Background: KMN (KNL/MIS12/NDC80) is a kinetochore constituent essential for chromosome movements in mitosis.
Results: KMN protein specifies the kinetochore localization of SKAP. SKAP regulates spindle microtubule dynamics for accurate chromosome movements.
Conclusion: The MIS13-SKAP interaction links kinetochore structural components to dynamic microtubule plus-ends.
Significance: MIS13-SKAP interaction governs chromosome dynamics and stability in mitosis.
Keywords: Chromosomes, Kinetochore, Microtubules, Mitosis, Molecular Dynamics, EB1, Mis13, SKAP, Chromosome Oscillation, Microtubule Plus-end
Abstract
Chromosome segregation in mitosis is orchestrated by the dynamic interactions between the kinetochore and spindle microtubules. Our recent study shows that mitotic motor CENP-E cooperates with SKAP to orchestrate an accurate chromosome movement in mitosis. However, it remains elusive how kinetochore core microtubule binding activity KMN (KNL1-MIS12-NDC80) regulates microtubule plus-end dynamics. Here, we identify a novel interaction between MIS13 and SKAP that orchestrates accurate interaction between kinetochore and dynamic spindle microtubules. SKAP physically interacts with MIS13 and specifies kinetochore localization of SKAP. Suppression of MIS13 by small interfering RNA abrogates the kinetochore localization of SKAP. Total internal reflection fluorescence microscopic assays demonstrate that SKAP exhibits an EB1-dependent, microtubule plus-end loading and tracking in vitro. Importantly, SKAP is essential for kinetochore oscillations and dynamics of microtubule plus-ends during live cell mitosis. Based on those findings, we reason that SKAP constitutes a dynamic link between spindle microtubule plus-ends and mitotic chromosomes to achieve faithful cell division.
Introduction
The faithful chromosome segregation during mitosis is orchestrated by dynamic interaction between spindle microtubules and the kinetochore, a protein supercomplex assembled at the chromosome centromere. Kinetochore acts as a structural platform for linking the centromere to spindle microtubules and functions as a signaling hub coordinating chromosome attachment and the metaphase to anaphase transition (1). The spindle assembly checkpoint senses kinetochore-microtubule attachment and/or tension at the kinetochore and delays the onset of anaphase until all chromosomes have successfully achieved stable, bi-oriented attachments (2).
In the past decade, our understanding on the molecular mechanism of dynamic kinetochore-microtubule interactions has achieved significant progress (3). As we know, multiple proteins function together at the kinetochore; they contribute to the establishment and/or regulation of dynamic kinetochore-microtubule interactions by forming attachment sites for spindle microtubules. One conserved protein network composed of KNL-1, Mis12 complex, and Ndc80 complex (KMN network) constitutes the core microtubule binding site at the kinetochore (4). When the KMN network is compromised, a severe kinetochore-microtubule attachment defect occurs. Another group of kinetochore proteins, including Clasp1/2, Clip170, and Lis1, can also interact with microtubules, but their main functions seem not to promote stable kinetochore-microtubule attachment. When knocking down these proteins via siRNA treatment, the number of attached microtubules did not decrease and sometimes even increased. However, in these siRNA-treated cells, the cell cycle was blocked at prometaphase, and perfect chromosome alignment could not be achieved. Further detailed examinations showed that the dynamics of attached microtubule plus-end were repressed in these cells, and oscillation behaviors of paired kinetochores did not coordinate with each other (3, 5). In addition, mitotic kinesin CENP-E also interacts with microtubules and is essential for chromosome alignment (6–9). Through tethering microtubules to the kinetochore, CENP-E is involved in chromosome movements and segregation from prometaphase to anaphase.
An early search for mitotic regulators has led to the identification of SKAP as a regulator for the anaphase onset (10). SKAP localizes at the outer kinetochore and is essential for chromosome alignment (10). SKAP forms a complex with Astrin, a spindle- and kinetochore-related protein (11). Both SKAP and Astrin exhibit microtubule activity (12). A recent study speculates that the SKAP·Astrin complex localizes and regulates microtubule plus-ends (13). Our investigation of protein networks interacting with CENP-E has led to the identification of SKAP-CENP-E interaction (14). Our study showed that SKAP cooperates with CENP-E to orchestrate kinetochore-microtubule interaction for faithful chromosome segregation (14). Interestingly, suppression of CENP-E does not abolish the kinetochore localization of SKAP (14), raising interest in the identification of the kinetochore constituent(s) that specifies SKAP localization to the kinetochore.
To delineate the molecular network underlying SKAP interaction with the kinetochore and spindle microtubule, we carried out a yeast two-hybrid screen for SKAP-interacting proteins at the kinetochore. Our biochemical characterization indicates that SKAP interacts with MIS13, and this interaction specifies the kinetochore localization of SKAP. Our present study has established that SKAP is a microtubule plus-end tracking protein in vitro and essential for normal kinetochore oscillation and microtubule plus-end dynamics in real-time mitosis. Our studies revealed mechanistic action of SKAP in mitosis, which functions as a linker protein connecting outer kinetochore with dynamic microtubule plus-ends.
MATERIALS AND METHODS
Yeast Two-hybrid Assay
Yeast two-hybrid assays were performed as described previously (15, 16). Briefly, SKAP cDNA was inserted into the BamHI-EcoRI sites of pGBKT7 vector to create a fusion with amino acids 1–147 of the Gal4 DNA-binding domain (BD).3 The resultant BD-SKAP was transformed into strain AH109 along with different recombinant plasmids expressing a Gal4 activation domain in fusion with different kinetochore proteins, respectively. The co-transformed yeast was grown up on SD plates with X-α-Gal but lacking Leu, Trp, His, and Ade.
Cell Culture
HeLa cells, from the American Type Culture Collection (Manassas, VA) were maintained as subconfluent monolayers in Dulbecco's modified Eagle's medium (Invitrogen) with 10% FBS (Hyclone, Logan, UT) and 100 units/ml penicillin plus 100 μg/ml streptomycin (Invitrogen).
Plasmids and Recombinant Protein Production
The full-length SKAP mRNA was amplified as described previously (14). GFP-tagged SKAP full-length and deletion truncations were cloned into pEGFP-C2 (Clontech). Bacterial expression constructs of SKAP were cloned into pGEX-5X-3 (GE Healthcare), pET-28a (Novagen), and pMal-C2 vector (New England Biolabs, Beverly, MA). All plasmid constructs were sequenced for verification.
Expression and Purification of Recombinant Proteins
Purification of recombinant proteins was carried out as described previously (14). Briefly, the GST fusion protein in bacteria in the soluble fraction was purified by using glutathione-agarose chromatography, whereas MBP-tagged protein was purified using Amylose beads.
In Vitro Pull-down Assay
Purified MBP-SKAP full-length and deletion mutants were used as affinity matrix to absorb GST-Mis13 protein. These MBP fusion protein-bound Amylose beads were incubated with GST-Mis13-expressing bacteria cell lysate for 1 h at 4 °C, respectively. After incubation, the beads were washed three times with PBS containing 0.25% Triton X-100 and once with PBS and then boiled in 1× SDS-PAGE sample buffer. The bound proteins were separated on 10% SDS-polyacrylamide gel for Coomassie Blue staining and transferred onto nitrocellulose membrane for Western blotting using GST antibody.
Purified GST-SKAP full-length and deletion mutants were used as affinity matrix to absorb MBP-Mis13 protein. These GST fusion protein-bound Sepharose beads were incubated with purified MBP-Mis13 fusion proteins for 1 h at 4 °C, respectively. After incubation, the beads were washed three times with PBS containing 0.25% Triton X-100 and once with PBS and then boiled in 1× SDS-PAGE sample buffer. The bound proteins were separated on 8% SDS-polyacrylamide gel for Coomassie Blue staining and transferred onto nitrocellulose membrane for Western blotting using MBP antibody.
Immunoprecipitation
pEGFP-C2 vector- or GFP-Mis12/Mis13/Mis14 plus 3× FLAG-SKAP-co-expressing 293T cells were lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.1% Triton X-100) on ice individually. Different lysates were clarified by centrifugation (12,000 rpm for 20 min at 4 °C) and then incubated with anti-FLAG M2 affinity beads (Sigma) at 4 °C for 4 h, respectively. After an extensive wash, the beads were boiled in 1× SDS-PAGE sample buffer for 5 min, and the bound proteins were separated on 10% SDS-polyacrylamide gel for transferring onto nitrocellulose membrane for Western blotting using GFP antibody.
To test if SKAP forms a cognate complex with EB1, mitotic cell lysates were prepared, clarified, and incubated with SKAP mouse IgG prebound Protein G-agarose beads as described previously (14). After an extensive wash, Protein G beads were boiled in SDS-PAGE sample buffer followed by Western blotting analyses to probe for SKAP and EB1.
Antibodies
Both rabbit and mouse antibodies against SKAP were generated using full-length recombinant proteins from bacteria using a standard protocol as described previously (17). Antibodies against Mis13 were generated as described previously (18). Anti-tubulin antibody (DM1A) was purchased from Sigma. Anti-Hec1 antibody was purchased from Abcam (Cambridge, MA).
siRNA Treatment
The siRNA sequence used for silencing of SKAP is 5′-AGGCTACAAACCACTGAGTAA-3′ (siRNA 1) or a SMARTpool (L-022219-00; Thermo Fisher Scientific; siRNA 2). Hec1, Mis12, Mis13, and CENP-E siRNA were reported previously (15, 19, 20). As a control, either a duplex targeting cyclophilin or scramble sequence was used (17, 21). The oligonucleotide RNA duplexes were synthesized by Dharmacon Research, Inc. (Boulder, CO).
HeLa cells were grown on 150-mm2 culture flasks at 37 °C with 10% CO2, and mitotic cells were collected by mitotic shake-off. Two hours after replating mitotic cells into 24-well plates with or without coverslips, cells were transfected with various siRNA oligonucleotide duplexes as described above using Lipofectamine 2000 (Invitrogen).
Immunofluorescence Microscopy
HeLa cells were grown on coverslips in 24-well plates (Corning Glass) and transfected with siRNA and/or plasmids using Lipofectamine 2000. Single thymidine block and release treatment was used to synchronize HeLa cell. For the immunofluorescence assay, HeLa cells were rinsed for 1 min with PHEM buffer (100 mm PIPES, 20 mm HEPES, pH 6.9, 5 mm EGTA, 2 mm MgCl2, and 4 m glycerol) and permeabilized for 1 min with PHEM buffer plus 0.1% Triton X-100. Then extracted HeLa cells were fixed in freshly prepared 4% paraformaldehyde in PHEM buffer and rinsed three times in 1× PBS. HeLa cells seeded on coverslips were blocked with 0.05% Tween 20 in 1× PBS (1× TPBS) with 1% bovine serum albumin (Sigma) and then incubated with various primary antibodies in a humidified chamber for 1 h at room temperature and then washed three times in 1× TPBS. Primary antibodies were visualized by fluorescein-conjugated secondary antibodies. DNA was stained with DAPI (Sigma).
For analysis of cold-stable microtubules, fixation was done as described previously (22). Briefly, HeLa cells were incubated for 15 or 30 min on ice in L-15 medium (Invitrogen) with 20 mm HEPES at pH 7.3 and then fixed for 10 min at room temperature with PTEMF buffer (3.7% formaldehyde in 100 mm PIPES, pH 6.8, 10 mm EGTA, 1 mm MgCl2, and 0.2% Triton X-100).
Images were taken at identical exposure times within each experiment, acquired as 24-bit RGB images, and processed in Adobe Photoshop. Images shown in the same panel have been identically scaled. Measurement of kinetochore intensities was performed in ZEN software (Carl Zeiss) on non-deconvolved images. Quantification of kinetochore intensities was performed as described previously (23). Essentially, a circular region with fixed diameter was centered on each kinetochore, and unless indicated otherwise, ACA intensity was measured in the same region and used for normalization after subtraction of background intensity measured outside the cell.
Kinetochore Tracking Assay
The kinetochore tracking experiment was performed as described (24). Briefly, HeLa cells were grown on a glass bottom culture dish (MatTek) at 37 °C with 10% CO2 after various transfections with siRNA or scrambled (control) oligonucleotides; HeLa cells were then transiently transfected with EGFP-CENP-B followed by synchronization. Then images were collected using a DeltaVision DVI microscopy system built on an Olympus IX-70 inverted microscope base as described previously (7, 25). For imaging, time points were acquired every 5 s for 5 min with a ×100, 1.35 numerical aperture objective lens on the imaging system (DeltaVision Core, Applied Precision) fitted with a 37 °C environmental chamber. Final images were processed using DeltaVision Softworx software. To characterize sister kinetochore dynamics over time, we monitored the sister kinetochore center position along the normal to the metaphase plate (spindle axis), and the position of the metaphase plate was calculated by setting a plane to the computed kinetochore positions. The autocorrelation ability of paired sister kinetochore movements along the metaphase plate yielded sister kinetochore oscillations.
GFP-Tubulin Photoactivation in Live Mitosis
The experiment was performed as described previously (26–28). Briefly, HeLa cells were grown on a glass bottom culture dish (MatTek) at 37 °C with 10% CO2 after various transfections with siRNA or scrambled (control) oligonucleotides; HeLa cells were transiently transfected with PA-GFP-tubulin and cherry-H2B followed by synchronization, and then mitotic cells were identified by DIC microscopy. Several pulses from a 405-nm diffraction-limited laser on LSM710 NLO (Carl Zeiss) were used to photoactivate an area of <2 μm2 within the spindle as described previously. Images were acquired with ×63, 1.4 numerical aperture objectives on a LSM710 laser-scanning microscope, and images were collected every 30 s.
Total Internal Reflection Fluorescence Microscopic Analyses
The microtubule plus-end tracking experiment was performed as described recently (29) with some modifications. The GMPCPP MT seeds were prepared by polymerizing 30 μm tubulin (at a bovine tubulin/rhodamine-tubulin/biotin-tubulin ratio of 30:2:1) in the presence of 1 mm GMP-CPP (Jena Bioscience) at 37 °C for 40 min. The seeds were then centrifuged and resuspended in BRB80 buffer (80 mm K-PIPES, pH 6.8, 2 mm MgCl2, 1 mm EGTA). These seeds were sheared with a 25-gauge needle before they were used to generate short seeds.
Flow chambers were prepared as described previously (30). Chambers were coated with 10% monoclonal anti-biotin antibody (Sigma) followed by blocking with 5% Pluronic F-127 (Sigma). After a brief wash, sheared MT seeds (125 nm) were added into the chamber. Tubulin polymerization mixture (30 μm tubulin in total containing 1:30 rhodamine-labeled bovine tubulin in BRB80, 50 mm KCl, 5 mm DTT, 1.25 mm Mg-GTP, 0.25 mg/ml κ-casein, 0.15% methylcellulose (Sigma), an oxygen-scavenging system, and +TIPs) was introduced into the chamber to initiate polymerization. Unless stated otherwise, the final concentrations of +TIPs were 250 nm EB1 and 250 nm N-SKAP or its SXIP mutant recombinant proteins. The temperature was kept at 25 °C. Images were collected with a superresolution microscope configured on an ELYRA system (Carl Zeiss). The laser intensities were kept at a low level to avoid photobleaching. For +TIPs tracking assays, 1 frame was taken per second. Plus-end tracking of GFPN-SKAP and its SXIP mutant was analyzed using kymographs in ZEN software (Carl Zeiss).
RESULTS
C-terminal Region of SKAP Determines Its Kinetochore Localization
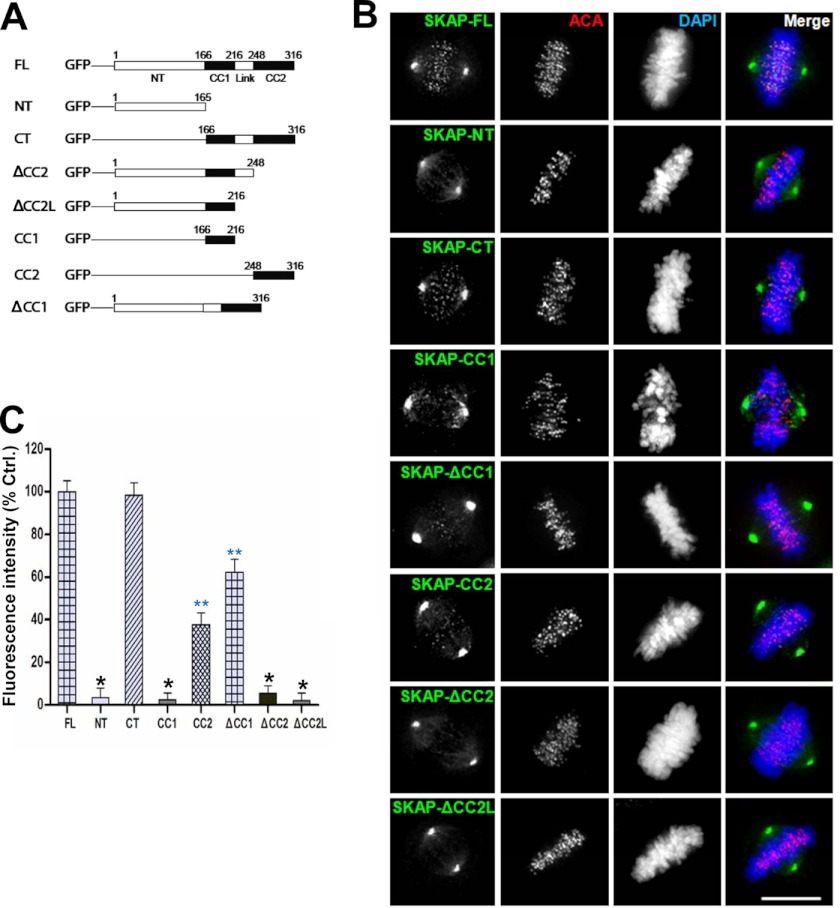
Our recent study revealed that SKAP cooperates with CENP-E in achieving accurate chromosome congression in mitosis (14). To delineate the mechanism underlying SKAP kinetochore localization and its regulation, we constructed a series of GFP-tagged deletion mutants of SKAP (as illustrated in Fig. 1A) and examined their localization in HeLa cells. As shown in Fig. 1B, full-length (FL) SKAP distributed to the spindle microtubules and kinetochores, which is consistent with previously published results (10, 12–14). The C-terminal region of SKAP, which contains two short coiled-coil regions, also distributed to the kinetochore. Furthermore, we noted that the CC2 fragment, which is a smaller region in CT deletion, had a weak kinetochore localization signal (Fig. 1B). However, the NT, ΔCC2, ΔCC2L, and CC1 deletion mutants failed to localize to kinetochore. In addition, deletion of CC1 attenuated the localization of SKAP to the kinetochore (Fig. 1B). Quantitative analyses of kinetochore relative fluorescence signal indicated that GFP-tagged CT deletion of SKAP had a fluorescence intensity similar to that of full-length, and deletion CC1 had a greater than 30% reduction judged by the fluorescence intensity at the kinetochore, whereas the kinetochore signals for SKAP-NT, ΔCC2, ΔCC2L, and CC1 deletion mutants were virtually undetectable based on the survey of 100 positively transfected HeLa cells (Fig. 1C; *, p < 0.001). In addition, the kinetochore signals for CC2 and ΔCC1 were significantly reduced (Fig. 1C; **, p < 0.01). Given the fact that deletion of either CC1 or CC2 significantly perturbed SKAP localization to the kinetochore (p < 0.01), we conclude that both coiled-coil domains of SKAP are responsible for its kinetochore targeting.
FIGURE 1.
Mapping the kinetochore binding region of SKAP. A, schematic representation of GFP-tagged SKAP full-length and various deletion mutants. CC1 and CC2, coiled-coil domain 1 and 2, respectively. B, representative images of metaphase HeLa cells expressing GFP-tagged full-length and different deletion mutants of human SKAP. 24 h after transfection, HeLa cells were fixed and then stained for ACA (red) and DNA (blue). Scale bar, 10 μm. C, quantification of kinetochore signal intensity of GFP-tagged SKAP full-length and different deletion mutants. Bars, mean ± S.E. (error bars) of three independent experiments. In each experiment, 20 cells were measured (≥15 kinetochores/cell; *, p < 0.001; **, p < 0.01).
KMN Is Essential for the Kinetochore Localization of SKAP
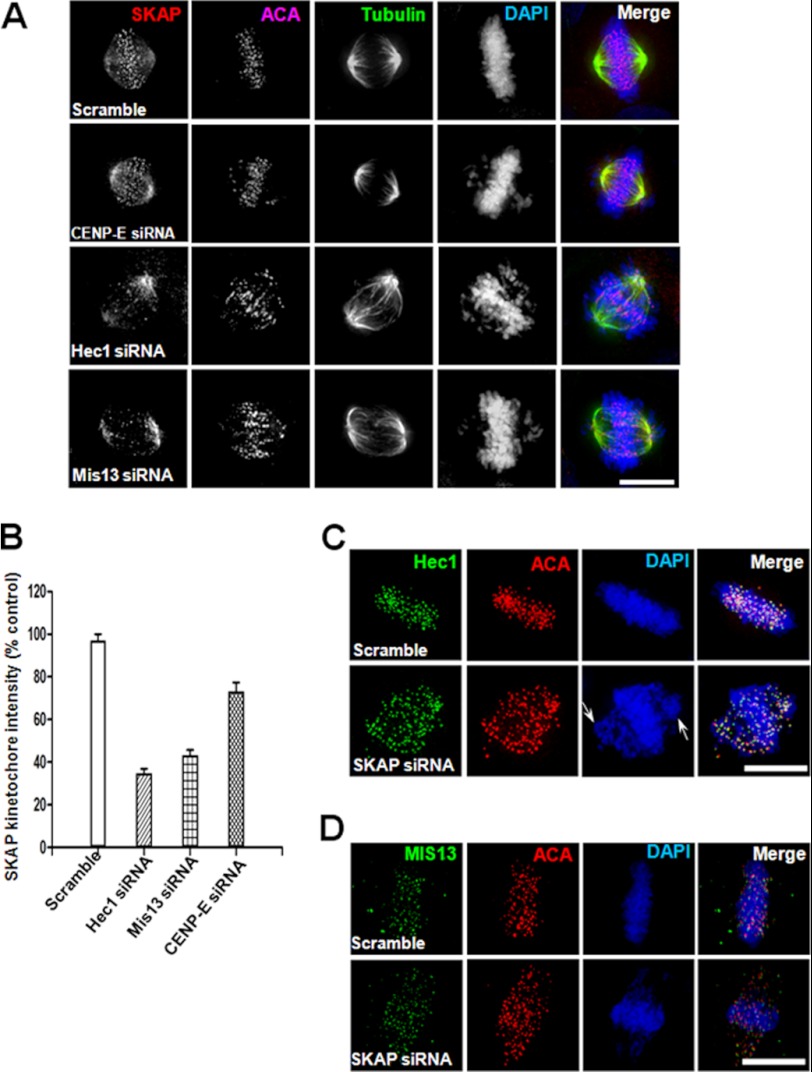
To understand the precise function of SKAP in kinetochore-microtubule interactions, we sought to probe for the kinetochore element(s) responsible for SKAP localization. To this end, we carried out an siRNA screen of kinetochore core constituents, such as KMN proteins. Previous studies have established that the NDC80 complex is indispensable for establishing microtubule-kinetochore attachments (e.g. see Refs. 1–3). Because the KMN network is one of the major structure platforms of the outer kinetochore and the core microtubule binding site, we examined the kinetochore signal of SKAP in HeLa cells in which individual KMN components were suppressed by siRNA treatment (4, 31). Preliminary experiments and a literature search have eliminated possible off-target effects of those siRNAs used in this study. In general, we employed two distinct siRNAs to targets (e.g. see Refs. 14–16).4 As shown in Fig. 2A, the level of SKAP at the kinetochore decreased significantly in the absence of Hec1, and loss of Mis13 also caused a sharply decreased signal of SKAP at kinetochores compared with that of control cells. However, when CENP-E is suppressed, the kinetochore signal of SKAP was slightly affected, which is consistent with our previous observation (14).
FIGURE 2.
Ndc80 complex and Mis12 complex are required for the kinetochore localization of SKAP. A, representative images of metaphase HeLa cells transfected with CENP-E, Hec1, or Mis13 siRNA for 36 h followed by fixation and then labeled with antibodies against SKAP (red), tubulin (green), ACA (purple), and DNA (blue). Scale bar, 10 μm. B, quantification of kinetochore signal intensity of SKAP. Bars, mean ± S.E. (error bars) of three independent experiments. In each experiment, 20 cells were measured (≥15 kinetochores/cell). C and D, representative images of metaphase HeLa cells transfected with SKAP siRNA for 36 h followed by fixation and then labeled with antibodies against Hec1 or Mis13 (green), ACA (red), and DNA (blue). Scale bar, 10 μm.
Quantitative analyses of relative fluorescence signal of SKAP at kinetochores demonstrated that depletion of Hec1 and Mis13 caused a 63 and 56% reduction in fluorescence intensity, respectively (Fig. 2B; p < 0.01). Therefore, the KMN network is required for recruitment of SKAP onto kinetochore, but not vice versa, because the protein levels of Hec1 or Mis13 at kinetochores were not affected when knocking down SKAP (Fig. 2, C and D; p > 0.05). Thus, both the Ndc80 complex and Mis12 complex are essential for the kinetochore localization of SKAP but not the contrary.
SKAP Physically Interacts with Mis12 Complex Components
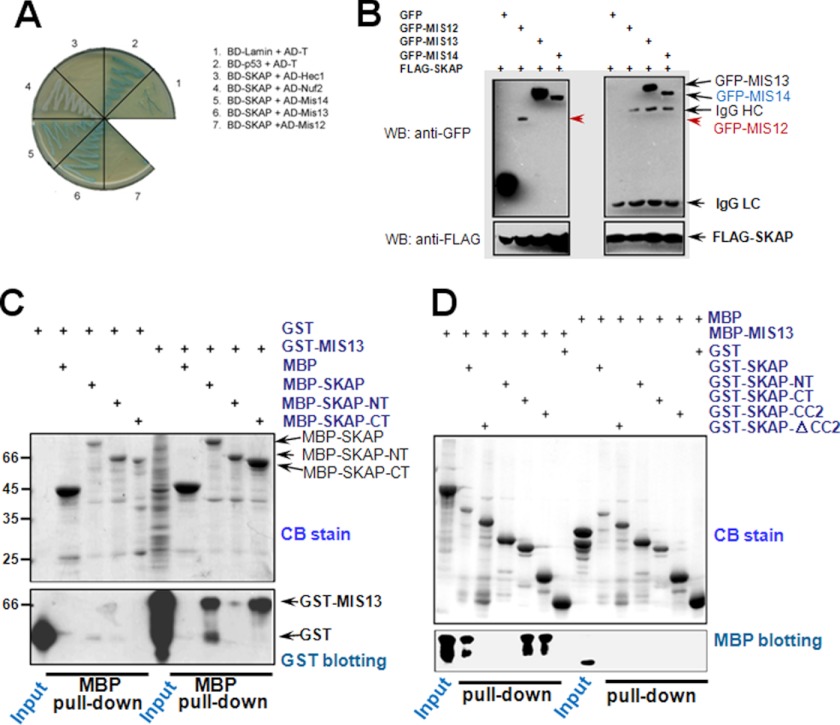
Given the dependence of SKAP localization to kinetochore on KMN components (e.g. see Fig. 2), we sought to examine if SKAP physically interacts with any components of the KMN complex. To this end, we carried out a yeast two-hybrid assay to screen for SKAP-interacting partners using the protocol established in our laboratory (e.g. see Refs. 15 and 16). Specifically, all candidate kinetochore protein cDNAs were cloned into AD vectors and co-transfected with BD-SKAP, followed by growth selection. As shown in Fig. 3A, co-transformation of BD-SKAP with AD-Mis13/Mis14 could rescue the transcription of the MEL1 reporter gene in the yeast strain, whereas no complementary effect was observed between BD-SKAP and AD-Hec1/Nuf2/Mis12. Thus, our yeast genetic screen suggested that Mis13 and Mis14 are potential interacting partners physically bound to SKAP.
FIGURE 3.
SKAP physically interacts with the components of Mis12 complex. A, yeast two-hybrid test of BD-SKAP against different AD fusion constructs. The image shows the growth of co-transformed yeast on the plate with X-α-Gal but lacking leucine, tryptophan, histidine, and adenine. B, GFP-Mis12/Mis13/Mis14 or pEGFP-C2 plus 3×FLAG-SKAP were co-transfected into 293T cells individually, followed by FLAG immunoprecipitations and Western blotting (WB) with anti-FLAG or anti-GFP antibodies. C, purified MBP-SKAP proteins (full-length, SKAP-NT, and SKAP-CT) were incubated with GST-Mis13-expressing bacteria cell lysates, respectively. After an extensive wash, the samples were analyzed by Coomassie Blue (CB) staining or anti-GST antibody Western blotting. D, purified GST-SKAP full-length protein and its deletion mutant protein were incubated with purified MBP-Mis13 protein, respectively. After an extensive wash, the samples were analyzed by Coomassie Blue staining or anti-MBP antibody Western blotting.
To confirm the interactions between SKAP and Mis13/14 observed in our yeast two-hybrid assay and to test whether SKAP forms a complex with Mis13/14, we carried out an immunoprecipitation assay using lysates of 293 cells previously co-transfected with GFP-Mis12/Mis13/Mis14 and FLAG-SKAP plasmids to express tagged proteins. The transfected cells were then collected at 36 h after transfection, followed by preparation of soluble cell lysates as previously described (e.g. (14, 18)). After the clarification centrifugation, FLAG-M2 affinity beads were added to cell lysates for capture of FLAG-SKAP and its interacting components. Immunoblotting with anti-FLAG antibody demonstrated a successful precipitation of SKAP, and immunoblotting with anti-GFP antibody verified that Mis12 complex components were co-precipitated with SKAP (Fig. 3B), whereas GFP was not recovered from the FLAG immunoprecipitates, showing the specific formation of SKAP·Mis12 complex. Because the interaction between Mis13 and SKAP is much stronger than that between Mis14 and SKAP (Fig. 3B), Mis13 was then studied as the main interacting partner for SKAP. This is also consistent with our previous conclusion because Mis12 and Mis13 are thought to be the major components responsible for linking Mis12 complex with other kinetochore components (18).
To define the concrete domain(s) required for their interaction, we carried out an MBP pull-down assay using full-length MBP-SKAP and its NT and CT deletion mutants as an affinity matrix to absorb GST-Mis13-expressing bacteria cell lysate, respectively. Immunoblotting with anti-GST antibody confirmed a direct interaction between the SKAP C-terminal region and Mis13 in vitro (Fig. 3C), and to further verify the minimum region of SKAP interacting with Mis13, different GST-tagged SKAP deletion mutants were purified and used as an affinity matrix to incubate with purified MBP-Mis13. As shown in Fig. 3D, immunoblotting with anti-MBP antibody showed that the SKAP-CC2 deletion mutant is the minimum Mis13-interacting region, which is consistent with the previously mentioned fact that this deletion mutant is also the minimum region for the kinetochore localization of SKAP. Thus, we concluded that SKAP directly interacts with Mis13 through its CC2 domain.
SKAP Governs Spindle Microtubule Dynamics and Accurate Kinetochore Attachment
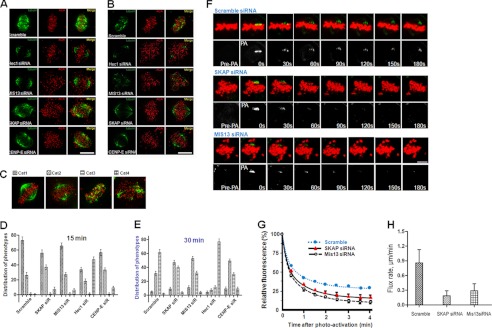
Our previous results suggested that SKAP is located at the interface between the kinetochore and dynamic spindle microtubule interface and is involved in a stable attachment of spindle microtubules to the kinetochore (14). The aforementioned experiments show that SKAP interacts with components of the KMN protein complex (e.g. see Fig. 3). To examine the precise function of SKAP in regulating the stability and/or dynamics of kinetochore-microtubule attachment, we sought to assess the kinetochore microtubule (K-fiber) stability in the absence of SKAP using a cold treatment protocol (7, 10). Briefly, after 15-min cold treatment (which depolymerizes all non-K-fibers, such as astral microtubules) or even longer cold treatment up to 30 min (which depolymerizes unstable K-fibers), microtubules were stained with tubulin antibody and imaged. As shown in Fig. 4A, most K-fibers of scramble siRNA transfected cells remain intact after exposure to cold treatment. The stability of K-fibers is sustained even after a longer exposure to cold (Fig. 4B). Consistent with the literature, suppression of the Ndc80 complex component Hec1 or the Mis12 complex component Mis13 dramatically reduced the K-fiber stability as judged by a minimized number of K-fibers after 15-min cold treatment (Fig. 4A). However, the K-fiber stability in Mis13-depleted cells was less severe compared with that of Hec1-suppressed cells. In the cells lacking SKAP or CENP-E, the K-fibers remained apparent but much altered in the 15-min treatment group (Fig. 4A) and were further destabilized after 30-min treatment (Fig. 4B), suggesting that SKAP does not determine the kinetochore-microtubule capture but facilitates a stable kinetochore microtubule attachment (14).
FIGURE 4.
SKAP regulates microtubule plus-end dynamics. A and B, HeLa cells were transfected with control and SKAP-, CENP-E-, Mis13-, and Hec1-specific siRNA, respectively. 36 h after transfection, HeLa cells were fixed after incubation at 4 °C for 15 min (A) or 30 min (B) before labeling with anti-α-tubulin antibody (green), ACA serum (red), and DAPI (DNA; blue). Scale bar, 10 μm. C, examples of spindle configurations seen in the SKAP-suppressed cells. A total of four categories were assigned. D and E, statistical analyses of spindle stability of SKAP-suppressed cells in response to cold treatments. Note that knockdown of SKAP and Mis13 did not cause the collapse of the mitotic spindle, whereas Hec1 suppression resulted in spindle collapse in the presence of cold. F, examples of time lapse fluorescent images of metaphase HeLa cells in the presence of scramble, SKAP, or Mis13 siRNA before (Pre-PA) and at the indicated times after activation (Post-PA) of GFP-tubulin fluorescence. The half-life (t½) of photoactivated GFP-tubulin from the aforementioned groups was then calculated and expressed as mean ± S.E. (error bars). G, statistic analyses of normalized fluorescence intensity over time after photoactivating kinetochore microtubule from the aforementioned three groups in F (n = 15 cells/group). H, quantification of kinetochore microtubule flux rate in HeLa cells transfected with SKAP, Mis13, or scramble control siRNA. Note that depletion of SKAP significantly reduces kinetochore microtubule flux (*, p < 0.01 compared with control cells).
To further assess the mechanisms of SKAP function in spindle plasticity, we conducted a whole population survey on the spindle stability challenged by cold treatment after siRNA-mediated protein knockdown. In general, we found four types of major spindle configuration in cold-treated HeLa cells after specific kinetochore protein suppression, as shown in Fig. 4C. Category 1 (Cat1) represents normal spindle by which cold stable microtubules were terminated at the kinetochore marked by ACA. Category 2 (Cat2) represents unstable spindle in which the bipolar spindle was shortened. Category 3 (Cat3) represents perturbed spindle in which kinetochore microtubules were fragmented but chromosomes appeared near the equator. Category 4 (Cat4) represents collapsed spindle as chromosome scattered around. The statistic analyses show that suppression of Hec1 perturbed bipolar spindle stability because there was a significant increase in category 4 count in the 15-min cold treatment (Fig. 4D). After prolonged exposure to cold treatment for 30 min, virtually no kinetochore microtubules were intact (Fig. 4E; p < 0.001).
In the Mis13-suppressed cells, the numbers of category 2 and 3 spindles increased with a concurrent decrease of category 1 upon cold treatment for 30 min (Fig. 4E). Interestingly, the number of collapsed spindles (category 4) in Mis13-suppressed cells was virtually unaltered with extended cold treatment. A similar scenario was observed in CENP-E-depleted cells and SKAP-suppressed cells, suggesting that the Ndc80 complex bears direct microtubule binding and potent stabilization activity, consistent with the literature (4, 18, 19).
If SKAP regulates the dynamic interaction of spindle microtubule with kinetochore core, suppression of SKAP would reduce the rate of spindle microtubule plus-end dynamics. To test this hypothesis, we sought to examine the microtubule dynamics using photoactivated GFP-tubulin to follow the growth of microtubule plus-ends in the various experimental conditions because the rates of pole-ward microtubule flux and tubulin turnover at plus-ends are usually measured to monitor the dynamics of kinetochore-attached spindle microtubules. Aliquots of HeLa cells were co-transfected with Cherry-H2B and PA-GFP-tubulin, to allow us to simultaneously follow mitotic chromosome and the respective spindle microtubule.
To monitor the rate of pole-ward microtubule flux, the spindle microtubules near the kinetochore were flushed with UV light to activate GFP and to allow tracking fluorescence-activated GFP-tubulin signal to be monitored (27, 32, 33); to monitor tubulin turnover and to describe tubulin dynamics at microtubule plus-ends, the time-dependent decrease in fluorescence intensity of the activated region was quantified and calculated (expressed as the half-life, t½). The pole-ward flux in either SKAP-depleted or Mis13-suppressed cells was much slower compared with scramble siRNA-transfected cells (0.86 ± 0.27 μm/min, Fig. 4H; *, p < 0.01), suggesting that SKAP promotes microtubule plus-end dynamics. Consistent with this, the measurement demonstrated that the half-time for tubulin turnover at the plus-ends became slower in SKAP-depleted cells (Fig. 4F). The t½ for control transfected cells is 35.3 ± 2.7 s, whereas t½ for SKAP-suppressed and Mis13-suppressed cells is 21.7 ± 2.6 and 17.3 ± 2.9 s, respectively. Statistical analyses of normalized fluorescence intensity over time after photoactivating kinetochore microtubule from the aforementioned three groups support the notion that SKAP stabilizes kinetochore microtubule attachment (Fig. 4G; n = 15 cells). Thus, we concluded that SKAP regulates spindle microtubule plus-end dynamics.
SKAP Orchestrates Kinetochore Oscillation and Accurate Alignment at the Equator
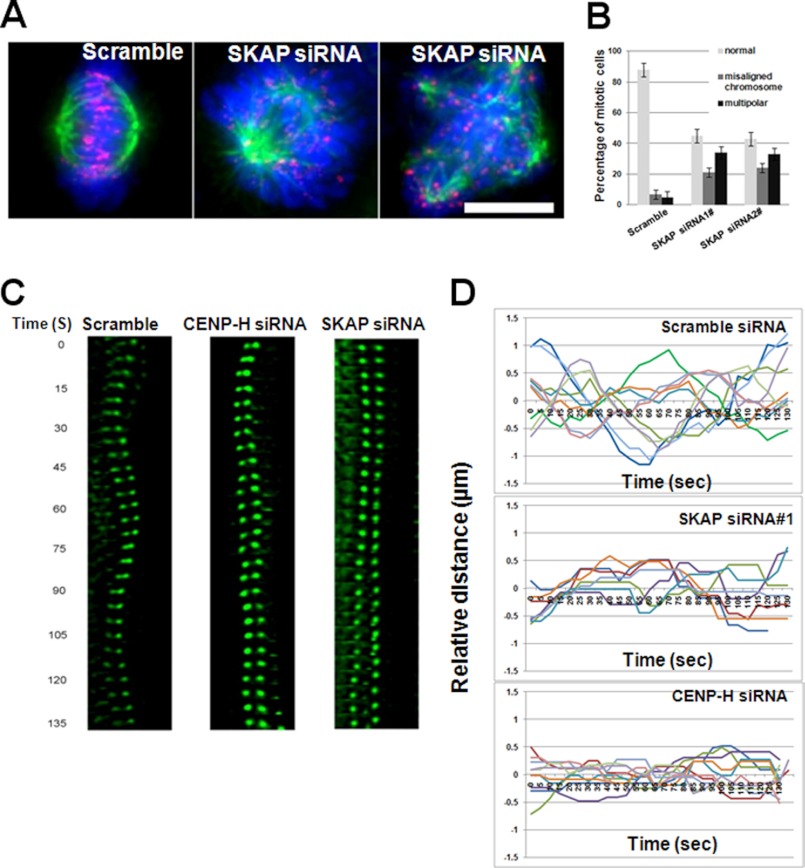
Accurate chromosome alignment is required for equal segregation of sister chromatids in anaphase. To achieve chromosome alignment, bi-oriented paired sister kinetochores generate force to drag chromosomes through regulation of attached microtubule plus-end dynamics. Kinetochore-binding microtubules processively alternate their state between growth and shrinkage, inducing regular kinetochore oscillation along the spindle axis. Consistent with recent publications (10, 12–14), knocking down SKAP by two independent siRNAs caused severe mitotic defects and arrest due to the failure to achieve metaphase chromosome alignment and satisfaction of the spindle assembling checkpoint. The ratio of multipolar spindle is also significantly higher in SKAP siRNA-transfected cells than that of control siRNA-transfected cells based on statistic analyses (Fig. 5, A and B; p < 0.01). This finding, to some extent, can be explained by the above mentioned fact that SKAP does not contribute to the formation of stable kinetochore-microtubule attachment but is involved in the regulation of dynamics of attached spindle microtubules. A high temporal resolution real-time examination of chromosome/kinetochore behavior in a small time window approximating metaphase is useful to illustrate this issue. In HeLa cells, GFP-CENP-B was overexpressed to mark kinetochores, and the GFP dots were imaged at high temporal resolution. In control cells, paired sister kinetochores gave an oscillated curve (Fig. 5C). In SKAP-depleted cells, paired sister kinetochores resulted in a flat curve, and as a positive control, CENP-H-depleted cells were analyzed in the same way, and the resulting curve suggested that loss of CENP-H disrupts sister kinetochore oscillations, which is consistent with a recent report (24) (Fig. 5C). To present these data in a vivid way, the motion path was portrayed, and the distance of sister kinetochores was plotted to the metaphase plate over time for these three groups (Fig. 5D). Those plots summarized from more than 30 cells show that suppression of SKAP minimizes the chromosome oscillatory ability. Those studies support the notion that SKAP constitutes a dynamic link between kinetochore core and spindle microtubule plus-ends.
FIGURE 5.
SKAP is required for accurate sister kinetochore oscillation. A, depletion of SKAP results in chromosome alignment errors. Immunofluorescence staining of control and SKAP siRNA-treated cells was conducted for ACA (red), tubulin (green), and DNA (blue). Scale bar, 10 μm. B, statistical analyses of phenotypes of SKAP-suppressed cells using two different siRNAs. C, HeLa cells expressing GFP-CENP-B were imaged 36 h after transfection of the indicated siRNAs (scramble, SKAP, and CENP-H); time is given in seconds. Paired sister kinetochores are color-coded to illustrate their dynamics. D, motion paths of color-coded sister kinetochores to the metaphase plate under various experimental conditions (transfection of scramble, CENP-H, and SKAP siRNAs individually) were traced during the given time. A kinetochore oscillatory kymograph is presented over time (seconds).
SKAP Is a Novel Microtubule Plus-end Tracking Protein Essential for Chromosome Dynamics
Recent Ndc80 complex crystal structure suggests that the Ndc80/Nuf2 head binds to microtubules at the interdimer interface at an angle of 60° (34). It has remained elusive how kinetochore core KMN complex links to dynamic kinetochore microtubule plus-ends as this dynamic interface modulates the assembly activities of K-fibers and facilitates motion along the kinetochore microtubules.
To examine whether SKAP could constitute an interface between the dynamic plus-end and KMN, we carried out computational analysis of SKAP for structural motifs and functional modules. Interestingly, SKAP contains a conserved SXLP motif within a basic Pro/Ser-rich domain at its N terminus (14), suggesting that SKAP bears EB1 binding activity. To test this possibility, we carried out a pull-down assay and validated that SKAP indeed binds to EB1 in vitro (14). Our immunoprecipitation assay further demonstrates that EB1 forms a cognate complex with SKAP in HeLa cells (14). If SKAP is an EB1-binding protein, SKAP should track microtubule plus-ends in an EB1-dependent manner.
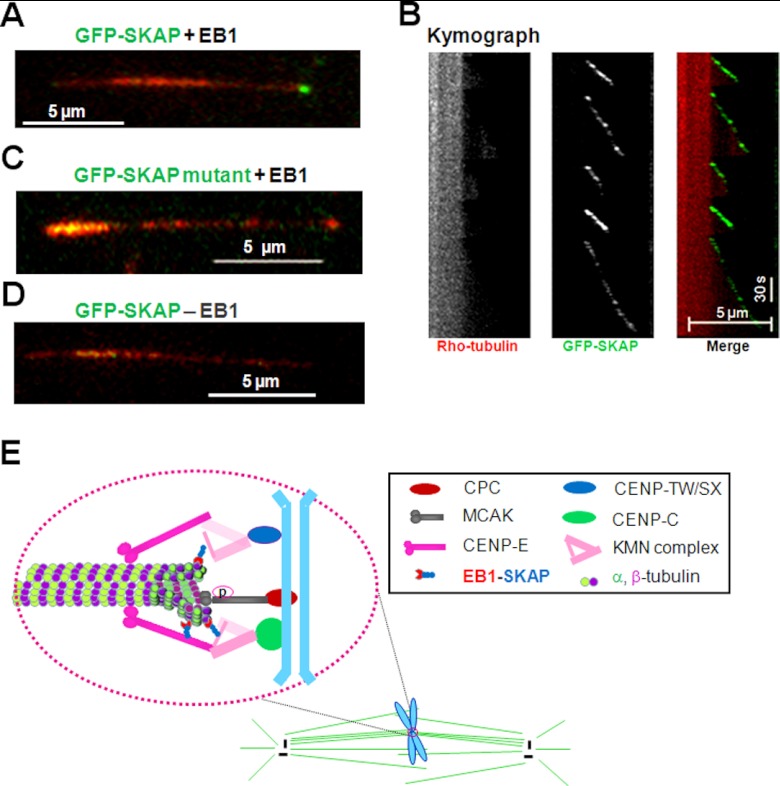
To determine if the SXLP motif of SKAP mediates the plus-end tracking activity on microtubules, a TIRF-based microscopic assay was established as illustrated in Xia et al. (40). As shown in Fig. 5A, in the presence of EB1, GFP-SKAP tracked the growing ends of MTs (Fig. 6A). Analyses of a kymograph of the SKAP tracking behavior reveals that SKAP rides only on the tip of growing microtubules but dissociates from shrinking microtubule (Fig. 6B). In contrast, almost no plus-end tracking was observed when the LP motif of SKAP was mutated to asparagines (Fig. 6C). These results indicated that the SXLP motif is responsible for linking SKAP to growing microtubule plus-ends in an EB1-dependent manner because SKAP failed to track microtubule plus-end when EB1 was absent. Thus, we conclude that SKAP is a novel EB1-dependent microtubule plus-end tracking protein that functions in chromosome dynamics during mitosis.
FIGURE 6.
SKAP constitutes a dynamic interface between KMN and growing microtubule plus-ends. A, TIRFM analysis shows that SKAP tracks microtubule plus-ends. The growing microtubule plus-end tracking of SKAP depends on EB1 in vitro. B, the corresponding kymograph shows that SKAP orchestrates the microtubule dynamics because SKAP is specifically located in the growing ends. C, validation of plus-end tracking of SKAP depends on its EB1-binding motif SXLP because mutation of LP into NN abolishes its tracking and end binding activity in vitro. D, validation of plus-end tracking of SKAP depends on EB1 because the absence of EB1 fails to support SKAP tracking activity in vitro. E, the faithful progression of mitosis is orchestrated by dynamic interaction between spindle microtubules and the kinetochore; it ensures the inheritance of genetic materials. The conserved KMN network, including KNL-1, Mis12 complex, and Ndc80 complex, constitutes the core microtubule binding site of the kinetochore. The CENP-C·CENP-T complex helps to target KMN to the centromere. The direct binding of SKAP to Mis13 and plus-end tracking protein EB1 links the outer kinetochore to microtubule plus-ends. Together with some other proteins, such as CENP-E, CLASP1, and MCAK, SKAP modulates kinetochore-microtubule attachment, sister kinetochore oscillations, and microtubule dynamics and regulates spindle plasticity.
DISCUSSION
The kinetochore is a complex molecular machine to power chromosome movements and orchestrates the checkpoint signaling during cell division cycle progression. Proper alignment and segregation of sister chromatids during mitosis is the most important event in eukaryotic cells; it depends on the dynamic interaction between kinetochore and spindle microtubules. To build effective kinetochore-microtubule attachment, it must satisfy two criteria: one is to maintain the dynamic flux of kinetochore microtubules, and the other is to secure the dynamic link between kinetochore and microtubules. Numerous microtubule-associated proteins play very important roles here. One such typical example is the KMN network proteins, which are composed of three components: Mis12 complex, KNL-1, and Ndc80 complex, all of which together form the major structure platform of the outer kinetochore and the core microtubule binding site. This is crucial for kinetochore assembly and chromosome segregation. Recently, it has been reported that kinetochore-localized microtubule plus-end proteins, such as Clasp1/2, Clip170, and Lis1, form a new class of regulator governing kinetochore-microtubule attachment and microtubule dynamics (3, 5). Our studies revealed that SKAP interacts directly with Mis13 through its C-terminal region and interacts with EB1 via its N-terminal SXLP motif, which adds SKAP to this list of dynamic regulators. Because Mis13 is a component of the Mis12 complex, the direct binding of SKAP to Mis12 complex provides a new link between the structural determinant of kinetochore core and dynamic microtubule plus-ends.
Our present study demonstrates that the N-terminal region of SKAP associates with spindle microtubules, although it is not sufficient to localize to the kinetochore. It implies that the function of SKAP can be separated into two parts according to its secondary structure characteristics. The C-terminal region of SKAP is responsible for its kinetochore localization, and the N-terminal region orchestrates the microtubule binding activity and functions as a regulator of microtubule plus-end dynamics.
Consistent with previous publications (e.g. see Ref. 14), SKAP participates in faithful mitotic progression. Therefore, we further investigated its role in mediating kinetochore-microtubule attachment and microtubule plus-end dynamics. We found that loss of SKAP causes severe chromosome alignment defects but still maintains a large proportion of the stable kinetochore-microtubule attachment. This is because the number of cold-stable K-fibers is minimally affected during depletion of SKAP (Fig. 4A). However, we found that loss of SKAP dramatically perturbs the normal sister kinetochore oscillations, which indicates that suppression of SKAP down-regulates microtubule plus-end dynamics. Moreover, we demonstrated that loss of SKAP causes a decreased pole-ward flux velocity of spindle microtubules. All of these findings suggest that instead of ensuring stable kinetochore-microtubule attachment, SKAP acts as a critical regulator for microtubule plus-end dynamics during cell division.
It was recently reported that Astrin and SKAP form a complex responsible for recruiting CLASP1 and Kif2b, respectively (12, 35). Our study is consistent with a recent report regarding the role of the Astrin·SKAP complex in chromosome alignment (13). However, the currently available evidence provides little well defined information regarding the molecular basis underlying SKAP localization to kinetochore and its precise role in the dynamic interface of kinetochore-microtubule. Given the fact that both CLASP1 and Kif2b contribute to the dynamics of kinetochore microtubule plus-ends, it would be of great interest to delineate how SKAP interacts with Astrin, CLASP1, and Kif2b in mitotic chromosome dynamics. Global quantification of protein expression indicates that EB1 is significantly more abundant than any of the plus-end tracking proteins containing the SXLP motif (36), which raises an interesting question as to whether multiple SXLP motif-containing proteins, such as CLASP1, Kif2b, and SKAP, function synergistically at the same kinetochore microtubule or a different microtubule of the same kinetochore. It is also possible that SKAP may participate in several protein complexes at the kinetochore, which cooperate in regulating kinetochore microtubule dynamics and plasticity. The recent advances in superresolution imaging and single molecule studies also provide insights into the molecular mechanisms underlying kinetochore interaction with the spindle microtubule plus-ends of spindle microtubules (e.g. see Refs. 37 and 38). Those techniques allow real-time tracking of the movements of individual molecules and open exciting new paths to delineate the spatiotemporal dynamics of kinetochore-microtubule interactions.
The kinetochore machinery is highly dynamic, both in the remodeling of a subset structure and turnover of its components during cell division cycle. The kinetochore plasticity and macromolecular assembly dynamics are orchestrated by cell cycle machinery through protein covalent modifications, such as phosphorylation, acetylation, and methylation (39, 40, 41). It will be very challenging and exciting to visualize and illustrate the precise mechanism of action of the aforementioned protein modifications in orchestrating dynamic kinetochore assembly at the individual molecular level relative to the global consequences of chromosome stability in cell division.
In summary, we show that SKAP interacts with Mis13 and that this interaction is essential for temporal regulation of kinetochore microtubule dynamics. Our present study refines the current working model of kinetochore molecular function. As illustrated in Fig. 6E, the KMN network, including the Ndc80 complex, KNL1, and the Mis12 complex, forms the core kinetochore-microtubule attachment site at outer the kinetochore. The KMN complex anchors to the centromere via interaction with CENP-C and CENP-T complex (42, 43). SKAP links the outer kinetochore KMN via its interaction with Mis13 to microtubule plus-ends via plus-end tracking protein EB1 during chromosome congression to the equator. It is likely that SKAP forms other functional protein complexes, such as CENP-E, with which SKAP cooperates to ensure the stability of kinetochore microtubules via lateral interactions. To facilitate microtubule growth, MCAK depolymerase activity is suppressed by phosphorylation. Further molecular delineation of SKAP function and regulation will uncover the underlying regulatory basis for temporal control of centromere plasticity during mitosis and the defective processes underlying aneuploidy.
This work was supported, in whole or in part, by National Institutes of Health Grants DK56292, CA164133, G12RR03034, UL1 RR025008, and CA132389. This work was also supported by Chinese 973 Project Grants 2010CB912103, 2012CB917204, 2012CB945002, and 2002CB713700; Chinese Academy of Science Grant KSCX2-YW-H-10; Anhui Province Key Project Grant 08040102005; International Collaboration Grant 2009DFA31010; Chinese Natural Science Foundation Grants 90508002, 91129714, 31071184, 81270466, 90913016, MOE20113402130010, Chinese Postdoctoral Fellowship 2012M510210; and the Fundamental Research Funds for Central Universities (WK2060190018 and WK2340000021).
X. Wang and Y. Huang, unpublished observation.
- BD
- DNA-binding domain
- AD
- activation domain
- NT
- N-terminal
- CT
- C-terminal
- MBP
- maltose-binding protein
- GMP-CPP
- Guanosine-5′-[(α,β)-methyleno]triphosphate.
REFERENCES
- 1. Welburn J. P., Cheeseman I. M. (2008) Toward a molecular structure of the eukaryotic kinetochore. Dev. Cell 15, 645–655 [DOI] [PubMed] [Google Scholar]
- 2. Musacchio A., Salmon E. D. (2007) The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 3. Cheeseman I. M., Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 4. Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 [DOI] [PubMed] [Google Scholar]
- 5. Maiato H., DeLuca J., Salmon E. D., Earnshaw W. C. (2004) The dynamic kinetochore-microtubule interface. J. Cell Sci. 117, 5461–5477 [DOI] [PubMed] [Google Scholar]
- 6. Yen T. J., Li G., Schaar B. T., Szilak I., Cleveland D. W. (1992) CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 359, 536–539 [DOI] [PubMed] [Google Scholar]
- 7. Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 8. McEwen B. F., Chan G. K., Zubrowski B., Savoian M. S., Sauer M. T., Yen T. J. (2001) CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell 12, 2776–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kapoor T. M., Lampson M. A., Hergert P., Cameron L., Cimini D., Salmon E. D., McEwen B. F., Khodjakov A. (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311, 388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang L., Seki A., Fang G. (2009) SKAP associates with kinetochores and promotes the metaphase-to-anaphase transition. Cell Cycle 8, 2819–2827 [DOI] [PubMed] [Google Scholar]
- 11. Thein K. H., Kleylein-Sohn J., Nigg E. A., Gruneberg U. (2007) Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 178, 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmidt J. C., Kiyomitsu T., Hori T., Backer C. B., Fukagawa T., Cheeseman I. M. (2010) Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J. Cell Biol. 191, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunsch A. K., Linnane E., Barr F. A., Gruneberg U. (2011) The Astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol. 192, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y., Wang W., Yao P., Wang X., Liu X., Zhuang X., Yan F., Zhou J., Du J., Ward T., Zou H., Zhang J., Fang G., Ding X., Dou Z., Yao X. (2012) CENP-E kinesin interacts with SKAP protein to orchestrate accurate chromosome segregation in mitosis. J. Biol. Chem. 287, 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu D., Ding X., Du J., Cai X., Huang Y., Ward T., Shaw A., Yang Y., Hu R., Jin C., Yao X. (2007) Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J. Biol. Chem. 282, 21415–21424 [DOI] [PubMed] [Google Scholar]
- 16. Zhu M., Wang F., Yan F., Yao P. Y., Du J., Gao X., Wang X., Wu Q., Ward T., Li J., Kioko S., Hu R., Xie W., Ding X., Yao X. (2008) Septin 7 interacts with centromere-associated protein E and is required for its kinetochore localization. J. Biol. Chem. 283, 18916–18925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H., Hu X., Ding X., Dou Z., Yang Z., Shaw A. W., Teng M., Cleveland D. W., Goldberg M. L., Niu L., Yao X. (2004) Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279, 54590–54598 [DOI] [PubMed] [Google Scholar]
- 18. Yang Y., Wu F., Ward T., Yan F., Wu Q., Wang Z., McGlothen T., Peng W., You T., Sun M., Cui T., Hu R., Dou Z., Zhu J., Xie W., Rao Z., Ding X., Yao X. (2008) Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 283, 26726–26736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kline S. L., Cheeseman I. M., Hori T., Fukagawa T., Desai A. (2006) The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173, 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding X., Yan F., Yao P., Yang Z., Wan W., Wang X., Liu J., Gao X., Abrieu A., Zhu T., Zhang J., Dou Z., Yao X. (2010) Probing CENP-E function in chromosome dynamics using small molecule inhibitor syntelin. Cell Res. 20, 1386–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., Yao X. (2004) NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J. Biol. Chem. 279, 20049–20057 [DOI] [PubMed] [Google Scholar]
- 22. Lampson M. A., Kapoor T. M. (2005) The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7, 93–98 [DOI] [PubMed] [Google Scholar]
- 23. Dou Z., von Schubert C., Körner R., Santamaria A., Elowe S., Nigg E. A. (2011) Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PLoS One 6, e18793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amaro A. C., Samora C. P., Holtackers R., Wang E., Kingston I. J., Alonso M., Lampson M., McAinsh A. D., Meraldi P. (2010) Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J., Wang Z., Jiang K., Zhang L., Zhao L., Hua S., Yan F., Yang Y., Wang D., Fu C., Ding X., Guo Z., Yao X. (2009) PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J. Biol. Chem. 284, 23059–23071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhai Y., Borisy G. G. (1994) Quantitative determination of the proportion of microtubule polymer present during the mitosis-interphase transition. J. Cell Sci. 107, 881–890 [DOI] [PubMed] [Google Scholar]
- 27. Cimini D., Wan X., Hirel C. B., Salmon E. D. (2006) Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16, 1711–1718 [DOI] [PubMed] [Google Scholar]
- 28. Bakhoum S. F., Genovese G., Compton D. A. (2009) Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 19, 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dixit R., Barnett B., Lazarus J. E., Tokito M., Goldman Y. E., Holzbaur E. L. (2009) Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. U.S.A. 106, 492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dixit R., Ross J. L. (2010) Studying plus-end tracking at single molecule resolution using TIRF microscopy. Methods Cell Biol. 95, 543–554 [DOI] [PubMed] [Google Scholar]
- 31. Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., 3rd, Oegema K., Desai A. (2004) A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18, 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitchison T. J. (1989) Polewards microtubule flux in the mitotic spindle. Evidence from photoactivation of fluorescence. J. Cell Biol. 109, 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhai Y., Kronebusch P. J., Borisy G. G. (1995) Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 131, 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J. G., De Wulf P., Salek M., Rappsilber J., Moores C. A., Salmon E. D., Musacchio A. (2008) Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Manning A. L., Bakhoum S. F., Maffini S., Correia-Melo C., Maiato H., Compton D. A. (2010) CLASP1, Astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 29, 3531–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 37. McIntosh J. R., Grishchuk E. L., Morphew M. K., Efremov A. K., Zhudenkov K., Volkov V. A., Cheeseman I. M., Desai A., Mastronarde D. N., Ataullakhanov F. I. (2008) Fibrils connect microtubule tips with kinetochores. A mechanism to couple tubulin dynamics to chromosome motion. Cell 135, 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao X., Fang G. (2009) Visualization and orchestration of the dynamic molecular society in cells. Cell Res. 19, 152–155 [DOI] [PubMed] [Google Scholar]
- 39. Chu L., Zhu T., Liu X., Yu R., Bacanamwo M., Dou Z., Chu Y., Zou H., Gibbons G. H., Wang D., Ding X., Yao X. (2012) SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J. Mol. Cell Biol. 4, 331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., Yao X. (2012) EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16564–16569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chu Y., Yao P. Y., Wang W., Wang D., Wang Z., Zhang L., Huang Y., Ke Y., Ding X., Yao X. (2011) Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell. Biol. 3, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishino T., Takeuchi K., Gascoigne K. E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I. M., Fukagawa T. (2012) CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148, 487–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tao Y., Jin C., Li X., Qi S., Chu L., Niu L., Yao X., Teng M. (2012) The structure of the FANCM-MHF complex reveals physical features for functional assembly. Nat. Commun. 3, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]