Background: Catalytic mechanism for acetyltransferase activity and its regulation in M. tuberculosis GlmU (GlmUMtb) is not addressed comprehensively.
Results: We present these and features unique to GlmUMtb.
Conclusion: Unique features include a short helix that presents Trp-460 for substrate binding, distinctive acetyl-CoA conformation and regulation upon PknB-mediated phosphorylation.
Significance: These insights may be exploited for designing selective inhibitors against GlmUMtb.
Keywords: Acetyl Coenzyme A, Crystal Structure, Phosphorylation, Protein Kinases, Tuberculosis, GlmU, Acetyltransferase
Abstract
N-Acetyl-glucosamine-1-phosphate uridyltransferase (GlmU), a bifunctional enzyme involved in bacterial cell wall synthesis is exclusive to prokaryotes. GlmU, now recognized as a promising target to develop new antibacterial drugs, catalyzes two key reactions: acetyl transfer and uridyl transfer at two independent domains. Hitherto, we identified GlmU from Mycobacterium tuberculosis (GlmUMtb) to be unique in possessing a 30-residue extension at the C terminus. Here, we present the crystal structures of GlmUMtb in complex with substrates/products bound at the acetyltransferase active site. Analysis of these and mutational data, allow us to infer a catalytic mechanism operative in GlmUMtb. In this SN2 reaction, His-374 and Asn-397 act as catalytic residues by enhancing the nucleophilicity of the attacking amino group of glucosamine 1-phosphate. Ser-416 and Trp-460 provide important interactions for substrate binding. A short helix at the C-terminal extension uniquely found in mycobacterial GlmU provides the highly conserved Trp-460 for substrate binding. Importantly, the structures reveal an uncommon mode of acetyl-CoA binding in GlmUMtb; we term this the U conformation, which is distinct from the L conformation seen in the available non-mycobacterial GlmU structures. Residues, likely determining U/L conformation, were identified, and their importance was evaluated. In addition, we identified that the primary site for PknB-mediated phosphorylation is Thr-418, near the acetyltransferase active site. Down-regulation of acetyltransferase activity upon Thr-418 phosphorylation is rationalized by the structures presented here. Overall, this work provides an insight into substrate recognition, catalytic mechanism for acetyl transfer, and features unique to GlmUMtb, which may be exploited for the development of inhibitors specific to GlmU.
Introduction
Tuberculosis is a major threat to human health. For therapeutic intervention, enzymes involved in the formation of the cell wall of Mycobacterium tuberculosis are promising targets, due to the complex and specialized nature of the cell wall, which consists mainly of lipopolysaccharides and mycolic acid.
GlmU, a prokaryotic enzyme conserved both in Gram-positive and Gram-negative bacteria, is involved in the biosynthesis of UDP-N-acetylglucosamine 1-phosphate (UDP-GlcNAc)4 (1). It is a bifunctional protein with two independent active sites catalyzing acetyl transfer and uridyl transfer reactions on glucosamine 1-phosphate (GlcN-1-P). It synthesizes two key intermediates of cell wall biosynthesis pathways, viz. N-Acetylglucosamine 1-phosphate (GlcNAc-1-P) and UDP-GlcNAc. The acetyl transfer reaction catalyzed by the C-terminal domain involves the transfer of the acetyl group from acetyl-CoA to the amine group of GlcN-1-P to produce GlcNAc-1-P (2). GlcNAc-1-P then diffuses to the N-terminal domain of the protein, where it is activated to form UDP-GlcNAc by the transfer of UMP from UTP. UDP-GlcNAc, the end product, is utilized for synthesizing lipopolysaccharide and peptidoglycan components of the cell wall. Given this important role for GlmU, it is not surprising that it is essential for the growth of M. tuberculosis and Mycobacterium smegmatis (3, 4). Hence, understanding the catalytic mechanisms employed by GlmUMtb would be valuable toward the discovery of inhibitors targeting cell wall biosynthesis in mycobacteria. Elucidating the acetyl transfer reaction catalyzed by the C-terminal domain is the focus of the work presented here.
Several structures of GlmU and their homologues were determined to understand the reactions catalyzed by this enzyme. The structures of GlmU from Escherichia coli (GlmUEc) (5, 6) and Streptococcus pneumoniae (GlmUSp) (7) bound to acetyl-CoA at the C-terminal acetyl transferase active site are available. Determining the structures of GlmUMtb bound to substrates of the acetyl transfer reaction was important. Attempts to obtain acetyl-CoA bound crystals of GlmUMtb by co-crystallization or by soaking acetyl-CoA with the apo crystals were unsuccessful (8, 9) until UDP-GlcNAc was supplied along with. Here, we present two structures of GlmUMtb, one in complex with acetyl-CoA and another in complex with CoA and GlcN-1-P. These allowed identifying likely catalytic residues and together with mutational data, we infer a reaction mechanism for the acetyl transfer reaction. Additionally, features unique and highly conserved in the Mycobacteriaceae family could be noted from these studies. Of these, a short helix present at the C-terminal extension provides a key tryptophan, which appears critical for acetyl-CoA binding. Besides, an uncommon mode of binding of the substrate acetyl-CoA was identified to be a feature unique to GlmU from mycobacteria. In addition, we identified that the major target site on GlmUMtb where PknB phosphorylates is Thr-418; we address how this phosphorylation down-regulates acetyltransferase activity. Overall, a better understanding of the reaction mechanism for acetyl transfer and its regulation is important. This and the unique features reported here, would aid elucidating structure-function relationships in the enzyme, which we anticipate may be gainfully employed for drug design.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Restriction/modification enzymes were obtained from New England Biolabs. Cloning and expression vectors pENTR/directional TOPO cloning kit (Invitrogen), pQE2 (Qiagen), were purchased from the respective sources. Oligonucleotide primers and analytical grade chemicals were purchased from Sigma. [γ-32P]ATP (6000Ci/mmol) was purchased from PerkinElmer Life Sciences. Malachite green phosphate assay kit (POMG-25H) was purchased from BioAssay System (Gentaur).
Cloning, Mutagenesis, and Purification of E. coli and M. tuberculosis GlmU
Cloning of GlmUMtb was described previously (10). Site-directed mutants were generated by PCR amplifications using specific primers (supplemental table S1) followed by cloning into pQE-2 expression vector. Nucleotide sequences of all the clones generated were verified by DNA sequencing. Expression and purification of GlmUMtb-WT and GlmUMtb mutants as described in a previous work (9).
Acetyl and Uridyltransferase Assays
Uridyltranferase assays were performed in an 8-μl volume containing 25 mm HEPES buffer (pH 7.6), 10 mm MgCl2, 1 mm DTT with 0.5 mm GlcNAc-1-P and 1 mm UTP, 0.04 units of thermostable inorganic pyrophosphatase and various concentrations of GlmUMtb or GlmUMtb mutants for 30 min at 30 °C. Reactions were terminated by incubating at 65 °C for 30 min, and the product formed was estimated by malachite green phosphate assay kit according to the manufacturer's instructions. The acetyltransferase activity of GlmUMtb was monitored in a coupled assay, and the end product was detected with the help of malachite green assay kit. Briefly, the 8-μl reaction acetyltransferase assay mix contained 25 mm HEPES buffer (pH 7.6), 10 mm MgCl2, 1 mm DTT with 1 mm GlcN-1-P, 1 mm acetyl-CoA. Subsequent to 30 min incubation at 30 °C, reactions were terminated at 65 °C for 30 min. The product, GlcNAc-1-P, was estimated by performing coupled uridyltransferase assay in a reaction containing 0.04 units of thermostable inorganic pyrophosphatase, 1 mm UTP, and 20 pmol of GlmUMtb:1–352 deletion mutant (10), which was estimated with the help of malachite green phosphate assay kit.
Determining the Kinetic Parameters (Km and Vmax) for the Acetyltransferase Activity
To determine the kinetic parameters, either acetyl-CoA or GlcN-1-P was varied keeping the other substrate at 1 mm. Concentration of the enzyme GlmUMtb or the active site mutants used for the reaction was decided based on the range finding experiments. The reactions were performed for 30 min at 30 °C followed by termination by heating it at 65 °C for 30 min. The product GlcNAc-1-P formed was measured by coupling this reaction with uridyltransferase assay as described above. Unit activity of the enzyme was defined as the μm of product formed/min/pmol of enzyme. GraphPad prism software was used for non-linear regression analysis while evaluating the Km and Vmax values.
Protein Crystallization
Crystals were obtained by mixing 400 nl of 15 mg/ml GlmUMtb, 5 mm acetyl-CoA, 5 mm MgCl2, 5 mm UDP-GlcNAc with 400 nl of 18% PEG 3350, 0.1 m Tris-Cl, pH 8.5, and 2% tacsimate in a sitting drop vapor diffusion set up. Crystals appeared in 4–5 days and grew to a size suitable for diffraction in 7–8 days. Hereafter, these crystals, obtained in the presence of acetyl-CoA, will be referred as GlmUMtb(AcCoA). Crystals in the presence of CoA and GlcN-1-P were obtained in two ways. First was by soaking GlmUMtb(AcCoA) crystals in 5 mm GlcN-1-P, 5 mm MgCl2, 5 mm UDP-GlcNAc, 5 mm acetyl-CoA, 18% PEG 3350, 0.1 m Tris-Cl, pH 8.5, and 2% tacsimate. The second method employed was co-crystallizing GlmUMtb with 5 mm GlcNAc-1-P, 5 mm MgCl2, 5 mm UDP-GlcNAc, 5 mm CoA in the conditions mentioned for obtaining GlmUMtb(AcCoA) crystals. Hereafter, these crystals will be referred as GlmUMtb(CoA:GlcN-1-P). For collecting data sets, crystals were transferred to a cryoprotectant solution containing 22% ethylene glycol in mother liquor for ∼5 s and were then snap frozen and stored in liquid nitrogen.
Data Collection and Processing
X-ray diffraction data were collected for GlmUMtb(AcCoA) and GlmUMtb(CoA:GlcN-1-P) crystals at Department of Biotechnology supported facility, Beam line BM14 of European Synchrotron Radiation Facility (Grenoble, France). A complete data set for GlmUMtb(AcCoA) was collected at 2.33 Å. Similarly, complete datasets for GlmUMtb(CoA:GlcN-1-P) crystals were collected at 1.98 Å. The data sets were indexed and scaled with XDS (11). Because the structure of GlmUMtb bound to UDP-GlcNAc (Protein Data Bank code 3DJ4) was available, molecular replacement technique was employed to obtain phase information, and consequently, the structures were determined. For this, the search model, the indexed data sets for GlmUMtb(AcCoA) and GlmUMtb(CoA:GlcN-1-P) were provided to the Auto-Rickshaw server (12). The partially refined models obtained from the Auto-Rickshaw server for GlmUMtb(AcCoA) and GlmUMtb(CoA:GlcN-1-P) were completed by several rounds of manual model building in COOT (13) and were refined with Refmac (14). The positive electron density at the active site was modeled with appropriate ligand molecules acetyl-CoA/CoA, UDP-GlcNAc, and/or GlcN-1-P. After several rounds of refinement, the final structures were examined for proper geometry and deposited in the Protein Data Bank (codes 3ST8 and 3SPT). The final statistics are given in Table 1.
TABLE 1.
X-ray data collection and refinement statistics
| GlmUMtb(AcCoA) | GlmUMtb(CoA:GlcN-1-P) | |
|---|---|---|
| Data collection | ||
| Space group | H32 | H32 |
| Cell dimensions | a = 110.19, b = 110.19, and c = 361.29 Å; α = β = 90°, γ = 120° | a = 110.312, b = 110.312, and c = 360.537; α = β = 90°, γ = 120° |
| Resolution (Å) | 2.33 (2.33–2.47) | 1.98 (1.98–2.10) |
| Rsym or Rmerge (%) | 6.6 (29.3) | 5.8 (22.3) |
| I/σI | 24.97 (5.54) | 29.7 (6.89) |
| Completeness (%) | 99.8 (99.0) | 99.8 (99.4) |
| Redundancy | 7.34 | 7.35 |
| Refinement | ||
| Resolution (Å) | 19.68–2.33 | 19.71–1.98 |
| No. of reflections | 35,443 | 57,748 |
| Rwork/Rfree | 0.192/0.223 | 0.1674/0.202 |
| No. of Atoms | ||
| Protein | 3471 | 3479 |
| Ligand/ion | 93 | 107 |
| Water | 212 | 529 |
| B-factors | ||
| Protein | 31.05 | 24.13 |
| Ligand/ion | 28.7 | 24.53 |
| Water | 39.1 | 41.45 |
| r.m.s.d.a | ||
| Bond lengths (Å) | 0.0257 | 0.034 |
| Bond angles | 2.3533° | 2.678° |
a r.m.s.d., root mean square deviation.
In Vitro Kinase Assays and Phosphopeptide Mapping
In vitro kinase assays were performed by incubating 5 pmol of PknB-KD (1–330 amino acids) and 25 pmol of GlmUMtb or GlmUMtb-T418A in a 30-μl reaction in 1× kinase buffer (25 mm HEPES (pH 7.6), 10 mm MnCl2, 1 mm DTT) and 10 μCi of [γ-32P]ATP for 30 min at 30 °C. Samples were resolved on 10% SDS-PAGE, transferred to nitrocellulose membranes, and subjected to autoradiography. Bands corresponding to the phosphorylated GlmUMtb were excised and digested with trypsin at 37 °C. Peptide mapping were carried out as described previously (10, 15, 16). For identification of phosphorylation site using mass spectrometry, in vitro kinase assays were carried out using 50 pmol of PknB-KD and 50 pmol of GlmUMtb in a 200-μl reaction in 1× kinase buffer containing 1 mm ATP for 1 h at 30 °C.
Identification of PknB Target Phosphorylation Site
In vitro-phosphorylated GlmUMtb was resolved on SDS-PAGE, and Coomassie-stained GlmUMtb bands were excised for in gel digestion. Gel pieces were treated three times with destaining solution (25 mm ammonium bicarbonate in 50% acetonitrile) at 37 °C for 30 min. Protein samples were reduced with 5 mm TCEP followed by alkylation with 50 mm iodoacetamide for 1 h. Gel pieces were digested overnight with 1 μg of trypsin gold (Promega) followed by zip tip purification (Millipore). The dried peptides were dissolved in 5% acetonitrile containing 0.1% formic acid. The tryptic peptide samples (∼500 ng) were separated for each experiment via Thermo Scientific Proxeon nano LC using a C18 analytical column (Newobjective), at a flow rate of 300 nl/min for 90 min. The running method consisted of acetonitrile gradient of 5–40% for 60 min, 80% for 10 min, and finally 90% for 20 min using acetonitrile containing 0.1% formic acid. The LTQ Orbitrap velos performed a full MS scan (RP 60,000) followed by 19 data-dependent MS/MS scans with detection of the fragment ions in the linear ion trap. Target values were 1e6 for full FT-MS scans and 1e4 for ion trap MSn scans. Supplemental activation was used for all ETD MSn scans. Data analysis was performed using Proteome Discoverer software suite (version 1.3). For the search engine SEQUEST, the peptide precursor mass tolerance was set to 10 ppm, and fragment ion mass tolerance was set to 0.8 Da. Carbamidomethylation on cysteine residues was used as fixed modification, and oxidation of methionine and phosphorylation of serine, threonine, and tyrosine was used as variable modifications. Spectra were queried against the M. tuberculosis UniProt database. The spectra were also queried against a decoy database using a target false discovery rate of 1% for strict and 5% for relaxed conditions.
RESULTS
Structure of GlmUMtb(AcCoA) Complex
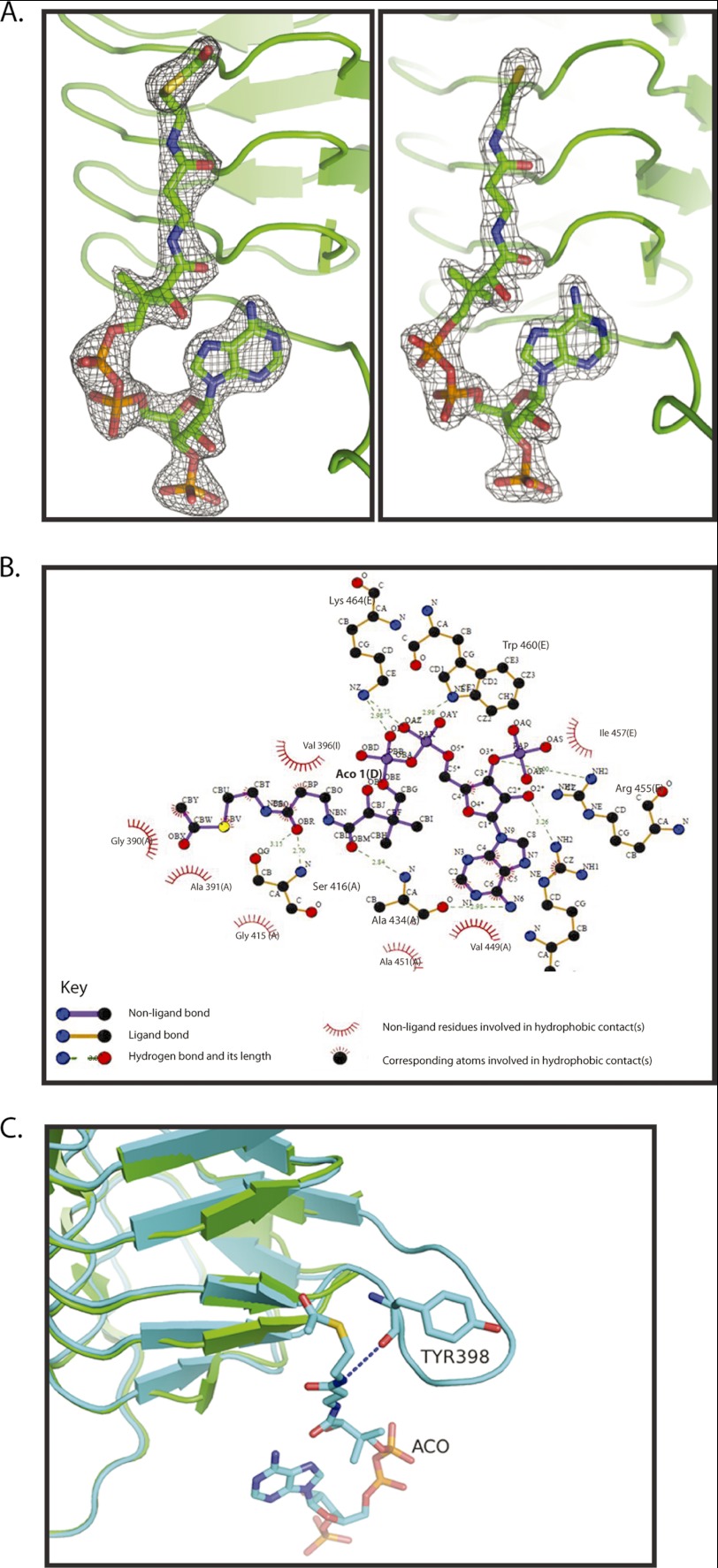
In a previous study, we identified a unique C-terminal extension in GlmUMtb, whose deletion led to an abrogated acetyltransferase activity (10). To further understand these observations, the structures of GlmUMtb bound to substrates/product at the acetyltransferase site were believed to be important. Previous attempts to obtain acetyl-CoA bound crystals by co-crystallization or by soaking apo crystals with acetyl-CoA were unsuccessful (8, 10). The structures of GlmUEc and GlmUSp bound to acetyl-CoA (at the acetyltransferase active site) revealed that the uridyltransferase active site was simultaneously occupied by UDP-GlcNAc, the product of the uridyl transfer reaction. It is likely that UDP-GlcNAc stabilizes the N-terminal domain, whereas acetyl-CoA binds the C-terminal active site. Anticipating such stabilization, attempts were made to crystallize GlmUMtb with acetyl-CoA, in the presence of UDP-GlcNAc. Indeed, crystals were obtained using this strategy, and the structure of GlmUMtb bound to acetyl-CoA, referred to as GlmUMtb(AcCoA), was determined to a resolution of 2.33 Å. An analysis of this structure shows the conserved two-domain architecture of GlmU, one monomer per asymmetric unit, and a trimeric quaternary structure known for GlmU proteins. A comparison of GlmUMtb(AcCoA) with the apo structure of GlmUMtb suggests the following. Distinctly, at the acetyltransferase active site formed by the C-terminal domains of the three monomers, a clear unbiased electron density suggestive of the bound acetyl-CoA was noticeable; acetyl-CoA could be easily modeled into this density (Fig. 1A). As expected, the uridyltransferase active site shows a clear and unbiased electron density corresponding to UDP-GlcNAc that was supplied during crystallization.
FIGURE 1.
Acetyl-CoA binding to GlmUMtb. A, electron density for the ligands in the structures of GlmUMtb(AcCoA) and GlmUMtb(CoA:GlcN-1-P). In the 2Fo − Fc maps, contoured at 2σ, electron density corresponding to acetyl-CoA (left) and CoA (right) are shown. B, interactions made by acetyl-CoA with residues contributed by the three monomers of the trimer in the structure GlmUMtb(AcCoA) are shown. Letters in parentheses represent the chain identifier of the protein molecule in the trimer. All interactions within 4 Å from acetyl-CoA are shown. Interactions were plotted with LIGPLOT. C, structural superposition of GlmUMtb apo (green) and GlmUMtb(AcCoA) (cyan) structures is shown. Interaction of Tyr-398 with the backbone amino group of acetyl-CoA leads to the ordering of the loop in GlmUMtb(AcCoA) structure.
A careful analysis of the structure of GlmUMtb(AcCoA) reveals that hydrogen bonds stabilize the polar groups of acetyl-CoA and hydrophobic interactions stabilize its large carbon chain. A two-dimensional representation of these interactions, plotted with LIGPLOT (17), is shown in Fig. 1B. Briefly, the amine group of the adenine ring of acetyl-CoA hydrogen bonds with the backbone oxygen of Ala-434. The hydrophobic part of the ring is stabilized by Ala-451 and Val-449. Oxygen O3* attached to C3* of the ribose ring is stabilized by the amino group of Arg-455. Backbone oxygen, OBR, is stabilized by hydrogen bonding to OG of Ser-416. In addition to this, Gly-390 and Val-396 provide hydrophobic interactions to the aliphatic carbon chain of the pentathenyl group of acetyl-CoA. The amine groups from Trp-460 and Lys-464 stabilize the backbone phosphate oxygen of acetyl-CoA. These interactions seem critical. Deleting the C-terminal tail (i.e. residues 457–495) of GlmUMtb that provides these residues abolishes all acetyltransferase activity (see below).
A structural superposition of GlmUMtb structures in the presence and absence of acetyl-CoA clearly reveals that a loop disordered in the apo structure becomes ordered upon acetyl-CoA binding (Fig. 1C). This conformational change appears significant as it contributes Tyr-398 to stabilize acetyl-CoA. The backbone oxygen of Tyr-398 interacts with the oxygen atom of the acetyl and amino group of acetyl-CoA. This interaction stabilizes the loop and leads to its ordering.
Ground State Structure Bound to Both of the Substrates Allows Identifying Residues Likely Catalyzing the Acetyltransferase Reaction
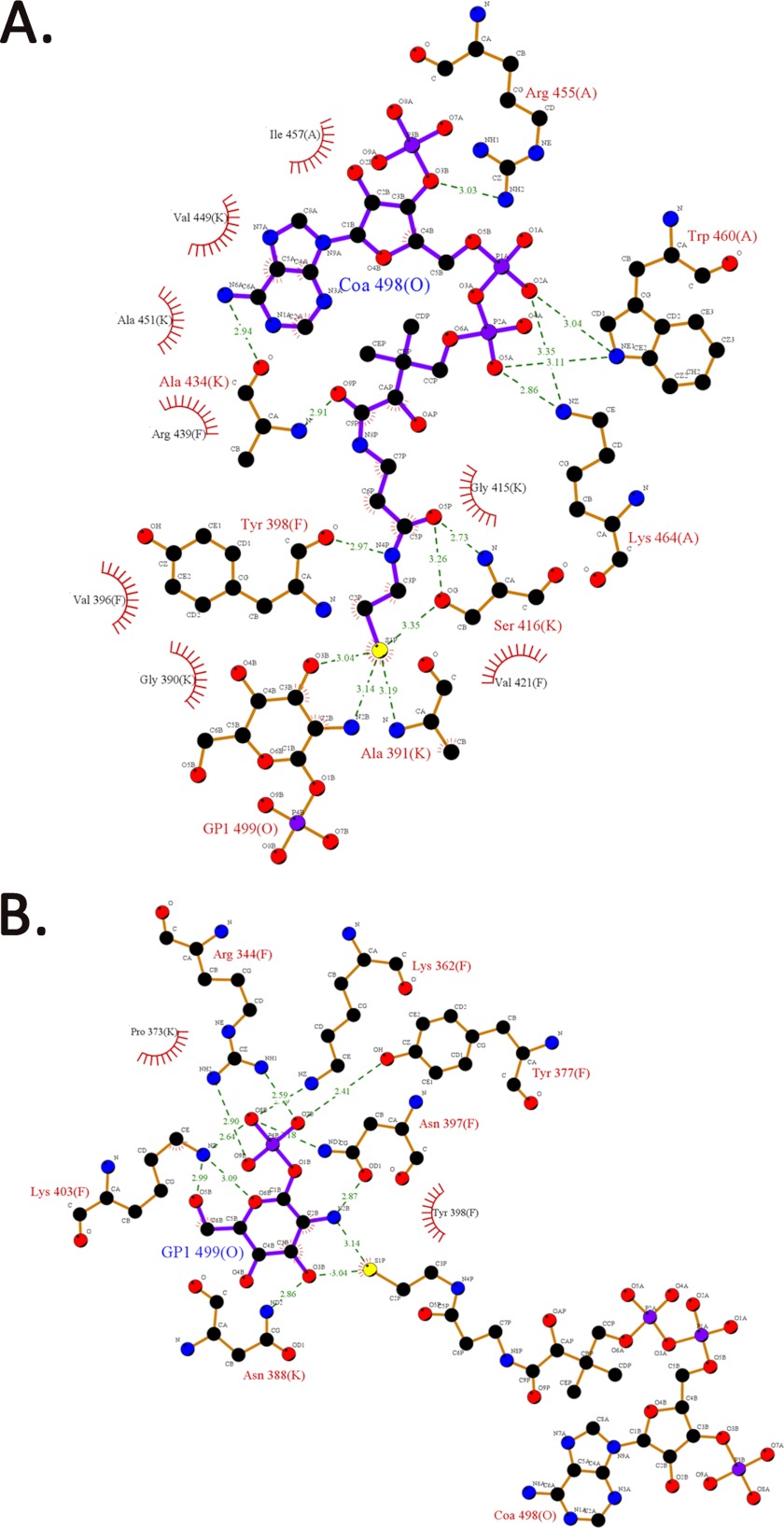
To obtain the structure bound to both the substrates of the acetyltransferase reaction, GlmUMtb(AcCoA) crystals were soaked with the second substrate GlcN-1-P, and the structure was determined to a resolution of 1.98 Å. The objective of these attempts was to understand acetyl group transfer from acetyl-CoA to GlcN-1-P, which leads to the formation of the product GlcNAc-1-P. Therefore, we anticipated that the active site would contain either the substrates or the product. Surprisingly, the electron density present at the active site did not reveal the presence of the acetyl moiety, and it depicted the presence of only CoA and GlcN-1-P. However, this structure closely mimics the ground state and is referred GlmUMtb(CoA:GlcN-1-P). We also co-crystallized GlmUMtb with UDP-GlcNAc, CoA, and GlcNAc-1-P, and the structures are identical (see “Experimental Procedures”). In the active site of GlmUMtb(CoA:GlcN-1-P), all interactions stabilizing the carbon backbone, ribose sugar, and adenine base of CoA are similar to those in GlmUMtb(AcCoA) structure, the only exception being four new interactions that the sulfur atom of CoA makes with the backbone nitrogen of Ala-391, OG of Ser-416, and N2B and O3B of GlcN-1-P (Fig. 2).
FIGURE 2.
Interactions between CoA, GlcN-1-P, and the three monomers of the trimer in the structure of GlmUMtb(CoA:GlcN-1-P). A, all of the interactions for CoA are similar to that made by acetyl-CoA in GlmUMtb(AcCoA) structure, except the thiol group of CoA, which forms four new interactions with the hydroxyl group of Ser-416, backbone oxygen of Ala-391, and O1B and N2B groups of GlcN-1-P. B, the phosphate group of GlcN-1-P is stabilized by Lys-403, Arg-344, Lys-362, Tyr-377, and Asn-397, whereas the oxygen and amine groups are stabilized by polar interactions from the protein side chains.
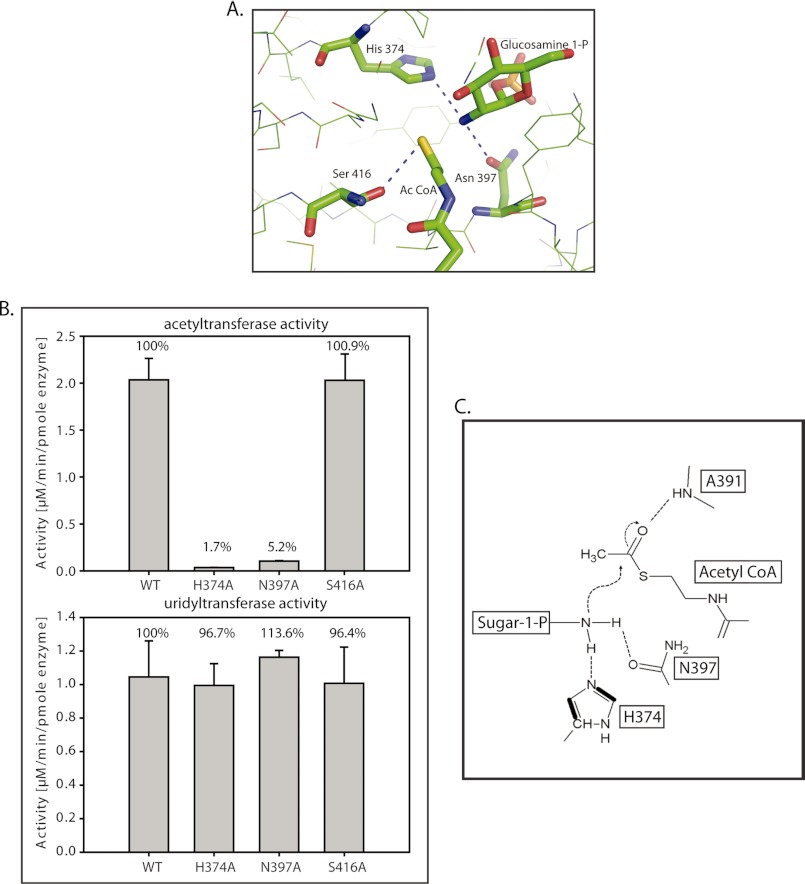
Despite the absence of the acetyl group of acetyl-CoA, this structure mimics the ground state and allows identifying residues likely to be important for catalysis. His-374, Asn-397, Ala-391, and Ser-416 were identified as probable candidate residues, as these were within 3.5 Å to the reacting groups, i.e. the amine of GlcN-1-P and thiol group of CoA (Fig. 3A). As the backbone amine, and not the side chain atom(s) of Ala-391 is involved in the interaction, it could not be mutated. The rest were individually mutated to alanine and mutant proteins were expressed, purified, and analyzed on SDS-PAGE to assess their purity (supplemental Fig. S1). Because the mutations were in the active site of the acetyltransferase domain, one would expect uridyltransferase activity to be unaffected. This was used as an internal positive control. In agreement, the uridyltransferase activity of GlmUMtb wild type (called GlmUMtb-WT henceforth) and the mutants was found comparable (Fig. 3B). Additionally, the acetyltransferase activity by mutants H374A and N397A was almost abolished and that by S416A was unchanged (Fig. 3B). To further understand the effect of these mutations, we have determined the kinetic parameters for both the substrates (Table 2). Indeed, Km and Vmax values obtained for S416A mutant were comparable with those obtained for GlmUMtb-WT, indicating that Ser-416 neither plays a role in catalysis nor in substrate binding. For the mutants N397A and H374A, Km values were similar to that observed for GlmUMtb-WT, whereas the Vmax values were greatly compromised. We speculate that both Asn-397 and His-374 are required for catalysis. Based on these and the prevailing mechanistic inferences based on the studies on GlmUSp and GlmUEc, we believe that the catalytic mechanism, as discussed later, would involve His-374, Asn-397, and Ala-391 (Fig. 3C).
FIGURE 3.
Residues participating in the acetyl transfer reaction. A, the acetyltransferase active site of GlmUMtb reveals probable catalytic residues His-374, Asn-397, and Ser-416, which are highly conserved. Backbone amide of Ala-391 that interacts with the acetyl group is not shown. B, active site mutants were generated, and their acetyltransferase activities were assayed. The activities of H374A and N397A are almost abolished. However, the activity of S416A is not affected significantly. Uridyltransferase activities of the proteins were used as a control. C, a schematic of the proposed acetyltransferase reaction mechanism in GlmUMtb (see text). The amino group activated by His-374 and Asn-397 launches a nucleophilic attack on the carbonyl carbon of acetyl-CoA. Simultaneously, the resultant negative charge on the carbonyl oxygen (oxyanion) is stabilized by the amide backbone of Ala-391.
TABLE 2.
Kinetic parameters (Km and Vmax) for the acetyltransferase activity of GlmUMtb and active site mutants
Kinetic parameters were determined as described under “Experimental Procedures.” S.D. values were calculated using the data obtained from three independent experiments.
| Protein | Km | Vmax | Relative Vmax/Km |
|---|---|---|---|
| μm | μm/min/pmol enzyme | % | |
| Acetyl CoA | |||
| GlmUMtb | 355.0 ± 45.9 | 5.9 ± 0.8 | 100 |
| GlmU-H374A | 300.0 ± 0.05 | 0.05 ± 0.01 | 0.96 |
| GlmU-N397A | 355.1 ± 38.9 | 0.13 ± 0.04 | 2.18 |
| GlmU-S416A | 360.1 ± 20.5 | 5.6 ± 2.2 | 92.39 |
| Glucosamine-1-P | |||
| GlmUMtb | 353.3 ± 64.6 | 7.5 ± 1.3 | 100 |
| GlmU-H374A | 300.0 ± 0.6 | 0.05 ± 0.02 | 0.8 |
| GlmU-N397A | 282.5 ± 20.7 | 0.08 ± 0.03 | 1.4 |
| GlmU-S416A | 370.0 ± 44.3 | 6.8 ± 2.0 | 86.7 |
Acetyl-CoA Adopts Different Conformations when Bound to GlmUMtb and GlmUEc
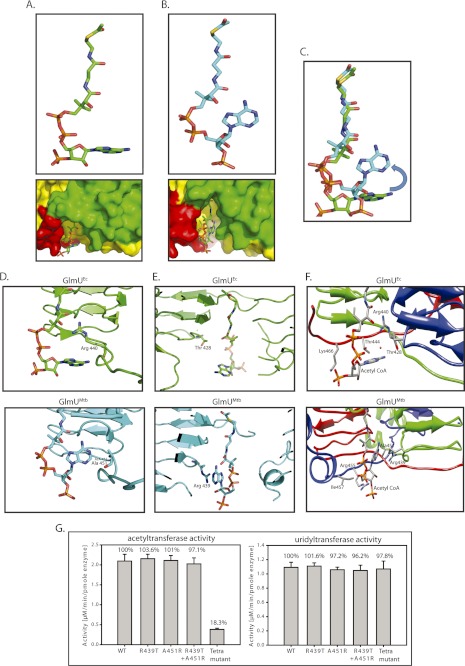
Structural superposition of GlmUMtb(AcCoA) and GlmUEc[AcCoA] (Protein Data Bank codes 3SPT and 2OI5, respectively) reveals a significant difference in the conformation of the adenine ring of acetyl-CoA (Fig. 4, A–C). For ease of presentation, we term the conformations of acetyl-CoA bound to GlmUEc as L conformation, whereas that bound to GlmUMtb as U conformation based on the shape adopted by it (Fig. 4, A and B, respectively). Careful analysis of both structures revealed that two key residues that interact with acetyl-CoA are interchanged in the two enzymes. How may these substitutions give rise to the observed differences, i.e. the L and U conformations, in acetyl-CoA conformation? It may be reasoned as follows. One of these residues, Arg-440 in GlmUEc interacts with the backbone oxygen atoms of acetyl-CoA, which appears to hinder the adenine ring from bending onto the carbon backbone of acetyl-CoA, and restricts it to the L conformation. In GlmUMtb, this arginine is replaced by an alanine. Ala, unlike Arg, cannot restrict the adenine ring similarly and therefore allows it to adopt a U conformation (Fig. 4D). Also, in GlmUMtb, the U conformation appears to be further stabilized due to cation-π interactions from Arg-439, which in GlmUEc is substituted by Thr-428, whose shorter side chain cannot provide a similar stabilization (Fig. 4E).
FIGURE 4.
Different conformations of acetyl-CoA bound to GlmUEc and GlmUMtb. A, GlmUEc displays an L confirmation of acetyl-CoA with the adenine base laying flat. A surface representation (see below) shows the adenine base, and backbone phosphates of acetyl-CoA bound to GlmUEc are exposed to the solvent, and the backbone phosphates interacts with the water molecules. B, acetyl-CoA bound to GlmUMtb displays a U conformation with a bent adenine base, whose amine groups interact with the oxygens of the backbone. A surface representation (see below) is shown with the three monomers colored green, red, and yellow. The adenine base of acetyl-CoA bound to GlmUMtb is buried at the interface of two monomers of the trimer. The backbone phosphates interact with Trp-460 and Lys-454 contributed by the tail of the third monomer of the trimer (see Fig. 6C). C, superposition of the U and L conformations of acetyl-CoA. It shows a ∼90° bend at the adenine base on the backbone carbon chain in the U conformation. D, comparing the two conformations, Arg-440 in GlmUEc interacts with the backbone oxygens of acetyl-CoA, which prevents the adenine base from bending onto itself. On the other hand, Ala-451 at an analogous position in GlmUMtb is less bulky and hence would not hinder the base from bending. E, the U conformation of acetyl-CoA in GlmUMtb is further stabilized by Arg-439, which provides cation-π interaction to the adenine base. No such stabilizing interaction is seen in GlmUEc, as Thr-428Ec substitutes Arg-439Mtb here. F, comparing the structures of GlmUMtb and GlmUEc two other substitutions viz. Ile-457 to Lys and Arg-455 to Thr were also noted. Together, all of these were believed to reverse the U conformation to an L conformation. G, acetyltransferase activities for mutants R439T, A451R, double mutant (R439T, A451R) and a tetra mutant (A451R,R439T,I457K,R455T) are shown. Uridyltransferase assays were used as internal controls.
To inquire whether the substitutions presumed to provide a different binding conformation to acetyl-CoA in GlmUMtb are uniquely conserved in Mycobacteriaceae family, the sequences of GlmU homologues were aligned. Indeed, the substitutions responsible for the U conformation are restricted to Mycobacterium genus and some members of Actinobacteria phyla, whereas those responsible for the L conformation, as in GlmUEc, are conserved in all of the other homologues (supplemental Fig. S3). The structures of acetyl-CoA, bound to different protein structures present in the Protein Data Bank, were analyzed using the ValiGurl server. From this, it was apparent that the U conformation is atypical; as of all of the observed conformations, only eight had a root mean square deviation ≤2 Å (see supplemental Fig. S4 and supplemental Table 2). To ascertain the importance of the aforesaid residues in acetyltransferase activity, mutations A451R and R439T were created in GlmUMtb, both independently and simultaneously, to mimic the corresponding residues in GlmUEc. From the structures, it appeared that the different nature of these substitutions would possibly result in an altered conformation of the adenine base of acetyl-CoA moiety. However, neither the single mutants A451R and R439T nor the double mutant (A451R + R439T) affected the acetyltransferase activity significantly (Fig. 4G). A careful analysis of the structures brought out two more substitutions that were suspected to be important for the U/L conformation of acetyl-CoA. These are I457K and R455T (Fig. 4F). When these mutations were included to create a tetra mutant (A451R, R439T, I457K, and R455T), a marked reduction in acetyltransferase activity was noted (Fig. 4G). Perhaps all of these residues collectively provide the unique U or L conformation to acetyl-CoA. Crystal structures of these mutants would evaluate the possible reversal of acetyl-CoA conformation at the active site, but attempts to crystallize these mutants were unfortunately not successful. Interestingly, the difference between GlmUMtb and GlmUEc in the C-terminal tail lies only in these four residues: Ala-451, Arg-439, Ile-457, and Arg-455. Overall, the tail region of GlmUMtb appears to play a significant role, perhaps in providing the U conformation to acetyl-CoA. The absence of activity in the tetra mutant likely indicates an importance of the U conformation for GlmUMtb.
A Short Helix Governs the Interaction of Trp-460 with Acetyl-CoA
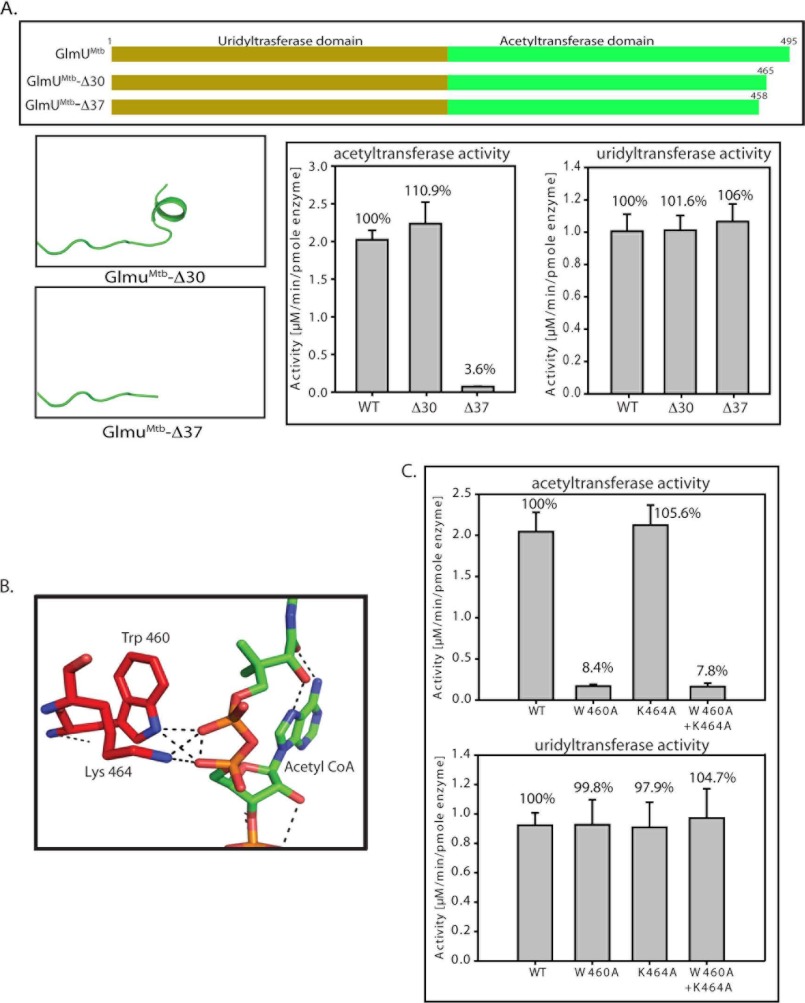
Previously, we reported that a 30-residue extension at the C terminus of GlmUMtb is crucial for its activity (10). Regrettably, upon revisiting the data, the mutant based on which this inference was drawn, actually had a 37-, and not 30-, amino acid deletion in the C terminus. (This mutant is hence referred GlmUMtbΔ37.) We generated another mutant, where the C-terminal 30 amino acids was deleted and designated it as GlmUMtbΔ30. The control experiments suggested that the uridyltransferase activities of these mutants were comparable with GlmUMtb-WT (Fig. 5A). Interestingly, deleting 30 residues at the C-terminal region had little impact on the acetyltransferase activity, whereas a further deletion of seven residues, i.e. Δ37, significantly affects the activity (Fig. 5A). The mutant Δ37 is devoid of the region (458 to 464) that forms a short helix at the C terminus and this region appears important for the acetyltransferase activity. Furthermore, we find that Trp-460 and Lys-464 are provided by this region, which interact with the backbone phosphate oxygens of the acetyl-CoA in GlmUMtb(AcCoA) structure (Fig. 5B). To address the role of these residues in acetyltransferase activity, we mutated Trp-460 and Lys-464 to alanines; both independently and simultaneously. It is apparent from the assays that although K464A is as active as GlmUMtb-WT, for W460A and the double mutant (K464A+W460A) acetyltransferase activity is significantly compromised (Fig. 5C). These studies implicate Trp-460 to be an important element in stabilizing acetyl-CoA binding.
FIGURE 5.
Tail truncation experiments identify residues important for acetyltransferase activity. A, acetyltransferase assays show that a complete loss in activity is seen only for Δ37, whereas Δ30 shows an activity comparable with the wild type protein. The right panel depicts the unchanged uridyltransferase activities for the mutants. B, structure analysis of GlmUMtb(AcCoA) structure revealed that Trp-460 and Lys-464 lie in this region and provide important interactions with the backbone phosphate of acetyl-CoA. C, W460A mutant displays complete loss in acetyltransferase activity, whereas K464A does not. The loss in activity of the double mutant (W460A + K464A) reinforces the importance of Trp-460. The control uridyltransferase assays are shown on the right.
Thr-418 Is the Major Phosphorylation Site
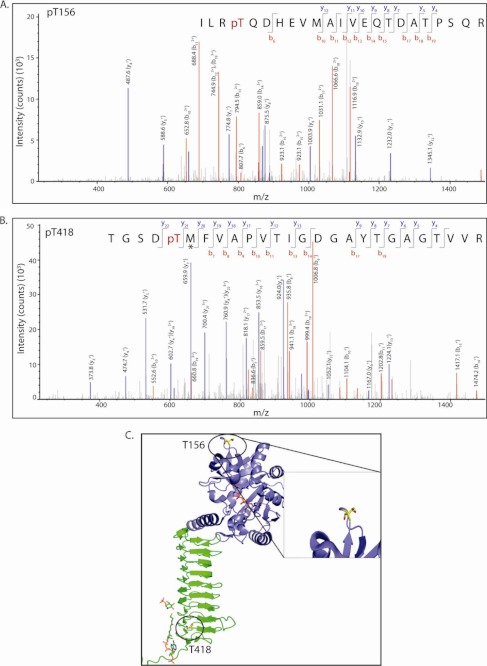
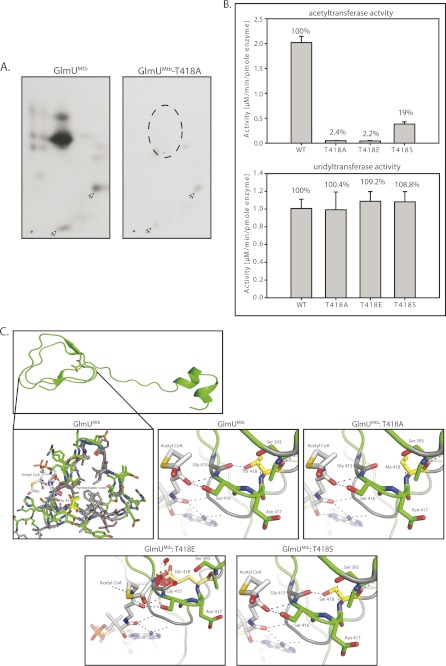
GlmUMtb is phosphorylated by the kinase PknB (7). This phosphorylation down-regulates acetyltransferase activity of GlmUMtb, leaving its uridyltransferase activity unaffected. To identify PknB target sites on GlmUMtb, we resorted to high-resolution mass spectrometry analysis of in vitro-phosphorylated protein. LC-MS analysis showed the presence of two phosphorpeptides with precursor m/z 906.76 and 880.40 corresponding to phosphorylated mass of triply charged semitryptic peptide from residues 153–175 and tryptic peptide from residues 413–439, respectively. MS/MS analysis of these two precursors unambiguously identified Thr-156 (Fig. 6A) and Thr-418 to be the target phosphorylation sites (Fig. 6, A and B). Analysis of peptide spectrum matches of all the identified GlmUMtb phosphopeptides in multiple LC-MS runs indicated that phosphorylation on Thr-418 is ∼5 times more abundant compared with Thr-156. Peptide map analysis of in vitro phosphorylated PknB showed two major spots, which disappear when Thr-418 is mutated to alanine (Fig. 7A). However, minor spots could be detected, both in wild type and the mutant, which may correspond to Thr-156 phosphorylation. Comparison of intensities of various spots also suggests Thr-418 to be the most abundant phosphorylation site on GlmUMtb (Fig. 7A).
FIGURE 6.
Identification of PknB-mediated phosphorylation site. LC-MS/MS data showing collision-induced fragmentation mass spectra identifying two phosphopeptides in GlmUMtb. pT indicates the site of phosphorylation. An asterisk indicates methionine oxidation. A, MS/MS spectrum of precursor m/z 906.76 (+3) and MH+: 2718.29111 Da, of semitryptic phosphopeptide ILR(pT)QDHEVMAIVEQTDATPSQR. Unambiguous location of the intact phosphate group on Thr-156 was evident by the observation of the “b” ion series containing b6, b10, b11, b12, b13, b14, b15, b17, b18, and b19. B, MS/MS spectrum of precursor m/z 880.40 (+3) and MH+: 2639.20804 Da of phosphopeptide TGSD(pT)MFVAPVTIGDGAYTGAGTVVR. Unambiguous location of the intact phosphate group on Thr-418 was apparent by the observation of the “y” series ion y22 and “b” ion series containing b7, b8, b9, b10, b11, b13, b14, b17, and b19. C, the location of the two threonines Thr-156 and Thr-418 that are phosphorylated are shown (circled) here. Thr-156 is solvent exposed and far from the active sites. Thr-418, the primary site, is closer to the acetyltransferase active site.
FIGURE 7.
The effect of phosphorylation of GlmUMtb by PknB. A, in vitro-phosphorylated GlmUMtb or mutants were digested with trypsin and the resulting phosphopeptides were mapped by two-dimensional resolution on thin layer chromatography. B, Thr-418 was mutated to glutamate (phospho-Thr mimic) and to alanine (phosphorylation negative). Acetyltransferase activities of GlmUMtb-WT and the phospohomutants are shown in the left panel. Corresponding uridyltransferase activities are shown in the right panel. C, the acetyltransferase domain, which consists of the β-helix, has a highly hydrophobic core (gray residues) and a few polar residues. Thr-418 is shown in yellow. Hydroxyl group of Thr-418 makes a hydrogen bond with the backbone oxygen of Gly-415. Mutation of Thr-418 to alanine (T418A) would disrupt this interaction (black arrow), whereas mutation to a glutamate (T418E) would introduce steric clashes in the region as shown by red patches. However, mutation to serine (T418S) would preserve the polar interaction with the backbone of Gly-415. The hydroxyl and backbone amine group of Ser-416 interacts with the backbone oxygen of the acetyl-CoA. It is expected that perturbing these interactions would affect the positioning of the acetyl group for catalysis.
The acetyltransferase activity of T418E mutant that mimics a phosphorylated Thr, was severely compromised as compared with GlmUMtb-WT protein (Fig. 7B). Surprisingly, the activity of T418A mutant was also severely compromised, suggesting an essential role for the side chain oxygen (Fig. 7B). Mutating Thr-418 to serine preserved some activity. Analysis of the structures of GlmUMtb reveals that the region near Thr-418 lies in a hydrophobic core of the LβH of the acetyltransferase domain and forms a hydrogen bond with the backbone carbonyl oxygen of Gly-415 (Fig. 7C). As the region is highly hydrophobic, an imbalance in charge would destabilize the region. The mutation T418A will not satisfy the interaction with Gly-415. The introduction of a bulky negatively charged phosphate group due to phosphorylation of Thr-418 will cause a steric clash and disturb the stabilizing interactions (Fig. 7C). This would further destabilize the interactions between the adjacent stacks of the β-helix. As these stacks provide stabilizing interactions to acetyl-CoA, their destabilization will result in the loss of acetyltransferase activity.
DISCUSSION
The acetyl transfer reaction catalyzed by GlmU is as follows.
Acetyl-CoA + GlcN-1-P ⇄ GlcNAc-1-P + CoA
As seen from the structures of GlmUMtb, the substrate acetyl-CoA binds at the trimeric interface. Its stabilization by interactions from all three monomers implicates an importance of trimerization for the acetyltransferase activity. Acetyl-CoA is similarly stabilized in GlmUEc and GlmUSp as well (5, 18). Here, in an attempt to obtain the crystals of GlmUMtb in the product bound form of acetyltransferase reaction, 1) GlmUMtb(AcCoA) crystals were soaked in GlcN-1-P; and 2) GlmUMtb was co-crystallized with GlcNAc-1-P and CoA. Although both revealed the presence of GlcN-1-P at the active site, they yielded incomplete information in not depicting electron density for the acetyl group. This can be due to the presence of catalytically active pairs of the acetyltransferase reaction in the reaction mixture, as the reverse reaction will lead to the presence of the reactants and products of the reaction, both, at any given time. If CoA and GlcN-1-P have more affinity for the active site as compared with acetyl-CoA and GlcNAc-1-P, then due to the presence of all the four components in the reaction mixture, CoA and GlcN-1-P would preferentially bind the active site.
Although the acetyl group could not be positioned in the active site, the GlmUMtb(CoA:GlcN-1-P) structure enabled the identification of His-374, Asn-397, Ala-391, and Ser-416 as potential catalytic residues (Fig. 2). Acetyltransferase assays using H374A, N397A, and S416A mutants, revealed that mutating His-374 and Asn-397, but not Ser-416, affects catalytic activity. For the GlmUMtb-WT and the mutants, kinetic constants were determined separately for the two substrates acetyl-CoA and GlcN-1-P (Table 2). The Vmax for H374A and N397A is drastically reduced as compared with GlmUMtb-WT. Furthermore, the Km values remain unchanged for both acetyl-CoA and GlcN-1-P, signifying that the mutants do not affect binding of the substrates. In the structure, the amine group of GlcN-1-P is only 3.14 Å away from the thioester group of CoA. Based on enzyme kinetics for GlmUMtb-WT and the mutants, we propose a catalytic mechanism where, the amine group of GlcN-1-P is activated for nucleophilic attack onto the carbonyl carbon of acetyl-CoA to form the products GlcNAc-1-P and CoA, as shown in a schematic (Fig. 3C). We also infer that a conserved histidine (His-374 in GlmUMtb) deprotonates the amine group. Besides, the nucleophilicity of the nitrogen (of the amine group) is further enhanced due to an H-bond interaction with the side chain oxygen (OD1) of Asn-397 (Fig. 3C). The activated amino group then makes a nucleophilic attack on the carbonyl carbon of acetyl-CoA, which is electrophilic. Simultaneously, this leads to the generation of an oxyanion on the carbonyl oxygen. The additional negative charge on the carbonyl oxygen is stabilized by an H-bond with the backbone amide of a conserved alanine Ala-391, as shown in Fig. 3. This mechanism is in line with those proposed for GlmUEc and GlmUSp (6, 7). Furthermore, it was suggested that the acetyltransferase reaction follows an SN2 mechanism (6). In this mechanism, the amino group of the GlcN-1-P is believed to be activated by a conserved histidine (His-374 in case of GlmUMtb). Although a role for histidine in catalysis was clearly suggested, the role of Asn-397 has not been addressed satisfactorily. Olsen et al. (6) only suggest that this Asn makes an interaction with the amine group (of GlcN-1-P) and do not mention its involvement in catalysis. However, for GlmUSp, the role of this Asn has been interpreted in providing a stable binding of acetyl-CoA (7). This view is negated by the kinetics data (Table 2) and the analysis of the crystal structures presented here. This apart, in the case of GlmUEc, a conserved serine was also believed to orient itself to stabilize the oxyanion group. However, upon mutating the analogous Ser-416 of GlmUMtb, a significant difference (in activity) was not observed (Fig. 3B, Table 2). In agreement with our data, mutational analysis carried out on the basis of amino acid residue conservation in the carboxyl terminal region of GlmUMtb also showed importance for His-374 and Trp-460 in catalysis (19).
Another intriguingly different aspect is the conformation of acetyl-CoA/CoA bound to GlmUMtb and that bound to GlmUEc/GlmUSp. An analysis of the conformations of the acetyl-CoA present in the Protein Data Bank revealed that only eight entries have an acetyl-CoA conformation as that found in GlmUMtb, suggesting this conformation is atypical in nature (root mean square deviation < 2 Å, supplemental Fig. S4 and supplemental Table S2). However, it is not uncommon for small molecules/ligands to adopt a wide range of conformations by varying only a few rotatable bonds. These changes in binding conformation may have a biological significance, including differences in reaction kinetics by homologous enzymes. Usually, catalytic mechanisms adopted by homologous enzymes from different species are conserved and the conformation adopted by substrates may not be identical. In these active sites, the residues near the reaction center are conserved to preserve the nature of the reaction mechanism. However, some that are away from the reaction center contribute to substrate binding. Differences in these, (among homologues) may manifest as differences in substrate binding (20). The residues interacting with the acetyl group of acetyl-CoA in GlmUMtb are largely conserved among GlmU homologues, except for Arg-439, Ala-451, Arg-455, and Ile-457 near the adenine base of acetyl-CoA. As indicated in Fig. 4, D–F, in GlmUEc, residues analogous to these are Thr-428, Arg-440, Thr-444, and Lys-446, respectively. Analysis of crystal structures suggested that the nature of these substitutions might significantly alter the conformation of the adenine base of acetyl-CoA moiety. We reasoned that these are likely responsible for the U or L conformations of acetyl-CoA seen in GlmUMtb and GlmUEc. However, upon mutating Ala-451 to Arg and Arg-439 to Thr, either independently in single mutants or simultaneously in a double mutant, did not affect the acetyltransferase activity (Fig. 4G). Interestingly, when additional mutations Ile-457 to Lys and Arg-455 to Thr were also included to create a tetra mutant, acetyltransferase activity was significantly affected (Fig. 4G). However, these experiments do not clarify whether the conformation of acetyl-CoA is altered from U to L in the tetra mutant. GlmUEc and GlmUMtb exhibit a large difference in their basal activities, with GlmUEc being ∼6–8 fold more efficient compared with its GlmUMtb counterpart. This study raises a possibility that the observed difference in activity may be associated with the distinct U/L conformations of acetyl-CoA. However, further investigations are required to evaluate the same.
Besides these differences, differential regulation operates in GlmUMtb. In an earlier report (10), we showed that PknB phosphorylates a threonine residue in the C-terminal domain of GlmUMtb, which in turn down-regulates the acetyltransferase activity. Here, with the help of mass spectrometry, we identified Thr-156 and Thr-418 to be the target sites on GlmUMtb. Peptide mapping analysis and peptide spectrum matches from multiple LC-MS runs suggested Thr-418 to be the most abundant phosphorylation site (Fig. 6, A and B). An analysis of the structure of GlmUMtb revealed Thr-156 to be present in a surface-exposed loop and away from the uridyltransferase active site (Fig. 6C). This is in line with the observation that the uridyltransferase activity is unaffected upon PknB-mediated phosphorylation. However, Thr-418 is important in providing stability to the left-handed β-helix: the T418A mutant, whose acetyltransferase activity is completely abolished, reflects this importance (Fig. 7, C and D). Based on the structure of GlmUMtb(AcCoA), it is possible to explain the effect of phosphorylation at Thr-418 on the acetyltransferase activity. It is evident that the introduction of a highly charged and bulky phosphate group on Thr-418 would destabilize the hydrophobic region immediately surrounding it (Fig. 7D). A T418E mutant generated to mimic the phosphorylated threonine corroborates this as it shows a complete abolishment of acetyltransferase activity (Fig. 7C). Preliminary florescence binding assays, albeit qualitative, suggest that phosphorylation does not inhibit acetyl-CoA binding (supplemental Fig. S5). Thr-418 lies in an important region that is directly involved in making polar contacts with the backbone oxygen of acetyl-CoA via the backbone amine group of Gly-415. By affecting the precise positioning of the acetyl group, phosphorylation appears to regulate acetyl transfer.
Another regulation of the acetyltransferase activity is due to the unique C-terminal extension in GlmUMtb. It contributes a short helix, which presents an important residue Trp-460 for acetyl-CoA binding. Crystal structure analysis and biochemical studies reiterate its importance. Zhang et al. (8), by modeling acetyl-CoA into the apo structure of GlmUMtb, suggested an interaction of Trp-460 and the substrate. Crystal structures of GlmUHi (from Haemophilus influenzae) bound with acetyltransferase inhibitors were reported recently (21). These depict an important contribution by the equivalent Trp-449 in inhibitor binding. Interestingly, drug resistant strains of H. influenzae carry a mutation at Trp-449, which results in the loss of this π-stacking interaction with the inhibitors.
Overall, GlmUMtb forms an ideal drug target to develop antitubercular drugs. The synthesis of UDP-GlcNAc takes place via a different route in eukaryotes than that in prokaryotes. In eukaryotes, acetyl transfer occurs on GlcN-6-P instead of GlcN-1-P. Also, the acetyltransferase and uridyltransferase reactions are catalyzed by two different enzymes, which show little homology to GlmUMtb (22–24). Perhaps, all of these are suggestive that targeting the acetyltransferase domain to inhibit the action of GlmUMtb may be a reasonable strategy. Identifying structural differences is necessary for the design of selective inhibitors against GlmUMtb. The U conformation of acetyl-CoA found in GlmUMtb and the active site residues that make this possible can be a target site. Besides, we also identify that a small, one-and-a-half-turn helix in the C-terminal extension, a feature unique to GlmUMtb, and a conserved tryptophan (Trp-460) are important contributors to the acetyl transfer reaction. Altering these interactions or the conformation of the short helix could be a viable strategy.
Supplementary Material
Acknowledgments
We gratefully acknowledge the use of the BM14 beam line at European Synchrotron Radiation Facility (Grenoble, France) for x-ray data collection under the program supported by Department of Biotechnology, India. We acknowledge the use of National Institute of Immunology mass spectrometry facility set up with the support provided by Department of Biotechnology, India.

This article contains supplemental Tables S1 and S2 and Figs. S1–S5.
The atomic coordinates and structure factors (codes 3ST8 and 3SPT) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- UDP-GlcNAc
- UDP-N-acetylglucosamine 1-phosphate
- KD
- knockdown
- GlmU
- N-acetyl-glucosamine-1-phosphate uridyltransferase
- SN2
- bimolecular nucleophilic substitution reaction
- GlcN-1-P
- glucosamine 1-phosphate
- GlcNAc-1-P
- N-acetylglucosamine 1-phosphate
- GlmUEc
- GlmU from Escherichia coli
- GlmUSp
- GlmU from Streptococcus pneumoniae
- GlmUMtb
- GlmU from Mycobacterium tuberculosis.
REFERENCES
- 1. Anderson M. S., Raetz C. R. (1987) Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J. Biol. Chem. 262, 5159–5169 [PubMed] [Google Scholar]
- 2. Mengin-Lecreulx D., van Heijenoort J. (1994) Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 176, 5788–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 4. Zhang W., Jones V. C., Scherman M. S., Mahapatra S., Crick D., Bhamidi S., Xin Y., McNeil M. R., Ma Y. (2008) Expression, essentiality, and a microtiter plate assay for mycobacterial GlmU, the bifunctional glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase. Int. J. Biochem. Cell Biol. 40, 2560–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olsen L. R., Roderick S. L. (2001) Structure of the Escherichia coli GlmU pyrophosphorylase and acetyltransferase active sites. Biochemistry 40, 1913–1921 [DOI] [PubMed] [Google Scholar]
- 6. Olsen L. R., Vetting M. W., Roderick S. L. (2007) Structure of the E. coli bifunctional GlmU acetyltransferase active site with substrates and products. Protein Sci. 16, 1230–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sulzenbacher G., Gal L., Peneff C., Fassy F., Bourne Y. (2001) Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture. J. Biol. Chem. 276, 11844–11851 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Z., Bulloch E. M., Bunker R. D., Baker E. N., Squire C. J. (2009) Structure and function of GlmU from Mycobacterium tuberculosis. Acta Crystallogr. D Biol. Crystallogr. 65, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verma S. K., Jaiswal M., Kumar N., Parikh A., Nandicoori V. K., Prakash B. (2009) Structure of N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) from Mycobacterium tuberculosis in a cubic space group. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parikh A., Verma S. K., Khan S., Prakash B., Nandicoori V. K. (2009) PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J. Mol. Biol. 386, 451–464 [DOI] [PubMed] [Google Scholar]
- 11. Kabsch W. (2010a). XDS. Acta. Cryst. D66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2005) Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an x-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 61, 449–457 [DOI] [PubMed] [Google Scholar]
- 13. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 15. Boyle W. J., van der Geer P., Hunter T. (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 16. Khan S., Nagarajan S. N., Parikh A., Samantaray S., Singh A., Kumar D., Roy R. P., Bhatt A., Nandicoori V. K. (2010) Phosphorylation of enoyl-acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J. Biol. Chem. 285, 37860–37871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 18. Kostrewa D., Brockhaus M., D'Arcy A., Dale G. E., Nelboeck P., Schmid G., Mueller F., Bazzoni G., Dejana E., Bartfai T., Winkler F. K., Hennig M. (2001) X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 20, 4391–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Y., Yu W., Zheng Q., Xin Y., Ma Y. (2012) Identification of amino acids involved in catalytic process of M. tuberculosis GlmU acetyltransferase. Glycoconj. J. 29, 297–303 [DOI] [PubMed] [Google Scholar]
- 20. Nicklaus M. C., Wang S., Driscoll J. S., Milne G. W. (1995) Conformational changes of small molecules binding to proteins. Bioorg. Med. Chem. 3, 411–428 [DOI] [PubMed] [Google Scholar]
- 21. Buurman E. T., Andrews B., Gao N., Hu J., Keating T. A., Lahiri S., Otterbein L. R., Patten A. D., Stokes S. S., Shapiro A. B. (2011) In vitro validation of acetyltransferase activity of GlmU as an antibacterial target in Haemophilus influenzae. J. Biol. Chem. 286, 40734–40742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mio T., Yabe T., Arisawa M., Yamada-Okabe H. (1998) The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases. Gene cloning, protein expression, and catalytic mechanism. J. Biol. Chem. 273, 14392–14397 [DOI] [PubMed] [Google Scholar]
- 23. Wang-Gillam A., Pastuszak I., Stewart M., Drake R. R., Elbein A. D. (2000) Identification and modification of the uridine-binding site of the UDP-GalNAc (GlcNAc) pyrophosphorylase. J. Biol. Chem. 275, 1433–1438 [DOI] [PubMed] [Google Scholar]
- 24. Wang-Gillam A., Pastuszak I., Elbein A. D. (1998) A 17-amino acid insert changes UDP-N-acetylhexosamine pyrophosphorylase specificity from UDP-GalNAc to UDP-GlcNAc. J. Biol. Chem. 273, 27055–27057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.