Background: HoxA10 target genes include CDX4 and FGF2, and HOXA10 is a Cdx4 target gene.
Results: Transcription of newly identified, β-catenin-binding cis elements in the CDX4 and HOXA10 promoters is regulated by Fgf2-dependent activity of β-catenin.
Conclusion: Positive feedback between CDX4 and HOXA10 involves β-catenin and Fgf2.
Significance: Understanding cross-regulation between HoxA10, Cdx4, and β-catenin provides insights into leukemogenesis.
Keywords: Gene Expression, Gene Transcription, General Transcription Factors, Homeobox, Leukemia
Abstract
HoxA10 is a homeodomain transcription factor that is involved in maintenance of the myeloid progenitor population and implicated in myeloid leukemogenesis. Previously, we found that FGF2 and CDX4 are direct target genes of HoxA10 and that HOXA10 is a Cdx4 target gene. We also found that increased production of fibroblast growth factor 2 (Fgf2) by HoxA10-overexpressing myeloid progenitor cells results in activation of β-catenin in an autocrine manner. In this study, we identify novel cis elements in the CDX4 and HOXA10 genes that are activated by β-catenin in myeloid progenitor cells. We determine that β-catenin interacts with these cis elements, identifying both CDX4 and HOXA10 as β-catenin target genes in this context. We demonstrate that HoxA10-induced CDX4 transcription is influenced by Fgf2-dependent β-catenin activation. Similarly, Cdx4-induced HOXA10 transcription is influenced by β-catenin in an Fgf2-dependent manner. Increased expression of a set of Hox proteins, including HoxA10, is associated with poor prognosis in acute myeloid leukemia. Cdx4 contributes to leukemogenesis in Hox-overexpressing acute myeloid leukemia, and increased β-catenin activity is also associated with poor prognosis. The current studies identify a molecular mechanisms through which increased expression of HoxA10 increases Cdx4 expression by direct CDX4 activation and by Fgf2-induced β-catenin activity. This results in Cdx4-induced HoxA10-expression, creating a positive feedback mechanism.
Introduction
Hox proteins are homeodomain transcription factors that are highly conserved from Drosophila to humans (1). In humans and mice, HOX genes are found in groups on four different chromosomes with 9–11 genes in each group (referred to as HOXA-D). During embryogenesis, HOX genes control organogenesis and are activated 5′ to 3′ in a cranial to caudal manner (1). During definitive hematopoiesis, HOX genes are activated 5′ to 3′ in a temporal manner with Hox1 to -4 highly expressed in hematopoietic stem cells (HSC)2 and Hox7 to -11 expressed in committed progenitor cells (2).
Consistent with this expression pattern, HoxB3 and -B4 are involved in HSC maintenance. Overexpression of these proteins in bone marrow cells results in HSC expansion in vitro and development of a myeloproliferative neoplasm in vivo in murine transplantation studies (3, 4). In contrast, overexpression of HoxA9 or A10 in bone marrow cells expands the common granulocyte/monocyte progenitor (GMP) population in vitro and results in a myeloproliferative neoplasm in vivo (5–9). In murine recipients of HoxA10-overexpressing bone marrow, this myeloproliferative neoplasm progresses to acute myeloid leukemia (AML), suggesting that HoxA10 is sufficient for GMP expansion, but acquisition of additional mutations is necessary for differentiation block and AML (10, 11).
Clinical correlative studies implicate Hox proteins in human leukemia. Increased and dysregulated expression of a set of Hox proteins (HoxB3, -B4, and -A7-11) is found in poor prognosis leukemias (12–14). This includes leukemias with translocations involving the MLL (mixed lineage leukemia) gene, referred to as 11q23 leukemias (15–18). Expression of this set of Hox proteins is also found in association with other recurring chromosomal translocations and in a poor prognosis subset with normal karyotype. Increased expression of Hox proteins in leukemia is often associated with dysregulated expression of other homeodomain transcription factors, including Meis and Cdx proteins.
Cdx proteins are of particular interest because they function as regulators of the HOX locus. Cdx4 influences expression of a number of HOX genes, including the genes encoding HoxB3, -B4, and -A7-11 (19). Cdx4 expression has been shown to contribute to Hox-induced leukemogenesis in murine models and to bypass the requirement for Mll for hematopoiesis (20). Overexpression of Cdx4 in murine bone marrow results in a myeloproliferative neoplasm that progresses to AML in vivo, and co-overexpression of Cdx4 and HoxB4 in ES cells induces long term repopulating activity in murine transplantation experiments (19, 21).
In previous studies, we identified CDX4 as a HoxA10 target gene using a high throughput, chromatin immunoprecipitation-based screening approach (22). We also found that HOXA10 is a direct target gene for Cdx4, indicating positive feedback between these genes. Other investigators identified Cdx4 as a downstream target for canonical Wnt signaling. These studies identified several β-catenin-Lef binding consensus sequences in the proximal 1.1 kb of the murine CDX4 promoter that contributed to promoter activity in transfection studies using embryonic carcinoma cells (23).
In other previous studies, we identified FGF2 as a HoxA10 target gene (24). We found that overexpression of HoxA10 in primary murine myeloid progenitor cells or myeloid cell lines increases Fgf2 expression and results in Fgf2-dependent activation of β-catenin in an autocrine manner. We also found that Fgf2-induced β-catenin activity contributes to the increased response of HoxA10-overexpressing cells to cytokine-induced proliferation (i.e. cytokine hypersensitivity) (24).
Therefore, the current studies investigate the hypothesis that HoxA10 influences CDX4 transcription both directly and via Fgf2-induced activation of β-catenin. In addition, we identified HOXA10 as a novel β-catenin target gene, indicating another positive feedback mechanism for HoxA10 expression. Because Hox expression and β-catenin activity are both associated with poor prognosis in human AML, these studies are of interest for unraveling the complex interactions between HOX genes, β-catenin, and adverse events in human disease.
MATERIALS AND METHODS
Plasmids
Protein Expression Vectors
The cDNA representing the major transcript of human HoxA10 was obtained from C. Largman (University of California, San Francisco) (25, 26). The cDNA for human Cdx4 was obtained by reverse transcription and PCR from U937 cells. The sequence of this cDNA was compared with the sequence in GenBankTM. Both were subcloned into the pSRα vector for expression in mammalian cell lines and the MSCV vector for generation of retrovirus (per manufacturer's instructions; Stratagene, La Jolla, CA). The cDNA for β-catenin was obtained from Origene Technologies Inc. (Rockville, MD).
shRNA Expression Vectors
HoxA10- and Cdx4-specific shRNAs and scrambled control sequences were designed with the assistance of the Promega Web site (Madison, WI). Double-stranded oligonucleotides representing the complementary sequences separated by a hairpin loop were subcloned into the pLKO.1puro vector (a gift from Dr. Kathy Rundell, Northwestern University, Chicago). Several sequences were tested, and the most efficient sequences were used in combination. Negative controls for these experiments were matched shRNAs with rearranged sequences (bp swapping or scrambled controls). Vectors to express an shRNA specific to mouse and human β-catenin were obtained from OriGene (OriGene, Rockville, MD).
CDX4 and HOXA10 Reporter Vectors
Various sequences from the CDX4 or HOXA10 5′-flanks were amplified by PCR from chromatin extracted from the human U937 myeloid cell line. The fragments were sequenced to ensure identity with the ENSEMBL database. CDX4 5′-flanking sequences were subcloned into the pGL3-E reporter vector, and HOXA10 5′-flanking sequences were subcloned into the pGL3-basic reporter vector (Promega). Additional constructs were generated in the pGL3-promoter vector with three copies of the −1020 to −1045 bp (β-catenin-binding) sequence from the CDX4 promoter or with two copies of the −800 to −825 bp (β-catenin-binding) sequence from the HOXA10 promoter (as described under “Oligonucleotides”).
Oligonucleotides
Oligonucleotides were custom synthesized by MWG Biotech (Piedmont, NC). Oligonucleotides used for EMSA were as follows: −1020 to −1045 bp of CDX4 promoter (wild type, 5′-TGTCTTTTTCAACAATTGGAGTTTT-3′; β-catenin-binding mutant, 5′-TGTCTTTTTCAAGTTTTGGAGTTTT-3′), −809 to −829 bp of HOXA10 promoter (wild type, 5′-TCTTTCCATCAAGGCTGGCA-3′; mutant, 5′-TCTTTCCTAGAAGGCTGGCA-3′), a consensus sequence for β-catenin/Tcf binding (5′-AAGATCAAAGGGGGTA-3′; mutant, 5′-GGTAAGGCCAAAGGG-3′; from FOPFlash and TOPFlash vectors, respectively). Oligonucleotides used for real time PCR to quantify mRNA expression were as follows (listed 5′ to 3′): Cdx4 (forward, CGAGAAGACTGGAGCGTGTA; reverse, CTGTAGTCGGTCGAGCAGAA), HoxA10 (forward, ACACTGGAGCTGGAGAAGGA; reverse, TCACTTGTCTGTCCGTGAGG).
Myeloid Cell Line Culture
The human myelomonocytic leukemia cell line U937 (28) was obtained from Andrew Kraft (Hollings Cancer Center, University of South Carolina, Charleston, SC). Cells were maintained as described (29).
Primary Murine Bone Marrow Studies
Animal studies were performed according to a protocol approved by the Animal Care and Use Committees of Northwestern University and the Jesse Brown Veterans Affairs Medical Center.
Bone Marrow Harvest and Culture
Bone marrow mononuclear cells were obtained from the femurs of WT or HoxA10−/− C57/BL6 mice (30). Sca1+ cells were separated using the Miltenyi magnetic bead system (Miltenyi Biotechnology, Auburn, CA). Bipotential myeloid progenitor cells were cultured (2 × 105 cells/ml) for 48 h in DMEM supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, 10 ng/ml murine GM-CSF (R&D Systems Inc., Minneapolis, MN), and 10 ng/ml murine recombinant IL-3 (R&D Systems Inc.). Cells were either maintained in GM-CSF + IL-3 for 48 h or differentiated over 48 h in 10 ng/ml G-CSF (granulocyte) or 10 ng/ml murine M-CSF (monocyte).
Bone Marrow Retroviral Transduction
Retrovirus (∼107 pfu/ml) was generated with the HoxA10/MSCV or control MSCV plasmid using the Phoenix cell packaging line according to the manufacturer's instructions (Stratagene, La Jolla, CA). Bone marrow mononuclear cells were cultured for 24 h in 10 ng/ml IL-3, 10 ng/ml GM-CSF, and 100 ng/ml SCF. Cells were transduced by incubation with retroviral supernatant in the presence of Polybrene (6 μg/ml) as described (10). Transduced cells were selected for 48 h in puromycin and differentiated with M-CSF or G-CSF (20 ng/ml).
Quantitative Real-time PCR
RNA was isolated using TRIzol reagent (Invitrogen) and tested for integrity by denaturing gel electrophoresis. Primers were designed with Applied Biosystems software, and real-time PCR was performed using SYBR Green according to the “standard curve” method. Results were normalized to 18S (for mRNA) or input chromatin (for co-precipitated chromatin).
Chromatin Immunoprecipitation
U937 cells were briefly treated with formaldehyde to generate DNA-protein cross-links. Cell lysates were sonicated to generate chromatin fragments of ∼200 bp and immunoprecipitated with a β-catenin-specific antibody (Abcam, Cambridge, MA). Precipitated chromatin was recovered and analyzed by PCR using primer sets to amplify various sequences in the 5′-flank of CDX4 or HOXA10. Several batches of precipitated chromatin were combined for each experiment (31).
Myeloid Cell Line Transfections and Assays
Stable Transfectants
U937 cells were transfected by electroporation with equal amounts of β-catenin, HoxA10, or Cdx4 expression vector or empty vector control (using pcDNAamp) plus a vector with a neomycin phosphotransferase cassette (pSRα) (30 μg each). Stable pools of transfected cells were selected in G418 (0.5 mg/ml), and aliquots of cells were tested for HoxA10 and Cdx4 expression by Western blot and real-time PCR.
Other cells were transfected by electroporation with a lentiviral vector (pLKO.1puro) for expression of β-catenin-specific shRNAs or scrambled control shRNAs. Stable pools of transfected cells were selected in puromycin (1.2 μg/ml). In some experiments, U937 cells were stably co-transfected with a HoxA10 expression vector, Cdx4 expression vector, or empty vector control plus a vector to express β-catenin-specific shRNAs or scrambled control shRNAs. Co-transfectants were selected in both G418 and puromycin.
CDX4 and HOXA10 Reporter Assays
To identify β-catenin-binding cis elements in the CDX4 promoter, U937 cells were co-transfected with a luciferase reporter vector (pGL3-E) containing 1.4 kb, 1050 bp (with or without mutation of the β-catenin-binding consensus in this construct), 1025 bp, or 539 bp of CDX4 5′-flanking sequences or pGL3-E control (30 μg) and a vector to overexpress β-catenin (or empty vector control) (50 μg). In other experiments, cells were co-transfected with a luciferase reporter vector (pGL3-basic) with 1.0 kb, 825 bp (with or without mutation of the β-catenin-binding consensus in this construct), 772 bp, or 200 bp of the HOXA10 5′-flank or with pGL3-basic control (30 μg) and a vector to overexpress β-catenin or empty expression vector (50 μg).
In additional experiments, cells were co-transfected with a minimal promoter/luciferase reporter vector with multiple copies of either the β-catenin-binding cis element from the CDX4 promoter (−1025 to −1050 bp) or the β-catenin-binding cis element from the HOXA10 promoter (−800 to −825 bp) (30 μg) and vectors to overexpress β-catenin or β-catenin-specific shRNAs or relevant controls (empty vectors or vector with scrambled shRNA sequences) (50 μg). Cells were also transfected with a β-galactosidase reporter vector to control for transfection efficiency (CMV/β-gal).
For some studies, cells were transfected with TopFlash or FopFlash reporter vectors. TopFlash contains three copies of a consensus binding site for Tcf-Lef linked to a minimal promoter and a luciferase reporter. FopFlash is a similar construct but with a mutation that abolishes Tcf-Lef binding. The TopFlash and FopFlash reporter constructs were purchased from Millipore (Billerica, MA).
Western Blots
U937 or murine bone marrow cells were lysed by boiling in 2× SDS sample buffer. Lysate proteins (50 μg) were separated by SDS-PAGE and transferred to nitrocellulose. Western blots were serially probed with antibodies to β-catenin, HoxA10, Cdx4, and GAPDH or tubulin. Each experiment was repeated at least three times with different lysate proteins. A representative blot is shown.
In Vitro DNA-binding Assays
Electrophoretic Mobility Shift Assays
Oligonucleotide probes were prepared, and EMSAs were performed, as described (32, 33, 34). For binding reactions with β-catenin antibody, disruption of the complex (not supershift) was anticipated, based on the location of the epitopes used for antibody production. Binding competition studies were performed with double-stranded synthetic oligonucleotides. These oligonucleotides were used in dose titration experiments to determine the efficiency of binding competition. Data are presented for assays with a 200-fold molar excess of the competitor (standard for such studies; see also Refs. 34 and 35).
For all experiments, at least three batches of nuclear proteins were tested in at least two independent experiments. A representative study is shown. Integrity of the nuclear proteins and equality of protein loading were determined in EMSA with a probe representing a classical CCAAT box from the α-globin gene promoter.
Isolation of Nuclear Proteins
Nuclear proteins were extracted from U937 cells by the method of Dignam (35) with protease inhibitors, as described (29).
Proliferation Assays
U937 cells that were stably transfected with a vector to overexpress Cdx4 or empty vector control and β-catenin-specific shRNAs or scrambled shRNA control in various combinations were deprived of fetal calf serum for 24 h and then treated with a dose titration of fetal calf serum (0.1–10%). Cell proliferation was determined by incorporation of [3H]thymidine (for the last 8 h of incubation) according to standard techniques.
Genomic Sequence Analysis
Conserved genomic sequences and consensus sequences for β-catenin protein DNA binding were identified using VISTA software (Genomics Division of the Lawrence Berkley National Laboratory, Berkley, CA) (36–38).
Statistical Analysis
Statistical significance was determined by Student's t test and analysis of variance using SigmaPlot and SigmaStat software. In all of the graphs, error bars represent S.E.
RESULTS
β-Catenin Influences Expression of HoxA10 and Cdx4 in Myeloid Progenitor Cells
We previously found increased β-catenin protein (but not mRNA) in primary myeloid progenitor cells or myeloid leukemia cell lines that were overexpressing HoxA10 (24). This increase in β-catenin was dependent on HoxA10-induced production of Fgf2 by these cells and on activation of phosphoinositol 3-kinase (PI3K) (24). Previous investigators identified consensus sequences for β-catenin-Lef binding in the proximal murine CDX4 promoter (23). They found that these sequences were necessary for CDX4 promoter activity in reporter gene transfection experiments using a carcinoma cell line. Therefore, we investigated the impact of β-catenin on Cdx4 expression.
We were initially interested in establishing a connection between β-catenin and Cdx4 in myeloid progenitor cells. For these studies, we generated stable transfectants of the U937 myeloid cell line using a vector to express β-catenin, a vector to express β-catenin-specific shRNAs, or control vectors (empty expression vector plus a vector expressing scrambled shRNA sequences). U937 cells were chosen for these experiments because they represent the granulocyte/monocyte stage of differentiation (28). This stage is of interest because HoxA10 is maximally expressed in GMP during normal myelopoiesis. Also, the GMP stage of differentiation is most similar to the leukemia stem cell in AML (rather than the more immature HSC) (39).
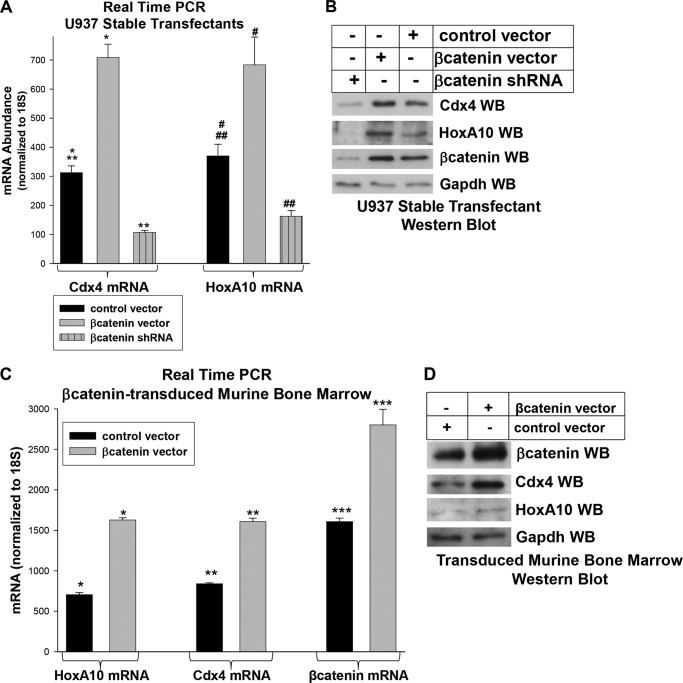
These cells were analyzed for Cdx4 mRNA by real-time PCR. We found that Cdx4 mRNA expression is increased by β-catenin overexpression and decreased by β-catenin knockdown in U937 cells (Fig. 1A). These changes in Cdx4 mRNA expression correlated with altered Cdx4 protein abundance in these cells (Fig. 1B). Because HOXA10 is a target gene for Cdx4 (22), we also determined HoxA10 mRNA and protein in these cells. As expected, HoxA10 mRNA (Fig. 1A) and protein (Fig. 1B) increased with β-catenin overexpression and decreased with β-catenin knockdown.
FIGURE 1.
β-Catenin increases Cdx4 and HoxA10 expression in myeloid progenitor cells. A, increased β-catenin expression increases Cdx4 and HoxA10 mRNA in myeloid cell lines. U937 cells were stably transfected with a vector to overexpress β-catenin, with an empty control vector, or with a vector to express β-catenin-specific shRNAs or scrambled control shRNA. Expression of Cdx4 and HoxA10 mRNA was determined by real-time PCR. *, **, *** and #, ##, or ###, statistically significant differences in gene expression (p < 0.001, n = 6). B, increased β-catenin expression increases Cdx4 and HoxA10 protein in myeloid cell lines. The stable transfectants referred to above were also analyzed for protein expression. Western blots (WB) of total cell lysates were probed with antibodies to β-catenin, Cdx4, HoxA10, and Gapdh (as a loading control). C, increased β-catenin expression increases Cdx4 and HoxA10 mRNA in primary bone marrow cells. Bone marrow cells were isolated from WT mice and transduced with a retroviral vector to express β-catenin or with control vector. Cells were cultured in GM-CSF, IL-3, and SCF, and CD34+ cells were separated. Expression of Cdx4, HoxA10, and β-catenin was determined by real-time PCR. *, **, or ***, statistically significant differences in gene expression (p < 0.001, n = 3). D, increased β-catenin expression increases Cdx4 and HoxA10 protein in primary bone marrow cells. The bone marrow cells described above were also analyzed for protein expression. Western blots of lysate proteins were probed with antibodies to Cdx4, HoxA10, β-catenin, and Gapdh (as a loading control). Error bars, S.E.
U937 is a leukemia cell line with multiple abnormalities in pathways that regulate growth and survival. Therefore, we also investigated the relationship between β-catenin and Cdx4 or HoxA10 using primary murine myeloid progenitor cells. For these studies, bone marrow was isolated from wild type C57BL6 mice, transduced with a retroviral vector to express β-catenin (or with empty vector control), and cultured in GM-CSF, IL-3, and SCF, and Cd34+ cells were isolated using antibody-conjugated magnetic beads (10, 22, 24). Our previous studies indicate that this procedure results in relative GMP enrichment (Sca1−CD34+CD38−Gr1−) (10). Cells were analyzed for expression of Cdx4, HoxA10, and β-catenin. We found that β-catenin overexpression increases Cdx4 and HoxA10 mRNA (Fig. 1C) and protein (Fig. 1D) in primary murine GMP, consistent with our studies in U937 cells.
β-Catenin Increases CDX4 Promoter Activity in Myeloid Progenitor Cells
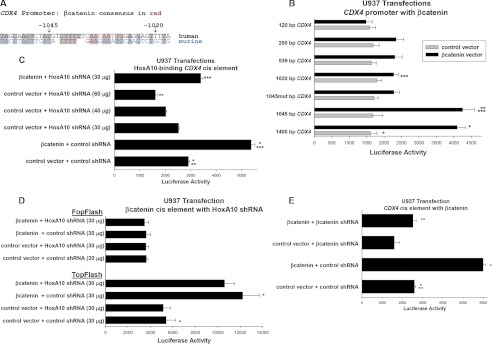
To determine the mechanism by which β-catenin increases Cdx4 expression in myeloid progenitor cells, we performed additional studies. We first examined the human CDX4 5′-flank for sequences corresponding to the β-catenin-Lef/Tcf DNA-binding consensus (5′-(A/T)(A/T)CAA(A/T)G-3′) and compared the results with analysis of the murine CDX4 5′-flank (40). We noted that only one of the three Lef/Tcf consensus sequences that were previously identified within the proximal 1.1 kb of the murine CDX4 promoter was conserved in the human promoter (−715 to −709 bp relative to the transcription start site) (23, 24). Promoter analysis identified a consensus sequence in the human CDX4 promoter between −1028 and −1035 that did not at first appear to be conserved in the murine promoter. However, closer analysis identified a Lef/Tcf consensus in the murine promoter in the same location but on the opposite strand (Fig. 2A).
FIGURE 2.
β-Catenin activates the CDX4 promoter in myeloid progenitor cells. A, sequence homology between the human and murine CDX4 promoters. A β-catenin-Lef DNA-binding consensus sequence in the human CDX4 promoter has a homologous sequence in the murine CDX4 promoter. Human sequence is in black, murine sequence is in blue, and the consensus sequence is in red. B, β-catenin overexpression activates the CDX4 promoter. U937 cells were co-transfected with a series of reporter gene constructs with sequence from the CDX4 5′-flank and a vector to overexpress β-catenin (or control vector). Transfectants were analyzed for luciferase reporter activity. *, **, or ***, statistically significant differences in reporter activity (p < 0.0001, n = 6). C, β-catenin overexpression increased activity of the HoxA10-binding CDX4 cis element in a HoxA10-dependent manner. U937 cells were co-transfected with a construct with three copies of the −139 to −145 bp sequence from the CDX4 promoter linked to a minimal promoter and reporter and a vector to overexpress β-catenin (or control vector) and a HoxA10-specific shRNA (or scrambled shRNA control vector). Transfectants were analyzed for luciferase reporter activity. *, **, or ***, statistically significant differences in reporter activity (p < 0.0001, n = 6). D, HoxA10 knockdown does not influence activation of a β-catenin-binding cis element. U937 cells were co-transfected with a construct with four copies of a β-catenin/Lef binding consensus sequence linked to a minimal promoter and reporter (TopFlash) or non-binding control vector (FopFlash), a vector to express β-catenin (or control vector), and a HoxA10-specific shRNA (or scrambled shRNA control vector). The HoxA10-specific shRNA vector was used at a level that did not alone influence activity of the −139 to −145 bp CDX4 cis element but blocked activation by overexpressed β-catenin. *, statistically significant differences (p < 0.001, n = 4). E, β-catenin activates a specific CDX4 cis element. U937 cells were co-transfected with a construct with three copies of the −1028 to −1035 bp CDX4 promoter sequence linked to a minimal promoter and a reporter and a vector to overexpress β-catenin (or control vector), a β-catenin-specific shRNA, both, or control vectors. Transfectants were analyzed for luciferase reporter activity. * or **, statistically significant differences in reporter activity (p < 0.0001, n = 6). Error bars, S.E.
We designed a series of constructs with various sequences from the human CDX4 5′-flank linked to a luciferase reporter (pGL3-basic). The 5′-end of these constructs was determined by the location of Lef/Tcf consensus sequences, and the 3′-end of all of the constructs was at the transcription start site. These constructs were co-transfected into U937 cells with a vector to express β-catenin or empty control vector. We found that overexpression of β-catenin significantly increased the activity of CDX4 promoter constructs in transfectants with constructs containing 1400 or 1045 bp of 5′-flank (154 ± 22% and 150 ± 27% increase, respectively, p < 0.001, n = 6 for each construct) (Fig. 2B). Reporter expression from constructs with between 1020 and 200 bp of CDX4 5′-flank were less affected by β-catenin overexpression (between 23 and 35% increase, p < 0.01, n = 6), and there was no change in reporter expression from constructs with 120 bp of CDX4 5′-flank. To determine if the consensus sequence at −1028 to −1035 was responsible for this activity, we generated a construct with −1045 bp of CDX4 5′-flank with a mutation in the Lef/Tcf consensus sequence. We found that this construct was only slightly activated by β-catenin overexpression in U937 transfectants (38 ± 8% increase, p < 0.001, n = 6) (Fig. 2B).
Constructs with at least 146 bp of CDX4 promoter include a HoxA10-binding cis element that we identified in previous studies (−139 to −146 bp) (22). If β-catenin increases Cdx4 expression, and Cdx4 increases HoxA10 expression, β-catenin would be anticipated to activate CDX4 transcription indirectly, via increased HoxA10. To investigate this hypothesis, U937 cells were co-transfected with an artificial promoter/reporter construct containing three copies of the HoxA10-binding cis element from the CDX4 promoter and vectors to express β-catenin- and HoxA10-specific shRNAs (or relevant control vectors) (22). We found that β-catenin overexpression increases activity of this cis element in a HoxA10-dependent manner (Fig. 2C). Specifically, these studies determined that expression of a HoxA10-specific shRNA blocked β-catenin-induced activity of the CDX4 cis element (Fig. 2C). This suggests that HoxA10 knockdown was blocking the indirect effects of overexpressed β-catenin. Alternatively, β-catenin might interact with this CDX4 cis element in a HoxA10-dependent manner. This possibility is explored below.
Reporter activity of empty pGL3-basic vector in U937 transfectants was <10% of the activity of constructs with CDX4 promoter sequences and was subtracted as background. Overexpression of β-catenin did not influence this minimal reporter activity.
We also performed control experiments to verify that expression of HoxA10-specific shRNA at this level did not influence activity of β-catenin protein (either endogenous or overexpressed). To do this, we used a construct with four copies of a consensus Lef/Tcf binding site linked to a minimal promoter and a luciferase reporter (TopFlash, versus non-binding, mutant FopFlash) (41). U937 cells were co-transfected with TopFlash or FopFlash control vector, a vector to express β-catenin (or control vector), and a vector to express HoxA10-specific shRNA (or scrambled control). We found that β-catenin-induced TopFlash activation was not influenced by HoxA10 knockdown under these conditions (Fig. 2D).
We next investigated the conserved β-catenin/Lef consensus sequence in the CDX4 promoter (−1028 to −1035 bp) for cis element activity using an artificial promoter construct with three copies of this sequence linked to a minimal promoter and a luciferase reporter. This construct was co-transfected into U937 cells with a vector to overexpress β-catenin, a β-catenin-specific shRNA, or both (or control vectors). We found that the activity of the CDX4 cis element-containing construct was increased by β-catenin overexpression and decreased by β-catenin knockdown (Fig. 2E). In contrast, β-catenin expression level had no influence on reporter activity from the control, minimal promoter construct, which was subtracted as background.
β-Catenin Increases HOXA10 Promoter Activity in Myeloid Progenitor Cells
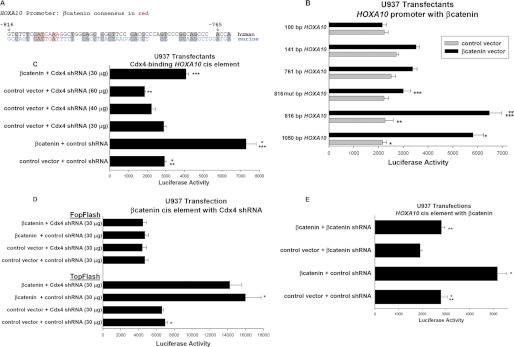
Analysis of the human HOXA10 promoter identified a previously unrecognized consensus sequences for β-catenin-Lef/Tcf binding (Fig. 3A). This Lef/Tcf binding consensus was conserved in the murine HOXA10 promoter but in the opposite orientation, similar to the cis elements in the human and murine CDX4 promoters that are influenced by β-catenin (see above) (22). Therefore, we considered the possibility that HOXA10 is also a β-catenin target gene.
FIGURE 3.
β-Catenin activates the HOXA10 promoter in myeloid progenitor cells. A, sequence homology between the human and murine HOXA10 promoters. A β-catenin-Lef DNA-binding consensus sequence in the human HOXA10 promoter has a homologous sequence in the murine HOXA10 promoter. The human sequence is in black, the murine sequence is in blue, and the consensus sequence is in red. B, β-catenin overexpression activates the HOXA10 promoter. U937 cells were co-transfected with a series of reporter gene constructs with sequence from the HOXA10 5′-flank and a vector to overexpress β-catenin (or control vector). Transfectants were analyzed for luciferase reporter activity. *, **, or ***, statistically significant differences in reporter activity (p < 0.002, n = 6). C, β-catenin overexpression increased activity of the Cdx4-binding HOXA10 cis element in a Cdx4-dependent manner. U937 cells were co-transfected with a construct with three copies of the −129 to −136 bp sequence from the HOXA10 promoter linked to a minimal promoter and reporter and a vector to overexpress β-catenin (or control vector) and a Cdx4-specific shRNA (or scrambled shRNA control vector). Transfectants were analyzed for luciferase reporter activity. *, **, or ***, statistically significant differences in reporter activity (p < 0.001, n = 6). D, Cdx4 knockdown does not influence activation of a β-catenin-binding cis element. U937 cells were co-transfected with a construct with four copies of a β-catenin/Lef binding consensus sequence linked to a minimal promoter and reporter (TopFlash) or non-binding control vector (FopFlash), a vector to express β-catenin (or control vector), and a Cdx4-specific shRNA (or scrambled shRNA control vector). The Cdx4-specific shRNA vector was used at a level that did not alone influence activity of the −129 to −136 bp HOXA10 cis element but blocked activation by overexpressed β-catenin. *, statistically significant differences (p < 0.001, n = 4). E, β-catenin activates a specific HOXA10 cis element. U937 cells were co-transfected with a construct with two copies of the −800 to −830 bp HOXA10 promoter sequence linked to a minimal promoter and a reporter and a vector to overexpress β-catenin (or control vector), a β-catenin-specific shRNA, both, or control vectors. Transfectants were analyzed for luciferase reporter activity. * or **, statistically significant differences in reporter activity (p < 0.01, n = 6). Error bars, S.E.
Based on the location of this putative β-catenin-binding cis element, a series of HOXA10 promoter/reporter constructs were generated. The 5′-end of these constructs was chosen based on the possible β-catenin/Lef consensus sequence, and the 3′-end of all of the constructs was the transcription start site. These constructs were co-transfected into U937 myeloid cells with a vector to overexpress β-catenin or empty control vector. We found that reporter gene activity from constructs with 1050 or 816 bp of HOXA10 5′-flank was significantly increased by β-catenin overexpression (increase of 169 ± 20% and 186 ± 25%, respectively, p < 0.001, n = 6 for each construct) (Fig. 3B). In contrast, β-catenin overexpression failed to exert this profound effect on constructs with 761 or 150 bp of HOXA10 5′-flank (34 ± 10% and 29 ± 12% increase, respectively; p < 0.01, n = 8) and exerted no effect on a construct with 100 bp (p = 0.7, n = 4). We found that mutation of the β-catenin/Lef consensus sequence abolished most of the effect of β-catenin overexpression on the 816-bp HOXA10 reporter construct (Fig. 3B).
The Cdx4-binding cis element in the HOXA10 promoter is at −129 to −136 bp (22). Therefore, this cis element is included in all of the constructs tested except for the 100-bp construct. If β-catenin activates the CDX4 promoter and Cdx4 activates HOXA10 transcription, we would expect indirect activation of the HOXA10 promoter by β-catenin via the Cdx4-binding cis element (as discussed above for the CDX4 promoter and HoxA10). To specifically explore the contribution of the Cdx4-binding cis element to β-catenin-induced HOXA10 promoter activity, U937 cells were co-transfected with an artificial promoter/reporter construct containing three copies of the −129 to −136 bp sequence from the HOXA10 promoter and vectors to express β-catenin- and Cdx4-specific shRNAs (or relevant control vectors) (22). We found that expression of a Cdx4-specific shRNA blocked the effect of β-catenin on this cis element, consistent with an indirect effect of β-catenin (Fig. 3C). The alternative possibility, that β-catenin interacts with this cis element in a HoxA10-dependent manner, is explored below.
We also performed control experiments to verify that expression of Cdx4-specific shRNA at this level did not influence the activity of β-catenin protein using the TopFlash β-catenin/Lef-binding reporter construct, as in the studies above. For these studies, U937 cells were co-transfected with TopFlash or FopFlash control vector, a vector to express β-catenin (or control vector), and a vector to express Cdx4-specific shRNA (or scrambled control). We found that β-catenin-induced TopFlash activation was not influenced by Cdx4 knockdown under these conditions (Fig. 3D).
To verify cis element activity of the potential β-catenin/Lef binding site in the HOXA10 promoter sequence, we generated a reporter construct with two copies of −803 to −809 bp sequence linked to a minimal promoter. This construct was co-transfected into U937 cells with vectors to express β-catenin, a β-catenin-specific shRNA, or both (or control vectors). We found that reporter activity was increased by β-catenin overexpression and decreased by knockdown, as anticipated (Fig. 3E).
β-Catenin Interacts with the CDX4 and HOXA10 Promoters
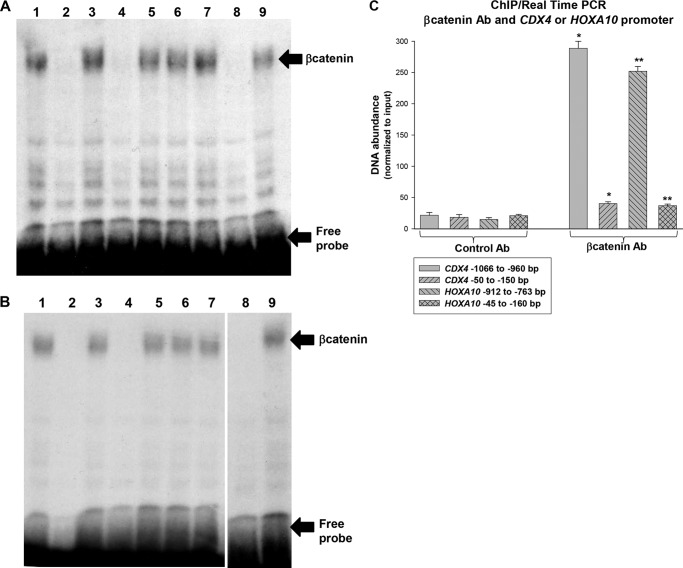
These studies suggest that β-catenin influences activity of the CDX4 and HOXA10 promoters via the Lef/Tcf consensus sequences that we identified above. To demonstrate this directly, we first performed in vitro DNA-binding assays. For these studies, radiolabeled, double-stranded synthetic oligonucleotide probes were generated with the −1020 to −1045 bp sequence from the CDX4 promoter or the −800 to −825 bp sequence from the HOXA10 promoter. The probes were incubated with nuclear proteins from U937 cells, and DNA-bound proteins were identified by EMSA.
We found that the CDX4 probe generated a specific low mobility band in this assay (Fig. 4A). Binding of this complex was competed for by an unlabeled molar excess of double-stranded homologous oligonucleotide or an oligonucleotide representing a β-catenin/Lef consensus (but not mutant control oligonucleotide). Binding of the low mobility complex was disrupted by preincubation with a β-catenin antibody. Similar studies were performed with the HOXA10 probe, and similar results were obtained (Fig. 4B).
FIGURE 4.
β-Catenin interacts with the CDX4 and HOXA10 promoters in myeloid cells. A, β-catenin interacts with the CDX4 promoter in vitro. EMSAs were performed with a double-stranded oligonucleotide probe representing −1020 to −1045 bp sequence from the CDX4 promoter and nuclear proteins from U937 cells. Some binding assays were preincubated with double-stranded oligonucleotide competitors. Lane 1, no competitor; lane 2, homologous oligonucleotide; lane 3, homologous oligonucleotide with mutation in the β-catenin-Lef consensus sequence; lane 4, another oligonucleotide with the β-catenin-Lef consensus sequence; lane 5, a mutant form of this oligonucleotide; lane 6, oligonucleotide representing the HoxA10 binding (−139 to −146 bp) CDX4 sequence; lane 7, irrelevant oligonucleotide from the CDX4 5′-flank; lane 8, β-catenin antibody; lane 9, control irrelevant antibody. B, β-catenin interacts with the HOXA10 promoter in vitro. EMSAs were performed with a double-stranded oligonucleotide probe representing −800 to −835 bp sequence from the HOXA10 promoter and nuclear proteins from U937 cells. Some binding assays were preincubated with double-stranded oligonucleotide competitors. Lane 1, no competitor; lane 2, homologous oligonucleotide; lane 3, homologous oligonucleotide with mutation in the β-catenin-Lef consensus sequence; lane 4, another oligonucleotide with the β-catenin-Lef consensus sequence; lane 5, a mutant form of this oligonucleotide; lane 6, oligonucleotide representing the Cdx4-binding (−129 to −136 bp) HOXA10 sequence; lane 7, irrelevant oligonucleotide from the HOXA10 5′-flank; lane 8, β-catenin antibody; lane 9, control irrelevant antibody. C, β-catenin interacts with the CDX4 and HOXA10 promoters in vivo. Chromatin was immunoprecipitated from U937 lysates with an antibody to β-catenin or control antibody. Co-precipitating chromatin was amplified with primer sets representing sequences from the CXD4 and HOXA10 promoters. * or **, statistically significant differences (p < 0.0001, n = 3). Error bars, S.E.
We also investigated β-catenin binding to these promoter sequences in vivo. For these studies, chromatin was sonicated prior to an average size of 200 bp and co-immunoprecipitated from U937 cells with an antibody to β-catenin (or irrelevant antibody control). Precipitated chromatin was amplified with primer sets to amplify the regions surrounding the β-catenin/Lef consensus sequences in the CXD4 and HOXA10 promoters. For these studies, non-precipitated chromatin was a positive control. We found specific interaction of β-catenin with these putative cis elements (Fig. 4C). Because β-catenin also influences the activity of a previously identified CDX4 cis element that binds HoxA10 (−136 to −146 bp) and a HOXA10 cis element that binds Cdx4 (−129 to −136 bp), we also tested in vivo binding of β-catenin to these promoter sequences. We found that β-catenin binding to these sequences was equivalent to background binding to irrelevant sequences (Fig. 4C). This suggested that β-catenin influences these cis elements indirectly by influencing HoxA10 and Cdx4 expression rather than by interacting with the cis elements in a HoxA10- or Cdx4-dependent manner.
Expression of β-Catenin, HoxA10, and Cdx4 Is Interdependent in Myeloid Cells
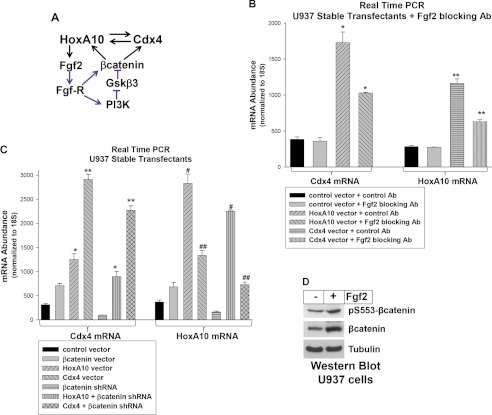
Our studies indicate that HoxA10 regulates CDX4, Cdx4 regulates HOXA10, and both genes are activated by β-catenin (22). We previously determined that HoxA10 activates transcription of the FGF2 gene (24) and that autocrine production of Fgf2 by HoxA10-overexpressing myeloid cells increases β-catenin activity (Fig. 5A) (24). Therefore, we investigated the role of Fgf2 in HoxA10-induced expression of Cdx4 and in Cdx4-induced HoxA10 expression. For these studies, U937 stable transfectants were generated with a vector to overexpress HoxA10 or Cdx4 or with empty control vector. Cells were treated with a blocking antibody to Fgf2 (versus control antibody) and analyzed for gene expression.
FIGURE 5.
Expression of HoxA10, Cdx4, and β-catenin is interdependent in myeloid leukemia cells. A, representation of influences of HoxA10, Cdx4, and β-catenin on each other. The schematic represents the interrelationship between HoxA10, Cdx4, and β-catenin. Black arrows, influence on gene transcription; purple arrows, signal transduction pathways. B, HoxA10 influences Cdx4 expression in an Fgf2-dependent manner, and Cdx4 influences HoxA10 expression in an Fgf2-dependent manner. U937 stable transfectants with vectors to express HoxA10 or Cdx4 or with empty vector control were analyzed after treatment with Fgf2-blocking antibody or control antibody. Real-time PCR was used to determine mRNA expression. * or **, statistically significant differences (p < 0.003, n = 3). C, HoxA10 influences Cdx4-expression in a β-catenin-dependent manner, and Cdx4 influences HoxA10 expression in a β-catenin-dependent manner. U937 stable transfectants were generated with vectors to express β-catenin, HoxA10, Cdx4, β-catenin-specific shRNAs, HoxA10 + β-catenin-specific shRNAs, Cdx4 + β-catenin-specific shRNAs, or control vectors. Cells were analyzed for mRNA expression by real-time PCR. *, **, #, or ##, statistically significant differences (p < 0.0001, n = 3). D, both total and serine 552-phosphorylated β-catenin are increased by Fgf2. U937 cells were treated with Fgf2, and cell lysate proteins were analyzed by Western blots serially probed with antibodies to Ser(P)-552 β-catenin, β-catenin, or tubulin (as a loading control). Error bars, S.E.
We found increased Cdx4 mRNA in HoxA10-overexpressing cells and increased HoxA10 mRNA in Cdx4-overexpressing cells, as anticipated (Fig. 5B). Consistent with our hypothesis, Fgf2 blocking antibody decreased Cdx4 in HoxA10-overexpressing cells and HoxA10 in Cdx4-overexpressing cells. We previously demonstrated the specificity and efficacy of blocking Fgf-R activation with this Fgf2-blocking antibody (24).
Based on these results, we next investigated whether β-catenin influences the effect of HoxA10 on Cdx4 expression or the effect of Cdx4 on HoxA10 expression. For these studies, U937 stable transfectants were generated with a vector to overexpress HoxA10 or Cdx4 (or with vector control) and a vector to express a β-catenin-specific shRNA (or scrambled control). Cells were analyzed for mRNA expression by real-time PCR.
We found that β-catenin knockdown decreased Cdx4 mRNA in HoxA10-overexpressing transfectants (Fig. 5C). β-Catenin knockdown also decreased Cdx4 mRNA in Cdx4-overexpressing transfectants, consistent with an effect of Cdx4-induced HoxA10 on transcription of the endogenous CDX4 gene. Similarly, the increase in HoxA10 in Cdx4-overexpressing transfectants was decreased by β-catenin knockdown, as was the increase in HoxA10 in cells transfected with a HoxA10 expression vector (Fig. 5C).
Fgf-R activation induces a specific phosphorylation event (Ser(P)-552) that increases β-catenin activity (42). We previously found increased total β-catenin protein in a PI3K-dependent manner in Fgf2-treated cells (24). To determine if total and phosphorylated β-catenin were both increased by Fgf2, we treated U937 cells with a dose of recombinant Fgf2 that we previously determined induces maximal proliferation (24). Cell lysate proteins were analyzed for total and phosphorylated β-catenin by Western blot. We found an equivalent increase in total and active β-catenin in Fgf2-treated cells in comparison with sham-treated cells (Fig. 5D).
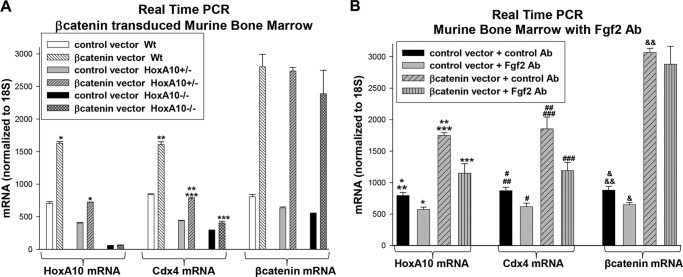
We also investigated these issues using bone marrow-derived primary myeloid progenitor cells. For these studies, we took advantage of a murine model with disruption of the HOXA10 gene (30). These mice are viable with decreased fertility and mild abnormalities in hematopoiesis. Bone marrow cells from WT, HoxA10+/−, and HoxA10−/− mice were transduced with a retroviral vector to express β-catenin or with empty control vector. Cells were cultured in GM-CSF, IL-3, and SCF, and CD34+ cells were isolated (GMP conditions). Cells were analyzed for expression of Cdx4, HoxA10, and β-catenin mRNA by real-time PCR.
We found that increased Cdx4 mRNA in β-catenin-transduced cells is abrogated by HoxA10 knock-out in a dose-dependent manner (Fig. 6A). We also found that β-catenin overexpression induces a greater increase in HoxA10 expression in WT bone marrow cells in comparison with HoxA10+/− cells (Fig. 6A). HoxA10 expression is not seen in any HoxA10−/− cells, as anticipated.
FIGURE 6.
Expression of HoxA10, Cdx4, and β-catenin is interdependent in primary murine myeloid progenitor cells. A, β-catenin influences Cdx4 expression in a HoxA10-dependent manner in primary bone marrow cells. Bone marrow cells were isolated from WT, HoxA10+/−, or HoxA10−/− mice and transduced with a retroviral vector to express β-catenin or with control vector. Cells were cultured in GM-CSF, IL-3, and SCF, and CD34+ cells were separated. Expression of Cdx4, HoxA10, and β-catenin was determined by real-time PCR. *, **, or ***, statistically significant differences in gene expression (p < 0.0001, n = 3). B, Fgf2 influences β-catenin-induced HoxA10 and Cdx4 expression in primary bone marrow cells. Some of the cells described above were treated with Fgf2-blocking antibody or control irrelevant antibody. Expression of Cdx4, HoxA10, and β-catenin was determined by real-time PCR. *, **, ***, #, ##, ###, &, &&, or &&&, statistically significant differences in gene expression (p < 0.002, n = 3). Error bars, S.E.
To investigate the role of Fgf2 in β-catenin-induced HoxA10 and Cdx4 expression, some transduced cells were treated with an Fgf2-blocking antibody (24). We found significantly decreased HoxA10 and Cdx4 expression in β-catenin-overexpressing cells that were treated with Fgf2-blocking antibody in comparison with control antibody (Fig. 6B). Expression was also decreased in control cells, as might be anticipated.
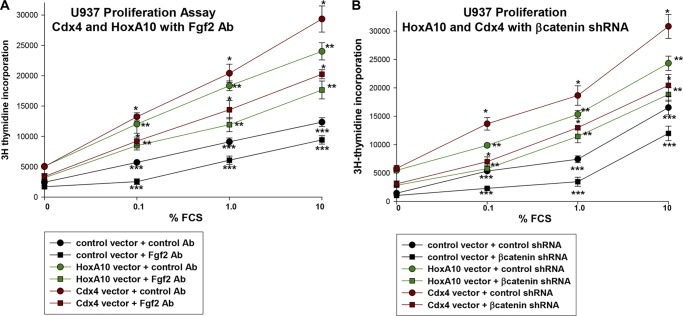
β-Catenin Influences Cytokine Hypersensitivity in Cdx4-overexpressing Myeloid Cells
In previous studies, we determined that HoxA10-overexpressing myeloid progenitor cells exhibit an increased proliferative response to hematopoietic cytokines (i.e. cytokine hypersensitivity) (10, 24). We also demonstrated that cytokine hypersensitivity in HoxA10-overexpressing myeloid cells was influenced by Fgf2 and β-catenin (24). Because Cdx4 increases expression of HoxA10, Cdx4-overexpressing cells would also be anticipated to exhibit Fgf2- and β-catenin-related cytokine hypersensitivity.
To test this hypothesis, Cdx4-overexpressing cells were compared with control vector-transduced cells (as a negative control) and HoxA10-overexpressing cells (as a positive control). Cells were assayed for proliferation in response to a dose titration of fetal calf serum (FSC; a source of cytokine in this study). To determine the effect of Fgf2 on proliferation, cells were treated with an Fgf2-blocking antibody or antibody control. We found that Cdx4-overexpressing cells exhibit an increased proliferative response to FCS in comparison with control cells (Fig. 7A). Consistent with our hypothesis, this was decreased by blocking Fgf2 (Fig. 7A).
FIGURE 7.
Cytokine hypersensitivity of Cdx4-overexpressing myeloid cells is influenced by Fgf2 and β-catenin. A, cytokine hypersensitivity of Cdx4-overexpressing myeloid cells is influenced by Fgf2. U937 cells were stably transfected with a vector to express Cdx4 or HoxA10 or with control vector. Cells were deprived of all cytokines for 24 h and stimulated with a dose titration of FCS with or without an Fgf2-blocking antibody. Proliferation was determined by incorporation of [3H]thymidine over 24 h. *, **, or ***, statistically significant differences in proliferation at a given FCS concentration. B, cytokine hypersensitivity of Cdx4-overexpressing myeloid cells is influenced by β-catenin. U937 cells were stably transfected with a vector to express Cdx4, HoxA10, or control vector plus a vector to express β-catenin-specific shRNAs or scrambled control shRNA. Cells were deprived of all cytokines for 24 h and stimulated with a dose titration of FCS. Proliferation was determined by incorporation of [3H]thymidine over 24 h. *, **, or ***, statistically significant differences in proliferation at a given FCS concentration. Error bars, S.E.
We next determined the impact of β-catenin on Cdx4-induced cytokine hypersensitivity. For these studies, proliferation assays were performed on U937 cells transfected with vectors to express Cdx4 or HoxA10 (or control vector) and a vector to express β-catenin-specific shRNAs (or scrambled control shRNA). We found that β-catenin knockdown decreases the proliferative response of Cdx4-overexpressing cells (Fig. 7B). β-Catenin knockdown also impaired cytokine hypersensitivity in HoxA10-overexpressing cells, consistent with our previous results (24).
DISCUSSION
Our previous studies determined that HoxA10 regulates transcription of CDX4 and Cdx4 regulates transcription of HOXA10 in a manner that results in positive feedback between these proteins (22). In other previous studies, we determined that HoxA10 regulates FGF2 transcription (24). We found that autocrine stimulation of HoxA10-overexpressing myeloid progenitor cells by Fgf2 activates β-catenin in a PI3K-dependent manner (24) and that this contributes to cytokine hypersensitivity of these cells. In the current studies, we demonstrate that β-catenin directly activates the CDX4 and HOXA10 promoters. We also find that β-catenin indirectly activates CDX4 in a HoxA10-dependent manner and HOXA10 in a Cdx4-dependent manner. Therefore, our studies identify another layer of complexity to cross-regulation of these homeodomain-containing proteins. Previous studies by other investigators determined that canonical Wnt signaling increases Cdx4 expression in a β-catenin-dependent manner (22). Other studies indicated that Fgf2 signaling influences Cdx4 expression in a PI3K-dependent manner (43). Therefore, our investigations integrate these two pathways via cross-talk between HoxA10 and Cdx4.
In our studies, we compared the human and murine CDX4 promoters to identify consensus sequences for β-catenin/Lef binding. Other investigators analyzed the murine CDX4 promoter and determined the impact of such consensus sequence on promoter activity in transfection experiments with an embryocarcinoma cell line (23). We found that one of the three β-catenin/Lef consensus sequences identified by these investigators was conserved in the human CDX4 promoter (at ∼−710 bp). However, we found that this conserved CDX4 cis element was not active in myeloid cells. This might suggest lineage specificity to activity of β-catenin/Lef-binding cis elements. Mechanisms for this might include the presence of lineage-specific cooperating or antagonistic transcription factors or epigenetic differences in myeloid progenitor cells versus carcinoma cells.
The β-catenin/Lef consensus sequence that we functionally defined in the CDX4 promoter is 5′ to all three previously characterized murine cis elements (23). However, this does not contradict their prior excellent work, because the studies by these investigators also indicated that there were more distal β-catenin-regulated CDX4 cis elements (23). Studies to better understand the lineage- and/or differentiation stage-specific regulation of the CDX4 promoter are ongoing in the laboratory.
Our studies represent the first identification of a HOX gene as a target for β-catenin. Because β-catenin is activated by Fgf2 in myeloid progenitor cells, this provides another positive feedback mechanism for HOXA10 transcription in HoxA10-overexpressing cells. Because β-catenin also activates CDX4, these studies identify molecular mechanisms with a multiplier effect. These mechanisms would be anticipated to sustain abnormal expression of HoxA10 and Cdx4 and further increase β-catenin activity. Similar to the human and murine CDX4 promoters, the β-catenin/Lef consensus sequences in the human and murine HOXA10 promoters were conserved but in the opposite orientation.
Increased expression of HoxA10 is associated with poor prognosis in human AML and results in myeloproliferation and AML in murine models. Increased expression of Cdx4 is also found in Hox-overexpressing human AML and contributes to leukemogenesis in murine models. Increased activity of β-catenin in leukemia stem cells is associated with resistance to drug therapy and poor prognosis. Fgf2 is critical for hematopoietic stem cell proliferation and survival and plays a role in lineage commitment (44). Fgf2 expression is increased in forms of human AML with increased HoxA10, and increased circulating Fgf2 has been documented in myeloid leukemias (45) (Oncomine, Research Edition, University of Michigan, Ann Arbor, MI). We previously identified functional connections between these proteins by demonstrating that cytokine hypersensitivity in HoxA10-overexpressing myeloid progenitor cells can be decreased by blocking Fgf2 or knocking down β-catenin. In the current studies, we show similar influences for Fgf2 and β-catenin on the proliferative response to cytokines in Cdx4-overexpressing myeloid cells.
Increased β-catenin activity could contribute to cytokine hypersensitivity in myeloid leukemia cells by increasing expression of β-catenin target genes, including cyclin D1 and c-Myc. However, our studies identify another potential mechanism, increased HoxA10 and Cdx4 with consequent dysregulation of the target genes for these homeodomain transcription factors. Cdx4 influences expression of other HOX genes, including HOXB3, -B4, and -A9 (19, 22). HoxA10 target genes that are involved in proliferation include genes encoding Fgf2, Tgfβ2, Mkp2, and β3 integrin (24, 27, 33, 34). Our studies suggest the possibility that disrupting this network might be a therapeutic approach to Hox-overexpressing AML. Because pharmacologic inhibitors to Fgf-R are under development, this approach is feasible.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-HL87717. This work was also supported by a Veterans Affairs Merit Review (to E. A. E.).
- HSC
- hematopoietic stem cell(s)
- GMP
- granulocyte/monocyte progenitor
- AML
- acute myeloid leukemia
- Ab
- antibody
- SCF
- stem cell factor.
REFERENCES
- 1. Acampora D., D'Esposito M., Faiella A., Pannese M., Migliaccio E., Morelli F., Stornaiuolo A., Nigro V., Simeone A., Boncinelli E. (1989) The human HOX gene family. Nucleic Acids Res. 17, 10385–10402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sauvageau G., Lansdorp P. M., Eaves C. J., Hogge D. E., Dragowska W. H., Reid D. S., Largman C. (1994) Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. U.S.A. 91, 12223–12227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thorsteinsdottir U., Kroon E., Jerome L., Blasi F., Sauvageau G. (2001) Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 21, 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sauvageau G., Thorsteinsdottir U., Eaves C. J., Lawrence H. J., Largman C., Lansdorp P. M., Humphries R. K. (1995) Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 9, 1753–1765 [DOI] [PubMed] [Google Scholar]
- 5. Calvo K. R., Sykes D. B., Pasillas M., Kamps M. P. (2000) Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol. Cell. Biol. 20, 3274–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lawrence H. J., Helgason C. D., Sauvageau G., Fong S., Izon D. J., Humphries R. K., Largman C. (1997) Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 89, 1922–1930 [PubMed] [Google Scholar]
- 7. Buske C., Feuring-Buske M., Antonchuk J., Rosten P., Hogge D. E., Eaves C. J., Humphries R. K. (2001) Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood 97, 2286–2292 [DOI] [PubMed] [Google Scholar]
- 8. Björnsson J. M., Andersson E., Lundström P., Larsson N., Xu X., Repetowska E., Humphries R. K., Karlsson S. (2001) Proliferation of primitive myeloid progenitors can be reversibly induced by HOXA10. Blood 98, 3301–3308 [DOI] [PubMed] [Google Scholar]
- 9. Thorsteinsdottir U., Mamo A., Kroon E., Jerome L., Bijl J., Lawrence H. J., Humphries K., Sauvageau G. (2002) Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 99, 121–129 [DOI] [PubMed] [Google Scholar]
- 10. Wang H., Lindsey S., Konieczna I., Bei L., Horvath E., Huang W., Saberwal G., Eklund E. A. (2009) Constitutively active SHP2 cooperates with HoxA10 overexpression to induce acute myeloid leukemia. J. Biol. Chem. 284, 2549–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Golub T. R., Slonim D. K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J. P., Coller H., Loh M. L., Downing J. R., Caligiuri M. A., Bloomfield C. D., Lander E. S. (1999) Molecular classification of cancer. Class discovery and class prediction by gene expression monitoring. Science 286, 531–537 [DOI] [PubMed] [Google Scholar]
- 12. Kawagoe H., Humphries R. K., Blair A., Sutherland H. J., Hogge D. E. (1999) Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia 13, 687–698 [DOI] [PubMed] [Google Scholar]
- 13. Armstrong S. A., Staunton J. E., Silverman L. B., Pieters R., den Boer M. L., Minden M. D., Sallan S. E., Lander E. S., Golub T. R., Korsmeyer S. J. (2002) MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30, 41–47 [DOI] [PubMed] [Google Scholar]
- 14. Camós M., Esteve J., Jares P., Colomer D., Rozman M., Villamor N., Costa D., Carrio A., Nomdedéu J., Montserrat E., Campo E. (2006) Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 66, 6947–6954 [DOI] [PubMed] [Google Scholar]
- 15. Guenther M. G., Jenner R. G., Chevalier B., Nakamura T., Croce C. M., Canaani E., Young R. A. (2005) Global and hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 102, 8603–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Milne T. A., Briggs S. D., Brock H. W., Martin M. E., Gibbs D., Allis C. D., Hess J. L. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 17. Ernst P., Mabon M., Davidson A. J., Zon L. I., Korsmeyer S. J. (2004) An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 14, 2063–2069 [DOI] [PubMed] [Google Scholar]
- 18. Horton S. J., Grier D. G., McGonigle G. J., Thompson A., Morrow M., De Silva I., Moulding D. A., Kioussis D., Lappin T. R., Brady H. J., Williams O. (2005) Continuous MLL-ENL expression is necessary to establish a “Hox Code” and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 65, 9245–9252 [DOI] [PubMed] [Google Scholar]
- 19. Davidson A. J., Ernst P., Wang Y., Dekens M. P., Kingsley P. D., Palis J., Korsmeyer S. J., Daley G. Q., Zon L. I. (2003) Cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature 425, 300–306 [DOI] [PubMed] [Google Scholar]
- 20. Bansal D., Scholl C., Fröhling S., McDowell E., Lee B. H., Döhner K., Ernst P., Davidson A. J., Daley G. Q., Zon L. I., Gilliland D. G., Huntly B. J. (2006) Cdx4 dysregulates Hox gene expression and generates acute myeloid leukemia alone and in cooperation with Meis1a in a murine model. Proc. Natl. Acad. Sci. U.S.A. 103, 16924–16929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Yates F., Naveiras O., Ernst P., Daley G. Q. (2005) Embryonic stem cell-derived hematopoietic stem cells. Proc. Natl. Acad. Sci. 102, 19081–19086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bei L., Huang W., Wang H., Shah C., Horvath E., Eklund E. (2011) HoxA10 activates CDX4 transcription and Cdx4 activates HOXA10 transcription in myeloid cells. J. Biol. Chem. 286, 19047–19064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pilon N., Oh K., Sylvestre J. R., Bouchard N., Savory J., Lohnes D. (2006) Cdx4 is a direct target of the canonical Wnt pathway. Dev. Biol. 289, 55–63 [DOI] [PubMed] [Google Scholar]
- 24. Shah C. A., Bei L., Wang H., Platanias L. C., Eklund E. A. (2012) HoxA10 regulates transcription of the gene encoding fibroblast growth factor 2 (FGF2) in myeloid cells. J. Biol. Chem. 287, 18230–18248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowney P., Corral J., Detmer K., LeBeau M. M., Deaven L., Lawrence H. J., Largman C. (1991) A human Hox 1 homeobox gene exhibits myeloid-specific expression of alternative transcripts in human hematopoietic cells. Nucleic Acids Res. 19, 3443–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takebe Y., Seiki M., Fujisawa J., Hoy P., Yokota K., Arai K., Yoshida M., Arai N. (1988) SR α promoter. An efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 8, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shah C. A., Wang H., Bei L., Platanias L. C., Eklund E. A. (2011) HoxA10 regulates transcription of the gene encoding transforming growth factor β 2 (TGFB2) in myeloid cells. J. Biol. Chem. 286, 3161–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larrick J. W., Fischer D. G., Anderson S. J., Koren H. S. (1980) Characterization of a human macrophage-like cell line stimulated in vitro. A model of macrophage functions. J. Immunol. 125, 6–12 [PubMed] [Google Scholar]
- 29. Eklund E. A., Jalava A., Kakar R. (1998) PU.1, interferon regulatory factor 1, and interferon consensus sequence-binding protein cooperate to increase gp91phox expression. J. Biol. Chem. 273, 13957–13965 [DOI] [PubMed] [Google Scholar]
- 30. Satokata I., Benson G., Maas R. (1995) Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature 374, 460–463 [DOI] [PubMed] [Google Scholar]
- 31. Oberley M. J., Farnham P. J. (2003) Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 371, 577–596 [DOI] [PubMed] [Google Scholar]
- 32. Eklund E. A., Jalava A., Kakar R. (2000) Tyrosine phosphorylation of HoxA10 decreases DNA-binding and transcriptional repression during IFNγ-induced differentiation of myeloid leukemia cell lines. J. Biol. Chem. 275, 20117–20126 [DOI] [PubMed] [Google Scholar]
- 33. Wang H., Lu Y., Huang W., Papoutsakis E. T., Fuhrken P., Eklund E. A. (2007) HoxA10 activates transcription of the gene encoding Mkp2 in myeloid cells. J. Biol. Chem. 282, 16164–16176 [DOI] [PubMed] [Google Scholar]
- 34. Bei L., Lu Y., Bellis S. L., Zhou W., Horvath E., Eklund E. A. (2007) Identification of a HoxA10 activation domain necessary for transcription of the gene encoding β3 integrin during myeloid differentiation. J. Biol. Chem. 282, 16846–16859 [DOI] [PubMed] [Google Scholar]
- 35. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dubchak I., Brudno M., Loots G. G., Mayor C., Pachter L., Rubin E. M., Frazer K. A. (2000) Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10, 1304–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., Pachter L. S., Dubchak I. (2000) VISTA. Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 [DOI] [PubMed] [Google Scholar]
- 38. Loots G. G., Ovcharenko I., Pachter L., Dubchak I., Rubin E. M. (2002) rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12, 832–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goardon N., Marchi E., Atzberger A., Quek L., Schuh A., Soneji S., Woll P., Mead A., Alford K. A., Rout R., Chaudhury S., Gilkes A., Knapper S., Beldjord K., Begum S., Rose S., Geddes N., Griffiths M., Standen G., Sternberg A., Cavenagh J., Hunter H., Bowen D., Killick S., Robinson L., Price A., Macintyre E., Virgo P., Burnett A., Craddock C., Enver T., Jacobsen S. E., Porcher C., Vyas P. (2011) Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 19, 138–152 [DOI] [PubMed] [Google Scholar]
- 40. van de Wetering M., Cavallo R., Dooijes D., van Beest M., van Es J., Loureiro J., Ypma A., Hursh D., Jones T., Bejsovec A., Peifer M., Mortin M., Clevers H. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88, 789–799 [DOI] [PubMed] [Google Scholar]
- 41. Kim K., Pang K. M., Evans M., Hay E. D. (2000) Overexpression of β-catenin induces apoptosis independent of its transactivation function with LEF-1 or the involvement of major G1 cell cycle regulators. Mol. Biol. Cell 11, 3509–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He X. C., Yin T., Grindley J. C., Tian Q., Sato T., Tao W. A., Dirisina R., Porter-Westpfahl K. S., Hembree M., Johnson T., Wiedemann L. M., Barrett T. A., Hood L., Wu H., Li L. (2007) PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat. Genet. 39, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keenan I. D., Sharrard R. M., Isaacs H. V. (2006) FGF signal transduction and the regulation of Cdx gene expression. Dev. Biol. 299, 478–488 [DOI] [PubMed] [Google Scholar]
- 44. Wilson E. L., Rifkin D. B., Kelly F., Hannocks M. J., Gabrilove J. L. (1991) Basic fibroblast growth factor stimulates myelopoiesis in long-term human bone marrow cultures. Blood 77, 954–960 [PubMed] [Google Scholar]
- 45. Aguayo A., Kantarjian H., Manshouri T., Gidel C., Estey E., Thomas D., Koller C., Estrov Z., O'Brien S., Keating M., Freireich E., Albitar M. (2000) Angiogenesis in acute and chronic leukemias and myelodysplastic syndromes. Blood 96, 2240–2245 [PubMed] [Google Scholar]