Background: Heparan sulfate (HS) regulates the transport and signaling activities of fibroblast growth factors (FGF).
Results: The molecular determinants of the interactions of FGFs and heparin were identified.
Conclusion: There are clear molecular specificities determining the interactions of FGFs with the polysaccharide.
Significance: The expansion of the FGFs in metazoan evolution parallels the diversification of the specificity of their interactions with heparin.
Keywords: Fibroblast Growth Factor (FGF), Glycosaminoglycan, Heparan Sulfate, Heparin, Heparin-binding Protein
Abstract
The functions of a large number (>435) of extracellular regulatory proteins are controlled by their interactions with heparan sulfate (HS). In the case of fibroblast growth factors (FGFs), HS binding determines their transport between cells and is required for the assembly of high affinity signaling complexes with their cognate FGF receptor. However, the specificity of the interaction of FGFs with HS is still debated. Here, we use a panel of FGFs (FGF-1, FGF-2, FGF-7, FGF-9, FGF-18, and FGF-21) spanning five FGF subfamilies to probe their specificities for HS at different levels as follows: binding parameters, identification of heparin-binding sites (HBSs) in the FGFs, changes in their secondary structure caused by heparin binding and structures in the sugar required for binding. For interaction with heparin, the FGFs exhibit KD values varying between 38 nm (FGF-18) and 620 nm (FGF-9) and association rate constants spanning over 20-fold (FGF-1, 2,900,000 m−1 s−1 and FGF-9, 130,000 m−1 s−1). The canonical HBS in FGF-1, FGF-2, FGF-7, FGF-9, and FGF-18 differs in its size, and these FGFs have a different complement of secondary HBS, ranging from none (FGF-9) to two (FGF-1). Differential scanning fluorimetry identified clear preferences in these FGFs for distinct structural features in the polysaccharide. These data suggest that the differences in heparin-binding sites in both the protein and the sugar are greatest between subfamilies and may be more restricted within a FGF subfamily in accord with the known conservation of function within FGF subfamilies.
Introduction
The fibroblast growth factors (FGFs)5 regulate many aspects of embryonic development and adult homeostasis. In humans and mice, there are 22 fgf genes encoding ligands and 5 fgfr genes encoding the cognate membrane receptors, whereas only 2 fgf genes and 1 fgfr gene have been identified in Caenorhabditis elegans, for example. This indicates that the FGF ligand-receptor system expanded significantly during evolution from primitive metazoa to vertebrates (1). The 22 human FGFs are divided into 7 subfamilies according to their sequence similarities, FGF-1 subfamily, FGF-4 subfamily, FGF-7 subfamily, FGF-8 subfamily, FGF-9 subfamily, FGF-11 subfamily, and FGF-19 subfamily, and these map to certain functional properties.
The stimulation of cell proliferation requires the formation of a ternary complex of the FGF, FGFR, and the heparan sulfate (HS) co-receptor (2, 3). This requirement for the co-receptor has since been demonstrated in vivo (4) and in structures of co-crystals of the ternary complex (5). Moreover, the interactions of FGFs with HS in extracellular matrices control their transport from source to target cell (6). HS and its experimental proxy heparin are glycosaminoglycans (GAGs) and have the same underlying disaccharide backbone, which is composed of a uronic acid, either α-l-iduronic or β-d-glucuronic acid linked 1,4 to α-d-glucosamine. The initial product of biosynthesis is a polysaccharide chain comprising 50–100 repeating disaccharides of 1,4-linked β-d-glucuronic acid and α-N-acetyl-d-glucosamine. This is then modified, initially by concomitant N-deacetylation/N-sulfation by N-sulfotransferases, which produces a series of clusters of N-sulfated glucosamines along the chain. These are the sites for subsequent enzyme action, which includes epimerization of glucuronic acid to iduronic acid, O-sulfation of C2 on iduronic acid, and of C6 and C3 on glucosamine (7). The resulting clustering of modifications results in HS chains possessing several distinct domains of varying sizes as follows: unsulfated (N-acetylated) domains and intermediate and transition (NAS) domains that have one glucosamine in two N-sulfated and sulfated domains in which every glucosamine is N-sulfated (S-domain). The latter resemble heparin but are generally less O-sulfated and are considered to contain the sites where proteins bind (7, 8).
There is biological evidence for the FGF ligand-receptor system possessing a high degree of specificity. For example, particular FGF ligand·FGFR complexes can be formed in situ only on HS from particular tissue compartments (9). In vitro, some specificity is apparent, particularly for FGFR preferences, e.g. FGF-7 subfamily and FGFR2b, although in general such preferences are fairly broad (1, 10, 11). The interactions of some FGFs with HS and its derivatives have been subjected to scrutiny in vitro, but their level of specificity is not clear. Indeed, it has been proposed that this specificity is largely at the level of HS versus other GAGs, although a greater degree of specificity might emerge at the level of the full ligand-receptor system (12). However, some detailed biophysical analyses suggest that distinct FGFs bind to heparin differently (13, 14). Given the expansion of FGFs in the course of evolutionary history and the central regulatory role of the HS co-receptor, it would seem likely that these are reflected in the degree of specificity of the FGF-HS interaction. To address this question, we have used a representative of each of five FGF subfamilies and interrogated their interactions with a library of polysaccharides. By employing a range of complementary techniques, binding parameters, Protect and Label, synchrotron radiation circular dichroism (SRCD), and differential scanning fluorimetry (DSF), a diversification of the specificity of FGF-heparin interactions and of secondary binding sites for heparin in the proteins has been identified.
EXPERIMENTAL PROCEDURES
FGF cDNA
cDNAs encoding FGF-1 (UniProt accession number P05230; residues 16–155) and FGF-2 (UniProt accession number P09038-2; residues 1–155) were cloned into vector pET-14b (Novagen, Merck). cDNAs encoding FGF-7 (Uniprot accession number P21781; residues 32–194) (shipped from the company undertaking gene synthesis, Eurofins Mwg, UK), FGF-9 (Uniprot accession number P31371; residues 1–208), FGF-18 (zFGF5) (UniProt accession number O76093; residues 28–207), and FGF-21 (Uniprot accession number Q9NSA1; residues 29–209) were inserted separately into a modified pET-24b vector (pETM-11, a kind gift of Dr Paul Elliott, University of Liverpool, UK), which contains sequence encoding a 6× histidine tag and a tobacco etch virus cleavage site (26 amino acids, MKHHHHHHPMSDYDIPTTENLYFQGA) at the N terminus of the inserted FGF.
Recombinant FGF Purification
pET-14b-FGF-1, pET-14b-FGF-2, pETM-11-FGF-9, pETM-11-FGF-18, and pETM-11-FGF-21 were transformed into Escherichia coli C41 (DE3) cells and expressed, as described previously (15–17). pETM-11-FGF-7 was transformed into E. coli BL21 (DE3) cells, expressed by 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h, and purified as described for FGF-18 (15). pETM-11-FGF-21 was transformed into C41 (DE3), grown, and induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside at 16 °C for 16 h. Cell pellets were sonicated in buffer A21 (0.15 m NaCl, 0.05 m Tris, pH 7.5) with 50 mm imidazole. After clarification by centrifugation, soluble protein was loaded onto a 3-ml ProbondTM nickel-chelating resin (Invitrogen) column. Bound proteins were eluted in the same buffer (buffer A21) with 500 mm imidazole and dialyzed against buffer B21 (10 mm Tris, 1 mm DTT, pH 8.0) with 50 mm NaCl, and then applied to a 1-ml HisTrap Q column (GE Healthcare), and bound proteins were eluted in buffer B21, with a gradient using up to 1 m NaCl. That the pure FGFs were correctly folded is supported by their binding to heparin affinity columns (FGF-1, FGF-2, FGF-7 FGF-9, and FGF-18) and that they stimulated all proliferation with the appropriate sensitivity (data not shown).
Sugars
The same batch of heparin (17 kDa average molecular mass, Celsus Lab, Cincinnati, OH) was used in all assays and for the production of modified derivatives and oligosaccharides. Porcine mucosal heparan sulfate, hyaluronic acid (HA), and chondroitin sulfate (CS C) were from Sigma; dermatan sulfate (DS) was from Iduron (Manchester, UK). Heparin oligosaccharides of degrees of polymerization (DP) DP2, DP4, DP6, DP8, DP10, and DP12 were produced, as described previously (18). A range of systematically modified heparin derivatives with different repeating units (D1–9) were prepared as described previously (Table 1) (19). Cation (Na+, K+, Ca2+, Cu2+, and Zn2+) forms of heparin were also prepared as described previously (20).
TABLE 1.
Nomenclature and structures of chemically modified heparin structures
The letter I stands for iduronate, and A stands for the amino sugar glucosamine.
| Analogue | Predominant repeat | IdoUA-2 | GlcN-6 | GlcN-2 | IdoUA-3 | GlcN-3a |
|---|---|---|---|---|---|---|
| 1 (heparin) | I2SA6SNS | SO3− | 3− | 3− | OH | OH |
| 2 | I2SA6SNAc | SO3− | 3− | COCH3 | OH | OH |
| 3 | I2OHA6SNS | OH | 3− | 3− | OH | OH |
| 4 | I2SA6OHNS | SO3− | OH | 3− | OH | OH |
| 5 | I2OHA6SNAc | OH | 3− | COCH3 | OH | OH |
| 6 | I2SA6OHNAc | SO3− | OH | COCH3 | OH | OH |
| 7 | I2OHA6OHNS | OH | OH | 3− | OH | OH |
| 8 | I2OHA6OHNAc | OH | OH | COCH3 | OH | OH |
| 9 | I2S,3SA6S3S,NS | SO3− | 3− | 3− | 3− | 3− |
a Numbers refer to the ring position of the carbon atoms.
Measurement of Binding Kinetics
Binding kinetics were measured in an IAsys (Farfield Group, Manchester, UK) optical biosensor, which reports responses in arc s (1 arc s = 1/3600°, 600 arc s = 1 ng of protein/mm2 sensor surface). Streptavidin (Promega, Southampton, UK) was immobilized on aminosilane surfaces (Farfield Group) using bis-sulfosuccinimidyl suberate (as the cross-link) (Thermo Scientific, Epsom, UK). Control experiments showed that FGF-1, FGF-7, and FGF-9 in buffer PBST (140 mm NaCl, 3.6 mm NaH2PO4, 12.9 mm Na2HPO4, 0.02% (w/v) NaN3, 0.02% (v/v) Tween 20, pH 7.2) did not bind to the unmodified surface or to a streptavidin surface. Oligosaccharides (DP8) were biotinylated at their reducing ends and immobilized on a streptavidin-derivatized surface as described previously (21, 22). Binding assays were performed as described for FGF-2 (21); kon was determined at lower concentrations of FGFs, and koff was measured at higher concentrations of FGFs with, in some instances, competing soluble heparin to avoid rebinding artifacts. Binding parameters of the association and dissociation phases were calculated using the nonlinear curve-fitting software provided with the instrument (Fastfit, Farfield). The off-rate constant (koff, equivalent to the dissociation rate constant, kd) was calculated from the dissociation phase, and the association rate constant (ka) was calculated from the values of kon determined at five to seven concentrations of FGFs. The equilibrium dissociation constant (KD) was calculated from the ratio of the dissociation and association rate constants (kd/ka) (21, 23).
Microscale Thermophoresis (MST), Measurements of Binding Kinetics
Binding kinetics of FGF-18 to DP8 were measured by Microscale Thermophoresis in a NanoTemperMonolith NT.115 instrument (NanoTemper Technologies, Munich, Germany), which detects changes in size, charge, and/or solvation induced by binding (24). The FGF-18 was labeled on a heparin (AF Heparin Beads, Tosoh Biosciences GmbH, Stuttgart, Germany) column to prevent the modification of lysine residues involved in heparin binding. Two nm FGF-18 was loaded onto a 20-μl heparin column and equilibrated with buffer M1 (17.9 mm Na2HPO4, 2.1 mm NaH2PO4, 150 mm NaCl, pH 7.8). The column was then washed three times with carbonate buffer (130 mm NaHCO3, 50 mm NaCl, pH 8.2). The heparin-bound protein was labeled with dye NT-647 (Nanotemper Technologies) (24, 25) at room temperature for 30 min. The labeled FGF-18 was eluted with buffer M2 (17.9 mm Na2HPO4, 2.1 mm NaH2PO4, 2 m NaCl, pH 7.8). Labeled proteins were concentrated and buffer exchanged to 0.1 μm in phosphate-buffered saline (2.7 mm KCl, 136 mm NaCl, 8.1 mm Na2HPO4, 1.5 mm KH2PO4, pH 7.3) with 0.02% Tween 20 (v/v) mixed (1:1, v/v) with 16 different concentrations of DP8 to form a titration gradient from 20 μg/ml to 0.61 ng/ml DP8. Protein sugar complexes were loaded onto hydrophilic MST-grade glass capillaries (NanoTemper Technologies), and changes in the size, charge, and/or solvation were measured by MST. Curve fitting and affinity KD calculation was done with the NanoTemper Analysis Software Version 1.2.210 (NanoTemper Technologies).
Protect and Label
Heparin-binding sites in FGF-1, FGF-7, FGF-9, and FGF-18 were identified by the “Protect and Label” approach, as described for FGF-2 (26), except that 1.5 nm FGF protein was used instead of 1.2 nm. Digested and biotinylated peptides were purified on a C18 ZipTip (Millipore Ltd., Watford, UK) (26) and then analyzed by tandem mass spectrometry. Up to 1 μg of biotinylated peptides was injected into a LTQ-Orbitrap Velos instrument (Thermo) using a nanoAcquity UPLC system (Waters Associates). Peptides were separated on a BEH300 C18 (75 μm × 250 mm, 1.7 μm) nanoAcquity UPLC column (Waters) using a 60-min linear gradient (5–35% (v/v) acetonitrile in 0.1% formic acid). Data acquisition was performed using a TOP-10 strategy where survey MS scans were acquired in the orbitrap (R = 30,000 at m/z 400), and up to 10 of the most abundant ions per full scan were fragmented by higher energy collision dissociation (HCD) and analyzed in the orbitrap (R = 7,500 at m/z 400).
Data analysis was performed using the Batch Tag tool of the Protein Prospector package Version 5.9.2 applying the following parameters: digest, chymotrypsin (FWYMEDLN); maximum missed cleavages, 5; possible modifications, acetyl (Lys), biotin (Lys), carbamidomethyl (Cys), carboxymethyl (Cys), oxidation (Met); parental ion tolerance, 10 ppm; fragment ion tolerance, 0.05 Da. The UniProt accession number of the protein analyzed was used as a research parameter (database, SwissProt 2011.01.11). Results were filtered using a peptide E-value <0.001 and SLIP score threshold for site assignment was set to 6 (cite PMID: 21490164).
SRCD Spectroscopy
SRCD analyses were performed on an Olis DSM20 monochromator (beam line B-23, Diamond Light Source) with quartz cuvettes of path lengths of 0.02 cm (178–260 nm). Samples included the following: heparin alone (1.4 mg/ml) and FGFs (FGF-1, FGF-2, FGF-7, FGF-9, FGF-18, and FGF-21) alone (0.5–1 mg/ml) or with a 5-fold molar excess of heparin were analyzed in CD buffer (15.3 mm Na2HPO4, 2.2 mm NaH2PO4, pH 7.5). Each set of data were based on the average of four scans. The program SELCON3 and database 3 were used to analyze the spectra for the secondary structures (27–32).
Differential Scanning Fluorimetry (DSF)
DSF was performed on a 7500 Fast Real PCR System (software version 1.4.0, Applied Biosystems), as described previously (15, 33). Different concentrations of heparin (0-500 μm) with a fixed concentration (5 μm) of FGF-7, FGF-9, or FGF-21 were tested for melting curves. Different glycosaminoglycan sugars (DP2–12, chemically modified derivatives D1–9 (Table 1), porcine mucosal heparan sulfate, HA, CS, DS, and cation-modified heparins), which were all used as 10 times concentrated stock solution (1.75 mg/ml) in HPLC grade water, were also tested with five different FGFs (FGF-1, FGF-2, FGF-7, FGF-9, and FGF-18) (5 μm). Experiments were run as described previously (15). First derivatives of the melting curves were calculated using Origin 7 (OriginLab Corp., Northampton, UK). At least two experiments each comprising triplicate wells were performed, but for the purpose of clarity the figures only show one melting curve or derivative per sample. Mean Tm and the standard error (S.E.) were calculated based on two repeats (6 data points). Data were normalized according to the formula: Tm x − Tm PBS/Tm hep-Tm PBS, where Tm x is the Tm of the protein sugar complex; Tm PBS is the Tm of protein itself, and Tm hep is the Tm of protein·heparin complex. The relative stability of FGFs in PBS was set as 0, and the relative stability of the full heparin effect was set as 1 (15).
Principal Component Analysis of DSF Data
Principal component analysis was performed using R (R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria). The data were analyzed as they were delivered, and no scaling was performed. The dendrograms were produced using statistical distance matrices generated from the component loadings.
RESULTS
To test possible relationships between the diversification of FGF ligands and their interactions with HS, a series of complementary measurements were used on representatives of five FGF subfamilies as follows: FGF-1 and FGF-2 (FGF-1 subfamily), FGF-7 (FGF-7 subfamily), FGF-9 (FGF-9 subfamily), FGF-18 (FGF-8 subfamily), and FGF-21 (FGF-19 subfamily). The biophysical determination of binding affinity and kinetics provides a quantitative insight into the specificity of an interaction. Because FGF-2, for example, possesses not only a canonical heparin-binding site, but also two other secondary sites, a Protect and Label structural proteomics approach (26) was used to identify heparin-binding surfaces in the FGFs. To determine whether heparin binding affected FGF structure, SRCD was used, because this accesses the solution structure of the protein and its complexes. DSF, by detecting thermal stabilization (15), identifies conformational change upon binding and allows the rapid screening of libraries of sugars to determine binding specificity.
Binding Parameters of FGFs to a Heparin-derived Octasaccharide (DP8)
Heparin-derived oligosaccharides of DP8 were chosen as a representative, because, according to previous results, FGFs bind this unit (15, 21). The level of binding of FGF-1, FGF-7, and FGF-9 to DP8 oligosaccharides immobilized through their reducing end was measured using an optical biosensor. As the concentration of FGFs increased, so did the observed level of binding (supplemental Figs. S1A–S9A). Analysis of the binding curves with a one-site model, revealed a random distribution of the data around the model (Figs. 1–9). A plot of the slope of initial rate against FGF concentration was fitted by a straight line (Figs. 1–9), indicating that binding was not limited by diffusion. Additionally, the data included most of the curve (≥90%) described by the single site model (supplemental Figs. S1H, S2J, S3I, S4I, S5I, S6H, S7H, S8I and S9H). These analyses showed that, at the concentrations used in this experiment, the interaction of oligosaccharides of DP8 with FGFs was monophasic, and there was no evidence for biphasic association kinetics. FGF-1 possessed a fast ka for immobilized DP8 (Table 2), which was indistinguishable from that of FGF-2 measured previously for DP8 (ka 2,000,000 ± 610,000 m−1 s−1) (21). The ka for FGF-7 was three times slower than that of FGF-1 (Table 2), whereas that for FGF-9 was even lower, 130,000 ± 17,000 m−1 s−1. The affinity of FGF binding to DP8 was largely the result of an altered value of ka. Thus, the highest affinity (KD) was measured for FGF-1, which was similar to that of FGF-2 to DP8 (21), whereas FGF-7 exhibited a substantially lower affinity of 140 ± 15 nm and FGF-9 the lowest KD 620 ± 340 nm. The affinity of these interactions calculated from the maximum extent of binding KD (equilibrium) was similar in each case to the value calculated from the kinetic parameters, indicating internal self-consistency (Table 2). FGF-18 was found to bind nonspecifically to both the underivatized amino silane surface and surfaces with streptavidin or neutravidin. Consequently, a solution approach, microscale thermophoresis, was used to measure its affinity for DP8 (supplemental Fig. S10). This indicated that FGF-18 had an affinity of 38 ± 12 nm for DP8, close to that of FGF-1 and FGF-2. When the affinity of FGF-2 was measured by microscale thermophoresis, the affinity was the same as when determined using an optical biosensor (result not shown).
FIGURE 1.
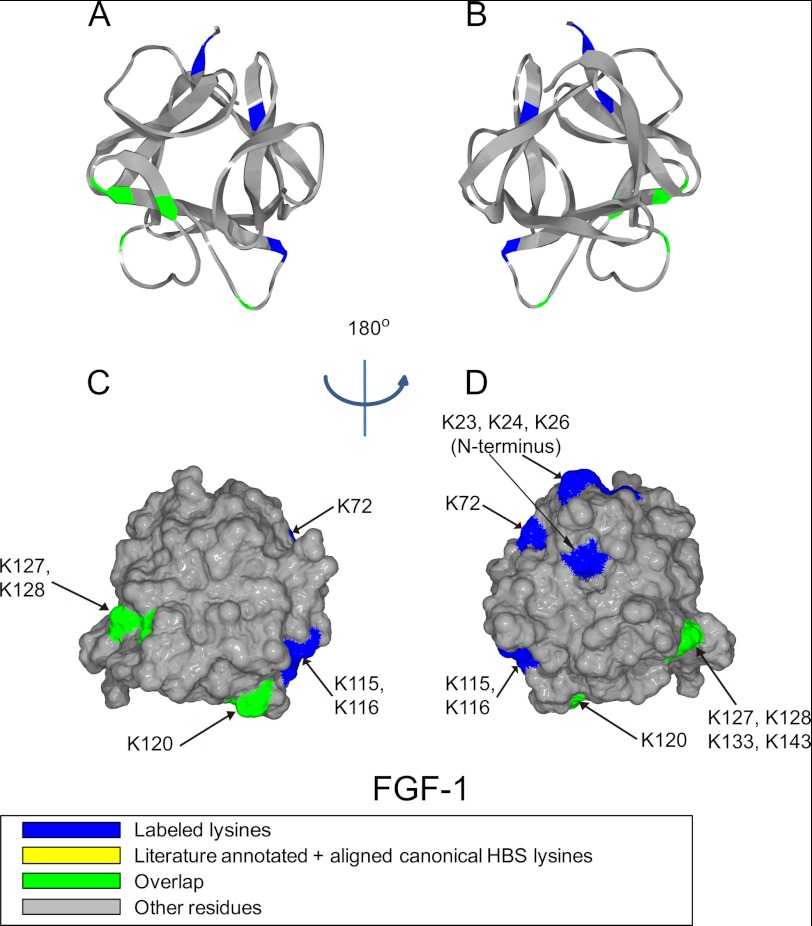
Position of biotinylated peptides in FGF-1 (residues 22–154) identified by structural proteomics mapped onto the predicted three-dimensional structure (PDB 2ERM (53)). Labeled peptides are colored in blue, and peptides overlapping with literature annotated and aligned canonical HBS lysines are colored in green (35, 36). A and B, ribbon diagram. C and D, corresponding molecular surface. FGF-1 is shown using schematic representation. B and D, 180° back view of A and C.
FIGURE 2.
Sequence alignment of FGFs by subfamily. The sequence alignment employed ClustalX2 (54). Secondary structures are from human FGF-1, rat FGF-7, human FGF-8b, and human FGF-9 crystal structures (39, 42, 43, 55, 56). Labeled lysines and literature reported lysines and arginines are colored in red, and the predicted lysines are colored in blue. The aligned HBSs are highlighted in red.
FIGURE 3.
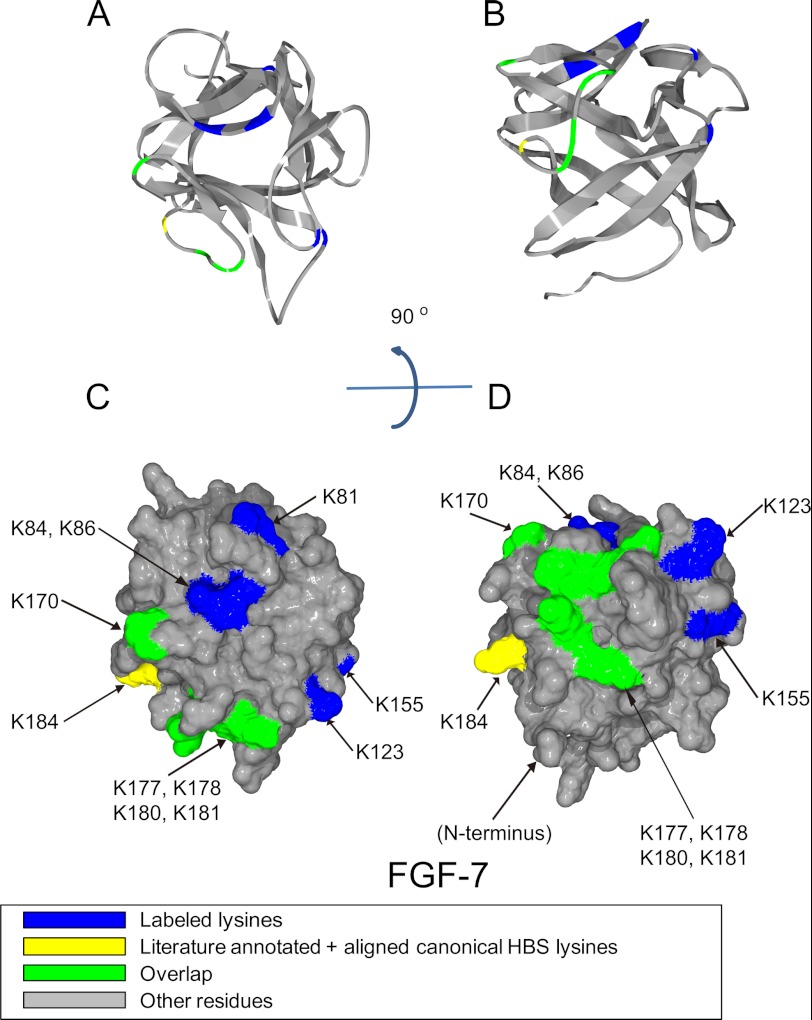
Position of biotinylated peptides in FGF-7 (residues 51–191) identified by structural proteomics mapped onto the predicted three-dimensional structure (rat FGF-7 3D PDB 1QQK (39)). Labeled peptides are colored in blue; literature annotated and aligned canonical HBS lysines are labeled in yellow, and peptides overlapping with literature annotated and aligned canonical HBS lysines are colored in green. A and B, ribbon diagram. C and D, corresponding molecular surface. FGF-7 is shown using schematic representation. B and D, 90° bottom view of A and C.
FIGURE 4.
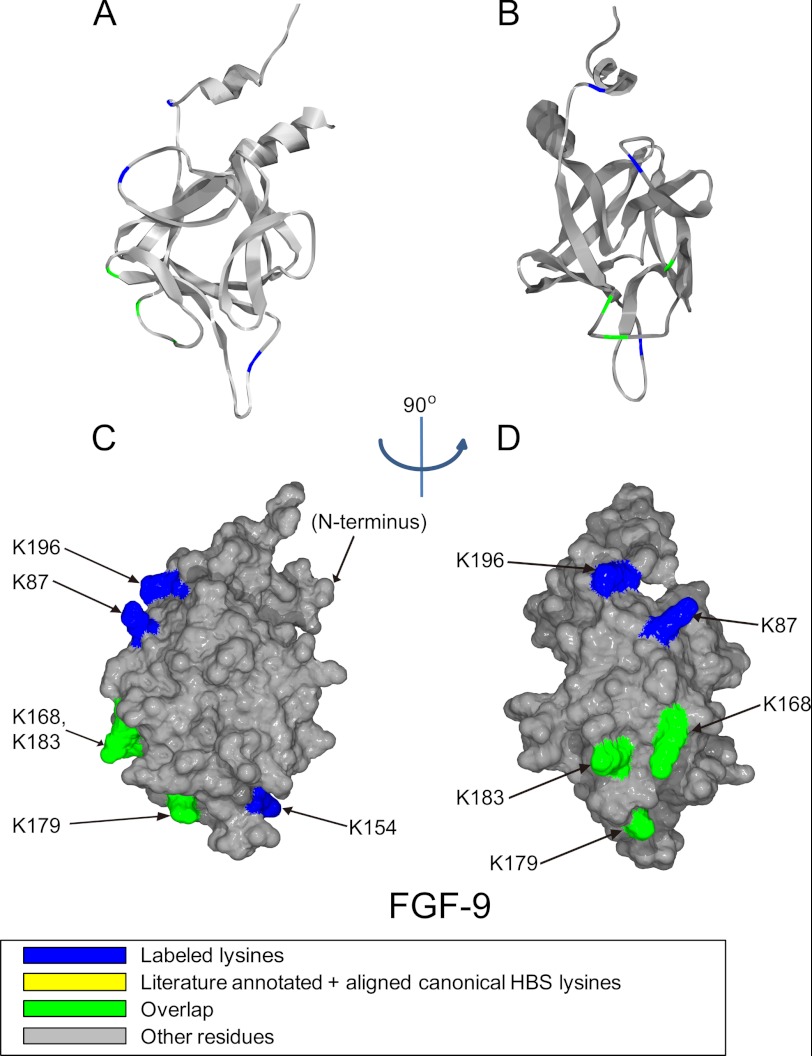
Position of biotinylated peptides in FGF-9 (residues 52–208) identified by structural proteomics mapped onto the predicted three-dimensional structure (PDB 1IHK (42)). Labeled peptides are colored in blue, and overlapped peptides are colored in green. A and B, ribbon diagram. C and D, corresponding molecular surface. FGF-9 is shown using schematic representation. B and D, 90° left view of A and C.
FIGURE 5.
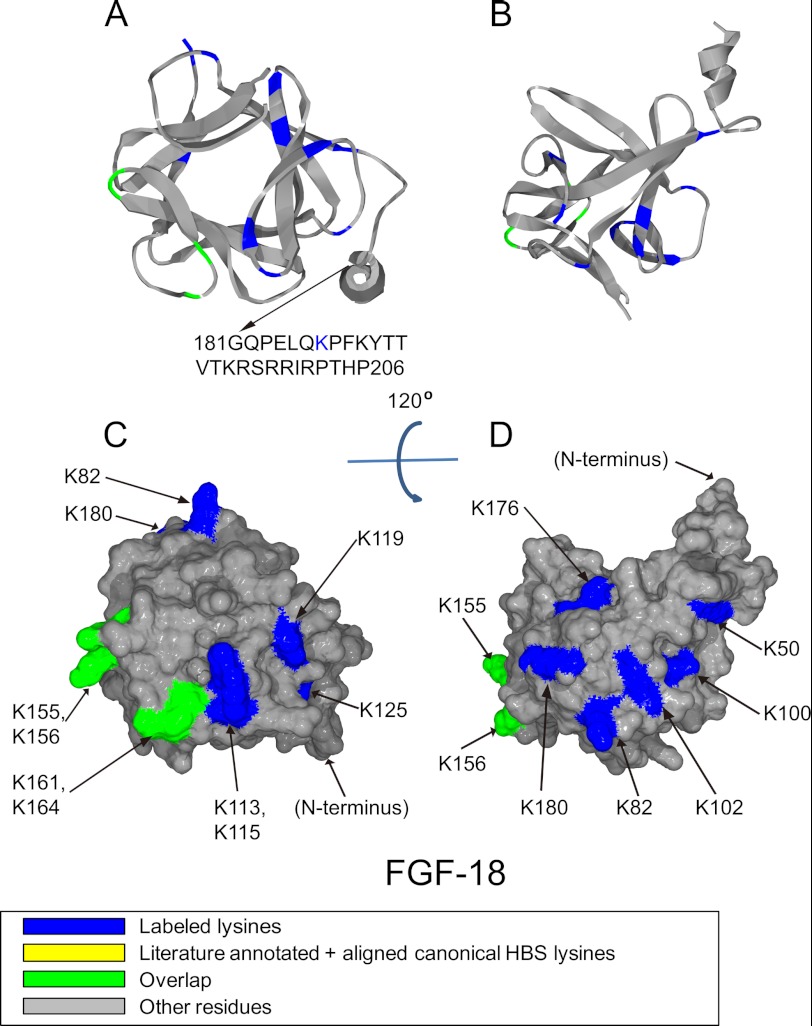
Position of biotinylated peptides in FGF-18 (residues 35–176) identified by structural proteomics mapped onto the predicted three-dimensional structure (modeled structure from FGF-8b PDB 2FDB (56) using software SPDBV (57). Labeled peptides are colored in blue, and peptides overlapping with literature annotated and aligned canonical HBS lysines are colored in green. Loop between Gly-70 and Arg-71 was missing, and residues after Gly-181 was not shown due to model (FGF-8b) differences compared with FGF-18. A and B, ribbon diagram. C and D, corresponding molecular surface. FGF-18 is shown using schematic representation. B and D, 120° top view of A and C.
FIGURE 6.
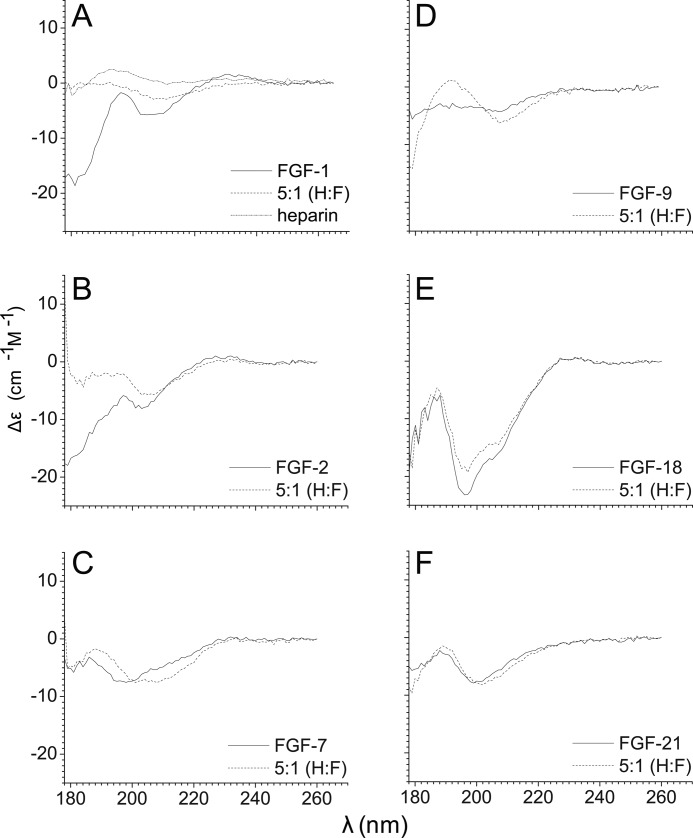
SRCD spectra of six FGFs and their complexes. SRCD spectra were recorded on beamline B-23 at Diamond Synchrotron between 180 and 260 nm of the FGFs (FGF-1, FGF-2, FGF-7, FGF-9, FGF-18, and FGF-21), in the absence or presence of heparin (molar ratio 1:5). A heparin control spectrum (1.4 mg/ml) was also recorded. A, FGF-1 and heparin spectra. B, FGF-2. C, FGF-7. D, FGF-9. E, FGF-18. F, FGF-21.
FIGURE 7.
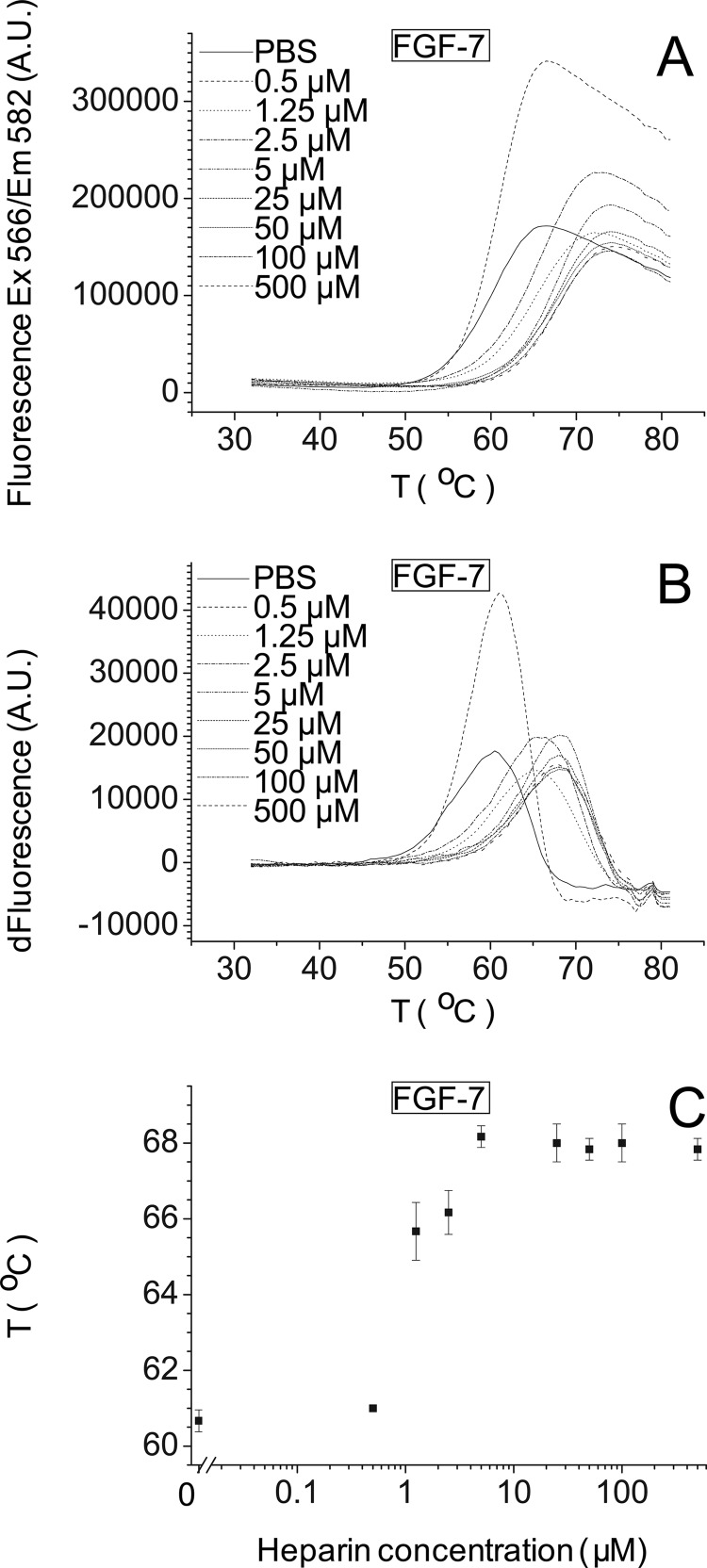
Stabilization effect of heparin on FGF-7. Differential scanning fluorimetry of 5 μm FGF-7 in the presence of different concentrations of heparin (“Experimental Procedures”). A, melting curve profiles of FGF-7 (5 μm) with a range of heparin concentrations (0–500 μm). B, first derivative of the melting curves in A. C, peak of the first derivative of the melting curves from B, which is the melting temperature, Tm (mean of triplicates ± S.E.).
FIGURE 8.
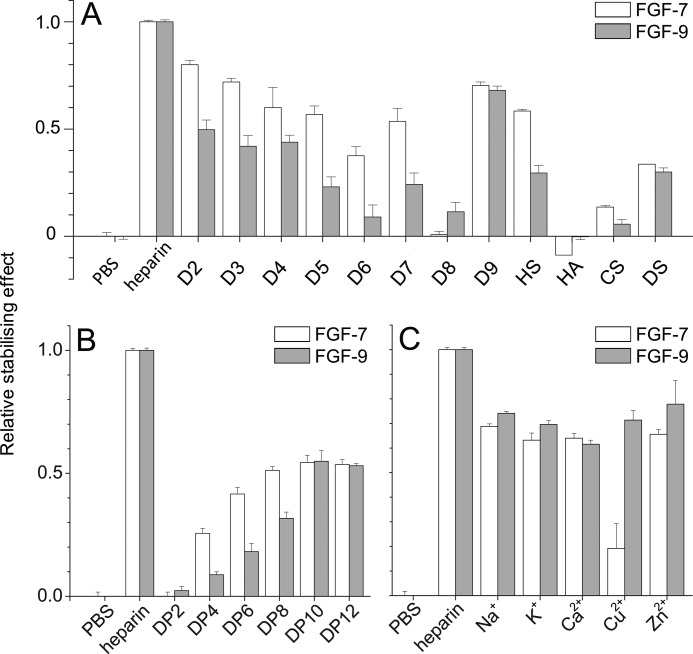
Differential scanning fluorimetry analysis of heparin derivatives on FGF-7 and FGF-9 reveals distinct dependence on substitution pattern. Differential scanning fluorimetry was performed with a range of heparin-based poly- and oligosaccharides with 5 μm protein and 175 μg/ml sugar. A, relative thermal stabilization effect of controls (PBS and heparin), chemically modified heparins (D2–D9), and other GAGs (HS, HA, CS, and DS). B, cation-modified heparin forms (20). C, heparin-derived oligosaccharides, ranging from DP2 to 12. Results are the mean of triplicates after normalization ± S.E., and an apparent absence of error bar is due to a small S.E.
FIGURE 9.
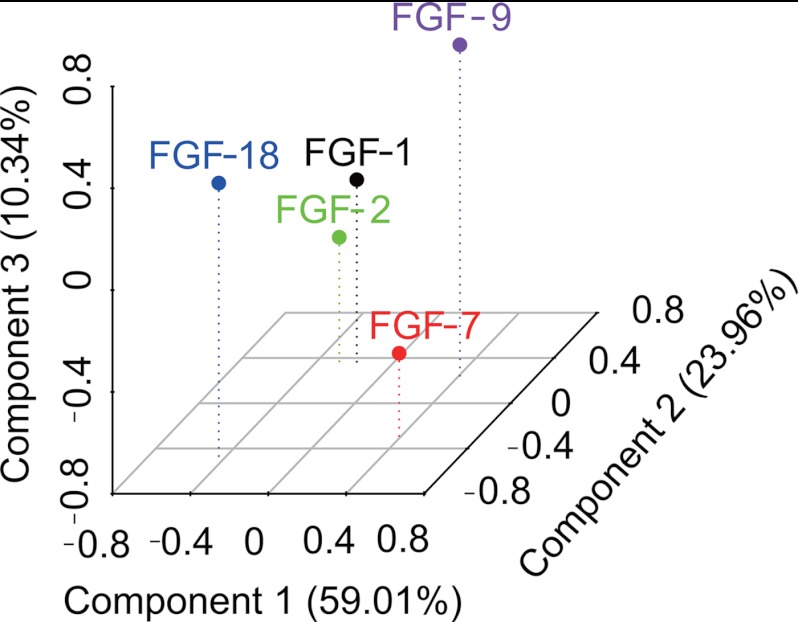
Identification of the structural specificity underlying the interactions of five FGFs with heparin. DSF was used to establish the relative affinity of each FGF for a library of modified heparins and heparin fragments of defined sizes. The binding specificity of the FGFs for the polysaccharides is revealed following principal component analysis by a combination of their interactions with heparin fragments and the modified heparin library. The three-dimensional plot (PC1 versus PC2 versus PC3) indicated that FGFs binding is affected by both substitution pattern and size of heparin represented by the first three components. PC1 explains 59.01%, PC2 explains 23.96%, and PC3 explains 10.34% of the variance.
TABLE 2.
Summary of binding data for FGF-1, FGF-7, and FGF-9
| FGF | kaa | rb | kdc | K>D(kinetic)d | KD (equilibrium)e |
|---|---|---|---|---|---|
| m−1 s−1 | s−1 | m−1 | m−1 | ||
| FGF-1 | 2,900,000 ± 140,000 | 0.92 | 0.084 ± 0.015 | 29 ± 5.5 | 60 ± 6.2 |
| FGF-7 | 870,000 ± 32,000 | 0.96 | 0.12 ± 0.012 | 140 ± 15 | 290 ± 49 |
| FGF-9 | 130,000 ± 17,000 | 0.91 | 0.030 ± 0.0055 | 240 ± 32 | 620 ± 340 |
| FGF-18f | 38 ± 12 |
a The S.E. is derived from the deviation of the data from a one-site model and was calculated by matrix inversion using the FastFit software provided with the instrument (see under “Experimental Procedures”). For three sets of values of kon, the resulting values for ka and their associated S.E. were combined.
b The correlation coefficient of the linear regression through the values for kon is shown.
c The kd is the mean ± S.E. of five values, obtained at high concentrations of FGF-7 and FGF-9.
d KD (kinetic) was calculated from the ratio of kd/ka, and the S.E. is the combined S.E. of the three kinetic parameters.
e KD (equilibrium) was calculated from the extent of the binding at, or near, equilibrium, and the S.E. is the combined value for three independent determinations of kd.
f FGF-18 and DP8 binding KD (equilibrium) was based on the data from MST (see under “Experimental Procedures”).
Identification of Heparin-binding Sites in FGFs
FGFs possess a canonical heparin-binding site toward their C terminus (5, 34, 35). However, other heparin-binding sites have been demonstrated in FGF-1 and FGF-2 (36, 37). Moreover, these sites have been successfully confirmed in FGF-2 by a Protect and Label analysis (26). In this approach, lysine side chains that remain exposed to solvent when the proteins bound to heparin are blocked with N-hydroxysuccinimide acetate. The protein is then eluted from heparin, and any protected lysines are labeled with N-hydroxysuccinimide-biotin; biotinylated peptides are identified by tandem mass spectrometry. Other amino acids involved in interactions with heparin (7) are not identified by this approach.
In this work, Protect and Label was used to identify heparin-binding sites (HBSs) in FGF-1, FGF-7, FGF-9, and FGF-18. High resolution tandem mass spectrometry allowed identification of the labeled peptides and, in most cases, unambiguous assignment of modified residues.
For FGF-1, the biotinylated peptides mapped to three areas on the protein surface (Fig. 1). One is the canonical binding site that extends from the loop between β-strands 9/10 to the loop between β-strands 11/12 and includes Lys-120, Lys-127–128, Lys-133, and Lys-143 (Figs. 1, A–D, and 2A and Table 3), in agreement with previous x-ray crystallographic studies (35, 36). Biotin-labeled Lys-115 and Lys-116, although close to the canonical binding site, are separated from it by an acidic amino acid Glu-119. This argues that this peptide forms part of an independent secondary binding site (Figs 1, A–D, and 2A and Table 3), which is supported by Lys-115 and Lys-116 being surrounded on other sides by acidic residues. The third area is near the N terminus and includes part of β-strand 1 and N-terminal to this Lys-23, Lys-24, and Lys-26 (Figs. 1, B and D, and 2A and Table 3), as well as Lys-72, which is in β-strand 5, but which is physically adjacent (Figs. 1, B and D, and 2A).
TABLE 3.
Summary of peptides identified by Protect and Label structure proteomics
Labeled peptides were identified by tandem mass spectrometry and analyzed by Protein Prospector package Version 5.9.2. Here, a summary of the peptides involved in the heparin-binding sites and the labeled position is provided. A full list of identified peptides is provided in the (supplemental Table S1).
| FGF | Peptide | Sequences | HBSa | Residuesb |
|---|---|---|---|---|
| FGF-1 | 1 | VGLK(biotin)K(biotin)NGSC(carboxymethyl)K(biotin)RGPRTHY GQ(carboxymethyl)K(biotin)AILFLPL | 1 | 124–150 |
| 2 | ISK(biotin/acetyl)K(biotin/acetyl)HAEK(biotin/acetyl)NWF | 2/1 | 113–123 | |
| 3 | K(biotin/acetyl)K(biotin/acetyl)PK(biotin/acetyl)LLY | 3 | 24–30 | |
| 4 | IK(biotin)STETGQYL | 3 | 71–80 | |
| FGF-7 | 1 | VALNQK(biotin)GIPVRGK(biotin/acetyl)K(biotin/acetyl)TK (biotin/acetyl)K(biotin)EQK(acetyl)TAHF | 1 | 165–188 |
| 2 | LAM(oxidation)NK(biotin)EGK(acetyl)LY | 1 | 119–128 | |
| 3 | ASAK(biotin)WTHNGGEMF | 1 | 152–164 | |
| 4 | YLRIDK(biotin/acetyl)RGK(biotin/acetyl)VK(biotin/acetyl)GTQE MK(acetyl)NNY | 4 | 76–95 | |
| FGF-9 | 1 | VALNK(biotin)DGTPREGTRTK(biotin)RHQK(biotin)F | 1 | 164–184 |
| 2 | HLEIFPNGTIQGTRK(biotin)DHSRF | 1 | 73–92 | |
| 3 | THFLPRPVDPDK(biotin)VPELY | 1 | 185–201 | |
| 4 | K(biotin)HVDTGRRY | 1 | 154–162 | |
| FGF-18 | 1 | TK(biotin)K(biotin)GRPRK(biotin)GPK(biotin)TRENQQDVHFM (oxidation) | 1 | 154–175 |
| 2 | C(carbamidomethyl)MNRK(biotin/acetyl)GK(biotin)LVGK | 1 | 109–129 | |
| (Biotin/acetyl)PDGTSK(biotin/acetyl)EC(carbamidomethyl)VF | ||||
| 3 | RIHVENQTRARDDVSRK(biotin)QL | 2 | 34–52 | |
| 4 | GRRISARGEDGDK(biotin)Y | 2 | 70–83 | |
| 5 | GSQVRIK(biotin/acetyl)GK(biotin/acetyl)ETEFYL | 2 | 94–108 | |
| 6 | M(oxidation)K(biotin/acetyl)RYPK(biotin/acetyl)GQPELQK (biotin/acetyl)PF | 2 | 175–189 |
In addition to its canonical binding site (HBS-1), FGF-2 has two other sites (26, 37, 38). Thus, FGF-2 could bind to heparin in the region 115YRSRKYSSWYVA126 (37, 38), which could be considered to form a secondary binding site, HBS-2. A third heparin-binding site (HBS-3) has been found at the N terminus of FGF-2 (26). According to this study, Lys-115- and Lys-116 in FGF-1 constitute the secondary corresponding binding site (HBS-2), which is at the same position as HBS-2 in FGF-2, but it has an extension in the form of β-5 Lys-72 (Fig. 2A). FGF-1 also has an HBS-3 at its N terminus at the same position as that of FGF-2 (Fig. 2A). Thus, based on these results, the FGF-1 subfamily can be proposed as having a conserved set of HBSs (Fig. 2A).
The structure of human FGF-7 has not yet been reported, so the rat FGF-7 crystal structure was used as a model (39). These two proteins differ by just 11 amino acids out of 163 (barring the first 31 amino acids of the secretory signal peptide), of which eight are similar. The aligned canonical binding site of FGF-7 contains Lys-170, Lys-177, Lys-178, Lys-180, Lys-181, and Lys-184. Work using the structure of the FGF-2·FGFR2c·heparin hexasaccharide co-complex (5) to model the heparin-binding site of FGF-7 suggests in contrast that only two lysines, Lys-180 and Lys-184, form the canonical heparin-binding site (14). The peptides identified here corresponded to the canonical binding site and contained all the previously identified amino acids, except for Lys-184 (Figs. 2B, 3, A–D, and Table 3). Moreover, another two lysine residues could be proposed as part of the canonical binding site as follows: one is in the middle of the sequence, contains Lys-123, and lies between strands β-6 and β-7 (Fig. 2B and Table 3); and the second lies in the loop between β-9 and β-10 (Lys-155, Figs. 2B, 3, A–D, and Table 3). In addition, to the canonical binding site, there may be another heparin-binding site in FGF-7 (Table 3), on the loop between β-2 and β-3 and part of β-3, as indicated by detection of biotinylated Lys-81, Lys-8, and Lys-86 (Figs. 2B, 3, A and C, and Table 3). This site is not found in FGF-1 (Fig. 2A) or FGF-2 (26), so it could be considered to form a distinct HBS, termed HBS-4. These binding lysines, Lys-81, Lys-84, and Lys-86, are on the top right side of the canonical site on the molecular surface (Fig. 3C). Together, these two sites form a “T”-shaped structure on the surface of FGF-7, uninterrupted by acidic residues, with the canonical binding forming the top of the T (Fig. 3C). However, Lys-123 and Lys-155 are a significant distance away from Lys-81, Lys-84, and Lys-86 (average 1.8 nm), so this T may not be a single large binding site but instead may accommodate HS in different orientations.
FGF-7 may have another binding site. Because only the binding lysines can be labeled, HBS that lack lysines will not be identified by Protect and Label, e.g. FGF-7 may have an HBS-3, as in FGF-1 and FGF-2 at the N terminus (Fig. 2, A and B), but the corresponding amino acids Arg-65, Arg-67, and Arg-68, cannot be identified by our approach.
FGF-9 has fewer labeled lysines (only six) compared with the other three FGFs. The overall heparin-binding site is located on one side of the protein, whereas the other side of the protein, despite being very basic and possessing lysines, appears to lack heparin-binding sites (Fig. 4, A–D). The alignment of the canonical binding site identifies Lys-168, Lys-179, and Lys-183, and these residues were all biotinylated (Fig. 2C and Table 3). Biotin-labeled Lys-87, between β-strand 3 and β-strand 4 (Fig. 2C and Table 3), and Lys-196 (Fig. 2C and Table 3), which is at the C terminus, are quite close to each other, and both of them are also close to the canonical binding site. Thus, these residues could be considered as an extension of the canonical binding site (Fig. 4, B and D). Lys-154 is on the loop area between β-strands 9 and 10 and can also be considered as a part of the canonical binding site (Fig. 4, A and C, and Table 3). Thus, the six labeled lysines identified here may form one large heparin-binding site, comprising the canonical site, extended by adjacent lysines. This is contributed to by sequences equivalent to HBS-1 and HBS-2, so in FGF-9 these sites may have merged (Fig. 2C). FGF-9 has Arg-62, Arg-63, and Arg-64, which cannot be identified by our strategy but may form HBS-3.
In FGF-18, Lys-155, Lys-156, Lys-161, and Lys-164 (Figs. 2D, 5, A and C, and Table 3) were all biotinylated, and these correspond to the canonical binding site identified by sequence alignment (Fig. 2D). Lys-113, Lys-115, Lys-119, and Lys-125, which were also labeled with biotin, are quite close to this and could also be considered to be part of it (Figs. 2D, 5, A and C, and Table 3). Residues corresponding to HBS-2 in FGF-1, Lys-100 and Lys-102, are also present in FGF-18 (Figs. 2, A and D, 5, B and D, and Table 3). However, the HBS-2 of FGF-18 has expanded into a much larger area, because another five lysines (Lys-50, Lys-82, Lys-176, Lys-180, and Lys-187), identified in FGF-18, are physically adjacent to Lys-100 and Lys-102, although in different parts of the linear sequence as follows: N terminus, loop 3/4, part of strand β-12 and the C terminus (Figs. 2D, 5, B and D, and Table 3). Lys-187 is not in the crystal structure of the protein. Nonetheless, because it is at the C terminus, it may also be close to these residues, which together would form a substantial HBS-2.
FGF-18 has a similar heparin-binding lysine as the HBS-3 of FGF-1 at its N terminus, Lys-50 (Table 3). However, Lys-50 is physically close to the extended HBS-2 of FGF-18. As a result, the expanded HBS-2 of FGF-18 is now adjacent to its HBS-3 identified by Lys-50, so these may operate as a single binding site. Thus, FGF-18 may possess a canonical binding site that is expanded by Lys-155, Lys-156, Lys-161, and Lys-164, and Lys-113, Lys-115, Lys-119 and Lys-125, and a single large secondary binding (HBS-2/3) to which contribute Lys-50, Lys-82, Lys-100, Lys-102, and also Lys-180, Lys-176, and Lys-187. The dimensions of a heparin chain, with its intrinsic flexibility and the added reach of appended sulfate groups, can readily span the protein surface even to bind amino acids that reside on opposite faces of the protein as seen in FGF-9 (Lys-87, Lys-168, Lys-183, Lys-17, and Lys-154) and FGF-18 (Lys-100 and Lys-176). This finding is similar to the situation seen with proteins possessing bipartite HBS, such as VEGF (7).
Circular Dichroism Spectroscopy Reveals Generally Distinct Secondary Structures among FGF Subfamily Members
To explore the structural changes of FGFs and their sugar complexes, SRCD was used. The heparin alone (1.4 mg/ml) control spectrum was first collected to confirm that it presented only a small signal, which would not affect the spectra of the FGF heparin complexes. The spectra of six FGFs and their heparin complexes were collected under the same conditions by SRCD (Fig. 6). The spectra of FGF-1, FGF-2, FGF-7, and FGF-9 typically include the β and random coil with little α-helix, which all have a peak around 190 nm (Fig. 6, A–D), and the secondary structure content is close to that observed in the corresponding structures determined by x-ray crystallography (39–42) or NMR spectroscopy (supplemental Table S2) (43, 44). Similarly to FGF-1 and FGF-2, which exhibited significant changes when the 1:5 molar ratio heparin was introduced, FGF-7, FGF-9, and FGF-18 also showed visible changes (Fig. 6, A–E). Nonetheless, the spectral changes, which related to the conformational change of the secondary structure, were different in these six FGFs. The spectra of the FGF-1 subfamily showed that, as heparin is introduced, the signals of FGF-1 and FGF-2 become more linear (Fig. 6, A and B), highlighting the similarity within this subfamily. The spectral features of FGF-7, FGF-18, and in particular FGF-9 became more pronounced in the presence of heparin. Only the spectrum of FGF-21 was virtually unaffected (Fig. 6F), agreeing with previous studies, confirming that FGF-21 binds to heparin only weakly (45, 46).
DSF and Heparin-dependent Thermostabilizing Effects on FGFs
DSF was developed previously to screen heparin binding specificity through the effects of binding on the thermal stabilities of FGF-1, FGF-2, and FGF-18 (15). DSF was employed here to determine the melting temperature of FGF-7, FGF-9, and FGF-21 and their sugar complexes to provide a data set across five FGF subfamilies. A range of heparin concentrations was tested against a fixed amount of FGFs (5 μm). The melting temperatures of FGF-7 and FGF-9 increased, as the concentration of heparin increased (Fig. 7A and supplemental Fig. S11A), whereas that of FGF-21 did not (supplemental Fig. S12A). Calculation of the first derivative (Fig. 7B and supplemental Figs. S11B and S12B) allowed the melting temperature to be determined at each concentration of polysaccharide (Fig. 7C and supplemental Figs. S11C and S12C) (see under “Experimental Procedures”). These data show that the Tm of FGF-7 progressively increased as the concentration of heparin increased from 0.5 μm (molar ratio FGF-7/heparin 10:1) to 5 μm (molar ratio FGF-7/heparin 1:1) and was then unchanged at higher concentrations of the sugar (Fig. 7C). In contrast, the effect of heparin on FGF-9 was apparent from a lower concentration (0.05 μm) of the polysaccharide (molar ratio FGF-9/heparin 100:1) and then progressively increased up to 5 μm of heparin (molar ratio FGF-9/heparin 1:1) (supplemental Fig. S11C) showing that heparin can cause a concentration-dependent thermal stabilization of FGF-7 and FGF-9, which is distinct for each of these FGFs. Furthermore, heparin influences the thermal stability of FGF-9 more than FGF-7. A range of heparin concentrations (0.5–500 μm) binding to a fixed concentration (5 μm) of FGF-21 was also tested by DSF (supplemental Fig. S12C) showing that heparin did not significantly influence the thermal stability of FGF-21, even at the highest concentrations (molar ratio of 100:1, heparin/FGF-21). The binding of FGF-21 to heparin cannot be measured by this method, in all likelihood because this interaction is too weak (45, 46); therefore, the His6 tags do not interact with heparin.
If we compare the Tm of these three FGFs (Fig. 7C and supplemental Figs. S11C and S12C and supplemental Table S3–S5) with that of FGF-1, FGF-2, and FGF-18 (15), FGF-2 (55.4 °C), FGF-9 (53.7 °C) and FGF-18 (56.9 °C) have a similar melting temperature; however, FGF-7 is more stable with a Tm at 60.7 °C, whereas FGF-1 (48.5 °C) and FGF-21 (48.2 °C) are less stable. After binding to heparin, FGF-2 has a dramatic increase of Tm (22.2 °C) (15), and FGF-9 is similar in this respect (19.1 °C) (supplemental Fig. S11C and supplemental Table S4), whereas the Tm of FGF-1 and of FGF-18 (15) increase to a lesser extent (FGF-1, 13.5 °C; FGF-18, 14.9 °C), and FGF-7 has the lowest increase (7.5 °C) (Fig. 7C and supplemental Table S3).
Characterization of the Thermostabilizing Effect of Different Polysaccharides on FGFs
To determine structures in the polysaccharide responsible for binding FGF-7 and FGF-9, we used full-length homogeneously modified heparin derivatives (Table 1) and libraries of oligosaccharides of various lengths bearing a range of different sulfation patterns. Three FGFs (FGF-1, FGF-2, and FGF-18) were tested only with cation-modified heparins and GAGs, because their binding to the other heparin derivatives has been published (15) (47). The experiments were all based on an approximate molar ratio of 1:1 protein to polysaccharide. At this ratio, the stabilization effect of heparin on both FGF-7 and FGF-9 was in the medium to high range, suitable for the measurement of binding specificity. The singly desulfated species of heparin had contrasting effects on FGF-7 and FGF-9. Singly desulfated heparins (D2, D3, and D4; Table 1) were all around 50% as effective as heparin in stabilizing FGF-9 (Fig. 8A). In contrast, for FGF-7 there is a trend of decreasing effectiveness from D2 (no N-S) to D3 (no 2-S) and D4 (no 6-S), and FGF-7 is less sensitive to the loss of a sulfate at any one position and to the loss of any two sulfates than FGF-9 (Fig. 8A). Interestingly, heparin lacking N- and 2-O-sulfates or 6-O- and 2-O-sulfates is as effective at stabilizing FGF-7 as heparin lacking just 6-O-sulfate, whereas heparin without N- and 6-O-sulfate is much less effective. Totally desulfated heparin is ineffective, whereas persulfated heparin is no more effective than the mono-desulfated species. Thus, FGF-7 clearly has a preference for N- and 6-O-sulfate and their conformational effects. FGF-9 shows a similar but less pronounced preference for N- and 6-O-sulfate when challenged with mono-sulfated heparins (D5, D6, and D7, Fig. 8A). However, totally desulfated heparin is as effective as heparin with just a 2-O-sulfate and persulfated heparin being less effective than native heparin. Thus, FGF-9 can be viewed as being somewhat more heavily influenced by the overall charge of the polysaccharide, but it still has a clear preference for N- and 6-O-sulfate groups and their conformational effect on heparin.
Other GAGs were also compared with the heparin compounds. HS exhibited 60% of the thermal stabilization of heparin for FGF-7, ∼30% for FGF-9, ∼50% for FGF-1 and FGF-2, and just ∼20% for FGF-18, which might reflect the relative abundance of suitable structures in HS for these FGFs (Fig. 8A and supplemental Fig. S13). HA and CS interacted only weakly with these FGFs, whereas DS was more effective, although it interacted more weakly than HS (Fig. 8A and supplemental Fig. S13). This may result from a strong preference for IdoUA residues, which are absent in HA and CS. The basic repeating disaccharide unit of heparin is a disaccharide termed DP2 (degree of polymerization 2). To understand the minimal length of oligosaccharide required to stabilize FGF-7 and FGF-9, various lengths of heparin oligosaccharides from DP2 to DP12 were tested by DSF. There is a trend of increasing effect from DP2 to DP12 observed, with FGF-7 and FGF-9 (Fig. 8B). This showed that the length of heparin oligomer is important for binding to these FGFs. Binding to FGF-7 was only apparent from DP4 and longer. As the length of the oligosaccharide increased from DP4 to DP8, there was a progressive increase in binding (Fig. 8B), but from DP8 to DP12, there was no measurable difference. FGF-9 in contrast clearly had a weak interaction with DP2, which then increased with oligosaccharide length up to a maximum at DP10 (Fig. 8B). Thus, FGF-9 appeared to require a larger structure than FGF-7 and was able to bind the shortest units. Both FGF-7 and FGF-9 were stabilized less effectively by DP12 than by heparin (Fig. 8B). Larger sugar structures may either bind differently or contain rare substructures that are particularly effective at binding these proteins.
The effect of the cation form of heparin was also tested using heparins in which cations had been exhaustively exchanged into the appropriate ion forms using cation exchange resin (20). Na+, K+, Ca2+, and Zn2+ forms of heparin had an ∼30% lower thermal stabilization effect on FGF-7 and FGF-9 compared with standard heparin (Fig. 8C). Interestingly, the Cu2+ form of heparin had a markedly reduced thermal stabilization effect on FGF-7 but not on FGF-9 (Fig. 8C). Cu2+ can cause the rigidification of the heparin chain (48), to which FGF-7 may be particularly sensitive. All the cation forms have a similar effect on FGF-2, which does not affect heparin binding (supplemental Fig. S13). Na+ and Cu2+ decrease binding to FGF-18 (∼50% of Na+ and ∼20% of Cu2+) (Fig. 8C). Na+, K+, Ca2+ forms of heparin have quite a similar effect on FGF-1, a reduction in binding (∼20%), but ∼30% for Zn2+ to FGF-1 compared with standard heparin (supplemental Fig. S13). FGF-1 binding to Cu2+ heparin could not be measured by this method as the melting curve became flat reflecting the documented effect of Cu2+ on FGF-1 (49).
The thermal stabilization effects on the five FGFs, of heparin fragments (DP2–12) and the modified heparin library (D1-D9), were analyzed by principal component analysis (PCA) (Fig. 9), showing distinct effects on FGF stabilization according to substitution pattern and size. PCA analysis of these spectra revealed that the first three principal components (PCs) described 93.34% of the variance and differentiated the FGFs (Fig. 9). According to the three-dimensional plot (Fig. 9), FGFs have binding specificities to these sugars. Although there is no simple relationship between substitution pattern and Tm, this is not unexpected, because removal of sulfate groups alters the overall conformation resulting in complex changes in structure (20). FGF-1 and FGF-2 bind to similar sugars, although it is known that their structural effects are distinct (47). This showed that there are similarities in subfamily members; however, they can still be separated by PC1 and PC2 (Fig. 9). PC1 differentiated the FGFs into three groups, consisting of FGF-18 and then FGF-1 and FGF-2, and FGF-7 and FGF-9; PC2 into three groups consisting of FGF-7 and FGF-18, FGF-1 and FGF-2, and FGF-9; and PC3 separated them into three groups, consisting of FGF-2 and FGF-7, FGF-1 and FGF-18, and FGF-9. PC1, PC2, and PC3 combined were able to differentiate all the FGFs (Fig. 9).
DISCUSSION
To address the specificity of the recognition of the HS co-receptor by FGFs, we have characterized this interaction from different perspectives as follows: the sites in the FGF that are recognized by the sugar, the sugar structures that are important for binding FGFs, and the effect of sugar binding on the FGF structure in solution.
Taking the present Protect and Label data (Figs. 1 and 3–5) together with those published previously (26), how some aspects of heparin binding may have evolved across the FGFs can be proposed. As expected from sequence alignments, Protect and Label identifies the consensus heparin-binding site, HBS-1, in all the FGFs (Figs. 1–5) (26). It is important to note that the Protect and Label technique only identifies lysine residues that are in contact with heparin for the duration of the protection step. Therefore, the lysines identified here and in a previous study (26) as bound to heparin represent real molecular contact points in the FGF·heparin complexes in solution. Because FGFs are small, single domain proteins and their lysine residues are exposed to solvent, labeled lysines are very unlikely to be protected due to intra-protein interactions resulting from conformational change induced by heparin binding. The fact that the biotinylation step is achieved on intact protein released from heparin under native conditions also argues strongly that the labeled lysines are contact points in the complex.
Our approach identifies the three documented HBSs in FGF-1 and FGF-2 (Table 3 and Fig. 2) (26, 34, 44) and the best characterized FGFs from the perspective of heparin binding. Thus, in the FGF-1 subfamily, the secondary HBS at β9 and the loop between β-strand 9 and 10 (HBS-2) and at the N terminus of the proteins (HBS-3) are conserved. Although the affinity of these secondary binding sites for heparin is modest (KD, 120 ± 50 μm (38)), there is evidence that they affect the activity of these FGFs. For example, deletion of HBS-3 at the N terminus of FGF-2 causes a small but significant reduction in the mitogenic activity of the growth factor (50). Together with the conservation of these sites within the subfamily, this argues that the secondary binding sites are functionally significant.
In FGF-7, a new secondary HBS, HBS-4, between β2 and 3, was identified. HBS-1 in FGF-7 has clearly expanded compared with the FGF-1 subfamily. The consequence is that the canonical HBS-1 together with HBS-4 form a large T-shaped patch on the surface of the protein. This indicates that FGF-7 may bind extended sulfated sequences in HS in a variety of orientations. This larger binding site in FGF-7 does not result in reduced stringency of interaction. On the contrary, FGF-7 has a slower association rate constant, ka, compared with FGF-1 and FGF-2, suggesting a requirement for a more precise alignment of the protein and particular sequences of saccharides if a molecular collision of the two is to result in a molecular complex and that electrostatic steering is less dominant in complex formation. Moreover, the observation that dodecasaccharides derived from heparin, DP12, persulfated heparin, and HS do not afford the same thermal stabilization as full-length heparin suggests that there may be rare structures in heparin that allow for a stronger interaction. These may encompass 3-O-sulfates on glucosamine, which have been shown to be critical for high affinity interactions of FGF-7 with heparin (51).
In FGF-18 the canonical HBS-1 has expanded considerably compared with the FGF-1 subfamily. Moreover, HBS-2 has also expanded, and as a consequence, these two sites form a single large HBS. This more extensive site requires a larger minimal oligosaccharide for binding than FGF-1, FGF-2, or FGF-7, with a DP10 accounting for the binding observed with heparin, and it will accept sugar sequences presenting a wide range of combinations of sulfate groups (Fig. 7) (15). This suggests that FGF-18 binds HS with quite a low stringency given an S-domain (or an NAS-S-NAS domain combination) of minimal length DP6.
Despite its very high basicity, FGF-9 has the most restricted HBS, consisting of just an extended HBS-1. However, compared with FGF-1 and FGF-2, this site does not accommodate the smaller oligosaccharides effectively. Moreover, as observed for FGF-7, the longer oligosaccharides, DP10 and DP12, do not bind FGF-9 as well as heparin. FGF-9 also has the slowest association rate constant, ka, of the FGFs. Together, these data indicate a mode of binding to heparin that is less dependent on electrostatic steering and require a reasonably precise alignment of HBS-1 and the polysaccharide, if a molecular collision is to give rise to a complex. The observation that, of all the FGFs, FGF-9 is the only one to significantly bind, albeit weakly, completely desulfated heparin supports the idea that electrostatic bonding contributes less to the interaction of FGF-9 with heparin. However, FGF-9 does not interact with HA, which has a different backbone to desulfated heparin (β1–3GlcNAc-GlcA versus β1–4GlcNAc-IdoUA). Given the conformational versatility of iduronate residues in heparin (52), this suggests that FGF-9 is sensitive to the conformation of the polysaccharide backbone and the uronic acid. Thus, as least some of the sulfation requirements observed for FGF-9 binding (Fig. 4) may be due to sugar conformation (53). The SRCD spectra show that the predominant solution structures of the FGFs are the combinations of β-sheets and unstructured regions, as would be expected. The same is true when the FGFs are bound to heparin, which acts as a proxy for the natural ligand, HS. However, regardless of whether the FGFs are bound to heparin or not, there is no common secondary structural form. Nevertheless, there are some similarities, most obviously between the closely related FGF-1 and FGF-2 (both members of FGF-1 subfamily). More surprising, however, is the similarity between the distantly related pair, FGF-7 and FGF-21, the latter of which binds heparin only weakly. This may indicate that these two FGFs converge to a similar secondary structure, despite their relatively low sequence similarity when heparin is bound. FGF-21 undergoes only a slight conformational change on binding heparin and may adopt a conformation very close to that required for complex formation and signaling before heparin binding. There is also some similarity between these two and FGF-9 with heparin bound, and together, these observations may indicate a common conformation for some heparin-bound FGFs. Because these three FGFs exhibit distinct receptor specificity (1, 10, 11), this suggests that the receptor specificity resides in their surface amino acids, rather than with the FGF conformation, and their different HBS characteristics might contribute as follows: FGF-9 possesses one HBS site (HBS-1, Figs. 2C and 4), whereas FGF-7 has two (HBS-1 and HBS-4, Figs. 2B and 3).
The present data demonstrate conservation of HBSs in the FGF-1 subfamily and considerable variation in both HBSs and in the sugar structures recognized across five FGF subfamilies. Alignment of the amino acid sequences of the FGFs suggests that the HBSs are conserved within subfamilies (Fig. 2). For example, all FGF-7 subfamily members display sequence conservation in both canonical and secondary HBSs (Fig. 2B). In agreement with this, a previous study identified Arg-186, Arg-193, and Lys-195 as part of the HBS of FGF-10 (13). Moreover, at the extension of HBS-1, all the FGF-7 subfamily members have lysines or arginines (FGF-3, Lys-102; FGF-22, Lys-99, and FGF-10, Lys-136) (Fig. 2B) at the same position as FGF-7, Lys-123. FGF-3, FGF-10, and FGF-22 may also have an HBS-4 like FGF-7, but with a lower affinity due to the presence of fewer basic residues, one arginine for FGF-3 and FGF-22 and two lysines for FGF-10 (Fig. 2B). Similar arguments for the conservation of both canonical and secondary HBSs can be made for the FGF-8 and FGF-9 subfamilies (Fig. 2, C and D).
The evolution of the FGF signaling system is closely related to the development of multicellular and complex life forms. In evolutionary terms, the increase in the demands on signaling required the diversification of the extent of signaling systems and their overall capacity. The FGFs have evolved by exploiting several features to provide additional diversity, while maintaining a conserved heparin-binding site, HBS-1. Alignments of the four invertebrate FGFs, Pyramus and Thisbe from Drosophila, and EGL-17 and LET-756 from C. elegans also revealed a probable HBS-1. In addition, Pyramus and Thisbe may also possess an HBS-2 equivalent to Lys-72 of FGF-1 and EGL-17 likewise at Lys-176 to Lys-187 of FGF-18. With the exception of EGL-17, they all have a potential HBS-3 at the same position as FGF-1, but all four apparently lack an HBS-4. Although the FGF family shows considerable variation in amino acid sequence, the resulting secondary structure has provided only moderate additional diversity. For example, there is evidence that several FGFs (FGF-7 and FGF-18, and to some extent FGF-9) adopt similar conformations on heparin binding, and their solution conformations therefore seem unlikely to determine their distinct receptor specificities. More likely, the identity of surface-exposed amino acids and the development of additional HBS (e.g. HBS-4 in FGF-7) provide the required additional diversity and hence increased signaling capacity. Whereas changes in HBS-1 could clearly have an impact on both the transport of FGFs (6) and the assembly of receptor complexes, the role of the secondary HBSs is less clear.
Thus, it would appear that in the course of the expansion of the FGF family, there has been a diversification of the specificity of FGF-heparin interactions and HBSs. However, selection pressure to maintain a degree of functional coherence within each FGF subfamily may have prevented their further divergence within subfamilies. These results highlight the central role in FGF function of HS binding, which has allowed a balance between diversification and conservation of HBSs during metazoan evolution.
Supplementary Material
Acknowledgment
We thank the Proteomics Core Facility at EMBL for technical assistance.
This work was supported in part by the Cancer and Polio Research Fund, the North West Cancer Research Fund, and Biotechnology and Biological Sciences Research Council.

This article contains supplemental Figs. S1–S13 and Tables S1–S5.
- FGF
- fibroblast growth factor
- HS
- heparan sulfate
- HBS
- heparin-binding site
- GAG
- glycosaminoglycan
- SRCD
- synchrotron radiation circular dichroism
- DSF
- differential scanning fluorimetry
- PMHS
- porcine mucosal heparan sulfate
- HA
- hyaluronic acid
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- DP
- heparin oligosaccharides of degree of polymerization
- MST
- microscale thermophoresis
- HCD
- higher collision decomposition
- PCA
- principal component analysis
- PC
- principal component.
REFERENCES
- 1. Ornitz D. M., Itoh N. (2001) Fibroblast growth factors. Genome Biol. 2, REVIEWS3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991) Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- 3. Rapraeger A. C., Krufka A., Olwin B. B. (1991) Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708 [DOI] [PubMed] [Google Scholar]
- 4. Lin X., Buff E. M., Perrimon N., Michelson A. M. (1999) Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development 126, 3715–3723 [DOI] [PubMed] [Google Scholar]
- 5. Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 6. Duchesne L., Octeau V., Bearon R. N., Beckett A., Prior I. A., Lounis B., Fernig D. G. (2012) Transport of fibroblast growth factor2 in the pericellular matrix is controlled by the spatial distribution of its binding sites in heparan sulfate. Plos. Biol. 10, e1001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ori A., Wilkinson M. C., Fernig D. G. (2008) The heparanome and regulation of cell function. Structures, functions, and challenges. Front. Biosci. 13, 4309–4338 [DOI] [PubMed] [Google Scholar]
- 8. Murphy K. J., Merry C. L., Lyon M., Thompson J. E., Roberts I. S., Gallagher J. T. (2004) A new model for the domain structure of heparan sulfate based on the novel specificity of K5 lyase. J. Biol. Chem. 279, 27239–27245 [DOI] [PubMed] [Google Scholar]
- 9. Chang Z., Meyer K., Rapraeger A. C., Friedl A. (2000) Differential ability of heparan sulfate proteoglycans to assemble the fibroblast growth factor receptor complex in situ. FASEB J. 14, 137–144 [DOI] [PubMed] [Google Scholar]
- 10. Ornitz D. M., Xu J., Colvin J. S., McEwen D. G., MacArthur C. A., Coulier F., Gao G., Goldfarb M. (1996) Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 271, 15292–15297 [DOI] [PubMed] [Google Scholar]
- 11. Zhang X., Ibrahimi O. A., Olsen S. K., Umemori H., Mohammadi M., Ornitz D. M. (2006) Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 281, 15694–15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) Interactions between heparan sulfate and proteins. The concept of specificity. J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rahmoune H., Chen H. L., Gallagher J. T., Rudland P. S., Fernig D. G. (1998) Interaction of heparan sulfate from mammary cells with acidic fibroblast growth factor (FGF) and basic FGF. Regulation of the activity of basic FGF by high and low affinity binding sites in heparan sulfate. J. Biol. Chem. 273, 7303–7310 [DOI] [PubMed] [Google Scholar]
- 14. Makarenkova H. P., Hoffman M. P., Beenken A., Eliseenkova A. V., Meech R., Tsau C., Patel V. N., Lang R. A., Mohammadi M. (2009) Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci. Signal. 2, ra55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uniewicz K. A., Ori A., Xu R., Ahmed Y., Wilkinson M. C., Fernig D. G., Yates E. A. (2010) Differential scanning fluorimetry measurement of protein stability changes upon binding to glycosaminoglycans. A screening test for binding specificity. Anal. Chem. 82, 3796–3802 [DOI] [PubMed] [Google Scholar]
- 16. Thompson S. M., Connell M. G., van Kuppevelt T. H., Xu R., Turnbull J. E., Losty P. D., Fernig D. G., Jesudason E. C. (2011) Structure and epitope distribution of heparan sulfate is disrupted in experimental lung hypoplasia. A glycobiological epigenetic cause for malformation? BMC Dev. Biol. 11, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ke Y., Wilkinson M. C., Fernig D. G., Smith J. A., Rudland P. S., Barraclough R. (1992) A rapid procedure for production of human basic fibroblast growth factor in Escherichia coli cells. Biochim. Biophys. Acta 1131, 307–310 [DOI] [PubMed] [Google Scholar]
- 18. Powell A. K., Ahmed Y. A., Yates E. A., Turnbull J. E. (2010) Generating heparan sulfate saccharide libraries for glycomics applications. Nat. Protoc. 5, 821–833 [DOI] [PubMed] [Google Scholar]
- 19. Yates E. A., Santini F., Guerrini M., Naggi A., Torri G., Casu B. (1996) 1H and 13C NMR spectral assignments of the major sequences of 12 systematically modified heparin derivatives. Carbohydr. Res. 294, 15–27 [DOI] [PubMed] [Google Scholar]
- 20. Rudd T. R., Guimond S. E., Skidmore M. A., Duchesne L., Guerrini M., Torri G., Cosentino C., Brown A., Clarke D. T., Turnbull J. E., Fernig D. G., Yates E. A. (2007) Influence of substitution pattern and cation binding on conformation and activity in heparin derivatives. Glycobiology 17, 983–993 [DOI] [PubMed] [Google Scholar]
- 21. Delehedde M., Lyon M., Gallagher J. T., Rudland P. S., Fernig D. G. (2002) Fibroblast growth factor-2 binds to small heparin-derived oligosaccharides and stimulates a sustained phosphorylation of p42/44 mitogen-activated protein kinase and proliferation of rat mammary fibroblasts. Biochem. J. 366, 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delehedde M., Lyon M., Vidyasagar R., McDonnell T. J., Fernig D. G. (2002) Hepatocyte growth factor/scatter factor binds to small heparin-derived oligosaccharides and stimulates the proliferation of human HaCaT keratinocytes. J. Biol. Chem. 277, 12456–12462 [DOI] [PubMed] [Google Scholar]
- 23. Duchesne L., Tissot B., Rudd T. R., Dell A., Fernig D. G. (2006) N-Glycosylation of fibroblast growth factor receptor 1 regulates ligand and heparan sulfate co-receptor binding. J. Biol. Chem. 281, 27178–27189 [DOI] [PubMed] [Google Scholar]
- 24. Jerabek-Willemsen M., Wienken C. J., Braun D., Baaske P., Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hadian K., Griesbach R. A., Dornauer S., Wanger T. M., Nagel D., Metlitzky M., Beisker W., Schmidt-Supprian M., Krappmann D. (2011) NF-κB essential modulator (NEMO) interaction with linear and Lys-63 ubiquitin chains contributes to NF-κB activation. J. Biol. Chem. 286, 26107–26117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ori A., Free P., Courty J., Wilkinson M. C., Fernig D. G. (2009) Identification of heparin-binding sites in proteins by selective labeling. Mol. Cell. Proteomics 8, 2256–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lees J. G., Miles A. J., Wien F., Wallace B. A. (2006) A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics 22, 1955–1962 [DOI] [PubMed] [Google Scholar]
- 28. Whitmore L., Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy. Methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 29. Whitmore L., Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lobley A., Whitmore L., Wallace B. A. (2002) DICHROWEB. An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18, 211–212 [DOI] [PubMed] [Google Scholar]
- 31. Sreerama N., Woody R. W. (1993) A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209, 32–44 [DOI] [PubMed] [Google Scholar]
- 32. Sreerama N., Venyaminov S. Y., Woody R. W. (1999) Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8, 370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niesen F. H., Berglund H., Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 34. Faham S., Hileman R. E., Fromm J. R., Linhardt R. J., Rees D. C. (1996) Heparin structure and interactions with basic fibroblast growth factor. Science 271, 1116–1120 [DOI] [PubMed] [Google Scholar]
- 35. DiGabriele A. D., Lax I., Chen D. I., Svahn C. M., Jaye M., Schlessinger J., Hendrickson W. A. (1998) Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature 393, 812–817 [DOI] [PubMed] [Google Scholar]
- 36. Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. (2000) Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 37. Baird A., Schubert D., Ling N., Guillemin R. (1988) Receptor- and heparin-binding domains of basic fibroblast growth factor. Proc. Natl. Acad. Sci. U.S.A. 85, 2324–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kinsella L., Chen H. L., Smith J. A., Rudland P. S., Fernig D. G. (1998) Interactions of putative heparin-binding domains of basic fibroblast growth factor and its receptor, FGFR-1, with heparin using synthetic peptides. Glycoconj. J. 15, 419–422 [DOI] [PubMed] [Google Scholar]
- 39. Ye S., Luo Y., Lu W., Jones R. B., Linhardt R. J., Capila I., Toida T., Kan M., Pelletier H., McKeehan W. L. (2001) Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs. Biochemistry 40, 14429–14439 [DOI] [PubMed] [Google Scholar]
- 40. Eriksson A. E., Cousens L. S., Weaver L. H., Matthews B. W. (1991) Three-dimensional structure of human basic fibroblast growth factor. Proc. Natl. Acad. Sci. U.S.A. 88, 3441–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Romero A., Pineda-Lucena A., Giménez-Gallego G. (1996) X-ray structure of native full-length human fibroblast-growth factor at 0.25-nm resolution. Eur. J. Biochem. 241, 453–461 [DOI] [PubMed] [Google Scholar]
- 42. Plotnikov A. N., Eliseenkova A. V., Ibrahimi O. A., Shriver Z., Sasisekharan R., Lemmon M. A., Mohammadi M. (2001) Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition. J. Biol. Chem. 276, 4322–4329 [DOI] [PubMed] [Google Scholar]
- 43. Moy F. J., Seddon A. P., Böhlen P., Powers R. (1996) High resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry 35, 13552–13561 [DOI] [PubMed] [Google Scholar]
- 44. Lozano R. M., Pineda-Lucena A., Gonzalez C., Angeles Jiménez M., Cuevas P., Redondo-Horcajo M., Sanz J. M., Rico M., Giménez-Gallego G. (2000) 1H NMR structural characterization of a nonmitogenic, vasodilatory, ischemia-protector, and neuromodulatory acidic fibroblast growth factor. Biochemistry 39, 4982–4993 [DOI] [PubMed] [Google Scholar]
- 45. Harmer N. J., Pellegrini L., Chirgadze D., Fernandez-Recio J., Blundell T. L. (2004) The crystal structure of fibroblast growth factor (FGF) 19 reveals novel features of the FGF family and offers a structural basis for its unusual receptor affinity. Biochemistry 43, 629–640 [DOI] [PubMed] [Google Scholar]
- 46. Goetz R., Beenken A., Ibrahimi O. A., Kalinina J., Olsen S. K., Eliseenkova A. V., Xu C., Neubert T. A., Zhang F., Linhardt R. J., Yu X., White K. E., Inagaki T., Kliewer S. A., Yamamoto M., Kurosu H., Ogawa Y., Kuro-o M., Lanske B., Razzaque M. S., Mohammadi M. (2007) Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 27, 3417–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rudd T. R., Uniewicz K. A., Ori A., Guimond S. E., Skidmore M. A., Gaudesi D., Xu R., Turnbull J. E., Guerrini M., Torri G., Siligardi G., Wilkinson M. C., Fernig D. G., Yates E. A. (2010) Comparable stabilization, structural changes, and activities can be induced in FGF by a variety of HS and non-GAG analogues. Implications for sequence-activity relationships. Org. Biomol. Chem. 8, 5390–5397 [DOI] [PubMed] [Google Scholar]
- 48. Rudd T. R., Skidmore M. A., Guimond S. E., Guerrini M., Cosentino C., Edge R., Brown A., Clarke D. T., Torri G., Turnbull J. E., Nichols R. J., Fernig D. G., Yates E. A. (2008) Site-specific interactions of copper(II) ions with heparin revealed with complementary (SRCD, NMR, FTIR, and EPR) spectroscopic techniques. Carbohydr. Res. 343, 2184–2193 [DOI] [PubMed] [Google Scholar]
- 49. Landriscina M., Bagalá C., Mandinova A., Soldi R., Micucci I., Bellum S., Prudovsky I., Maciag T. (2001) Copper induces the assembly of a multiprotein aggregate implicated in the release of fibroblast growth factor 1 in response to stress. J. Biol. Chem. 276, 25549–25557 [DOI] [PubMed] [Google Scholar]
- 50. Seno M., Sasada R., Kurokawa T., Igarashi K. (1990) Carboxyl-terminal structure of basic fibroblast growth factor significantly contributes to its affinity for heparin. Eur. J. Biochem. 188, 239–245 [DOI] [PubMed] [Google Scholar]
- 51. Wang F., Kan M., McKeehan K., Jang J. H., Feng S., McKeehan W. L. (1997) A homeo-interaction sequence in the ectodomain of the fibroblast growth factor receptor. J. Biol. Chem. 272, 23887–23895 [DOI] [PubMed] [Google Scholar]
- 52. Ferro D. R., Provasoli A., Ragazzi M., Torri G., Casu B., Gatti G., Jacquinet J. C., Sinay P., Petitou M., Choay J. (1986) Evidence for conformational equilibrium of the sulfated l-iduronate residue in heparin and in synthetic heparin mono- and oligosaccharides. NMR and force-field studies. J. Am. Chem. Soc. 108, 6773–6778 [Google Scholar]
- 53. Canales A., Lozano R., López-Méndez B., Angulo J., Ojeda R., Nieto P. M., Martín-Lomas M., Giménez-Gallego G., Jiménez-Barbero J. (2006) Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue. FEBS J. 273, 4716–4727 [DOI] [PubMed] [Google Scholar]
- 54. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 55. Blaber M., DiSalvo J., Thomas K. A. (1996) X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry 35, 2086–2094 [DOI] [PubMed] [Google Scholar]
- 56. Olsen S. K., Li J. Y., Bromleigh C., Eliseenkova A. V., Ibrahimi O. A., Lao Z., Zhang F., Linhardt R. J., Joyner A. L., Mohammadi M. (2006) Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 20, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer. An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.