Background: Low affinity autoreactive B cells are activated by co-engagement of the BCR and TLRs.
Results: Endogenous RNAs that are associated with autoantigens are ligands for TLR7 and TLR3.
Conclusion: Sequence and structure of RNA determines recognition by TLRs.
Significance: B cells are activated by RNA delivered through the BCR, and the sequences of RNAs define their stimulatory capacity as TLR ligands.
Keywords: Antibodies, Autoimmune Diseases, Autoimmunity, Immunology, RNA, Toll-like Receptors (TLR), B Cell
Abstract
The key step in the activation of autoreactive B cells is the internalization of nucleic acid containing ligands and delivery of these ligands to the Toll-like Receptor (TLR) containing endolysosomal compartment. Ribonucleoproteins represent a large fraction of autoantigens in systemic autoimmune diseases. Here we demonstrate that many uridine-rich mammalian RNA sequences associated with common autoantigens effectively activate autoreactive B cells. Priming with type I IFN increased the magnitude of activation, and the range of which RNAs were stimulatory. A subset of RNAs that contain a high degree of self-complementarity also activated B cells through TLR3. For the RNA sequences that activated predominantly through TLR7, the activation is proportional to uridine-content, and more precisely defined by the frequency of specific uridine-containing motifs. These results identify parameters that define specific mammalian RNAs as ligands for TLRs.
Introduction
Systemic autoimmune diseases such as systemic lupus erythematosus (SLE),2 systemic sclerosis (SSc), Sjögren's syndrome, and polymyositis are characterized by autoantibodies reactive with nuclear or subcellular organelles. The targeted autoantigens are commonly components of macromolecular complexes involved in RNA assembly, processing, or quality control. Intriguingly, particular autoantibody reactivities commonly associate with specific disease subcategories (1, 2). For example, SLE patients often make autoantibodies against nucleosome (DNA, histone) or splicesome components (SmD), SSc patients make autoantibodies reactive with nucleolar proteins (topoisomerase), and polymyositis patients make autoantibodies against aminoacyl tRNA synthetases (Jo-1) (2–4). The circumstances that expose such autoantigens to the immune system and promote a breach in B cell tolerance are incompletely understood, but most likely involve detection by innate immune receptors, such as the Toll-like receptor (TLR) family. Previous studies from our group and others have implicated TLR7 and TLR8 in the detection of mammalian RNA and RNA-binding autoantigens (5–7). Numerous reports involving TLR7-deficient mice have pointed to a critical role for TLR7 in a wide range of murine models of SLE (8, 9). However, the RNA motifs and/or structures that most effectively engage these receptors in the context of various autoimmune diseases are still unclear. A more precise delineation of these motifs is likely to provide important insights into the etiology of systemic autoimmune diseases.
The initial studies that identified ssRNA as the ligand for TLR7 showed that only certain viral RNA sequences stimulated PBMCs, and that polyuridylic acid (polyU), but no other single base polyribonucleotide-activated DCs (10, 11). These observations raised the question as to whether TLR7 recognizes uridine-rich RNA in general or has sequence specificity. One study found that TLR7 stimulation by RNA was defined only by the presence of uridine ribonucleotides, while another study found that the order of the bases differentially affected TNF-α and IFN-α production (12, 13). Recent follow-up work by Vollmer's group has defined a TLR7 stimulating motif that is dependent on the sequence surrounding a key uridine but does not need to be uridine-rich (14). Endogenous RNA sequences, such as those derived from the U1 snRNP, are stimulatory ligands for TLR7 (15). U1 RNA has also been shown to activate TLR3 when delivered to epithelial cells (16). A significant limitation of all of these studies is that they all examined short, often sulfur-stabilized, RNA oligonucleotides delivered to cells by conjugation with cationic lipids, such as DOTAP. The mode of delivery is important in determining the specificity of TLRs, as it has been shown that DNA devoid of CG dinucleotides only activates when delivered by DOTAP, but not via spontaneous uptake nor by Fc receptor mediated uptake of immune complexes (ICs) (17). Also, while it has been shown that B cells can be activated by synthetic dsRNA through a TRIF-dependent pathway, and that influenza virus activates B cells through a MyD88-dependent pathway, the parameters that define the stimulatory capacity of RNAs have not been examined in B cells (18, 19).
The nucleic acid sensing TLRs, including TLR3, -7, and -9, are sequestered in endosomal/lysosomal compartments in immune cells (20, 21). Compartmentalization is thought to prevent inappropriate detection of self-nucleic acids (21). In order for autoantigens to gain access to the TLR compartments, they require directed transport, which in the case of B cells is provided by the B cell receptor (BCR) (22). Normally topologically excluded autoantigens, including ribonucleoproteins, become exposed on the cell surface during apoptosis (23, 24), and failure to appropriately remove this cell debris can provide ligands for autoreactive B cells. To examine TLR reactivity to mammalian RNAs initially detected by the BCR, we used B cells from an autoreactive BCR site-directed transgenic line designated AM14 (25, 26). AM14 B cells recognize endogenous IgG2a and can be activated by natural or defined ICs that incorporate mammalian DNAs or RNAs (5, 22, 27). We previously used AM14 B cells to show that TLR9 is able to discriminate between CG-rich and CG-poor mammalian DNAs, despite a paucity of the “canonical motif” described for bacterial DNA (28). Here, we show that endogenous RNA sequences are capable of stimulating through TLR7 and TLR3, depending on their self-complementarity. Further, we show that both the frequency and the sequence context of uridines, determine the degree to which an RNA can activate murine B cells through TLR7.
EXPERIMENTAL PROCEDURES
Mice
AM14 mice have been described previously (26). AM14 mice were crossed to TLR7-deficient mice (TLR7 KO) provided by Dr. Shizuo Akira, and backcrossed 8 generations to the BALB/c background. AM14 mice were also crossed to FcγRIIB KO BALB/c background mice (Jackson). Unc93b1 deficient (3d) mice were a generous gift from Dr. B. Beutler. TLR3/TLR7 double deficient mice were kindly provided by Drs. D. Golenbock and R. Gazzinelli. All mice were maintained at the Boston University School of Medicine Laboratory Animal Sciences Center or University of Massachusetts Medical School Department of Animal Medicine, in accordance with the regulations of the American Association for the Accreditation of Laboratory Animal Care.
B Cell Stimulation and Analysis
B cells were positively selected with CD45R/B220 magnetic particles (BD Biosciences). B cells purity was validated by FACS and determined to be >98%. B cells were cultured in RPMI/5% heat-inactivated FCS and stimulated with: 1 μg/ml CpG (s-oligodeoxynucleotide (ODN) 1826, Coley Pharmaceuticals); 300 ng/ml CL097 (InvivoGen); 10 μg/ml BWR4, an RNA-specific mAb provided by Dr. D. Eilat (Hadassah University Hospital, Jerusalem, Israel); defined 1D4/RNA ICs consisting of 300 ng/ml Biotin-RNA fragments (see below) combined with 7.5 μg/ml of the IgG2a anti-biotin mAb, 1D4 (Biolegend); or defined Fab/RNA or Fab/DNA ICs consisting of 300 ng/ml of biotinylated nucleic acids, 1–3 μg/ml streptavidin, and 1.5 μg/ml biotinylated Fab fragment of goat anti-mouse IgM. Defined ICs were initially prepared in a small volume of PBS in the presence of 10 units/μl of RNasin (Promega) (28), and then diluted to the working concentration in RPMI/FCS. The effect of type I IFN was determined by adding 300 U/ml IFN-α or IFN-β (PBL) to the cells. B cells were stimulated in 96-well plates at a final concentration of 2 × 106 cells/ml for 24 h, then pulsed for 6 h with [3H]thymidine (Amersham Biosciences). Incorporation of [3H]thymidine was quantified via a liquid scintillation beta counter (Trilux 1450 MicroBeta, PerkinElmer).
Cytokine ELISA
Cytokine secretion by B cells stimulated in vitro in the presence of 50 ng/ml BLyS (Human Genome Sciences), were measured at 48 h. Plates were coated with anti-murine IL-6 (BD Bioscience) at 1 μg/ml or anti-murine RANTES (RND systems) at 2 μg/ml for 16 h at 4 °C. Samples were added for 4 h and detected with biotin-anti-murine IL-6 (BD Bioscience) or biotin- anti-murine RANTES at 0.5 μg/ml. Plates were developed with streptavidin-HRP (BD Bioscience) and tetramethylbenzidine liquid substrate system (Sigma-Aldrich). Standard curves were generated using recombinant murine IL-6 (BD Bioscience) sensitive to 125 ng/ml and recombinant murine RANTES (RND Systems) sensitive to 62.5 ng/ml.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was extracted from AM14 and AM14 TLR7 KO B cells, reverse transcribed, and analyzed by qPCR. Data are the means ± S.E. of seven independent experiments done in triplicate. Standard commercial TaqMan probes were used for TLR3, TLR7, MyD88, and GAPDH (Applied Biosystems). Samples were normalized to GAPDH and represented as fold change over medium control using the ΔΔCT method previously described
In Vitro Transcribed RNA
Templates for in vitro transcribed RNAs were linearized DNA plasmids with the sequence downstream of a T7 or an SP6 promoter. The templates were cloned by our laboratory (Alu1, mt-R-loop, r11, and r13), or were generous gifts of Dr. S. Wolin (Y4, Y5), Dr. L. Gehkre (HCV 3′-UTR and SS1 (29)), or Dr. P. J. Utz and Dr. Ger Pruijn. RNA was generated by in vitro transcription using RiboMAX SP6 and T7 Large Scale RNA Production Systems (Promega), or DuraScribe T7 kit (Epicenter Biotechnologies). Biotinylated RNAs were made by substituting a fraction of the nucleotides with biotin-16-aaCTP or biotin-16-aa-UTP (TriLink) (see supplemental Table S1); T7 RNA polymerase has been previously shown to efficiently accept biotinylated bases (30). Other modified bases, 2′-Ome-U and pseudo-U (TriLink) and 2′-F-dU and 2′-F-dC (epicenter) were incorporated for further analyses. DNA templates were removed using RNase-free DNase at 1 unit/μg of template (Promega), and RNA was isolated using RNeasy columns (Qiagen), according to the manufacturer's recommendations. RNA concentration was determined by spectrophotometry using a nanodrop spectrophotometer (Thermo). RNA integrity, biotinylation, and concentration were further validated by visualization with ethidium bromide by gel electrophoresis.
RESULTS
Defined RNA ICs Activate AM14 B Cells
AM14 B cells express a low affinity receptor for IgG2a and do not respond to monomeric IgG2a or IgG2a-bound protein ICs. However, IgG2a mAbs that bind DNA, or RNA, or associated proteins induce a robust proliferative response, dependent on TLR9 or TLR7, respectively (5, 22). As no exogenous sources of nucleic acid were added to these cultures, the actual autoantigen in these “natural” ICs is, of necessity, derived from cell debris, or the B cells themselves, and thus ill-defined. We have previously shown that the RNA reactive mAb, BWR4, can bind undefined mammalian RNA ligands to form ICs that effectively activate AM14 B cells through a TLR7-dependent mechanism (5). BWR4 can also bind defined RNA fragments, as shown by EMSA for an Alu RNA fragment (Fig. 1A). Alu RNAs are short, high copy number, repetitive genetic elements, which can be transcribed during times of cellular stress (31). Since Alu RNAs are transcribed by RNA polymerase II, they will not be capped. The scAlu RNAs (including Alu1, tested here) are shorter sequences that lack the adenine-rich regions (32). We found that the AM14 B cell response to BWR4 could be enhanced by precombining BWR4 with Alu RNA or other RNAs that had been previously described as TLR7 ligands, including VSV genome, polyU, and ssRNA40 (Fig. 1B and supplemental Fig. S1, A and B). However, the relatively modest increase indicated that this would not be the optimal method for parsing differences in the stimulatory capacity of specific autoantigen-associated RNAs. Moreover, BWR4 preferentially binds A-C-U-containing sequences (33), which could further confound data obtained with this reagent. Therefore, we used an approach similar to one used previously to compare dsDNA fragments by constructing ICs consisting of an anti-biotin mAb, 1D4, and biotinylated dsDNA fragments (28).
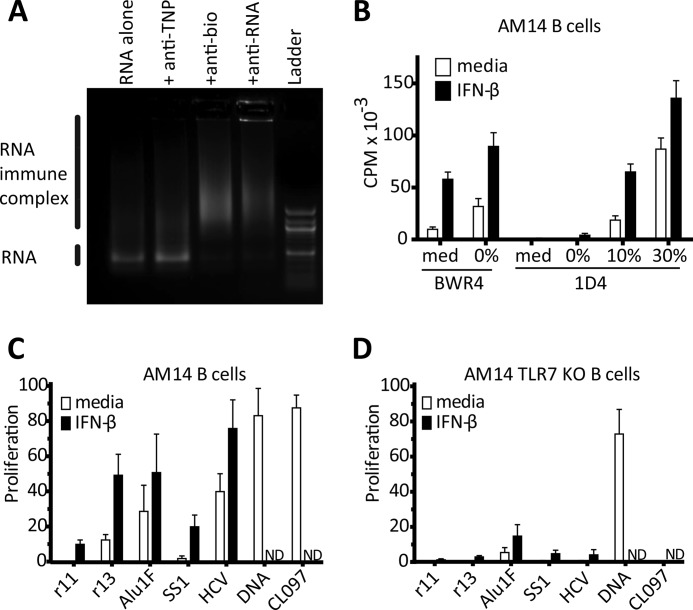
FIGURE 1.
Uridine-rich RNAs activate AM14 B cells, when delivered as ICs, through a TLR7-dependent manner. A, in vitro transcribed bio-Alu1 RNA, alone or premixed with the indicated mAbs, was electrophoresed in a 1% agarose gel and subsequently visualized with ethidium bromide. B, AM14 B cells were stimulated with BWR4 or 1D4 alone (med) or premixed with bio-Alu1 RNA transcribed in the presence of 0, 10, or 30% biotinylated cytodine bases, in the absence □ or presence ■ of IFN-β. Proliferation was quantified by [3H]thymidine incorporation. Average ± S.E. is shown, n = 3. C and D, AM14 B cells, or AM14 TLR7-deficient B cells were stimulated with 1D4 premixed with the indicated bio-RNAs (with 8 biotins for every 100 nt) or stimulated with 1D4 and bio-DNA, or the small molecule ligand CL097 and in the absence □ or presence ■ of IFN-β. Proliferation is measured by [3H]thymidine uptake, and plotted as the percent of the response to CpG-B. Average ± S.E. is shown, n = 3.
Biotinylated-RNA fragments (Bio-RNAs) were generated by incorporating biotinylated RNA bases into our in vitro transcription reactions. These bio-RNAs formed ICs when combined with either BWR4 or 1D4, but not with an irrelevant anti-hapten mAb, as shown by EMSA (Fig. 1A). Importantly, 1D4 alone fails to activate AM14, but 1D4 + bio-Alu1 ICs induced a robust proliferative response (Fig. 1B). A relatively high degree of biotin incorporation and high levels of anti-biotin antibody are needed to make stimulatory RNA ICs (Fig. 1B and supplemental Fig. S1C). Presumably the antibody is both protecting the RNA from degradation as well as delivering it to the appropriate TLR-associated compartment. In a more physiological setting, RNA-binding proteins could serve a similar role, protection of RNA and binding to the BCR of an autoreactive B cell. Optimal activation of B cells by natural TLR7 requires exogenous type I IFN priming, and similarly, the stimulation by Alu RNA containing RNA ICs is enhanced by type I IFN-priming (Fig. 1A and supplemental Fig. S1B, and Ref. 34). Therefore this strategy recapitulates B cell activation by natural RNA ICs (such as BWR4) and provides a method for the precise comparison of endogenous RNA sequences.
Uridine-rich RNAs Activate B Cells in a TLR7-dependent Manner
We first used defined RNA ICs to compare uridine-rich and uridine-poor RNAs. Uridine-poor (r11: 17% U), and uridine-rich (r13: 42% U) biotinylated, in vitro transcribed, RNAs were generated from mammalian cloned sequences available in the laboratory (28). We also generated viral RNA sequences from the 3′-untranslated region of hepatitis C virus (HCV 3′-UTR: 52% U) and a uridine-poor section of the same virus (SS1: 18% U) (29). Both the uridine-rich endogenous sequence RNA and the uridine-rich viral sequence induced robust B cell proliferation, whereas the uridine-poor RNAs barely activated the cells at all (Fig. 1C). The responses to the Alu1, HCV 3′-UTR, and r13 RNAs were highly dependent on TLR7, as these ICs did not activate AM14 TLR7 KO B cells, similar to the small molecule TLR7 ligand CL097 (Fig. 1, C and D). As expected, the AM14 response to a bio-dsDNA IC was TLR7 independent (Fig. 1, C and D). These data demonstrate that uridine-rich RNAs are effective TLR7 ligands when taken up by B cells through a physiologically relevant mechanism.
RNAs from Major RNP Autoantigens Activate Autoreactive B Cells
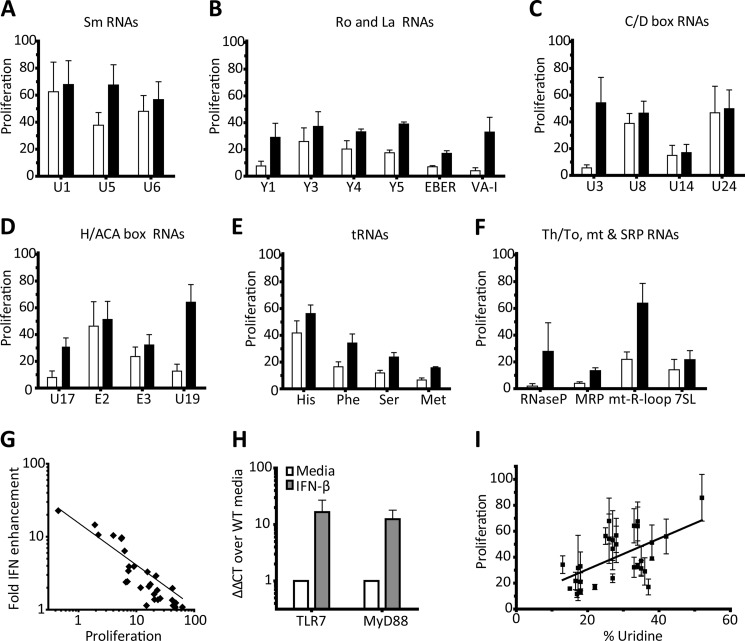
Structural RNAs, associated with RNPs that function to process immature RNAs to their mature forms, are some of the most abundant RNAs in the cell (35). In addition to their critical function, these RNPs are common targets of autoantibodies. It follows that these structural RNAs provide adjuvant like activity that can promote the activation of low affinity autoreactive B cells. To test this assumption, we generated a panel of biotinylated, in vitro transcribed RNAs corresponding to mammalian RNP-associated sequences. The RNAs were tested, as RNA ICs, for their capacity to activate AM14 B cells in the presence or absence of type I IFNs. RNAs associated with the canonical SLE autoantigen Sm were quite stimulatory, even without type I IFN priming (Fig. 2A). Ro associated Y-RNAs (Y1, Y3, Y4, and Y5) were somewhat less stimulatory, and La-associated RNAs (EBER and VA-I) only induced proliferation with type I IFN-priming (Fig. 2B). RNAs involved in splicing and post-transcriptional modifications of ribosomal RNAs that contain the highly conserved C/D or H/ACA boxes stimulated AM14 B cells to varying degrees; for example, the fibrillarin-associated RNA U8 induced a strong response even in the absence of IFN (Fig. 2, C and D). tRNA His, the RNA component of the prevalent autoantigen in polymyositis, was more stimulatory than the other tRNAs that occasionally are more rarely targeted by autoimmune diseases (Fig. 2E). The RNA from the Th/To antigen, RNase MRP, was not very stimulatory, but the mitochondrial R-loop (the stable RNA transcript at the heavy strand origin of replication) was quite stimulatory (Fig. 2F). Overall, these data demonstrate that many, but not all of the RNAs associated with known RNP autoantigens are likely to engage endosomal TLRs. In general, the less stimulatory RNAs showed the greatest fold enhancement by type I IFN priming (Fig. 2G).
FIGURE 2.
The response of AM14 B cells to endogenous, RNP-associated RNA sequences is variably enhanced by type I IFN. A–F, AM14 B cells were stimulated with 1D4 or premixed with the indicated bio-RNAs, in the absence □ or presence ■ of IFN-β. Proliferation was quantified by [3H]thymidine incorporation, and is shown as the percent of the response to CpG-B. Average ± S.E. is shown, A and C, n = 4, B and D–F n = 3. G, proliferation of the samples from A–F in the absence of interferon plotted against the fold enhancement in the presence of IFN-β. Log-log regression line Y = 10(−0.590×log X +1.164), R2 = 0.8543. H, qRT-PCR for TLR7 and MyD88 from resting and 1 h IFN-primed AM14 B cells. Data represented as ΔΔCT fold change over media, relative to GAPDH. Average ± S.E. is shown, n = 5. I, proliferation of the samples from A–F in the presence of IFN, plotted against % of uridine content in each fragment. Linear regression line, R2 = 0.3206, p = 0.0011.
We and others have previously shown that type I IFN leads to increased TLR7 expression, and that IFN increases BCR signaling (28, 34, 36). IFN priming increases the expression of TLR7 and MyD88, which most likely accounts for the increased sensitivity of IFN-primed B cells to suboptimal TLR7 ligands (Fig. 2H).
Somewhat unexpectedly, the stimulatory capacity of the RNA fragments did not correlate particularly well with uridine content (Fig. 2I). The base content and length of our RNAs, ranging from 73–877 nt, with an average length of 215 nt, are summarized in supplemental Table S1. These data show that certain endogenous sequences from RNAs associated with known autoantigens can serve as strong TLR ligands, since they robustly stimulate autoreactive B cells, even in the absence of exogenous type I IFN.
RNAs from Major RNP Autoantigens Activate Autoreactive B Cells Independently of TLR7
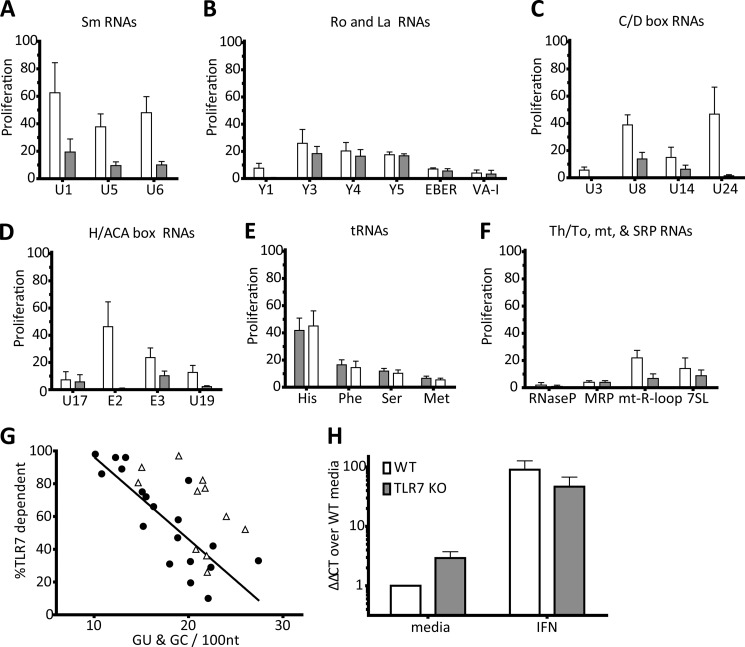
To determine to what extent AM14 B cell activation by our panel of defined RNA ICs was TLR7-dependent, we tested our ICs on TLR7-deficient AM14 B cells. In general, RNA ICs stimulated AM14 B cells more effectively than AM14 TLR7 KO B cells (Fig. 3, A–F), with a few notable exceptions. Ro associated Y-RNAs induced TLR7-independent B cell proliferation, as did the tRNAs (Fig. 3, B and E). Additionally, U1 RNA, of the U1–70KDa autoantigen, and U8, were partially TLR7 independent (Fig. 3, A and C). While it was possible that these RNAs formed exceedingly large ICs that were capable of activating AM14 B cells through a TLR-independent mechanism, we have found that large protein complexes that do not incorporate nucleic acids routinely fail to activate AM14 B cells. Alternatively, activation by these ICs depended on a pattern recognition receptor other than TLR7.
FIGURE 3.
Endogenous RNP-associated RNA sequences with regions of sequence complementarity can activate AM14 B cells independently of TLR7. A–F, AM14 □ or AM14 TLR7-deficient ■ B cells were stimulated with 1D4 premixed with the indicated bio-RNAs. Proliferation is plotted as the percent of the response to CpG-B. Average ± S.E. is shown, A and C, n = 4, B and D–F, n = 3. G, percent of proliferation dependent of TLR7 from A–F, plotted against the number of base-paired guanines predicted by MFOLD. Stimulatory RNAs (●) were compared for the linear regression, R2 = 0.6575, p < 0.0001, weakly stimulatory (<10% Proliferation on WT) RNAs (Δ) were not included in this analysis. H, qRT-PCR for TLR3 from resting and 1 h IFN-primed AM14 □ or AM14 TLR7 KO ■ B cells. Data represented as ΔΔCT fold change over media, relative to GAPDH. Average ± S.E. is shown, n = 3.
TLR3 is an endosomal receptor that recognizes dsRNA (37). While all of the RNAs that we tested are made as single-stranded in vitro transcribed RNAs, some are of sufficient length and self-complementarity to potentially form partial double-stranded structures. The stability of RNA secondary structure is an active area of research, and the rules defining optimal stable structures are still being determined. Nonetheless, we examined the potential for secondary structure by running our RNA sequences through the mFOLD folding prediction software, and the number of base-paired guanines was enumerated (38). The extent of secondary structure, as enumerated by the number of paired-Gs per length of RNA, was inversely proportional to the TLR7 dependence of the stimulation (Fig. 3G). This indicates that these TLR7 independent RNAs could have more stable double-stranded secondary structures, and therefore might stimulate through the dsRNA receptor TLR3. To support this idea, we used qRT-PCR to demonstrate that TLR7 KO B cells express TLR3 (Fig. 3H). TLR3 expression was slightly higher in resting TLR7 KO B cells than in TLR7-sufficient B cells, and expression level was in both cases enhanced by the addition of type I IFN (Fig. 3H). The correlation between TLR7 independent activity and more self-complementary suggested that these mammalian RNA sequences could function as TLR3 ligands.
TLR3 Is the Second Receptor for Highly Structured RNAs
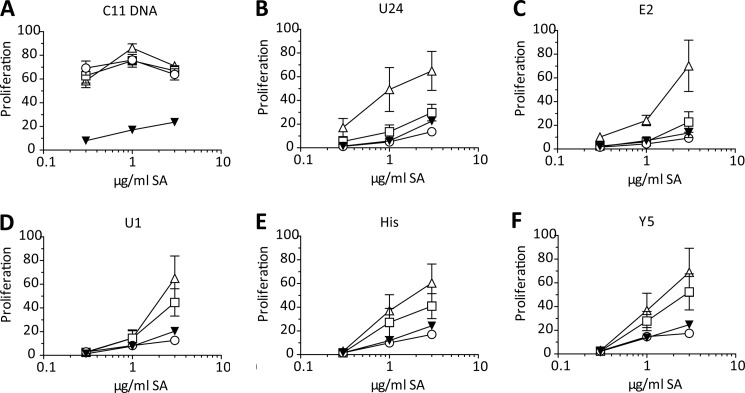
TLR3 is often overlooked in the context of autoimmune disease because dsRNA is considered to be a viral product. Interestingly, TLR3 ligands have been used to induce type I IFN in models of SSc (39). To be able to test the role of TLR3 in non-transgenic B cells, we developed a second method for targeting nucleic acid ICs to the BCR; bio-RNAs (or bio-dsDNAs) were coupled to biotinylated-anti-IgM Fab fragments with streptavidin to make reagents that could target the BCR but not cause extensive cross-linking of the receptor. Fab-nucleic acid ICs activated WT and AM14 B cells with comparable titrations and dependence on TLR7, demonstrating that activation of B cells by RNA delivered to TLRs via the BCR is a general feature of B cells (Fig. 4 and supplemental Fig. S2). We found that these Fab-nucleic acid ICs failed to activate unc93B-deficient B cells, and therefore activation most likely depends on endosomal TLRs (Fig. 4). As expected, Fab DNA IC, that incorporate the CG-rich bio-dsDNA fragment, clone 11, activated TLR3/7 double KO B cells as well as WT B cells, but failed to activate UNC93B-deficient cells. This shows that activation by Fab DNA ICs remains TLR9-dependent (Fig. 4A). Furthermore, the RNA fragments previously shown to activate predominantly through TLR7 as 1D4/bio-RNA ICs (Fig. 3), were also highly TLR7 dependent in the form of Fab RNA ICs (Fig. 4, B and C). Importantly, the RNA fragments that induced predominantly TLR7-independent stimulation of AM14 B cells failed to stimulate TLR3/7 double deficient B cells (Fig. 4, D–F). We therefore concluded that the residual TLR7-independent activity of these RNAs was due to TLR3.
FIGURE 4.
Endogenous RNP-associated RNA sequences with regions of sequence complementarity can activate B cells through TLR3. A–F, WTΔ, TLR7 KO □, TLR3/7 dKO ▾, or unc93b3d ο B cells were stimulated with the indicated Fab ICs. Proliferation was quantified by [3H]thymidine incorporation and plotted as the percent of the response to CpG-B. Average ± S.E. is shown, n = 7.
TLR3-dependent RNAs Elicit IL-6 in the Absence of Exogenous Type I IFN
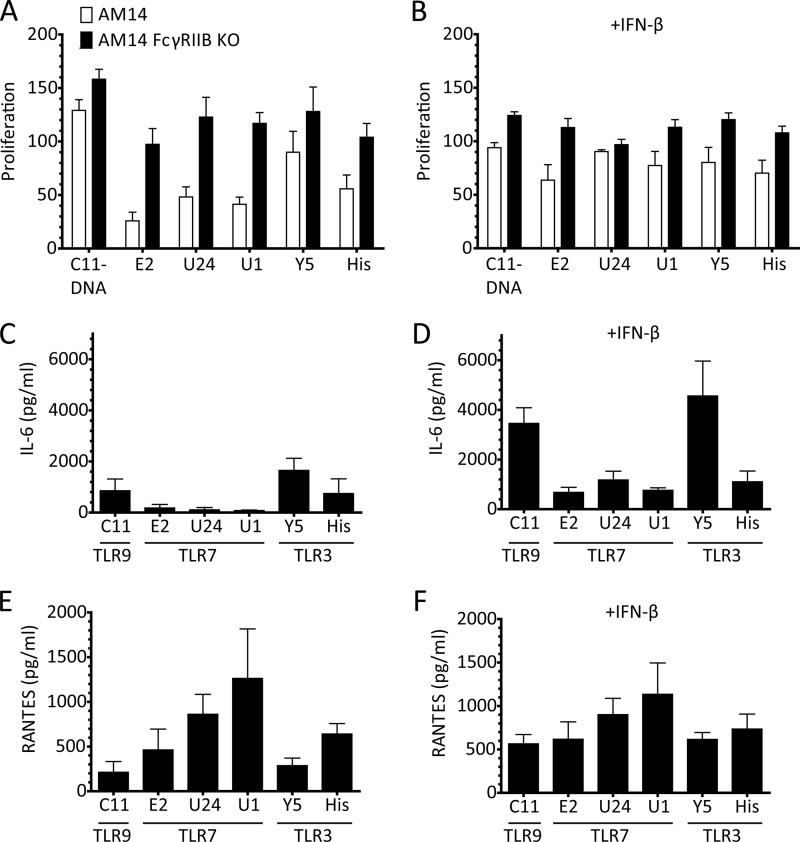
Since TLR3 and TLR7 signal through different pathways, we sought to determine whether RNA ligands for TLR7 and for TLR3-elicited different responses. We evaluated both AM14 and AM14 FcγRIIB-deficient (R2KO) B cells, as the R2KO cells have previously been shown to be more responsive to spontaneous RNA ICs (40). We found that the AM14 R2KO B cells also proliferate better in response to defined RNA ICs, especially with regard to the TLR7-dependent RNAs (Fig. 5A). The suboptimal TLR7 response of the R2+ B cells was enhanced by the addition of type I IFN (Fig. 5B). Because the R2KO cells proliferated comparably to both the TLR7 and TLR3 ligands in both the presences and absence of type I IFN, we examined these culture supernatants for cytokine production. The two highly TLR3-dependent RNAs, Y5, and His, elicited IL-6, even without type I IFN priming, whereas the RNAs with a strong TLR7 component only made IL-6 when provided with exogenous IFN-β (Fig. 5, C and D). By contrast, the TLR7-dependent RNAs made high levels of RANTES in the absence of type I IFN, and RANTES production was not substantially increased by type I IFN priming (Fig. 5, E and F). These data are consistent with the notion that in B cells the TLR3-TRIF pathway is more directly connected to the production of type I IFN than the TLR7-MyD88 pathway.
FIGURE 5.
TLR3-dependent RNAs make IL-6 independently of IFN priming. A and B, proliferation of AM14 □ and AM14 FcγRIIB KO ■ B cells were stimulated with with 1D4 or premixed with the indicated bio-RNAs, in the absence (A) or presence (B) of IFN-β. Proliferation was quantified by [3H]thymidine incorporation, and is shown as the percent of the response to CpG-B. Average ± S.E. is shown, n = 4. C–F, cytokine production of AM14 FcγRIIB KO B cells in the absence (C and E) or presence (D and F) of IFN-β. The TLRs through which the given ligands stimulate are noted under the name of the nucleic acid. Cytokine production was measured by ELISA, and the average ± S.E. of four independent experiments is shown.
Uridines Are the Key Determinant in Stimulatory TLR7 Ligands
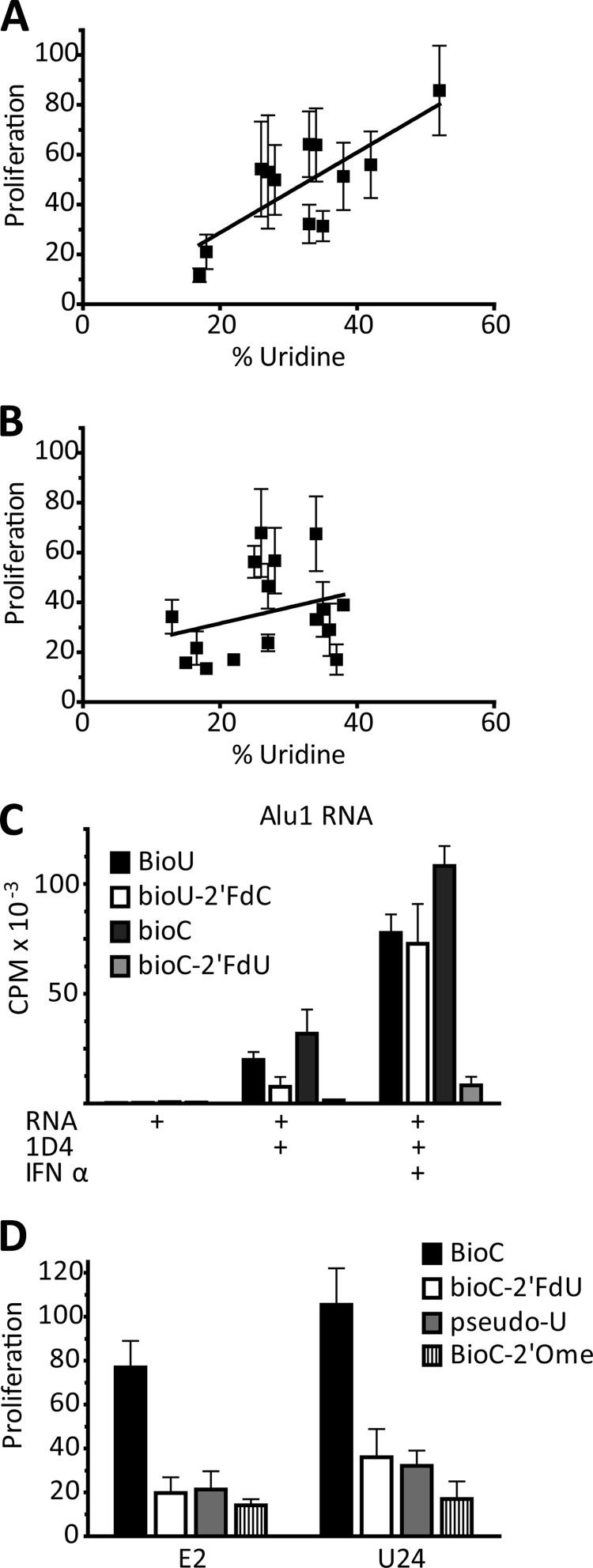
We next applied our RNA IC strategy to the further examination of sequence specificity of TLR7. As shown above, the uridine content of our RNA fragments did not correlate well with capacity to activate AM14 B cells (Fig. 3G). However, if we exclude the RNAs with substantial TLR7 independent activity from the analysis, then a significant correlation between TLR7-dependent stimulation and uridine content of the RNA ligands is revealed (Fig. 6A). Conversely, if we compare the uridine content of the excluded RNAs to their stimulatory capacity, there is no correlation (Fig. 6B). To determine the contribution of uridine more precisely, we made variants of the highly TLR7-dependent RNA, Alu1, and substituted either the cytosines or the uridines with 2′-fluoro-substituted bases, using either Bio-U or Bio-C for 1D4-mediated BCR delivery. Incorporation of Bio-C or Bio-U did not affect stimulatory capacity of the RNA ICs, presumably because the 16 carbon linker that attaches the biotin to the base keeps the biotin at sufficient distance to not block recognition by the TLR (Fig. 6C). By contrast, fluorine substitution of the 2′-OH of the ribose of the uridines (2′-FdU) abolished the stimulatory capacity of the RNA ICs, whereas the same substitution on cytosine (2′-FdC) had a minimal effect (Fig. 6C). To validate the potential physiological relevance of this finding, we took another two highly TLR7 dependent RNAs and made additional uridine substitutions. In addition to the artificial 2′-FdU, we substituted with a 2′O-methylated sugar, and the isomerized base psuedouridine. All of these uridine modifications significantly abrogated AM14 stimulatory activity (Fig. 6D). These data demonstrate that there is a strict requirement for intact uridine, both at the level of the base and the sugar, for RNA to effectively engage TLR7 in B cells.
FIGURE 6.
Uridines are an essential determining factor in RNA activation through TLR7. A, proliferation in the presence of IFN of the samples from Fig. 3, A–F, plotted against % uridine content of the fragment; analysis includes RNAs that are >75% TLR7 dependent. Linear regression, R2 = 0.5840, p = 0.0038. B, proliferation in the presence of IFN of the samples from Fig. 3, A–F, plotted against % uridine content of the fragment; analysis includes RNAs that are <75% TLR7 dependent for stimulation. Linear regression line, R2 = 0.02927, p = 0.5115. C, Bio-Alu 1 RNA ICs activation of AM14 B cells ± IFN-β; RNAs transcribed with BioU or Bio-C, and/or 2′FdC or 2′FdU as indicated. Proliferation is measured by [3H]thymidine incorporation. Average ± S.E. is shown, n = 3. D, Bio-E2 and Bio-U24 RNA IC activation of AM14 B cells; RNAs transcribed with Bio-C and either 2′-FdU, pseudouridine (pseudo-U), or 2′-O-methyl-uridine (2′Ome) substitutions. Proliferation is plotted as the percent of the response to CpG-B. Average ± S.E. is shown, n = 3.
The Sequence Specificity of TLR7 Is Determined by the Motif in Which the Uridines Reside
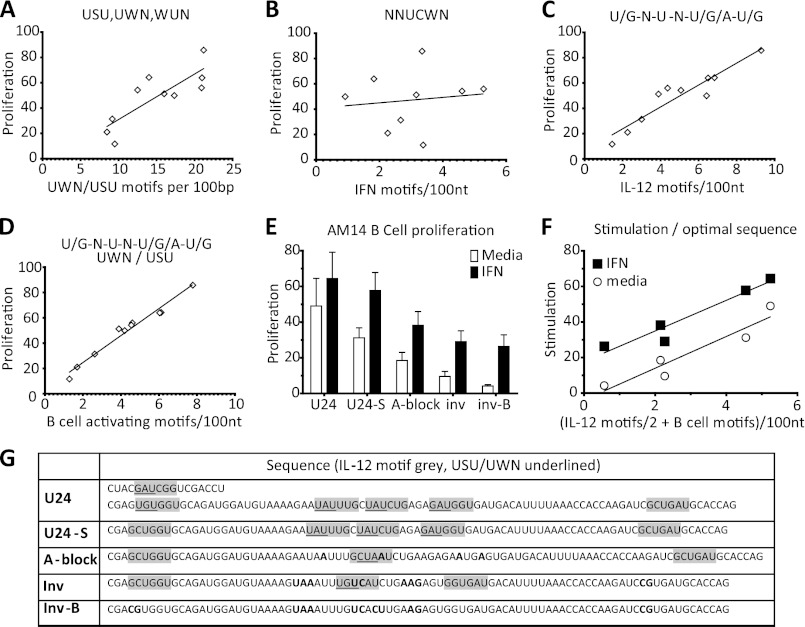
The strong dependence on intact uridines in the stimulatory TLR7 ligands does not rule out the possibility for even finer specificity of the receptor. To determine whether the context of the uridine was also an important determinant for TLR7 activation, we searched for 3-mers that were overrepresented in some of the more stimulatory TLR7-dependent sequences (E2, U24, HCV3′UTR), relative to less stimulatory sequences (U17, r11). The consensus stimulatory 3-mer that emerged from this analysis is defined as a uridine adjacent to another uridine or adenine (W), or one nucleotide away from another uridine (U-S-U; S is C or G). The frequency of these triplets correlated with greater stimulatory activity (Fig. 7A). A recent report by Vollmer and co-workers showed that the substitution of individual bases around a single key uridine in a stimulatory RNA motif could greatly diminish the capacity of their RNAs to induce PBMCs to make cytokine (14). They identified two motifs. The first motif, NNUCWN, was associated with optimal IFN-α production. The frequency of this motif did not correlate with the B cell stimulatory capacity of our RNAs (Fig. 7B). The second motif, KNUNDK (K is U or G, and D is any but C), optimally elicited IL-12. There was a strong correlation between the frequency of this motif and TLR7-dependent B cell stimulatory capacity as defined by our assays (Fig. 7C). By limiting the IL-12 motif to only include sequences where the central uridine also fulfills our U-W-N/U-S-U triplet requirement, we identified a more optimal motif for the activation of murine B cells through TLR7. The new motif is the IL-12 motif, KNUNDK, but with the central U being adjacent to a weak base or one space away from another U in either direction. If we screen for the frequency of this B cell stimulating motif in our TLR7-dependent RNAs, the frequency of this motif explains over 95% of the variance in stimulation by TLR7 dependent RNA ICs (Fig. 7D). To validate this analysis, we made modified versions of one of the more TLR7-dependent stimulatory mammalian RNAs, U24. We started with a shortened version of U24, U24-S, which eliminated the first 2 B cell activating motifs, and replaced them with one initial IL-12 motif. If we then inserted adenines to disrupt all of the B cell stimulating motifs (A-block), but not two of the IL-12 motifs, or made inversions to accomplish the same effect (invert), the stimulatory capacity is slightly reduced (Fig. 7E). If we then further modify the RNA to disrupt both the B cell stimulating motifs and the IL-12 motifs (invert-B), then the stimulation is reduced further (Fig. 7E). These sequences are shown in Fig. 7G. Analysis of these four RNAs shows that stimulatory capacity correlated with the frequency of both IL-12 and B cell activating motifs, whether or not the cells were primed with type I IFN (Fig. 7F). It is also important to note that these modified RNA sequences are still comparably uridine-rich. These data demonstrate that TLR7 has a sequence preference beyond simply uridine density. The optimal motif for recognition of RNA by TLR7 in B cells differs substantially from the motif associated with maximal type I IFN production in PBMCs.
FIGURE 7.
A specific uridine-containing motif determines the stimulatory capacity of RNA ligands for TLR7. A, proliferation, in the presence of IFN, of the >90% TLR7-dependent samples from Fig. 3, A–F plotted against the frequency of USU/UWN 3-mers. Linear regression, R2 = 0.6771, p = 0.0035. B, proliferation, in the presence of IFN, of the >90% TLR7 dependent samples from Fig. 3, A–F plotted against NNUCWN (IFN) motifs. Linear regression, R2 = 0.01623, p = 0.7440. C, proliferation, in the presence of IFN, of the >90% TLR7-dependent samples from Fig. 3, A–F plotted against frequency of KNUNDK (IL-12) motif. Linear regression, R2 = 0.8838, p < 0.0001. D, proliferation, in the presence of IFN, of the >90% TLR7-dependent samples from Fig. 3, A–F plotted against frequency of KNUNDK & USU/UWN (B cell activating) motif. Linear regression, R2 = 0.9710, p < 0.0001. E, U24-bio RNA, and sequence modifications, with 1D4 stimulation of AM14 B cells, in the absence □ or presence ■ of IFN-β. Proliferation is measured by [3H]thymidine uptake, and plotted as the percent of the response to CpG-B. Average ± S.E. is shown, n = 3. F, proliferation from E plotted against the frequency of B cell stimulating motifs + half the IL-12 motifs that do not fulfill the USU/UWN motif. Linear regression for media: R2 = 0.8964, p = 0.0146. Linear regression for IFN: R2 = 0.9242, p = 0.0091. G, sequences of RNAs analyzed in E and F.
DISCUSSION
Autoantibodies reactive with particular RNPs have used as a diagnostic marker for rheumatic diseases for many years, but the importance of innate immune receptors in breaking tolerance to these self-antigens has only been appreciated more recently. It is now clear that TLR7 plays a critical role in the pathogenesis of SLE, and that TLR7 is required for the generation of anti-RNP autoantibodies in multiple murine models of SLE (9). We now show that RNAs associated with SLE-targeted RNPs effectively engage TLR7 and/or TLR3. Importantly, many of these RNAs are so stimulatory that they activate autoreactive B cells without exogenous type I IFN priming. Such RNAs may play a key role in the early events associated with the loss of B cell tolerance, the production of autoantibodies, and the subsequent formation of ICs. Such ICs could then contribute to the activation of dendritic cell subsets that produce type I IFNs and other proinflammatory cytokines that prime autoreactive B cells to respond to RNPs associated with less stimulatory RNAs. It has been shown that serum levels of anti-Ro, anti-La, anti-Sm, and anti-U1-RNP together correlate with expression of type I IFN inducible genes in SLE patients, and the presence of anti-Sm and anti-U1-RNP autoantibodies correlates with clinical manifestations of disease (41, 42). RNPs and RNP ICs most likely play a critical role in both the initiation of inflammation and propagation of tissue damage in systemic autoimmune diseases, through TLR recognition of their RNA.
TLR detection of endogenous RNA is prevented in part by modifications that silence the capacity of RNA to stimulate TLR7 (Fig. 5 and Ref. 43, 44). Many of the RNA fragments tested in the current study are derived from small RNAs that are normally not extensively modified. One major exception is the tRNAs, which routinely undergo extensive post-transcriptional modifications. The two major instances where unmodified tRNA sequences could become available to the immune system are as pre-tRNAs, and as B2-like SINE RNAs, which are thought to be ancient duplications of tRNA genes. Similar to the Alu RNAs, B2-like SINEs can also be induced by cellular stress (45). Accumulation and availability of unmodified RNAs may therefore serve as a danger signal leading to immune activation. When dead cells are not appropriately cleared by phagocytic cells in an autoimmune prone individual, autoreactive B cells can become activated in response to these signals. Since SLE patients may have defects in the clearance of apoptotic debris and autoantigens can become exposed on the surface of apoptotic blebs, the availability of unmodified RNAs to the immune system may contribute to the activation of autoreactive B cells and the initiation of autoimmune disease.
Autantibodies reactive with RNPs are found in autoimmune diseases other than SLE. Many of the self-complementary stimulatory RNAs analyzed in the current study are associated with such conditions. For example, the Jo-1 autoantigen (histidyl-tRNA synthetase), targeted in patients with polymyositis, has been shown to stimulate the innate immune system (46). TLR3 activation in muscle cells appears to play a role in polymyositis. In the current study we found His-tRNA to be a TLR3 ligand, thereby causally connecting the Jo-1 antigen to disease pathogenesis (Ref. 47 and Fig. 4D). Bacterial tRNAs can elicit IFN-α production through TLR7 in DCs, but we find that B cell activation by tRNAs is mostly TLR7 independent. The difference could be explained by the difference between RNA motifs for optimal B cell proliferation and for optimal IFN-α production from DCs (48, 49) and Fig. 7). SSc is an autoimmune disease that targets the vasculature, and major autoantibody targets in that disease include U1-RNP and nucleolar-associated proteins such as fibrillarin. Poly I:C is a TLR3 ligand and poly(I:C) administration has been used to model of SSc. This report demonstrates that self-complementary mammalian sequence RNAs can act as both TLR7 and TLR3 ligands. U1 RNA has been independently shown to be a TLR7 or a TLR3 ligand, and here we demonstrate that it can act as both (15, 16 and Figs. 3A and 4C). The fibrillarin-associated RNA, U3, was almost completely TLR7 dependent, while another, U8, had a substantial TLR7-independent component (Fig. 3C). Exogenous synthetic TLR3 ligands can exacerbate glomerulonephritis and sialoadenitis in the context of murine models of SLE and Sjögren's syndrome, respectively (50, 51). Since many of the TLR3 reactive RNAs we tested are also TLR7 ligands, it is reasonable to assume that there is synergy between TRIF and MyD88 pathways, and TLR3 could be an important co-factor for TLR7 driven autoimmune responses. Both TLR7 and TLR3 ligands are most likely present in a variety of autoantigen macromolecular structures. Whether there is a role for TLR3 in spontaneous murine models of SLE is unclear. The only model that has been closely examined for TLR3 is the MRL.lpr, where TLR3 deficiency did not ameliorate disease, but this model is dominated by DNA autoantibodies (52). On the other hand, B cells containing a genetic susceptibility locus from the New Zealand Black autoimmune prone strain are hyperresponsive to TLR3 ligands (53). A better understanding of the role played by TLR7 and TLR3 in murine models and in patients will be useful when paired with autoantibody profiles to identify which TLR pathways are most relevant as therapeutic targets.
Prior to this study, the direct activation of B cells by TLR7 ligands had not been examined. We have now identified a uridine-associated stimulatory motif. The current report explains the controversy as to whether TLR7 recognized any uridine-rich RNA or has sequence specificity. While a variety of sequences with as few as one central uridine can fulfill the optimal motif identified here, KNUNDK (UWN/USU) (e.g. GAUCAG), many uridine-dense sequences, including a string of six consecutive uridines, also fulfill the motif. Therefore highly uridine-rich RNAs are likely to make good TLR7 ligands, because they are more likely to contain uridine in the context of the correct motif. Further study is needed to determine whether the sequence of RNA ligands defines the interaction with TLR7 directly or indirectly by allowing for a more optimal secondary structure of the RNA. These data would lead to the prediction that an RNP associated with unmodified ssRNA stimulatory motifs would make a good TLR7 ligand. This is the case in the pristane model of SLE, where Su (also called Ago2), is a frequent autoantibody target. Ago2 binds a wide range of micro-RNAs, including RNAs that have the optimal B cell activating motif UGUGUG, overrepresented in an Ago2-binding screen (54). Pristane-injected TLR7-deficient mice do not make Su-reactive autoantibodies (55). Intriguingly, the optimal motif for murine B cells is most similar to a motif previously shown to induce IL-12, and not to the motif for optimal IFN-α production in human PBMCs (Fig. 6 and Ref. 14). Cell-specific optimal motifs may reflect cell-associated differences in TLR endolysomal trafficking and divergent conditions, such as pH, or TLR-associated proteins may influence RNA/TLR7 interactions.
The sequence specificity of TLR7 has most likely been evolutionarily selected to confer resistance to viral pathogens, with self-RNA recognition as a counter-balancing force. The optimal stimulatory uridine-containing motif (B cell-activating motif) is present in the highly conserved 3′-UTRs of negative sense ssRNA viruses, including Ebola, Rabies, VSV, Measles, Sendai, and RSV, all of which have been previously identified as TLR7 ligands (56). The hypothesis of molecular mimicry proposes that viral infection can promote the onset of autoimmune disease by driving the production antibodies specific for viral epitopes that cross-react with self-antigens. Another possible explanation for the connection between viral infection and autoimmune disease is that viral infections lead to elevated levels of type I IFN, which in turn lowers the activation threshold for TLR7. Elevated levels of type I IFN are associated with more severe SLE. TLR7 detection of endogenous RNAs in B cells is normally avoided by multiple layers of protection. For B cells to respond to TLR7 ligands, TLR7 expression needs to be up-regulated, either in response to a strong stimulus, or through type I IFN priming (34). The data in this report show that RNAs from clinically relevant RNPs are able to activate resting, autoreactive B cells, when delivered to the endolysosomal TLRs by the BCR. The reason that certain RNPs are frequent autoantibody targets is therefore due to the RNA component acting as an adjuvant through either TLR7 or TLR3.
Supplementary Material
Acknowledgments
We thank Patricia Busto for the production of mAbs, and Gregory Viglianti and Kerstin Kiefer for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AR05-256 and AR035230.

This article contains supplemental Table S1 and Figs. S1 and S2.
- SLE
- systemic lupus erythematosus
- SSc
- systemic sclerosis
- TLR
- Toll-like receptor
- polyU
- polyuridylic acid
- IC
- immune complex
- BCR
- B cell receptor.
REFERENCES
- 1. Tan E. M., Cohen A. S., Fries J. F., Masi A. T., McShane D. J., Rothfield N. F., Schaller J. G., Talal N., Winchester R. J. (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 25, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 2. Lerner M. R., Steitz J. A. (1979) Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. U.S.A. 76, 5495–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris M. L., Rosen A. (2003) Autoimmunity in scleroderma: the origin, pathogenetic role, and clinical significance of autoantibodies. Current Opin. Rheumatol. 15, 778–784 [DOI] [PubMed] [Google Scholar]
- 4. Levine S. M., Raben N., Xie D., Askin F. B., Tuder R., Mullins M., Rosen A., Casciola-Rosen L. A. (2007) Novel conformation of histidyl-transfer RNA synthetase in the lung: the target tissue in Jo-1 autoantibody-associated myositis. Arthritis Rheum. 56, 2729–2739 [DOI] [PubMed] [Google Scholar]
- 5. Lau C. M., Broughton C., Tabor A. S., Akira S., Flavell R. A., Mamula M. J., Christensen S. R., Shlomchik M. J., Viglianti G. A., Rifkin I. R., Marshak-Rothstein A. (2005) RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J. Exp. Med. 202, 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrat F. J., Meeker T., Gregorio J., Chan J. H., Uematsu S., Akira S., Chang B., Duramad O., Coffman R. L. (2005) Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 202, 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yasuda K., Richez C., Maciaszek J. W., Agrawal N., Akira S., Marshak-Rothstein A., Rifkin I. R. (2007) Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J. Immunol. 178, 6876–6885 [DOI] [PubMed] [Google Scholar]
- 8. Christensen S. R., Shupe J., Nickerson K., Kashgarian M., Flavell R. A., Shlomchik M. J. (2006) Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity 25, 417–428 [DOI] [PubMed] [Google Scholar]
- 9. Green N. M., Marshak-Rothstein A. (2011) Toll-like receptor driven B cell activation in the induction of systemic autoimmunity. Semin Immunol. 23, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. (2004) Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529 [DOI] [PubMed] [Google Scholar]
- 11. Diebold S. S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. (2004) Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 [DOI] [PubMed] [Google Scholar]
- 12. Diebold S. S., Massacrier C., Akira S., Paturel C., Morel Y., Reis e Sousa C. (2006) Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur. J. Immunol. 36, 3256–3267 [DOI] [PubMed] [Google Scholar]
- 13. Forsbach A., Nemorin J. G., Montino C., Müller C., Samulowitz U., Vicari A. P., Jurk M., Mutwiri G. K., Krieg A. M., Lipford G. B., Vollmer J. (2008) Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 180, 3729–3738 [DOI] [PubMed] [Google Scholar]
- 14. Jurk M., Chikh G., Schulte B., Kritzler A., Richardt-Pargmann D., Lampron C., Luu R., Krieg A. M., Vicari A. P., Vollmer J. (2011) Immunostimulatory potential of silencing RNAs can be mediated by a non-uridine-rich toll-like receptor 7 motif. Nucleic Acid Therap. 21, 201–214 [DOI] [PubMed] [Google Scholar]
- 15. Vollmer J., Tluk S., Schmitz C., Hamm S., Jurk M., Forsbach A., Akira S., Kelly K. M., Reeves W. H., Bauer S., Krieg A. M. (2005) Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J. Exp. Med. 202, 1575–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sadik C. D., Bachmann M., Pfeilschifter J., Mühl H. (2009) Activation of interferon regulatory factor-3 via toll-like receptor 3 and immunomodulatory functions detected in A549 lung epithelial cells exposed to misplaced U1-snRNA. Nucleic Acids Res. 37, 5041–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yasuda K., Richez C., Uccellini M. B., Richards R. J., Bonegio R. G., Akira S., Monestier M., Corley R. B., Viglianti G. A., Marshak-Rothstein A., Rifkin I. R. (2009) Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. J. Immunol. 183, 3109–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marshall-Clarke S., Downes J. E., Haga I. R., Bowie A. G., Borrow P., Pennock J. L., Grencis R. K., Rothwell P. (2007) Polyinosinic acid is a ligand for toll-like receptor 3. J. Biol. Chem. 282, 24759–24766 [DOI] [PubMed] [Google Scholar]
- 19. Marshall-Clarke S., Tasker L., Buchatska O., Downes J., Pennock J., Wharton S., Borrow P., Wiseman D. Z. (2006) Influenza H2 haemagglutinin activates B cells via a MyD88-dependent pathway. Eur. J. Immunol. 36, 95–106 [DOI] [PubMed] [Google Scholar]
- 20. Matsumoto M., Funami K., Tanabe M., Oshiumi H., Shingai M., Seto Y., Yamamoto A., Seya T. (2003) Subcellular localization of Toll-like receptor 3 in human dendritic cells. J. Immunol. 171, 3154–3162 [DOI] [PubMed] [Google Scholar]
- 21. Nishiya T., DeFranco A. L. (2004) Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J. Biol. Chem. 279, 19008–19017 [DOI] [PubMed] [Google Scholar]
- 22. Leadbetter E. A., Rifkin I. R., Hohlbaum A. M., Beaudette B. C., Shlomchik M. J., Marshak-Rothstein A. (2002) Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 416, 603–607 [DOI] [PubMed] [Google Scholar]
- 23. Radic M., Marion T., Monestier M. (2004) Nucleosomes are exposed at the cell surface in apoptosis. J. Immunol. 172, 6692–6700 [DOI] [PubMed] [Google Scholar]
- 24. Casciola-Rosen L. A., Anhalt G., Rosen A. (1994) Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J. Exp. Med. 179, 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hannum L. G., Ni D., Haberman A. M., Weigert M. G., Shlomchik M. J. (1996) A disease-related rheumatoid factor autoantibody is not tolerized in a normal mouse: implications for the origins of autoantibodies in autoimmune disease. J. Exp. Med. 184, 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sweet R. A., Christensen S. R., Harris M. L., Shupe J., Sutherland J. L., Shlomchik M. J. (2010) A new site-directed transgenic rheumatoid factor mouse model demonstrates extrafollicular class switch and plasmablast formation. Autoimmunity 43, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Viglianti G. A., Lau C. M., Hanley T. M., Miko B. A., Shlomchik M. J., Marshak-Rothstein A. (2003) Activation of autoreactive B cells by CpG dsDNA. Immunity 19, 837–847 [DOI] [PubMed] [Google Scholar]
- 28. Uccellini M. B., Busconi L., Green N. M., Busto P., Christensen S. R., Shlomchik M. J., Marshak-Rothstein A., Viglianti G. A. (2008) Autoreactive B cells discriminate CpG-rich and CpG-poor DNA and this response is modulated by IFN-α. J. Immunol. 181, 5875–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uzri D., Gehrke L. (2009) Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 83, 4174–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fenn B. J., Herman T. M. (1990) Direct quantitation of biotin-labeled nucleotide analogs in RNA transcripts. Anal. Biochem. 190, 78–83 [DOI] [PubMed] [Google Scholar]
- 31. Liu W. M., Chu W. M., Choudary P. V., Schmid C. W. (1995) Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 23, 1758–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chu W. M., Liu W. M., Schmid C. W. (1995) RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res. 23, 1750–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eilat D., Fischel R. (1991) Recurrent utilization of genetic elements in V regions of antinucleic acid antibodies from autoimmune mice. J. Immunol. 147, 361–368 [PubMed] [Google Scholar]
- 34. Green N. M., Laws A., Kiefer K., Busconi L., Kim Y. M., Brinkmann M. M., Trail E. H., Yasuda K., Christensen S. R., Shlomchik M. J., Vogel S., Connor J. H., Ploegh H., Eilat D., Rifkin I. R., van Seventer J. M., Marshak-Rothstein A. (2009) Murine B cell response to TLR7 ligands depends on an IFN-β feedback loop. J. Immunol. 183, 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gesteland R. F., Atkins J. F. (1993) The RNA World: the Nature of Modern RNA Suggests a Prebiotic RNA World, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Braun D., Caramalho I., Demengeot J. (2002) IFN-α/β enhances BCR-dependent B cell responses. Int. Immunol. 14, 411–419 [DOI] [PubMed] [Google Scholar]
- 37. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 38. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farina G. A., York M. R., Di Marzio M., Collins C. A., Meller S., Homey B., Rifkin I. R., Marshak-Rothstein A., Radstake T. R., Lafyatis R. (2010) Poly(I:C) drives type I IFN- and TGFβ-mediated inflammation and dermal fibrosis simulating altered gene expression in systemic sclerosis. J. Invest. Dermatol. 130, 2583–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Avalos A. M., Uccellini M. B., Lenert P., Viglianti G. A., Marshak-Rothstein A. (2010) FcγRIIB regulation of BCR/TLR-dependent autoreactive B-cell responses. Eur. J. Immunol. 40, 2692–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hua J., Kirou K., Lee C., Crow M. K. (2006) Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 54, 1906–1916 [DOI] [PubMed] [Google Scholar]
- 42. Villarreal G. M., Drenkard C., Villa A. R., Slor H., Shafrir S., Bakimer R., Shoenfeld Y., Alarcón-Segovia D. (1997) Prevalence of 13 autoantibodies and of the 16/6 and related pathogenic idiotypes in 465 patients with systemic lupus erythematosus and their relationship with disease activity. Lupus 6, 425–435 [DOI] [PubMed] [Google Scholar]
- 43. Karikó K., Buckstein M., Ni H., Weissman D. (2005) Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 [DOI] [PubMed] [Google Scholar]
- 44. Tluk S., Jurk M., Forsbach A., Weeratna R., Samulowitz U., Krieg A. M., Bauer S., Vollmer J. (2009) Sequences derived from self-RNA containing certain natural modifications act as suppressors of RNA-mediated inflammatory immune responses, Int. Immunol. 21, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li T., Spearow J., Rubin C. M., Schmid C. W. (1999) Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene 239, 367–372 [DOI] [PubMed] [Google Scholar]
- 46. Soejima M., Kang E. H., Gu X., Katsumata Y., Clemens P. R., Ascherman D. P. (2011) Role of innate immunity in a murine model of histidyl-transfer RNA synthetase (Jo-1)-mediated myositis. Arthritis Rheum. 63, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tournadre A., Lenief V., Eljaafari A., Miossec P. (2012) Immature muscle precursors are a source of interferon-β in myositis: role of Toll-like receptor 3 activation and contribution to HLA class I up-regulation. Arthritis Rheum. 64, 533–541 [DOI] [PubMed] [Google Scholar]
- 48. Jockel S., Nees G., Sommer R., Zhao Y., Cherkasov D., Hori H., Ehm G., Schnare M., Nain M., Kaufmann A., Bauer S. (2012) The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209, 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gehrig S., Eberle M. E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W. M., Erdmann V. A., Sprinzl M., Bec G., Keith G., Dalpke A. H., Helm M. (2012) Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Patole P. S., Pawar R. D., Lichtnekert J., Lech M., Kulkarni O. P., Ramanjaneyulu A., Segerer S., Anders H. J. (2007) Coactivation of Toll-like receptor-3 and -7 in immune complex glomerulonephritis. J. Autoimmun. 29, 52–59 [DOI] [PubMed] [Google Scholar]
- 51. Nandula S. R., Scindia Y. M., Dey P., Bagavant H., Deshmukh U. S. (2011) Activation of innate immunity accelerates sialoadenitis in a mouse model for Sjogren's syndrome-like disease. Oral Dis. 17, 801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Christensen S. R., Kashgarian M., Alexopoulou L., Flavell R. A., Akira S., Shlomchik M. J. (2005) Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J. Exp. Med. 202, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Loh C., Pau E., Chang N. H., Wither J. E. (2011) An intrinsic B-cell defect supports autoimmunity in New Zealand black chromosome 13 congenic mice. Eur. J. Immunol. 41, 527–536 [DOI] [PubMed] [Google Scholar]
- 54. Leung A. K., Young A. G., Bhutkar A., Zheng G. X., Bosson A. D., Nielsen C. B., Sharp P. A. (2011) Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 18, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee P. Y., Kumagai Y., Li Y., Takeuchi O., Yoshida H., Weinstein J., Kellner E. S., Nacionales D., Barker T., Kelly-Scumpia K., van Rooijen N., Kumar H., Kawai T., Satoh M., Akira S., Reeves W. H. (2008) TLR7-dependent and FcγR-independent production of type I interferon in experimental mouse lupus. J. Exp. Med. 205, 2995–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Forsbach A., Nemorin J. G., Völp K., Samulowitz U., Montino C., Müller C., Tluk S., Hamm S., Bauer S., Lipford G. B., Vollmer J. (2007) Characterization of conserved viral leader RNA sequences that stimulate innate immunity through TLRs. Oligonucleotides 17, 405–417 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.