Background: Kindlin-3 is a novel integrin activator with unclear mechanism.
Results: Calpain cleaves Kindlin-3 at Y-373. Cleavage-resistant mutant Y373N Kindlin-3 promotes cell adhesion but hinders migration by altering the pattern of interaction with β integrins.
Conclusion: Kindlin-3 cleavage by calpain controls dynamics of integrin complexes.
Significance: A novel mechanism regulating kindlin-dependent integrin functions in hematopoietic cells is identified.
Keywords: Calcium Intracellular Release, Calpain, Cell Migration, Integrins, Protein Degradation, Cell Adhesion, Kindlin-3
Abstract
Integrin activation on hematopoietic cells is essential for platelet aggregation, leukocyte adhesion, and transmigration through endothelium and extracellular matrix into inflamed tissues. To migrate through matrix, leukocyte integrin adhesion complexes undergo dynamic changes. Here we show that Kindlin-3, a main activator and binding partner of integrins in hematopoietic cells, can be cleaved by calpain in an activation-dependent manner. This calpain-mediated cleavage occurs in platelets and leukocytes as well as in endothelial cells. We determined the calpain I cleavage site in Kindlin-3 at tyrosine 373 in the N-terminal part of Kindlin-3 pleckstrin homology domain. Expression of the calpain-resistant Y373N mutant of Kindlin-3 promotes stronger cell adhesion to extracellular matrix under flow as well as to activated endothelium. In contrast, Y373N mutation in Kindlin-3 hinders cell migration. Mechanistically, calpain-resistant Y373N mutant of Kindlin-3 exhibited an activation-independent association with β integrin cytoplasm domain. Thus, cleavage of Kindlin-3 by calpain controls the dynamics of integrin-Kindlin-3 interaction and as a result, integrin-dependent adhesion and migration of hematopoietic cells. This represents a novel mechanism regulating reversibility of integrin adhesion complexes in leukocytes, which, in turn, is critical for their successful transmigration through the extracellular matrix.
Introduction
Integrins are a major class of extracellular matrix (ECM)4 receptors responsible for cell adhesion and migration (1). These functions of integrins are regulated by multiple mechanisms including affinity and avidity modulation and cellular localization (2). Integrin activation is achieved by a series of inside-out signaling events culminating in the binding of two key integrin activators, Talin head domain and Kindlin, to integrin cytoplasmic domain. The sequence of interactions between the cytoplasmic domain of the β3 integrin and its activators appears to be well defined and tightly controlled to transform integrin to a high affinity state and ensure strong adhesion to ECM. However, tight adhesion to the substrate might not permit effective cell movement; therefore, the strength and stability of integrin-ECM bonds require dynamic regulation by a distinct mechanism. As a result, there is a constant turnover of integrin-containing adhesion complexes in migrating cells.
Kindlin is the most recently identified integrin activator, and the presence of at least one Kindlin isoform is required to achieve integrin activation in any given cell type. In cells of hematopoietic origin, including platelets and immune cells, Kindlin-3 is the main functional isoform required for integrin-mediated responses. These responses include, but are not limited to, platelet adhesion and aggregation, leukocyte adhesion to ECM and to activated endothelium, and their subsequent transmigration to the sites of inflammation. Lack of Kindlin-3 causes severe bleeding and immune disorder in humans (3, 4). The mechanisms underlying Kindlin-mediated integrin activation are only now being explored. It is clear that the key event in Kindlin-mediated integrin activation is Kindlin-3 binding to the membrane-distal NXXY/F motif in the β cytoplasmic tails, which serves as a prerequisite for affinity modulation of the integrin (2, 5). It remains unclear, however, how Kindlin-3 function can be regulated during dynamic integrin-mediated processes such as cell adhesion and cell migration.
One of the possible mechanisms regulating the stability of integrin-ECM complexes is the activation of calpain cysteine proteases by calcium influx (6). There are 14 known human calpain paralogs, defined by the presence of a protease domain similar to that found in μ-calpain (CAPN1) and m-calpain (CAPN2) (7). Protein cleavage by calpains has been proposed to serve as an important regulator of cytoskeletal remodeling and cell migration (6). A number of focal adhesion (FA)-associated proteins can be cleaved by calpain proteases, often producing opposite outcomes on FA stability and cell migration (8–12). Calpain-mediated cleavage of several proteins, including focal adhesion kinase (FAK) and Talin, leads to the destabilization of FA and increased cell migration (9, 10). The effect of Paxillin cleavage, in contrast, was reported to be inhibitory for cell migration (8).
Because Kindlin-3 is a crucial modulator of integrin affinity in hematopoietic cells (3, 4, 13), we assessed the mechanisms by which its interaction with β3 cytoplasmic domain is controlled and regulated. We show that calpain cleaves Kindlin-3 at Y373 in response to cell stimulation and/or an increase in intracellular calcium in platelets, leukocytes, and endothelial cells. This cleavage regulates the dynamics of the interaction between β integrin subunit and Kindlin-3, thereby the integrin-mediated cell adhesion and migration are controlled and regulated.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Chemicals were purchased from Sigma if not stated otherwise. The rabbit anti-Kindlin-3 (N-12) antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and the rabbit anti-URP2 antibody was purchased from Abcam (Cambridge, MA). Calpain I enzyme was purchased from Biovision, Inc. (Milpitas, CA). The rabbit anti-Kindlin-3 antibodies were described previously (14). The in-house mouse antibody was produced by using full-length bacterial expressed Kindlin-3 as antigen in the Hybridoma core (Cleveland Clinic). The RGD peptide, c(Arg-Gly-Asp-dTyr-Lys), the selective protease-activated receptor 1 (PAR1) agonist with sequence of SFLLRN-NH2, and the selective protease-activated receptor 4 (PAR4) agonist with sequence of AYPGKF-NH2, were purchased from the American Peptide Company (Sunnyvale, CA).
Plasmids
The pGEX-4T-1 vector was purchased from GE Healthcare. The pcDNA3.1-V5-His vector was from Invitrogen. Expression plasmid constructs, including pEGFP-C2-FERMT3, were described previously (14). Long accurate PCR was used for mutagenesis, by using the Pfu enzyme (New England Biolabs) and pEGFP-FERMT3 or pcDNA3-FERMT3-V5 plasmids as template. Primers used are as following: FERMT3_Y373N forward, AAGCTGACCCTGAAGGGCAACCGCCAACACTGGGTGGTGTTCAAG, and FERMT3_Y373N reverse, CTTGAACACCACCCAGTGTTGGCGGTTGCCCTTCAGGGTCAGCTT.
Cell Culture
K562, THP-1, MEG01, HL-60, and HEK293 cells were purchased from ATCC (Manassas, VA). Primary cultures of HUVECs were provided by Dr. Paul DiCorleto (Cleveland Clinic), as we described previously (15). Human T lymphocyte isolation and culture followed the method described (16).
Platelet Isolation
Informed consent was obtained from donors in accordance with the Declaration of Helsinki. Human venous blood was drawn from healthy donors into acid-citrate-dextrose (ACD; 85 mm trisodium citrate, 65 mm citric acid, and 111 mm d-glucose, pH 4.6) solution containing prostaglandin I2, (1 μg/ml of ACD). Gel-filtered platelets were isolated as described previously (17). Platelet counts were assessed by a CellometerTM Auto-M10 from Nexcelom Bioscience (Lawrence, MA).
Liquid Chromatography-Mass Spectrometry (LC/MS)
Endogenous Kindlin-3 was immunoprecipitated from HUVECs or other types of cells which were described previously (14). Polyacrylamide gels were stained with Coomassie Blue for band visualization. Protein bands were cut and digested with trypsin and followed with LC/tandem MS analysis. Peptides were identified via mapping to the Mascot database. The experiments were done in the Proteomics Laboratory (Cleveland Clinic).
Adhesion to Fibronectin under Flow
K562 cells were transfected with GFP-conjugated wild type (WT) Kindlin-3, the Y373N mutant, or vector alone, and stable cell lines were selected with G418. Vena8 Fluoro biochips (Cellix) were coated with 100 μg/ml fibronectin. The cells were pretreated with PMA (200 nm) for 10 min at a concentration of 2 × 106 cells/ml and injected into the channels using a syringe pump and operated by Flow Assay software at a flow rate of 20 dynes/cm2. Nonadherent cells were then removed by growth medium perfusion, and fluorescent images of adhered cells were acquired at ×200 magnification.
Transwell Migration Assay
Three-μm Transwell inserts (Corning) were coated with 100 μg/ml fibronectin with or without 100 μm RGD or α5β1 blocking antibody (clone AB 1950; EMD Millipore). 2 × 105 cells in serum-free medium containing 0.1% BSA were added to the upper chamber. The cells were allowed to migrate toward 20% FCS for 14 h. Following incubation, cells from the upper chamber were removed, and images of the migrated GFP fluorescent cells were acquired at ×200 magnification.
Adhesion Assay of HL-60 Neutrophil-like Cells to HUVEC Monolayers
HUVECs were grown on 6-well plates and when confluent were incubated with 10 ng/ml TNF-α for 4 h. HL-60 cells were incubated in RPMI-160 medium containing the cell-permeable fluorescent indicator 1 μg/ml Cell Trace Calcein red-orange AM (C-34851; Invitrogen) for cells with pEGFP-Y373N mutant FERMT3 overexpression, or 1 μg/ml Calcein AM green (C-3100MP; Invitrogen) for cells with pEGFP-Y373 wild type FERMT3 overexpression, for 15 min at 37 °C and 5% CO2. After such incubation, the HL-60 cells were treated with or without 20 μm ALLM for 15 min at 37 °C and 5% CO2. 4×105 cells of Y373 (green) and Y373N (red) with same type treatment were added to the same well of the TNF-α activated HUVEC monolayer in 6-well plate. After 30 min, cells were washed with RPMI-160 medium three times, each time for 2 min. Micrographs of adhered cells were taken at ×100 magnification.
Immunostaining, Immunoprecipitation, and Immunoblotting
All procedures were performed using standard protocol. Nondenaturing conditions were employed for immunoprecipitation experiments (18).
Calpain Cleavage Site Prediction
Reference Kindlin-3 protein sequence was downloaded from the NCBI Entrez Gene database, and predicted at the calpain web site by using given algorithms (19) or alternative methods (20). Related sequences were downloaded and aligned by using ClustalW (21). These sequences include high quality mammals Kindlin-3 sequences (22) and other paralogs. Briefly, related sequences were downloaded from the Ensembl Release 67, May, 2012. Of 48 FERMT3 orthologs which encode Kindlin-3, 28 nonambiguous sequences from distinct species including fish, reptile, and mammals, were chosen for analysis. These species were Homo sapiens, Pan troglodytes, Gorilla gorilla, Pongo abelii, Pteropus vampyrus, Tursiops truncates, Equus caballus, Sus scrofa, Otolemur garnettii, Myotis lucifugus, Macaca mulatta, Canis familiaris, Loxodonta africana, Ochotona princeps, Cavia porcellus, Mus musculus, Rattus norvegicus, Anolis carolinensis, Latimeria chalumnae, Oreochromis niloticus, Gasterosteus aculeatus, Takifugu rubripes, Danio rerio, and Oryzias latipes. Especially, two zebrafish (D. rerio) FERMT3 sequences were chosen because there are two genes encoding for Kindlin-3. The final aligned sequences, in which the Y373 site was localized in the center, ranging from amino acid residue 365 to 383, were plotted with WebLogo (23).
Image Analysis
image analysis in this study was developed within the ImageJ (National Institutes of Health) platform with related Plugins.
Statistical Analysis
Each experimental point represents the mean ± S.D. for triplicate determinations of a representative experiment performed at least three times. Statistical analyses were made using two-way ANOVA. For all hypotheses, the significance level was 0.05. When multiple comparisons were made, a Bonferroni correction to the significance criterion for each test was made.
RESULTS
Kindlin-3 Is Cleaved by Calpain in Various Cells and in Vitro
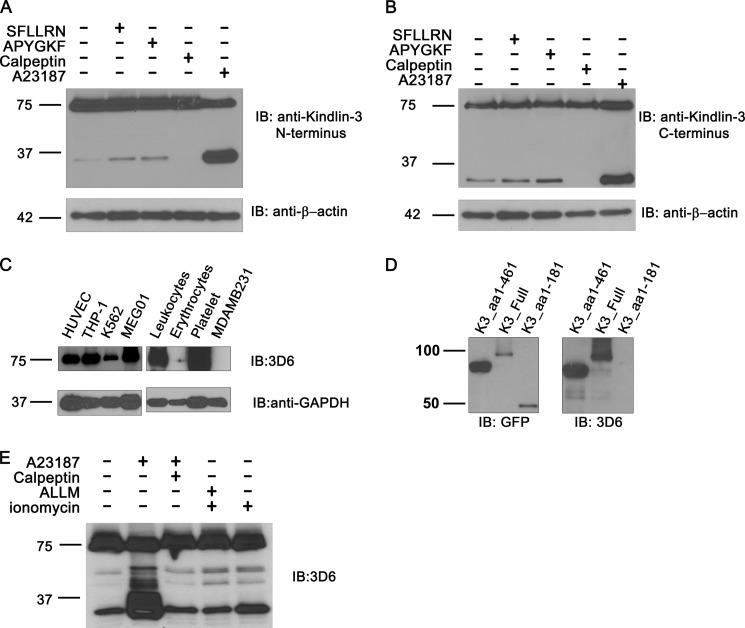
First, we assessed the possibility of Kindlin-3 cleavage in response to stimulation with agonists that induce integrin activation. In platelets, treatment with PAR agonists or calcium ionophore resulted in accumulation of two Kindlin-3 fragments of 37 kDa (Fig. 1A) and 33 kDa (Fig. 1B). The 37-kDa fragment is recognized by antibodies to the N-terminal part of Kindlin-3, whereas the 33-kDa fragment is recognized by an antibody against the C-terminal part. Based on the molecular mass, these two fragments roughly accounted for the full-length protein. The strongest cleavage was observed upon stimulation with calcium ionophore, which is known to serve as a strong inducer of calpain activity (Fig. 1A). Small amounts of Kindlin-3 cleavage products were also detected in unstimulated platelets, however they disappeared after treatment with calpeptin, a known calpain inhibitor (Fig. 1A, B).
FIGURE 1.
Proteolysis of Kindlin-3 in platelets is calpain-dependent. A and B, platelets isolated from healthy donors were activated with PAR-1 ligand SFLLRN (100 μm), PAR-4 ligand APYGKF (20 μm), or calcium ionophore A23187 (1 μm) for 15 min, or calpeptin (10 μm) as indicated, and then lysed and immunoblotted (IB) with two anti-Kindlin 3 antibodies recognizing the N terminus (A) or the C terminus (B). Loading control was β-actin. C, novel 3D6 antibody detects endogenous Kindlin-3 in selected cell lines and primary blood cells, i.e. leukocytes, erythrocytes, and platelets as indicated. The loading control is GAPDH, with distinct level in different cells. D, the 3D6 antibody recognizes an epitope on the C-terminal half of the protein (amino acids 181–461). The whole lysates of HEK293 cells expressing EGFP fusion products of Kindlin-3 deletion mutants as indicated were lysed and blotted with anti-EGFP (left) or 3D6 antibody (right). E, the Kindlin-3 cleavage in platelets induced by calcium ionophore A23187 and ionomycin is calpain/cysteine protease-dependent. The platelets were treated with calcium ionophore A23187 (1 μm) or ionomycin (1 μm) for 15 min as indicated in the presence or absence of calpeptin (10 μm) or ALLM (10 μm), and the lysates were immunoblotted with the 3D6 antibody.
We have developed a new monoclonal antibody to Kindlin-3 (clone 3D6) which very efficiently recognized full-lengthKindlin-3 in various hematopoietic cell lines, primary leukocytes, platelets and HUVECs (Fig. 1C). We mapped the epitope to the residues 181–461 (Fig. 1D). The antibody also recognized the 33-kDa band representing the previously observed C-terminal cleavage product of Kindlin-3 in platelet lysates. The amount of this product increased dramatically after platelet activation with calcium ionophore A23187 and moderately upon activation with ionomycin (Fig. 1E).
When platelets were pretreated with calpeptin prior to the calcium ionophore stimulation, the amount of Kindlin-3 cleavage product was reduced by ∼10-fold, indicating that the cleavage depends on calpain activity (Fig. 1E). Similar inhibitory effect was observed after pretreatment with ALLM, a more general cysteine protease inhibitor (Fig. 1E).
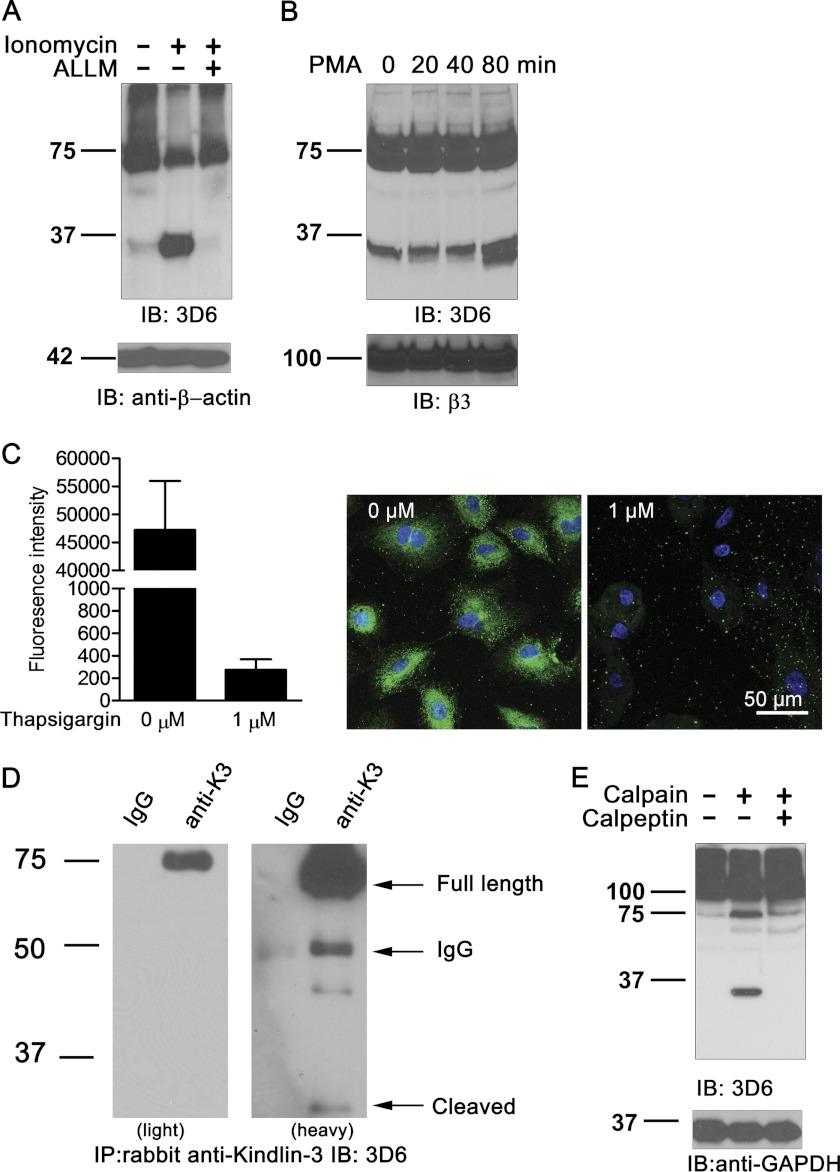
Next we examined Kindlin-3 cleavage in monocytic leukemia cell line THP-1 (Fig. 2A) and in HUVECs (Fig. 2B). Treatment of THP-1 cells with ionomycin promoted Kindlin-3 cleavage as indicated by the presence of 33-kDa band and 65% reduction in the amount of full-length Kindlin-3 (Fig. 2A). The cleavage was blocked when cells were pretreated with cysteine protease inhibitor ALLM (Fig. 2A).
FIGURE 2.
Calpain-dependent cleavage of Kindlin-3 in leukocytes, endothelium, and in purified system. A, THP-1 cells were treated with ionomycin (1 μm) for 15 min in the presence or absence of ALLM (10 μm) or remain untreated, and the lysates were assayed by immunoblotting (IB) with the 3D6 anti-Kindlin-3 antibody. Loading control was β-actin. B, HUVECs were treated with 200 nm PMA for the indicated periods of time (in minutes), and the lysates were assayed by immunoblotting with the 3D6 antibody. Results of immunoblotting for β3 integrin are shown as a control. C, Kindlin-3 degradation in HUVEC is triggered by calcium mobilization. HUVEC were treated with thapsigargin (1 μm for 30 min at 37 °C) as indicated, fixed, and immunostained with the rabbit anti-human Kindlin-3 antibodies. Means of fluorescence intensity ± S.D. (error bars; left) and representative images (right) are shown. D, cleavage of Kindlin-3 in unstimulated K562 cells. Left, light exposure; right, heavy exposure. The C-terminal specific rabbit anti-human antibody was used for immunoprecipitation (IP), and the 3D6 antibody was used for subsequent immunoblotting. E, in vitro cleavage of Kindlin-3 by the recombinant calpain I. Lysates of HEK293 cells expressing the N-terminal EGFP fusion product of Kindlin-3 were treated with recombinant calpain I for 1 h or remained untreated in the presence or absence of calpain inhibitor calpeptin (10 μm) as indicated. Immunoblotting was performed with the 3D6 anti-Kindlin-3 antibody. Loading control was GAPDH.
Similar to hematopoietic cells and cell lines, stimulation of HUVECs with PMA resulted in a time-dependent increase of the 33-kDa cleavage product of Kindlin-3 (Fig. 2B). Although the process of Kindlin-3 cleavage in HUVECs was substantially slower than in platelets, the amount of 33-kDa product increased by ∼5-fold after 80 min of treatment (Fig. 2B). The most dramatic cleavage of Kindlin-3 was achieved by the treatment of endothelial cells with thapsigargin, known to mobilize intracellular calcium (Fig. 2C). In this case, 30 min after the treatment, full-length Kindlin-3 was hardly detectable in HUVECs (Fig. 2C).
In all cell types tested, there was detectable cleavage of Kindlin-3 in unstimulated cells; however, the degree of this cleavage differed depending on the cell type. The cleavage occurred at the lowest level in resting platelets (Fig. 1A) and at the highest level in unstimulated endothelial cells represented by HUVECs (Fig. 2B). In certain cell types expressing Kindlin-3 at the levels substantially lower compared with platelets (Fig. 1C), the cleavage products can only be detected by immunoprecipitation of Kindlin-3 with subsequent anti-Kindlin Western blotting, when longer exposures were used (Fig. 2D for K562 cells).
To demonstrate directly the cleavage of Kindlin-3 by calpain, N-terminal EGFP-Kindlin-3 fusion protein was overexpressed in HEK293 cells, and the cell lysates were incubated with recombinant calpain I in the presence or absence of its inhibitor, calpeptin (Fig. 2D). As evident from Fig. 2D, the 33-kDa C-terminal fragment can only be observed in calpain-treated sample, confirming the identity of the enzyme responsible for the cleavage of endogenous Kindlin-3.
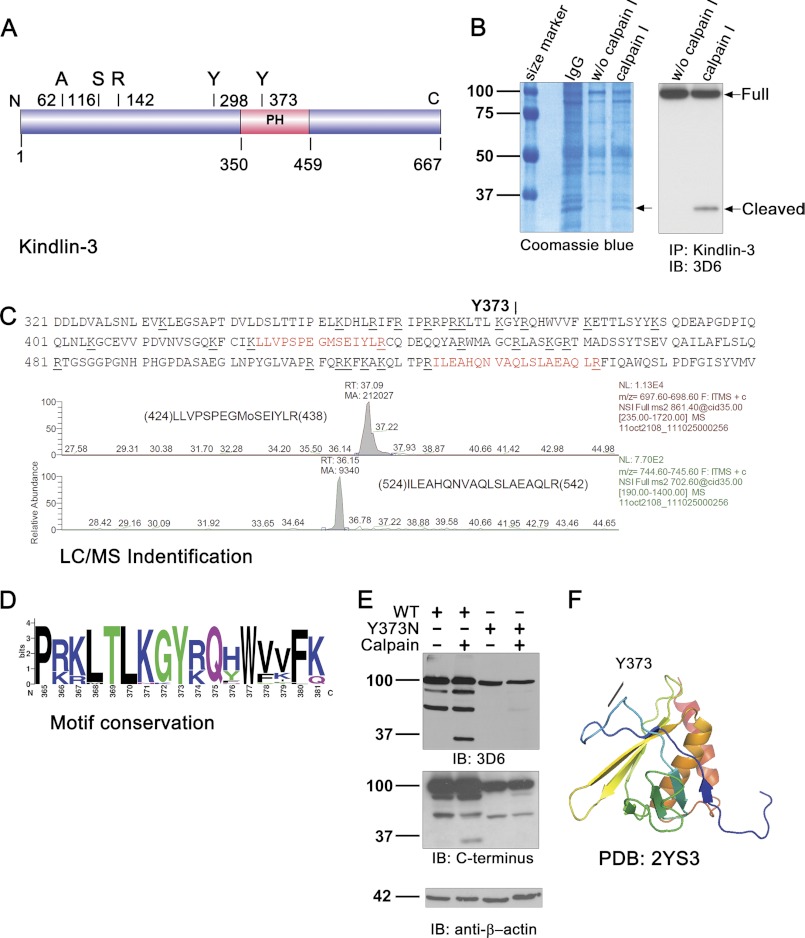
Kindlin-3 Is Cleaved by Calpain at Y373
With several algorithms (as described under “Experimental Procedures”), at least five potential calpain cleavage sites can be predicted within Kindlin-3 protein sequence (Fig. 3A). To identify the actual site in Kindlin-3, immunoprecipitated EGFP-Kindlin-3 fusion was treated with recombinant calpain I in vitro, and the resulting mixture was resolved by SDS-PAGE, followed by Coomassie staining (Fig. 3B, left panel). Further, both the full-length and the cleaved 33-kDa fragment were confirmed by immnoblotting for mouse 3D6 antibody followed by immunoprecipitation with rabbit C-terminal antibody using Kindlin-3-overexpressed HEK293 cell lysates after calpain I digestion. The 33-kDa band, which was calpain I-dependent, was excised and subjected to LC/tandem MS analysis and mapped to the Mascot database. Most of N-terminal peptides identified in this fragment were positioned near the potential cleavage site at tyrosine 373 (Fig. 3C). We analyzed the conservation of Kindlin-3 profile in 28 vertebrates spanning from zebrafish to human. The surrounding sequence TLKGY has been highly conserved (Fig. 3D) with the full conservation of the Y373 site, suggesting that Y373 is an important and essential function constraint amino acid residue site required for vertebrates. Analysis of the structural characteristics of Kindlin-3 PH domain revealed that Y373 is positioned on the solvent-exposed loop of Kindlin-3 and, therefore, might be readily accessible for the enzyme (Fig. 3F). Altogether, the size of Kindlin-3 fragment and the result of mass spectroscopy suggest that calpain cleaves Kindlin-3 at tyrosine 373.
FIGURE 3.
Calpain cleaves Kindlin-3 at tyrosine 373. A, schematic represents predicted calpain cleavage sites. Five predicted cleavage sites were labeled with amino acid residues, and the region of the PH domain is also highlighted in red. B, electrophoresis profile of EGFP-Kindlin-3 fusion protein sample treated or untreated with recombinant calpain I prior to immunoprecipitation (IP) with the C-terminal rabbit antibody is shown. The SDS-PAGE gel was stained with Coomassie Blue (left). In parallel, samples were resolved by SDS-PAGE and immunoblotted (IB) with the 3D6 antibody (right). Arrows indicate cleaved fragments that were analyzed by LC/MS. C, LC/MS data support the putative cleavage site at tyrosine 373. Spectrum of the two peptides identified from the fragment shown in B identifies it as C-terminal to the Y373 putative calpain cleavage site. D, motif conservation analysis of sequence surrounding the Y373 calpain cleavage site in 28 Kindlin-3 amino acid sequences spanning from zebrafish to the human is shown. The Y373 site is highly conserved. E, the Y373N substitution prevents Kindlin-3 cleavage by calpain. The EGFP-tagged Kindlin-3 wild type and Y373N mutant were expressed in HEK293 cell line. The cell lysates were treated with recombinant calpain I as indicated. The samples were separated by SDS-PAGE and probed by immunoblotting with the 3D6 antibody and C terminus-specific rabbit antibody. Loading control was β-actin. F, amino acid Y373 was mapped to solution structure of the PH domain of Kindlin-3 from human (Protein Data Bank code, 2YS3). The Y373 amino acid residue is localized in a solvent-exposed loop which is easily assessable for proteases.
To confirm further the cleavage site at Y373 within Kindlin-3, this tyrosine was mutated to asparagine. WT and Y373N mutant forms of Kindlin-3 were treated with recombinant calpain I. Based on results of Western blotting with two distinct mouse and rabbit antibodies, calpain was able to cleave WT but not Y373N mutant Kindlin-3 to generate 37-kDa cleavage fragment (Fig. 3E).
Calpain-resistant Kindlin-3 Supports Stronger Cell Adhesion but Hinders Cell Migration
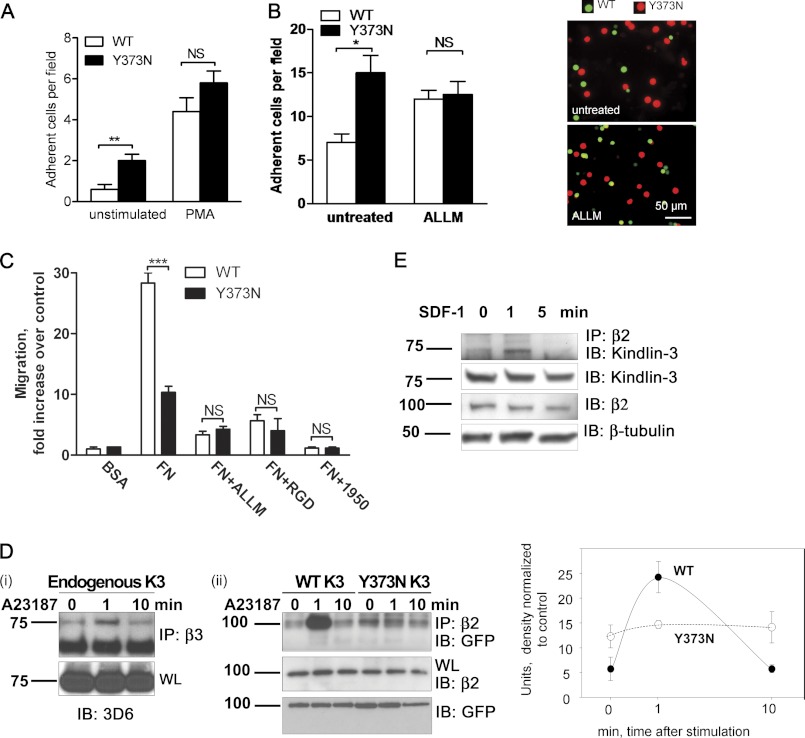
Next, we assessed whether a cleavage-resistant form of Kindlin-3 is able to alter integrin-mediated cell functions. To determine this, WT and Y373N mutant forms of Kindlin-3 were stably expressed in K562 cells (4). Overall levels of Kindlin-3 expression did not differ between the two cell lines (data not shown). The strength of integrin adhesion to extracellular matrix was tested under flow conditions (20 dyn/cm2). K562 cells expressing Y373N mutant Kindlin-3 adhered at least 3-fold stronger under flow conditions compared with WT (Fig. 4A). It appears that Kindlin-3 cleavage occurred in these cells without additional stimulation (Fig. 2D). Treatment with a strong agonist such as PMA further stimulated cell adhesion in both WT- and Y373N mutant-expressing cells (Fig. 4A). Adhesion of Y373N mutant-expressing cells remained higher, although the difference was not significant for PMA-treated cells (Fig. 4A).
FIGURE 4.
Calpain-resistant mutant of Kindlin-3 conferred an increased cell adhesion but limits cell migration through altered integrin binding. A, K562 cells express either EGFP wild type or Y373N mutant. The cells were tested in adhesion under flow in both resting and activated (100 nm PMA) conditions in Vena8 Fluoro biochips (Cellix) coated with fibronectin. 100 nm PMA pretreatment for 10 min where indicated was followed by perfusion through the channels using a syringe pump operated at a flow rate of 20 dynes/cm2. **, p < 0.01, n = 5. Error bars, S.D.; NS, not significant. B, HL-60 neutrophil-like cell adhesion shown on activated endothelial cells monolayer. HL-60 cells stably expressing either the wild type or the Y373N Kindlin-3 mutant were differentially labeled either with green (wild type) or red (mutant) calcein. Cells were mixed in 1:1 ratio and plated on TNFα preactivated HUVEC monolayer, as described under “Experimental Procedures.” The cells were pretreated with 20 μm ALLM for 15 min at 37 °C as indicated. Right, representative images of untreated and ALLM treated cells. Left, quantification from three fields of view, adhesion of cells expressing wild type Kindlin-3 is >1-fold higher. *, p < 0.05. C, Transwell migration of K562 cells shown expressing EGFP fusion of either the wild type (WT) or the Y373N Kindlin-3 mutant in response to serum gradient with a series of treatments or coatings. BSA was used as a control. FN, fibronectin; ALLM was calpain inhibitor; RGD peptide blocked integrins; 1950(AB1950), anti-α5β1 blocking antibody. Data are shown with quantification of the migration assay. ***, p < 0.001, two-way ANOVA test. D, i, interaction of endogenous Kindlin-3 with β3 integrin is time-dependent. THP-1 cells were stimulated with 1 μm A23187 for indicated time (1 or 10 min) or remained unstimulated (0) and lysed. Integrin was immunoprecipitated (IP) with anti-β3 integrin antibody followed by Western blotting (IB) with 3D6 anti-Kindlin 3 antibody (upper panel). Lower panel shows results of Western blotting of whole lysate (WL) with 3D6 anti-Kindlin-3 antibody. ii, interaction of Kindlin-3 wild type or Y373N mutants is expressed as the EGFP fusion proteins with endogenous β2 integrins in HL-60 cells. Cells were stimulated with 1 μm A23187 for indicated periods of time or remained unstimulated. β2 integrin was immunoprecipitated from the whole cell lysates, and the samples were probed with anti-GFP antibody. Right, quantification of results shown in ii. Closed circles represent WT Kindlin-3, and open circles represent Y373N Kindlin-3. Band densities were normalized to corresponding loading controls and are shown as means ± S.D. (error bars) from three experiments. E, Kindlin-3 cleavage and its interaction with β2 integrin in human T lymphocytes stimulated with 10 ng/ml of SDF-1 for indicated periods of time are shown. Left, results of immunoprecipitation of β2 integrin and subsequent Western blotting. From top to bottom, binding of endogenous Kindlin-3 detected with 3D6 antibody; full-length Kindlin-3 in cell lysates; β2 integrin in immunoprecipitates; β-tubulin as a loading control.
The key function of hematopoietic cells in vivo is their adhesion to the activated endothelium which precedes their transmigration. Inhibition of calpain is known to slow down disassembly of adhesion complexes, thereby affecting both adhesion and transmigration of cells through endothelium (8, 12). Accordingly, neutrophil-like HL-60 cells stably expressing either WT or Y373N Kindlin-3 were labeled with green or red calcein, respectively, and added to the HUVECs stimulated with TNFα. As evident from Fig. 4B, adhesion of Y373N cells was at least 2-fold higher compared with WT cells. Treatment with calpain inhibitor resulted in an increased number of attached WT cells. Importantly, WT and Y373N cells adhered similarly when treated with ALLM to inhibit calpain (Fig. 4B). Thus, the mutant Y373N Kindlin-3 has no effect on cell adhesion when calpain is inactive. Altogether, these data suggest that calpain cleavage of Kindlin-3 might negatively regulate integrin-dependent cell adhesion.
Because strong adhesion often antagonizes cell migration, we next measured migratory responses of K562 cells expressing WT and Y373N Kindlin-3 mutant. As shown in Fig. 4C, the number of migrated cells expressing WT Kindlin-3 was at least 3-fold higher compared with Y373N Kindlin-3-expressing cells, whereas no difference was observed on BSA. This migratory response was integrin-dependent as indicated by its inhibition with RGD peptide and anti-integrin blocking antibody (Fig. 4C). Similar to previous reports (12), treatment of cells with calpain inhibitor ALLM diminished migration of cells expressing WT Kindlin-3. Importantly, no difference in migratory responses of WT and Y373N mutant cells were observed when calpain was inactivated by ALLM (Fig. 4C). This suggests that effect of Y373N mutation in Kindlin-3 on cell migration is calpain-dependent. Thus, Kindlin-3 cleavage by calpain limits cell adhesion but promotes migratory responses.
Calpain Cleavage Regulates Kindlin-3 Binding to β Integrin Subunit
Direct interaction between integrin cytoplasmic domain and Kindlin-3 was shown to be a prerequisite for integrin activation. Based on a recent report, Kindlin-3 binds to α4β1 integrin in an activation-dependent and transient fashion, reaching maximum binding at 1 min after activation with agonist (24). Likewise, Kindlin-3 was reported to forma complex with β2 integrin in an activation-dependent fashion (25). Similarly, we show that β3 integrin is able to exhibit a similar binding profile with endogenous Kindlin-3 (Fig. 4D). Whereas limited binding was observed in the unstimulated cells, activation with agonist for 1 min produced a severalfold increase in β3 integrin-Kindlin-3 binding, which returned to the basal level at the 10-min time point (Fig. 4D). Thus, Kindlin-3 is able to interact with integrins in an activation-dependent and transient manner. Next, we compared the ability of WT and the cleavage-resistant form of Kindlin-3 to interact with the β subunit of integrin in an activation-dependent fashion. To discriminate from endogenous Kindlin-3, we expressed both WT and Y373N mutant Kindlins as GFP-containing fusion proteins in HL-60 cells. As shown in Fig. 4Dii, limited binding of WT Kindlin-3 to β2 integrin was observed in unstimulated cells. Activation of cells with calcium ionophore for 1 min resulted in a significant increase of interaction between the WT Kindlin-3 and the β2 integrin. This binding, however, was transient because it returned to the original level 10 min after stimulation, thereby resembling the binding pattern of endogenous Kindlin-3 (Fig. 4Di). In contrast, Y373N mutant Kindlin-3 exhibited binding to β2 integrin in the absence of cell stimulation, and this binding was not activation-dependent because it remained at the similar levels 1 and 10 min after A2387 treatment. Thus, the calpain-resistant form of Kindlin-3 does not follow an activation-dependent pattern of interaction with integrins.
Importantly, we were able to show that stimulation of human T lymphocytes with physiological agonist stromal-derived factor-1 (SDF-1) also induced the complex between β2 integrin and Kindlin-3. Although the interaction was substantially weaker compared with one stimulated by calcium ionophore, it exhibited a similar activation-dependent transient pattern (Fig. 4E). This interaction was observed 1 min after stimulation with SDF-1 and diminished at the later time point (5 min after treatment). Stimulation with SDF-1 also promoted cleavage of Kindlin-3 in a time-dependent manner resulting in accumulation of 33-kDa fragment. In sum, cleavage of Kindlin-3 with calpain seems to regulate activation-dependent interaction between Kindlin-3 and the β subunit of integrin.
DISCUSSION
The main conclusions from this study are the following. Kindlin-3 is cleaved by calpain in agonist-activated cells ranging from platelets to leukocytes and endothelium. The cleavage occurs at Y373. Mutation Y373N renders Kindlin-3 resistant to calpain cleavage in vitro and in vivo. Y373N Kindlin-3 mutant promoted stronger cell adhesion to ECM and to activated endothelium compared with WT Kindlin-3 while impeding cell migration. Mechanistically, Y373N mutation impairs ability of Kindlin-3 to interact with the β subunit of integrin in an activation-dependent and reversible fashion. Thus, cleavage of Kindlin-3 by calpain controls the dynamics of integrin adhesion complexes and as a result, integrin-dependent adhesion and migration of hematopoietic cells. This represents a novel mechanism permitting reversibility of integrin-matrix interactions in leukocytes which is critical for successful transmigration of these cells through ECM.
Calpain has been proposed to serve as a key regulator of integrin-mediated functions due to its ability to cleave β3 integrin itself as well as one of the key integrin-binding partners and activators, Talin (6). It appears that to achieve a balanced turnover of adhesion complexes, both Talin and Kindlin need to be cleaved by calpain. In addition to Talin (10), other integrin-binding partners, Paxillin and FAK, have been previously shown to serve as calpain substrates (8, 9). FAK cleavage destabilizes cell adhesion and increases focal adhesion turnover (26). In contrast, Paxillin cleavage promotes cell adhesion and consequently limits migration (8). Our results implicate calpain cleavage of Kindlin-3 in the regulation of integrin adhesion complex stability as demonstrated by increased adhesion and decreased migration of cells expressing calpain-resistant mutant. This scenario is somewhat similar to Talin and FAK, where calpain-mediated proteolysis promoted cell migration thereby antagonizing the consequences of Paxillin cleavage. Whereas calpain cleavage in leukocytes enables the dynamic regulation and reversibility of integrin adhesion complexes, its function in platelets, where integrin-ECM bonds need to be durable to ensure thrombus stability, might be different. Calpain I accounts for 80% of the protease activity in platelets and its knock-out increases bleeding time due to impaired clot formation and retraction, implicating calpain in postligand binding events (27). At the same time, in the calpain I-null mouse model, enhanced platelet spreading on collagen and fibrinogen-coated surfaces is consistent with compromised platelet aggregation and clot retraction despite normal tail bleeding times (28). In platelets, a substantial fraction of Kindlin-3 seems to be cleaved by calpain. This creates the possibility that after its cleavage, Kindlin-3 might be replaced at the cytoplasmic domain of integrins by other proteins, linking integrins more firmly to the cytoskeleton to ensure stability of platelet aggregates. The cleavage might affect interactions with other proteins which are dependent on the N-terminal part of Kindlin-3 and, as a result, disrupt the optimal sequence of binding events. In endothelial cells which express two Kindlins, Kindlin-3 and -2, the scenario might be more complicated. In a short term, the blockade of Kindlin-3 cleavage by calpain might delay dissociation of Kindlin-3 from activated integrin and as a result, might slow down turnover of integrin adhesion complexes, eventually slowing down dynamic processes such as migration. Moreover, it appears that after initial cleavage by calpain, Kindlin-3 levels in endothelium decrease substantially, possibly due to Kindlin degradation. This might further impair integrin function and cell movement. However, HUVECs express another Kindlin, Kindin-2, which together with Kindlin-3 controls integrin-dependent adhesion and cell migration. How these cleavage events are balanced remains to be assessed in the future. In sum, cleavage with calpain might promote the process of removal of Kindlin-3 from activated integrin to free binding sites for other integrin partners.
The repertoire of potential binding partners for Kindlin-3 seems to be of particular interest, especially considering the promiscuous nature of the PH domain. However, these studies are still in the early stages of development. The main binding partner shown to interact with endogenous Kindlin-3 is the cytoplasmic domain of β subunit of integrin. We demonstrate that the Y373N mutation that renders Kindlin-3 resistant to calpain cleavage affects transient and activation-dependent pattern of interaction between Kindlin-3 and β integrin. Although this seems to be the most plausible mechanism underlying the functional differences between WT and Y373N mutant Kindlin-3, we cannot exclude the possibility that this mutation might affect interactions with other unknown or less characterized partners of Kindlin-3. As such, it has been proposed that the PH domain of Kindlins interacts with ARF1 and/or ARF6 (5).
Because calpain-mediated proteolysis is known to be controlled by multiple mechanisms, including tyrosine phosphorylation of its substrate, it is likely that other molecules, e.g. tyrosine kinases and adaptors, might actively participate in the regulation of the complex between integrin and Kindlin-3. Together, it seems that calpain cleavage of integrin-binding partners including Kindlin-3 allows formation of highly dynamic adhesion complexes where the key players can be rearranged to permit an optimal adhesive and migratory response.
Acknowledgments
We thank Dr. Belinda Willard and Earl Poptic for assistance with the proteomics study; Dr. John W. Peterson for assistance with image analysis; and Drs. Elizabeth Klipfell and Youngwoong Kim and Emelye Crehore for assistance with manuscript proofreading.
This work was supported, in whole or in part, by National Institutes of Health Grants HL073311 (to T. V. B.), HL071625 (to T. V. B.), HL077213 (to E. A. P.), and 2P01HL073311–06 (to T. V. B. and E. A. P.).
- ECM
- extracellular matrix
- EGFP
- enhanced green fluorescent protein
- FAK
- focal adhesion kinase
- HUVEC
- human umbilical vein endothelial cell
- PAR
- protease-activated receptor
- PH
- pleckstrin homology
- PMA
- phorbol myristate acetate
- SDF-1
- stromal cell-derived factor-1.
REFERENCES
- 1. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2. Moser M., Legate K. R., Zent R., Fässler R. (2009) The tail of integrins, talin, and kindlins. Science 324, 895–899 [DOI] [PubMed] [Google Scholar]
- 3. Svensson L., Howarth K., McDowall A., Patzak I., Evans R., Ussar S., Moser M., Metin A., Fried M., Tomlinson I., Hogg N. (2009) Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat. Med. 15, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malinin N. L., Zhang L., Choi J., Ciocea A., Razorenova O., Ma Y. Q., Podrez E. A., Tosi M., Lennon D. P., Caplan A. I., Shurin S. B., Plow E. F., Byzova T. V. (2009) A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat. Med. 15, 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Malinin N. L., Plow E. F., Byzova T. V. (2010) Kindlins in FERM adhesion. Blood 115, 4011–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Franco S. J., Huttenlocher A. (2005) Regulating cell migration: calpains make the cut. J. Cell Sci. 118, 3829–3838 [DOI] [PubMed] [Google Scholar]
- 7. Huang Y., Wang K. K. (2001) The calpain family and human disease. Trends Mol. Med. 7, 355–362 [DOI] [PubMed] [Google Scholar]
- 8. Chan K. T., Bennin D. A., Huttenlocher A. (2010) Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK). J. Biol. Chem. 285, 11418–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cortesio C. L., Boateng L. R., Piazza T. M., Bennin D. A., Huttenlocher A. (2011) Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. J. Biol. Chem. 286, 9998–10006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Du X., Saido T. C., Tsubuki S., Indig F. E., Williams M. J., Ginsberg M. H. (1995) Calpain cleavage of the cytoplasmic domain of the integrin β3 subunit. J. Biol. Chem. 270, 26146–26151 [DOI] [PubMed] [Google Scholar]
- 11. Franco S. J., Rodgers M. A., Perrin B. J., Han J., Bennin D. A., Critchley D. R., Huttenlocher A. (2004) Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 6, 977–983 [DOI] [PubMed] [Google Scholar]
- 12. Svensson L., McDowall A., Giles K. M., Stanley P., Feske S., Hogg N. (2010) Calpain 2 controls turnover of LFA-1 adhesions on migrating T lymphocytes. PLoS One 5, e15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moser M., Bauer M., Schmid S., Ruppert R., Schmidt S., Sixt M., Wang H. V., Sperandio M., Fässler R. (2009) Kindlin-3 is required for β2 integrin-mediated leukocyte adhesion to endothelial cells. Nat. Med. 15, 300–305 [DOI] [PubMed] [Google Scholar]
- 14. Bialkowska K., Ma Y. Q., Bledzka K., Sossey-Alaoui K., Izem L., Zhang X., Malinin N., Qin J., Byzova T., Plow E. F. (2010) The integrin co-activator Kindlin-3 is expressed and functional in a non-hematopoietic cell, the endothelial cell. J. Biol. Chem. 285, 18640–18649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Byzova T. V., Kim W., Midura R. J., Plow E. F. (2000) Activation of integrin αVβ3 regulates cell adhesion and migration to bone sialoprotein. Exp. Cell Res. 254, 299–308 [DOI] [PubMed] [Google Scholar]
- 16. Pellat-Deceunynck C., Jego G., Harousseau J. L., Vié H., Bataille R. (1999) Isolation of human lymphocyte antigens class I-restricted cytotoxic T lymphocytes against autologous myeloma cells. Clin. Cancer Res. 5, 705–709 [PubMed] [Google Scholar]
- 17. Ma Y., Ashraf M. Z., Podrez E. A. (2010) Scavenger receptor BI modulates platelet reactivity and thrombosis in dyslipidemia. Blood 116, 1932–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., protocol 4, pp. 18.60–18.68, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19. DuVerle D. A., Ono Y., Sorimachi H., Mamitsuka H. (2011) Calpain cleavage prediction using multiple kernel learning. PLoS One 6, e19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Z., Ma Q., Cao J., Gao X., Ren J., Xue Y. (2011) GPS-PUP: computational prediction of pupylation sites in prokaryotic proteins. Mol. Biosyst. 7, 2737–2740 [DOI] [PubMed] [Google Scholar]
- 21. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lindblad-Toh K., Garber M., Zuk O., Lin M. F., Parker B. J., Washietl S., Kheradpour P., Ernst J., Jordan G., Mauceli E., Ward L. D., Lowe C. B., Holloway A. K., Clamp M., Gnerre S., Alföldi J., Beal K., Chang J., Clawson H., Cuff J., Di Palma F., Fitzgerald S., Flicek P., Guttman M., Hubisz M. J., Jaffe D. B., Jungreis I., Kent W. J., Kostka D., Lara M., Martins A. L., Massingham T., Moltke I., Raney B. J., Rasmussen M. D., Robinson J., Stark A., Vilella A. J., Wen J., Xie X., Zody M. C., Broad Institute Sequencing Platform and Whole Genome Assembly Team, Baldwin J., Bloom T., Chin C. W., Heiman D., Nicol R., Nusbaum C., Young S., Wilkinson J., Worley K. C., Kovar C. L., Muzny D. M., Gibbs R. A., Cree A., Dihn H. H., Fowler G., Jhangiani S., Joshi V., Lee S., Lewis L. R., Nazareth L. V., Okwuonu G., Santibanez J., Warren W. C., Mardis E. R., Weinstock G. M., Wilson R. K., Delehaunty K., Dooling D., Fronik C., Fulton L., Fulton B., Graves T., Minx P., Sodergren E., Birney E., Margulies E. H., Herrero J., Green E. D., Haussler D., Siepel A., Goldman N., Pollard K. S., Pedersen J. S., Lander E. S., Kellis M. (2011) A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hyduk S. J., Rullo J., Cano A. P., Xiao H., Chen M., Moser M., Cybulsky M. I. (2011) Talin-1 and kindlin-3 regulate α4β1 integrin-mediated adhesion stabilization, but not G protein-coupled receptor-induced affinity up-regulation. J. Immunol. 187, 4360–4368 [DOI] [PubMed] [Google Scholar]
- 25. Kliche S., Worbs T., Wang X., Degen J., Patzak I., Meineke B., Togni M., Moser M., Reinhold A., Kiefer F., Freund C., Förster R., Schraven B. (2012) CCR7-mediated LFA-1 functions in T cells are regulated by 2 independent ADAP/SKAP55 modules. Blood 119, 777–785 [DOI] [PubMed] [Google Scholar]
- 26. Sieg D. J., Hauck C. R., Schlaepfer D. D. (1999) Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J. Cell Sci. 112, 2677–2691 [DOI] [PubMed] [Google Scholar]
- 27. Azam M., Andrabi S. S., Sahr K. E., Kamath L., Kuliopulos A., Chishti A. H. (2001) Disruption of the mouse μ-calpain gene reveals an essential role in platelet function. Mol. Cell. Biol. 21, 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuchay S. M., Wieschhaus A. J., Marinkovic M., Herman I. M., Chishti A. H. (2012) Targeted gene inactivation reveals a functional role of calpain-1 in platelet spreading. J. Thromb. Haemost. 10, 1120–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]