Background: Proton leak through UCP2 is modulated by glutathionylation, and UCP2 modulates GSIS.
Results: Glutathionylation of UCP2 amplifies GSIS from β cells. Matrix ROS activates UCP2-desensitizing GSIS.
Conclusion: Reversible glutathionylation of UCP2 aids in regulating GSIS.
Significance: Findings enhance our understanding of the role of redox circuits in the modulation of GSIS.
Keywords: Glutathionylation, Insulin Secretion, Reactive Oxygen Species (ROS), Redox Signaling, Uncoupling Proteins
Abstract
The role of reactive oxygen species (ROS) in glucose-stimulated insulin release remains controversial because ROS have been shown to both amplify and impede insulin release. In regard to preventing insulin release, ROS activates uncoupling protein-2 (UCP2), a mitochondrial inner membrane protein that negatively regulates glucose-stimulated insulin secretion (GSIS) by uncoupling oxidative phosphorylation. With our recent discovery that the UCP2-mediated proton leak is modulated by reversible glutathionylation, a process responsive to small changes in ROS levels, we resolved to determine whether glutathionylation is required for UCP2 regulation of GSIS. Using Min6 cells and pancreatic islets, we demonstrate that induction of glutathionylation not only deactivates UCP2-mediated proton leak but also enhances GSIS. Conversely, an increase in mitochondrial matrix ROS was found to deglutathionylate and activate UCP2 leak and impede GSIS. Glucose metabolism also decreased the total amount of cellular glutathionylated proteins and increased the cellular glutathione redox ratio (GSH/GSSG). Intriguingly, the provision of extracellular ROS (H2O2, 10 μm) amplified GSIS and also activated UCP2. Collectively, our findings indicate that the glutathionylation status of UCP2 contributes to the regulation of GSIS, and different cellular sites and inducers of ROS can have opposing effects on GSIS, perhaps explaining some of the controversy surrounding the role of ROS in GSIS.
Introduction
Blood glucose homeostasis is highly dependent on pancreatic β cell mitochondrial energetics (1). When blood glucose levels rise, β cells import and oxidize glucose to support ATP synthesis. Glucose oxidation is coupled to the formation of an electrochemical gradient across the mitochondrial inner membrane, referred to as protonmotive force (PMF),4 which is then used to drive ATP production. The increase in cytoplasmic ATP/ADP deactivates KATP channels prompting Ca2+ uptake and subsequent release of insulin (2). The importance of coupled oxidative phosphorylation in insulin release can be illustrated by the use of chemical uncouplers that depolarize the PMF impeding ATP synthesis and glucose-stimulated insulin release (3). Hence, effective glucose-stimulated insulin secretion (GSIS) is reliant on efficient coupling of glucose catabolism to ATP synthesis. (4, 5).
For over 15 years, researchers have been trying to assign physiological functions to mitochondrial uncoupling protein 2 (UCP2), which was discovered and named based on its sequence homology to UCP1, a protein in brown adipose tissue that, when active, dissipates the PMF preventing ATP production. However, unlike UCP1, inducible proton leak through UCP2 does not play a thermogenic function but rather a cell signaling role (6). UCP2 activity and expression are associated with negative regulation of insulin secretion. Knock-out of UCP2 expression or inhibition of UCP2 function with genipin increases GSIS from pancreatic islets, observations that have been replicated in INS-1E cells (7–9). The putative signaling function of UCP2 is thought to stem from its capacity to control mitochondrial ROS emission (reviewed in Ref. 10). For instance, using a cell-specific UCP2 deletion mouse model, Robson-Doucette et al. (9) demonstrated that β cell UCP2 has little effect on mitochondrial ATP production, but it significantly contributes to the control of mitochondrial ROS production, which in turn regulates GSIS. In support of this, various reports have shown that exposing β cells (either insulinoma cells or in pancreatic islets) to low amounts of superoxide (O2˙̄, generated artificially with menadione) or H2O2 stimulates insulin release (reviewed in Refs. 9, 11–14). Furthermore, Leloup et al. (15) showed that the induction of ROS emission from the electron transport chain stimulates insulin release to the same degree as glucose-mediated ATP production. Glucose metabolism has also been shown to increase intracellular ROS levels in rat islets, Min6 (mouse β cell line), and INS-1 832/13 cells (rat β cell line), conditions associated with GSIS (9, 12).
In addition to the regulation of GSIS-amplifying ROS signals, ROS are also important regulators of UCP2 function itself (1). In a series of publications, Brand and co-workers (16, 17) showed that proton leak through the uncoupling proteins is acutely controlled by ROS. As there is a non-Ohmic relationship between PMF and mitochondrial ROS production, even minor increases in uncoupling cause significant decreases in mitochondrial ROS emission when PMF is high (18, 19). Recently, Affourtit et al. (8) showed that proton leak through UCP2 decreases GSIS by diminishing ROS production. UCP2 is well known to regulate mitochondrial ROS production in many tissues and cell types (reviewed in Ref. 20). However, as discussed above, ROS also activate GSIS. It is therefore paradoxical that mitochondrial ROS amplify GSIS and also activate UCP2, a negative regulator GSIS. One potential explanation is that the cellular location of ROS genesis is important in controlling GSIS.
Reversible glutathionylation involves the formation of a disulfide linkage between a protein thiol and glutathione. This post-translational modification is required to modulate protein function in response to fluctuations in cell redox state (21). Recently, our group showed that reversible glutathionylation is required to modulate proton leak through UCP2 and UCP3 but not UCP1 (6, 22). Small nontoxic increases in ROS deglutathionylate UCP2- and UCP3-activating proton leak, thereby diminishing mitochondrial ROS emission through a negative feedback loop. Conversely, glutathionylation deactivates leak through these proteins. We have established that reversible glutathionylation of UCP2 and UCP3 is required to acutely control mitochondrial ROS production (23).
Using Min6 cells as a model system, we set out to determine whether reversible glutathionylation of UCP2 plays a signaling role during GSIS. Pharmacological induction of glutathionylation with diamide (100 μm), a powerful glutathionylation catalyst, inhibited proton leak through UCP2 and increased GSIS. These observations were confirmed in pancreatic islets. Intriguingly, the treatment of cells with H2O2 (10 μm) had a dual effect amplifying GSIS yet activating proton leak through UCP2. Using paraquat, a superoxide-generating bipyridine that accumulates in mitochondria, we found that matrix ROS actually inhibits GSIS by activating the UCP2 leak. Hence, our results show that glutathionylation of UCP2 deactivates proton leak and amplifies GSIS. We also demonstrate that the impact of ROS on GSIS depends on the ROS location. The implications of ROS signaling in the matrix versus the cytoplasm are also discussed.
MATERIALS AND METHODS
Cell Culture and Treatment
Min6 insulinoma cells were routinely cultured in T75-cm2 flasks on plastic and maintained in high glucose (25 mm) Dulbecco's modified Eagle's medium (DMEM; 4 mm glutamine, 1 mm pyruvate) containing 10% fetal bovine serum (FBS), 2% antibiotics/antimycotics, and 50 μm β-mercaptoethanol. Medium was changed every 2 days, and cells were split every 4 days. For cell splitting, medium was aspirated, and the cell monolayer was treated with full strength trypsin (Invitrogen) for 1 min at 37 °C. Trypsin was then deactivated with 3 volumes of medium, and cells were pelleted by centrifugation. The pellet was then resuspended in medium and split into new cultures. For experiments, cells were diluted to ∼80,000 cells/ml in clear or black 96-well plates or in Seahorse tissue culture plates 2 days prior to experimentation. For all paraquat assays, cells were treated with paraquat (0–500 μm) for 18 h prior to experimentation. On the day of the experiments, medium was aspirated; cells were washed once with Krebs-Ringer buffer (KRB: 128 mm NaCl, 4.8 mm KCl, 1.2 mm KH2PO4, 1.2 mm MgSO4, 2.5 mm CaCl2, 10 mm HEPES, 0.1% (w/v) BSA, pH 7.4, 5 mm NaHCO3 added fresh on the day of experiments) and incubated for 1 h at 37 °C in KRB containing 1 mm glucose. Cells were then incubated for 1 h at 37 °C in KRB containing 1 or 25 mm glucose and supplemented with either diamide (0–1000 μm), hydrogen peroxide (H2O2; 0–100 μm or 5 mm as a control for ROS and cytotoxicity assays), or biotinylated glutathione ethyl ester (BioGEE, 1 mm). Following the incubation, the final assay medium was removed and stored at 4 °C for insulin release determinations.
UCP2 Knockdown
Following 1 day of growth, cells were treated with Polybrene (2 μg/ml, Santa Cruz Biotechnology) with either UCP2 shRNA (shUCP2; Santa Cruz Biotechnology) or scrambled (control, shCtl; Santa Cruz Biotechnology) shRNA lentiviral particles (5000 infectious viral particles/ml, Santa Cruz Biotechnology) for 48 h. Cultures were re-supplemented with fresh medium devoid of lentiviral particles or Polybrene and incubated for an additional 24 h in a medium, including puromycin (1 μg/ml, Santa Cruz Biotechnology). Transduced cells were then lifted and treated accordingly for experimentation. The cell transduction protocol was performed according to the manufacturer's instructions and was optimized to minimize cell death and ensure maximum UCP2 knockdown.
Animals and Islet Isolation
The loxUCP2 mice were a gift from Dr. Bradford Lowell (24). β cell-specific UCP2 deletion was accomplished by crossing loxUCP2 mice with rat insulin promoter-driven Cre recombinase (RIPCre) mice (The Jackson Laboratory, Bar Harbor, ME). Mice were genotyped using standard PCR of ear notch DNA. RIPCre mice were chosen as controls for experimentation because RIPCre and floxed mice (mice that express the floxed Ucp2 gene without Cre) gave similar results. All mice (10–13 weeks old) were age- and sex-matched and maintained on a 129J-C57BL/6-mixed background. A mixture of male and female mice were used for experiments. All animal experiments were approved by the University of Toronto Animal Care Committee, and animals were handled according to the guidelines of the Canadian Council of Animal Care.
Islets were isolated from anesthetized mice by perfusing the pancreas via the pancreatic duct with a solution of collagenase type-V (0.8 mg/ml) (Sigma) in RPMI 1640 medium (11.1 mm glucose) supplemented with 1% v/v penicillin/streptomycin (Invitrogen) and 1% v/v l-glutamine (Invitrogen). The pancreas was digested for 17 min at 37 °C before the addition of RPMI 1640 medium (11.1 mm glucose) supplemented with 10% v/v FBS, 1% v/v penicillin/streptomycin (Invitrogen), and 1% v/v l-glutamine (Invitrogen) to stop the digestion.
Insulin Release
Assay medium was collected from Min6 cell cultures that were incubated in the various conditions described above. Insulin levels were tested using the Cisbio HTRF insulin assay kit (CisBio Bioassays, Bedford, MA), as described by the manufacturer's instructions. Results were normalized to total cellular protein levels, which were determined using the Bradford assay.
Islet GSIS was tested as described previously (9) with a few modifications. Islets were washed with ice-cold KRB and preincubated at 37 °C in 2.8 mmol/liter glucose (±10 μm diamide) for 1 h. Tubes were put in ice water, and the 2.8 mmol/liter glucose buffer was replaced with ice-cold 2.8 or 16.7 mmol/liter glucose KRB (±10 μm diamide) and incubated for 30 min at 37 °C. All insulin values were corrected to DNA content. Data were represented as the fold change in insulin release relative to islets treated with 0 μm diamide.
In Situ Monitoring of Cellular Bioenergetics
Characteristics of Min6 cellular bioenergetics were determined using the Seahorse extracellular flux analyzer (Seahorse Bioscience, North Billerica, MA). Cells were washed once with warmed PBS and then incubated for 30 min at 37 °C and ambient CO2 in HCO3-free DMEM containing 1 mm glucose, 4 mm glutamine, 1 mm pyruvate (pH 7.4). Diamide (100 μm) or H2O2 (10 μm) was injected immediately followed by a 5-min incubation and the measurement of resting oxygen consumption rate (OCR). Cells were then treated sequentially with glucose (25 mm final concentration), oligomycin (0.2 μg/ml), and antimycin A (1 μm) to test the impact of diamide or H2O2 on glucose-stimulated, proton leak-dependent, and extra-mitochondrial respiration, respectively. Mitochondrial respiration was calculated by subtracting extra-mitochondrial respiration values. Corrected OCR values were normalized to total protein/well using the Bradford assay. Changes in respiration rates relative to resting respiration values were determined by expressing the percent change in OCR relative to resting OCR.
ROS Measurements
Total cellular and matrix levels of ROS were measured using dichlorofluorescein diacetate (DCFHDA; 20 μm) and MitoSOX (20 μm), respectively. Reagent concentrations and durations of exposure were optimized prior to assays. Cells were loaded with DCFHDA or MitoSOX during glucose starvation (1 h of incubation in KRB containing 1 mm glucose), washed twice with KRB, and then incubated for 1 h in 1 or 25 mm glucose. Following the incubation, cells were washed twice with PBS. DCFHDA and MitoSOX fluorescence was measured at excitation/emission wavelengths of 480/530 nm and 514/585 nm. Cells not exposed to DCFHDA or MitoSOX were used to test background fluorescence. Results were normalized to protein content/well using Bradford assay and background fluorescence.
TMRE Fluorescence
Mitochondrial membrane potential was measured in situ using TMRE under nonquench mode conditions. Following cell starvation, cells were incubated for 1 h at 37 °C in KRB containing 5 mm NaHCO3, 1 or 25 mm glucose, and diamide or H2O2 and 10 nm TMRE. Incubation medium was then removed; cells were washed twice with PBS, and fluorescence was measured at excitation/emission wavelengths of 548/573 nm. Cells not incubated in TMRE were used to assess background fluorescence. Results were normalized to protein content/well using the Bradford assay and background fluorescence.
Cytotoxicity Assays
The effect of diamide on Min6 cell viability was tested using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Calbiochem) and propidium iodide (PI; Sigma) assays (25). Min6 cells were starved of glucose and then incubated in KRB containing 5 mm NaHCO3, 25 mm glucose, diamide, and MTT assay reagent. Cells exposed to 5 mm H2O2 served as the control. MTT assays were conducted as described in the manufacturer's instructions. For PI assays, following the 1 h of incubation of cells with diamide and 25 mm glucose, cells were washed once with PBS and then incubated for 10 min in PI diluted in PBS (10 μg/ml). Cells were then washed twice, and plates were read at excitation/emission wavelengths of 530/615 nm. PI results were normalized to protein content as determined by the Bradford assay.
Glutathione Pool and ATP/ADP
For GSH/GSSG, GSH bound to protein, and ATP/ADP were measured using HPLC as described previously (20, 25). For GSH and GSSG levels, cells were trypsinized, washed once with PBS, and then diluted to 0.5 mg/ml in ice-cold 1% (w/v) trifluoroacetic acid (TFA; 1% w/v), meta-phosphoric acid solution and incubated on ice for 10 min. Precipitate was removed by centrifugation for 10 min at 12,000 × g at 4 °C. The supernatant was collected and stored at −80 °C. The protein pellet was treated with 1 n KOH for 20 min at room temperature; the base was neutralized with perchloric acid, and the precipitate was removed by centrifugation. The pellet was then discarded, and the supernatant was collected for analysis. On the day of the experiments, samples were injected into an Agilent 1100 Series HPLC equipped with a Pursuit C18 column (150 × 4.6 mm, 5 μm, Agilent Technologies). For proper separation of GSH and GSSG, a flow rate of 1 ml/min was used with a mobile phase consisting of 0.1% (w/v) TFA and HPLC-grade methanol in a 90:10 ratio. GSH and GSSG were detected using an Agilent UV-visible variable wavelength detector operating at 215 nm. Retention times were confirmed by injecting GSH and GSSG standards. GSH and GSSG were quantified using Agilent Chemstation software.
For ATP and ADP level determinations, cells were collected, washed once with PBS, and then diluted to 0.5 mg/ml in a 0.5% (v/v) solution of perchloric acid and incubated on ice for 10 min. Following removal of the protein precipitate by centrifugation, the sample was injected into a C18 hydrophilic reverse phase column operating at a flow rate of 0.7 ml/min (Synergy Hydro-RP; 4 μm; 250 × 4.6 mm, Phenomenex). The mobile phase was 20 mm KH2PO4, pH 2.9. ATP and ADP were detected at 254 nm for nucleotides. Quantification was performed by injecting varying amounts of ATP and ADP.
Western Blotting
Min6 cells were trypsinized, washed once with PBS, and lysed on ice in RIPA buffer. Protein was diluted in Laemmli buffer and then electrophoresed on SDS-polyacrylamide denaturing gels. Upon completion, gel slabs were removed and equilibrated in transfer buffer, and then proteins were electroblotted onto nitrocellulose membranes. Membranes were blocked for 1 h with 5% (w/v) nonfat skim milk and then probed overnight with anti-UCP2 antibodies (N-19, Santa Cruz Biotechnology), glutaredoxin-1 (Grx1, Abcam), or glutaredoxin-2 (Grx2, Abcam). Probing for succinate dehydrogenase (Santa Cruz Biotechnology) served as the loading control. For overnight incubation, antibodies were diluted in Tris-buffered saline, 2% (v/v) Tween 20. Bands were visualized by incubating membranes for 1 h with anti-goat or anti-mouse HRP-conjugated secondary antibodies followed by a 5-min incubation in chemiluminescent substrate (ECL kit, Thermo Scientific).
UCP2 Glutathionylation Status
The glutathionylation status of UCP2 was determined using BioGEE (Invitrogen) and immunoblotting, as described previously (17). Min6 cells were pre-starved with KRB solution containing 1 mm glucose and then treated with 1 mm glucose or 25 mm glucose + 1 mm BioGEE with or without 10 μm H2O2 for 1 h at 37 °C. Cells were washed once with PBS and lysed in RIPA buffer containing protease inhibitors and 50 mm N-ethylmaleimide (to deactivate any unbound BioGEE and block unmodified thiol residues). Protein lysate was diluted to 2 mg/ml and incubated overnight under constant agitation in streptavidin beads at 4 °C to elute proteins modified with BioGEE. A more deglutathionylated protein would bind more BioGEE. BioGEE-modified proteins were eluted by centrifugation (150 × g for 5 min at 4 °C); the supernatant was placed on ice, and the pellet was then treated with 4 m urea (in PBS, pH 7.4) to detach the BioGEEylated proteins from the streptavidin. The solution was centrifuged (150 × g for 5 min at 4 °C); the pellet discarded, and the resulting supernatant was placed on ice. Protein samples were then subjected to electrophoresis and then immunoblotted for the presence of UCP2 under reducing conditions, as described above. The amount of protein used in the initial elution (2 mg/ml) was loaded as an input control.
Immunoblot Detection of the Glutathionylated Proteome
Min6 cells treated with BioGEE were lysed and electrophoresed under nonreducing conditions. The amount of protein modified by BioGEE was detected by immunoblot using avidin-HRP antibody as described previously (17). Briefly, blocked membranes were washed twice with TBS-T and then incubated for 1 h in the dark in avidin-HRP antibody diluted in blocking solution (1:500, Abcam). Bands were visualized with chemiluminescent reagent (ECL kit, Thermo Scientific).
Statistical Analysis
Student's t tests were performed with Microsoft Excel software. One-way ANOVA with Fisher's protected least significant difference post hoc test or ANOVA repeated measures with Student's t tests were performed with StatView software. All values were expressed as means ± S.E.
RESULTS
Low Doses of H2O2 Amplify GSIS
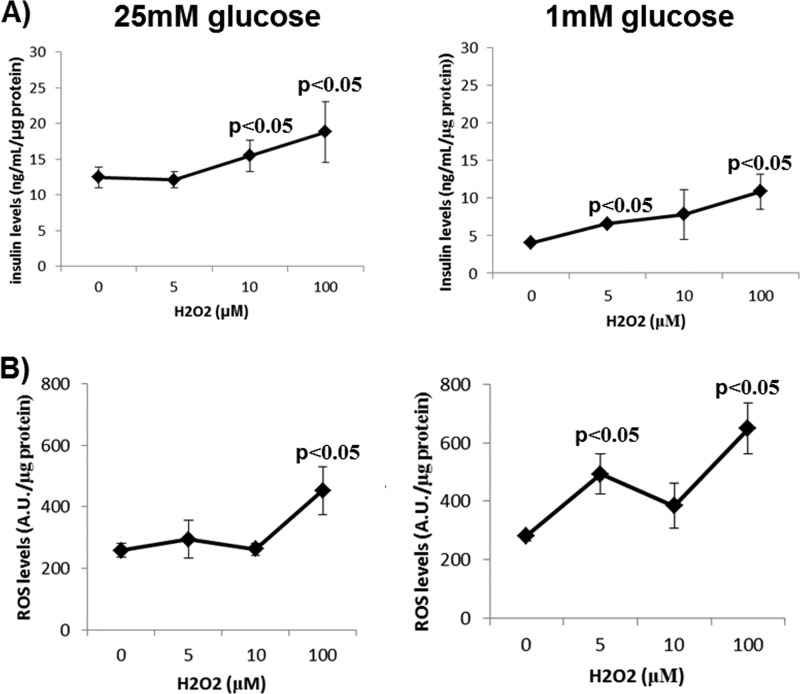
The role of ROS in enhancing GSIS is paradoxical in the sense that ROS can increase insulin release but also activate UCP2, which is known to negatively regulate GSIS. To confirm that low doses of ROS enhance GSIS, Min6 cells, a mouse insulinoma cell line that expresses UCP2 (supplemental Fig. 1), were treated with H2O2 (0–100 μm) under glucose-starved (1 mm) or -replete (25 mm) conditions. Although we did not observe an increase in GSIS when Min6 cells were exposed to 5 μm H2O2, providing cells with 10 μm did enhance GSIS (Fig. 1A), which is consistent with previous observations (12). Exposure of glucose-energized cells to 100 μm H2O2 increased GSIS further. However, cells exposed to 100 μm H2O2 also had high cellular ROS levels, indicating that the latter increase in extracellular insulin may be due to oxidative stress and cell damage (Fig. 1B). We also determined whether H2O2 could stimulate insulin release under low glucose (1 mm) conditions. H2O2 dose-dependently increased insulin release under these conditions (Fig. 1A), and this correlated with increases in cellular ROS levels (at H2O2 levels as low as 5 μm) (Fig. 1B). Hence, it would appear that, at least in Min6 cells, ROS alone may not be sufficient to stimulate insulin release in the absence of glucose but is required to amplify insulin release under glucose-replete conditions.
FIGURE 1.
Effect of different doses of H2O2 on insulin release and ROS levels in Min6 cells exposed to 25 or 1 mm glucose. A, insulin release. Cells were washed once with KRB, starved for 1 h, and then exposed to KRB containing either 25 or 1 mm glucose with H2O2 for 1 h. Following the incubation, the assay medium was collected and tested for insulin content. Data were normalized to total cell protein content/well. B, ROS levels. Cells were washed once with KRB, starved for 1 h in the presence of DCFHDA, and then exposed to KRB containing 25 or 1 mm glucose with H2O2 for 1 h. The assay medium was then removed, and DCFHDA fluorescence was measured. Data were normalized to total cell protein content/well and background fluorescence. n = 4, mean ± S.E., one-way ANOVA with Fisher's protected least significant difference post hoc test. A.U., absorption units.
Induction of Glutathionylation Amplifies GSIS
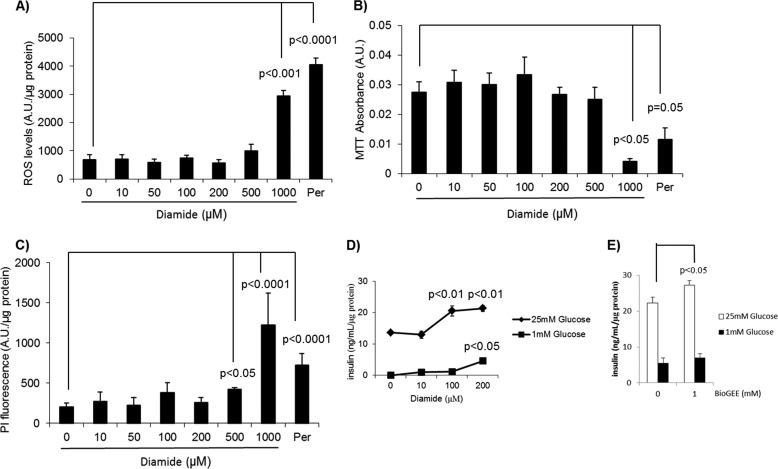
It is clear that redox circuits, through ROS signaling, modulate GSIS (26, 27). Conjugation of GSH to exposed thiol residues, a covalent modification referred to as glutathionylation, also plays a part in redox signaling (28). Indeed, changes in the glutathionylated proteome coincide with alterations in ROS levels that control many mitochondrial and cellular functions, including aerobic metabolism, signaling, and cell division (29, 30). We used diamide, a commonly employed glutathionylation catalyst, to investigate the effect of glutathionylation on insulinoma cell physiology and GSIS (6, 31). First, we tested the toxicity of diamide in Min6 cells energized for 1 h with 25 mm glucose because diamide can induce mitochondrial permeability transition and cell death (32). A concentration of 1000 μm diamide was required to induce significant increases in ROS and decreases in cell redox potential (Fig. 2, A and B). PI fluorescence, an index of cell death, increased upon exposure of Min6 cells to ≥500 μm diamide (Fig. 2C). Acute treatment of Min6 cells with 10 μm did not increase GSIS (Fig. 2D). However, exposure of Min6 cells to 100 and 200 μm diamide amplified GSIS (Fig. 2D). These concentrations did not induce cell death (Fig. 2, A–C). In glucose-starved cells, 100 μm diamide did not stimulate insulin release. However, a significant increase in insulin release was observed when glucose-starved cells were exposed to 200 μm diamide (Fig. 2D). The lower concentration of diamide required at high glucose concentrations to amplify GSIS suggests that in glucose-replete conditions UCP2 is in a more deglutathionylated state. We confirmed the GSIS-amplifying effect of glutathionylation using BioGEE, a cell-permeable glutathione molecule tagged to a biotin group. Exposure of glucose-replete Min6 cells to 1 mm BioGEE amplified GSIS (Fig. 2E).
FIGURE 2.
Impact of diamide on Min6 cell viability and physiology. Min6 cells were washed with KRB, starved for 1 h with KRB containing 1 mm glucose, and then incubated for 1 h in KRB containing 20 mm glucose and varying amounts of diamide (0–1000 mm). Exposure to H2O2 (Per; 5 mm) served as the control. A, ROS levels determined with 20 μm DCFHDA. For ROS measurements, cells were loaded with DCFHDA prior to exposure to 25 mm glucose and diamide. ROS levels were then measured fluorometrically and normalized to protein and background fluorescence. B, assessment of the reductive cellular environment. Cells were exposed simultaneously to MTT and diamide/glucose and then tested for the amount of reduced tetrazolium. Values were normalized to background cellular absorption. C, measurement of cell death using propidium iodide (PI). Following exposure to diamide/glucose, cells were treated for 10 min with 1 μg/ml PI diluted in PBS. Amount of cell death was then tested fluorometrically. Values were normalized to background fluorescence and amount of protein. n = 6, mean ± S.E., one-way ANOVA with Fisher's protected least significant difference post hoc test. D, impact of diamide on insulin release. For insulin release determinations, Min6 cells were washed once with KRB, starved for 1 h, and then treated for 1 h with KRB containing 25 mm glucose and different amounts of diamide. The supernatant was then collected and tested for insulin content. n = 4, mean ± S.D. one-way ANOVA with Fisher's protected least significant difference post hoc test. E, effect of BioGEE on insulin release. Min6 cells were washed once with KRB, starved for 1 h, and then treated for 1 h with KRB containing 25 mm glucose and BioGEE (1 mm). n = 4, mean ± S.E., Student's t test. A.U., absorption units.
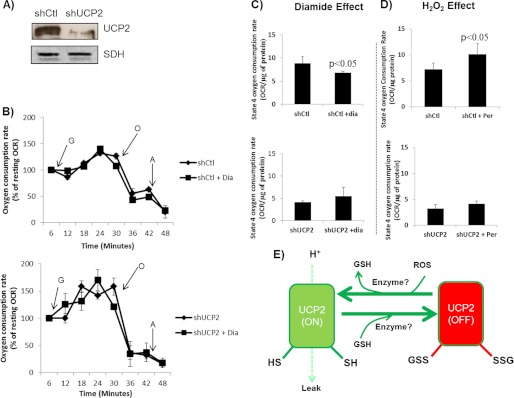
The glutathionylation status of UCP2 and UCP3 dictates the degree of proton leak through either protein, which in turn modulates ROS emission from mitochondria (22). Because mitochondrial bioenergetics play a central role in GSIS and UCP2 is known to negatively regulate this process, we tested the effect of diamide and H2O2 on proton leak-dependent respiration in intact Min6 cells transduced with either scrambled shRNA (control; shCtl) or shRNA directed against UCP2 (UCP2 knock down; shUCP2). Transduction of Min6 cells with shUCP2 lentiviral particles decreased UCP2 protein levels by ∼81% (Fig. 3A). Cells were incubated in diamide (100 μm) or H2O2 (10 μm) for 5 min, and then resting, glucose-stimulated state 4 (proton leak-dependent) and extramitochondrial respirations were tested. A summary of the Seahorse XF24 trace generated during the experiment is provided in Fig. 3B. Diamide inhibited and H2O2 activated UCP2 proton leak. In the shUCP2 Min6 cells, the contribution of proton leak-dependent respiration was reduced by ∼30%. This observation is consistent with previous reports that have shown that UCP2 makes a significant contribution to total respiration in insulinoma cells (20). As shown in Fig. 3C, diamide treatment decreased proton leak-dependent respiration in a UCP2-dependent fashion, suggesting glutathionylation inhibits proton leak through UCP2. However, acute treatment with H2O2 (10 μm) had the opposite effect. H2O2 treatment increased proton leak in cells transduced with control shRNA, suggesting that in the presence of small amounts of H2O2 respiration is less coupled (Fig. 3D). We also used TMRE to measure the effect of diamide on mitochondrial membrane potential. TMRE measurements revealed that diamide treatment increased mitochondrial membrane potential in Min6 cells in a UCP2-dependent fashion (supplemental Fig. 1). Conversely, H2O2 (10 μm) decreased mitochondrial membrane potential in a UCP2-dependent fashion. Overall, these results indicate that glutathionylation and ROS work in tandem to deactivate and activate UCP2-mediated uncoupling, respectively, in Min6 cells (Fig. 3E).
FIGURE 3.
ROS and glutathionylation activate and deactivate, respectively, proton leak in a UCP2-dependent manner in Min6 cells. Min6 cells were transduced with short hairpin control (shCtl) or UCP2 (shUCP2) lentiviral particles and then tested for the effect of H2O2 (0 or 10 μm) and diamide (100 μm) on bioenergetics using the Seahorse XF24 analyzer. A, immunoblot detection of UCP2 in Min6 cells transduced with either shCtl or shUCP2. Succinate dehydrogenase (SDH) served as the loading control. B, summary of the method for determining the impact of diamide on Min6 cell bioenergetics using the XF24 analyzer. Following the injection of diamide, resting respiration was tested; this was then followed by the injection of glucose (G, 25 mm), oligomycin (O, 0.13 μg/ml), and antimycin A (A, 2 μm). All values were expressed as a percentage of resting respiration. n = 4, mean ± S.E. C, effect of diamide on absolute oligomycin-induced state 4 respiration rates in shCtl and shUCP2 Min6 cells. n = 4, mean ± S.E., Student's t test. D, effect of H2O2 on absolute oligomycin-induced state 4 respiration rates in shCtl and shUCP2 Min6 cells. Determinations were performed as described in B. n = 4, mean ± S.E., Student's t test. E, summary of the effect of reversible glutathionylation on proton leak through UCP2.
Assessment of GSIS in UCP2 Knockdown Cells Exposed to Diamide or H2O2
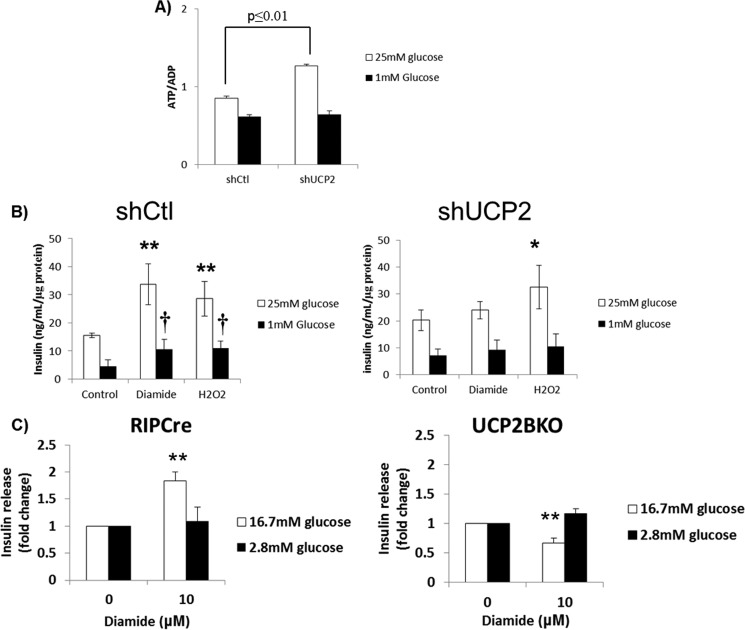
The evidence provided above is contradictory because 10 μm H2O2 amplified GSIS but also activated leak through UCP2. We next tested the impact of UCP2 knockdown on GSIS and UCP2-dependent ROS-mediated amplification of GSIS. Loss of UCP2 led to a small but significant increase in ATP/ADP, consistent with the notion that UCP2 negatively regulates GSIS by changing mitochondrial coupling efficiency (Fig. 4A). Diamide treatment amplified GSIS in Min6 cells transduced with control shRNA (Fig. 4B). This increase was blunted in cells knocked down for UCP2. In comparison, H2O2 treatment (10 μm) increased GSIS in both the shCtl and shUCP2 cells indicating that the amplification of GSIS with H2O2 is independent of UCP2 (Fig. 4B). In glucose-starved cells, diamide and H2O2 also promoted insulin release but only in the cells transduced with control shRNA (Fig. 4B). The observation that diamide augments GSIS by preventing leak through UCP2 prompted us to also test if diamide could have similar effects in mouse islets. Treatment of islets from RIPCre mice with 10 μm diamide led to a significant increase in glucose-stimulated insulin release (Fig. 4C). These effects were blunted in islets from mice with β cell-specific UCP2 deletion (UCP2BKO) exposed to 16.7 mm glucose (Fig. 4C). In fact, treatment of islets from UCP2BKO mice with 10 μm diamide led to a significant decrease in glucose-stimulated insulin release. No changes in insulin secretion were observed in RIPCre or UCP2BKO islets exposed to 2.8 mm glucose (Fig. 4C). These results reveal that diamide is also able to amplify GSIS from mouse pancreatic islets, and this amplification is UCP2-specific. Overall, our results indicate that the pharmacological induction of UCP2 glutathionylation amplifies GSIS in both insulinoma cells and mouse islets.
FIGURE 4.
UCP2 is required for the diamide-mediated regulation of insulin release. A, UCP2 knockdown increases the ATP/ADP ratio in Min6 cells. Min6 cells transduced with either short hairpin control (shCtl) or UCP2 (shUCP2) lentiviral particles were starved and then incubated for 1 h in KRB containing 25 or 1 mm glucose. ATP and ADP were detected as described under “Materials and Methods.” n = 3, mean ± S.E., Student's t test. B, UCP2 knockdown abolishes the diamide-mediated increase in insulin release. Min6 cells transduced with either shCtl or shUCP2 were starved and then incubated for 1 h in KRB containing 25 or 1 mm glucose with either diamide (100 μm) or H2O2 (10 μm). Media were then collected and tested for insulin release. Data were normalized to total cellular protein/well. n = 4, mean ± S.E., one-way ANOVA with Fisher's protected least significant difference post hoc test. * and ** denotes p ≤ 0.05 and 0.01, respectively, when compared with the 25 mm glucose control. † denotes p ≤ 0.05 when compared with the 1 mm glucose control. C, diamide modulates glucose-stimulated insulin release in a UCP2-dependent manner. Islets from control (RIPCre) and pancreas-specific UCP2 knock-out (UCP2BKO) mice were treated with diamide (0 and 10 μm) and then tested for insulin release as described under “Materials and Methods.” Insulin release was measured following a 1-h exposure to high glucose (16.7 mm) or low glucose (2.8 mm) conditions. n = 4, mean ± S.E., Student's t test. ** denotes p ≤ 0.01 when compared with 0 μm diamide control.
Impact of Glucose Energization on Glutathione Pools, Total Cellular ROS, and Mitochondrial ROS
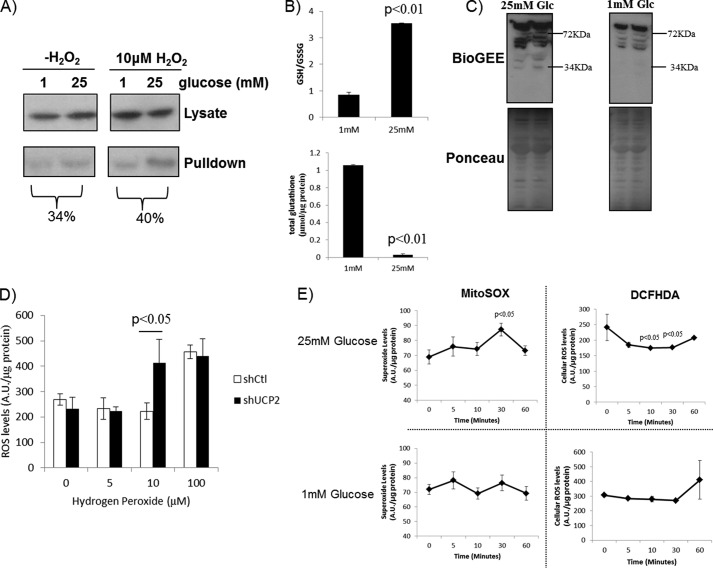
The observation that glutathionylation impedes UCP2-mediated proton leak in Min6 cells prompted us to test the glutathionylation status of UCP2 in cells exposed to low or high glucose conditions for 60 min. UCP2 was less glutathionylated in cells exposed to 25 mm glucose signifying UCP2 activation (Fig. 5A). Treatment with 10 μm H2O2 increased UCP2 pulldown from cells treated with 25 mm glucose. There appeared to be an increase in UCP2 enrichment in cells treated with 1 mm glucose and 10 μm H2O2, but the difference was small. These results illustrate that UCP2 is less glutathionylated following exposure to 25 mm glucose, which can be enhanced by a brief H2O2 treatment. We also measured the expression levels of glutaredoxin (Grx) 1 and Grx2. Grx1 is expressed in the cytosol and intermembrane space of mitochondria, whereas Grx2 is found in the matrix. Both enzymes display thiol transferase activity and are, to date, the most well characterized enzymes involved in (de)glutathionylation (21). Small increases in Grx1 and Grx2 protein levels were observed in Min6 cells energized with 25 mm glucose (supplemental Fig. 1). The observation that glucose energization deglutathionylated UCP2 prompted us to measure cellular GSH/GSSG and the total amount of GSH associated with the proteome following a 1-h exposure to either 1 or 25 mm glucose. Incubation of cells in 25 mm glucose increased the GSH/GSSG ratio substantially in comparison with cells incubated in 1 mm glucose (Fig. 5B). It is important to point out that despite this increase, the GSH/GSSG ratio was extremely oxidized (∼1 and ∼3.5 in cells treated with 1 and 25 mm glucose, respectively). This is in contrast to the values generated by Pi et al. (12), who found that the GSH/GSSG was actually quite reduced (∼50–70) in rat INS-1 (832/13) cells. In comparison, mouse islets exposed to 20 mm glucose have a GSH/GSSG of ∼10 (33). To our knowledge GSH/GSSG in Min6 cells has never been reported. Exposure to 25 mm glucose, conversely, substantially decreased the total amount of GSH associated with the proteome (Fig. 5B). This change in the glutathionylated proteome was confirmed by detecting the amount of BioGEE bound to the proteome using avidin-HRP and immunoblot (Fig. 5C). Immunoblotting revealed several proteins at various molecular weights were modified by BioGEE, including a faint band at ∼34 kDa (which corresponds to the approximate molecular mass of UCP2) (Fig. 5C). This would indicate that upon glucose exposure, cell proteins become deglutathionylated. The oxidized nature of the GSH pool prompted us to determine whether UCP2 was required to protect Min6 cells from an oxidative challenge. Knockdown of UCP2 did not increase total cell ROS levels in cells exposed to high glucose conditions (Fig. 5D). However, treatment with 10 μm H2O2 increased cellular ROS levels only in cells knocked down for UCP2 (Fig. 5D). We next decided to test the following: 1) total cellular ROS levels and 2) mitochondrial ROS levels in cells treated with 1 or 25 mm glucose over a 1-h period. Interestingly, exposure to 25 mm glucose led to a gradual increase in matrix ROS (Fig. 5E, top left panel). A small but significant increase in mitochondrial matrix ROS (measured with MitoSOX) was observed after a 30-min incubation, which then decreased at 60 min (Fig. 5E). This change in matrix ROS from 30 to 60 min is most likely associated to some extent with the activation of UCP2 proton leak (UCP2 was more deglutathionylated after a 60-min incubation in 25 mm glucose). No changes in mitochondrial matrix ROS were observed in cells treated with 1 mm glucose. We also measured cellular ROS changes using DCFHDA over the 60-min period (note that the MitoSOX and DCFHDA measurements were performed as separate experiments). Intriguingly, exposure to 25 mm glucose over the 60-min period significantly decreased total cellular ROS (Fig. 5E). In contrast, ROS levels did not change in cells exposed to 1 mm glucose.
FIGURE 5.
Effect of glucose energization on glutathione redox in Min6 cells. A, Min6 cells were starved for 1 h and then incubated in 25 or 1 mm glucose in the absence or presence of 10 μm H2O2 +1 mm BioGEE. Cells were then lysed in RIPA containing 25 mm N-ethylmaleimide, and BioGEE-tagged proteins were enriched using streptavidin beads. UCP2 was detected by immunoblot. B, HPLC analysis of GSH/GSSG ratio and total GSH associated with proteins in Min6 cells exposed to 25 or 1 mm glucose. Cells were starved, incubated in KRB containing 25 or 1 mm glucose, and then lysed with 0.1% (v/v) trifluoroacetic acid/methanol solution (90:10). Supernatant was then injected into the HPLC. For GSH associated with protein (total glutathione associated with proteome), protein was treated with KOH and then the resulting supernatant was injected into the HPLC. GSH and GSSG retention times were confirmed and quantified by injecting standard solutions. n = 3, mean ± S.E., Student's t test. C, immunodetection of BioGEE-modified proteins in Min6 cell extract. Cells were treated with BioGEE and lysed, and the amount of BioGEE-tagged protein was detected with avidin-HRP. D, UCP2 knockdown increases cellular ROS levels following H2O2 challenge. Min6 cells transduced with either short hairpin control (shCtl) or UCP2 (shUCP2) lentiviral particles were loaded with DCFHDA (20 μm) during cell starvation, washed with KRB, and then incubated for 1 h in 25 or 1 mm glucose with H2O2 (0–100 μm). Cells were then measured for DCFHDA fluorescence. Data were normalized to total cell protein/well and background fluorescence. n = 4, mean ± S.E., Student's t test. E, time course analysis of ROS production in mitochondria (MitoSOX) or total cell (DCFHDA) following exposure to 25 or 1 mm glucose. Amount of ROS was then detected following various incubation times. Data were normalized to total protein levels. n = 4, mean ± S.E., one-way ANOVA with Fisher's protected least significant difference post hoc test. A.U., absorption units.
Matrix ROS Activates UCP2 That Impedes GSIS
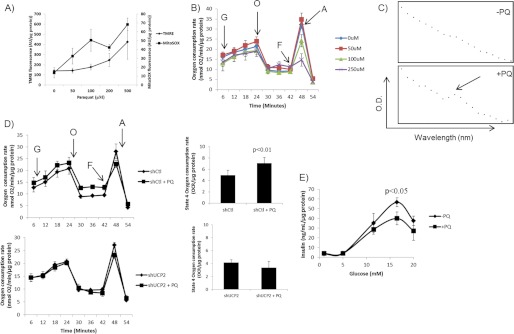
We then determined whether matrix ROS, generated using paraquat, were able to activate leaks in a UCP2-dependent manner and whether the activation of UCP2 leaks impeded GSIS. Paraquat is a superoxide-generating bipyridine that accumulates in mitochondria in a membrane potential-dependent manner (34) and can be a useful tool to artificially increase mitochondrial matrix superoxide. We first performed a number of dose response assays to identify the appropriate paraquat concentration that can be used to increase superoxide without disrupting mitochondrial function. We had tried a 1-h pretreatment, but no changes in mitochondrial ROS were observed (data not shown). Therefore, cells were preloaded with paraquat for 18 h prior to each assay. MitoSOX and TMRE measurements (performed separately) revealed that 50 μm paraquat induced a sharp increase in matrix O2˙̄ but did not disrupt mitochondrial membrane potential (Fig. 6A). Higher concentrations of paraquat led to further increases in matrix ROS but also progressively altered TMRE fluorescence (Fig. 6A). We next performed Seahorse XF24 analyses on the impact of paraquat on mitochondrial energetics. As shown in Fig. 6B, paraquat (up to 250 μm) did not have any effect on resting or glucose-stimulated respiration. Paraquat had a dose-dependent effect on maximal respiration (stimulated by FCCP). Exposure to 100 and 250 μm paraquat induced a steady decline in the FCCP response. In fact, 250 μm paraquat abolished the FCCP effect entirely. However, 50 μm paraquat had no effect on the FCCP response (Fig. 6B). Treatment with 50 μm paraquat did however increase respiration associated with proton leak. Hence, 50 μm paraquat is the optimal concentration to simulate ROS production in the matrix of Min6 mitochondria without detectable disruption of metabolism. In addition, we confirmed that paraquat was accumulating in mitochondria. Min6 mitochondria were collected, solubilized with 1% maltoside, treated with dithionite, and then tested for the presence of paraquat using a UV-visible scan as described by Cochemé and Murphy (34) (Fig. 6C). A peak between 500 and 650 nm was observed only in digested mitochondria from paraquat-treated cells. Because 50 μm paraquat increased the leak in Min6 cells, we decided to test if this increase was UCP2-dependent. As shown in Fig. 6D, paraquat increased proton leak only in cells transduced with scrambled shRNA. In contrast, knockdown of UCP2 abrogated the proton leak-activating effect of paraquat. Because paraquat activated the UCP2 leak, we tested if paraquat was able to impede GSIS. Paraquat treatment led to a small but significant decrease in GSIS when cells were treated with 16.5 mm glucose (Fig. 6E).
FIGURE 6.
Matrix ROS activates leak through UCP2 that impedes GSIS. To simulate production of superoxide in the matrix without inhibiting the respiratory complexes, Min6 cells were incubated for 18 h with paraquat (PQ). A, PQ-mediated increases in matrix ROS are dose-dependent. Cells were pre-loaded with either MitoSOX (20 μm) or TMRE (10 nm) and energized for 1 h with 25 mm glucose. Fluorescent signals were then detected and compared to determine whether PQ generates superoxide in the matrix and whether PQ uptake has an effect on mitochondrial membrane potential. Data were normalized to total protein per well. B, impact of PQ on Min6 mitochondrial bioenergetics. Following an assessment of resting respiration, respiration rates in cells treated to PQ (0–250 μm) were tested following exposure to glucose (G; 25 mm), oligomycin (O; 0.13 μg/ml), FCCP (F; 2 μm), and antimycin A (A; 2 μm). Data were normalized to total protein per well. n = 4, mean ± S.E. C, PQ accumulates in mitochondria. Following exposure of Min6 cells to 0 or 50 μm PQ, mitochondria were isolated, lysed, and treated with dithionite. PQ was detected by UV-visible scan from 500 to 700 nm. D, PQ activates UCP2 proton leak. Min6 cells transduced with either short hairpin control (shCtl) or UCP2 (shUCP2) lentiviral particles and treated with or without PQ (50 μm) were sequentially treated with glucose (G; 25 mm), oligomycin (O; 0.13 μg/ml), FCCP (F; 2 μm), and antimycin A (A; 2 μm). Impact of PQ on UCP2-dependent proton leak is summarized to the right of the bioenergetic data. All data were normalized to total protein per well. n = 4, mean ± S.D. Student's t test. E, PQ impedes GSIS. Min6 cells were starved and then treated with different amounts of glucose (1–20 mm) for 1 h. Insulin levels in the incubation medium were normalized to total protein amounts/well. n = 4, mean ± S.E. Student's t test.
DISCUSSION
By dissipating the PMF and decreasing mitochondrial coupling efficiency and ROS emission, UCP2 can diminish GSIS. These observations have made UCP2 a potential target for treatment of type 2 diabetes mellitus (1, 7). However, the mechanisms governing UCP2 function have remained elusive. Our group has recently shown that reversible glutathionylation controls leak through UCP2 and UCP3 (6, 22). In this study, we show that reversible glutathionylation of UCP2 plays an important signaling role in GSIS. Pharmacological induction of glutathionylation with diamide augmented GSIS from insulinoma cells and islets in a UCP2-dependent manner. Conversely, ROS had the opposite effect, but we found that the site of ROS production can have different effects on GSIS. Indeed, matrix ROS, produced by paraquat, activated leak through UCP2. The ROS-mediated activation of leak through UCP2 is associated with the deglutathionylation of the protein. Indeed, in this study we provided evidence that glucose metabolism, which increases matrix ROS, deglutathionylates UCP2. These effects were amplified by co-treatment with H2O2. We have previously established that ROS is able to deglutathionylate UCP3 (6). ROS-mediated UCP3 deglutathionylation does not proceed spontaneously and requires the presence of a cellular environment. Concentrations of H2O2 in the millimolar range cannot deglutathionylate UCP3 in vitro, but H2O2 in micromolar amounts deglutathionylates UCP3 in intact cells (6). Although we still have not identified the enzyme that mediates this process, it is clear that ROS deglutathionylates UCP2 (and UCP3) with the aid of an as yet to be identified enzyme. Activation of leak subsequently decreased the GSIS signal. Intriguingly, extracellular ROS amplified GSIS indicating different sources of ROS have different cellular effects, e.g. matrix ROS blunts GSIS, whereas cytosolic/extracellular ROS amplifies GSIS. Hence, insulin-secreting cells most likely rely on a number of redox circuits to modulate GSIS.
Reversible glutathionylation is emerging as an important post-translational modification required to modulate protein function in response to changes in redox status (30). Regulation of protein function by reversible glutathionylation is especially relevant to mitochondrial energetics because mitochondria are a significant source of ROS and have an environment that promotes reversible glutathionylation reactions (35). Despite the importance of mitochondrial energetics in insulin release from pancreatic β cells, the role of glutathionylation in modulating insulin release has never been tested. In this study, we observed that multiple cellular proteins can be modified by glutathionylation, and the extent of this modification depends on the amount of extracellular glucose. To this end, UCP2 was less glutathionylated when Min6 cells were exposed to high glucose. This was amplified by co-incubation in H2O2. UCP2 is known to partake in the negative regulation of GSIS. However, the observation that the glutathionylation state of other proteins changes in response to glucose metabolism would suggest that redox environment plays an important role in modulating insulin release in general. Redox biology was recently suggested to play a key role in insulin signaling and release (36). Incubation in high glucose substantially decreased the glutathionylated proteome of cells, which was matched by an increase in GSH/GSSG. The increase in GSH/GSSG is most likely due to the liberation of GSH from surrounding proteins and increased NADPH production from the hexose monophosphate pathway. Despite this increase in GSH/GSSG, the glutathione pool was highly oxidized especially when cells were starved. β cells have relatively low anti-oxidative enzyme expression and therefore may exhaust their GSH pools quite rapidly (37). In fact, insulinoma cells need to be cultured in the presence of powerful reducing agents, like β-mercaptoethanol, to maintain a reduced GSH pool and by extension their β cell-like properties (38–40). In this study, UCP2 became deglutathionylated following a 60-min glucose treatment. Using intact primary thymocytes, we previously showed that pharmacological induction of glutathionylation with diamide deactivated proton leak through UCP2 (6). Similar observations were made herein with Min6 cells. This effect was dependent on the presence of UCP2 (e.g. the leak was not responsive to diamide in UCP2 knockdown cells). Conversely, the deactivation of leak with diamide enhanced the GSIS signal, an effect that was UCP2-dependent. This result indicates that control of leak through UCP2 by reversible glutathionylation is required to control GSIS.
ROS are well recognized “amplifiers” of GSIS. Pi et al. (12) were the first to show that doses of H2O2 as low as 5 μm amplified GSIS. Providing β cells with redox cycling molecules like menadione also increases GSIS (13). Other studies have used electron transport chain inhibitors (e.g. antimycin A or rotenone) to artificially enhance cell ROS levels and stimulate GSIS (15). In our opinion, however, use of electron transport chain inhibitors should be avoided when investigating mitochondrial ROS signaling (especially in β cells) because these drugs also compromise ATP production and induce cell death quite rapidly through uncontrolled ROS production and impaired ATP genesis (41–43). Here, we demonstrate that low doses of H2O2 amplify GSIS but can also induce proton leak through UCP2. The induction of proton leak with ROS is consistent with work published previously by our group, as well as by others (6, 16, 22, 44). The dual effect of ROS on GSIS prompted us to test if matrix-generated ROS had a different effect on GSIS than ROS provided in the extracellular medium. To simulate matrix ROS production, we used paraquat, which generates superoxide by cycling between an oxidized and semi-reduced state (34) and has been used extensively to study oxidative stress in a wide array of biological systems (45, 46). After a battery of assays, we established that exposure to 50 μm paraquat for 18 h was sufficient to induce an increase in matrix ROS production without appreciably perturbing Min6 cell bioenergetics. We also decided to use paraquat because, in comparison with other drugs that generate ROS by inhibiting the electron transport chain (e.g. rotenone or antimycin A), it takes advantage of complex I activity to generate ROS. At 50 μm paraquat we were able to activate proton leak in a UCP2-dependent fashion. Paraquat induced a small but significant inhibitory effect on GSIS, indicating that matrix ROS impedes GSIS and overall that ROS produced in different cellular compartments can have very different effects on insulin secretion.
Glucose metabolism in β cells has been reported to robustly increase ROS levels, which then are thought to amplify GSIS (11). Glucose energization induced a small but significant increase in mitochondrial matrix ROS. In contrast, glucose metabolism led to an immediate suppression of total cell ROS. This latter observation coincides with several other studies reporting that glucose metabolism actually decreases cellular ROS levels (47, 48). In fact, evidence generated in other studies attributes this decline in cell ROS to increased glucose flux through the hexose monophosphate shunt pathway that provides a bulk of the NADPH to a cell (47). In our study, we also observed that GSH/GSSG increased following glucose energization, which coincided with the deglutathionylation and activation of UCP2, a key protein involved in controlling cell ROS levels. The decrease in total cell ROS (from 5–60 min of incubation) and the gradual increase in matrix ROS would suggest that two separate pools of ROS in the cell are being modulated following glucose energization. For example, the gradual increase in matrix ROS due to glucose metabolism would trigger the dissipation of PMF by UCP2 proton leak, which is required to desensitize the insulin secretion signal. It is also important to note that an increase in proton leak through UCP2 would also diminish the emission of ROS from mitochondria, which is also a putative amplifier of GSIS (8). Mitochondrial ROS levels were observed to gradually increase during the initial 30 min following glucose provision and then drop back down at the 60-min mark. Hence, the location of ROS genesis determines its impact on GSIS (e.g. extracellular ROS activates although matrix ROS inhibits) and that activation of UCP2 by matrix ROS is used to eventually dissipate the GSIS signal.
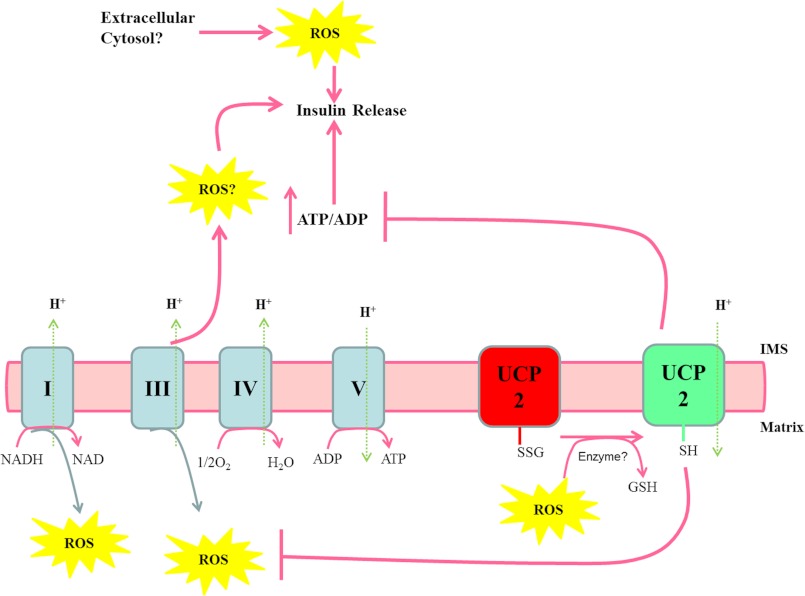
UCP2 is expressed in many tissues and has been linked to a number of cell signaling functions (10, 49). In previous studies, we provided novel evidence that UCP2 is controlled by glutathionylation (6). Here, we have shown that reversible glutathionylation of UCP2 can play a signaling role in GSIS. Matrix ROS activated UCP2 proton leak, thereby decreasing GSIS, whereas pharmacological glutathionylation of UCP2 enhanced GSIS. These observations are completely consistent with our previous publications (6, 22). A glucose-dependent metabolically driven redox circuit is most likely required to regulate insulin release (summarized in Fig. 7). The reducing equivalents derived from glucose oxidation drive mitochondrial synthesis of ATP, which then signals insulin secretion. However, if the provision of reducing equivalents exceeds the oxidative capacity of the mitochondrial respiratory chain, matrix ROS levels increase. We propose that following glucose uptake, UCP2 remains glutathionylated to allow the establishment of a PMF, which drives ATP production and insulin release. Progressive increases in glucose metabolism increase matrix ROS, and the latter induces deglutathionylation of UCP2, thereby activating proton leak and decreasing insulin release. What are the implications of our finding that extracellular ROS enhanced GSIS? Perhaps this is indicative of another source of ROS that is required to activate a completely separate redox circuit dedicated to GSIS amplification. Hence, it is important to consider contributions from the cytosol (e.g. NADPH oxidase) or the intermembrane space of mitochondria. UCP2 activation by matrix ROS could very well prevent ROS emission from mitochondria, thus diminishing the GSIS signal. Indeed, such a scenario would be consistent with previous observations that ROS self-regulate their own production by activating the uncoupling proteins (8, 16). Thus, it appears that ROS-mediated redox signaling in β cells is highly ordered and complex.
FIGURE 7.
Matrix ROS, glutathionylation of UCP2, and the control of GSIS. Following glucose catabolism, reducing equivalents enter complex I and complex II of the electron transport chain. The liberated electrons are transferred through complexes III and IV to the terminal electron acceptor O2. The energetically favorable transfer of electrons to O2 is coupled to the genesis of PMF, which is then used to drive complex V (ATP synthase). ATP is exported into the cytosol from the mitochondria increasing the ATP/ADP ratio, which induces insulin release. Over time with progressive glucose metabolism and the decrease in available ADP, PMF increases, as does matrix ROS generation from complexes I and III. The increase in matrix ROS leads to the deglutathionylation of UCP2 and the activation of proton leak. Proton leak decreases PMF and ATP/ADP, thus hindering further insulin release. SSG, protein-glutathione adduct; IMS, intermembrane space.
Supplementary Material
This work was supported in part by Canadian Institutes of Health Research Canadian Institutes of Health Research (to M. E. H.).

This article contains supplemental Fig. 1.
- PMF
- protonmotive force
- ROS
- reactive oxygen species
- GSIS
- glucose-stimulated insulin secretion
- TMRE
- tetramethylrhodamine ethyl ester
- ANOVA
- analysis of variance
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI
- propidium iodide
- PQ
- paraquat
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- DCFHDA
- dichlorofluorescein diacetate
- BioGEE
- biotinylated glutathione ethyl ester
- OCR
- oxygen consumption rate.
REFERENCES
- 1. Lowell B. B., Shulman G. I. (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307, 384–387 [DOI] [PubMed] [Google Scholar]
- 2. Affourtit C., Brand M. D. (2008) On the role of uncoupling protein-2 in pancreatic beta cells. Biochim. Biophys. Acta 1777, 973–979 [DOI] [PubMed] [Google Scholar]
- 3. MacDonald M. J., Fahien L. A. (1990) Insulin release in pancreatic islets by a glycolytic and a Krebs cycle intermediate. Contrasting patterns of glyceraldehyde phosphate and succinate. Arch. Biochem. Biophys. 279, 104–108 [DOI] [PubMed] [Google Scholar]
- 4. Malmgren S., Nicholls D. G., Taneera J., Bacos K., Koeck T., Tamaddon A., Wibom R., Groop L., Ling C., Mulder H., Sharoyko V. V. (2009) Tight coupling between glucose and mitochondrial metabolism in clonal beta-cells is required for robust insulin secretion. J. Biol. Chem. 284, 32395–32404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spacek T., Santorová J., Zacharovová K., Berková Z., Hlavatá L., Saudek F., Jezek P. (2008) Glucose-stimulated insulin secretion of insulinoma INS-1E cells is associated with elevation of both respiration and mitochondrial membrane potential. Int. J. Biochem. Cell Biol. 40, 1522–1535 [DOI] [PubMed] [Google Scholar]
- 6. Mailloux R. J., Seifert E. L., Bouillaud F., Aguer C., Collins S., Harper M. E. (2011) Glutathionylation acts as a control switch for uncoupling proteins UCP2 and UCP3. J. Biol. Chem. 286, 21865–21875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang C. Y., Parton L. E., Ye C. P., Krauss S., Shen R., Lin C. T., Porco J. A., Jr., Lowell B. B. (2006) Genipin inhibits UCP2-mediated proton leak and acutely reverses obesity- and high glucose-induced beta cell dysfunction in isolated pancreatic islets. Cell Metab. 3, 417–427 [DOI] [PubMed] [Google Scholar]
- 8. Affourtit C., Jastroch M., Brand M. D. (2011) Uncoupling protein-2 attenuates glucose-stimulated insulin secretion in INS-1E insulinoma cells by lowering mitochondrial reactive oxygen species. Free Radic. Biol. Med. 50, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robson-Doucette C. A., Sultan S., Allister E. M., Wikstrom J. D., Koshkin V., Bhattacharjee A., Prentice K. J., Sereda S. B., Shirihai O. S., Wheeler M. B. (2011) Beta-cell uncoupling protein 2 regulates reactive oxygen species production, which influences both insulin and glucagon secretion. Diabetes 60, 2710–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mailloux R. J., Harper M. E. (2011) Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic. Biol. Med. 51, 1106–1115 [DOI] [PubMed] [Google Scholar]
- 11. Gray J. P., Heart E. (2010) Usurping the mitochondrial supremacy. Extramitochondrial sources of reactive oxygen intermediates and their role in beta cell metabolism and insulin secretion. Toxicol. Mech. Methods 20, 167–174 [DOI] [PubMed] [Google Scholar]
- 12. Pi J., Bai Y., Zhang Q., Wong V., Floering L. M., Daniel K., Reece J. M., Deeney J. T., Andersen M. E., Corkey B. E., Collins S. (2007) Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes 56, 1783–1791 [DOI] [PubMed] [Google Scholar]
- 13. Heart E., Palo M., Womack T., Smith P. J., Gray J. P. (2012) The level of menadione redox-cycling in pancreatic β-cells is proportional to the glucose concentration. Role of NADH and consequences for insulin secretion. Toxicol. Appl. Pharmacol. 258, 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacDonald M. J. (1991) Stimulation of insulin release from pancreatic islets by quinones. Biosci. Rep. 11, 165–170 [DOI] [PubMed] [Google Scholar]
- 15. Leloup C., Tourrel-Cuzin C., Magnan C., Karaca M., Castel J., Carneiro L., Colombani A. L., Ktorza A., Casteilla L., Pénicaud L. (2009) Mitochondrial reactive oxygen species are obligatory signals for glucose-induced insulin secretion. Diabetes 58, 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Echtay K. S., Roussel D., St-Pierre J., Jekabsons M. B., Cadenas S., Stuart J. A., Harper J. A., Roebuck S. J., Morrison A., Pickering S., Clapham J. C., Brand M. D. (2002) Superoxide activates mitochondrial uncoupling proteins. Nature 415, 96–99 [DOI] [PubMed] [Google Scholar]
- 17. Echtay K. S., Murphy M. P., Smith R. A., Talbot D. A., Brand M. D. (2002) Superoxide activates mitochondrial uncoupling protein 2 from the matrix side. Studies using targeted antioxidants. J. Biol. Chem. 277, 47129–47135 [DOI] [PubMed] [Google Scholar]
- 18. Korshunov S. S., Skulachev V. P., Starkov A. A. (1997) High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 416, 15–18 [DOI] [PubMed] [Google Scholar]
- 19. Parker N., Affourtit C., Vidal-Puig A., Brand M. D. (2008) Energization-dependent endogenous activation of proton conductance in skeletal muscle mitochondria. Biochem. J. 412, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Affourtit C., Brand M. D. (2008) Uncoupling protein-2 contributes significantly to high mitochondrial proton leak in INS-1E insulinoma cells and attenuates glucose-stimulated insulin secretion. Biochem. J. 409, 199–204 [DOI] [PubMed] [Google Scholar]
- 21. Gallogly M. M., Mieyal J. J. (2007) Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 22. Mailloux R. J., Adjeitey C. N., Xuan J. Y., Harper M. E. (2012) Crucial yet divergent roles of mitochondrial redox state in skeletal muscle versus brown adipose tissue energetics. FASEB J. 26, 363–375 [DOI] [PubMed] [Google Scholar]
- 23. Mailloux R. J., Harper M. E. (2012) Mitochondrial proticity and ROS signaling: lessons from the uncoupling proteins. Trends Endocrinol. Metab. 23, 451–458 [DOI] [PubMed] [Google Scholar]
- 24. Kong D., Vong L., Parton L. E., Ye C., Tong Q., Hu X., Choi B., Brüning J. C., Lowell B. B. (2010) Glucose stimulation of hypothalamic MCH neurons involves K(ATP) channels, is modulated by UCP2, and regulates peripheral glucose homeostasis. Cell Metab. 12, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mailloux R. J., Harper M. E. (2010) Glucose regulates enzymatic sources of mitochondrial NADPH in skeletal muscle cells; a novel role for glucose-6-phosphate dehydrogenase. FASEB J. 24, 2495–2506 [DOI] [PubMed] [Google Scholar]
- 26. Pi J., Collins S. (2010) Reactive oxygen species and uncoupling protein 2 in pancreatic β-cell function. Diabetes Obes. Metab. 12, 141–148 [DOI] [PubMed] [Google Scholar]
- 27. Rani S., Mehta J. P., Barron N., Doolan P., Jeppesen P. B., Clynes M., O'Driscoll L. (2010) Decreasing Txnip mRNA and protein levels in pancreatic MIN6 cells reduces reactive oxygen species and restores glucose-regulated insulin secretion. Cell. Physiol. Biochem. 25, 667–674 [DOI] [PubMed] [Google Scholar]
- 28. Mieyal J. J., Chock P. B. (2012) Post-translational modification of cysteine in redox signaling and oxidative stress. Focus on S-glutathionylation. Antioxid. Redox Signal. 16, 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sabens Liedhegner E. A., Gao X. H., Mieyal J. J. (2012) Mechanisms of altered redox regulation in neurodegenerative diseases–focus on S-glutathionylation. Antioxid. Redox Signal. 16, 543–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooper A. J., Pinto J. T., Callery P. S. (2011) Reversible and irreversible protein glutathionylation. Biological and clinical aspects. Expert Opin. Drug Metab. Toxicol. 7, 891–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shelton M. D., Chock P. B., Mieyal J. J. (2005) Glutaredoxin. Role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid. Redox Signal. 7, 348–366 [DOI] [PubMed] [Google Scholar]
- 32. Kushnareva Y. E., Sokolove P. M. (2000) Prooxidants open both the mitochondrial permeability transition pore and a low conductance channel in the inner mitochondrial membrane. Arch. Biochem. Biophys. 376, 377–388 [DOI] [PubMed] [Google Scholar]
- 33. Kitiphongspattana K., Khan T. A., Ishii-Schrade K., Roe M. W., Philipson L. H., Gaskins H. R. (2007) Protective role for nitric oxide during the endoplasmic reticulum stress response in pancreatic beta-cells. Am. J. Physiol. Endocrinol. Metab. 292, E1543–E1554 [DOI] [PubMed] [Google Scholar]
- 34. Cochemé H. M., Murphy M. P. (2008) Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 283, 1786–1798 [DOI] [PubMed] [Google Scholar]
- 35. Hurd T. R., Costa N. J., Dahm C. C., Beer S. M., Brown S. E., Filipovska A., Murphy M. P. (2005) Glutathionylation of mitochondrial proteins. Antioxid. Redox Signal. 7, 999–1010 [DOI] [PubMed] [Google Scholar]
- 36. Fisher-Wellman K. H., Neufer P. D. (2012) Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol. Metab. 23, 142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robertson R. P., Harmon J., Tran P. O., Tanaka Y., Takahashi H. (2003) Glucose toxicity in beta-cells. Type 2 diabetes, good radicals gone bad, and the glutathione connection. Diabetes 52, 581–587 [DOI] [PubMed] [Google Scholar]
- 38. Koshkin V., Wang X., Scherer P. E., Chan C. B., Wheeler M. B. (2003) Mitochondrial functional state in clonal pancreatic beta-cells exposed to free fatty acids. J. Biol. Chem. 278, 19709–19715 [DOI] [PubMed] [Google Scholar]
- 39. Asfari M., Janjic D., Meda P., Li G., Halban P. A., Wollheim C. B. (1992) Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology 130, 167–178 [DOI] [PubMed] [Google Scholar]
- 40. Janjic D., Wollheim C. B. (1992) Effect of 2-mercaptoethanol on glutathione levels, cystine uptake, and insulin secretion in insulin-secreting cells. Eur. J. Biochem. 210, 297–304 [DOI] [PubMed] [Google Scholar]
- 41. Park W. H., Han Y. W., Kim S. H., Kim S. Z. (2007) An ROS generator, antimycin A, inhibits the growth of HeLa cells via apoptosis. J. Cell. Biochem. 102, 98–109 [DOI] [PubMed] [Google Scholar]
- 42. Porterfield D. M., Corkey R. F., Sanger R. H., Tornheim K., Smith P. J., Corkey B. E. (2000) Oxygen consumption oscillates in single clonal pancreatic beta-cells (HIT). Diabetes 49, 1511–1516 [DOI] [PubMed] [Google Scholar]
- 43. Mertz R. J., Worley J. F., Spencer B., Johnson J. H., Dukes I. D. (1996) Activation of stimulus-secretion coupling in pancreatic beta-cells by specific products of glucose metabolism. Evidence for privileged signaling by glycolysis. J. Biol. Chem. 271, 4838–4845 [DOI] [PubMed] [Google Scholar]
- 44. MacLellan J. D., Gerrits M. F., Gowing A., Smith P. J., Wheeler M. B., Harper M. E. (2005) Physiological increases in uncoupling protein 3 augment fatty acid oxidation and decrease reactive oxygen species production without uncoupling respiration in muscle cells. Diabetes 54, 2343–2350 [DOI] [PubMed] [Google Scholar]
- 45. Mason R. P. (1990) Redox cycling of radical anion metabolites of toxic chemicals and drugs and the Marcus theory of electron transfer. Environ. Health Perspect. 87, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bus J. S., Gibson J. E. (1984) Paraquat. Model for oxidant-initiated toxicity. Environ. Health Perspect. 55, 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rebelato E., Abdulkader F., Curi R., Carpinelli A. R. (2011) Control of the intracellular redox state by glucose participates in the insulin secretion mechanism. PLoS ONE 6, e24507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ammon H. P., Grimm A., Lutz S., Wagner-Teschner D., Händel M., Hagenloh I. (1980) Islet glutathione and insulin release. Diabetes 29, 830–834 [DOI] [PubMed] [Google Scholar]
- 49. Mailloux R. J., Harper M. E. (2012) Mitochondrial proticity and ROS signaling. Lessons from the uncoupling proteins. Trends Endocrinol. Metab. 23, 451–458 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.