Background: Huntingtin is a large nuclear protein with no previously identified nuclear localization signal.
Results: Huntingtin has a PY-NLS that is recognized by karyopherin β1 and β2.
Conclusion: The huntingtin NLS has pathway redundancy and has a unique intervening sequence.
Significance: This work provides insight into the mechanism and regulation of huntingtin nuclear entry.
Keywords: Huntington Disease, Neurodegeneration, Neurodegenerative Diseases, Nuclear Translocation, Stress, Importin β, Karyopherin β1, Karyopherin β2, Transportin
Abstract
Among the known pathways of protein nuclear import, the karyopherin β2/transportin pathway is only the second to have a defined nuclear localization signal (NLS) consensus. Huntingtin, a 350-kDa protein, has defined roles in the nucleus, as well as a CRM1/exportin-dependent nuclear export signal; however, the NLS and exact pathway of import have remained elusive. Here, using a live cell assay and affinity chromatography, we show that huntingtin has a karyopherin β2-dependent proline-tyrosine (PY)-NLS in the amino terminus of the protein. This NLS comprises three consensus components: a basic charged sequence, a downstream conserved arginine, and a PY sequence. Unlike the classic PY-NLS, which has an unstructured intervening sequence between the consensus components, we show that a β sheet structured region separating the consensus elements is critical for huntingtin NLS function. The huntingtin PY-NLS is also capable of import through the importin/karyopherin β1 pathway but was not functional in all cell types tested. We propose that this huntingtin PY-NLS may comprise a new class of multiple import factor-dependent NLSs with an internal structural component that may regulate NLS activity.
Introduction
One of the most fundamental differences between a eukaryotic and prokaryotic cell is the evolution of the nucleus, which provides an isolated and protected environment for the cellular genetic information. Nuclear import and export of proteins are strictly controlled, allowing for more complex functions and thus providing greater opportunity for biological diversity (1). Proteins smaller than 50–60 kDa may enter the nucleus by passive diffusion (2), but larger proteins must be accompanied into the nucleus through 1 of at least 14 nuclear transport pathways. These pathways are often used for smaller proteins as well, because they provide more rapid and strict control over import and export than is possible by diffusion alone (3). Facilitated diffusion across the nuclear pores is mediated by nuclear import proteins termed karyopherins or importins (4). Despite identification of every importin/karyopherin family member in the mammalian proteome, alternative mechanisms for nuclear entry that do not rely on these pathways also exist (5). The first isolated and best understood nuclear import pathway is mediated by karyopherin β1 via the adaptor protein karyopherin α. In this pathway, karyopherin α recognizes a well characterized classical nuclear localization signal (NLS)2 consensus consisting of either one basic rich cluster (monopartite) or two clusters separated by an intervening sequence (bipartite). These NLSs have now been identified in thousands of nuclear proteins (6). Multiple isoforms of karyopherin α are expressed differentially across cell types, allowing tissue specific regulation of NLS function (7).
A second NLS consensus has been defined for karyopherin β2-mediated import (8, 9). The karyopherin β2 type NLS, or “PY-NLS” has an invariant proline-tyrosine (PY) sequence, a conserved arginine, and a basic or hydrophobic sequence 8–13 amino acids upstream of the PY (8). This class of NLS has been described as unstructured and assumes a binding conformation only when interacting with the inner helical face of the huntingtin, EF3, PP2A, mTor (HEAT)-rich karyopherin β2 protein (10). The PY-NLS is often found in proteins involved in mRNA trafficking (9) and has recently been described as a key regulator of import to the neuronal primary cilium (11). Compared with typical karyopherin α/β1-dependent NLSs, the PY-NLS is relatively rare, although an increasing number have been identified since the discovery of its consensus.
Huntingtin is a HEAT-rich protein of 350 kDa that has known functions in both the nucleus and the cytoplasm and shuttles between the two compartments (12). Previously, we described a typical consensus CRM1-dependent, leptomycin B-sensitive nuclear export signal in the carboxyl terminus of huntingtin. Although several putative sequences were tested, a typical karyopherin α/β1-type NLS was not found (13). This led to the hypothesis that huntingtin itself may be capable of nuclear import because HEAT repeats are a common structural element between huntingtin and the karyopherin family. However, this prediction was later disproved when we noted by fluorescence recovery after photobleaching, that a fragment of huntingtin between amino acids 81 and 586 containing few HEAT repeats was still capable of active nuclear transport (14).
Here, we identify an active NLS sequence between amino acids 174 and 207 of huntingtin that interacts with karyopherin β2 both in a novel live cell assay and by affinity chromatography. The huntingtin NLS contains all of the necessary epitopes to be recognized by karyopherin β2: an upstream basic region, or “B” site, and a downstream arginine-proline-tyrosine (R-X5-PY) “A” site (8). However, the space between the A and B sites of this sequence far exceeds the length limits defined by the consensus. Furthermore, a functional redundancy appears to exist whereby karyopherin β1 is also capable of mediating import through the same NLS motif. CD spectroscopy and mutational analysis have identified a proline-flanked structured intervening sequence (IVS) between the A and B sites of the huntingtin NLS that is critical to its function. This sequence may also provide clues regarding regulation of huntingtin nuclear import. In isolation, the IVS exhibits independent targeting to the cytoplasm. Coupled with the observation that huntingtin NLS activity varies substantially among cell types, we predict that the IVS has the ability to negatively regulate huntingtin NLS function in a cell type-specific manner by a unique competitive or allosteric mechanism.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
All heterologous GFP-β-galactosidase fusions were created using the phm830 cloning vector (13, 15). Synthetic DNA oligonucleotides were generated bearing overhangs at either end corresponding to SacII and XbaI restriction sites (MOBIX, McMaster University). These oligonucleotides were then annealed to produce double-stranded DNA and ligated into digested phm830 vector. A similar cloning strategy was implemented for mCherry-M9M (EcoRI, BamHI) and mCerulean-Htt NLS (EcoRI, SacII), along with the various NLS mutants. The base cloning vectors for these constructs was p-mCherry-C1 and p-mCerulean-C1 (BD Biosciences/Clontech).
Htt 1–465-eYFP was generated by deletion of the sequence encoding amino acids 466–586 from Htt 1–586-eYFP, cloned as described previously (14). This product was created by performing inverse PCR using primers flanking the region of deletion. The resulting PCR product was then phosphorylated at the 5′ OH using T4 Polynucleotide Kinase (New England Biolabs) followed by blunt end ligation to recircularize the vector. Full or partial deletion of the NLS sequence was achieved by the same method. Point mutations of the NLS sequence were introduced to Htt 1–465-eYFP by either QuikChange II XL site-directed mutagenesis (Stratagene) or the inverse PCR method. The inverse PCR method is similar to the deletion method described above, but alternatively the point mutation is introduced as an overhang on the forward primer. mCherry-karyopherin β2(541–890) was created by PCR of the karyopherin β2 fragment and ligation into the p-mCherry-C1 vector using the restriction sites SmaI and XhoI. mCherry-karyopherin β1(256–876) was cloned in the same way, except using the restriction sites XhoI and SacII. All restriction enzymes and PCR polymerases were purchased from New England Biolabs unless otherwise stated.
Tissue Culture
Conditionally immortalized STHdh Q7/Q7 striatal progenitor cells (a kind gift from M. E. MacDonald) were originally derived from a wild-type (7 glutamine) knock-in transgenic HD mouse containing a humanized Exon 1 and G418 resistance cassette. To maintain the expression of the transgene and of the SV40 large T antigen, the cells were grown under G418 selection at 33 °C in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Invitrogen). The HEK293 cell line was grown at 37 °C (5% CO2) in α-minimal essential medium containing 10% fetal bovine serum.
Microscopy
All widefield imaging was performed using a Nikon TE200 inverted widefield epifluorescence microscope and plan apochromat 60× oil immersion objective. The imaging platform controlling the scope was NIS elements 3.1. All filter sets and dichroic filters were from Semrock (Rochester, NY), and the filter wheel was from Sutter Instruments (Novato, CA). The light source was a 175-W xenon lamp (Sutter Instruments), with ND2 or ND4 filters. Images were acquired using a Hamamatsu Orca ER digital camera (Hamamatsu Photonics). The confocal imaging was performed using a Leica DMI6000B with a Leica TCS SP5 scanner with acoustic optical beam splitter and a 100-mW 488-nm argon ion laser line. The objective used was a 63× 1.3 NA differential interference contrast glycerol HCX Plan-Apochromat, and the detector was a 2× steady-state photomultiplier tubes. The microscope was controlled using Leica Application Suite Advanced Fluorescence software.
Transfections and Quantitative Analysis
Both the STHdh Q7/Q7 and the HEK293 cells were transfected with either Exgen 500 (Fermentas) or Turbofect (Fermentas) and imaged ∼20–24 h after transfection. To determine the percent nuclear fluorescence, ∼50–100 cells were imaged per construct and quantified using Simple PCI (Compix). To ensure unbiased data collection, the cells were co-transfected with mCherry, and cells were selected under the red channel before being imaged in the green channel, in a method of unbiased imaging data acquisition described elsewhere (16). Nuclear and cytoplasmic intensities were collected by manually defining the nuclear and cytoplasmic regions in each image and then collecting blank areas of equal size to represent the image background. The intensities of the defined regions were then measured using the Simple PCI measurement tool. Percent nuclear fluorescence was calculated using the equation % nuclear fluorescence = ((nuclear fluorescence − background)/(whole cell fluorescence − background))/100.
Statistical Analysis
All statistical analysis was performed using SigmaPlot 11.0. Determination of statistical significance was calculated as deemed most appropriate by the software, where for normal data it employed Student's t test, and for nonnormal data, the Whitney-Mann method.
NLS Peptide Affinity Chromatography
Wild-type and mutant (K177A/K178A) huntingtin NLS peptides were diluted in ddH20 to create 20 mg/ml stocks. The peptides were diluted to a final concentration of 1 mg/ml in 500 μl of peptide coupling buffer (50 mm Tris, 5 mm Na-EDTA, pH 8.5) and then coupled to 30 μl of resin according to the manufacturer's instructions (Thermo Scientific). After the coupling reaction, the resin was equilibrated in 500 μl of binding buffer (0.1 m NaCl, 10 mm HEPES, pH 7.6, 10% glycerol). STHdh Q7/Q7 lysates were prepared in 1% Nonidet P-40 lysis buffer (0.15 m NaCl, 0.05 m Tris, pH 8, 1% Nonidet P-40) and diluted into 500 μl of binding buffer. One confluent plate was lysed per affinity column. The coupled resin was mixed end-over-end with diluted lysate at 4 °C before loading into a 200-μl pipette tip column stuffed with glass wool. The columns were washed four times with 200 μl of binding buffer and then 60-μl volumes of 0.2, 0.3, and 0.4 m NaCl in 10 mm HEPES, pH 7.6, and 10% glycerol. The bound proteins were eluted in 60 μl of 0.5 m NaCl. All affinity chromatography was conducted at 4 °C.
SDS-PAGE and Western Blot Analysis
Equal amounts of each elution were run on a 12% SDS-polyacrylamide gel, transferred to PVDF membrane (PALL), and blocked with 5% (w/v) milk in TBST. The membrane was blotted with anti-karyopherin β2 (D45; Abcam) at 1/2000. Rabbit anti-mouse HRP-conjugated secondary antibody (ab97046; Abcam) was used at 1/25,000. Blots were developed with Immobilon Western Chemiluminescent Substrate (Millipore) and imaged on a DNR Microchemi Western blot imager.
Circular Dichroism Spectroscopy
Huntingtin NLS Peptides (NH2-LYKEIKKNGAPRSLRAALWRFAELAHLVRPQKCRPY-COOH) were purchased from New England Peptide and reconstituted in PBS (10 mm NaH2PO4, 0.14 m NaCl, 1 mm EDTA, pH 7.4). The peptides were further diluted in PBS to 0.25 mg/ml for CD spectroscopy analysis in an Aviv CD spectrometer model 215 (Aviv Instruments). The data were collected at 25 °C in millidegrees and then converted to mean residue ellipticity []λ in degree/cm2 per dmol through the formula (/molarity) × (no. of residues) × pathlength. After the spectrum was generated, the numbers were fed into the online CD spectroscopy analysis software, K2D Neural Network (17). This tool allows for interpretation of CD spectra by systematically comparing them with a large library of spectra from other proteins or peptides of known structure.
RESULTS
Huntingtin Contains a Highly Conserved PY-NLS Sequence Defined by Amino Acids 174–207
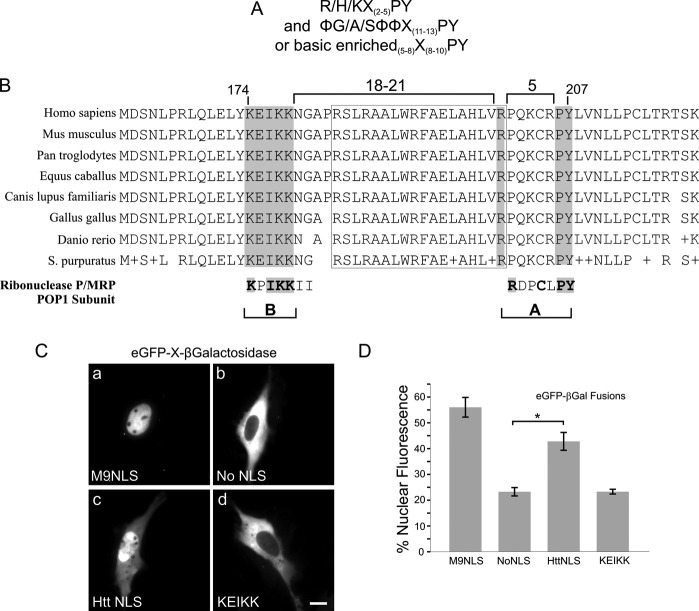
An NLS consensus for karyopherin β2-mediated nuclear transport has been elucidated (9). Termed the PY-NLS, this sequence is defined by an R-X5-PY motif, 8–13 amino acids downstream from either a hydrophobic or basic rich region (Fig. 1A). We noted the presence of a strictly conserved arginine and downstream PY motif within huntingtin 81–586, a region previously shown by our laboratory to exhibit nuclear import activity (Fig. 1B). In our first attempt to delineate the boundaries of the NLS, we identified a hydrophobic sequence that closely matched the rules of the consensus, falling only 8 amino acids upstream from the PY (LWRFAEL). When tested, however, this fragment failed to demonstrate NLS activity (data not shown). We next proceeded to examine sequences further upstream and located a basic rich region (KEIKK) 27 amino acids away from the PY. To assess whether or not this fragment possessed NLS activity we implemented a classic live cell in vivo NLS assay, in which huntingtin fragments were fused to the reporter proteins GFP and β-galactosidase, creating a triple fusion larger than 120 kDa (15). Because this far exceeds the diffusion limit of the nuclear pore, and neither GFP nor β-galactosidase has an endogenous NLS, nuclear import is only achieved only by fusion of an active NLS sequence. Activity was assayed by quantification of the percent nuclear fluorescence compared with the total cell fluorescence within a population of 50–100 transfected cells/data set. In STHdh cells, nuclear import activity of the PY-NLS sequence (20) was found to be dependent upon the KEIKK basic sequence located upstream (Fig. 1, B–D) in addition to the PY region (Fig. 1, C and D). The KEIKK sequence alone was not sufficient to mediate nuclear entry of the reporter (Fig. 1C, c versus d). Thus, the nuclear localization sequence of huntingtin strictly requires amino acids 174–207, falling outside the length limits of the defined PY-NLS consensus (Fig. 1A) (9). However, despite this key difference, the sequence still contains individual elements of the consensus that are strikingly similar to other PY-NLSs, particularly those motifs found within the POP1 subunit of P/MRP ribonuclease (Fig. 1B).
FIGURE 1.
Huntingtin amino acids 174–207 contain elements of a karyopherin β2 basic PY NLS. The defined consensus for karyopherin β2-type NLSs is shown in A, alongside sequence alignments of the huntingtin NLS displaying full conservation of the basic rich KEIKK region, arginine 200, and proline-tyrosine PY 206/207 (boxed in gray) in B. The 18-amino acid intervening sequence flanked by proline turns, highlighted by a boxed outline, is also conserved. Below, the PY-NLS of ribonuclease P/MRP POP1 subunit is shown, with highly similar A and B sites. C, huntingtin 174–207 was cloned in-frame with GFP and β-galactosidase, a >120-kDa reporter for live cell expression. Sample images show huntingtin NLS as a GFP-β-galactosidase fusion, compared with no NLS, M9 hnRNPA1 NLS, and the KEIKK sequence. D, quantification of data in C, n = ∼50 cells quantified per construct. *, p < 0.01. Error bars, S.E.
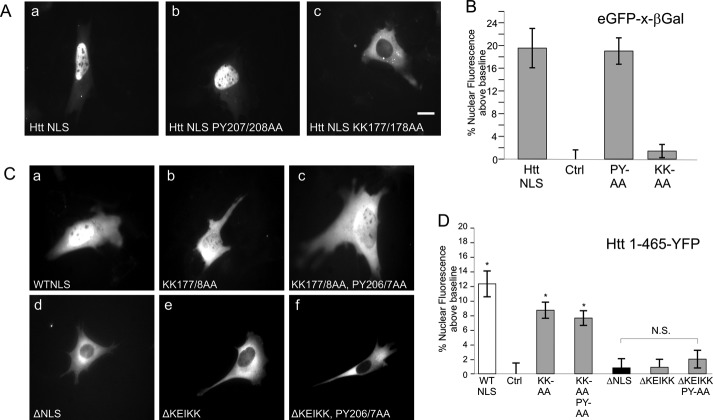
Using the same GFP-β-galactosidase fusion assay in STHdh cells, we next explored the relative importance of individual NLS residues to import activity. Mutation of the PY residues at 207/208 had no effect, whereas mutation of lysines 177 and 178 within the basic rich region abrogated nuclear import activity (Fig. 2A, a–c, quantified in Fig. 2B). All residues were converted to alanines. We examined similar mutations in the basic region and PY residues in the context of huntingtin 1–465 as a carboxyl-terminal eYFP fusion (Fig. 2C). In this context, K177A/K178A mutation reduced, but did not eliminate nuclear entry, whereas mutation of the PY residues had no effect (Fig. 2C, b and c). Similar results have previously been observed when PY mutations were made to the hnRNPA1 M9 NLS (9). Complete abrogation of nuclear entry was observed upon deletion of the entire NLS sequence from 174 to 207 (Fig. 2Cd). However, the same loss of nuclear import could be achieved by deletion of the KEIKK basic rich sequence alone (Fig. 2C, e and f). Although the KEIKK sequence is clearly necessary for NLS activity, it is not sufficient to mediate nuclear import in a heterologous nuclear transport assay, indicating that this is not a typical basic karyopherin α/β1-type NLS (Fig. 1Cd) To ensure that the observed changes to NLS activity after mutation are not due to gross alterations of expression, we quantified the average exposure time in each of our data sets. Although the exposures varied greatly from image to image in all our data sets, the average fluorescence intensity did not change appreciably and showed no correlation with high or low levels of nuclear import (data not shown).
FIGURE 2.
Huntingtin PY-NLS basic region is critical for NLS activity in vivo. A, positive control of GFP-Htt NLS-β-galactosidase, compared with P207A/Y208A and K177A/K178A mutants, quantified in B. Ctrl is GFP-β-galactosidase, which has been quantified and then set as base line to 0. C and D, analysis of huntingtin NLS mutants in the (Q17)1–465-eYFP context. n = ∼50 cells quantified per construct. *, p < 0.01. Error bars, S.E.
Huntingtin PY-NLS Activity Is Cell Type-specific and Employs Multiple Nuclear Import Pathways
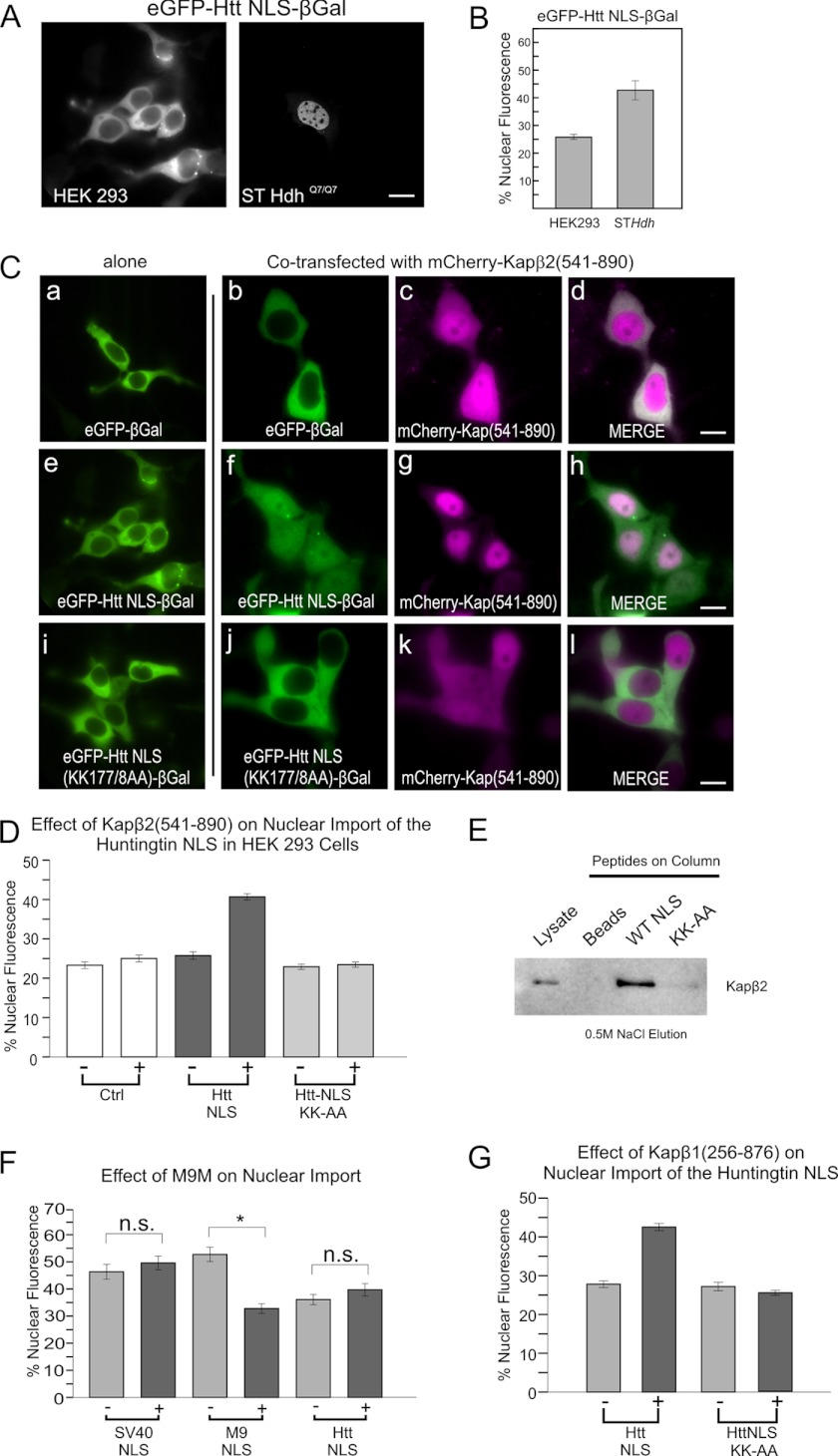
During the course of these experiments we observed that the huntingtin PY-NLS sequence exhibited low nuclear import levels in human embryonic kidney HEK293 cells (Fig. 3, A and B). This was despite the detection of similar levels of karyopherin β2 by Western blotting (data not shown). Thus, for unknown reasons, we had found a nonpermissive cell line for huntingtin NLS nuclear entry. To determine whether karyopherin β2 is capable of mediating nuclear transport of the huntingtin NLS in live cells we attempted to drive nuclear entry in the HEK293 cell line. A fragment of karyopherin β2 (residues 541–890) lacking the RanGTP/GDP binding domain (18) was fused to mCherry and overexpressed in the HEK293 cells (Fig. 3C). Without the Ran interaction domain, we predicted that karyopherin β2 would be incapable of cargo release once across the nuclear pore complex. Co-expression of karyopherin β2(541–890) with GFP-huntingtin NLS-β-galactosidase resulted in nuclear entry of the reporter. However, it did not promote nuclear entry of GFP-β-galactosidase alone (Fig. 3C, a–d versus e–h). Consistent with the earlier work in STHdh mouse cells, this nuclear entry could be abrogated with the K177A/K178A mutation (Fig. 3C, i–l, quantified in 3D). As further confirmation that the huntingtin NLS interacts with karyopherin β2, we performed affinity column chromatography of STHdh cell lysate using huntingtin NLS and mutant (K177A/K178A) peptides. After washing the columns with increasing concentrations of NaCl (up to 0.4 m), the bound proteins were eluted at 0.5 m NaCl. Karyopherin β2 was found to associate with the wild-type huntingtin NLS peptide, but not the K177A/K178A mutant. The beads alone did not interact with karyopherin β2 (Fig. 3E). Together, these findings suggest that huntingtin NLS activity can be mediated by karyopherin β2. To assess the specificity of huntingtin NLS for the karyopherin β2 pathway, we tested the effect of overexpressing a karyopherin β2 NLS inhibitor, M9M (Fig. 3F). M9M is a synthetic chimera of the A and B sites from two different PY-NLS sequences that produces a motif with higher than physiological affinity for karyopherin β2. This increased affinity prevents M9M release from karyopherin β2, even in the presence of RanGTP in the nucleus (18). In our live cell assay, mCherry-M9M overexpression inhibited nuclear import of the hnRNPA1 M9 karyopherin β2-dependent PY-NLS but had no effect on the huntingtin PY-NLS or the classical SV40 NLS (Fig. 3F).
FIGURE 3.
Huntingtin PY-NLS utilizes the karyopherin β2/β1 import pathways. A, comparison of huntingtin NLS-mediated nuclear entry in human HEK293 versus mouse STHdh cells, quantified in B. C, overexpression of mCherry-karyopherin β2(541–890) (ΔRan binding) restoring wild-type NLS activity in HEK 293 cells (f–h), but not K177A/K178A mutant NLS (j–l). D, quantification of 3C. E, affinity chromatography of STHdh cell lysates on huntingtin NLS peptide-coupled columns. Column elutions at 0.5 m NaCl were separated by SDS-PAGE and Western blotted for karyopherin β2 (anti-Kapβ2,D45). Lysate lane contains 1/20 of column input. Beads indicates the beads-alone column. F, comparison of the SV40 NLS, M9 NLS, and huntingtin NLS in response to overexpression of M9M peptide competitor to karyopherin β2. G, overexpression of karyopherin β1(256–876) (ΔRan binding) increasing nuclear entry via the huntingtin NLS, but not a K177A/K178A mutant NLS. All NLSs are in GFP-β-galactosidase context in STHdh cells unless otherwise stated. For all quantification, n = ∼50 cells per construct. *, p < 0.01. Error bars, S.E.
The observation that M9M does not inhibit nuclear import of the huntingtin NLS suggests that it may not be restricted to recognition by karyopherin β2 alone. To test this hypothesis, we overexpressed a similar Ran-independent fragment of karyopherin β1 (19) in the HEK293 assay and found that it could also restore huntingtin NLS activity. This occurred without the need to overexpress a karyopherin α adaptor protein (Fig. 3G). This suggests that aside from being capable of nuclear entry via karyopherin β2, the huntingtin NLS can also utilize the karyopherin β1 pathway, but via a karyopherin β1 direct mechanism, and not the classical α/β1 interactions.
The Huntingtin PY-NLS Contains an 18-Mer β-Sheet Sequence That Is Critical for NLS Activity
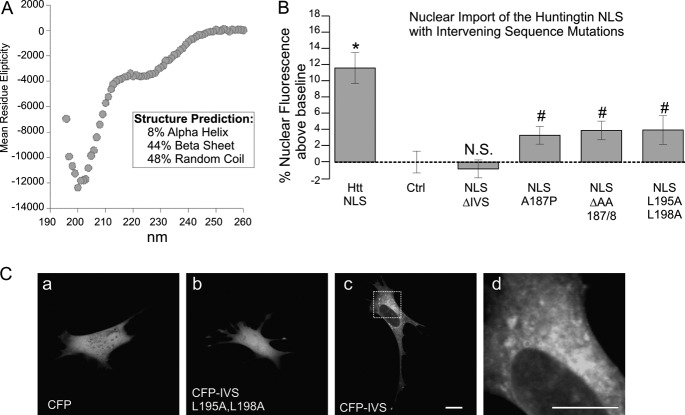
A unique aspect of the huntingtin PY-NLS is the 27-amino acid intervening sequence between the basic region and the proline/tyrosine of the A site defined for PY-NLSs. An 18-amino acid sequence within this region is flanked by two proline residues (Fig. 1B), suggesting that it may be a structured element lying between two proline turns. To test this hypothesis, we synthesized a synthetic peptide corresponding to the 18-amino acid sequence and performed CD spectroscopy at physiological salt concentrations with a protein concentration of 0.25 mg/ml (Fig. 4A). Using a neural two-dimensional network algorithm (17), the secondary structure was calculated to be 44% β sheet. Similar results were seen when the entire PY-NLS 174–207 peptide was assayed (data not shown). When an A187P or ΔAA(187,188) mutation was made at the center of the IVS (Fig. 4B), NLS activity was lost in context of GFP-β-galactosidase, despite the fact that these mutations fall at a considerable distance from the consensus elements. This indicates that structure is critical to huntingtin NLS nuclear import activity.
FIGURE 4.
Huntingtin NLS 18-amino acid intervening sequence has a β sheet structure and cytoplasmic targeting activity. A, CD spectroscopy of the huntingtin IVS peptide, NH2-PRSLRAALWRFAELAHLVRP-COOH, showing 44% β sheet characteristics. B, effect of IVS mutants on huntingtin NLS activity in the context of eGFP-X-β-galactosidase. Note the single proline substitution A187P, ΔAA(187,188) deletion, and double leucine substitution L195A/L198A. C, comparison of nuclear localization of CFP alone (a), CFP-IVS (c) and CFP-IVS L195A/L198A (b) in STHdh cells. Leucines 195 and 198 are critical for targeting to cytoplasm. Area in panel c is magnified in panel d to highlight targeting to cytoplasmic structures. n = ∼70 cells per construct. #, p values that are statistically significant compared with the wild-type huntingtin NLS (p < 0.01). *, p values that are statistically significant compared with Ctrl (p < 0.01). N.S. is nonsignificant compared with control. Error bars, S.E.
The Huntingtin PY-NLS Intervening Sequence Has Cytoplasmic Targeting Activity
To determine whether the structured PY-NLS intervening sequence has intrinsic biological activity or is only behaving as a spacer element, we fused it to an mCerulean blue fluorescent protein for expression in live cells. When expressed alone, mCerulean blue (CFP) localizes to both the cytoplasm and the nucleus because its molecular mass falls below the diffusion limit of the nuclear pore complex (Fig. 4Aa). AFP proteins can even be trimerized in tandem and still diffuse across the nuclear pore complex (20). Fusion of the IVS to CFP only increases the molecular mass by a few kDa; therefore, it would be expected to display the same diffuse nuclear and cytoplasmic localization. However, in STHdh cells, this fusion was found to target the cytoplasm. To establish that the targeting of the CFP-IVS fusion was not due solely to insolubility of the small peptide, we created a L195A/L198A double mutation of similar hydrophobicity. However, the interaction appeared to be specific because the mutation led to a loss of cytoplasmic targeting (Fig. 4C, a–d). Because the L197A/L198A mutation disrupted cytoplasmic targeting, we anticipated that it might allow functional separation of the counteractive cytoplasmic targeting of the intervening sequence and the nuclear targeting of the NLS. However, even this structurally subtle mutation inhibited the activity of the NLS (Fig. 4B). We noted that the hydrophobic residues and spacing in the IVS could potentially fit the consensus of a CRM1-dependent nuclear export signal (21). However, the sequence appeared to be to targeted to specific cytoplasmic structures (Fig. 4Cd), inconsistent with a nuclear export signal, and localization was not affected by leptomycin B treatment (data not shown). Thus, the huntingtin PY-NLS intervening sequence appears to have independent targeting capacity to cytoplasmic factors, an activity that may sterically regulate the PY-NLS functional interactions with karyopherins.
DISCUSSION
The defined PY-NLS consensus was derived in vitro by establishing important interaction residues between NLSs and karyopherin β2 by x-ray co-crystal and isothermal calorimetry studies (9). We have discovered a PY-NLS in the huntingtin protein that is both highly evolutionarily conserved and active in heterologous fusion nuclear import assays in live cells. Although components of the huntingtin PY-NLS conform to the sequence requirements of the consensus, including the A site (R/H/KX(2–5)-PY) and the B site (the upstream basic rich region) (9), it diverges in terms of the spacing of these epitopes. Whereas the consensus defines 8–10 amino acids between the PY motif and the basic rich region, the intervening sequence of the huntingtin PY-NLS is 27 amino acids (from the proline-tyrosine to the upstream basic region). CD spectroscopy analysis reveals another significant inconsistency with typical PY-NLSs. Whereas typical PY-NLSs are found in unstructured regions, it appears that the huntingtin PY-NLS contains structured elements within its IVS. These two important findings define the huntingtin NLS as a novel subtype of structured PY-NLS. To fit the binding pocket of karyopherin β2 or β1, this longer PY-NLS may require the formation of secondary structure to align the critical epitopes for import factor interaction in three-dimensional space. This is consistent with the ability of the consensus elements of the PY-NLS to bind karyopherin β2 in an energetically independent manner (10).
HEK293 cells do not permit huntingtin PY-NLS function, despite the presence of karyopherin β2 and β1. This suggests that there may be an additional factor in this cell line that competitively inhibits nuclear entry by binding to the PY-NLS, possibly through the intervening sequence. Consistent with this hypothesis, the intervening sequence has independent cytoplasmic targeting ability in vivo. These data present a caveat for the interpretation of studies that use HEK293s as a model system to examine huntingtin nuclear import and toxicity. If huntingtin is able to activate or repress nuclear entry through the IVS it may explain how huntingtin nuclear localization is regulated in different cell types throughout the body. Some differentiated cells may restrict the ability of huntingtin to enter the nucleus after development. Indeed, classic studies of huntingtin localization in human brain have shown a neuronal cell type variance in huntingtin nuclear localization (22, 23).
Consistent with the concept of an additional class of karyopherin β1- and β2-dependent regulated NLSs, recently, the HIV-1 Rev protein was shown to import into the nucleus by both karyopherin β1-direct (24) and karyopherin β2 pathways, with a binding factor that regulates these NLS-transport factor interactions (25). Other proteins, such as the c-Jun oncoprotein, have been seen to interact with both karyopherin β1 and β2 as well as other import factors in vitro, via a basic stretch of amino acids, but without the PY epitope of a karyopherin β2-dependent NLS (26, 27). In the case of c-Jun, regulation of the karyopherin β1/β2 NLS was demonstrated to depend upon steric hindrance by homodimerization (28). The mRNA export factor, NXF1 contains a PY-NLS, but this signal can enter the nucleus through redundant pathways that include karyopherins β1, β2, importin α, and importins 4 and 11 (29).
Huntingtin has an essential and critical role in neurogenesis during development and a role at the mitotic spindle apparatus in cell division (29–31). Thus, it is likely that the nuclear import of such a critical factor has evolved to not rely solely on one import pathway. Furthermore, by utilizing a karyopherin β1 direct pathway, the huntingtin NLS would not depend on the karyopherin α adaptor family and thus not be limited by their cell type-specific expression (30). Consistent with the karyopherin β2 import factor pathway for huntingtin nuclear entry, in mouse brain, karyopherin β2 is highly expressed in regions of neurogenesis and circadian clock regulation (31). Disruption in circadian rhythms has been noted in both HD patients and HD mouse models (32).
Our mutational analysis in the 1–465 fragment of huntingtin concludes that this PY-NLS sequence is the only NLS within the amino terminus of huntingtin. This fragment contains the expanded polyglutamine tract and comprises regions seen in proteolytic fragments of huntingtin implicated in HD pathology (33–35). The identification of huntingtin intervening sequence binding proteins, and their possible regulation of huntingtin nuclear entry, are the subject of further study.
Acknowledgment
We thank Dr. Yuh Min Chook (University of Texas, Southwestern Medical Center, Dallas, TX) for invaluable advice and reagent support.
This work was supported by the Huntington Society of Canada, CHDI Inc., the Krembil Foundation, and the Canadian Institutes of Health Research Operating Grant MOP-119391 (to R. T.).
- NLS
- nuclear localization signal
- HEAT
- huntingtin, EF3, PP2A, mTor
- IVS
- intervening sequence.
REFERENCES
- 1. Chook Y. M., Blobel G. (2001) Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715 [DOI] [PubMed] [Google Scholar]
- 2. Terry L. J., Shows E. B., Wente S. R. (2007) Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 [DOI] [PubMed] [Google Scholar]
- 3. Cullen B. R. (2003) Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28, 419–424 [DOI] [PubMed] [Google Scholar]
- 4. Ribbeck K., Görlich D. (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20, 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wagstaff K. M., Jans D. A. (2009) Importins and beyond: non-conventional nuclear transport mechanisms. Traffic 10, 1188–1198 [DOI] [PubMed] [Google Scholar]
- 6. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 8. Chook Y. M., Süel K. E. (2011) Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., Chook Y. M. (2006) Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Süel K. E., Gu H., Chook Y. M. (2008) Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 6, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dishinger J. F., Kee H. L., Jenkins P. M., Fan S., Hurd T. W., Hammond J. W., Truong Y. N., Margolis B., Martens J. R., Verhey K. J. (2010) Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and RanGTP. Nat. Cell Biol. 12, 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Truant R., Atwal R. S., Burtnik A. (2007) Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease. Prog. Neurobiol. 83, 211–227 [DOI] [PubMed] [Google Scholar]
- 13. Xia J., Lee D. H., Taylor J., Vandelft M., Truant R. (2003) Huntingtin contains a highly conserved nuclear export signal. Hum. Mol. Genet. 12, 1393–1403 [DOI] [PubMed] [Google Scholar]
- 14. Atwal R. S., Xia J., Pinchev D., Taylor J., Epand R. M., Truant R. (2007) Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 16, 2600–2615 [DOI] [PubMed] [Google Scholar]
- 15. Sorg G., Stamminger T. (1999) Mapping of nuclear localization signals by simultaneous fusion to green fluorescent protein and to β-galactosidase. BioTechniques 26, 858–862 [DOI] [PubMed] [Google Scholar]
- 16. Voss T. C., Demarco I. A., Day R. N. (2005) Quantitative imaging of protein interactions in the cell nucleus. BioTechniques 38, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greenfield N. J. (2006) Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cansizoglu A. E., Lee B. J., Zhang Z. C., Fontoura B. M., Chook Y. M. (2007) Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 14, 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vetter I. R., Arndt A., Kutay U., Görlich D., Wittinghofer A. (1999) Structural view of the Ran-importin β interaction at 2.3 Å resolution. Cell 97, 635–646 [DOI] [PubMed] [Google Scholar]
- 20. Seibel N. M., Eljouni J., Nalaskowski M. M., Hampe W. (2007) Nuclear localization of enhanced green fluorescent protein homomultimers. Anal. Biochem. 368, 95–99 [DOI] [PubMed] [Google Scholar]
- 21. Bogerd H. P., Fridell R. A., Benson R. E., Hua J., Cullen B. R. (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16, 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoogeveen A. T., Willemsen R., Meyer N., de Rooij K. E., Roos R. A., van Ommen G. J., Galjaard H. (1993) Characterization and localization of the Huntington disease gene product. Hum. Mol. Genet. 2, 2069–2073 [DOI] [PubMed] [Google Scholar]
- 23. Sapp E., Schwarz C., Chase K., Bhide P. G., Young A. B., Penney J., Vonsattel J. P., Aronin N., DiFiglia M. (1997) Huntingtin localization in brains of normal and Huntington's disease patients. Ann. Neurol. 42, 604–612 [DOI] [PubMed] [Google Scholar]
- 24. Truant R., Cullen B. R. (1999) The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol. Cell. Biol. 19, 1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gu L., Tsuji T., Jarboui M. A., Yeo G. P., Sheehy N., Hall W. W., Gautier V. W. (2011) Intermolecular masking of the HIV-1 Rev NLS by the cellular protein HIC: novel insights into the regulation of Rev nuclear import. Retrovirology 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nikolaev I., Cochet M. F., Felenbok B. (2003) Nuclear import of zinc binuclear cluster proteins proceeds through multiple, overlapping transport pathways. Eukaryot. Cell 2, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waldmann I., Wälde S., Kehlenbach R. H. (2007) Nuclear import of c-Jun is mediated by multiple transport receptors. J. Biol. Chem. 282, 27685–27692 [DOI] [PubMed] [Google Scholar]
- 28. Malnou C. E., Salem T., Brockly F., Wodrich H., Piechaczyk M., Jariel-Encontre I. (2007) Heterodimerization with Jun family members regulates c-Fos nucleocytoplasmic traffic. J. Biol. Chem. 282, 31046–31059 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Z. C., Satterly N., Fontoura B. M., Chook Y. M. (2011) Evolutionary development of redundant nuclear localization signals in the mRNA export factor NXF1. Mol. Biol. Cell 22, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamei Y., Yuba S., Nakayama T., Yoneda Y. (1999) Three distinct classes of the α-subunit of the nuclear pore-targeting complex (importin-α) are differentially expressed in adult mouse tissues. J. Histochem. Cytochem. 47, 363–372 [DOI] [PubMed] [Google Scholar]
- 31. Sato M., Mizoro Y., Atobe Y., Fujimoto Y., Yamaguchi Y., Fustin J. M., Doi M., Okamura H. (2011) Transportin 1 in the mouse brain: appearance in regions of neurogenesis, cerebrospinal fluid production/sensing, and circadian clock. J. Comp. Neurol. 519, 1770–1780 [DOI] [PubMed] [Google Scholar]
- 32. Petersén A., Hult S., Kirik D. (2009) Huntington's disease: new perspectives based on neuroendocrine changes in rodent models. Neurodegener. Dis. 6, 154–164 [DOI] [PubMed] [Google Scholar]
- 33. Mende-Mueller L. M., Toneff T., Hwang S. R., Chesselet M. F., Hook V. Y. (2001) Tissue-specific proteolysis of Huntingtin (htt) in human brain: evidence of enhanced levels of N- and C-terminal htt fragments in Huntington's disease striatum. J. Neurosci. 21, 1830–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., Qin Z. H., Aronin N., DiFiglia M. (2001) Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gafni J., Hermel E., Young J. E., Wellington C. L., Hayden M. R., Ellerby L. M. (2004) Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J. Biol. Chem. 279, 20211–20220 [DOI] [PubMed] [Google Scholar]