Background: The mechanism of marine elastin degradation is unclear.
Results: A novel M23 metalloprotease pseudoalterin from a marine bacterium degraded elastin by cleaving both the glycyl bonds and the peptide bonds involved in cross-linking.
Conclusion: Pseudoalterin adopts a novel elastolytic mechanism different from other M23 metalloproteases.
Significance: The results shed light on the mechanism of marine elastin degradation.
Keywords: Elastin, Electron Microscopy (EM), Metalloprotease, Protein Cross-linking, Protein Purification, M23 Metalloprotease, Cross-linking Domain, Deep-sea Sediment, Elastolytic Mechanism
Abstract
Elastin is a common insoluble protein that is abundant in marine vertebrates, and for this reason its degradation is important for the recycling of marine nitrogen. It is still unclear how marine elastin is degraded because of the limited study of marine elastases. Here, a novel protease belonging to the M23A subfamily, secreted by Pseudoalteromonas sp. CF6-2 from deep-sea sediment, was purified and characterized, and its elastolytic mechanism was studied. This protease, named pseudoalterin, has low identities (<40%) to the known M23 proteases. Pseudoalterin has a narrow specificity but high activity toward elastin. Analysis of the cleavage sites of pseudoalterin on elastin showed that pseudoalterin cleaves the glycyl bonds in hydrophobic regions and the peptide bonds Ala–Ala, Ala–Lys, and Lys–Ala involved in cross-linking. Two peptic derivatives of desmosine, desmosine-Ala-Ala and desmosine-Ala-Ala-Ala, were detected in the elastin hydrolysate, indicating that pseudoalterin can dissociate cross-linked elastin. These results reveal a new elastolytic mechanism of the M23 protease pseudoalterin, which is different from the reported mechanism where the M23 proteases only cleave glycyl bonds in elastin. Genome analysis suggests that M23 proteases may be popular in deep-sea sediments, implying their important role in elastin degradation. An elastin degradation model of pseudoalterin was proposed, based on these results and scanning electron microscopic analysis of the degradation by pseudoalterin of bovine elastin and cross-linked recombinant tropoelastin. Our results shed light on the mechanism of elastin degradation in deep-sea sediment.
Introduction
Elastin is used in diverse tissues that require elasticity and as such are insoluble proteins that are abundant in all higher organisms, including marine animals (1). On this basis, elastin represents an important component of marine organic nitrogen, and its degradation cannot be ignored for marine nitrogen recycling.
The formation of elastin is classically characterized by four stages as follows: tropoelastin synthesis and secretion, association through coacervation, deposition on microfibrils, and cross-linking (2). Tropoelastin is the precursor of elastin whose sequence contains two major types of domains, hydrophobic and hydrophilic regions. The hydrophobic domains are rich in nonpolar residues, Gly, Val, and Pro, whereas the classical hydrophilic domains participate in cross-linking and are mostly composed of Lys and Ala (1). Secreted tropoelastin molecules are ∼15-nm asymmetric monomers that aggregate on the cell surface to form spherules through coacervation by hydrophobic interactions (3–6). In vitro observation of coacervated recombinant tropoelastin has demonstrated that this association is a noncovalent process that can by reversed by simple changes in environmental conditions such as temperature and salt (7). These spherules deposit on microfibrils for further coacervation and alignment promoted by microfibrillar proteins (2). Lysine residues on the massed tropoelastin spherules are oxidized and polymerized to bi-, tri-, and tetrafunctional cross-links, including desmosine (DES)2 and isodesmosine (IDE), to form native elastin networks with the participation of lysyl oxidase (8). Various sizes of tropoelastin aggregates before being cross-linked have been identified in vitro, ranging from ∼200 nm to 2–6 μm (2, 5, 9, 10). However, the precise details of elastin assembly in vivo are still unclear.
Human tropoelastin is translated from a single gene and spliced to multiple isoforms. Exon 26A is a unique domain in human elastin that is usually spliced out in healthy elastic tissue and is occasionally retained under damaged elastin conditions (11). Tropoelastin SHELdelta26A corresponding to amino acid residues 27–724 of GenBankTM entry AAC98394 (gi182020) is an isoform of synthetic human elastin without domain 26A. Tropoelastin SHELdelta26A can reversibly associate by coacervation at suitable temperature, concentration, NaCl concentration, and pH (12). Additionally, chemical cross-linker bis(sulfosuccinimidyl) suberate (BS3) has been used to cross-link the SHELdelta26A tropoelastin to study the structural features of tropoelastin monomer (10).
Because of their insoluble and highly cross-linked nature, elastin is resistant to most proteases, except for a limited number of elastases. Although some serine proteases and metalloproteases from terrestrial bacteria are reported to be elastases, there are very few studies on marine elastase-producing bacteria and elastases (13, 14). For this reason, the mechanism of marine elastin degradation remains unclear.
The metalloproteases of family M23 are divided into two subfamilies, M23A and M23B. Thus far, three metalloproteases are known for family M23A, six in M23B and one that is not assigned to a subfamily; these proteases are all from terrestrial bacteria (15–23). Proteases in this family are designated by their lysis of other organisms for nutrition and infection (23, 24). M23B subfamily proteases are additionally proposed to be required in cell separation (25, 26). The metalloproteases of family M23 are all endopeptidases that are synthesized as precursors and are activated extracellularly. Because they are not autoprocessed during maturation, it is difficult to heterogeneously express the M23 proteases, which somewhat limits their biochemical analysis. These proteases contain a zinc ion, and the active site residues occur in HXXXD and HXH motifs. They are all β proteins containing two stacked half-β-barrels, and the active site is at the end of the larger C-terminal barrel (27). All the M23 proteases have a preference for cleavage of glycyl bonds (23, 28–32). For this reason, elastin is recognized as a preferred substrate for the M23 proteases because there are ∼33% glycine residues in the tropoelastin sequence (33). Nevertheless, the elastolytic mechanism of the M23 proteases is largely unclear. Studies on the specificity of the M23 proteases show that they cleave glycyl bonds, including Gly-Gly, Gly-Ala, Gly-Phe, Gly-Leu, Gly-His, and Gly-Trp, where Gly-Gly is the most preferred (15, 24, 28–31, 34).
It is generally assumed that except for glycyl bonds very few other peptide bonds can be cleaved by the M23 proteases. It was reported that β-lytic endopeptidase could cleave the Val-18–Cya-19 bond in oxidized insulin B chain and the N-acylmuramoyl-l-Ala bond between the cell wall glycoproteins (35), but the relevance of these isolated examples is unclear.
Pseudoalteromonas sp. CF6-2 is a protease-producing bacterium isolated from the deep-sea sediment in the Jiulong methane reef area off the southwest of the island of Taiwan (36). In this study, a novel M23 metalloprotease secreted by strain CF6-2, designated pseudoalterin, was purified and characterized. Moreover, the elastolytic mechanism of pseudoalterin was studied in detail by biochemical experiments and through SEM observations, thus revealing that pseudoalterin digests elastin effectively by a mechanism that is distinct from the reported M23 proteases. The results help to clarify the degradation mechanism of deep-sea sedimentary elastin.
EXPERIMENTAL PROCEDURES
Experimental Materials
Pseudoalteromonas sp. CF6-2 was isolated from the deep-sea sediment at a water depth of 2441 m at site 119° 30.060′E, 22° 0.316′N in the Jiulong methane reef area off the southwest of the island of Taiwan during the South China Sea Open Cruise of R/V Shiyan 3 (36). Escherichia coli DH5α was purchased from Novagen and cultivated at 37 °C on Luria-Bertani (LB) medium supplemented with ampicillin (0.1 mg/ml) for selection of transformants. Insoluble type I collagen fiber (bovine achilles tendon) was purchased from Worthington. α-casein, gamma globulin, elastinorcein, BS3, and bovine neck ligament elastin isoform 1 (SwissProt accession number P04985–1) (hereafter called bovine elastin) were purchased from Sigma, and gelatin was from Boston Biomedical Inc. DES was purchased from Merck.
Protease Purification
Pseudoalteromonas sp. CF6-2 was cultured at 15 °C for 48 h in the fermentation medium containing 0.2% (w/w) yeast extract, 0.3% (w/w) bovine elastin, 0.5 mm CaCl2, 0.5 mm Na2HPO4, and artificial seawater (pH 8.0). The culture was centrifuged at 10,000 × g, 4 °C for 10 min. The supernatant was dialyzed against Tris-HCl buffer (50 mm Tris-HCl (pH 9.0)) for desalination. The dialyzed sample then was loaded onto a DEAE-Sepharose Fast Flow column (Amersham Biosciences), and bound proteins were eluted with an increasing gradient NaCl (0–0.3 m). Fractions with protease activity were collected. The purified protease was analyzed by 12.5% SDS-PAGE, and was named pseudoalterin.
N-terminal Sequence Analysis
The purified pseudoalterin electrophoresed into SDS-polyacrylamide gel was transferred to a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad). The N-terminal sequence of pseudoalterin was obtained by Edman degradation with PROCISE491 (Applied Biosystems) at Beijing University (China), and the resulting sequence was ATFTMNLPWSQGYYY. This sequence was subjected to NCBI Blastp, which showed that it has a relatively high identity with staphylolysin, a protease in family M23A.
Gene Cloning
Two degenerate primers were designed, based on the N-terminal sequence of pseudoalterin and the conserved sequence in the catalytic domain of peptidases in family M23. The genomic DNA of Pseudoalteromonas sp. CF6-2 was extracted using a genomic extraction kit (Bioteke, China). Using the genomic DNA of Pseudoalteromonas sp. CF6-2 as template, PCR amplification was performed using EasyTaq DNA polymerase (TransGen Biotech, China) for 30 cycles consisting of 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 2 min. A 303-bp fragment was amplified and sequenced by Biosune Inc. (China). The thermal asymmetric interlaced PCR was then performed as described by Chen et al. (37), and the remainder of the gene was amplified. Through assembly, a 1212-bp open reading frame encoding pseudoalterin was obtained. Following verification by PCR, the sequence of this gene was submitted to GenBankTM under the accession number HQ005379.
Enzyme Assays and Protein Determination
The proteolytic activity on casein was determined at 25 °C in Tris-HCl buffer (50 mm (pH 9.0)) as described by He et al. (38). One unit of caseinolytic activity was defined as the amount of 1 μg of tyrosine that formed by enzyme catalysis in 1 min. Collagenolytic and elastolytic activities were assayed at 25 °C (39), and the proteolysis of gelatin was measured at 25 °C according to the method recommended by Worthington (40). One unit of elastinolytic activity was defined as the amount of enzyme that increased 0.01 unit of absorbance at 590 nm/min. For collagen and gelatin, 1 unit is defined as the release of 1 μmol of l-leucine equivalents from collagen in 5 h and from gelatin in 1 min. Fibrin degradation was determined electrophoretically (41). To assay the activity of pseudoalterin on gamma globulin, 50 μg of the substrate was treated with 0.6 μg of pseudoalterin at 25 °C for 0–120 min, and then the digested samples were subjected to SDS-PAGE. Proteolytic activity using the substrates furylacryloyl-Gly-Leu-NH2 and furylacryloyl-Gly-Phe-NH2 was detected using Feder's method (42). Bradford assays were used to determine protein concentrations (43) with bovine serum albumin (Sigma) as the standard.
Analysis of the Cleavage Sites of Pseudoalterin on Oxidized Insulin B Chain
Pseudoalterin was incubated with oxidized insulin B chain (1:40, by mass) in 50 mm Tris-HCl (pH 9.0) at 25 °C for 5, 10, and 15 min. 1% trifluoroacetic acid was added to terminate the reaction. The products were separated on a C18 column (Venusil MP C18, China) using a high performance liquid chromatography (HPLC) Shimadzu system with two LC-20AT pumps and an SPD-M20A spectrometer (44). Molecular masses of the hydrolytic peptides were analyzed by liquid chromatography-mass spectrometry (LC-MS) Ion Trap 6340 (Agilent), and the sequences of the peptides were obtained with MASCOT MS/MS Ion Research tools.
Light Microscope Observation of Bovine Elastin Hydrolysis by Pseudoalterin
Bovine elastin (10 mg) was incubated with pseudoalterin (10 μg) in 0.5 ml of 50 mm Tris-HCl (pH 9.0) at 25 °C with continuous stirring. Bovine elastin in the same buffer without pseudoalterin served as negative controls. After treatment for either 1 or 2 h, samples were assessed by inverted microscopy (Olympus IX71, Japan) at room temperature.
Cleavage Sites for Pseudoalterin on Bovine Elastin
Soluble products from pseudoalterin digestion of 10 mg of elastin were boiled for 10 min to terminate the reaction and loaded onto LC-MS to analyze the molecular masses of the released peptides. The sequences of these peptides were analyzed by MASCOT MS/MS Ion Research tools and ExPASy tools.
To confirm these cleavage sites, peptides GVGVAP, IGGGAGG, GIGLGP, PGAGARF, AAAKAA, and AAAAAKAAAK were synthesized by ChinaPeptides Co., Ltd. (China). Each of these peptides (5 μg) was mixed with 5 μg of pseudoalterin in 50 μl of 50 mm Tris-HCl (pH 9.0). After incubation at 25 °C for 600 min for GVGVAP, IGGGAGG, GIGLGP, and PGAGARF and 110 min for AAAKAA and AAAAAKAAAK, reactions were terminated by the addition of 1% trifluoroacetic acid. Molecular masses and sequences of all peptides were then determined as described above.
Detection of Desmosine Derivatives Released from Bovine Elastin by Pseudoalterin
Twenty milligrams of bovine elastin was mixed with 20 μg of pseudoalterin in 500 μl of 50 mm Tris-HCl (pH 9.0). The mixture was incubated at 25 °C for 36 h followed by addition of 500 μl of 4% sulfosalicylic acid to terminate the reaction and precipitate long peptides. After centrifugation at 10,000 × g for 10 min, the supernatant was lyophilized and redissolved in 50 μl of 0.01 m hydrochloric acid. DES and its derivatives in the sample were desalinated and detected using Ion Trap 6340 LC-MS (Agilent). DES standard was prepared in 0.01 m HCl at a final concentration of 2 mg/ml. The same treatment with BSA instead of bovine elastin served as controls.
Preparation, Degradation, and Observation of Cross-linked Recombinant Tropoelastin
Recombinant human tropoelastin isoform SHELdelta26A was purified as described previously (12, 45). Tropoelastin was dissolved to 10 mg/ml in 10 mm phosphate-buffered saline (PBS) (pH 7.4). Chemical cross-linker BS3 (10 mm) was added to the solution, which was spread thinly on aluminum foil and incubated at 37 °C for 30 min to cross-link. The cross-linked tropoelastin was washed with the PBS repeatedly, followed by hydrolysis with 0.1 mg/ml pseudoalterin for 10 and 15 min at 25 °C. The residual cross-linked layer was washed and dehydrated (10). The samples on the aluminum foil were spatter-coated with 5 nm of platinum and examined with a Hitachi FE-S4800 scanning electron microscope (SEM) at 5.0 kV.
SEM Observation of Bovine Elastin Degradation by Pseudoalterin
Five milligrams of bovine elastin was mixed with 10 μg of pseudoalterin in 200 μl of 50 mm Tris-HCl (pH 9.0), and the mixture was incubated at 25 °C with continuous stirring for different times. The same reaction system without pseudoalterin served as the negative control. After washing twice with deionized water, the insoluble mass was lyophilized, mounted on stubs, and examined by SEM as described above.
RESULTS
Purification and Physicochemical Characterization of the Protease Secreted by Pseudoalteromonas sp. CF6-2
The most abundant protease secreted by strain CF6-2 was purified to homogeneity from the culture supernatant, which resulted in 1.5-fold purification and a final yield of 33.7% (supplemental Table S1 and supplemental Fig. S1). SDS-PAGE analysis indicated an apparent molecular mass of about 19 kDa (supplemental Fig. S2). This purified protease was designated pseudoalterin. The specific activity of pseudoalterin toward elastinorcein at 25 °C in 50 mm Tris-HCl (pH 9.0) was quite high. The activity of pseudoalterin toward elastinorcein was detectable over a broad range from pH 7 to 11 with a maximal activity at pH 9.5, whereas the optimal temperature of pseudoalterin was 25 °C (supplemental Fig. S3 and Table 1). The half-life of pseudoalterin activity was 14.13 min at 35 °C, indicating a low thermostability (supplemental Fig. S4). Pseudoalterin appeared to require zinc ion for its hydrolytic activity as 1 mm 1,10-phenanthroline completely abolished its activity against elastinorcein. Addition of 2 mm Zn2+ inhibited the activity of pseudoalterin severely (Table 2), which is consistent with the behavior of other zinc-dependent metalloproteases (46).
TABLE 1.
Physicochemical characteristics of pseudoalterin from Pseudoalteromonas sp. CF6-2
| Characteristics | Results |
|---|---|
| Length of DNA sequence | 1212 bp |
| No. of amino acid residues in mature protein sequence | 173 |
| No. of amino acid residues in signal sequence | 22 |
| No. of amino acid residues in prosequence | 208 |
| Molecular mass of mature enzyme (sequence) | 19,372 Da |
| Isoelectric point (sequence) | 5.86 |
| Half-life at 30 °Ca | 30.84 min |
| Half-life at 35 °Ca | 14.13 min |
| Optimum pH with elastinorcein (30 °C)b | 9.5 |
| Optimum temperature with elastinorceinc | 25 °C |
a Pseudoalterin (0.1 mg/ml) was incubated at 25, 30, and 35 °C. Samples were taken at intervals for the activity assay. The half-life was the time that it took to eliminate 50% of the activity of pseudoalterin with elastinorcein at a given temperature.
b The optimum pH was determined by measuring the activities of pseudoalterin with elastinorcein (30 °C) in Na2HPO4/NaH2PO4 buffer at pH values ranging from 6.5 to 8, barbital sodium/HCl buffer at pH values ranging from 7.5 to 9.6, and NaHCO3/NaOH buffer ranging from 9.6 to 11.0.
c The optimum temperature was determined by measuring the activities of pseudoalterin with elastinorcein in 50 mm Tris buffer (pH 9.0) or in artificial sea water at temperatures ranging from 0 to 45 °C.
TABLE 2.
Effects of metal ions and inhibitors on the elastolytic activity of pseudoalterin
PAPMSF is p-amidinophenylmethylsulfonyl fluoride.
| Metal ion (2 mm) | Relative activitya | Metal ion (2 mm) | Relative activitya | Inhibitors | Residual activityb |
|---|---|---|---|---|---|
| % | % | % | |||
| Control | 100 | Ni2+ | 71.3 | Control | 100 |
| Ca2+ | 287.3 | Mn2+ | 64.5 | PAPMSF | 110.0 |
| Sr2+ | 228.3 | Cu2+ | 63.3 | Leuhistin | 92.9 |
| Mg2+ | 104.3 | Sn2+ | 63.2 | EDTA | 13.4 |
| K+ | 95.9 | Fe2+ | 49.1 | 1,10-Phenanthroline | 0 |
| Na+ | 94.4 | Zn2+ | 2.8 | ||
| Co2+ | 86.7 |
a Elastolytic activity of pseudoalterin was measured at 25 °C for elastinorcein. The activity of pseudoalterin without any metal ion was used as a control (100%). Activity was expressed as the activity of pseudoalterin with a metal ion relative to the activity with the control. The data are the means of three experiments, and the standard deviations were ≤5%.
b Pseudoalterin was incubated with each inhibitor at 25 °C for 20 min, and then the enzyme activity was measured at 25 °C for elastinorcein. The activity of pseudoalterin without any inhibitor was used as the control (100%). Activity was expressed as the activity of pseudoalterin treated with an inhibitor relative to the control activity. The data are the means of three experiments, and the standard deviations were ≤5%.
Gene Cloning and Sequence Analysis of Pseudoalterin
N-terminal sequence analysis suggested that pseudoalterin is probably a member of family M23. Based on the N-terminal sequence of pseudoalterin and the conserved sequence in the catalytic center of family M23 proteases, the complete gene was cloned by a combination of PCR and thermal asymmetric interlaced PCR. The open reading frame of this gene encodes the 403-amino acid precursor of pseudoalterin. This precursor contains a signal peptide sequence of 22 residues (Met-1 to Ala-22), as predicted by SignalP 3.0 (47). A propeptide of 208 residues (Gly-23 to Gln-230) between the signal peptide and mature pseudoalterin was determined according to the N-terminal sequence of the mature protein. The mature enzyme pseudoalterin includes 173 residues (Ala-231 to Arg-403) (supplemental Fig. S5). BLAST searching the NCBI Conserved Domain Database (CDD version 3.06; www.ncbi.nlm.nih.gov) indicated that pseudoalterin is a zinc metallopeptidase belonging to the peptidase M23 family. Pseudoalterin shows the highest identity (53%) to a hypothetical metalloprotease from Pseudoalteromonas tunicata D2 (accession number EAR27283). Among the characterized proteases, pseudoalterin shows the highest identity (39%) to staphylolysin from Pseudomonas aeruginosa PAO1 (accession number AAG05260), a protease of the M23A subfamily. The conserved residues in and around the active site of M23 proteases are also found in pseudoalterin (Fig. 1). These results all point to pseudoalterin as a novel metalloprotease of the M23A subfamily.
FIGURE 1.
Alignment of the sequence of mature pseudoalterin with those of the other M23 proteases. Staphylolysin (P14789) from Pseudomonas aeruginosa PAO1 belongs to M23A; β-lytic metalloprotease (P27458) from Aeromonas hydrophila belongs to M23A; lysostaphin (P10547) from Staphylococcus simulans belongs to M23B; Enterolysin (Q9F8B0) from Enterococcus faecalis belongs to M23B; and zoocin A (Q3K3Z6) from Streptococcus zooepidemicus belongs to M23B. Identical amino acids between pseudoalterin and other proteases are shaded in black. Similar amino acids are shaded in gray. Arrows denote the active site residues.
Substrate Specificity of Pseudoalterin
The substrate specificity of pseudoalterin for proteins and peptides was assayed and compared with that of pseudolysin, a known bacterial elastase in family M4 (48), and myroilysin, a characterized bacterial elastase in family M12 from marine sediment (13). Pseudoalterin had very high activity toward elastinorcein (528 units/mg), much higher than pseudolysin and myroilysin. Except for elastin, pseudoalterin showed little hydrolysis of any other proteins that were tested, including casein, type I collagen fibers, fibrin, and gamma-globulin (Table 3 and supplemental Fig. S6). Pseudoalterin displayed only slight activity for gelatin (Table 3).
TABLE 3.
Substrate specificity of pseudoalterin on various substrates
| Substrate | Activity (units/mg)a |
||
|---|---|---|---|
| Pseudoalterin | Myroilysinb | Pseudolysinb | |
| Casein | 0 | 8541 | 7061 |
| Elastinorcein | 527.9 | 243.3 | 91.1 |
| Gelatin | 12.0 | 4.5 | 5.0 |
| Bovine-insoluble type I collagen fiber | 4.9 | 3.8 | 0 |
| Gamma globulinc | 0 | – | – |
| Furylacryloyl-Gly-Leu-NH2d | 0 | 0 | 642 |
| Furylacryloyl-Gly-Phe-NH2d | 0 | 0 | 3,364 |
| Fibrinc | No hydrolysis | Hydrolyzes α, β, and γ | Hydrolyzes α, β, and γ |
| Oxidized insulin B chain | Gly-8↓Ser-9, Glu-13↓Ala-14, Leu-15↓Tyr-16, Tyr-16↓Leu-17, Leu-17↓Val-18, Cys-19↓Gly-20, Glu-21↓Arg-22, Gly-23↓Phe-24, Phe-24↓Phe-25, Lys-29↓Ala-30 | Asp-3↓Gln-4, His-5↓Leu-6, Leu-6↓Cys-7, Ser-9↓His-10, His-10↓Leu-11, Tyr-16↓Leu-17, Phe-25↓Tyr-26, Tyr-26↓Thr-27, Thr-27↓Pro-28, Lys-29↓Ala-30 | His-5↓Leu-6, His-10↓Leu-11, Ala-14↓Leu-15, Tyr-16↓Leu-17, Leu-17↓Val-18, Gly-23↓Phe-24, Phe-24↓Phe-25, Phe-25↓Tyr-26, Lys-29↓Ala-30 |
a Unless otherwise indicated, the values are specific activities (in units/mg) at 25 °C. The data are the means of three experiments, and the standard deviations were ≤5%.
b The data on the substrate specificity of myroilysin and pseudolysin are cited from a previous study (13) and the MEROPS Database.
c Hydrolysis of gamma globulin and fibrin by pseudoalterin was performed at 25 °C, and the results were analyzed by SDS-PAGE (supplemental Fig. S6). – indicates not done.
d The proteolytic activities with furylacryloyl-Gly-Leu-NH2 and furylacryloyl-Gly-Phe-NH2 were measured with Feder's method at 25 °C (42); the data are the values of kcat/Km (in m−1 s−1).
Those peptides that were released from the oxidized insulin B chain by pseudoalterin were separated and analyzed by HPLC and LC-MS (supplemental Fig. S7). The cleavage sites were analyzed by MASCOT MS/MS Ion Research tools (supplemental Table S2). As shown in Table 3, 10 cleavage sites were identified, among which two cleavage sites (Tyr-16↓Leu-17 and Lys-29↓Ala-30) were also reported for myroilysin and pseudolysin, whereas the cleavage site Gly-23↓Phe-24 was in accord with the specificity of the typical protease of family M23, β-lytic endopeptidase (35).
Observation of Bovine Elastin Degradation by Light Microscopy
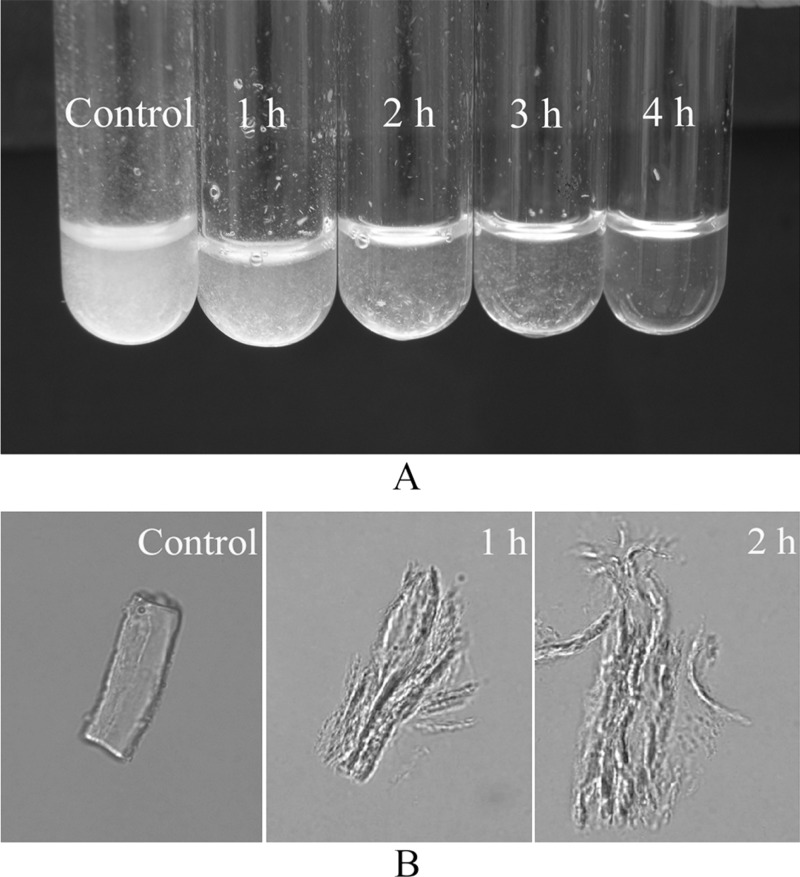
Fig. 2 revealed that pseudoalterin displayed high hydrolytic activity toward bovine elastin. When incubated at 25 °C, 10 mg of bovine elastin was visually degraded by 10 μg of pseudoalterin within 4 h (Fig. 2A). After treatment for 1 h, the elastin swelled, and numerous filaments were released, and these filaments were further degraded into smaller segments with further treatment time (Fig. 2B). These observations suggested that pseudoalterin was likely capable of disrupting the intermolecular cross-links in elastic fibers.
FIGURE 2.
Hydrolysis of bovine elastin by pseudoalterin. Ten milligrams of bovine elastin in 0.5 ml of 50 mm Tris-HCl (pH 9.0) containing 10 μg of pseudoalterin was incubated at 25 °C with continuous stirring. Bovine elastin in 50 mm Tris-HCl (pH 9.0) without pseudoalterin served as the negative control. A, macroscopic observation of the hydrolysis of bovine elastin by pseudoalterin. B, light microscopic observation of hydrolysis of bovine elastin by pseudoalterin after 1 and 2 h. Samples were photographed with an inverted microscope (Olympus IX71, Japan) at room temperature. Magnification is ×960.
Analysis of the Cleavage Sites of Pseudoalterin on Bovine Elastin
To further analyze the elastolytic mechanism of pseudoalterin, its cleavage sites on bovine elastin were analyzed (supplemental Fig. S8). Twenty seven peptides released from bovine elastin by pseudoalterin digestion were separated, and their molecular masses and sequences were identified. Based on the sequences of these peptides, 32 cleavage sites on bovine elastin were determined (supplemental Table S3). Among these cleavage sites, the P1 position is almost always occupied by Gly (31 sites), and the P1′ position is occupied by Gly, Ala, Leu, or Val (supplemental Table S4). Furthermore, a cleavage site Ala↓ Lys in the hydrophilic domain involved in cross-linking in elastin was also detected, which has not been reported for any other M23 protease.
To confirm the cleavage sites analyzed above, based on the sequence of bovine tropoelastin in the database (SwissProt accession number P04985-1), four peptides (GVGVAP, IGGGAGG, GIGLGP, and PGAGARF) in the hydrophobic regions and two typical peptides (AAAKAA and AAAAAKAAAK) in the hydrophilic regions were synthesized to assess their efficacy as potential substrates for pseudoalterin. LC-MS analysis revealed that all six synthetic peptides were digested by pseudoalterin. The cleavage sites were at Gly↓Val, Gly↓Gly, Gly↓Leu, Gly↓Ala, Ala↓Ala, Ala↓Lys, and Lys↓Ala (Fig. 3 and supplemental Fig. S9). This result confirmed the cleavage sites analyzed above and indicated that pseudoalterin can cleave the peptide bonds Ala-Ala and Lys-Ala in the hydrophilic regions in elastin.
FIGURE 3.
Analysis of the cleavage sites of pseudoalterin on the synthetic peptides. Each of these peptides (5 μg) was mixed with 5 μg of pseudoalterin in 50 μl of 50 mm Tris-HCl (pH 9.0). After incubation at 25 °C for 600 min for GVGVAP, IGGGAGG, GIGLGP, and PGAGARF and 110 min for AAAKAA and AAAAAKAAAK, the reaction was terminated by the addition of 1% trifluoroacetic acid. The molecular masses and sequences of peptides were analyzed by LC-MS and the ExPASy FindPept Tool, as shown in supplemental Fig. S9.
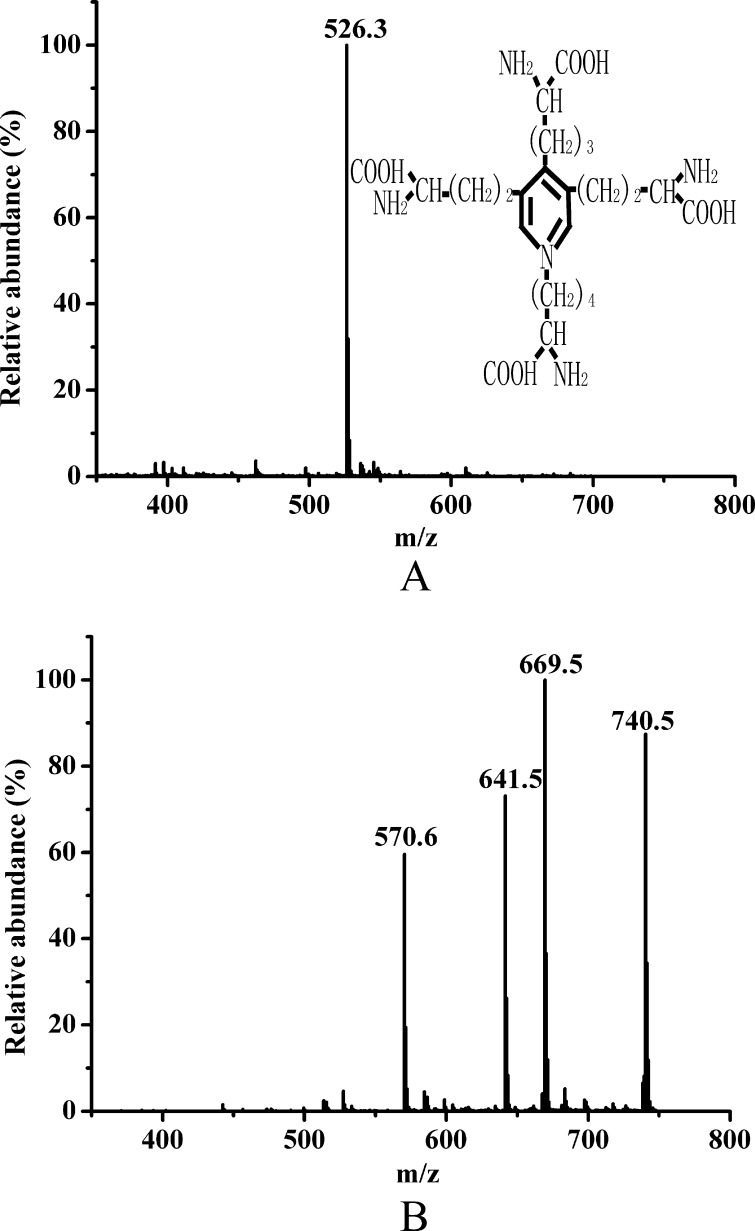
DES is a cross-link that is characteristic of elastin (8). If pseudoalterin can break down the cross-links in elastin by cleaving the peptide bonds Ala–Ala, Ala–Lys, and/or Lys–Ala in the hydrophilic regions, then DES and/or peptic derivatives of DES with one or more alanine residues, such as DES-Ala, DES-Ala-Ala, DES-Ala-Ala-Ala, or DES-Ala-Ala-Ala-Ala, would be released in the elastin hydrolysate. To test this hypothesis, elastin hydrolysates were analyzed by LC-MS. The m/z of the DES standard ([M + H]+) was 526.3 (Fig. 4A), which matched its molecular formula. Because the long peptides in the elastin hydrolysate were removed before LC-MS analysis, a subset of compounds was detected in the mass spectrum of the elastin hydrolysate. Among these peaks, two abundant ions were m/z 669.5 and m/z 740.5, representing the masses of DES-Ala-Ala and DES-Ala-Ala-Ala, respectively (Fig. 4B), indicating that DES-Ala-Ala and DES-Ala-Ala-Ala were released from bovine elastin by pseudoalterin. There were no counterparts in the negative control samples (data not shown).
FIGURE 4.
LC-MS analysis of peptic derivatives of DES in the elastin hydrolysate released by pseudoalterin. A, mass spectrum of DES standard. DES standard was prepared in 0.01 m HCl at a final concentration of 2 mg/ml, treated with an equal volume of 4% sulfosalicylic acid, and loaded onto LC-MS. B, mass spectrum of elastin hydrolysate. Twenty milligrams of bovine elastin was degraded by 20 μg of pseudoalterin at 25 °C for 36 h followed by addition of 500 μl of 4% sulfosalicylic acid to remove the long peptides. After centrifugation, the sample was lyophilized and redissolved in 50 μl of 0.01 m HCl and was then detected using the same method as described for the DES standard.
On this basis, the cleavage sites of pseudoalterin on bovine tropoelastin are shown in Fig. 5. These cleavage sites indicate that pseudoalterin can cleave the peptide bonds both in the hydrophilic regions and in the hydrophobic regions to effectively degrade insoluble elastin.
FIGURE 5.
Cleavage sites by pseudoalterin on bovine tropoelastin (SwissProt accession number P04985-1). Cleavage sites were determined by analysis of the sequences of peptides released by pseudoalterin from bovine elastin as shown in supplemental Table S3, and the cleavage sites of pseudoalterin on six peptides were synthesized according to the sequence of bovine tropoelastin as shown in Fig. 3. Hollow arrows indicate the cleavage sites that have not been identified for other M23 proteases. Solid arrows indicate the cleavage sites that have been reported for other M23 proteases. Gly-Leu bond was reported to be cleaved by Lysobacter β-lytic protease (23). Gly-Gly and Gly-Ala bonds were reported to be cleaved by both Achromobacter β-lytic protease (15) and staphylolysin (29).
SEM Observation of the Degradation of Cross-links between Recombinant Tropoelastin Spherules by Pseudoalterin
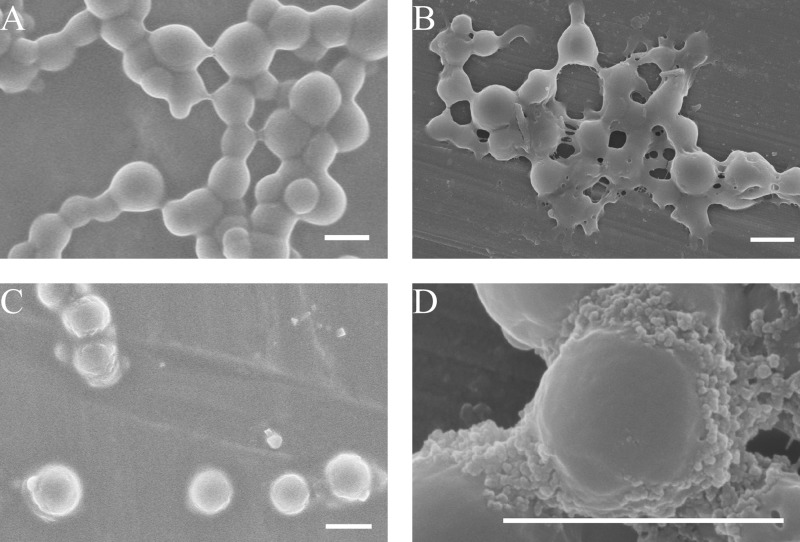
To model the degradation of elastin by pseudoalterin, human tropoelastin was expressed and purified and then chemically cross-linked in vitro. After cross-linking, the tropoelastin spherules were linked into clusters or filaments like beads on a string under SEM (Fig. 6A). When the cross-linked tropoelastin spherules were incubated with pseudoalterin for 10 min, physical contacts between the spherules dissipated or fractured (Fig. 6B). After 15 min of incubation, most clusters separated into single spherules (Fig. 6C), and some spherules began to reveal ∼15 nm substructures, i.e. the size of tropoelastin monomers (Fig. 6D). These observations revealed that pseudoalterin was capable of proteolytically attacking a synthetic elastin model where no other substrate was present other than that which contained target elastolytic sequences, regardless of the fact that it was cross-linked.
FIGURE 6.
Degradation of recombinant tropoelastin cross-linking clusters by pseudoalterin. A, 10 mg/ml tropoelastin following incubation with 10 mm BS3 in 10 mm PBS for 30 min. B–D, cross-linked product incubated with 0.1 mg/ml pseudoalterin for 10 min (B) and 15 min (C and D). Bars, 1 μm.
SEM Observation of Bovine Elastin Degradation by Pseudoalterin
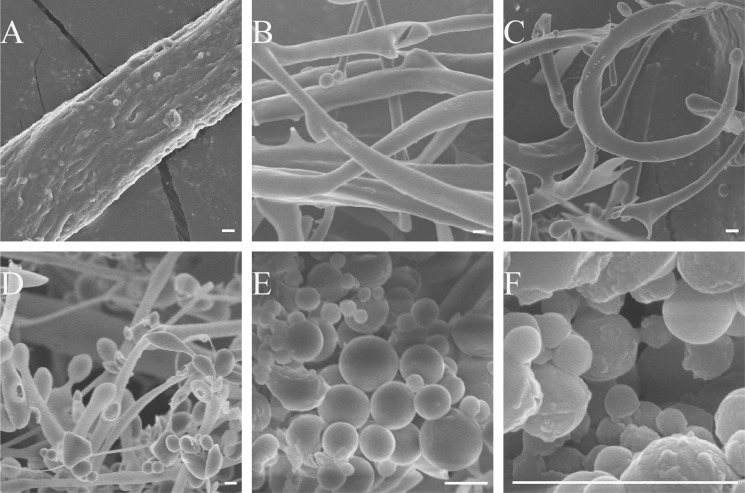
To further understand the degradation mechanism of pseudoalterin on elastin, the process was observed by SEM. Bovine elastin is composed of tightly arranged filaments with a diameter of 1–2 μm under SEM (Fig. 7A). After the fibers were incubated with pseudoalterin for 20 min, the filaments in the fiber appeared to separate (Fig. 7B), which was consistent with the light microscopic observations (Fig. 2B). After 40 min of incubation, these filaments completely separated and curled due to their increased flexibility. Meanwhile, droplets with the same diameter as the filaments began to appear at filamentous termini (Fig. 7C). More protein droplets formed with increasing incubation time (Fig. 7D). After 80 min, almost all the fibers had digested into nanometer- to micrometer-range spherules (Fig. 7E), which presumably then degraded into smaller peptides that were undetectable by SEM. The smallest detected spherules were ∼150 nm (Fig. 7F).
FIGURE 7.
SEM observation of elastin degradation by pseudoalterin. Pseudoalterin was added to bovine elastin at a ratio of 1:500 in 50 mm Tris-HCl (pH 9.0) at 25 °C with continuous stirring for 20 min (B), 40 min (C), 60 min (D), and 80 min (E and F). The same reaction system without pseudoalterin served as control (A). After washing twice with deionized water, the insoluble material was lyophilized and mounted on stubs, spatter-coated with 5 nm platinum, and examined with a Hitachi FE-S4800 at 5.0 kV. Bars, 1 μm.
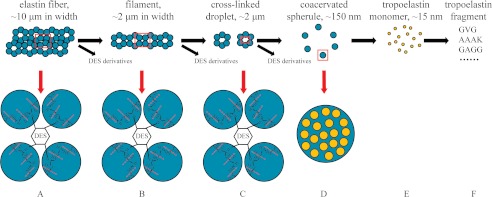
Based on SEM observations and the other results above, a model for the degradation mechanism of pseudoalterin on insoluble elastin is proposed (Fig. 8). Elastin is composed of cross-linked filaments (Fig. 8A). When elastin is treated with pseudoalterin, the cross-links between these filaments are initially broken down and result in separating filaments and the release of cross-links in the form of DES-Ala-Ala and DES-Ala-Ala-Ala (Fig. 8B). These filaments appear as cross-linked droplets and are further cut into single droplets due to the continuing digestion of cross-links (Fig. 8C). The droplets formed by cross-linked coacervated spherules are further digested, and the coacervated spherules are released when the peptide bonds connecting the cross-links between spherules are attacked (Fig. 8D). Subsequently, these spherules, which are a coacervate of damaged elastin fragments, are digested into scattered molecules (Fig. 8E), which are further hydrolyzed into small peptides due to the attack by pseudoalterin on Gly-Xaa sites in the hydrophobic regions (Fig. 8F).
FIGURE 8.
Schematic model for the elastin degradation mechanism of pseudoalterin. In this model, elastin is composed of filaments cross-linked by DES and IDE (A). When the fibers are partly hydrolyzed by pseudoalterin, single filaments and cross-links are released where the cross-links are attacked (B). Cross-links between protein droplets in the filaments are further attacked, and separated droplets and cross-linking molecules appear (C). The droplets then dissociate to release coacervated spherules due to destruction of the cross-links between the coacervation spherules (D). Spherules comprising coacervated and partly degraded elastin are digested into scattered damaged elastin species around the same size as tropoelastin (E), which are further hydrolyzed to yield small peptides due to the continued attack by pseudoalterin on Gly-Xaa sites in the hydrophobic regions (F). Blue balls, coacervate spherules; orange balls, tropoelastin monomers; red broken lines indicate the potential cleavage sites of pseudoalterin in the cross-linking regions of elastin.
DISCUSSION
Identification of the main microorganisms in deep sea sediment and the enzymes that they use for organic nitrogen hydrolysis is necessary to gain an understanding of the degradation and burial of particulate organic nitrogen in the seabed (49). However, the bacterial species and the kinds of proteases participating in particulate organic nitrogen hydrolysis process remain largely unknown. Analysis of the genomes of 21 bacteria isolated from deep-sea sediments and hydrothermal vents, which are in GenBankTM database, indicates that most of these bacteria could secrete one or several M23 family proteases (supplemental Table S5). This finding suggests that the M23 family proteases may be popular in deep sea sediments and may therefore play an important role in sedimentary particulate organic nitrogen degradation. However, the properties and functionality of marine M23 proteases have not been reported. In this study, the novel M23 protease pseudoalterin, secreted by the deep-sea sedimentary bacterium Pseudoalteromonas sp. CF6-2, was characterized, and its elastolytic mechanism was studied in detail.
Sequence analysis showed that pseudoalterin has the highest identity (39%) to the M23 protease staphylolysin from P. aeruginosa PAO1. However, in contrast to staphylolysin that slowly degrades insoluble elastin (18), pseudoalterin quickly degrades insoluble elastin. It is worth noting that pseudoalterin's activity on elastinorcein is much higher than that of pseudolysin, an elastase of family M4 from P. aeruginosa (48), and that of myroilysin, an elastase of family M12 from the deep-sea bacterium M. profundi D25 (13). This increased activity points to difference(s) in the elastolytic mechanism between pseudoalterin and staphylolysin as well as other M23 proteases.
Light microscopic observations indicated that pseudoalterin swells elastin and separates elastin into filaments, implying that this enzyme initially break downs cross-links between the filaments. This elastolytic pattern is quite different from that of myroilysin that cuts elastic fibers transversely at first (13). All reported M23 proteases show specificity for glycyl bonds (23, 28–32) and therefore only have the capacity to cleave in the hydrophobic regions in elastin. However, analysis of the cleavage sites of pseudoalterin on bovine elastin indicated that, in addition to glycyl bonds, the peptide bond Ala–Lys, which is found in elastin cross-linking domains, was also cleaved by pseudoalterin. Further analysis with six peptides synthesized according to the bovine tropoelastin sequence showed that pseudoalterin also cleaves the peptide bonds Ala–Ala and Lys–Ala, which are found in the hydrophilic elastin cross-linking domains. These results indicated that pseudoalterin cleaves not only glycyl bonds in the hydrophobic regions but also peptide bonds in elastin cross-linking domain in the hydrophilic regions during elastin degradation.
DES and IDE are the cross-linking molecules in the network of elastin. Detection in elastin hydrolysate of their peptic derivatives DES/IDE-A, DES/IDE-AA, DES/IDE-AAA, and DES/IDE-AAAA can provide direct evidence for the activity of pseudoalterin toward the peptide bonds on both sides of lysine residue in the hydrophilic domains. Several methodologies for monitoring DES/IDS have been developed in the last 2 decades, among which the LC-MS methods provide the best specificity and sensitivity (50). However, methods to monitor their peptic derivatives are rare. We aimed to detect DES or the peptic derivatives of DES in the elastin hydrolysate released by pseudoalterin by using LC-MS. Two molecular masses, 669.5 (m/z) and 740.5 (m/z), were detected in the hydrolysate, which was in complete accord with DES-Ala-Ala and DES-Ala-Ala-Ala. This finding indicated that pseudoalterin cleaves peptide bonds in the hydrophilic regions of elastin to release cross-links. Moreover, as a model system with defined components, degradation of pseudoalterin of the chemical cross-links between recombinant tropoelastin spherules was directly observed under SEM, and it provided strong evidence for the same conclusion. Furthermore, SEM observation of the degradation process by pseudoalterin on bovine elastin showed that pseudoalterin successively released filaments, droplets, and spherules from elastic fibers by destroying the cross-links at each structural level. Based on the results of biochemical experiments and SEM observations, a detailed mechanism for elastin degradation by pseudoalterin is proposed (Fig. 8).
In summary, in the novel elastolytic mechanism of pseudoalterin, a new M23 protease from deep sea bacterium Pseudoalteromonas sp. CF6-2 was studied. In addition to the cleavage of glycyl bonds in elastin as other M23 proteases, pseudoalterin also cleaves the peptide bonds in elastin's hydrophilic regions that include cross-links, leading to an ensuing rapid degradation of elastin. As the bacterial proteases of M23 may be common in marine sediments, as analyzed above (supplemental Table S5), the high activity of pseudoalterin toward elastin indicates the important role of M23 proteases in the degradation of the marine sedimentary elastin. Our results on the elastolytic mechanism of pseudoalterin are helpful for clarifying the degradation mechanism of deep-sea sedimentary elastin.
It is worth reiterating that, as stated in the Introduction, specific steps in the assembly of tropoelastin in vivo remain unclear. The degradation of bovine elastin by pseudoalterin supplies a new path to help investigate this system from the opposite direction. The smallest observed spherules indicate that ∼150 nm is the fundamental size in coacervation. Many researchers recognize 100–200-nm fibrillar intermediates in native and artificial elastin (51–54). At the late stage of cross-linked tropoelastin degradations that were seen here, particles were released that were similar to the size of the tropoelastin monomer. By using pseudoalterin to dissect components of the native elastic fiber, it could be speculated that tropoelastin monomers interconvert with ∼150-nm spherules by initial coacervation, then into 2–3-μm spheres accompanied by progressive cross-linking (3), and association with microfibrils where they are cross-linked into networked elastic fibers.
Supplementary Material
The work was supported by National Natural Science Foundation of China Grants 31025001, 31000034, 91228210, 31170055, 81271896, and 41276149, Hi-Tech Research and Development Program of China Grants 2011AA090703 and 2012AA092103, COMRA Program Grants DY125-15-T-05 and DY125-13-E-01, and Special Fund of China for Marine-scientific Research in the Public Interest Grant 201005032-6.

This article contains supplemental Figs. S1–S9, Tables S1–S5, and additional references.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) HQ005379.1.
- DES
- desmosine
- IDE
- isodesmosine
- SEM
- scanning electron microscope
- BS3
- bis (sulfosuccinimidyl) suberate.
REFERENCES
- 1. Wise S. G., Weiss A. S. (2009) Tropoelastin. Int. J. Biochem. Cell Biol. 41, 494–497 [DOI] [PubMed] [Google Scholar]
- 2. Yeo G. C., Keeley F. W., Weiss A. S. (2011) Coacervation of tropoelastin. Adv. Colloid. Interfac. SCI 167, 94–103 [DOI] [PubMed] [Google Scholar]
- 3. Kozel B. A., Rongish B. J., Czirok A., Zach J., Little C. D., Davis E. C., Knutsen R. H., Wagenseil J. E., Levy M. A., Mecham R. P. (2006) Elastic fiber formation. A dynamic view of extracellular matrix assembly using timer reporters. J. Cell. Physiol. 207, 87–96 [DOI] [PubMed] [Google Scholar]
- 4. Czirok A., Zach J., Kozel B. A., Mecham R. P., Davis E. C., Rongish B. J. (2006) Elastic fiber macro-assembly is a hierarchical, cell motion-mediated process. J. Cell. Physiol. 207, 97–106 [DOI] [PubMed] [Google Scholar]
- 5. Tu Y., Weiss A. S. (2010) Transient tropoelastin nanoparticles are early-stage intermediates in the coacervation of human tropoelastin whose aggregation is facilitated by heparan sulfate and heparin decasaccharides. Matrix Biol. 29, 152–159 [DOI] [PubMed] [Google Scholar]
- 6. Muiznieks L. D., Jensen S. A., Weiss A. S. (2003) Structural changes and facilitated association of tropoelastin. Arch. Biochem. Biophys. 410, 317–323 [DOI] [PubMed] [Google Scholar]
- 7. Clarke A. W., Arnspang E. C., Mithieux S. M., Korkmaz E., Braet F., Weiss A. S. (2006) Tropoelastin massively associates during coacervation to form quantized protein spheres. Biochemistry 45, 9989–9996 [DOI] [PubMed] [Google Scholar]
- 8. Debelle L., Tamburro A. M. (1999) Elastin. Molecular description and function. Int. J. Biochem. Cell Biol. 31, 261–272 [DOI] [PubMed] [Google Scholar]
- 9. Mithieux S. M., Tu Y., Korkmaz E., Braet F., Weiss A. S. (2009) In situ polymerization of tropoelastin in the absence of chemical cross-linking. Biomaterials 30, 431–435 [DOI] [PubMed] [Google Scholar]
- 10. Tu Y., Wise S. G., Weiss A. S. (2010) Stages in tropoelastin coalescence during synthetic elastin hydrogel formation. Micron 41, 268–272 [DOI] [PubMed] [Google Scholar]
- 11. Chen Z., Shin M. H., Moon Y. J., Lee S. R., Kim Y. K., Seo J. E., Kim J. E., Kim K. H., Chung J. H. (2009) Modulation of elastin exon 26A mRNA and protein expression in human skin in vivo. Exp. Dermatol. 18, 378–386 [DOI] [PubMed] [Google Scholar]
- 12. Wu W. J., Vrhovski B., Weiss A. S. (1999) Glycosaminoglycans mediate the coacervation of human tropoelastin through dominant charge interactions involving lysine side chains. J. Biol. Chem. 274, 21719–21724 [DOI] [PubMed] [Google Scholar]
- 13. Chen X. L., Xie B. B., Bian F., Zhao G. Y., Zhao H. L., He H. L., Zhou B. C., Zhang Y. Z. (2009) Ecological function of myroilysin, a novel bacterial M12 metalloprotease with elastinolytic activity and a synergistic role in collagen hydrolysis, in biodegradation of deep-sea high molecular weight organic nitrogen. Appl. Environ. Microbiol. 75, 1838–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyamoto K., Tsujibo H., Nukui E., Itoh H., Kaidzu Y., Inamori Y. (2002) Isolation and characterization of the genes encoding two metalloproteases (MprI and MprII) from a marine bacterium, Alteromonas sp. strain O-7. Biosci. Biotechnol. Biochem. 66, 416–421 [DOI] [PubMed] [Google Scholar]
- 15. Li S., Norioka S., Sakiyama F. (1998) Bacteriolytic activity and specificity of Achromobacter β-lytic protease. J. Biochem. 124, 332–339 [DOI] [PubMed] [Google Scholar]
- 16. Loewy A. G., Santer U. V., Wieczorek M., Blodgett J. K., Jones S. W., Cheronis J. C. (1993) Purification and characterization of a novel zinc proteinase from cultures of Aeromonas hydrophila. J. Biol. Chem. 268, 9071–9078 [PubMed] [Google Scholar]
- 17. Nilsen T., Nes I. F., Holo H. (2003) Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69, 2975–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peters J. E., Galloway D. R. (1990) Purification and characterization of an active fragment of the LasA protein from Pseudomonas aeruginosa. Enhancement of elastase activity. J. Bacteriol. 172, 2236–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schindler C. A., Schuhardt V. T. (1965) Purification and properties of lysostaphin-A lytic agent for Staphylococcus aureus. Biochim. Biophys. Acta 97, 242–250 [DOI] [PubMed] [Google Scholar]
- 20. Simmonds R. S., Naidoo J., Jones C. L., Tagg J. R. (1995) The streptococcal bacteriocin-like inhibitory substance, zoocin A, reduces the proportion of Streptococcus mutans in an artificial plaque. Microb. Ecol. Health Dis. 8, 281–292 [Google Scholar]
- 21. Whitaker D. R. (1965) Lytic enzymes of Sorangium sp. Isolation and enzymatic properties of the α- and β-lytic proteases. Can. J. Biochem. 43, 1935–1954 [DOI] [PubMed] [Google Scholar]
- 22. Zyskind J. W., Pattee P. A., Lache M. (1965) Staphylolytic substance from a species of Pseudomonas. Science 147, 1458–1459 [DOI] [PubMed] [Google Scholar]
- 23. Ahmed K., Chohnan S., Ohashi H., Hirata T., Masaki T., Sakiyama F. (2003) Purification, bacteriolytic activity, and specificity of β-lytic protease from Lysobacter sp. IB-9374. J. Biosci. Bioeng. 95, 27–34 [DOI] [PubMed] [Google Scholar]
- 24. Kessler E., Safrin M., Olson J. C., Ohman D. E. (1993) Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268, 7503–7508 [PubMed] [Google Scholar]
- 25. Uehara T., Dinh T., Bernhardt T. G. (2009) LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 191, 5094–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Möll A., Schlimpert S., Briegel A., Jensen G. J., Thanbichler M. (2010) DipM, a new factor required for peptidoglycan remodeling during cell division in Caulobacter crescentus. Mol. Microbiol. 77, 90–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rawlings N. D., Barrett A. J., Bateman A. (2010) MEROPS. The peptidase database. Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kessler E. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawling N. D., Woessner J. F., eds) 2nd Ed., pp. 998–1000, Elsevier, London [Google Scholar]
- 29. Kessler E., Ohman D. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawling N. D., Woessner J. F., eds) 2nd Ed., pp. 1001–1003, Elsevier, London [Google Scholar]
- 30. Kessler E., Safrin M., Abrams W. R., Rosenbloom J., Ohman D. E. (1997) Inhibitors and specificity of Pseudomonas aeruginosa LasA. J. Biol. Chem. 272, 9884–9889 [DOI] [PubMed] [Google Scholar]
- 31. Park P. W., Mecham R. P. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawling N. D., Woessner J. F., eds) 2nd Ed., pp. 1004–1005, Elsevier, London [Google Scholar]
- 32. Vessillier S., Delolme F., Bernillon J., Saulnier J., Wallach J. (2001) Hydrolysis of glycine-containing elastin pentapeptides by LasA, a metalloelastase from Pseudomonas aeruginosa. Eur. J. Biochem. 268, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 33. Rosenbloom J., Abrams W. R., Mecham R. (1993) Extracellular matrix 4. The elastic fiber. FASEB J. 7, 1208–1218 [PubMed] [Google Scholar]
- 34. Oza N. B. (1973) β-Lytic protease, a neutral sorangiopeptidase. Int. J. Pept. Protein Res. 5, 365–369 [PubMed] [Google Scholar]
- 35. Whitaker D. R., Roy C., Tsai C. S., Jurásek L. (1965) Lytic enzymes of Sorangium sp. A comparison of the proteolytic properties of the α- and β-lytic proteases. Can. J. Biochem. 43, 1961–1970 [DOI] [PubMed] [Google Scholar]
- 36. Zhou M. Y., Chen X. L., Zhao H. L., Dang H. Y., Luan X. W., Zhang X. Y., He H. L., Zhou B. C., Zhang Y. Z. (2009) Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China sea. Microb. Ecol. 58, 582–590 [DOI] [PubMed] [Google Scholar]
- 37. Chen X. L., Xie B. B., Lu J. T., He H. L., Zhang Y. (2007) A novel type of subtilase from the psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Catalytic and structural properties of deseasin MCP-01. Microbiology 153, 2116–2125 [DOI] [PubMed] [Google Scholar]
- 38. He H. L., Chen X. L., Li J. W., Zhang Y. Z., Gao P. J. (2004) Taste improvement of refrigerated meat treated with cold-adapted protease. Food Chem. 84, 307–311 [Google Scholar]
- 39. Zhao G. Y., Chen X. L., Zhao H. L., Xie B. B., Zhou B. C., Zhang Y. Z. (2008) Hydrolysis of insoluble collagen by deseasin MCP-01 from deep-sea Pseudoalteromonas sp. SM9913. Collagenolytic characters, collagen-binding ability of C-terminal polycystic kidney disease domain, and implication for its novel role in deep-sea sedimentary particulate organic nitrogen degradation. J. Biol. Chem. 283, 36100–36107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Worthington Biochemical Corp (1972) Worthington Enzyme Manual, pp. 43–45, Freehold, NJ [Google Scholar]
- 41. Datta G., Dong A., Witt J., Tu A. T. (1995) Biochemical characterization of basilase, a fibrinolytic enzyme from Crotalus basiliscus basiliscus. Arch. Biochem. Biophys. 317, 365–373 [DOI] [PubMed] [Google Scholar]
- 42. Feder J. (1968) A spectrophotometric assay for neutral protease. Biochem. Biophys. Res. Commun. 32, 326–332 [DOI] [PubMed] [Google Scholar]
- 43. Kruger N. J. (1996) in The Protein Protocols Handbook (Walker J. M., ed) pp. 15–20, Humana Press Inc., Totowa, NJ [Google Scholar]
- 44. Authier F., Metioui M., Fabrega S., Kouach M., Briand G. (2002) Endosomal proteolysis of internalized insulin at the C-terminal region of the B chain by cathepsin D. J. Biol. Chem. 277, 9437–9446 [DOI] [PubMed] [Google Scholar]
- 45. Martin S. L., Vrhovski B., Weiss A. S. (1995) Total synthesis and expression in Escherichia coli of a gene encoding human tropoelastin. Gene 154, 159–166 [DOI] [PubMed] [Google Scholar]
- 46. Salvesen G., Nagase H. (1989) in Proteolytic Enzymes: A Practical Approach (Beynon R. J., Bond J. S., eds) pp. 83–104, IRL Press at Oxford University Press, Oxford [Google Scholar]
- 47. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides. SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 48. Kessler E., Ohman D. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawling N. D., Woessner J. F., eds) 2nd Ed., pp. 401–409, Elsevier, London [Google Scholar]
- 49. Jørgensen B. B., Boetius A. (2007) Feast and famine–microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781 [DOI] [PubMed] [Google Scholar]
- 50. Albarbarawi O., Barton A., Lin Z., Takahashi E., Buddharaju A., Brady J., Miller D., Palmer C. N., Huang J. T. (2010) Measurement of urinary total desmosine and isodesmosine using isotope-dilution liquid chromatography-tandem mass spectrometry. Anal. Chem. 82, 3745–3750 [DOI] [PubMed] [Google Scholar]
- 51. Ushiki T. (2002) Collagen fibers, reticular fibers, and elastic fibers. A comprehensive understanding from a morphological viewpoint. Arch. Histol. Cytol. 65, 109–126 [DOI] [PubMed] [Google Scholar]
- 52. Kewley M. A., Steven F. S., Williams G. (1977) The presence of fine elastin fibrils within the elastin fiber observed by scanning electron microscopy. J. Anat. 123, 129–134 [PMC free article] [PubMed] [Google Scholar]
- 53. Bressan G. M., Castellani I., Giro M. G., Volpin D., Fornieri C., Pasquali Ronchetti I. (1983) Banded fibers in tropoelastin coacervates at physiological temperatures. J. Ultrastruct. Res. 82, 335–340 [DOI] [PubMed] [Google Scholar]
- 54. Ushiki T., Murakumo M. (1991) Scanning electron microscopic studies of tissue elastin components exposed by a KOH-collagenase or simple KOH digestion method. Arch. Histol. Cytol. 54, 427–436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.