Background: AtREM1.3 is involved in plant immune signaling.
Results: The N-terminal region of AtREM1.3 is intrinsically disordered.
Conclusion: N-terminal region facilitates protein interactions.
Significance: Remorins are dynamic signaling proteins with respect to structure and localization.
Keywords: Intrinsically Disordered Proteins, Membrane Proteins, Plant Biochemistry, Protein Phosphorylation, Protein-Protein Interactions, Signaling, Remorin
Abstract
The longstanding structure-function paradigm, which states that a protein only serves a biological function in a structured state, had to be substantially revised with the description of intrinsic disorder in proteins. Intrinsically disordered regions that undergo a stimulus-dependent disorder-to-order transition are common to a large number of signaling proteins. However, little is known about the functionality of intrinsically disordered regions in plant proteins. Here we investigated intrinsic disorder in a plant-specific remorin protein that has been described as a signaling component in plant-microbe interactions. Using bioinformatic, biochemical, and biophysical approaches, we characterized the highly abundant remorin AtREM1.3, showing that its N-terminal region is intrinsically disordered. Although only the AtREM1.3 C-terminal domain is essential for stable homo-oligomerization, the N-terminal region facilitates this interaction. Furthermore, we confirmed the stable interaction between AtREM1.3 and four isoforms of the importin α protein family in a yeast two-hybrid system and by an in planta bimolecular fluorescent complementation assay. Phosphorylation of Ser-66 in the intrinsically disordered N-terminal region decreases the interaction strength with the importin α proteins. Hence, the N-terminal region may constitute a regulatory domain, stabilizing these interactions.
Introduction
Intrinsically disordered regions are flexible and extended protein segments that have no ordered secondary structure under physiological conditions. However, this unstructured state is crucial for their biological function. Proteins harboring long stretches of intrinsic disorder (ID)3 have been shown to be key components of signal transduction cascades because these flexible regions confer high specificity and low affinity to protein-protein interactions (1).
Plants, as sessile organisms, require highly efficient regulatory circuits to modulate cellular signal transduction cascades. Plant-specific remorin proteins, which may act as molecular scaffolds regulating signal transduction, have been described to be highly phosphorylated and to associate with signaling compartments at the plasma membrane (2–4). These proteins evolved in all land plants, including ferns and mosses and constitute a multigene family that comprises six subgroups with no significant sequence similarity to other described proteins or domains (5). All remorins are characterized by the presence of a conserved C-terminal region (Pfam domain Remorin_C; PF03763) that encodes a predicted coiled-coil motif and a dynamic membrane-anchoring motif (6). Additionally, almost all groups harbor a highly diverse N-terminal region, which is predicted to be intrinsically disordered and to harbor most phosphorylation sites (7). However, its biological function has not been elucidated.
The best-studied remorins belong to group 1b. Potato StREM1.3 binds simple and complex galacturonides, cell wall-derived molecules that induce defense during pathogenesis (3, 8). Tomato SlREM1.2 interacts with the viral protein TGBp1, and its overexpression inhibits movement of potato virus X in leaves (9). Arabidopsis thaliana AtREM1.3 is a filament-forming, ubiquitously expressed, and highly abundant protein (5, 10). Furthermore, it is differentially phosphorylated upon treatment with bacterial elicitors and has been proposed to play a dynamic role as scaffold protein in plant innate immunity (11, 12). Therefore, AtREM1.3 is a prominent candidate to biochemically assess the presence of ID protein domains and their possible function in remorin signal transduction.
To increase our knowledge about the role of the remorin N-terminal region in plant signaling, we show the first structural characterization of AtREM1.3 N- and C-terminal domains by circular dichroism spectroscopy, limited proteolysis, and mass spectrometry, experimentally proving that the N terminus of AtREM1.3 is intrinsically disordered. Furthermore, the ID N-terminal region contributes to homo-oligomerization of AtREM1.3 and interaction with members of the cytosolic and nuclear importin α protein family, probably via an induced folding mechanism.
EXPERIMENTAL PROCEDURES
Bioinformatic Tools
Amino acid compositional analysis was performed using the Composition Profiler tool and the SwissProt version 51 database as background data set (13). The charge-hydrophobicity phase space was constructed using data of intrinsically disordered proteins described in literature (data taken partially from Ref. 14) and from randomly chosen natively folded proteins obtained from the Protein Data Bank. The mean net charge (〈R〉) and the mean hydrophobicity 〈H〉 were calculated using the FoldIndex server (15). The border between intrinsically disorder and folded proteins is described by the formula,
The intrinsic disorder profile was determined using the PONDR VL-XT algorithm (16–18).
Ab Initio Modeling, Molecular Graphics, and Protein Interaction Predictions
Ab initio modeling of AtREM1.3 was performed using the I-TASSER server (19, 20). Models for the N- and C-terminal regions were constructed independently and fused subsequently. Predictions of putative regions involved in protein interactions were performed using the PPI-Pred (University of Leeds) (21) and ProMate (Weizmann Institute) servers (22). Molecular graphics images were produced using the UCSF Chimera package (23).
Cloning
For protein expression, the A. thaliana AtREM1.3 coding sequence was PCR-amplified from a cDNA library with primers that encode a Strep-tag II (supplemental Table S1) and inserted into the expression vector pETDuet-1 (Novagen) via the NdeI and XhoI restriction sites, resulting in the vector pETDuet1_Strep_AtREM1.3 (supplemental Table S2). The truncation variants AtREM1.3-N and AtREM1.3-CΔ were cloned by the same strategy using primer pairs specified in the supplemental material (supplemental Table S1).
Fusion constructs for the yeast two-hybrid assays were created using Gateway (GW) technology (Invitrogen) according to the manufacturer's guidelines. Entry clones for AtREM1.3 and importin α isoforms were generated in the pENTR-D-modified vectors. The coding sequences were amplified from A. thaliana Col-0 cDNA using primers containing a stop codon and BsaI restriction sites specified in the supplemental material (supplemental Table S1). Insertion of the fragments was carried by cut-ligation as described previously (24), and positive clones were sequenced. Correct plasmids were used for in-frame delivery of genes into respective destination vectors by LR reaction (Invitrogen). The fusions with GAL4 activation (AD) and binding (BD) domains were performed using the GW-compatible destination vectors pGADT7::GW and pGBKT7::GW, respectively (supplemental Table S2). The fusions with YFPn and YFPc were performed using the GW-compatible plant expression destination vectors pAM-PAT-YFPn::GW and pAM-PAT-YFPc::GW, respectively (supplemental Table S2).
Site-directed Mutagenesis
The plasmid pETDuet1_Strep_AtREM1.3 was used as template to introduce the S66A and S66D site-directed mutations in the AtREM1.3 coding sequence. Mutations were generated using the primers indicated in supplemental Table S1 as described earlier (25). Template DNA was removed by DpnI digestion. Integrity of the mutants was verified by sequencing.
Protein Expression and Purification
Escherichia coli Rosetta pLacI harboring the expression plasmids was grown in LB medium supplemented with 1% (w/v) glucose, 100 μg/ml ampicillin, and 34 μg/ml chloramphenicol at 37 °C until A600 ∼0.6–0.7. Cells were induced at 28 °C with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 3–4 h. Induced cells were centrifuged, and the obtained pellet was resuspended in lysis buffer (50 mm Tris/HCl (pH 8.0), 200 mm NaCl, 1 mg/ml lysozyme (Sigma), 100 μg/ml DNase I (Sigma)) and incubated for 30 min at 4 °C. Cells were further disrupted using a French press (Aminco), and the cell debris was removed by ultracentrifugation for 30 min at 30,000 × g and 4 °C. The clear lysates were loaded onto the respective columns equilibrated in binding buffer (50 mm Tris/HCl (pH 8.0), 200 mm NaCl).
The Strep-tagged proteins were purified by a single-step affinity chromatography using a gravity flow Strep-Tactin-Sepharose column (IBA). The column was washed with binding buffer, and the Strep-tagged proteins were eluted with elution buffer (50 mm Tris/HCl (pH 8.0), 200 mm NaCl, 2.5 mm desthiobiotin). The fractions containing the protein were pooled and concentrated using an Amicon Ultra centrifugal filter (Millipore).
Far-UV Circular Dichroism (CD) Spectroscopy
Far-UV CD spectra were collected using a J-810 spectropolarimeter (Jasco) at 20°. The protein samples were extensively dialyzed against 10 mm Tris/HCl buffer, pH 8.0, and diluted to a 0.1 mg/ml final concentration. Spectra were collected from 185 to 260 nm using a 1-mm path length cell. Each spectrum is the average of three scans with a 1-nm bandwidth. Spectra manipulation was done using the spectra manager software (Jasco), and data analysis was performed using the CDSSTR program from the DichroWeb server (26–28). Folding was studied by incubation with increasing concentrations of 2,2,2-trifluoroethanol (TFE; >99.5%) (Sigma). TFE was added to the protein samples to final concentrations of 12.5–50% (v/v).
Proteinase K (PK) Digestion and Mass Spectrometry Sequencing
5 μg of AtREM1.3 protein variants were incubated in proteolysis buffer (50 mm Tris, pH 8.0, 5 mm CaCl) with increasing amounts of PK (50 pg, 500 pg, 5 ng, 50 ng, and 500 ng) for 30 min on ice. Reactions were stopped by adding SDS sample buffer and phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 5 mm and incubated for 5 min at 95 °C. Samples were loaded onto a 15% SDS-polyacrylamide gel and electrophoresed. Protein bands were excised and analyzed by mass spectrometry.
Transmission Electron Microscopy (TEM)
A 10-μl droplet of each protein sample (100 μg/ml) was applied to a carbon-coated Formvar nickel grid (300-mesh) (Plano) for 10 min and stained twice with 2% (w/v) uranyl acetate. Micrographs were collected on an FEI Morgani transmission electron microscope.
Yeast Two-hybrid Interaction Assay
Plasmids harboring GAL4 fusion proteins (supplemental Table S2) were transformed into the yeast strain pJ694A, which harbors the HIS3 reporter gene. The transformed yeast cultures were plated on to synthetic defined (SD; 0.67% yeast nitrogen base, 2% glucose, 2% Bacto-agar, and amino acid mix) plates without tryptophan and leucine (-LW) for 3 days at 28 °C. Transformants were tested for interactions on SD plates without the appropriate auxotrophic markers and in the presence of 10–20 mm 3-amino-1,2,4-triazole in different dilution series. β-Galactosidase activity of yeast strains was determined by the Miller method (29). 3–6 independent yeast transformants were grown overnight in 3 ml of SD−LW medium at 30 °C. Stationary phase cultures were diluted to A600 ∼0.2 in SD−LW medium and grown at 30 °C to A600 ∼0.5–0.8). Cells were lysed using SDS and chloroform. β-Galactosidase activity was assayed according to the Miller method (29). Expression of the different fusion proteins was detected by Western blot using a rat monoclonal α-HA-HRP antibody (Roche Applied Science) and a mouse monoclonal α-Myc antibody (Santa Cruz Biotechnology, Inc.) for AD and BD fusion proteins, respectively.
Statistical Analysis
Multiple comparisons among groups were performed in R version 2.15.1 (30) using a one-way analysis of variance followed by Tukey's honestly significant difference test (95% family-wise confidence level). Homogeneous subsets (samples not being different from each other, as indicated by the same letters on the bar plots (see Figs. 4 and 5)) were retrieved using the package multcompView. Samples with a p value of <0.05 were considered statistically different. Results are presented as means ± S.E.
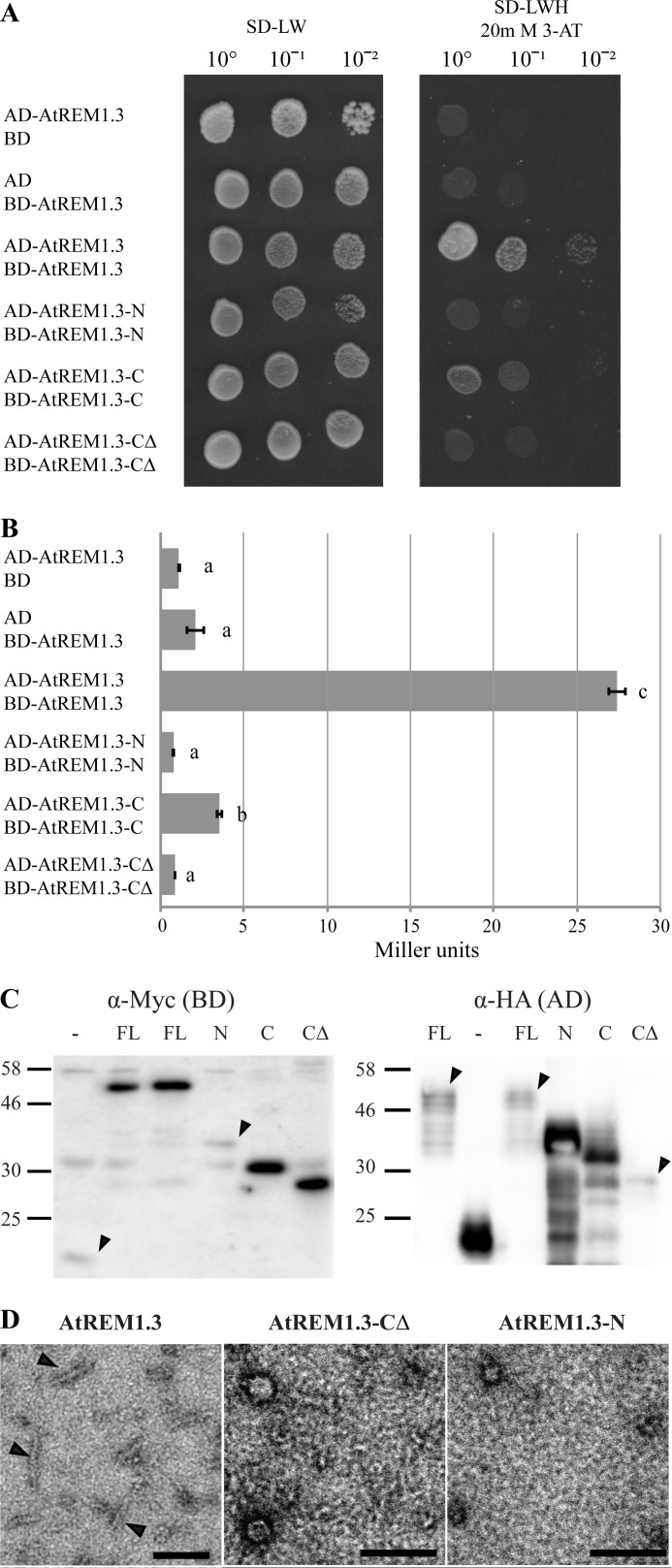
FIGURE 4.
Oligomerization assays for AtREM1.3 truncation variants. A, yeast two-hybrid interaction assay. Fusion proteins with GAL4-AD and -BD were expressed in yeast and assayed for their interaction on selective SD medium depleted in leucine (L), tryptophan (W), and histidine (H) and supplemented with 20 mm 3-amino-1,2,4-triazole to suppress residual histidine biosynthesis. Serial dilutions of yeast cells were grown to obtain single colonies. Empty pGBKT7::GW (BD) and pGADT7::GW (AD) were used as negative controls. B, quantitative β-galactosidase assay. β-Galactosidase activity was assayed using ortho-nitrophenyl-β-galactoside as substrate and measured in Miller units. Error bars, S.E. values from three independent biological replicates. Lowercase letters indicate different significance groups obtained by one-way analysis of variance. C, detection of fusion proteins expressed in yeast by Western blot. BD and AD fusion proteins were detected with α-Myc and α-HA monoclonal antibodies, respectively. Arrowheads, weakly expressed proteins. Molecular masses are indicated in kDa. FL, full-length AtREM1.3; N, AtREM1.3-N; C, AtREM1.3-C; CΔ, AtREM1.3-CΔ; −, empty vector control. D, recombinantly expressed and purified AtREM1.3 protein variants were investigated by TEM. Samples were negatively stained with 2% uranyl acetate. Arrowheads, filaments. Scale bar, 100 nm.
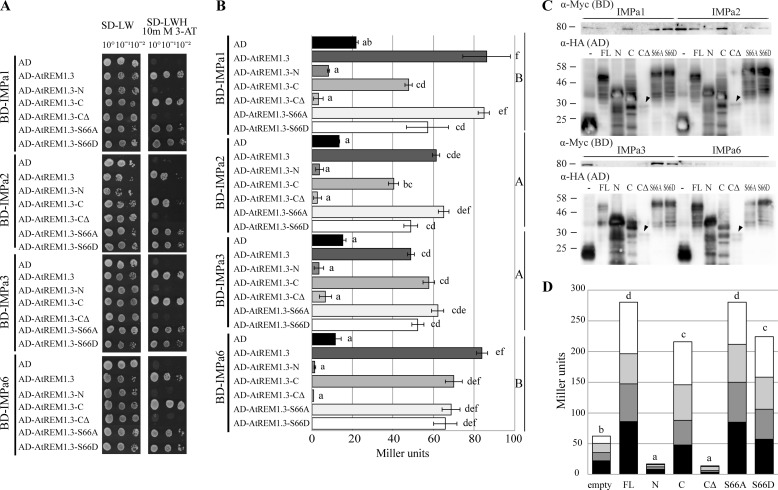
FIGURE 5.
ID N-terminal domain of AtREM1.3 facilitates interaction with importin α isoforms. A, yeast two-hybrid interaction assay. Importin α isoforms and AtREM1.3 truncation variants were fused to GAL4-BD and -AD, respectively. Fusion proteins were expressed in yeast and assayed for their interaction on SD−LWH selective medium supplemented with 10 mm 3-amino-1,2,4-triazole. Empty pGBKT7::GW (BD) and pGADT7::GW (AD) were used as negative controls. B, quantitative β-galactosidase assay. β-Galactosidase activity was assayed using ortho-nitrophenyl-β-galactoside as substrate and measured in Miller units. Error bars, S.E. values from 3–9 biological replicates. Lowercase and capital letters indicate different significance groups for each individual combination and for BD subgroups (importin α combination), respectively. C, detection of fusion proteins expressed in yeast by Western blot. BD and AD fusion proteins were detected with α-Myc and α-HA monoclonal antibodies, respectively. FL, full-length AtREM1.3; N, AtREM1.3-N; C, AtREM1.3-C; CΔ, AtREM1.3-CΔ; S66A, AtREM1.3-S66A; S66D, AtREM1.3-S66D; −, empty vector control. Arrowheads, weakly expressed proteins. Molecular masses are indicated in kDa. D, stacked column plot representation of yeast β-galactosidase activity grouped by AD domain fusion. Black, dark gray, light gray, and white bars, AtIMPa1-, AtIMPa2-, AtIMPa3-, and AtIMPa6-related activities, respectively. Lowercase letters indicate different significance groups.
Bimolecular Fluorescence Complementation and Confocal Microscopy
Agrobacterium infiltration of tobacco leaves was performed using Agrobacterium tumefaciens GV3101 carrying the constructs described before. Briefly, bacterial cultures were pelleted and resuspended in infiltration solution (10 mm MES-KOH, 10 mm MgCl2, 0.15 mm acetosyringone) to an A600 of 0.25. Cultures were mixed with an Agrobacterium strain harboring the silencing suppressor p19 (31). Nicotiana benthamiana leaves were infiltrated with the agrobacteria mix and analyzed 2 days postinfiltration. Imaging was performed with a spectral TCS SP5 MP confocal laser-scanning microscope (Leica Microsystems) with settings described earlier (2).
RESULTS
N-terminal Region of AtREM1.3 Is Intrinsically Disordered
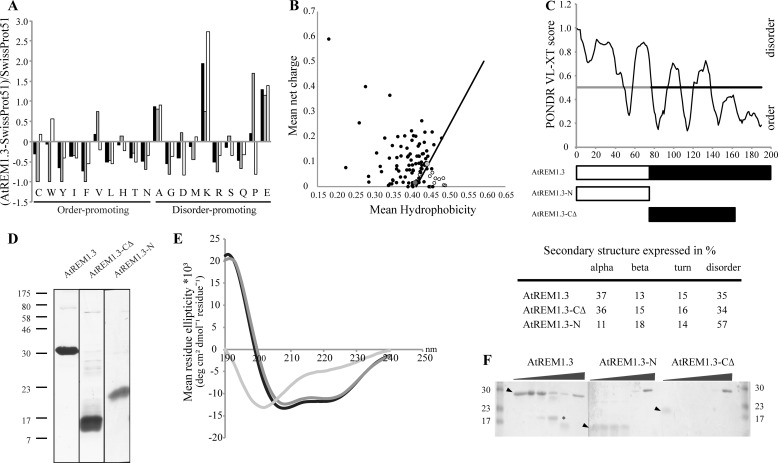
The N-terminal region (residues 1–77) of AtREM1.3 (At2g45820) shares no sequence similarity with any functionally characterized protein domain and is lowly conserved among remorin proteins. To gain insights into the structure-function characteristics of this region, we analyzed the AtREM1.3 amino acid composition using a composite profiler tool. This analysis shows that AtREM1.3 is depleted of order-promoting residues (Cys, Trp, Tyr, Ile, Phe, Leu, His, Thr, and Asn), whereas it is enriched in disorder-promoting residues (Ala, Lys, Pro, and Glu) compared with a set of random proteins (SwissProt version 51 database) (Fig. 1A). The abundance of charged residues (Lys and Glu) and the lack of hydrophobic residues (Trp, Tyr, and Phe) indicate a high net charge and a low mean hydrophobicity, respectively. Such a combination has been correlated with ID in proteins and thus can be used to discriminate intrinsically disordered from folded proteins (14). Investigation of AtREM1.3 in a charge-hydrophobicity phase space revealed a tendency toward ID, with AtREM1.3 being located in the border between order and disorder (Fig. 1B). Based on in silico analyses, this structural feature has recently been proposed to be a common feature of the remorin protein family (7). Indeed, a more detailed inspection of AtREM1.3 using the PONDR VL-XT predictor clearly indicates ID for the N-terminal region. In contrast, the C-terminal region is predicted to be mostly folded (Fig. 1C).
FIGURE 1.
The N-terminal region of AtREM1.3 is intrinsically disordered. A, composition profile of AtREM1.3 full-length (black), N-terminal (gray), and C-terminal (white) domains. Analysis was performed using the Composite Profiler tool and the SwissProt version 51 database as a background data set (13). Horizontal bars indicate order-promoting and disorder-promoting residues. B, charge-hydrophobicity phase space for AtREM1.3 (dark gray dot) and its N-terminal region (light gray dot). Black circles, intrinsically disordered proteins described in the literature (data taken partially from Ref. 14). White circles, natively folded proteins obtained from the Protein Data Bank. Solid black line, border between ID proteins and folded proteins, described by the formula, 〈R〉 = 2.785 〈H〉 − 1.151, where 〈R〉 and 〈H〉 are the mean net charge and the mean net hydrophobicity, respectively. C, PONDR VL-XT intrinsic disorder prediction and schematic representation of protein variants. White and black bars, N- and C-terminal regions, respectively. The gray line highlights the ID region. D, SDS-PAGE of AtREM1.3 and truncation variants. Proteins were separated on a 15% acrylamide gel and stained with Coomassie Brilliant Blue. Molecular masses of broad range protein marker (7–175 kDa) (New England Biolabs) are indicated in kDa. E, far-UV CD spectroscopy. CD spectra were recorded in 10 mm Tris/HCl buffer, pH 8.0, for the full-length AtREM1.3 (black), the AtREM1.3 N-terminal region (AtREM1.3-N; light gray), and the truncated AtREM1.3 C-terminal region (AtREM1.3-CΔ; dark gray) protein variants at a protein concentration of 0.1 mg/ml. The table below shows the secondary structure content expressed as a percentage. F, limited proteolysis with PK. Recombinantly expressed and purified AtREM1.3 and truncation variants (5 μg) were incubated with increasing amounts of PK (ratios 1:10−5, 1:10−4, 1:10−3, 1:10−2, and 1:10−1) for 30 min on ice. Fragments were resolved on a 15% SDS-PAGE. Arrowheads, non-digested protein. *, PK-resistant fragment that was subjected to mass spectrometry.
To experimentally verify this prediction, we recombinantly expressed and purified the full-length AtREM1.3(1–190) as well as truncated variants harboring the N-terminal region (AtREM1.3-N(1–77)) and part of the C-terminal region (AtREM1.3-CΔ(78–161)). The complete C-terminal region (residues 78–190) unfortunately could not be successfully expressed and purified. Interestingly, the C-terminal 28 residues of potato remorin StREM1.3 are responsible for plasma membrane attachment (6). Based on sequence comparisons, these 28 terminal residues are probably also required for AtREM1.3 plasma membrane attachment. Strep-tagged recombinant proteins were purified by affinity chromatography, and their purity (∼95%) was assessed by SDS-PAGE (Fig. 1D). The full-length protein as well as both truncation variants migrated abnormally in a denaturing acrylamide gel. Full-length AtREM1.3 (predicted mass, 22.8 kDa) ran as a globular 30-kDa protein, whereas AtREM1.3-N (predicted mass, 9.9 kDa) and AtREM1.3-CΔ (predicted mass, 11.5 kDa) ran at 23 and 17 kDa, respectively (Fig. 1D). The abnormally slow migration of AtREM1.3-N supports the presence of intrinsic disorder within this region (32, 33).
Next we used far-UV CD spectroscopy to analyze the secondary structure composition of the protein. The presence of α-helices is characterized by the presence of two distinct minima at 208 and 222 nm, as observed for the full-length AtREM1.3 (Fig. 1E). The percentage of α-helical structure in AtREM1.3-CΔ (36%) was comparable with values obtained for the full-length protein, indicating that the N-terminal region may partially gain helical structure upon protein oligomerization. In contrast, AtREM1.3-N showed low ellipticity at 190 nm and a minimum ellipticity at 200 nm, the characteristic pattern of a disordered conformation (57% random coil) (Fig. 1E, table).
To verify these findings by an independent method, we used limited proteolysis with PK, a protease that hydrolyzes flexible and therefore disordered regions. Incubation of purified AtREM1.3 with increasing concentrations of PK led to the accumulation of a stable fragment migrating at 17 kDa (Fig. 1F). Sequence determination by mass spectrometry of the proteolysis-resistant fragment resulted in the identification of residues 62–176, representing the majority of the C-terminal region. This suggests the presence of ID segments within the oligomerized full-length protein that are prone to PK cleavage. Furthermore, PK treatment of AtREM1.3-CΔ and AtREM1.3-N revealed that the ID N-terminal region is more susceptible to such a treatment (Fig. 1F). This supports our data obtained by CD spectroscopy and the idea that the N- and C-terminal regions constitute two independent domains.
The N-terminal Region Folds upon Incubation with TFE
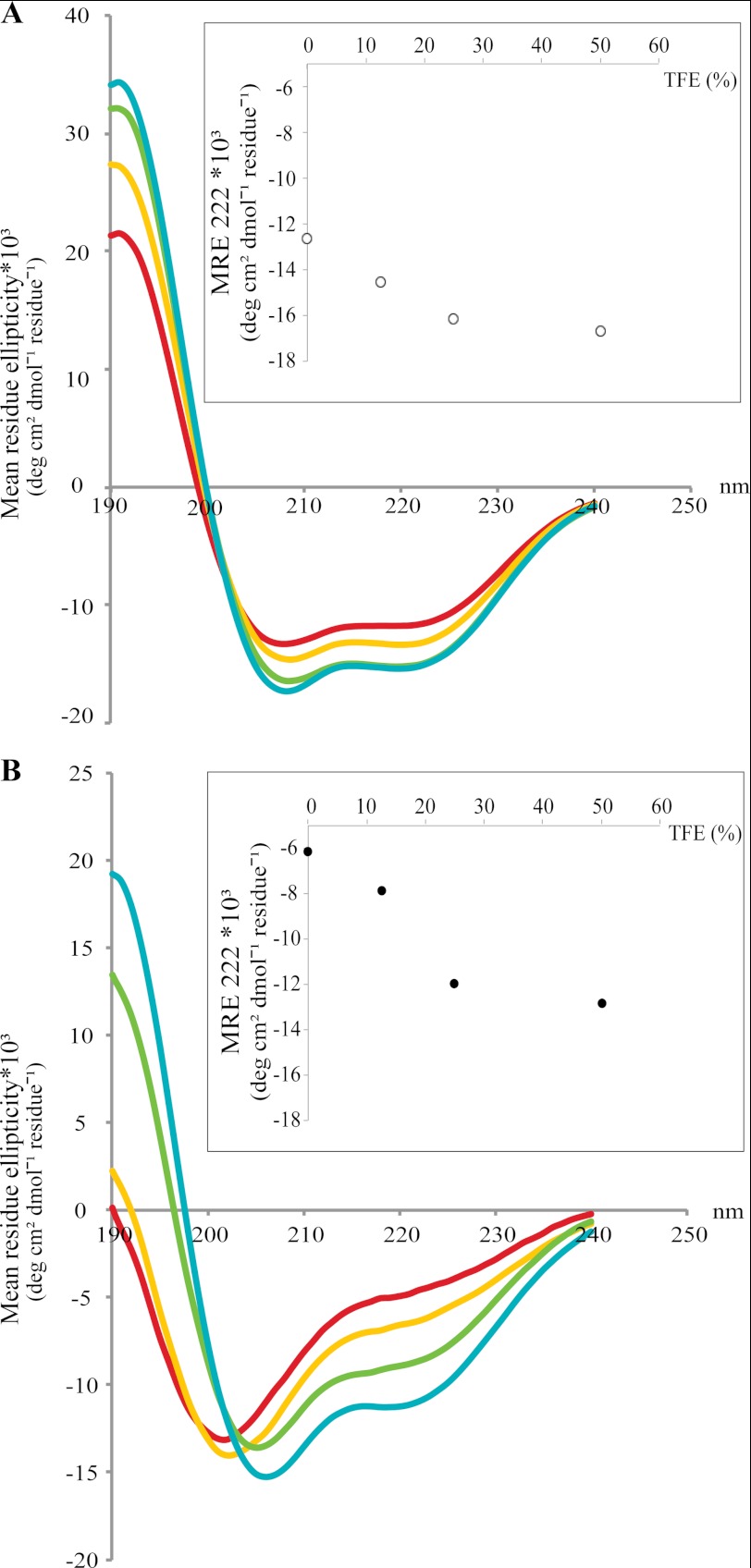
As ID regions often exhibit induced folding upon interaction with other proteins, we tested whether ordered structures can be induced within AtREM1.3 and AtREM1.3-N under conditions mimicking protein-protein interactions. Thus, we recorded structural changes by acquisition of far-UV CD spectra in the presence of TFE. This solvent resembles hydrophobic environments present in protein-protein interactions and therefore has been used to test the propensity of intrinsically disordered proteins to undergo an induced folding upon target binding (34). Incubation of the full-length protein with increasing TFE concentrations (0–50%) led to a moderate increase in α-helicity (Fig. 2A). A more pronounced effect was observed for AtREM1.3-N, indicated by the appearance of the distinct minima at 208 and 222 nm (Fig. 2B), with most of the disorder-to-order transition occurring at TFE concentrations of up to 25% (Fig. 2B, inset). These results clearly demonstrate folding propensity of the ID region and suggest α-helix formation upon interaction with binding partners.
FIGURE 2.
TFE induced folding of full-length AtREM1.3 and its N-terminal domain. Recombinantly expressed and purified full-length AtREM1.3 (A) and AtREM1.3-N (B) were subjected to far-UV CD spectroscopy with increasing TFE concentrations. The TFE percentage was as follows: 0% (red), 12.5% (yellow), 25% (green), and 50% (cyan). The mean residue ellipticity (MRE) at 222 nm versus TFE percentage is shown in the inset, illustrating that most of the disorder-to-order transition occurs up to 25% TFE.
The N-terminal Region of AtREM1.3 Is Required for AtREM1.3 Homo-oligomerization and Filament Formation in Vitro
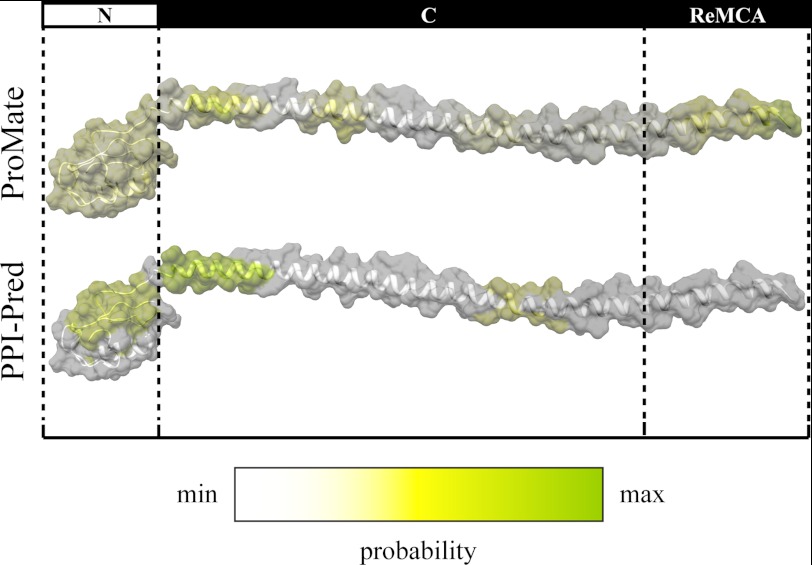
To predict if the ID N-terminal domain or other regions within the AtREM1.3 protein mediate protein interactions, we constructed an ab initio model of AtREM1.3 using the I-TASSER program. The model predicts an α-helical C-terminal domain and a vastly disordered N-terminal domain (Fig. 3), in accordance with the results obtained by CD and limited proteolysis. This model resembles a recently published homology model of the potato remorin StREM1.3, which also describes an α-helical C-terminal domain (6). Subsequently, we searched for regions within the model, which may be prone to mediate protein interactions using the PPI-Pred and ProMate programs. Both algorithms that are based on a support vector machine and probing of the surfaces of proteins using 10-Å radius spheres, respectively (21, 22), indicated that patches within the N- and C-terminal domains may contribute to protein interactions (Fig. 3).
FIGURE 3.
Ab initio model of AtREM1.3. The model was constructed using the I-Tasser program (19), and images were created with the UCSF Chimera package (23). Discontinuous lines mark the borders between the different protein regions. N, C, and ReMCA, N-terminal, C-terminal, and membrane-binding regions, respectively. Predictions of putative interaction surfaces were performed using the PPI-Pred (21) and ProMate programs (22). Interaction probability ranges from minimum (white) to maximum (green).
To verify these results experimentally we addressed whether the different domains contribute to oligomerization of AtREM1.3 using a yeast two-hybrid GAL4 system. In this system, the AD and the BD of the GAL4 transcription factor are split into two halves and fused to nuclear localization signals. Upon protein interaction in the nucleus, both domains get into proximity enabling the activation of a HIS3 reporter. This allows the yeast to complement a histidine auxotrophy and thus growth on histidine-depleted medium. Full-length AtREM1.3 as well as the AtREM1.3-N and AtREM1.3-CΔ variants were fused to the GAL4-AD and GAL4-BD. Additionally, a fusion protein comprising the complete C-terminal domain (AtREM1.3-C(78–190)) was constructed. In agreement with data reported for other remorin proteins (2), full-length AtREM1.3 homo-oligomerized (Fig. 4A). Additionally, only AtREM1.3-C (78–190) but neither AtREM1.3-N nor AtREM1.3-CΔ was able to interact with itself under these conditions (Fig. 4A). This indicates that residues critical for oligomerization reside within the C-terminal region and that the terminal 28 residues are essential for this interaction. To quantify the differences in interaction strength, we made use of a β-galactosidase activity assay. The β-galactosidase enzyme is encoded by the lacZ gene, which is transcriptionally activated by the GAL4 transcription factor. These experiments revealed that the ID N-terminal region also contributed to protein oligomerization, as shown by the significant reduction in the interaction strength of AtREM1.3-C compared with AtREM1.3 in the quantitative β-galactosidase assay (Fig. 4B). In all cases, expression of the respective fusion proteins was assessed by Western blot analyses to verify expression of the constructs (Fig. 4C).
To confirm these results by an independent method, we analyzed AtREM1.3 in vitro filament formation because this feature of group 1b remorins was reported earlier (10). For this, recombinantly expressed truncated variants were purified and inspected by TEM. As observed before, the full-length AtREM1.3 form filaments in vitro, whereas both AtREM1.3-N and AtREM1.3-CΔ truncation variants mostly form amorphous aggregates (Fig. 4D).
To get a more detailed understanding of this interaction, we studied the shape of the AtREM1.3 filaments using TEM, observing that a fraction of the filaments were twisted, which makes them not suitable for further analysis by high resolution cryo-EM. Additionally, we determined if AtREM1.3 filaments were amyloid in nature because several ID filament-forming proteins have this structural property (35). We performed a set of previously described experiments (36), which indicated the lack of any amyloid structure in AtREM1.3 filaments.
AtREM1.3 Interacts with Importin α Isoforms
It was previously shown that two group 2 remorin proteins from the legume plants Medicago truncatula (MtSYMREM1) and Lotus japonicus (LjSYMREM1) interact with at least three plasma membrane-resident receptor-like kinases (2, 37). In LjSYMREM1, this interaction is stabilized by the conserved C-terminal region, whereas two of the kinase domains phosphorylate residues in the putative ID N-terminal region of this protein (SYMREM1-N) (2). We currently hypothesize that the N-terminal region of SYMREM1 may be involved in the recruitment of cytoplasmic proteins to the plasma membrane. Because the intrinsically disordered N-terminal region of AtREM1.3 contributes to remorin oligomerization, we next asked if this region could also mediate interaction with other, non-membrane-resident proteins. To identify putative cytoplasmic interactors, we searched the Center for Cancer Systems Biology plant interactome database, which provides data from a high throughput yeast two-hybrid screen (38). Interestingly, these data not only confirmed homo- and hetero-oligomerization with itself and the closely related AtREM1.4 protein, respectively, but also yielded interactions with three importin α isoforms (AtIMPa1 (At3g06720), AtIMPa3 (At4g02150), and AtIMPa6 (At1g02690)). Importin α proteins are soluble receptors, which facilitate translocation of a broad spectrum of proteins across the nuclear envelope (reviewed in Refs. 39–41). Biologically, importin α proteins play roles in processes such as chromosome segregation, plant innate immunity, and nuclear envelope formation (42–45).
First, we verified these interactions between AtREM1.3 and importin α proteins independently. For this, AtIMPa1, AtIMPa3, and AtIMPa6 were amplified from a cDNA library and cloned into the above-mentioned DNA-binding domain yeast vectors. Indeed, all three IMPa proteins interacted with the full-length AtREM1.3 protein (Fig. 5A). Furthermore, we additionally found positive interaction with AtIMPa2 (At4g16143) (84% identity to AtIMPa1) (Fig. 5A), which did not interact with AtREM1.3 in the Center for Cancer Systems Biology Y2H screen. As described above, we additionally performed a β-galactosidase activity assay to quantify the interaction strength, showing that the interaction of AtREM1.3 with AtIMPa2 is comparable with the one with AtIMPa3 but weaker than with AtIMPa1 or AtIMPa6 (Fig. 5B). Interestingly, this does not correlate with the phylogenetic relation between importin α proteins (46).
The N-terminal Region of AtREM1.3 Facilitates Interaction with Importin α Isoforms
Next we addressed if the intrinsically disordered N-terminal region of AtREM1.3 mediates the interaction with IMPa proteins. For this, different truncated AtREM1.3 variants were co-expressed with AtIMPa1, AtIMPa2, AtIMPa3, and AtIMPa6 using the GAL4 system. Similar to AtREM1.3 homo-oligomerization, the C-terminal domain was found to be essential for the interaction with all tested importin α isoforms (Fig. 5A). However, the influence of the terminal 28 residues within this domain was difficult to assess because expression levels of the AD-AtREM1.3-CΔ construct were found to be highly reduced (Fig. 5C). Deletion of the N-terminal region led to a significant reduction in the interaction strength with AtIMPa1 and AtIMpa2 in the quantitative β-galactosidase assay (Fig. 5, B and D), indicating that the ID N-terminal region also contributed to interaction with the importin α proteins.
A prominent feature of remorin N-terminal regions is that they contain most of the phosphorylation sites of the highly phosphorylated remorin proteins (7). Across phylogenetic group 1b, Ser-66 (in AtREM1.3) is conserved and seems to be constitutively phosphorylated (7). Phosphoablative (AtREM1.3-S66A) and phosphomimetic (AtREM1.3-S66D) mutant variants of Ser-66 were created and used to assess the possible impact of phosphorylation within the ID N-terminal domain in the interaction with importin α proteins. Quantitative β-galactosidase assays showed a tendency to reduced interaction strength of AtIMpa1 and AtIMpa2 with the phosphomimetic variant. Interestingly, this reduction is comparable with the reduction observed in the variant devoid of the N-terminal domain (Fig. 5, B and D). In contrast, the phosphoablative variant showed no difference compared with the full-length protein. In all cases, expression of the respective fusion proteins was assessed by Western blot analyses (Fig. 5C). In summary, these experiments indicate that the ID N-terminal region of AtREM1.3 quantitatively contributes to the interaction with IMPa proteins and may thus serve regulatory functions.
Ectopic Expression of IMPa Proteins Leads to Subcellular Redistribution of AtREM1.3
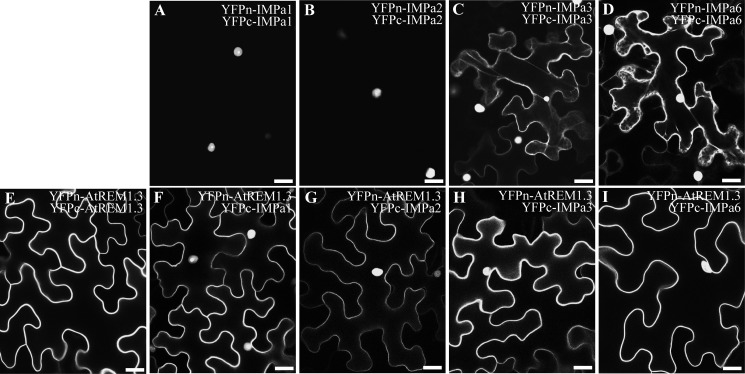
In a last set of experiments, we asked whether cellular translocation of at least one of the partners occurs during the interaction between nuclear IMPa proteins, and plasma membrane-resident AtREM1.3 indeed occurs in planta. To determine this, we used bimolecular fluorescence complementation, where the N- (YFPn) and C-terminal (YFPc) regions of YFP are individually fused to the proteins of interest and expressed in different combinations in N. benthamiana leaves for 2 days. The interaction between proteins results in the reassembly of the functional YFP protein and thus in fluorescence at the sites of interaction. Here we made use of the fact that both YFP halves still show a tendency to autoassemble a functional fluorophore during overexpression and thus potentially stabilize protein complexes. This allowed us to better visualize changes in cellular distributions of the proteins. Co-expression of IMPa1 and IMPa2 resulted in almost exclusive nuclear fluorescence (Fig. 6, A and B), whereas an additional cytosolic signal was observed for IMPa3 and IMPa6, indicated by distinct cytoplasmic strands that traverse the large central vacuole (Fig. 6, C and D). In contrast and as expected, the fluorescent signal of AtREM1.3 homo-oligomers resided exclusively in the periphery of the cell, indicating plasma membrane localization of the proteins (Fig. 6E). Interestingly, co-expression of AtREM1.3 with any of the four IMPa proteins resulted in a signal inside the nucleus and the plasma membrane (Fig. 6, F–I). In none of these cases were cytoplasmic strands observed, clearly demonstrating that the fluorescence signal did not derive from any YFP cleavage product. These data show for the first time that remorins can be translocated from the plasma membrane into the nucleus.
FIGURE 6.
AtREM1.3 and importin α isoforms are translocated and interact in planta. YFPn and YFPc were fused to the N terminus of AtREM1.3 and importin α proteins, and their subcellular localization and interaction were assessed by bimolecular fluorescence complementation in N. benthamiana leaf epidermal cells. IMPa1 (A) and IMPa2 (B) proteins interact mostly in the nucleus, whereas additional cytoplasmic localization was observed with IMPa3 (C) and IMPa6 (D), as indicated by cytoplasmic strands. AtREM1.3 homo-oligomerizes exclusively at the plasma membrane (E). However, co-expression with importin α proteins reconstitutes YFP fluorescence in the cell periphery and in the nucleus (F–I). Scale bars, 25 μm.
DISCUSSION
Plants require efficient signaling cascades to rapidly respond to environmental cues. It has been predicted that around 8% of the total A. thaliana proteome contains completely intrinsically disordered proteins. Moreover, it has been estimated that 29% of the A. thaliana proteome is composed of ID regions that are 50 residues or longer (47). However, only a few plant-signaling components have been experimentally characterized with respect to their ID. Examples of ID proteins are the A. thaliana ERD10 and ERD14 dehydrin proteins (48, 49), the N-terminal region of the A. thaliana Hy5 bZIP transcription factor involved in photomorphogenesis (50), and the Hordeum vulgare (barley) senescence-associated NAC transcription factor (33). In the case of the Alb3 membrane insertase (51) and the DELLA proteins (34), their ID regions mediate protein interactions.
Plant-specific remorin proteins have been proposed to be involved in plant-microbe cell signaling, where the group 2 symbiotic remorins LjSYMREM1 and MtSYMREM1 were shown to interact with the plasma membrane-resident symbiotic receptor-like kinases (NFR5, NFR1, and SYMRK in L. japonicus and NFP, LYK3, and DMI2 in M. truncatula) via their conserved and α-helical C-terminal regions (2, 37). Here we experimentally demonstrated that the N-terminal domain of the remorin protein AtREM1.3 is intrinsically disordered (Figs. 1 and 2). This experimentally proves previous predictions of ID in the N-terminal regions of diverse remorin proteins (7). A key functional advantage over structurally ordered proteins is that ID regions can bind a number of partners with high specificity and low affinity (1). This high flexibility possibly explains the lack of interaction of the N-terminal domain alone because stability is one of the requirements to detect interactions with yeast two-hybrid systems (Figs. 4 and 5). ID regions achieve high specificity when they undergo binding-induced folding. This is characterized by the presence of short rigid segments located within disordered regions termed molecular recognition features (52, 53). A potential molecular recognition feature is located in the ID N-terminal region, as indicated by the AtREM1.3 disorder profile, where a region of order within disorder is predicted (Fig. 1). Furthermore, this ordered segment is conserved among all group 1b remorins (7). TFE-induced formation of α-helical structure also indicates possible induced folding of the ID N-terminal domain of AtREM1.3 upon protein-protein interactions (Fig. 2) and therefore putative functional contributions to this process.
Such a hypothesis is supported by data obtained by fluorescence lifetime imaging microscopy and in vitro kinase assays that clearly indicate the involvement of the putative ID N-terminal region of LjSYMREM1 in the interaction with the receptor-like kinases (2). Similarly, deletion of the ID N-terminal region of AtREM1.3 results in reduced interaction strength with IMPa1 and IMPa2 (Fig. 5B). Furthermore, constitutive phosphorylation of Ser-66 decreases the interaction with importin α proteins, implying that phosphorylation of AtREM1.3 in its ID N-terminal domain could be a mechanism of controlling protein interactions with cytosolic proteins. These data are in agreement with previous reports (54–57), which describe that phosphorylation clusters preferentially within ID regions and often mediates protein interactions. For example, 14-3-3 proteins can discriminate between phosphorylated and non-phosphorylated partners (58). Interaction of a 14-3-3 protein with the RNA-binding K-homology splicing regulator protein (KSRP) is promoted by phosphorylation of the latter, which drives relocalization of KSRP into the nucleus, where it performs its mRNA degrading activity (59). In other cases, phosphorylation of residues within ID regions mediates stimulus-specific degradation, as in the case of plant GRAS transcription factors (60, 61).
Here we confirmed putative interactions between AtREM1.3 and members of the importin α family that were originally identified in a large unbiased yeast two-hybrid screen (38). Importin α proteins are soluble receptors involved in nuclear trafficking. In A. thaliana, this family comprises 9 members, some of which are involved in interaction with VirD2 and VirE2 bacterial T-complex proteins and mediate nuclear localization of these bacterial proteins (46). Moreover, AtIMPa3 (MOS6) plays a role in plant innate immunity because it is a suppressor of resistance responses and immunity to pathogens (42), further supporting possible roles of remorin proteins during plant-microbe interactions and innate immunity. Although the C-terminal domain of AtREM1.3 predominantly mediates stable oligomerization and interaction with importin α proteins, deletion of the ID N-terminal domain reduces significantly the strength of these interactions (Fig. 5). These data underline putative regulatory functions of ID domains in remorin proteins during protein-protein interactions.
Supplementary Material
Acknowledgments
We kindly thank Axel Strauss for help in statistical analyses, Petra Wendler (Gene Center of the Ludwig-Maximilians-University of Munich, Germany) for a contribution to the high-resolution TEM analysis, Marcus Fändrich and Senthil Kumar (Max Planck Research Unit for Enzymology of Protein Folding, Halle, Germany) for amyloid determination, and Susan Urbanus for critical reading of the manuscript.
This work was supported by Collaborative Research Center (Sonderforschungsbereich) SFB924 and Emmy Noether Program Grant OT423/2-1 funded by the Deutsche Forschungsgemeinschaft.

This article contains supplemental Tables S1 and S2.
- ID
- intrinsic disorder
- AD
- activation domain
- BD
- DNA-binding domain
- GW
- Gateway
- PK
- proteinase K
- SD
- synthetic defined
- TEM
- transmission electron microscopy
- TFE
- 2,2,2-trifluoroethanol.
REFERENCES
- 1. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 2. Tóth K., Stratil T. F., Madsen E. B., Ye J., Popp C., Antolín-Llovera M., Grossmann C., Jensen O. N., Schüssler A., Parniske M., Ott T. (2012) Functional domain analysis of the remorin protein LjSYMREM1 in Lotus japonicus. PLoS One 7, e30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reymond P., Kunz B., Paul-Pletzer K., Grimm R., Eckerskorn C., Farmer E. E. (1996) Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell 8, 2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Farmer E. E., Pearce G., Ryan C. A. (1989) In vitro phosphorylation of plant plasma membrane proteins in response to the proteinase inhibitor inducing factor. Proc. Natl. Acad. Sci. U.S.A. 86, 1539–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raffaele S., Mongrand S., Gamas P., Niebel A., Ott T. (2007) Genome-wide annotation of remorins, a plant-specific protein family. Evolutionary and functional perspectives. Plant Physiol. 145, 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perraki A., Cacas J. L., Crowet J. M., Lins L., Castroviejo M., German-Retana S., Mongrand S., Raffaele S. (2012) Plasma membrane localization of StREM1.3 remorin is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of PVX movement. Plant Physiol. 160, 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marín M., Ott T. (2012) Phosphorylation of intrinsically disordered regions in remorin proteins. Front. Plant Sci. 3, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reymond P., Grünberger S., Paul K., Müller M., Farmer E. E. (1995) Oligogalacturonide defense signals in plants. Large fragments interact with the plasma membrane in vitro. Proc. Natl. Acad. Sci. U.S.A. 92, 4145–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raffaele S., Bayer E., Lafarge D., Cluzet S., German Retana S., Boubekeur T., Leborgne-Castel N., Carde J. P., Lherminier J., Noirot E., Satiat-Jeunemaître B., Laroche-Traineau J., Moreau P., Ott T., Maule A. J., Reymond P., Simon-Plas F., Farmer E. E., Bessoule J. J., Mongrand S. (2009) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bariola P. A., Retelska D., Stasiak A., Kammerer R. A., Fleming A., Hijri M., Frank S., Farmer E. E. (2004) Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol. Biol. 55, 579–594 [DOI] [PubMed] [Google Scholar]
- 11. Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 12. Jarsch I. K., Ott T. (2011) Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol. Plant Microbe Interact. 24, 7–12 [DOI] [PubMed] [Google Scholar]
- 13. Vacic V., Uversky V. N., Dunker A. K., Lonardi S. (2007) Composition Profiler. A tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics 8, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uversky V. N., Gillespie J. R., Fink A. L. (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 15. Prilusky J., Felder C. E., Zeev-Ben-Mordehai T., Rydberg E. H., Man O., Beckmann J. S., Silman I., Sussman J. L. (2005) FoldIndex. A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21, 3435–3438 [DOI] [PubMed] [Google Scholar]
- 16. Romero, Obradovic, Dunker K. (1997) Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. Ser. Workshop Genome Inform. 8, 110–124 [PubMed] [Google Scholar]
- 17. Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., Dunker A. K. (2001) Sequence complexity of disordered protein. Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 18. Li X., Romero P., Rani M., Dunker A. K., Obradovic Z. (1999) Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform. Ser. Workshop Genome Inform. 10, 30–40 [PubMed] [Google Scholar]
- 19. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bradford J. R., Westhead D. R. (2005) Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics 21, 1487–1494 [DOI] [PubMed] [Google Scholar]
- 22. Neuvirth H., Raz R., Schreiber G. (2004) ProMate. A structure based prediction program to identify the location of protein-protein binding sites. J. Mol. Biol. 338, 181–199 [DOI] [PubMed] [Google Scholar]
- 23. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 24. Morbitzer R., Elsaesser J., Hausner J., Lahaye T. (2011) Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 39, 5790–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shenoy A. R., Visweswariah S. S. (2003) Site-directed mutagenesis using a single mutagenic oligonucleotide and DpnI digestion of template DNA. Anal. Biochem. 319, 335–336 [DOI] [PubMed] [Google Scholar]
- 26. Whitmore L., Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy. Methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 27. Whitmore L., Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lobley A., Whitmore L., Wallace B. A. (2002) DICHROWEB. An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18, 211–212 [DOI] [PubMed] [Google Scholar]
- 29. Miller J. H. (1974) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. R Core Team (2012) R. A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna [Google Scholar]
- 31. Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 [DOI] [PubMed] [Google Scholar]
- 32. Uversky V. N., Dunker A. K. (2010) Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kjaersgaard T., Jensen M. K., Christiansen M. W., Gregersen P., Kragelund B. B., Skriver K. (2011) Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J. Biol. Chem. 286, 35418–35429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun X., Jones W. T., Harvey D., Edwards P. J., Pascal S. M., Kirk C., Considine T., Sheerin D. J., Rakonjac J., Oldfield C. J., Xue B., Dunker A. K., Uversky V. N. (2010) N-terminal domains of DELLA proteins are intrinsically unstructured in the absence of interaction with GID1/gibberellic acid receptors. J. Biol. Chem. 285, 11557–11571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uversky V. N. (2008) Amyloidogenesis of natively unfolded proteins. Curr. Alzheimer Res. 5, 260–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Habicht G., Haupt C., Friedrich R. P., Hortschansky P., Sachse C., Meinhardt J., Wieligmann K., Gellermann G. P., Brodhun M., Götz J., Halbhuber K. J., Röcken C., Horn U., Fändrich M. (2007) Directed selection of a conformational antibody domain that prevents mature amyloid fibril formation by stabilizing Aβ protofibrils. Proc. Natl. Acad. Sci. U.S.A. 104, 19232–19237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lefebvre B., Timmers T., Mbengue M., Moreau S., Hervé C., Tóth K., Bittencourt-Silvestre J., Klaus D., Deslandes L., Godiard L., Murray J. D., Udvardi M. K., Raffaele S., Mongrand S., Cullimore J., Gamas P., Niebel A., Ott T. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Importin α. A multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 40. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) Classical nuclear localization signals. Definition, function, and interaction with importin α. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Merkle T. (2011) Nucleo-cytoplasmic transport of proteins and RNA in plants. Plant Cell Rep. 30, 153–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palma K., Zhang Y., Li X. (2005) An importin α homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15, 1129–1135 [DOI] [PubMed] [Google Scholar]
- 43. Askjaer P., Galy V., Hannak E., Mattaj I. W. (2002) Ran GTPase cycle and importins α and β are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell 13, 4355–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Loeb J. D., Schlenstedt G., Pellman D., Kornitzer D., Silver P. A., Fink G. R. (1995) The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. U.S.A. 92, 7647–7651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yano R., Oakes M. L., Tabb M. M., Nomura M. (1994) Yeast Srp1p has homology to armadillo/plakoglobin/β-catenin and participates in apparently multiple nuclear functions, including the maintenance of the nucleolar structure. Proc. Natl. Acad. Sci. U.S.A. 91, 6880–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhattacharjee S., Lee L. Y., Oltmanns H., Cao H., Veena, Cuperus J., Gelvin S. B. (2008) IMPa-4, an Arabidopsis importin α isoform, is preferentially involved in agrobacterium-mediated plant transformation. Plant Cell 20, 2661–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dunker A. K., Obradovic Z., Romero P., Garner E. C., Brown C. J. (2000) Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 11, 161–171 [PubMed] [Google Scholar]
- 48. Mouillon J. M., Gustafsson P., Harryson P. (2006) Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol. 141, 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kovacs D., Kalmar E., Torok Z., Tompa P. (2008) Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoon M. K., Shin J., Choi G., Choi B. S. (2006) Intrinsically unstructured N-terminal domain of bZIP transcription factor HY5. Proteins 65, 856–866 [DOI] [PubMed] [Google Scholar]
- 51. Falk S., Ravaud S., Koch J., Sinning I. (2010) The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J. Biol. Chem. 285, 5954–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oldfield C. J., Cheng Y., Cortese M. S., Romero P., Uversky V. N., Dunker A. K. (2005) Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry 44, 12454–12470 [DOI] [PubMed] [Google Scholar]
- 53. Mohan A., Oldfield C. J., Radivojac P., Vacic V., Cortese M. S., Dunker A. K., Uversky V. N. (2006) Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 [DOI] [PubMed] [Google Scholar]
- 54. Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., Dunker A. K. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chrivia J. C., Kwok R. P., Lamb N., Hagiwara M., Montminy M. R., Goodman R. H. (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859 [DOI] [PubMed] [Google Scholar]
- 56. Pawson T. (2004) Specificity in signal transduction. From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 [DOI] [PubMed] [Google Scholar]
- 57. Bustos D. M. (2012) The role of protein disorder in the 14-3-3 interaction network. Mol. Biosyst. 8, 178–184 [DOI] [PubMed] [Google Scholar]
- 58. Liang X., Van Doren S. R. (2008) Mechanistic insights into phosphoprotein-binding FHA domains. Acc. Chem. Res. 41, 991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Díaz-Moreno I., Hollingworth D., Frenkiel T. A., Kelly G., Martin S., Howell S., García-Mayoral M., Gherzi R., Briata P., Ramos A. (2009) Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14–3-3 binding. Nat. Struct. Mol. Biol. 16, 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fu X., Richards D. E., Ait-Ali T., Hynes L. W., Ougham H., Peng J., Harberd N. P. (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14, 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hussain A., Cao D., Cheng H., Wen Z., Peng J. (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J. 44, 88–99 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.