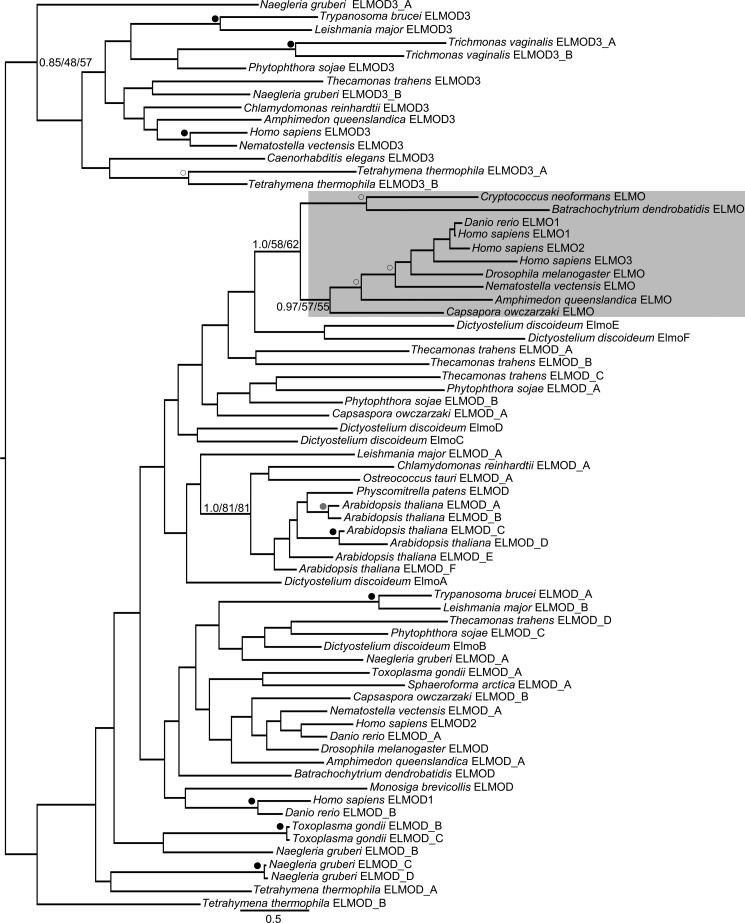
To examine the distribution of ELMO family members among eukaryotes, we sampled genomic databases that should represent the full span of eukaryotic diversity (36, 37). We identified ELMO domain-containing family members in species from every eukaryotic supergroup examined, suggesting that the family dates back to the LECA. We failed to identify homologs in Acanthamoeba castellanii and Takifugu rubripes, which may be a matter of incompleteness of the databases. However, we also failed to identify ELMOD homologs in the more complete databases of the parasitic Plasmodium vivax and Entamoeba histolytica. Although several fungi examined encoded one or more ELMO domain-containing family members, we did not identify any in the well studied yeasts S. cerevisiae and Schizosaccharomyces pombe. Within the human ELMO family there is a clear distinction between ELMO proteins (ELMOs) and ELMO domain proteins (ELMODs) in both their domain architecture (see Fig. 1) and function in cells (see above). To determine the distribution of each of these two functionally distinct subgroups, we performed a thorough phylogenetic analysis of the family of ELMO domain-containing proteins. A total of 71 genes from 24 taxa were used in a maximum likelihood analysis using PhyML and RAxML and a Bayesian analysis using MrBayes, as described under “Experimental Procedures.” These three methods produced similar phylogenetic trees represented by the best Bayesian topology in Fig. 2. A distinct, moderately supported clade (support values of 0.97/57/55) clustered various holozoan sequences with all three of the human ELMOs, suggesting that ELMOs represent a subfamily within the ELMODs. The tree also suggests that the ELMO subfamily includes fungi with support values of 1.0/58/62.
FIGURE 2.
ELMOs cluster into a distinct phylogenetic subfamily. Isolated ELMO domains from ELMO family members spanning the diversity of eukaryotic evolution were collected for phylogenetic analysis using MrBayes, PhyML, and RaxML, as described under “Experimental Procedures.” The tree shows the best Bayesian topology and is rooted to highlight the separation of the ELMOD3 and ELMO clusters. Support values for the relevant nodes are provided, and other nodes with strong (≥0.95/90/90), robust (≥0.9/80/80), or moderate (≥0.8/50/50) support are indicated with black, gray, and white dots, respectively. Gene names were assigned with either ELMO or ELMOD based on clustering with the human subfamilies except for genes with established nomenclature from human and D. discoideum. The ELMO subfamily is shaded. Scale bar indicates number of substitutions per site.
The clustering of ELMOs as a subfamily was further supported by the domain architecture of the proteins within this group. In addition to the ELMO domain, each protein contained the DUF3361 and PH domains found in each of the three human ELMOs, ELMO1–3. DUF3361 was not found outside of the ELMOD family, but we cannot determine whether the same is true of the ELMO-associated PH domain, due to the large number of PH domains and limited sequence conservation among them. The presence of the PH domain is strongly correlated with that of DUF3361 in the ELMO subfamily as 20 of 23 proteins clustering as ELMO homologs by phylogenetic analysis had both domains. This correlation and the divergence of the ELMO domain are of considerable interest and suggest a possible functional relationship between these domains. DUF3361 is found only in proteins within the ELMO subfamily with two notable exceptions. One of the five ELMOD family members (ElmoD_A) in the apusomonad Thecamonas trahens, a putative sister group to the Opisthokonta supergroup (50, 51), and one of six ELMOD family members (ElmoA) in the amoebozoan D. discoideum contains DUF3361 but no PH domain. These proteins did not cluster with the ELMO subfamily, but no strongly supported nodes separated them. Interestingly, however, the next closest, albeit unsupported, branches to the ELMO subfamily are also from T. trahens and D. discoideum. Together, these raise the possibility that ELMOs may also be found in amoebas and that their origin may lie within the Unikonts, i.e. the group of opisthokonts and amoebozoans. Regardless, due to the species limitation of the ELMO subfamily and because the majority of the remaining proteins (which span the breadth of sampled eukaryotic diversity) contain only the ELMO domain, we speculate that the remaining proteins represent the more ancient form of the ELMO domain-containing family and will refer to these proteins (those that did not resolve with the fungal or animal ELMOs) as ELMODs.
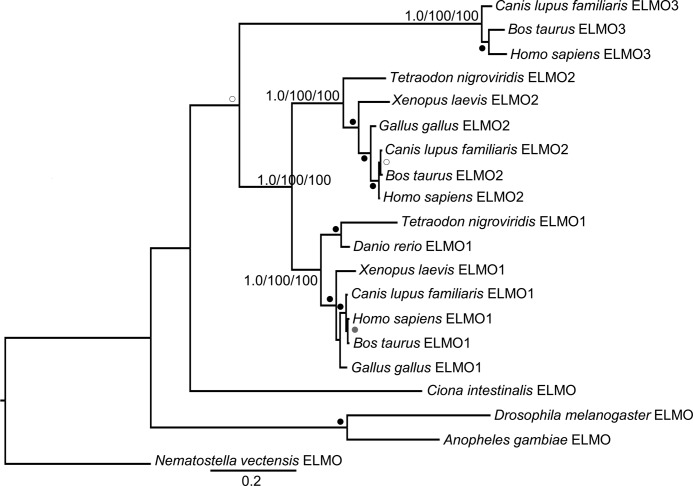
We were interested to see the resolution of the ELMOs, for which we have the most functional information, into a distinct phylogenetic group, so we further pursued this subfamily with a more specific analysis of the ELMO subfamily alone. Fungi and more basal metazoan lineages (e.g. insects and sponges) contained only a single ELMO subfamily protein, although in vertebrates, including mammals and fish, an expansion of the subfamily was observed. Thus, to further study this gene duplication, we sampled additional metazoan genomes and performed a phylogenetic analysis limited to metazoan ELMO proteins. This revealed three distinct and well supported clades classified as ELMO1, ELMO2, and ELMO3 based on three human ELMO sequences as landmarks (Fig. 3). The ELMO1 and ELMO2 clades are clearly defined and span the diversity of the sampled vertebrates. The clustering of ELMO3 more closely to the single ELMOs from basal metazoa, if taken at face value, would suggest that ELMO3, which is only found in mammals, is the more ancient form of the ELMO subfamily and that ELMO3 would have been lost multiple times in higher eukaryotes, including fish, amphibian, and bird lineages. However, it is also possible, and perhaps more likely, that the ELMO3 cluster is simply more divergent, and clustering with basal ELMO is the result of long branch attraction.
FIGURE 3.
ELMO subfamily is further divided into three distinct phylogenetic clades. Full-length sequences of ELMO subfamily members from metazoa were collected for phylogenetic analysis using MrBayes, PhyML, and RaxML. The tree shows the best Bayesian topology and is rooted on the basal organism of the analysis. Support values for the relevant nodes are provided, and other nodes with strong (≥0.95/90/90), robust (≥0.9/80/80), or moderate (≥0.8/50/50) support are indicated with black, gray, and white dots, respectively. Gene names were assigned ELMO1, ELMO2, or ELMO3 based on clustering with the three human ELMOs. Scale bar indicates number of substitutions per site.
In the large scale analysis of ELMOD-containing proteins (Fig. 2), we did not observe significant resolution within the backbone of the ELMODs in the phylogenetic analysis. Interestingly, although not well supported (0.85/48/57), human ELMOD3 resolved differently than human ELMOD1 and ELMOD2 in a clade that contains the diversity of eukaryotes sampled. Thus, the ELMOD3 clade may represent a deep evolutionary split and raises the possibility of two ancestral ELMOD proteins present in the LECA. We also recovered a robustly supported clade of plant ELMODs (1.0/81/81) and consistently observed an expansion of the ELMOD family in higher plants with six members in Arabidopsis. Despite the fact that ELMO proteins (ELMOs) were discovered first and are currently better understood functionally, our evidence that the ELMODs arose much earlier in eukaryotic evolution, are present in every eukaryotic supergroup sampled, are more divergent in sequence, and that the ELMOs are proposed to have diverged from them leads us to conclude that the most appropriate name of this family is ELMOD and not ELMO. This nomenclature also helps avoid confusion if we can refer to the ELMO clade within the ELMOD family and not within the ELMO family. We will adhere to this nomenclature below in efforts to minimize confusion.
Identification of the GAP Domain of the ELMODs
Our previous work indicated that human ELMOD2 exhibited GAP activity against Arl2, Arl3, and Arf1, and ELMOD1 was active against Arl2 using in vitro GAP assays, although we failed to detect Arl2 GAP activity for any of the three human ELMOs or ELMOD3 (3). Because the ELMO proteins per se appear to be lineage-specific and derived versions of the larger ELMOD family, and because all other members of that family are composed of the ELMO domain alone, we speculated that it was this domain that contained the GAP activity. We next aligned sequences of ELMO domains from all ELMODs identified by the unbiased sorting of the phylogenetic analysis in an effort to identify key residues within the GAP domain. The region with the highest level of conservation consisted of a stretch of 26 residues with 13 very highly conserved sites. This putative GAP domain of the ELMODs has the consensus sequence of WX3G(F/W)QX3PXTD(F/L)RGXGX3LX2L (Fig. 4). This consensus bears no obvious homology to the previously described Arf GAP or other Ras superfamily GAP domains. Of particular interest was the presence of a very highly conserved arginine residue toward the middle of the consensus sequence. A mechanism frequently used by GAPs of the Ras superfamily uses an arginine residue that is essential for efficient GAP activity known as the catalytic arginine. Thus, based upon sequence alignments of members of the ELMOD cluster, we have identified a putative GAP domain as well as an arginine residue that is predicted to be critical to GAP activity. When alignments included both ELMOD and ELMO proteins (Fig. 5), we note a much lower level of sequence conservation in this region of ELMOs, including the absence of the highly conserved arginine, identified in the ELMODs.
FIGURE 4.
Sequence alignment of the ELMODs reveals a highly conserved motif that includes a central arginine residue. Sequences representing the diversity of eukaryotic ELMO domains from the ELMODs were aligned using MUSCLE. The region with the highest level of conservation consisted of a stretch of 26 residues (gray shading) with 13 very highly conserved sites (bold type). The same consensus sequence was obtained when all ELMOD sequences (shown in Fig. 2) were used in the alignment. An asterisk indicates a conserved residue with two possible identities (e.g. F/W or F/L). Gene names were assigned with prefix abbreviations to indicate genus and species (see supplemental Table S1).
FIGURE 5.
Putative catalytic arginine and some of the other residues within the putative GAP domain of the ELMODs (top) are not conserved in members of the ELMO subfamily (bottom). The ELMO domains from representative members of opisthokont ELMODs and the ELMO subfamily were aligned using MUSCLE. The putative GAP motif of ELMOD is indicated by bold type in the sequences from both subfamilies to highlight the differences between ELMOD and ELMO in this region. Most notably, the putative catalytic arginine residue is not conserved in ELMO subfamily members. Consensus sequences of all ELMOD subfamily members analyzed in Fig. 2 are shown in the middle, and the effect on this consensus of including the ELMO subfamily members shown is included in the consensus at the bottom. Gene names were assigned with prefix abbreviations to indicate genus and species (see supplemental Table S1).
Arginine Mutations Affect the Biochemical GAP Activity of ELMODs in Vitro
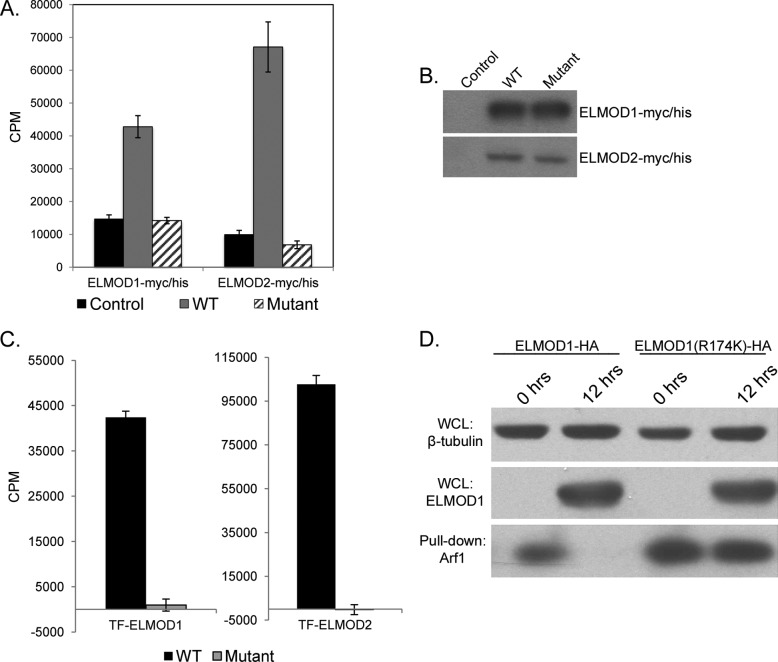
Recombinant human ELMOD2 was shown to have GAP activity against the Arf family members Arl2, Arl3, and Arf1 using an in vitro GAP assay, whereas ELMOD1 was only tested against Arl2 in the earlier study (3). We used a variation of this assay to test the importance of the putative catalytic arginine residue to the GAP activity of the ELMODs. Lysates from HeLa cells expressing ELMOD1 or ELMOD2 or their corresponding arginine mutants, tagged at the C terminus with c-Myc and His6 epitopes, were used as a source of GAP in the assays. HeLa lysates from cells expressing ELMOD1-myc/His or ELMOD2-myc/His had Arl2 GAP activity that was severalfold (∼3–5) higher than from mock-transfected HeLa cells (Fig. 6A). Note that the Arl2 GAP activity present in controls is not solely the result of endogenous Arl2 GAP activity. The substrate in the assay, [γ-32P]GTP, is susceptible to hydrolysis, e.g. by nucleotidases found in all cells. High concentrations of nonradioactive GTP and ATP are included in the GAP assay in efforts to minimize the contributions of this non-GAP-mediated hydrolysis to the background, control levels, but we know that suppression is incomplete (3). Thus, the fold stimulation of Arl2 GAP activity resulting from expression of ELMOD1-myc/His or ELMOD2-myc/His seen in HeLa cell lysates is likely considerably greater than the ∼3–5-fold increases shown in Fig. 6A, but for technical reasons a more accurate determination of fold increases in activity of total cell lysates is not possible in this assay.
FIGURE 6.
Mutation of the putative catalytic arginine residue reduces GAP activity of ELMOD1-myc/His and ELMOD2-myc/His in in vitro and cell-based assays. A, arginine mutants of ELMOD1-myc/His or ELMOD2-myc/His have lost activity in in vitro Arl2 GAP assays. HeLa cells were transiently transfected with no DNA (control) or plasmid encoding ELMOD1-myc/His, ELMOD2-myc/His, or their corresponding arginine mutants (ELMOD1(R174K)-myc/His or ELMOD2(R167K)-myc/His, respectively), and 16 h later cells were collected for assay. Whole cell lysates (20 μg of protein) were assayed for Arl2 GAP activity, as described under “Experimental Procedures.” Data shown are the averages from three independent assays, each performed in triplicate, and bars represent one standard deviation. B, wild type and arginine mutants are each expressed to the same levels. Immunoblots of HeLa lysates from control cells and those expressing ELMOD1-myc/His, ELMOD2-myc/His, or their corresponding arginine mutants show equal expression of wild type and mutant protein. Lysates from ELMOD1-myc/His cells (10 μg of total cell protein) and ELMOD2-myc/His cells (20 μg of total cell protein) were analyzed by immunoblot with ELMOD1 and ELMOD2 antibodies, respectively. Note that although each wild type/mutant pair expressed to the same levels, ELMOD1 expressed to roughly 10 times the levels seen for ELMOD2. For this reason, the immunoblot shown in the bottom panel was exposed for longer times than that shown on the top panel. C, arginine mutants of purified recombinant TF-ELMOD1 or TF-ELMOD2 have lost activity in in vitro Arl2 GAP assays. Purified proteins were assayed for Arl2 GAP activity, as described under “Experimental Procedures.” Data shown are from a single experiment representative of at least three repetitions performed in duplicate, and bars represent one standard deviation. D, ELMOD1-HA but not ELMOD1(R174K)-HA lowers levels of activated Arf1 pulled down from HeLa cell lysates. HeLa cells were previously transduced with lentiviruses carrying ELMOD1 proteins whose expression was induced by doxycycline (2 μg/ml for 0 or 12 h). Lysates were prepared, and the GST-GGA3 pulldown for activated Arf (Arf-GTP) was performed, as described under “Experimental Procedures.” HeLa whole cell lysate (WCL; 20 μg) was analyzed by immunoblot using antibodies specific for β-tubulin to show equal protein loading (top panel) and ELMOD1 (middle panel) to show equal expression of wild type and mutant proteins. The levels of Arf1 from pulldowns were determined by immunoblot with our Arf1-specific antibody (bottom panel).
To determine whether the most highly conserved, homologous arginines in ELMOD1 and ELMOD2 were important to Arl2 GAP activity, we generated plasmids directing expression of the conservative arginine to lysine mutants for ELMOD1(R174K) and ELMOD2(R167K) in pCDNA3.1-myc/His and for ELMOD1(R174K) in lentiviral vectors. Expression of ELMOD1(R174K)-myc/His or ELMOD2(R167K)-myc/His by transient transfection of HeLa cells led to similar levels of expression as the wild type proteins (Fig. 6B). We consistently have found that in the same vector(s) ELMOD1 is expressed to higher levels than is ELMOD2. When the cell lysates were assayed in the Arl2 GAP assay, we found that each of the arginine to lysine mutations was sufficient to completely ablate the increased GAP activity associated with ELMOD expression (Fig. 6A).
HeLa cells were also transduced with high titer stocks of lentiviruses that direct expression of ELMOD1 with a C-terminal HA epitope tag (ELMOD1-HA) or its arginine mutant, ELMOD1 (R174K)-HA, under control of the Tet-On promoter. In this case, controls were cells grown in the absence of doxycycline, and protein expression was achieved by exposure to 2 μg/ml doxycycline for 16 h. The levels of induced protein expression achieved were lower than those seen after transient transfections but were the same between ELMOD1-HA and ELMOD1 (R174K)-HA (Fig. 6). We found lower levels of Arl2 GAP activity in HeLa lysates with viral expression than transients, but again the mutation of arginine 174 to lysine was sufficient to eliminate the increased Arl2 GAP activity (data not shown). Thus, mutation of the putative catalytic arginine is sufficient to ablate Arl2 GAP activity of ELMOD1-myc/His, ELMOD2-myc/His, or ELMOD1-HA in in vitro assays.
To confirm that the assayed Arl2 GAP activities were the direct result of ELMOD protein and that mutation of the conserved arginine residues directly affected this activity, we generated purified recombinant protein stocks of ELMOD1 and ELMOD2 and their corresponding arginine mutants. Initial attempts to generate soluble recombinant stocks were unsuccessful as a number of constructs and fusion proteins were almost entirely insoluble. Fusion at the N terminus with the prokaryotic chaperone trigger factor (TF) increased solubility allowing for the expression and purification of milligram quantities of recombinant proteins. TF-ELMOD1 and TF-ELMOD2 exhibited detectable Arl2 GAP activity (Fig. 6C), but their specific activities were estimated to be less than 0.1% of ELMOD2 purified from bovine testes (3). Despite the low specific activity, the high solubility and ease of purification from bacteria allowed us to use relatively high concentrations of the TF fusion proteins in Arl2 GAP assays and without the concerns of other HeLa proteins contributing, directly or indirectly, to activities. No Arl2 GAP activity was detected for TF-ELMOD1 (R174K) or TF-ELMOD2 (R167K) (Fig. 6C). Thus, loss of Arl2 GAP activity is a direct consequence of mutation of the conserved arginine and not the result of some indirect effect.
The effects of mutation of the putative catalytic arginine residue were also examined using a cell-based assay for Arf GAP activity developed in the laboratory of James Casanova (University of Virginia). Briefly, a GST fusion of the Arf binding domain of the Arf-dependent adaptor GGA3 was used to selectively precipitate endogenous activated Arfs (Arf-GTP) from cell lysates to estimate the level of activated Arf under different conditions. We used the doxycycline-inducible lentiviral expression system for ELMOD1-HA or ELMOD1(R174K)-HA in HeLa cells to compare activated Arf levels of control cells to those in cells expressing ELMOD1-HA. Expression of ELMOD1-HA substantially reduced the levels of activated Arf1 that could be brought down with GST-GGA3 (Fig. 6D, bottom panel), consistent with the recombinant protein acting in cells as an Arf1 GAP. We used our Arf-specific antisera to test for specificity among the soluble Arfs and found that the expression of ELMOD1-HA led to clear decreases in the levels of activated Arf1 or Arf3, the most abundantly expressed Arfs in most cell lines, but technical concerns and lower levels of endogenous protein expression limited definitive answers as to effects on Arf4 or Arf5. We believe that the expressed ELMOD1 is acting directly on Arfs in cells but cannot rule out less direct mechanisms of action in reducing cellular Arf activities. Nevertheless, this is the first evidence to suggest that an ELMO domain-containing protein demonstrates GAP activity against Arfs in cells.
We next used the same assay and inducible lentiviral expression system to assess GAP activities from cell lysates expressing ELMOD1(R174K)-HA. Expression of ELMOD1(R174K)-HA to the same level as ELMOD1-HA had little to no effect on the amount of activated Arf1 brought down by GST-GGA3, in comparison with uninduced control lysates (Fig. 6D). Thus, the GAP activity of ELMOD1(R174K) mutants against purified recombinant Arl2 and endogenous Arf1 was considerably lower than that of tagged ELMOD1 against the same proteins in the same assays, suggesting that the putative catalytic arginine residue is critical for these activities.
Expression of ELMOD2-HA in lentivirus-infected HeLa cells with doxycycline induction had no effect on cellular levels of activated Arf in this assay. ELMOD2-HA was expressed to substantially lower levels than ELMOD1-HA. Thus, it is unclear whether expression levels of ELMOD2-HA were insufficient to detect a change in the levels of activated Arf, ELMOD2-HA did not localize to regions of the cell containing high levels of activated Arfs, or whether ELMOD2-HA did not exhibit Arf GAP activity when expressed in cells under these conditions.
ELMOD1-HA and ELMOD1(R174K)-HA Display Different Phenotypes When Expressed in HeLa Cells
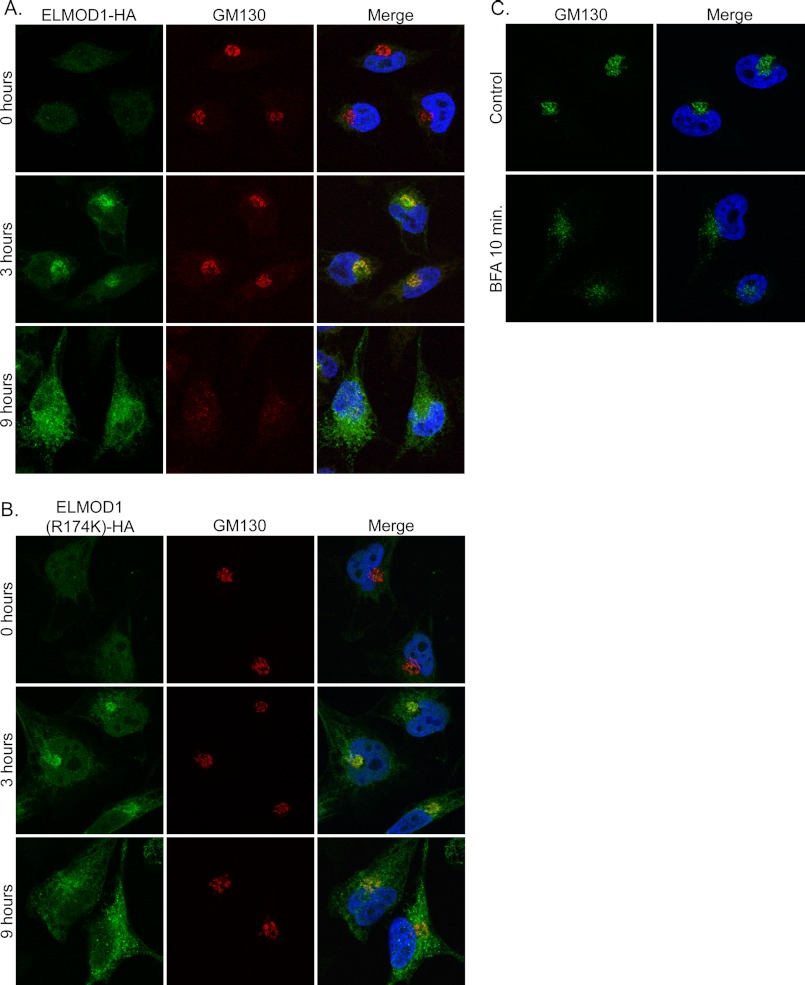
We next used our inducible lentiviral expression system in efforts to determine the cellular location of ELMOD1 (about which nothing is currently known) as well as effects of protein overexpression on cell functions. Human ELMOD1-HA expression was induced, and HeLa cells were fixed at the earliest times after detection of protein expression in an effort to minimize artifacts resulting from overexpression. We observed that at early times of expression (e.g. 3 h) there was a strong increase in the perinuclear staining with either our ELMOD1 rabbit polyclonal antibody or a commercial HA monoclonal antibody (Covance). Indeed, images obtained from these antibodies were superimposable, indicating that the induced protein was being identified by each. The perinuclear compartment to which ELMOD1-HA localized at early times of expression was identified as Golgi by double labeling with at least 15 markers of different organelles (e.g. LAMP1, EEA1, transferrin receptor, TOM20, calnexin, etc.). The most extensive overlap in staining was clearly between ELMOD1-HA and markers of the Golgi, e.g. GM130 (Fig. 7A, middle row).
FIGURE 7.
Overexpression of ELMOD1-HA alters Golgi morphology, but this phenotype is absent for ELMOD1(R174K)-HA. Expression of ELMOD1-HA (A) or ELMOD1(R174K)-HA (B) was induced in lentivirus-infected HeLa cells with doxycycline (2 μg/ml) for the given times. Note that ELMOD1-HA localizes to the Golgi at early times (3 h; middle row) and disrupts Golgi morphology at later times (9 h; bottom row) of expression, but although localization is retained, the effect on Golgi morphology is lost with ELMOD1(R174K)-HA. C, uninfected cells were treated with 10 μm BFA for 10 min as indicated. Cells were fixed and stained, as described under “Experimental Procedures,” using antibodies directed against ELMOD1 and GM130, a Golgi marker. Representative cells are shown for each condition.
The same procedure was carried out in HeLa cells expressing ELMOD1(R174K)-HA, and we saw the same pattern of Golgi localization at early times (∼3–5 h) of protein expression (Fig. 7B, middle panel). Thus, mutation of arginine 174 did not visibly alter the localization of ELMOD1 to the Golgi at early times in its expression. At later times (>6 h), we noted that localization of both ELMOD1-HA and ELMOD1(R174K)-HA to the Golgi became less evident as the staining throughout the cell increased. At these and later times of expression of the two ELMOD1 proteins, the staining of Golgi markers began to diverge in appearance as the Golgi itself was altered by expression of ELMOD1-HA but not the R174K mutant.
Because Arfs also localize to the Golgi, and we found earlier that ELMOD1 acts on Arfs in cells, we speculated that its presence at that organelle might be responsible for changes in the organelle itself. To determine whether expression of ELMOD1 had an effect on membranes of the secretory pathway, we performed double labeling for ELMOD1 and markers of membrane-bound organelles at different times after expression. Although most of these markers displayed no evident differences, the Golgi was strongly perturbed in cells expressing ELMOD1-HA. GM130 staining was more diffuse and peripheral (Fig. 7A, bottom panel). This was also true of other Golgi markers examined, including β-COP, Giantin, and TGN46 and consistent with the conclusion that ELMOD1-HA expression caused the breakdown of cis-, medial-, and trans-Golgi compartments. This effect was somewhat reminiscent of the effects of BFA (Fig. 7C), a specific inhibitor of some Arf GEFs, on the Golgi that result from the lowering of cellular Arf-GTP levels. There were subtle differences in the phenotypes of BFA treatment and expression of ELMOD1-HA. The early Golgi marker GM130 adopts a more punctate phenotype with BFA treatment, whereas GM130 staining appears more diffuse with some tubular elements with expression of ELMOD1-HA (compare Fig. 7, B, bottom row, to C). The effects of ELMOD1-HA expression on Golgi morphology is also similar to that reported from overexpression of the Arf GAP, ArfGAP1 (52, 53), which is consistent with our evidence (see above) that ELMOD1-HA lowers activated Arf levels in cells.
Finally, we investigated the ability of ELMOD1(R174K)-HA to alter Golgi morphology. At comparable times and levels of expression to ELMOD1-HA, we found that the arginine mutant had no discernible effect on the Golgi or the staining of GM130, TGN46, or β-COP (Fig. 7B, bottom panel). No effect on Golgi morphology was observed even at times of ELMOD1(R174K)-HA expression up to 48 h. Thus, arginine 174 of ELMOD1 is critical to GAP activity in vitro, Arf GAP activity in cells, and the changes in Golgi integrity and recruitment to the Golgi of the Arf-dependent adaptor COPI when overexpressed in cultured cells. These data are consistent with Arg-174 of ELMOD1 acting as a catalytic arginine in GAP reactions and with Arfs being potential targets of ELMOD1 in cells and/or ELMOD1 being an effector of Arfs in cells.
We emphasize that the staining of ELMOD1-HA and ELMOD1(R174K)-HA at the Golgi was transient and that the R174K mutant is deficient in effects on Golgi morphology and associated proteins. However, at later times of expression, both proteins were seen accumulating in quite distinct structures that could not be identified initially but that were striking in appearance (see below). These results led us to define to the extent technically possible with existing reagents the location of endogenous ELMOD1 and exogenously expressed ELMOD1-HA.
Cellular Localization of Endogenous and Epitope-tagged ELMOD1
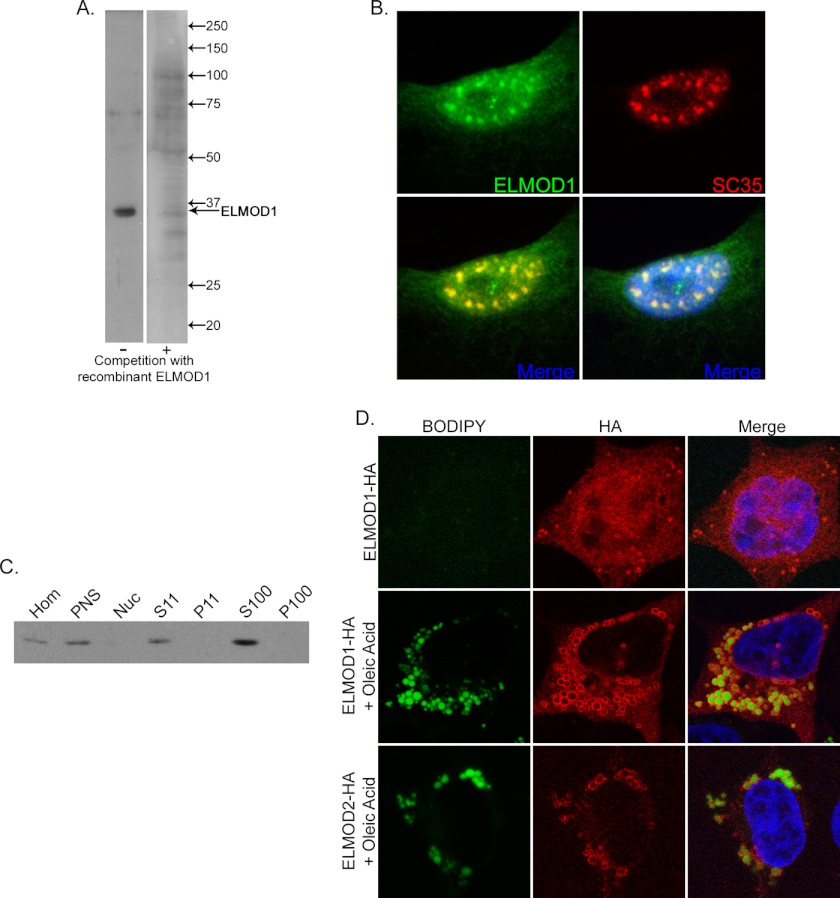
Using rabbit polyclonal antibodies raised against recombinant human ELMOD1, we investigated the cellular localizations of endogenous and epitope-tagged exogenous ELMOD1 in cultured cells. The only staining we describe in this report is that which is seen with immune but not preimmune serum (from the same rabbit) and is eliminated or strongly reduced upon prior incubation of the primary antibody with excess antigen in immunoblots and with immunofluorescence (Fig. 8A). In several cell lines (including HeLa, COS-7, and, most prominently, NRK cells), there was strong nuclear staining for endogenous ELMOD1 with distinct puncta that co-stained with a marker of nuclear speckles, SC35 (Fig. 8B). No other specific staining was detected for endogenous ELMOD1, but cellular fractionation experiments from NRK cells revealed that ELMOD1 is predominantly found in the 100,000 × g supernatant, consistent with it being a soluble protein (Fig. 8C). By comparison of purified recombinant ELMOD1 to total cell lysates or cell fractions, we have noted that ELMOD1 is expressed to very low levels in each of the cell lines tested, limiting our ability to identify all cellular localizations of endogenous ELMOD1 by indirect immunofluorescence or cell fractionation. We estimate that our ELMOD1 antibody can detect as little as a few nanograms of ELMOD1 in a lane of an immunoblot and reacts with cell lines derived from human, mouse, rat, and monkey, but detection in whole cell homogenates is weak at best. Thus, in these cell lines, ELMOD1 is typically expressed to less than 0.01% of total cell protein.
FIGURE 8.
Cellular localization of endogenous ELMOD1 and exogenous ELMOD1-HA or ELMOD2-HA. A, rabbit polyclonal antibody directed against ELMOD1 detects a single prominent band in immunoblots that is effectively competed with antigen. Total cell lysate from NRK cells (20 μg) was analyzed by immunoblot using our ELMOD1 antibody with (right) and without (left) competition with antigen. B, NRK cells were fixed and permeabilized as described under “Experimental Procedures” before developing with antibodies to ELMOD1 (top left panel) or SC35 (top right panel), a marker for nuclear speckles. C, endogenous ELMOD1 is predominantly found in the supernatant (S100) after centrifugation at 100,000 × g. NRK cells were lysed with a Dounce homogenizer, as described under “Experimental Procedures,” in the absence of detergent before being subjected differential centrifugation. Equal protein (20 μg) from each fraction was analyzed by immunoblot using ELMOD1 antibody. Hom, homogenate; PNS, post-nuclear supernatant; Nuc, nucleus. D, ELMOD1-HA localizes to ring-like structures seen throughout the cytosol and predominantly after 9 h of induction (2 μg/ml doxycycline) or longer (top row; 16 h of induction). Small droplets can be seen with staining for HA in the top panel, but co-staining with BODIPY is not evident due to its rapid photobleaching. ELMOD1-HA-positive ring structures increase in size and number with treatment of cells with oleic acid (300 μm, 6 h), which induces biosynthesis of lipid droplets (middle panel). Note that the increased staining of BODIPY is easily captured after oleate treatment (middle row, left panel). ELMOD2-HA also localizes to oleic acid induced lipid droplets (bottom row). Cells were fixed in paraformaldehyde and stained with antibody specific for the HA epitope and with BODIPY 493/503 to stain lipid droplets, as described under “Experimental Procedures.”
When expressed using our lentiviral expression system in HeLa cells, ELMOD1-HA localized predominantly to the Golgi at early time points (Fig. 7A, middle row). At later time points, ELMOD1-HA adopted a more diffuse reticular staining pattern reminiscent of ER localization (Fig. 7A, bottom row), but co-staining with several markers of the ER (including calnexin and BiP) did not display extensive overlap (data not shown). At similar times of protein expression, ELMOD1-HA also localized to ring-like structures throughout the cytosol. After failing to find markers of the secretory or endocytic pathways that also stained these structures, we suspected that they might be lipid droplets, which are found in every cell line but differ in size and abundance with conditions. Lipid droplets increase in size and number upon treatment of cells with oleic acid (300 μm), which promotes their biosynthesis (see Fig. 8D) (48). We found that oleic acid treatment of HeLa cells increased the size and number of ELMOD1-HA-positive ring structures and that these were also stained by the lipid droplet dye, BODIPY 493/503 (e.g. see Fig. 8D, middle row). BODIPY 493/503 staining of cells that were not treated is more variable due to the rapid photobleaching of the dye but is strongly increased by oleic acid. Note that although ELMOD1-HA staining was limited to the surface of the droplets, BODIPY 493/503 stains neutral lipids and thus more of the interior of the lipid droplet. It is clear from Fig. 8D that the ring structures identified by the presence of ELMOD1 are positive for BODIPY 493/503. Finally, to confirm that ELMOD1-HA was specifically localizing to lipid droplets, we purified lipid droplets from HeLa cells, using a previously established procedure (48). ELMOD1-HA was found to be enriched in this preparation as detected by immunoblotting of purified lipid droplets (data not shown). Despite the strong evidence supporting association of ELMOD1-HA with lipid droplets, we were unable to detect endogenous ELMOD1 in purified lipid droplet preparations from HeLa cells or by immunofluorescence with our ELMOD1 antibodies (data not shown), possibly due to low levels of endogenous protein expression. No changes in the number or size of lipid droplets were evident upon expression of ELMOD1-HA or ELMOD2-HA either with or without oleate treatment of HeLa cells.
Because several proteomics studies of purified lipid droplets detected ELMOD2 as a constituent of these organelles (54, 55), we also tested whether ELMOD2 localized to lipid droplets. Expression of ELMOD2-HA using our inducible lentiviral expression system showed strong localization to lipid droplets that was further stimulated by treatment of cells with oleic acid (Fig. 8D, bottom row). Endogenous ELMOD2 was also detected by immunoblot of purified lipid droplets using our ELMOD2 antibody (data not shown), but we did not observe staining of endogenous ELMOD2 on lipid droplets of fixed HeLa cells by immunofluorescence. We were further limited in our ability to test for ELMOD2 on lipid droplets in other cell lines because our ELMOD2 antibody appears to be specific to the human and monkey (e.g. COS-7 cells) proteins.
In attempts to provide mechanistic insight into the subcellular localization of ELMOD1 in cells, we performed ELMOD1 knockdown in NRK cells or HeLa cells. We constructed a total of six different pSUPER-based plasmids and purchased the Dharmacon Smart Pool siRNAs directed against rat or human ELMOD1. These assays were complicated by the decrease in levels of ELMOD1 protein that correlated with the cells approaching confluence, more readily seen in HeLa than NRK cells. However, none of these reagents gave consistent knockdown of ELMOD1 protein.
DISCUSSION
In this study, we identified and performed initial characterization of the GAP domain of ELMODs, a novel group of GAPs with wider specificity for Arf family GTPases than previously described for the family of Arf GAPs (27). A highly conserved arginine residue within the GAP domain was critically important to the GAP activity of ELMOD1 and ELMOD2 in vitro and for the phenotype of epitope-tagged ELMOD1 expression in HeLa cells. Studies in HeLa cells of exogenous epitope-tagged protein provide initial evidence of a biological function for ELMOD1 as an Arf family GAP at the Golgi, more likely involving Arfs than Arl2, based upon the known cellular localizations and functions of these GTPases. Homology searching identified ELMOD-containing proteins in genomes from taxa spanning the diversity of eukaryotes. Phylogenetic analysis of these proteins suggested that the majority of the protein family consists of the ELMODs for which homologs were found in the breadth of sampled eukaryotic diversity. By contrast, the ELMOs, for which we currently have the most functional information, represent a distinct subfamily, most likely restricted to the supergroup Opisthokonta. Thus, the ELMODs represent the more ancient form of the family and were likely present in the LECA, whereas the ELMO subfamily is a much more recent evolutionary development. We also describe localization of endogenous and overexpressed ELMOD1 in a few cultured cell lines. The diversity of cellular locations for ELMOD1 is consistent with the diversity of actions of both Arfs and Arls and supports the hypothesis that it functions in cells as a GAP and potential effector for one or more Arf family members. These findings offer initial insights into an evolutionarily conserved family of proteins that is predicted to provide the basis for further characterization of their biological functions.
We used the unbiased sorting of the Arf family GAP ELMODs and segregation of the ELMOs, which lack the Arf family GAP activities, in our phylogenetic analysis and sequence alignments to identify the putative GAP domain within the ELMODs. A single region of very high conservation was identified with a consensus sequence WX3G(F/W)QX3PXTD(F/L)RGXGX3LX2L. A notable feature of this region was the presence of a very highly conserved arginine residue that was not conserved in the ELMO subfamily. All of the known Arf family GAPs and many, but not all, other GAPs of small regulatory GTPases use a mechanism known as a catalytic arginine residue to confer GAP activity. The arginine side chain stabilizes the transition state through interactions with the γ-phosphate of the bound GTP and can increase rates of hydrolysis by several orders of magnitude (28). Consequently, mutation of this arginine residue is generally sufficient to reduce GAP activity by several orders of magnitude. We used a variety of in vitro and cell-based assays to determine the importance of the conserved arginine residue. The conserved arginine residue was essential for efficient in vitro GAP activity of ELMOD1 and ELMOD2, as even a minimal Arg → Lys mutation was sufficient to lower GAP activities to undetectable levels. These findings support our hypothesis that the conserved region is the GAP domain and offer initial characterization of the ELMOD GAP domain. Highly charged arginine residues are likely to be on the protein surface and not required for protein folding. The findings that the point mutant is expressed to the same levels and localizes as the wild type protein are consistent with the point mutant folding properly, and thus the loss of cellular activities is consistent with the loss of the biochemical GAP activity. Loss of in vitro Arl2 GAP activity of the point mutants of recombinant TF-ELMOD1 and TF-ELMOD2 more directly indicates that the conserved arginine is critical for the biochemical GAP activity and precludes the possibility that our observations in HeLa lysates were the result of indirect effects. Nevertheless, confirmation that ELMODs use the catalytic arginine mechanism of GTP hydrolysis described for other GAPs or that Arg-174 of ELMOD1 or Arg-167 of ELMOD2 are directly involved in catalysis will require more detailed enzymatic and structural work. We speculate that the drastically lower specific activity of TF-ELMOD1 and TF-ELMOD2 compared with epitope-tagged protein expressed in HeLa lysates (<0.1%) is due to the lack of an obligate binding partner(s). ELMOD2 required detergent addition at late stages in its purification from bovine testes, to resolve it from other proteins, and the GAP activity became less stable under these conditions (3). Preliminary data from quantitative co-immunoprecipitation experiments of ELMOD1-HA in HeLa cells using stable isotope labeling reproducibly showed enrichment of the nonopioid σ receptor 1 (SIGMAR1) to near stoichiometric levels.4 Members of the ELMO subfamily from several species also require DOCK180 obligate binding partners for their activity in vitro and in cells (1, 2, 5, 14). Thus, it is likely that our problems with solubility and with reduced GAP activity of recombinant ELMODs are the result of a missing obligate binding partner, although preliminary testing of co-expression with SIGMAR1 failed to remedy this situation.
ELMOD2 was the first mammalian GAP identified for any of the Arl proteins that make up the bulk of the Arf family of regulatory GTPases. Of particular interest was that ELMOD2 (3) and ELMOD1 (this study) possess GAP activity for both Arls and Arfs, offering the first evidence for cross-talk between these two functionally distinct groups of regulatory GTPases. Because every other known Arf GAP uses the canonical arginine finger Arf GAP domain and presumably catalytic mechanism, the GAP domain of the ELMODs represents both a novel Arf GAP domain and a founding member of a novel group of Arl GAPs. Only one other protein, RP2, has been found to share Arl3 GAP activity (24, 56, 57), and ELMODs do not share any extensive sequence homology or similarity and, thus, likely represent functionally and structurally distinct Arf family GAPs. We note that the Arl2-binding partner cofactor D and the RP2 homolog cofactor C have been implicated as GAPs for tubulin at the completion of the regulated folding reaction (58, 59) and that the binding of Arl2 can modulate this step, but this is a quite different process from the regulation of signaling GTPase by its GAPs.
At early times of ELMOD1-HA expression in HeLa cells using our lentivirus-inducible expression system, ELMOD1-HA localized predominantly to the Golgi. Shortly thereafter, the morphology of the cis-, medial-, and trans-Golgi compartments were altered. This phenotype was reminiscent of the overexpression of the well characterized Golgi Arf GAP, ArfGAP1, and of treatment with cells with BFA, an inhibitor of Arf activation. Although Golgi morphology is disrupted in cells treated with BFA and in cells expressing ELMOD1-HA, the two phenotypes are distinct from one another. In BFA-treated cells, GM130 staining adopted a more punctate staining pattern, although GM130 staining was more diffuse with some tubular elements when ELMOD1-HA was expressed. These differences are consistent with the two proposed mechanisms of BFA and ELMOD1-HA as inhibitors of Arf GEFs and as an Arf GAP, respectively. Because of the previously established roles for Arfs in maintenance of Golgi integrity (e.g. as seen by effects of BFA) together with our findings that ELMOD1-HA lowers cellular levels of activated Arf in HeLa cells (using the GST-GGA3 pulldown assay), these data suggest a role for ELMOD1 at the Golgi as an Arf GAP. The model for the function of ELMOD1 in cells as an Arf family GAP is further supported by studies of the catalytic arginine mutant ELMOD1(R174K)-HA in the same assays. Expression of ELMOD1(R174K)-HA had no effect on Golgi morphology and did not affect cellular levels of activated Arf. These data suggest that the disruption of Golgi morphology by ELMOD1 is tightly linked to its Arf GAP activity, and thus, loss of Golgi morphology is the result of an overabundance of GAP activity at the Golgi. However, this model is weakened by the incomplete testing of substrate specificities of ELMODs as Arf family GAPs and the roles of other family members (including but not limited to Arl1, Arl3, Arl5, Arl8, and Arfrp1) in maintenance of Golgi morphology. The model for the function of ELMOD1 as an Arf GAP at the Golgi is further limited by the fact that it is based solely on results from protein overexpression. Attempts to localize endogenous ELMOD1 to the Golgi by indirect immunofluorescence have been unsuccessful, although it is unclear whether this is due to low levels of ELMOD1 expression in cultured mammalian cells and/or to limits of sensitivity of our ELMOD1 antibody. In addition, although we have found ELMOD1 to possess Arf1 GAP activity both in vitro and in cultured cells, we have not yet tested it for GAP activity against several other Arf family members that are known to localize to the Golgi. Characterization of the specificities, functions, and locations of ELMODs as GAPs and potential effectors (60) for Arf family members will clearly require additional testing.
ELMOD protein homologs were identified in nearly all genomes examined, a sampling that spans eukaryotic diversity. This extent suggests that the protein family is ancient and builds on the emerging conclusion of a sophisticated membrane traffic system in the LECA (34, 35). Indeed, ELMOD homologs are more conserved in their distribution than several other ancient but patchily distributed proteins recently described (61, 62) and reinforces the idea that the proteins likely serve an important cellular function. By contrast, the ELMOs were only found in the Opisthokonta supergroup, with the open possibility of homologs in the apusomonads and amoebozoa. Regardless of the precise point of origin, this distribution suggests that ELMOs were a more recent evolutionary development and evolved from the more ancient ELMODs.
Our phylogenetic analyses offered some enlightening information regarding the contrasting functions of the human ELMOs, which function as heterodimeric unconventional Rac1 GEFs, and ELMODs, which are Arf family GAPs. Because ELMOs cluster away from ELMODs in a distinct phylogenetic subfamily, the contrasting functions of the ELMOs and ELMODs may be linked to divergent evolution of the ELMO domain itself, which is the only region of homology between the ELMOs and ELMODs. However, we note that the Rac GEF activity associated with ELMOs is actually found within its binding partner, Dock180, and paralogs, and thus the biochemical function of the ELMO domain in the ELMO subfamily has no known function(s). The presence of DUF3361 is consistent with the divergence of the ELMO domain from its ancestral ELMOD as it is found almost exclusively in ELMO subfamily members. Thus, DUF3361 may also play an important role in defining the functions of the ELMO family members and may be functionally linked to the ELMO domain in these proteins. We speculate that ELMOs lost the biochemical GAP activity of their ancestral ELMODs but may have retained the ability to bind Arf family GTPases. If so, they may be effectors of Arf family signaling. This is currently being tested in our laboratory with the development of protein interaction assays that are independent of GAP activity. In this model, ELMOs would be predicted to have adopted a more predominant role as effectors of GTPases, whereas the ELMODs retained the biochemical GAP activity. In many cases, a GAP for a given GTPase is able to function as both a terminator of GTPase signaling through its GAP activity and as an effector of GTPase signaling to mediate downstream signaling (60). We speculate that the divergent evolution of ELMO from ELMODs was the result of a specialization in the effector function of these two tasks.
Whether ELMODs function as effectors or terminators of Arf family signaling, or both, requires additional definition of their cellular roles. Only a few clues to this end have appeared in the literature. The one ELMOD gene in flies (Drosophila melanogaster) emerged from a screen of genes whose mutations alter cytokinesis (64), and human ELMOD2 is implicated as a gene linked to idiopathic pulmonary fibrosis (65, 66). Two studies in D. discoideum also reported that ElmoA regulates actin dynamics and forms a complex with the Dock180 ortholog DockD and Rac1 (29, 30). It is unclear whether this protein is an ELMO or ELMOD homolog. It fails to cluster with the other ELMOs but is not separated from that clade by any supported nodes. It has the domain architecture of ELMOs, but it possesses the catalytic arginine of ELMOD proteins. Therefore, this protein may provide the first indication of a functional overlap between the two classes of ELMO domain-containing proteins. A recent study in mice also linked hearing and balance deficits to two different naturally occurring mutations in ELMOD1 (67). Further analysis of these mice revealed morphological defects in the actin-based mechanoelectrical signaling organelles, stereocilia, in both inner and outer hair cells. Stereocilia morphology appeared normal in newborn mice but progressively worsened with age. Thus, ELMOD1 is essential for the maintenance of stereocilia in hair cells. Although the function of ELMODs in stereocilia is intriguing and has significant clinical relevance to deafness, ELMODs arose in evolution well before these highly specialized organelles. Thus, we expect to find other fundamental functions of ELMODs in more ubiquitous biological processes. Further experimentation is necessary to determine the function of ELMOD1 in these processes. Thus, the function of the ELMOs in regulating actin dynamics and cell motility may be relevant to ELMOD proteins. ElmoA may also be a useful tool to link the Arf family GAP activity of the ELMODs to the cellular roles of the ELMOs because it has both the conserved GAP domain of the ELMODs and interacts with binding partners essential to the function of ELMOs. We speculate that ELMOs may be Arf family GTPase effectors and similarly that ELMODs may be found capable of regulating actin and cell motility.
Finally, we provide initial data on the localization of endogenously and exogenously expressed ELMOD1 in a few cultured cell lines. Although data on the endogenous protein provide more reliable answers as to the biological contexts, the low levels of endogenous protein expression limited our ability to define locations for ELMOD1. Endogenous ELMOD1 quite prominently stains nuclear speckles, as evidenced by its co-localization with the marker SC35, although its function there is particularly obscure as a result of the lack of specific localization of any Arf family members to such structures and the limited functional information linked to these structures. A previous study (68) has found that Dock180 and members of the mammalian ELMO subfamily are found predominantly in the nucleus, and immunofluorescence images are consistent with localization to nuclear speckles, although this was not specifically examined in the previous study. Arl2 antibodies also stain the nucleus (although not specifically speckles), and there is one report of a role for Arl2 and BART to act inside the nucleus in STAT signaling (69). Cell fractionation experiments suggest that the majority of endogenous ELMOD1 is soluble. Thus, it is likely that there is a free soluble pool of ELMOD1 that is recruited to its sites of action (e.g. Golgi, nuclear speckles, lipid droplets) in a manner that may parallel the recruitment of one of its proposed substrate GTPases, Arf, which cycles on and off membranes depending on the bound nucleotide (70). When epitope-tagged ELMOD1-HA is exogenously expressed in cells, it first localizes to the Golgi and then adopts a more disperse reticular staining pattern throughout the cell that is reminiscent of ER staining, although we were unable to find a marker of the ER with extensive overlap. This ER-like staining pattern was not the result of disruption of the Golgi by ELMOD1-HA overexpression because ELMOD1(R174K)-HA, which does not disrupt the Golgi, adopts a similar staining pattern at later times of overexpression. Thus, the ER may be a site of localization and function for ELMOD1. ELMOD1-HA also localized to lipid droplets by immunofluorescence and was detected in purified lipid droplets by immunoblot. It is not clear whether endogenous ELMOD1 localizes to lipid droplets although it was not detected in our purified HeLa lipid droplet preparation and is also absent from the published proteome of lipid droplet preparations (at least one of which was from HeLa cells). Localization to both ER and lipid droplets would be consistent with the proposal that the ER is the site of new lipid droplet formation (71). The reason for the discrepancies between the localizations of endogenously and exogenously expressed ELMOD1 remains unclear. Exogenously expressed ELMOD1 (WT or epitope-tagged) does not appear to localize specifically to nuclear speckles or to the nucleus, and we have failed to localize endogenous ELMOD1 to any other organelle. Our polyclonal ELMOD1 antibody may not be sensitive enough to recognize other localizations of the endogenous protein in immunofluorescence experiments given the low levels of expression of endogenous ELMOD1. ELMOD2-HA also localized to lipid droplets by immunofluorescence when overexpressed, and endogenous ELMOD2 was detected in purified lipid droplets by immunoblot (Fig. 8C) and in published proteomes (54, 55). Several Arf family GTPases have been shown to localize to and are important for the formation of lipid droplets, including Arf1 (72, 73), Arfrp1 (74), and Arl4A (63). Thus, even though our initial testing for effects of ELMOD1 or ELMOD2 overexpression on lipid droplet number or size was negative, we predict that functions for these GAPs will likely be found and will involve these or other Arf family members.
Together, we conclude that ELMOD1 and ELMOD2, like the members of the Arf family GTPases that serve as substrates for their GAP activities, are ancient cell regulators found at multiple sites in eukaryotic cells, and we predict that they act on a more diverse array of Arf family GTPases than do the Arf GAPs, making their functional dissection even more difficult but no less important.
Supplementary Material
Acknowledgments
We thank James Olzmann (Stanford University) for helpful discussions and sharing data on the proteomes of different lipid droplet preparations and assistance in purifying lipid droplets from cells and Alex Schlacht (University of Alberta) for help in the phylogenetic analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM090158 (to R. A. K.) and Grant P30-NS055077 from NINDS (to the Viral Vector and Microscopy Cores of the Emory Neuroscience Core Facilities). This work was also supported by American Heart Association Grant 09PRE2140029 (to M. P. E.), Natural Sciences and Engineering Research Council of Canada, and an Alberta Innovates Technology Futures Discovery grant and New Faculty award (to J. B. D.).

This article contains supplemental Table 1.
M. East and R. A. Kahn, unpublished observations.
- ELMOD
- cell engulfment and motility domain
- Arf
- ADP-ribosylation factor
- Arl
- Arf-like protein
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- LECA
- last eukaryotic common ancestor
- PH
- pleckstrin homology
- BFA
- brefeldin A
- TF
- trigger factor
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., Ravichandran K. S. (2002) Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4, 574–582 [DOI] [PubMed] [Google Scholar]
- 2. Lu M., Kinchen J. M., Rossman K. L., Grimsley C., deBakker C., Brugnera E., Tosello-Trampont A. C., Haney L. B., Klingele D., Sondek J., Hengartner M. O., Ravichandran K. S. (2004) PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struct. Mol. Biol. 11, 756–762 [DOI] [PubMed] [Google Scholar]
- 3. Bowzard J. B., Cheng D., Peng J., Kahn R. A. (2007) ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 282, 17568–17580 [DOI] [PubMed] [Google Scholar]
- 4. Chung S., Gumienny T. L., Hengartner M. O., Driscoll M. (2000) A common set of engulfment genes mediates removal of both apoptotic and necrotic cell corpses in C. elegans. Nat. Cell Biol. 2, 931–937 [DOI] [PubMed] [Google Scholar]
- 5. Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., Nishiwaki K., Walk S. F., Nemergut M. E., Macara I. G., Francis R., Schedl T., Qin Y., Van Aelst L., Hengartner M. O., Ravichandran K. S. (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 [DOI] [PubMed] [Google Scholar]
- 6. Wu Y. C., Tsai M. C., Cheng L. C., Chou C. J., Weng N. Y. (2001) C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell 1, 491–502 [DOI] [PubMed] [Google Scholar]
- 7. Zhou Z., Caron E., Hartwieg E., Hall A., Horvitz H. R. (2001) The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev. Cell 1, 477–489 [DOI] [PubMed] [Google Scholar]
- 8. Elliott M. R., Ravichandran K. S. (2010) ELMO1 signaling in apoptotic germ cell clearance and spermatogenesis. Ann. N. Y. Acad. Sci. 1209, 30–36 [DOI] [PubMed] [Google Scholar]
- 9. Elliott M. R., Zheng S., Park D., Woodson R. I., Reardon M. A., Juncadella I. J., Kinchen J. M., Zhang J., Lysiak J. J., Ravichandran K. S. (2010) Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature 467, 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shimazaki A., Tanaka Y., Shinosaki T., Ikeda M., Watada H., Hirose T., Kawamori R., Maeda S. (2006) ELMO1 increases expression of extracellular matrix proteins and inhibits cell adhesion to ECMs. Kidney Int. 70, 1769–1776 [DOI] [PubMed] [Google Scholar]
- 11. Yang C., Sorokin A. (2011) Up-regulation of fibronectin expression by COX-2 is mediated by interaction with ELMO1. Cell. Signal. 23, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komander D., Patel M., Laurin M., Fradet N., Pelletier A., Barford D., Côté J. F. (2008) An α-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell 19, 4837–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sévajol M., Reiser J. B., Chouquet A., Pérard J., Ayala I., Gans P., Kleman J. P., Housset D. (2012) The C-terminal polyproline-containing region of ELMO contributes to an increase in the lifetime of the ELMO-DOCK complex. Biochimie 94, 823–828 [DOI] [PubMed] [Google Scholar]
- 14. Grimsley C. M., Kinchen J. M., Tosello-Trampont A. C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O., Ravichandran K. S. (2004) Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 279, 6087–6097 [DOI] [PubMed] [Google Scholar]
- 15. Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. (2006) Nomenclature for the human Arf family of GTP-binding proteins. ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donaldson J. G., Jackson C. L. (2011) ARF family G proteins and their regulators. Roles in membrane transport, development, and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nie Z., Randazzo P. A. (2006) Arf GAPs and membrane traffic. J. Cell Sci. 119, 1203–1211 [DOI] [PubMed] [Google Scholar]
- 18. Zhou C., Cunningham L., Marcus A. I., Li Y., Kahn R. A. (2006) Arl2 and Arl3 regulate different microtubule-dependent processes. Mol. Biol. Cell 17, 2476–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y., Wei Q., Zhang Y., Ling K., Hu J. (2010) The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189, 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Larkins C. E., Aviles G. D., East M. P., Kahn R. A., Caspary T. (2011) Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell 22, 4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burd C. G., Strochlic T. I., Gangi Setty S. R. (2004) Arf-like GTPases. Not so Arf-like after all. Trends Cell Biol. 14, 687–694 [DOI] [PubMed] [Google Scholar]
- 22. Graham D. J. (2011) Visual perception. Lightness in a high dynamic range world. Curr. Biol. 21, R914–R916 [DOI] [PubMed] [Google Scholar]
- 23. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins. Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Boil. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 24. Veltel S., Gasper R., Eisenacher E., Wittinghofer A. (2008) The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat. Struct. Mol. Biol. 15, 373–380 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y. W., Huang C. F., Huang K. B., Lee F. J. (2005) Role for Gcs1p in regulation of Arl1p at trans-Golgi compartments. Mol. Biol. Cell 16, 4024–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cukierman E., Huber I., Rotman M., Cassel D. (1995) The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 270, 1999–2002 [DOI] [PubMed] [Google Scholar]
- 27. Kahn R. A., Bruford E., Inoue H., Logsdon J. M., Jr., Nie Z., Premont R. T., Randazzo P. A., Satake M., Theibert A. B., Zapp M. L., Cassel D. (2008) Consensus nomenclature for the human ArfGAP domain-containing proteins. J. Cell Biol. 182, 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheffzek K., Ahmadian M. R., Wittinghofer A. (1998) GTPase-activating proteins. Helping hands to complement an active site. Trends Biochem. Sci. 23, 257–262 [DOI] [PubMed] [Google Scholar]
- 29. Isik N., Brzostowski J. A., Jin T. (2008) An Elmo-like protein associated with myosin II restricts spurious F-actin events to coordinate phagocytosis and chemotaxis. Dev. Cell 15, 590–602 [DOI] [PubMed] [Google Scholar]
- 30. Para A., Krischke M., Merlot S., Shen Z., Oberholzer M., Lee S., Briggs S., Firtel R. A. (2009) Dictyostelium Dock180-related RacGEFs regulate the actin cytoskeleton during cell motility. Mol. Biol. Cell 20, 699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brzostowski J. A., Fey P., Yan J., Isik N., Jin T. (2009) The Elmo family forms an ancient group of actin-regulating proteins. Commun. Integr. Biol. 2, 337–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adl S. M., Simpson A. G., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., James T. Y., Karpov S., Kugrens P., Krug J., Lane C. E., Lewis L. A., Lodge J., Lynn D. H., Mann D. G., McCourt R. M., Mendoza L., Moestrup O., Mozley-Standridge S. E., Nerad T. A., Shearer C. A., Smirnov A. V., Spiegel F. W., Taylor M. F. (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451 [DOI] [PubMed] [Google Scholar]
- 33. Field M. C., Gabernet-Castello C., Dacks J. B. (2007) Reconstructing the evolution of the endocytic system. Insights from genomics and molecular cell biology. Adv. Exp. Med. Biol. 607, 84–96 [DOI] [PubMed] [Google Scholar]
- 34. Elias M. (2010) Patterns and processes in the evolution of the eukaryotic endomembrane system. Mol. Membr. Biol. 27, 469–489 [DOI] [PubMed] [Google Scholar]
- 35. Dacks J. B., Field M. C. (2007) Evolution of the eukaryotic membrane-trafficking system. Origin, tempo, and mode. J. Cell Sci. 120, 2977–2985 [DOI] [PubMed] [Google Scholar]
- 36. Walker G., Dorrell R. G., Schlacht A., Dacks J. B. (2011) Eukaryotic systematics. A user's guide for cell biologists and parasitologists. Parasitology 138, 1638–1663 [DOI] [PubMed] [Google Scholar]
- 37. Burki F., Okamoto N., Pombert J. F., Keeling P. J. (2012) The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc. Biol. Sci. 279, 2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abascal F., Zardoya R., Posada D. (2005) ProtTest. Selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 [DOI] [PubMed] [Google Scholar]
- 40. Guindon S., Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 41. Stamatakis A. (2006) RAxML-VI-HPC. Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- 42. Ronquist F., Huelsenbeck J. P. (2003) MrBayes 3. Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 43. Cavenagh M. M., Whitney J. A., Carroll K., Zhang C., Boman A. L., Rosenwald A. G., Mellman I., Kahn R. A. (1996) Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 271, 21767–21774 [DOI] [PubMed] [Google Scholar]
- 44. Cuthbert E. J., Davis K. K., Casanova J. E. (2008) Substrate specificities and activities of AZAP family Arf GAPs in vivo. Am. J. Physiol. Cell Physiol. 294, C263–C270 [DOI] [PubMed] [Google Scholar]
- 45. Bowzard J. B., Sharer J. D., Kahn R. A. (2005) Assays used in the analysis of Arl2 and its binding partners. Methods Enzymol. 404, 453–467 [DOI] [PubMed] [Google Scholar]
- 46. Valnes K., Brandtzaeg P. (1985) Retardation of immunofluorescence fading during microscopy. J. Histochem. Cytochem. 33, 755–761 [DOI] [PubMed] [Google Scholar]
- 47. Austin C., Boehm M., Tooze S. A. (2002) Site-specific cross-linking reveals a differential direct interaction of class 1, 2, and 3 ADP-ribosylation factors with adaptor protein complexes 1 and 3. Biochemistry 41, 4669–4677 [DOI] [PubMed] [Google Scholar]
- 48. Brasaemle D. L., Wolins N. E. (2006) Isolation of lipid droplets from cells by density gradient centrifugation. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.15 [DOI] [PubMed] [Google Scholar]
- 49. Brummelkamp T. R., Bernards R., Agami R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 50. Katz L. A., Grant J., Parfrey L. W., Gant A., O'Kelly C. J., Anderson O. R., Molestina R. E., Nerad T. (2011) Subulatomonas tetraspora nov. gen. nov. sp. is a member of a previously unrecognized major clade of eukaryotes. Protist. 162, 762–773 [DOI] [PubMed] [Google Scholar]
- 51. Sebé-Pedrós A., Roger A. J., Lang F. B., King N., Ruiz-Trillo I. (2010) Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl. Acad. Sci. U.S.A. 107, 10142–10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu S., Roth M. G. (2002) Casein kinase I regulates membrane binding by ARF GAP1. Mol. Biol. Cell 13, 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Randazzo P. A., Hirsch D. S. (2004) Arf GAPs. Multifunctional proteins that regulate membrane traffic and actin remodeling. Cell. Signal. 16, 401–413 [DOI] [PubMed] [Google Scholar]
- 54. Hodges B. D., Wu C. C. (2010) Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 51, 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bouchoux J., Beilstein F., Pauquai T., Guerrera I. C., Chateau D., Ly N., Alqub M., Klein C., Chambaz J., Rousset M., Lacorte J. M., Morel E., Demignot S. (2011) The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biol. Cell 103, 499–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kühnel K., Veltel S., Schlichting I., Wittinghofer A. (2006) Crystal structure of the human retinitis pigmentosa 2 protein and its interaction with Arl3. Structure 14, 367–378 [DOI] [PubMed] [Google Scholar]
- 57. Veltel S., Kravchenko A., Ismail S., Wittinghofer A. (2008) Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett. 582, 2501–2507 [DOI] [PubMed] [Google Scholar]
- 58. Tian G., Bhamidipati A., Cowan N. J., Lewis S. A. (1999) Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the α/β-tubulin heterodimer. J. Biol. Chem. 274, 24054–24058 [DOI] [PubMed] [Google Scholar]
- 59. Bhamidipati A., Lewis S. A., Cowan N. J. (2000) ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. East M. P., Kahn R. A. (2011) Models for the functions of Arf GAPs. Semin. Cell Dev. Biol. 22, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hirst J., Barlow L. D., Francisco G. C., Sahlender D. A., Seaman M. N., Dacks J. B., Robinson M. S. (2011) The fifth adaptor protein complex. PLos Biol. 9, e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koumandou V. L., Klute M. J., Herman E. K., Nunez-Miguel R., Dacks J. B., Field M. C. (2011) Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J. Cell Sci. 124, 1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Patel M., Chiang T. C., Tran V., Lee F. J., Côté J. F. (2011) The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J. Biol. Chem. 286, 38969–38979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Echard A., Hickson G. R., Foley E., O'Farrell P. H. (2004) Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14, 1685–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pulkkinen V., Bruce S., Rintahaka J., Hodgson U., Laitinen T., Alenius H., Kinnula V. L., Myllärniemi M., Matikainen S., Kere J. (2010) ELMOD2, a candidate gene for idiopathic pulmonary fibrosis, regulates antiviral responses. FASEB J. 24, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 66. Hodgson U., Pulkkinen V., Dixon M., Peyrard-Janvid M., Rehn M., Lahermo P., Ollikainen V., Salmenkivi K., Kinnula V., Kere J., Tukiainen P., Laitinen T. (2006) ELMOD2 is a candidate gene for familial idiopathic pulmonary fibrosis. Am. J. Hum. Genet. 79, 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson K. R., Longo-Guess C. M., Gagnon L. H. (2012) Mutations of the mouse ELMO domain-containing 1 gene (Elmod1) link small GTPase signaling to actin cytoskeleton dynamics in hair cell stereocilia. PLoS ONE 7, e36074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yin J., Haney L., Walk S., Zhou S., Ravichandran K. S., Wang W. (2004) Nuclear localization of the DOCK180/ELMO complex. Arch. Biochem. Biophys. 429, 23–29 [DOI] [PubMed] [Google Scholar]
- 69. Muromoto R., Sekine Y., Imoto S., Ikeda O., Okayama T., Sato N., Matsuda T. (2008) BART is essential for nuclear retention of STAT3. Int. Immunol. 20, 395–403 [DOI] [PubMed] [Google Scholar]
- 70. Kahn R. A. (1991) Fluoride is not an activator of the smaller (20–25 kDa) GTP-binding proteins. J. Biol. Chem. 266, 15595–15597 [PubMed] [Google Scholar]
- 71. Kalantari F., Bergeron J. J., Nilsson T. (2010) Biogenesis of lipid droplets–how cells get fatter. Mol. Membr. Biol. 27, 462–468 [DOI] [PubMed] [Google Scholar]
- 72. Nakamura N., Akashi T., Taneda T., Kogo H., Kikuchi A., Fujimoto T. (2004) ADRP is dissociated from lipid droplets by ARF1-dependent mechanism. Biochem. Biophys. Res. Commun. 322, 957–965 [DOI] [PubMed] [Google Scholar]
- 73. Nakamura N., Banno Y., Tamiya-Koizumi K. (2005) Arf1-dependent PLD1 is localized to oleic acid-induced lipid droplets in NIH3T3 cells. Biochem. Biophys. Res. Commun. 335, 117–123 [DOI] [PubMed] [Google Scholar]
- 74. Hommel A., Hesse D., Völker W., Jaschke A., Moser M., Engel T., Blüher M., Zahn C., Chadt A., Ruschke K., Vogel H., Kluge R., Robenek H., Joost H. G., Schürmann A. (2010) The ARF-like GTPase ARFRP1 is essential for lipid droplet growth and is involved in the regulation of lipolysis. Mol. Cell. Biol. 30, 1231–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.