Background: For Kv channels, only crystal structures for the open state are available.
Results: Using LRET, we determined the movement of the S4-S5 linker during gating in KvAP channels.
Conclusion: A small displacement of the S6 by only 4 Å is sufficient for closing of the Kv channels.
Significance: We provide the first Kv channel closed state model based on cytosolic restraints.
Keywords: Electrophysiology, Fluorescence Resonance Energy Transfer (FRET), Gating, Molecular Dynamics, Potassium Channels, LRET, Internal Gate, Lipid Regulation
Abstract
Voltage-gated ion channels are responsible for the generation of action potentials in our nervous system. Conformational rearrangements in their voltage sensor domains in response to changes of the membrane potential control pore opening and thus ion conduction. Crystal structures of the open channel in combination with a wealth of biophysical data and molecular dynamics simulations led to a consensus on the voltage sensor movement. However, the coupling between voltage sensor movement and pore opening, the electromechanical coupling, occurs at the cytosolic face of the channel, from where no structural information is available yet. In particular, the question how far the cytosolic pore gate has to close to prevent ion conduction remains controversial. In cells, spectroscopic methods are hindered because labeling of internal sites remains difficult, whereas liposomes or detergent solutions containing purified ion channels lack voltage control. Here, to overcome these problems, we controlled the state of the channel by varying the lipid environment. This way, we directly measured the position of the S4-S5 linker in both the open and the closed state of a prokaryotic Kv channel (KvAP) in a lipid environment using Lanthanide-based resonance energy transfer. We were able to reconstruct the movement of the covalent link between the voltage sensor and the pore domain and used this information as restraints for molecular dynamics simulations of the closed state structure. We found that a small decrease of the pore radius of about 3–4 Å is sufficient to prevent ion permeation through the pore.
Introduction
Voltage-gated potassium channels (Kv) play a central role in the physiology of all excitable cells and are particularly involved in the repolarization of action potentials in neuronal and muscle cells (1). Defects in Kv channels may cause severe conditions such as cardiac arrhythmias, epilepsy, muscular ataxia, or congenital deafness (2). Function of Kv channels is tightly regulated through communication between its two main components: the voltage sensor domain and the pore domain. Kv channels are symmetrical tetramers formed by four (mostly) identical monomers, each consisting of six transmembrane helices (S1–S6). The first four α-helices of each monomer (S1–S4) yield to the formation of four peripheral voltage sensor domains, whereas the collective assembly of the last helices (S5 and S6) constitutes the central pore. Upon depolarization of the cell membrane, the S4 segments undergo a conformational change driven by the positively charged arginines in the electric field (3–6). This conformational change leads to pore opening in a process called electromechanical coupling.
Electromechanical coupling has been intensively studied in recent years (reviewed in Ref. 7). Voltage sensor and pore are covalently linked by the S4-S5 linker. This amphipathic helix also anneals to the C-terminal end of S6 (S6T) of the same and neighboring subunit (8–14). The link to the S6T is essential for the voltage sensor-controlled pore opening, during which the internal gate formed by the S6 is widened. Despite the considerable effort in recent years to study the electromechanical coupling in Kv channels, no structural information is available on the conformational changes on the cytosolic face of the channels, the site of electromechanical coupling. The existing models built using molecular mechanics techniques rely mainly on the open state crystal structure and experimentally defined restraints on the extracellular accessible part of the channel. The problem in obtaining biophysical information from the cytosolic face of the channel is that labeling methods, typically using thiol-reactive chemistry, are not practical for channels expressed in cells or Xenopus oocytes as too many cysteines are available in the cytosol (15). Spectroscopic methods, on the other hand, were impeded by the inability to control the voltage in solution or in small vesicles. Here, we overcame this problem by controlling the state of the channel by varying the lipid composition. It has been described earlier that the voltage dependence of Kv channel opening shifts with differently charged headgroups of the surrounding lipid matrix (16–23). We made use of the positively charged lipid DOTAP3 (1,2-dioleoyl-3-trimethylammonium-propane) to switch the KvAP channel into its resting-closed position. This allowed us to carry out luminescence resonance energy transfer measurements (24–26) in both open and closed conformation. We determined distances of residues all along the S4-S5 linker of KvAP-dimers reconstituted in lipid vesicles to determine the position of the S4-S5 linker in both conformations and reconstructed the movement of the cytosolic face of the channel in molecular dynamics simulations.
EXPERIMENTAL PROCEDURES
Molecular Biology
KvAP coding sequence starting from methionine 14 was inserted in the vector pQE70 (Qiagen) between restriction sites SphI and BglII, with a thrombin cleavage site between the C-terminal hexahistidine tag and the channel. The only endogenous cysteine residue of KvAP was replaced with a serine (C247S) by site-directed mutagenesis (Stratagene). Two KvAP inserts (I and II) were linked together by a stretch of 5 amino acids (QQQQS) to form the dimer. Insert I had a cysteine mutation in the S4-S5 linker, whereas insert II was cysteine-less. The first 8 residues of insert II were deleted.
Purification of KvAP Dimers
KvAP dimers were expressed in Escherichia coli M15 strain cell cultures (Qiagen) grown in LB medium with 100 μg/ml ampicillin and 25 μg/ml kanamycin. At A600 = 0.6, channel expression was induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside. At this time, 50% glycerol and 10 mm BaCl2 were also added to the medium, and the temperature was lowered to 25 °C. After 3–5 h, bacterial cells were harvested and lysed by pressure at 15,000 p.s.i. with an EmulsiFlex-C5 (Avestin). 1 mm PMSF was added to prevent protein cleavage. The membranes were isolated by centrifugation at 200,000 × g and solubilized with 20% decyl-maltoside (Anatrace) for 1 h at 4 °C. The sample was bound to a metal affinity column (Talon Superflow, Clontech) and washed with 10 mm imidazole (Sigma). Sample was maintained in a 200 mm NaCl, 50 mm KCl, 50 mm Hepes, and 0.25% decyl-maltoside, pH 7.4, buffer. The cysteines were reduced with 10 mm tris(2-carboxytheyl)phosphine (Pierce) before elution with 400 mm imidazole. Sample was concentrated (Amicon 30K, Millipore) to 2–5 mg/ml and run on SDS-PAGE to confirm the presence of dimer and absence of monomer by GelCode Blue staining and immunoblotting.
Labeling
For the donor-only condition (DO), sample was mixed overnight at 4 °C with a 5-fold excess (mol:mol) of a terbium chelate complex (LanthaScreen, Invitrogen, or synthesized in-house, based on previous protocols (27, 28)). For the donor and acceptor condition, the terbium chelate complex and an organic fluorophore (Alexa Fluor® 488 C5 maleimide, tetramethylrhodamine-5-maleimide, fluorescein-5-maleimide, all Invitrogen; Atto465M, ATTO-TEC GmbH) were mixed with proportions that allowed a final efficacy of labeling of 1:1. Excess of dyes was removed by successive washes and centrifugations in a concentrator with a cutoff weight of 50 kDa (Amicon, Millipore). Background labeling of KvAP cysteine-less dimer was 10–14 times lower than specific labeling.
Channel Reconstitution in Lipid Vesicles
DPhPC (1,2-diphytanoyl-sn-glycero-3-phosphocholine) or DOTAP (Avanti Polar Lipids) small unilamellar vesicles were prepared by vigorous sonication. The lipids were mixed with the sample in a ratio of ∼1:4 protein:lipid (w/w) at a final lipid concentration of 10 mg/ml, well vortexed (2–5 min), shortly sonicated, and rocked for 1 h at room temperature. To remove the detergent, sample was loaded in a dialysis device (Slide-A-Lyzer MINI 20K MWCO, Pierce) and kept at 4 °C shielded from light. Dialysis buffer (450 mm KCl, 10 mm HEPES at pH 7.4) was exchanged twice a day for at least 5 days.
Electrophysiology Recordings
Pure DPhPC, pure DOTAP (Avanti Polar Lipids), or a 3:4 DPhPC:DOTAP mixture (w/w) were dissolved in decane at 25 mg/ml to form planar lipid bilayers over an aperture (250-μm diameter) in a polymer partition that separated the internal and external solutions (150 mm KCl, 10 mm HEPES, pH 7.4). Vesicles containing the labeled channels were shortly sonicated and fused to the voltage-clamped bilayer. The elicited macroscopic currents were recorded using an Axiovert 1D amplifier (Axon Instruments) and registered using GPatch (UCLA, Department of Anesthesiology). Holding potential was −100 mV, and depolarizing pulses ranged from −100 to +100 mV in 10-mV increments, with a waiting time of 90 s between each pulse to allow channels to recover from inactivation.
LRET Measurements
The setup was based on a Zeiss Axiovert 200 microscope. Light at 337 nm from a pulsed 3-milliwatt N2-laser (Spectra-Physics) was directed in a wide-field illumination onto a drop of vesicles containing the labeled channels on a quartz coverslip. The light from the vesicles was collected using a 1.25 NA 40× glycerol immersion quartz objective (Partec) and filtered with a bandpass emission filter. Light was detected by a photon counter (Laser Components). Before each measurement, the samples were vortexed and centrifuged. Analysis of the lifetime decays was done with an exponential fitting program (MATLAB). Distances were determined using the following relations
where τDO is the time constant of the donor lifetime decay in absence of acceptor (donor-only), and τSE is the time constant decay of the sensitized emission of the acceptor.
The distance between the donor and the acceptor was determined using
where R0 is the distance at which 50% energy transfer occurs. The R0 values for the different pairs of fluorophores were calculated from experimentally determined spectra based on the spectral overlap of the donor and the acceptor.
Molecular Dynamics Simulations
The Kv1.2/2.1 chimera structure (Protein Data Bank (PDB) code 2R9R) was used as a template for model generation. Using the CHARMM27 force field for proteins (29), the initial structure was placed in a generalized born implicit solvent membrane model (30), minimized, and briefly simulated (50 ps) with flat-bottomed harmonic restraints of 10 kcal/mol to mimic the experimental results discussed herein. The resulting “closed” structure was then reconstituted into DPPC bilayers using the CHARMM-GUI web service (31) and equilibrated by gradually releasing protein backbone restraints over 450 ps. A potential of −500 mV was applied to the closed system with NAMD 2.8 (32) and the CHARMM27 and CHARMM 36 (33) force fields for proteins and lipids, respectively. The system was simulated for 100 ns with harmonic restraints applied to linker atoms throughout. Temperature was held constant at 303.15 K with Langevin dynamics, and pressure was held constant using the Langevin Nosé-Hoover method. Water was represented with the TIP3P water model, and high frequency hydrogen vibrations were constrained with the SHAKE algorithm. Aspartate, glutamate, and arginine residues were reparameterized to recently published values (34). Additional harmonic restraints between the α-carbons of Ile-230 and Tyr-267, Ile-230 and Arg-290, and Ile-177 and Arg-290 were added to the closed system in accordance with previously published work (35, 36). To compare our closed state model with the open structure, the Kv1.2/2.1 chimera crystal structure was also simulated in a DPPC bilayer built by the CHARMM-GUI web service. The system was simulated at 0 mV following an identical protocol; however, the system was neither restrained nor reparameterized.
To create a final model of the closed state, one subunit from the closed simulation was used to create a four-fold symmetric model. This tetramer was reinserted in a generalized born implicit membrane and minimized with flat-bottomed harmonic restraints of 50 kcal/mol on the linker atoms, as well as lighter harmonic restraints of 10 kcal/mol along the linker backbone to promote helicity. The protein was again reconstituted into a DPPC bilayer, solvated with TIP3 water molecules, and re-equilibrated with 10 kcal/mol backbone helical restraints applied to the linker and the bottom of S4, as well as the aforementioned flat-bottomed 50 kcal/mol harmonic linker restraints in accordance with our luminescence resonance energy transfer (LRET) measurements.
RESULTS
KvAP Dimers Can Be Gated by the Lipid Environment
LRET is a powerful spectroscopic tool to determine atomic-scale distances within a protein (24–26, 37, 38). Here, we used LRET to measure the position of the S4-S5 linkers of KvAP in the closed and the open state. To do so, a series of cysteine mutations of several positions along the S4-S5 linker (Phe-137–Asp-146) was expressed and purified to allow site-directed labeling with fluorophores attached to a maleimide linker (see “Experimental Procedures”).
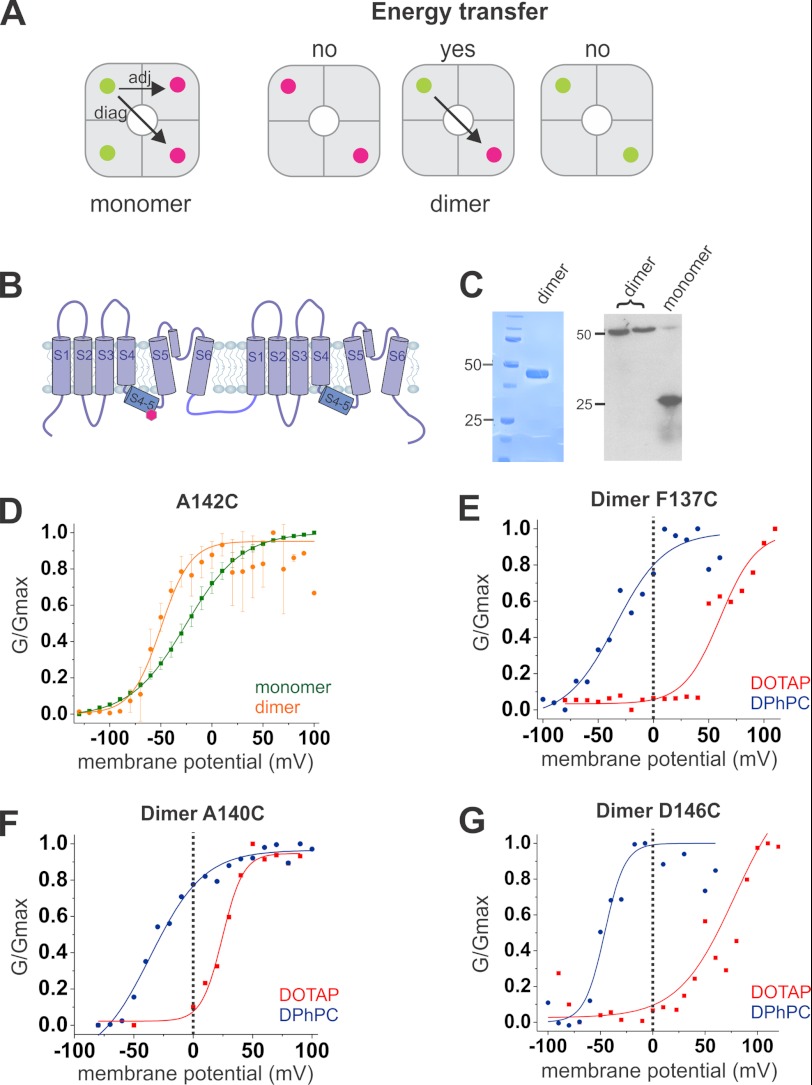
In a homotetrameric protein, and hence four identical binding sites, stoichiometric labeling would give us any combination of donor and acceptor pairs and two different distances (adjacent and diagonal), requiring a complex analysis of the signals (Fig. 1A) (see Ref. 25). To circumvent these difficulties, we constructed a dimer of dimers, where two subunits are linked by their intracellular C- and N-terminal ends (Fig. 1B and see “Experimental Procedures”). Dimer formation was confirmed by SDS-polyacrylamide gel electrophoresis and Western blot analysis (Fig. 1C). Liu et al. (39) demonstrated that such a construct assembles with identical monomers at diagonal positions. A single cysteine per dimer is then inserted by directed mutagenesis into the S4-S5 linker, allowing the specific binding of only two fluorophores per tetramer at identical positions within two opposite monomers (39). The dimer of dimers labeled with a 1:1 ratio of donor and acceptor will thus contain either two identical labels (only donors or acceptors) or one FRET pair (each one donor and one acceptor). Only those proteins that are labeled with a FRET pair will prompt energy transfer and show sensitized emission (acceptor emission upon donor excitation), leading to a single distance per protein (Fig. 1A). After purification and specific labeling, KvAP dimer channels are functionally reconstituted in lipid vesicles (Fig. 1D and see “Experimental Procedures”).
FIGURE 1.
KvAP dimer and response to lipid environment changes. A, a tetramer formed of monomers leads to four possible fluorophore binding sites, with the possibility of two energy transfer distances between the donor (green) and the acceptor (fuchsia): a smaller distance between two adjacent monomers (adj) and a larger one between two opposite monomers (diag). Use of a dimer of dimers leads to only two binding sites, located on opposite monomers. Only one combination of fluorophores will lead to an LRET signal; no energy transfer will occur between two donors or two acceptors. B, the dimer of KvAP is formed by two monomers linked together by an intracellular loop. C, dimer expression was confirmed by SDS-PAGE (left) and immunoblotting (right). D–G, vesicles containing the channels are fused to a planar lipid bilayer. D, normalized conductance in function of the potential of the dimer (orange) confirmed that it retained its voltage dependence in a similar way as the monomer (green). E–G, the midpoint activation potential of a N-terminal position (E), a middle position (F), and a C-terminal position (G) of the S4-S5 linkers in the DOTAP environment (red) is shifted to more positive potentials, and the channels are mostly closed at 0 mV, whereas in a phospholipid environment (DPhPC, blue), they are mostly opened at 0 mV.
The investigation of voltage-gated ion channels with spectroscopic methods has always been hindered by the lack of control over the membrane potential. Also, our LRET measurements are performed on KvAP channels reconstituted into liposomes in the absence of a membrane potential. Recently, more and more evidence indicated that the lipid environment has an important influence on channel behavior (17, 40–42). In particular, Schmidt et al. (18–20) showed that the state of the KvAP channel is dependent on the lipid environment. In typical phospholipid mixtures, the channels are open in the absence of a membrane potential. Adding increasing amounts of the cationic DOTAP to the membrane, however, shifts the voltage dependence to more positive potentials such that the open probability at 0 mV approaches zero at a DOTAP content of 67% (19). Here, we made use of this lipid dependence to control the state of the channel during our LRET recordings. The channels were reconstituted into vesicles composed of DPhPC or DOTAP where KvAP channels reside in the open and closed state, respectively.
Due to the key role that the S4-S5 linker fulfills in electromechanical coupling, mutations in this region might influence channel function. Thus, we had to ensure that the labeled mutants were functional and that they followed the correct voltage dependence under both conditions (Fig. 1, D–G). To this end, the reconstituted channels were fused into planar lipid bilayers formed of DPhPC and DOTAP, respectively. All dimer mutants, except A144C, expressed functionally with no significant differences when compared with wild type KvAP. A145C did not show a clear pattern in the Western blot analysis, and the cysteine in I141C was weakly accessible. Both mutants were thus excluded from further analysis. As predicted, at 0 mV, all other mutants were mostly open and closed when reconstituted in DPhPC and DOTAP, respectively (Fig. 1, E–G). Channels in pure DOTAP vesicles fused in a pure DOTAP bilayer were functional, but the bilayer was very unstable. For this reason, bilayers of mixed lipids (25% DPhPC + 75% DOTAP) were used with DOTAP vesicles, still leading to closed channels at 0 mV. Interestingly, the nature of the bilayer seemed to have less influence than the vesicle composition, suggesting a strong interaction between the voltage sensor and the lipids that it came into contact with first.
LRET Reveals Position of S4-S5 Linker in Both Closed and Open State
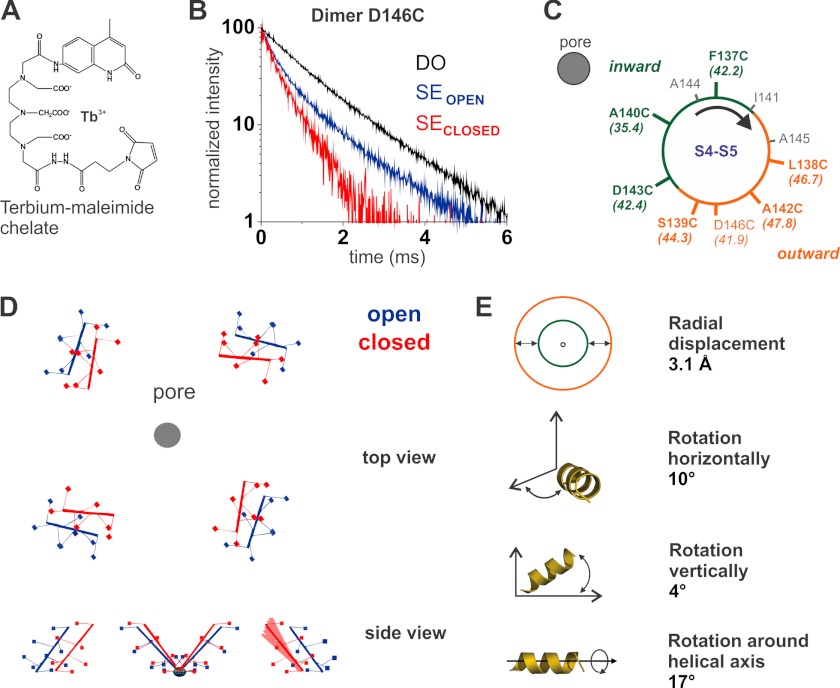
Identical samples of labeled KvAP dimer mutants reconstituted in lipid vesicles were used for the bilayer experiments and the LRET measurements (Fig. 2). A lanthanide (Tb3+) complex (Fig. 2A) was used as the donor whose characteristic long lifetime in the excited state (∼1 ms) allowed detecting its temporal decay with high accuracy. More importantly, its fully isotropic emission evaded any inaccuracy due to the orientation between donor and acceptor typical for fluorescence-based RET (24). The comparison of the lifetimes in the absence and presence of the acceptor reflects the efficiency of the energy transfer (see “Experimental Procedures”); if the acceptor is very close to the donor (closed state), then the energy transfer will be strong and the lifetime short and vice versa (Fig. 2B). We obtained cross-pore distances for seven positions along the S4-S5 linker in the open and closed position from LRET measurements on dimers reconstituted in DPhPC or in DOTAP, respectively (Table 1). The distances varied between 4 and 10 Å for each position between the different lipid conditions, i.e. between the closed and the open state. The open state distances, except S139C, correlated well with the distances from the corresponding positions in the Kv1.2/2.1 chimera x-ray structure (PDB: 2R9R, Ref. 43), indicating that our data reflect well the native conformation of the S4-S5 linker but also that the structures of Kv1.2/2.1 and KvAP in this region coincide (Table 1). We ensured that the distances were independent of the specific donor-acceptor pair by assessing distances with different acceptor fluorophores for the various positions (Table 2).
FIGURE 2.
S4-S5 linker movement during gating determined by LRET measurements. A, structure of the lanthanide complex used as donor. B, acceptor-sensitized emission (SE) is shown for the D146C dimer reconstituted in DOTAP (red) and in DPhPC (blue). τSE = 285 μs for the closed state (red), and τSE = 602 μs for the open state (blue). In black, the control of donor in the absence of acceptor is shown (τDO = 1.17 ms). C, residues of the S4-S5 linker of KvAP are projected on a helical wheel with an α-helical periodicity. The smallest experimental distances are from the residues on the inward side of the helix (green) with respect to the pore, and the largest ones, form the outward side (orange). D, the top view of the movement of the S4-S5 linker shows the radial displacement and the horizontal rotation it undergoes from the open state (blue) to the closed state (red). The side view shows the vertical rotation. One subunit of the closed state is superposed with all possible helix orientations where the residues remain within the error margins of the experimental distances. From these variations, we can estimate the error for each parameter: radial distance to the pore, ± 0.1 Å; angle with respect to the helix axis, ± 4°; deviation from horizontal plane, ± 4°; angle within horizontal plane, ± 1°. E, the three-dimensional movement of the S4-S5 linker is decomposed into four components.
TABLE 1.
Distances (Å) between opposite S4-S5 linkers
Cross-pore distances (Å) for seven residues along the S4-S5 linker of a KvAP dimer were determined by LRET in the open and the closed state. Data are the average of at least n = 5 for each distance with S.D. as the error. Distances for the corresponding positions of Kv1.2/2.1 chimera were measured from the crystal structure (2R9R) in the open state and served as a comparison reference (in italics).
| Position | Closed | Open | Kv 1.2/2.1 | Position |
|---|---|---|---|---|
| F137C | 33.4 ± 2.7 | 42.2 ± 0.3 | 42.9 | G309C |
| L138C | 42.8 ± 0.9 | 46.7 ± 0.6 | 49.8 | L310C |
| S139C | 40.2 ± 0.4 | 44.3 ± 1.4 | 52.4 | Q311C |
| A140C | 30.1 ± 0.5 | 35.4 ± 0.1 | 38.0 | I312C |
| A142C | 37.9 ± 1.5 | 47.8 ± 0.3 | 48.3 | G314C |
| D143C | 33.2 ± 1.1 | 42.4 ± 0.1 | 39.8 | L315C |
| D146C | 35.9 ± 0.3 | 41.9 ± 0.6 | 44.4 | K318C |
TABLE 2.
Comparison of distances (Å) obtained with different acceptors
Distances (Å) between two opposite residues were obtained with different pairs of fluorophores without any significant variations. Numbers in parentheses indicate the R0 (Å) of the dyes used. The table shows an example of one experiment for four different positions in the S4-S5 linker of KvAP.
| Residue | Dye |
|||
|---|---|---|---|---|
| ATTO 465 (30.6) | Fluorescein (42.7) | Alexa Fluor 488 (43.4) | TMR (50.8) | |
| F137C | 45.3 | 42.6 | ||
| A142C | 47.9 | 47.5 | ||
| D143C | 42.3 | 42.5 | ||
| D146C (O) | 49.3 | 48.5 | 50.6 | |
| D146C (C) | 36.3 | 35.9 | ||
The distances among the different residues along the S4-S5 linker vary significantly but with a periodicity of 3–4 residues. These variations are caused by the helical structure of the S4-S5 linker and its orientation with respect to the pore. When projected onto a helical wheel, we observed that those residues pointing toward the central pore yielded the smallest cross pore distances, and vice versa (Fig. 2C). Following this logic, the residues Phe-137, Ala-140, and Asp-143 are located toward the pore, whereas residues Leu-138, Ser-139, and Ala-142 are oriented outward. The distance determined for the residue Asp-146 does not fit perfectly the helical wheel, possibly because this residue is located at the C-terminal end of the linker, in the transition into the S5 segment. Based on these distance restraints and assuming an ideal α-helical conformation of the linker, we were able to predict the position and orientation of the helix in space relative to the pore in both open and closed state and to reconstruct the movement of the S4-S5 linker during gating (Fig. 2D).
When decomposed into four independent components (Fig. 2E), we observed a radial displacement of 3.1 Å, in combination with rotations of the S4-S5 linker around its own axis (17°), horizontally (10°), i.e. in the membrane plane, and (less marked) vertically (4°). Fig. 2D displays the displacement of the S4-S5 linker during gating. The rotation is consistent with current models postulating a rotation of the S4 helix (44, 45). According to our model, the S4 rotation seems to translate into a rotation of the S4-S5 linker. Also, the rather small shift radially away from the central axis is in agreement with predictions on the movement of the S4 helix (34).4 The channels are closed in the DOTAP environment, indicating that such a small movement is sufficient to allow and block ion conduction. In other words, limited linker displacement can open and close the internal gate of the Kv channels.
Atomistic Model Reveals Closing of the Pore by 3–4 Å Displacement
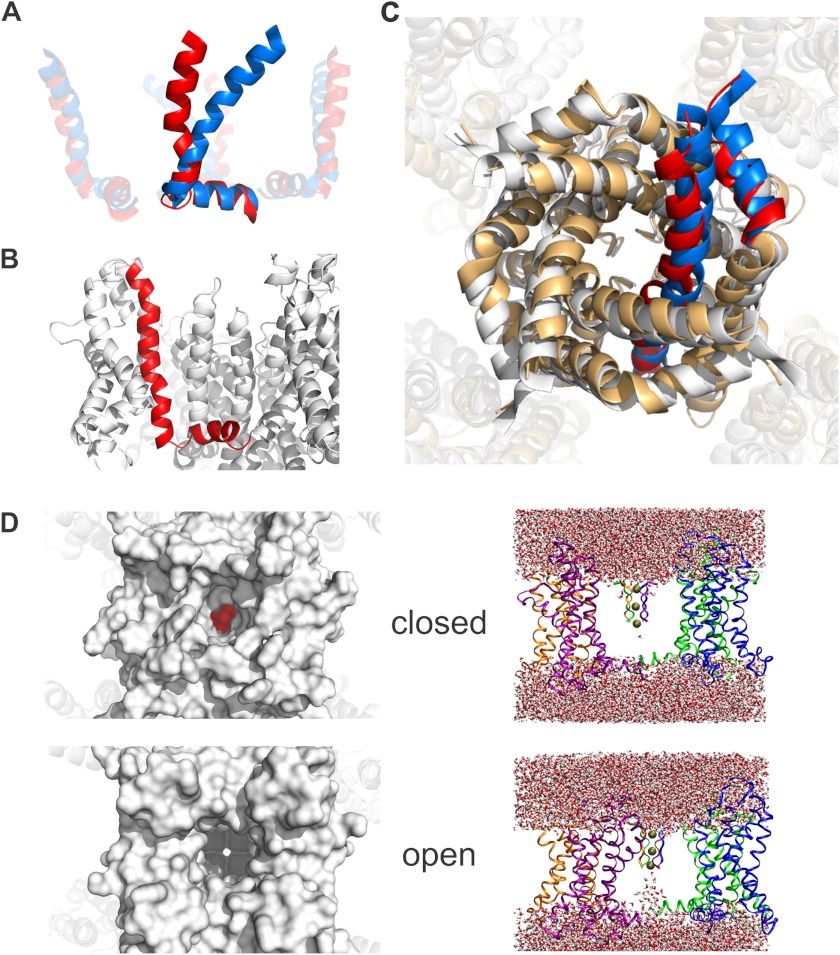
Our “rigid helix” model assumes that the S4-S5 linker is a rigid, perfect α-helix, which does not necessarily have to be the case. In addition, the above model does not yet give any information about S4-S5 linker movements that are invariant in the diameter, i.e. rotations around the central axis of the channel and vertical translations of the entire linker. We also wanted to answer the question as to how the displacement of the S4-S5 linker leads to pore opening and closing. We therefore built a closed state model satisfying our experimentally obtained distances using molecular dynamics simulations. Starting with the open state structure of Kv1.2/2.1 (43), we used our closed state data as restraints and applied an electric field over the membrane. To more rapidly reach the resting state conformation of the voltage sensor domain, we used additional restraints that were previously reported (35, 36) (for details, see “Experimental Procedures”). Under these conditions, the voltage sensor transited from the open state into its resting state and closed the ion-conducting pore (Fig. 3A).
FIGURE 3.
Closed state model satisfying LRET restraints. A, from the open (blue) to the closed (red) state, the S4 helix undergoes a translocation of 9 Å along its axis together with a rotation of 37° relative to the S4-S5 linker and of 24° around its own axis. B, in its resting state, the lower S4 maintains a 310-helical conformation. The S4-S5 linker is slightly bent. C, the 4 Å radial displacement of the S4-S5 linker is transferred to the C-terminal part of the S6, therefore closing the internal gate. White, the open state crystal structure (2R9R); light orange, the closed state model. One subunit is highlighted in blue (open) and red (closed). It shows the small displacement that is sufficient for pore closing. D, the water-filled column is interrupted in the bottom part of the pore when the internal gate is closed in our model (top) in comparison with the open state (bottom). For clarity, only the voltage sensors, linkers, and selectivity filter are shown. On the left, the view into the pore of closed model and open structure are shown. The pore is sealed by the ring of isoleucines Ile-398 (highlighted in red) in the closed model, whereas the selectivity filter is freely accessible in the open crystal structure (2R9R).
During the gating movement, the S4 helix was pulled downward by about 9 Å and rotated by 24° (Fig. 3A). The S4-S5 linker was moved inward, which pushed the S6 into the central pore by about 4 Å (Fig. 3C). As speculated, the S4-S5 linker did not remain rigid but was “bent” slightly by the C-terminal end of S4. The angle between S4 and the linker increased from 67° to 104°, and the horizontal tilt of the S4-S5 linker was close to the prediction above (9°). The S4 had a 310-helical conformation C-terminal to Arg-296 (Fig. 3B).
The movement of the S6 helix was less pronounced than expected previously. Therefore, we verified that the pore was indeed closed. Fig. 3D shows the water molecules in the central pore in the open (bottom) and closed (top) conformation. In the closed conformation, the water column is disrupted at a hydrophobic ring at position Ile-398 just above the PVP motif (Fig. 3D, left; highlighted in red), indicating that the channel is indeed closed and ion conduction is interrupted. Thus, a small radial displacement of only 3–4 Å is sufficient to push the internal gate of Kv channels into a closed state and prevent ion conduction.
DISCUSSION
In this study, we varied the composition of the lipid environment to control the state of Kv channels in the absence of a membrane potential. Using positively charged lipids modified the electric field around the voltage sensor such that the channel remains in its closed state even in the absence of a membrane potential. The lipid-induced closed state is likely to resemble the voltage-induced one; membranes composed of DOTAP have been shown to have a very high surface potential (47), suggesting that it exerts a similar electrostatic effect on the voltage sensor as the electric field. This is further supported by the finding that negatively charged polyunsaturated fatty acids develop the opposite effect (conductance-voltage relation (GV) shifted to more negative potentials), which is mainly of electrostatic nature (21, 22). Zheng et al. (42) also demonstrated that the lipid- and voltage-induced conformations are equivalent.
The use of lipids to control voltage-gated ion channels facilitated the use of spectroscopic methods for this class of membrane proteins. Interestingly, when fusing KvAP from vesicles into planar lipid bilayers, the composition of the lipid vesicles had a stronger influence than the composition of the bilayer itself. This suggests a tight interaction between protein and its immediate lipid environment and may have significant implications on lipid-induced regulation of the channels.
Using LRET, we succeeded in determining the position of the S4-S5 linker in both the open and the closed states with Å resolution and obtained a model for Kv channels in their resting state satisfying these and previous restraints. This is, to our knowledge, the first closed state model based on restraints from the cytosolic face of the channel. To enter the closed state, the S4-S5 linker underwent a radial translation by 3–4 Å. It rotated around its own axis by 17° and tilted 4° vertically and 10° horizontally (Fig. 2E).
We also analyzed how the S4 helix in our model was displaced with respect to the open state structure because S4 helix and S4-S5 linker movements are tightly linked. In an interpolation between the open state crystal structure and our closed state model, the S4 is vertically translocated by 9 Å down along its axis and tilts with respect to the S4-S5 linker from 67° in the open structure to 104° in our resting state model (Fig. 3A). These movements of S4 are accompanied by a rotation around its own axis by 24°. The rotation ensures that the arginines keep toward the lumen of the voltage sensor domain, whereas the translation downward brings the fourth arginine below the Phe-233. The parameters predicted for the S4 movement are consistent with previous models. Values for the vertical displacement of S4 vary between 6 and 10 Å (44, 45) and 15 Å (34). The angle between the S4-S5 linker was proposed to change by 40° from 60° to 100° (45) in accordance with our experimental data.
The C-terminal portion of the S4 maintained a 310-helical conformation starting with the arginine Arg-296, as observed in the crystal structure of the Kv1.2/2.1 chimera (43). The extent of the 310-helix coincided with those observed in the crystal structure and other closed state models (48), despite the proposed transition from a 310- to an α-helix, or an extension of the 310-helix in the resting state (43, 45, 49, 50). The extension of the 310-region, however, might occur upon longer simulation times, as suggested by Bjelkmar et al. (50).
The more important aim of our study, however, was to define the movement of the S4-S5 linker and the mechanism by which this movement controls the internal gate of the Kv channels. Here, two features of the S4-S5 linker movement give significant insight into the gating process. First, the S4-S5 linker, and with it the internal gate (S6), are radially displaced by only 3–4 Å, and second, the S4-S5 linker is slightly bent in the resting state. The movement of the S4-S5 linker is initiated by the displacement of the S4 helix described above. In particular, the tilt and the rotation of S4 are translated into a similar rotation and a bend in the S4-S5 linker. The relatively small radial displacement of the S4-S5 linker is translated to closing of the internal gate by 4 Å (Fig. 3C). The interaction is mediated by annealing of the S4-S5 linker to the C-terminal end of S6 (S6T (9, 10, 11, 13, 14, 51)). The S4-S5 linker “presses” against the S6 at the level of its PVP motif, and accordingly, the pore closes with a “hydrophobic seal” by pushing the isoleucine 398 residues into the pathway, excluding most water molecules from the central cavity in the closed state (Fig. 3D).
If such a small displacement of S6 is sufficient to interrupt the water-filled column in the ion-conducting pathway, it raises the question whether all four S6 helices have to be in the closed position or whether a single S6 in the closed position is sufficient to disrupt the water pathway. The movement of the four voltage sensor domains in Kv channels is supposed to occur independently followed by the cooperative opening of the pore (52). This implies that the energy generated by the movement of the first three voltage sensors has to be stored in the system before the final opening step. The flexibility and bending of the S4-S5 linker could potentially provide the required “elasticity” to the system. Disruption of the conduction pathway by the transition of a single voltage sensor, and with it of one S6, would offer an alternative explanation for the observed cooperativity. In this case, the actual conformational changes for the other three subunits may already have occurred when the last S6 opens and allows access to the pore. However, this would contradict the suggested correlation of subconductance levels with partial opening (53); thus, this question will have to be addressed in direct measurements as performed for instance in KcsA (46).
CONCLUSION
In this study, we determined the movement of the S4-S5 linker during the gating process and established that a small radial movement of 4 Å is sufficient to control the cytosolic pore gate. Controlling the state of a voltage-gated potassium channel via its lipid environment opens up the possibility to investigate these channels with any spectroscopic method in both open and closed state even in the absence of a membrane potential.
Acknowledgments
We thank Mireille Marsolais and Michel Brunette for technical assistance and Drs. William Skene and Matthieu Starck for help in synthesizing terbium chelate. The molecular dynamics simulations were performed at the Swiss National Supercomputing Center (CSCS). GÉPROM is a research group funded by the Fonds de la recherche en santé du Québec (FRSQ).
This work was funded by the Canadian Institutes for Health Research Grant MOP-102689 (to R. B.), the Canadian Foundation for Innovation (to R. B.), and the Swiss National Science Foundation (SNF-Professorship Number 118928) (to S. B).
Q. Li, S. Wanderling, P. Somponspisut, and E. Perozo, submitted for publication.
- DOTAP
- 1,2-dioleoyl-3-trimethylammonium-propane
- DPhPC
- 1,2-diphytanoyl-sn-glycero-3-phosphocholine
- DPPC
- dipalmitoylphosphatidylcholine
- LRET
- lanthanide-based resonance energy transfer
- DO
- donor-only
- SE
- sensitized emission.
REFERENCES
- 1. Hille B. (2001) Ion Channels of Excitable Membranes, 3rd Ed., Sinauer, Sunderland, MA [Google Scholar]
- 2. Ashcroft F. (1999) Ion Channels and Disease, pp. 97–122, Academic Press, New York [Google Scholar]
- 3. Liman E. R., Hess P., Weaver F., Koren G. (1991) Voltage-sensing residues in the S4 region of a mammalian K+ channel. Nature 353, 752–756 [DOI] [PubMed] [Google Scholar]
- 4. Papazian D. M., Timpe L. C., Jan Y. N., Jan L. Y. (1991) Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature 349, 305–310 [DOI] [PubMed] [Google Scholar]
- 5. Aggarwal S. K., MacKinnon R. (1996) Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron 16, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 6. Seoh S. A., Sigg D., Papazian D. M., Bezanilla F. (1996) Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron 16, 1159–1167 [DOI] [PubMed] [Google Scholar]
- 7. Blunck R., Batulan Z. (2012) Mechanism of electromechanical coupling in voltage-gated potassium channels. Front Pharmacol. 3, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long S. B., Campbell E. B., Mackinnon R. (2005) Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 9. Lu Z., Klem A. M., Ramu Y. (2001) Ion conduction pore is conserved among potassium channels. Nature 413, 809–813 [DOI] [PubMed] [Google Scholar]
- 10. Lu Z., Klem A. M., Ramu Y. (2002) Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J. Gen. Physiol. 120, 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Labro A. J., Raes A. L., Grottesi A., Van Hoorick D., Sansom M. S., Snyders D. J. (2008) Kv channel gating requires a compatible S4-S5 linker and bottom part of S6, constrained by non-interacting residues. J. Gen. Physiol. 132, 667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barghaan J., Bähring R. (2009) Dynamic coupling of voltage sensor and gate involved in closed-state inactivation of Kv4.2 channels. J. Gen. Physiol. 133, 205–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Batulan Z., Haddad G. A., Blunck R. (2010) An intersubunit interaction between S4-S5 linker and S6 is responsible for the slow off-gating component in Shaker K+ channels. J. Biol. Chem. 285, 14005–14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haddad G. A., Blunck R. (2011) Mode shift of the voltage sensors in Shaker K+ channels is caused by energetic coupling to the pore domain. J. Gen. Physiol. 137, 455–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blunck R., Starace D. M., Correa A. M., Bezanilla F. (2004) Detecting rearrangements of shaker and NaChBac in real-time with fluorescence spectroscopy in patch-clamped mammalian cells. Biophys. J. 86, 3966–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu X. P., Erichsen D., Börjesson S. I., Dahlin M., Amark P., Elinder F. (2008) Polyunsaturated fatty acids and cerebrospinal fluid from children on the ketogenic diet open a voltage-gated K channel: a putative mechanism of antiseizure action. Epilepsy Res. 80, 57–66 [DOI] [PubMed] [Google Scholar]
- 17. Xu Y., Ramu Y., Lu Z. (2008) Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature 451, 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt D., Cross S. R., MacKinnon R. (2009) A gating model for the archeal voltage-dependent K+ channel KvAP in DPhPC and POPE:POPG decane lipid bilayers. J. Mol. Biol. 390, 902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmidt D., Jiang Q. X., MacKinnon R. (2006) Phospholipids and the origin of cationic gating charges in voltage sensors. Nature 444, 775–779 [DOI] [PubMed] [Google Scholar]
- 20. Schmidt D., MacKinnon R. (2008) Voltage-dependent K+ channel gating and voltage sensor toxin sensitivity depend on the mechanical state of the lipid membrane. Proc. Natl. Acad. Sci. U.S.A. 105, 19276–19281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Börjesson S. I., Elinder F. (2011) An electrostatic potassium channel opener targeting the final voltage sensor transition. J. Gen. Physiol. 137, 563–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Börjesson S. I., Hammarström S., Elinder F. (2008) Lipoelectric modification of ion channel voltage gating by polyunsaturated fatty acids. Biophys. J. 95, 2242–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blunck R., Scheel O., Müller M., Brandenburg K., Seitzer U., Seydel U. (2001) New insights into endotoxin-induced activation of macrophages: involvement of a K+ channel in transmembrane signaling. J. Immunol. 166, 1009–1015 [DOI] [PubMed] [Google Scholar]
- 24. Selvin P. R. (2002) Principles and biophysical applications of lanthanide-based probes. Annual review of biophysics and biomolecular structure 31, 275–302 [DOI] [PubMed] [Google Scholar]
- 25. Richardson J., Blunck R., Ge P., Selvin P. R., Bezanilla F., Papazian D. M., Correa A. M. (2006) Distance measurements reveal a common topology of prokaryotic voltage-gated ion channels in the lipid bilayer. Proc. Natl. Acad. Sci. U.S.A. 103, 15865–15870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Posson D. J., Ge P., Miller C., Bezanilla F., Selvin P. R. (2005) Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature 436, 848–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J., Selvin P. R. (1999) Thiol-reactive luminescent chelates of terbium and europium. Bioconjugate chemistry 10, 311–315 [DOI] [PubMed] [Google Scholar]
- 28. Ge P., Selvin P. R. (2003) Thiol-reactive luminescent lanthanide chelates: part 2. Bioconjugate chemistry 14, 870–876 [DOI] [PubMed] [Google Scholar]
- 29. MacKerell A. D., Jr., Bashford D., Bellott M., Dunbrack R. L., Jr., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., III, Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 30. Im W., Feig M., Brooks C. L., 3rd. (2003) An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophys. J. 85, 2900–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jo S., Kim T., Iyer V. G., Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 [DOI] [PubMed] [Google Scholar]
- 32. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jensen M. Ø., Jogini V., Borhani D. W., Leffler A. E., Dror R. O., Shaw D. E. (2012) Mechanism of voltage gating in potassium channels. Science 336, 229–233 [DOI] [PubMed] [Google Scholar]
- 35. Campos F. V., Chanda B., Roux B., Bezanilla F. (2007) Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc. Natl. Acad. Sci. U.S.A. 104, 7904–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin M. C., Abramson J., Papazian D. M. (2010) Transfer of ion binding site from ether-a-go-go to Shaker: Mg2+ binds to resting state to modulate channel opening. J. Gen. Physiol. 135, 415–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cha A., Snyder G. E., Selvin P. R., Bezanilla F. (1999) Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 402, 809–813 [DOI] [PubMed] [Google Scholar]
- 38. Sandtner W., Bezanilla F., Correa A. M. (2007) In vivo measurement of intramolecular distances using genetically encoded reporters. Biophys. J. 93, L45–L47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y. S., Sompornpisut P., Perozo E. (2001) Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Biol. 8, 883–887 [DOI] [PubMed] [Google Scholar]
- 40. Ramu Y., Xu Y., Lu Z. (2007) Inhibition of CFTR Cl− channel function caused by enzymatic hydrolysis of sphingomyelin. Proc. Natl. Acad. Sci. U.S.A. 104, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramu Y., Xu Y., Lu Z. (2006) Enzymatic activation of voltage-gated potassium channels. Nature 442, 696–699 [DOI] [PubMed] [Google Scholar]
- 42. Zheng H., Liu W., Anderson L. Y., Jiang Q. X. (2011) Lipid-dependent gating of a voltage-gated potassium channel. Nature communications 2, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 44. Vargas E., Bezanilla F., Roux B. (2011) In search of a consensus model of the resting state of a voltage-sensing domain. Neuron 72, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yarov-Yarovoy V., DeCaen P. G., Westenbroek R. E., Pan C. Y., Scheuer T., Baker D., Catterall W. A. (2012) Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. U.S.A. 109, E93–E102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blunck R., McGuire H., Hyde H. C., Bezanilla F. (2008) Fluorescence detection of the movement of single KcsA subunits reveals cooperativity. Proc. Natl. Acad. Sci. U.S.A. 105, 20263–20268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zuidam N. J., Barenholz Y. (1997) Electrostatic parameters of cationic liposomes commonly used for gene delivery as determined by 4-heptadecyl-7-hydroxycoumarin. Biochim. Biophys. Acta 1329, 211–222 [DOI] [PubMed] [Google Scholar]
- 48. Khalili-Araghi F., Jogini V., Yarov-Yarovoy V., Tajkhorshid E., Roux B., Schulten K. (2010) Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys. J. 98, 2189–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Villalba-Galea C. A., Sandtner W., Starace D. M., Bezanilla F. (2008) S4-based voltage sensors have three major conformations. Proc. Natl. Acad. Sci. U.S.A. 105, 17600–17607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bjelkmar P., Niemelä P. S., Vattulainen I., Lindahl E. (2009) Conformational changes and slow dynamics through microsecond polarized atomistic molecular simulation of an integral Kv1.2 ion channel. PLoS Comput. Biol. 5, e1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Long S. B., Campbell E. B., Mackinnon R. (2005) Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309, 903–908 [DOI] [PubMed] [Google Scholar]
- 52. Zagotta W. N., Hoshi T., Aldrich R. W. (1994) Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J. Gen. Physiol. 103, 321–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chapman M. L., VanDongen A. M. (2005) K channel subconductance levels result from heteromeric pore conformations. J. Gen. Physiol. 126, 87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]