Background: How TGF-β signaling pathway is modulated by intracellular trafficking is not fully understood.
Results: Tollip interacts with Smad7 and TGF-β receptors and negatively regulates cellular signaling and function of TGF-β.
Conclusion: Tollip modulates intracellular trafficking and degradation of TGF-β receptors.
Significance: Tollip is a novel modulator of TGF-β signaling pathway.
Keywords: Endosomes, Protein Degradation, Signal Transduction, SMAD Transcription Factor, Transforming Growth Factor β (TGFβ), Tollip
Abstract
Upon activation, TGF-β type I receptor (TβRI) undergoes active ubiquitination via recruitment of E3 ligases to the receptor complex by Smad7. However, how ubiquitination of TβRI is coupled to intracellular trafficking, and protein degradation remains unclear. We report here that Tollip, an adaptor protein that contains both ubiquitin-associated domains and endosome-targeting domain, plays an important role in modulating trafficking and degradation of TβRI. Tollip was previously demonstrated to possess a functional role in modulating the signaling of interleukin-1 and Toll-like receptors. We identify here that Tollip interacts with Smad7, a major modulatory protein involved in the negative regulation of TGF-β signaling. Overexpression of Tollip antagonizes TGF-β-stimulated transcriptional response, Smad2 phosphorylation, and epithelial-mesenchymal transition. Tollip also interacts with ubiquitinated TβRI, and such interaction requires ubiquitin-associated domains of Tollip. The interaction and intracellular colocalization of Tollip with TβRI is enhanced by Smad7. Overexpression of Tollip accelerates protein degradation of activated TβRI. In addition, Tollip alters subcellular compartmentalization and endosomal trafficking of activated TβRI. Collectively, our studies reveal that Tollip cooperates with Smad7 to modulate intracellular trafficking and degradation of ubiquitinated TβRI, whereby negatively regulates TGF-β signaling pathway.
Introduction
Members of the transforming growth factor-β (TGF-β) superfamily play essential roles in a variety of physiological and biological processes including embryonic development, tumorigenesis, inflammation, differentiation, and apoptosis (1). The canonical signaling events induced by TGF-βs start by binding of the ligands to the cognate type II receptor that then recruits type I receptor to form the heteromeric complex where TGF-β type I receptor (TβRI)3 is activated by phosphorylation in its GS domain (2). The activated TβRI phosphorylates and activates receptor-restrictive Smads (i.e. Smad2 and Smad3 for TGF-β signaling) which form a heterocomplex with the common Smad, Smad4. The activated Smad complex is translocated into the nucleus, where it regulates the expression of the target genes (1, 3). Smad7, consisting of MH1 and MH2 domains, is one of the key negative regulators of TGF-β signaling pathway. Smad7 antagonizes TGF-β signaling through multiple mechanisms, such as by interfering with the recruitment of receptor-restrictive Smads into the type I receptor upon ligand activation (4) by inducing degradation of TβRI receptor via recruitment of E3 ubiquitin ligases such as Smurfs (5, 6) and by recruiting protein phosphatase 1 (PP1) to the receptor complex (7). The other important inhibitory Smad of TGF-β family is Smad6 (8), which together with Smad7 fall into the inhibitory Smad subfamily.
Endocytosis of cell surface receptors is an important regulatory event in signal transduction. Like most other cell surface proteins, TGF-β receptors undergo endocytosis and degradation upon activation (9). The internalization process of TGF-β receptors takes place mainly through two distinct endocytic pathways including clathrin- and caveolin-mediated pathways. The clathrin-mediated endocytosis targets receptors to the early endosomes in which the receptors may continue their signaling activity (9). The internalized receptors can be sorted to late endosomes and targeted to lysosomes for degradation (10). On the other hand, the lipid-raft-mediated caveolar internalization contains the Smad7-Smurf2 complex and facilitates receptor degradation (9).
Tollip (Toll-interacting protein) was initially identified as an adaptor protein involved in the signaling of interleukin-1 (IL-1) receptor (11). It is a small protein consisting of 274 residues in humans and contains a C2 (protein kinase C conserved region 2) domain in the central region. One of the major functions of the C2 domain is to anchor Tollip to the sorting endosomes (12). Tollip also contains a CUE (coupling of ubiquitin to endoplasmic reticulum degradation) domain in the C terminus, and this domain functions as an interaction motif for ubiquitinated proteins (13). In the N terminus Tollip contains a Tom1 binding domain (TBD) that is involved in the interaction with Tom1, a protein that binds ubiquitinated proteins (14). Tollip may recruit Tom1 and subsequently clathrin onto the sorting endosomes (12, 14). Via its endosome-targeting domain and ubiquitin binding domains, Tollip is required for intracellular trafficking of IL-1 receptor and the ubiquitination-dependent degradation of the receptor (15). In addition, Tollip can interact with IL-1 receptor-associated kinase 1 (IRAK-1) before IL-1β stimulation and suppress the kinase activity of IRAK-1 (11, 16). Overexpression of Tollip also impairs signaling pathways downstream of NF-κB and JNK upon Toll-like receptor activation (16–18). In agreement with the observations that Tollip plays a crucial role in the signaling of IL-1 receptor and Toll-like receptors, the mice depleted of Tollip have dysregulated inflammatory cytokine production in response to IL-1β and lipopolysaccharide (19).
In the present study we identified that Tollip is able to interact with Smad7. Functionally, Tollip suppresses TGF-β signaling via cooperation with Smad7. Smad7 facilitates recruitment of Tollip to TβRI. Subsequently, Tollip preferentially associates with ubiquitinated TβRI and accelerates degradation of activated TβRI. These findings, therefore, uncover a novel function of Tollip in the regulation of TGF-β signaling pathway.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Human Tollip cDNA and its truncation mutants were subcloned into the mammalian expression vector pRc/CMV (Invitrogen) with a FLAG epitope tag added to the N terminus by a PCR-based method. The GFP-tagged Tollip was constructed by subcloning into pEGFPc1 (Clontech, Mountain View, CA) to fuse with an enhanced green fluorescence protein at the N terminus. The human Smad7 (including its truncation mutants) and Smad2 were subcloned into the mammalian expression vector pCS2+MT with six Myc tags at the N terminus. The constructs of Smurf1, TβRI, and its constitutively active mutant were gifts from Dr. Yeguang Chen (Tsinghua University, Beijing, China). All clones were confirmed by DNA sequencing. The Tollip short hairpin RNA (shRNA) and Smad7 shRNA constructs were generated using a lentiviral system as previously reported (20, 21). In short, the annealed small interfering RNA (siRNA) cassettes with targeting sequences of AGCCATGGAGGACATGTTC for human Tollip or AGGTCACCACCATCCCCA for human Smad7 were inserted into the pBS-SKII-hU6 vector downstream of the hU6 promoter. The siRNA expression cassette was then subcloned into the FG12 vector and confirmed by DNA sequencing. The FG12 plasmid containing Tollip or Smad7 shRNA was directly used in cell transfection to silence expression of endogenous Tollip or Smad7.
Antibodies and Reagents
The antibodies were purchased as follows: rabbit anti-phospho-Smad2, anti-Tollip and anti-Snail antibodies from Cell Signaling Technology (Danvers, MA); anti-Myc, anti-HA, anti-total Smad2, anti-Smad7, anti-vimentin, and anti-ubiquitin antibodies from Santa Cruz Biotechnology (Santa Cruz, CA); mouse anti-FLAG antibody from Sigma; rabbit anti-TβRI and anti-GM130 antibodies from Abcam (Cambridge, MA); rabbit anti-EEA1 antibody from BD Biosciences; Alexa Fluor 488 donkey anti-mouse IgG, Alexa Fluor 546 goat anti-mouse, rabbit IgG, and Hoechst 33342 from Molecular Probes (Eugene, OR); Cy-5-labeled goat anti-mouse and rabbit IgG from Jackson ImmunoResearch (Baltimore Pike, PA).
Cell Culture and Transfection
HEK293T cells, HepG2 cells, HeLa cells, and mouse embryonic fibroblasts were cultured at 37 °C in 5% CO2 in DMEM containing 10% FBS. Transient transfection was performed using PolyJet™ DNA in Vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD) for HepG2 cells and mouse embryonic fibroblasts, calcium phosphate precipitation or polyethyleneimine method for HEK293T and HeLa cells.
Luciferase Reporter Assay, Immunoprecipitation, Immunoblotting, and Immunofluorescence
The reporter assay was performed as previously described (22). Briefly, 12 h after transfection, the cells were serum-starved for 16 h before harvesting. We used a luciferase assay kit (KenReal, Shanghai, China) to measure the luciferase activity with a luminometer (Berthold Technologies, Bad Wildbad, Germany). Reporter activity was normalized to co-transfected Renilla. The experiments were repeated in triplicate. Immunoprecipitation was performed as previously described (22). Briefly, cells were washed 3 times and then lysed for 30 min at 4 °C. The homogenates were centrifuged for 20 min at 12,000 rpm at 4 °C. The supernatant was added with antibody and mixed overnight at 4 °C. Protein A/G plus agarose was added for 4 h at 4 °C. The immunoprecipitated was washed three times in cell lysis buffer followed by Western blotting analysis. For the two-step co-immunoprecipitation (23), the protein complexes were eluted with lysis buffer containing 250 mm NaCl and 100 μg/ml 3×FLAG peptide (Sigma) for 3 h at 4 °C. The second immunoprecipitation was performed using 150 μl of eluate and 350 μl of lysis buffer containing 464 mm NaCl and 2 μg of anti-HA antibody for 3 h. Immunoblotting and immunofluorescence was performed as previously described (22).
RNA Isolation and Quantitative RT-PCR
The cells were lysed in TRIzol reagent (Invitrogen). Total RNA was purified according to the manufacturer's instructions. Quantitative real-time PCR was done with the SYBR Green PCR system (Applied Biosystems, Foster City, CA) using actin as an internal control for normalization. Primers used for each gene are listed as follows: 5′-CAACCTCGTCATGTCCTAC-3′ and 5′-GCTGGTACACTGTTGGCATC-3′ for human Tollip, 5′-CTCCTGCTGTGCAAAGTGTT-3′ and 5′-GATTCACAGCAACACAGCCT-3′ for human Smad7, 5′-GATCATTGCTCCTCCTGAGC-3′ and 5′-ACTCCTGCTTGCTGATCCAC-3′ for β-actin, 5′-ACACCCCCTGTTGGTGTCTTT-3′ and 5′-TGTATGTGGCAATGCGTTCTC-3′ for human E-cadherin, 5′-AGCCAACCTTAACTGAGGAGT-3′ and 5′-GGCAAGTTGATTGGAGGGATG-3′ for human N-cadherin, 5′-GTCCGCAGTCTTACGAGGAG-3′ and 5′-GCTTGAGGGTCTGAATCTTGCT-3′ for human Twist, 5′-AGATGCATATTCGGACCCAC-3′ and 5′-CCTCATGTTTGTGCAGGAGA-3′ for human Slug, 5′-GATGATGAATGCGAGTCAGATGC-3′ and 5′-ACAGCAGTGTCTTGTTGTTGT-3′ for human Zeb1, and 5′-GCGATGGTCATGCAGTCAG-3′ and 5′-CAGGTGGCAGGTCATTTTCTT-3′ for human Zeb2.
Protein Degradation Analysis
Twenty-four hours after transfection, HEK293T cells were treated with 50 μg/ml cycloheximide and harvested at various time points. The cell lysate was subjected to immunoblotting.
Subcellular Fractionation with Continuous Iodixanol Gradients
Subcellular fractionation was performed using a method as previously reported (24, 25). In short, the cells were washed with cold PBS and homogenized in buffer (20 mm Hepes, pH 7.4, 1 mm EDTA, 250 mm sucrose with protease and phosphatase inhibitors) 20 times with a 22-gauge needle and 5 times with a 26 gauge needle. The nuclei were pelleted at 3000 × g for 10 min at 4 °C. After measurement of protein concentration, a 10-20-30% continuous gradient was generated by mixing postnuclear supernatant 1:1 with 60% iodixanol (Opti-Prep, Axis-Shield PoC, Oslo, Norway) and layered under 1.2 ml of 20% iodixanol with 1.2 ml of 10% iodixanol at the top of the centrifuge tubes. The samples were centrifuged at 260,000 × g for 3 h at 4 °C.
Statistical Analysis
Statistical significance was assessed by Student's t test.
RESULTS
Interaction of Tollip with Smad7
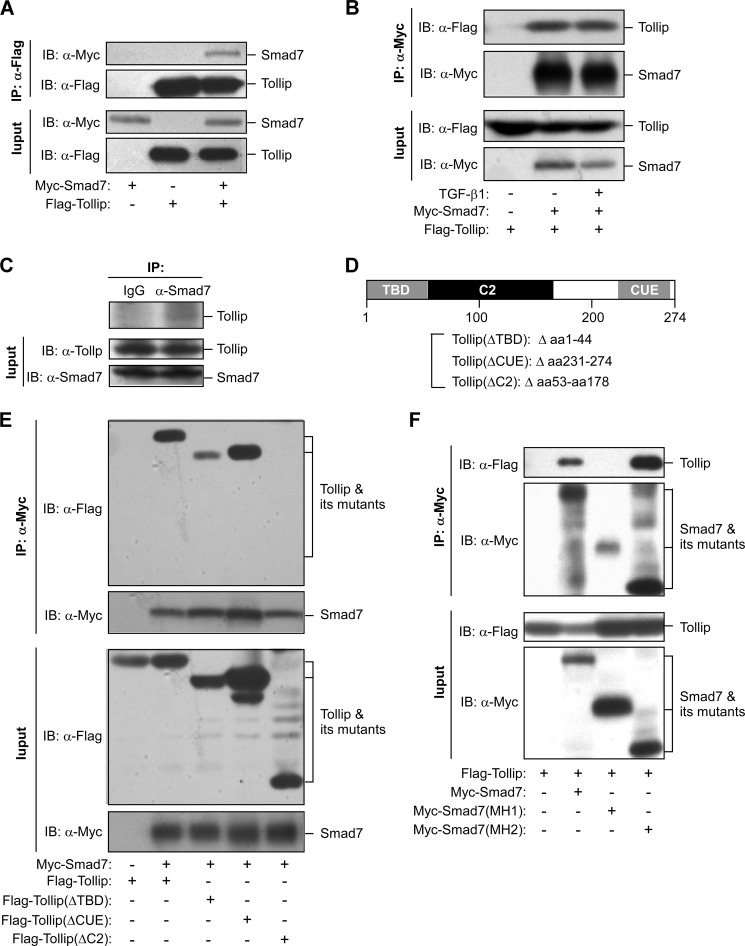
We originally used a yeast two-hybrid screening to identify proteins that interact with full-length Smad7 and found that Tollip was one of the Smad7-interacting proteins.4 We confirmed the interaction between Smad7 and Tollip by co-immunoprecipitation assays. When Smad7 and Tollip were co-expressed in HEK293T cells, they could interact with each other (Fig. 1A). The interaction of Tollip with Smad7 was not affected by TGF-β1 treatment (Fig. 1B). In addition, we found that endogenous Tollip could interact with endogenous Smad7 in HEK293T cells (Fig. 1C). On the other hand, Tollip only interacted with Smad7 but not Smad2 (supplemental Fig. S1). We next investigated the functional domain(s) of Tollip implicated in the interaction with Smad7. Tollip contains three functional domains, TBD, C2, and CUE (coupling of ubiquitin to endoplasmic reticulum degradation), as shown in Fig. 1D (13, 14). Smad7 bound efficiently to the full-length Tollip and the mutants with either TBD or CUE being deleted (Fig. 1E). However, no interaction could be detected between Smad7 and Tollip deleted of the C2 domain (Fig. 1E). These results indicate that the C2 domain of Tollip is required for the interaction of Tollip with Smad7. In addition, we analyzed which domain of Smad7 is involved in its interaction with Tollip. We demonstrated that the MH2 domain of Smad7 was sufficient to mediate the interaction with Tollip (Fig. 1F). Collectively, these data reveal that the C2 domain of Tollip is able to specifically interact with the MH2 domain of Smad7.
FIGURE 1.
Interaction of Tollip and Smad7. A, association of overexpressed Tollip with overexpressed Smad7 is shown. HEK293T cells were transfected with the plasmids as indicated. At 48 h post-transfection, the cell lysate was used in immunoprecipitation (IP) and immunoblotting (IB) using the antibodies as indicated. B, the interaction between Tollip and Smad7 was not affected by TGF-β1 treatment. HEK293T cells were transfected with the plasmids as indicated and then treated with TGF-β1 (2 ng/ml) for 2 h before being harvested for IP and IB. C, interaction of endogenous Tollip with endogenous Smad7 is shown. HEK293T cell lysate was subjected to IP with control rabbit IgG or anti-Smad7 antibody followed by IB with an anti-Tollip antibody. D, shown is a diagram depicting the functional domains of Tollip and the mutant Tollip used in the study. E and F, shown is identification of the functional domains involved in the interaction between Tollip and Smad7. Different mutants of Tollip and Smad7 were expressed in HEK293T cells, and coimmunoprecipitation assay were used to identify the region(s) involved in the protein-protein interaction.
Tollip Inhibits TGF-β Signaling
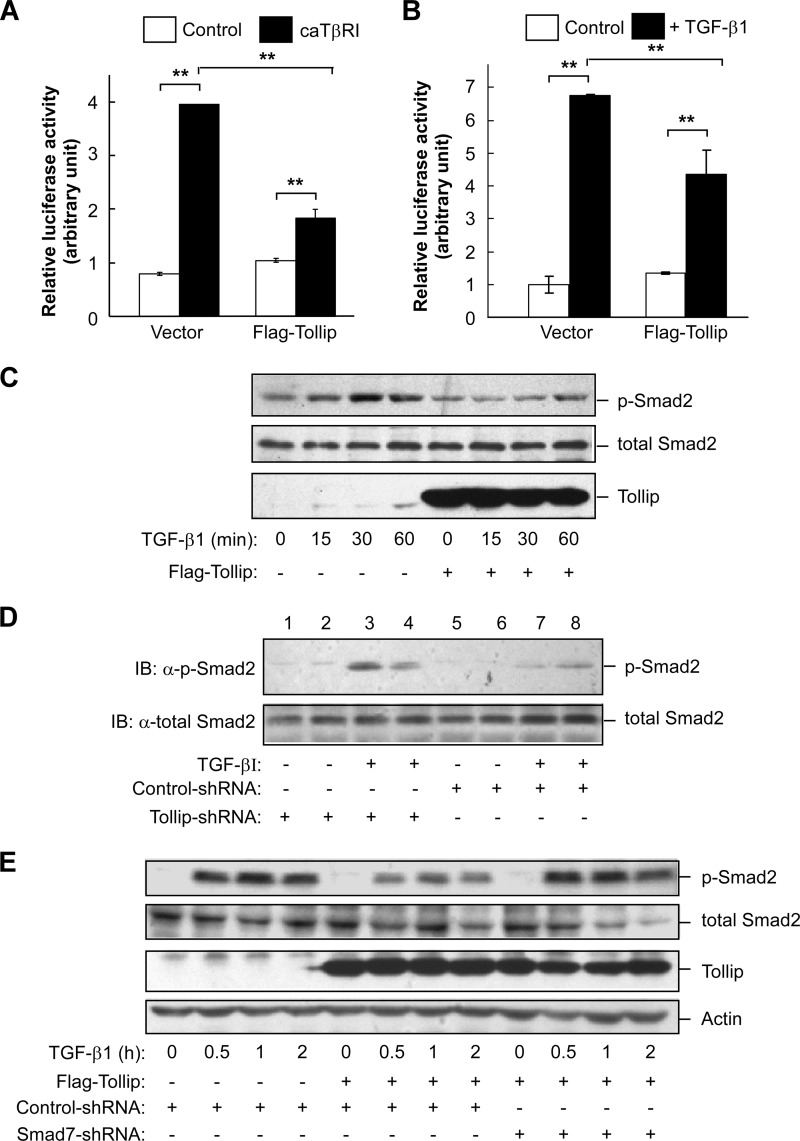
To assess the functional role of Tollip on TGF-β signaling, we first examined the effect of Tollip overexpression on TGF-β-induced transcriptional response using a luciferase reporter that contained four repeats of Smad binding elements as previously described (26). As expected, we found that overexpression of the constitutively active TGF-β type I receptor (caTβRI) was able to stimulate the luciferase reporter in both HEK293T and HepG2 cells (Fig. 2A and supplemental Fig. S2). However, overexpression of Tollip in these two cell lines could significantly reduce caTβRI-mediated activation of the luciferase reporter (Fig. 2A and supplemental Fig. S2). Consistently, TGF-β1-stimulated transcriptional response was lessened by Tollip overexpression in HEK293T cells (Fig. 2B). Taken together, these results indicate that Tollip is inhibitory to TGF-β-mediated signaling pathway.
FIGURE 2.
Tollip inhibits TGF-β signaling. A and B, Tollip suppresses TGF-β-induced transcriptional response. HEK293T cells were transfected with a luciferase reporter containing four repeats of Smad binding element, Renilla-luciferase, Tollip, and constitutively active TβRI (caTβRI, A) as indicated. Luciferase activity was measured 28 h after transfection (A). The cells were treated with TGF-β1 (2 ng/ml) for 24 h before luciferase assay (B). Each experiment was performed in triplicate. The data are presented as the mean ± S.D. after being normalized to Renilla activity. ** indicates p < 0.01 as comparison between the groups as indicated by Student's t test. C, Tollip attenuates TGF-β-induced Smad2 phosphorylation. HepG2 cells were transfected with the plasmids as indicated and then treated with 2 ng/ml TGF-β1 for various times. The cell lysate was used in immunoblotting using the antibody as indicated. D, Tollip knockdown increases TGF-β-stimulated Smad2 phosphorylation. HepG2 cells were transfected with nonspecific shRNA or Tollip-specific shRNA and then treated with TGF-β1 (2 ng/ml) for 1 h. Total cell lysate was subjected to immunoblotting (IB) with the antibodies as indicated. The knockdown efficiency is shown in supplemental Fig. S3A. E, Smad7 is required for suppressing TGF-β signaling by Tollip. HepG2 cells were transfected with nonspecific shRNA or Smad7-specific shRNA in the presence or absence of Tollip expression. The cells were then treated with TGF-β1 (2 ng/ml) for different length of time before being harvested for immunoblotting. The knockdown efficiency of Smad7 is shown in supplemental Fig. S3B.
To further confirm the inhibitory activity of Tollip on TGF-β signaling, we stimulated HepG2 cells with TGF-β1 in the presence or absence of Tollip overexpression and analyzed TGF-β1-mediated phosphorylation of Smad2. Although TGF-β1 treatment could markedly induce phosphorylation of Smad2 in these cells, overexpression of Tollip profoundly suppressed TGF-β-induced phosphorylation of Smad2 (Fig. 2C). Consistently, the TGF-β1-induced Smad2 phosphorylation was enhanced by knockdown of Tollip in HepG2 cells (Fig. 2D). On the other hand, knockdown of Smad7 could abrogate the inhibitory effect of Tollip on TGF-β1-induced Smad2 phosphorylation (Fig. 2E), indicating that the inhibitory effect of Tollip on TGF-β signaling is dependent on Smad7. The effectiveness of using shRNA to silence Tollip and Smad7 in these cells was confirmed by real time RT-PCR and Western blotting analyses (supplemental Fig. S3).
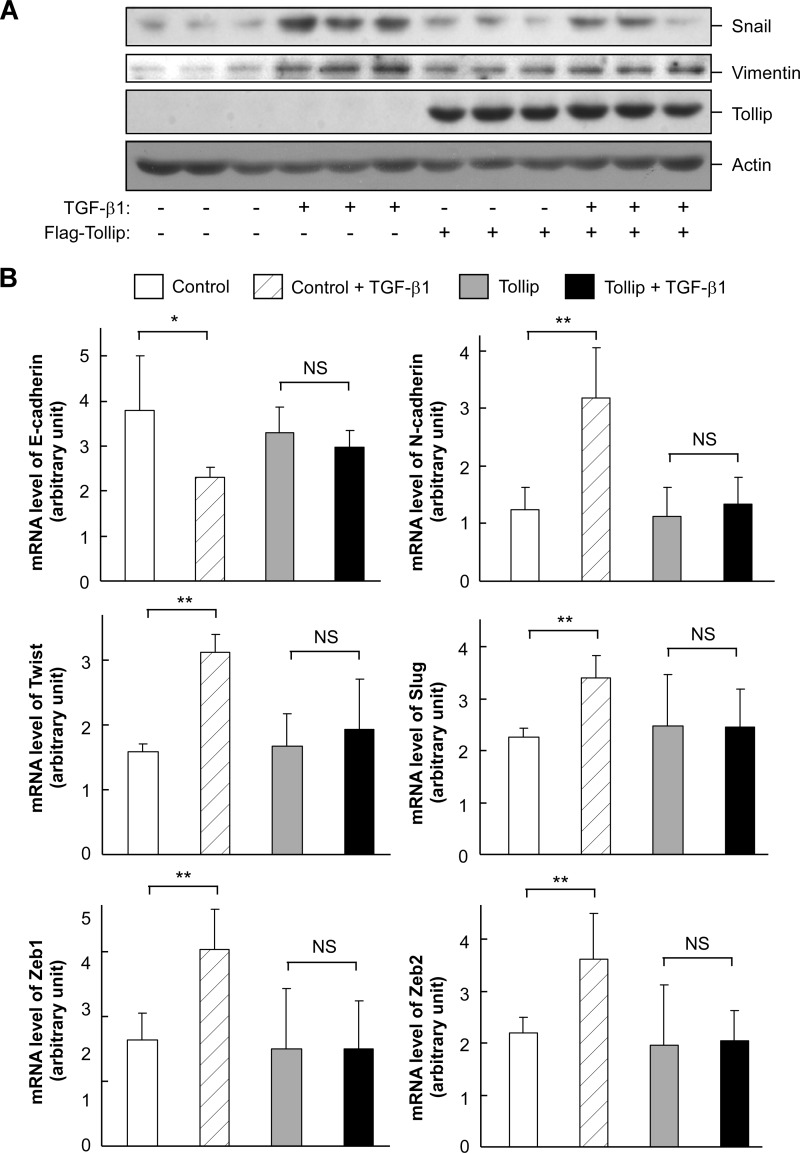
To analyze the potential effect of Tollip on TGF-β action, we examined whether Tollip could alter TGF-β-induced epithelial-mesenchymal transition in HepG2 cells. As expected, TGF-β1 treatment could elevate the protein levels of epithelial-mesenchymal transition markers including Snail and vimentin (Fig. 3A). At the mRNA level, TGF-β1 treatment could significantly reduce E-cadherin expression while increasing the expression levels of N-cadherin, Twist, Slug, Zeb1, and Zeb2 (Fig. 3B). However, Tollip overexpression could attenuate TGF-β1-induced alterations of these epithelial-mesenchymal transition markers (Fig. 3). Collectively, these data indicate that overexpression of Tollip has a negative effect on TGF-β signaling and function.
FIGURE 3.
Tollip attenuates TGF-β-induced epithelial-mesenchymal transition. HepG2 cells were transfected with FLAG-tagged Tollip as indicated and treated with TGF-β1 (2 ng/ml) for 24 h. The cell lysate was used in immunoblotting (A) and quantitative RT-PCR (B). The data are presented as mean ± S.D. ** indicates p < 0.01 by Student's t test. NS stands for non-significant.
Tollip Interacts with TGF-β Type I Receptor in a Smad7-dependent Manner
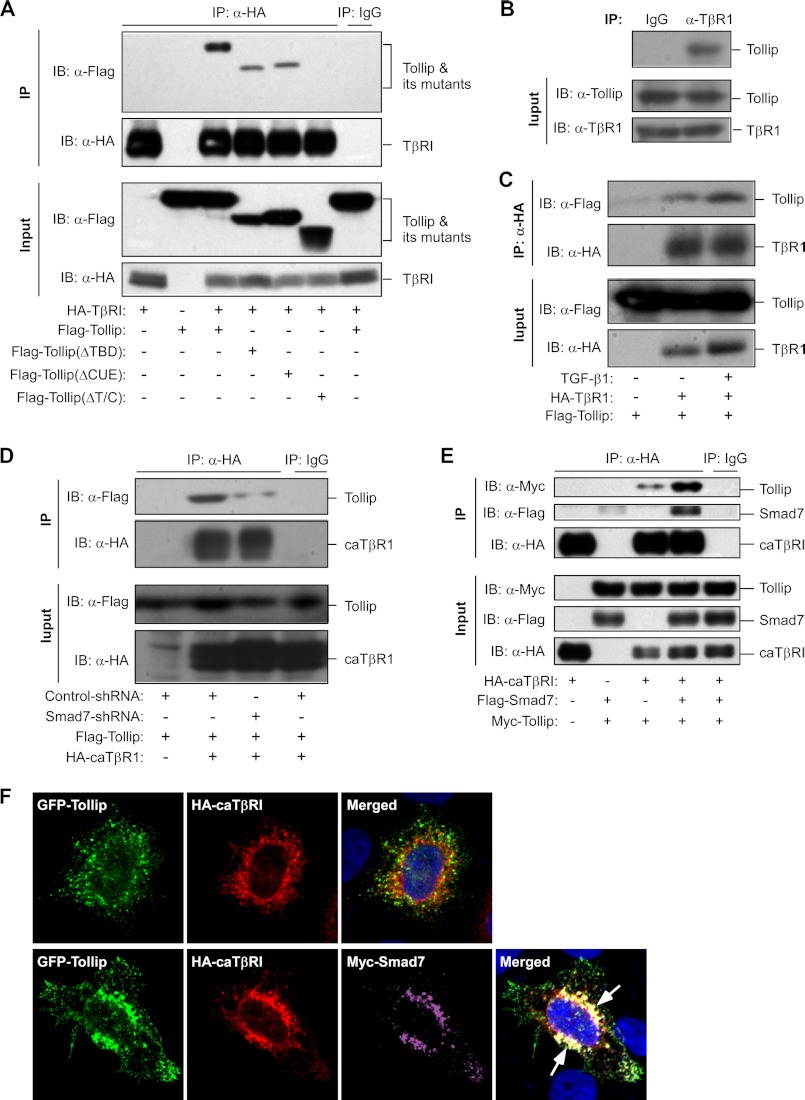
To investigate the molecular mechanism underlying the inhibitory effect of Tollip on TGF-β signaling, we analyzed the interaction between Tollip and TβRI. Using a co-immunoprecipitation assay, we found that Tollip could efficiently bind to TβRI (Fig. 4A). Deletion of either TBD or CUE domain had no obvious effect on the interaction between Tollip and TβRI (Fig. 4A). However, deletion of both the TBD and CUE domains led to complete loss of the interaction capacity of Tollip with TβRI (Fig. 4A). We also found that endogenous Tollip could interact with endogenous TβRI in HepG2 cells (Fig. 4B). Interestingly, TGF-β1 treatment appeared to elevate the interaction between Tollip and TβRI (Fig. 4C), indicating that Tollip preferentially binds activated TβRI.
FIGURE 4.
Tollip interacts with TβRI in a Smad7-dependent manner. A, interaction between overexpressed Tollip with TβRI is shown. HEK293T cells were transfected with the plasmids as indicated. At 48 h post-transfection, the cell lysate was used in immunoprecipitation (IP) and immunoblotting (IB) using the antibodies as indicated. B, interaction of endogenous Tollip with endogenous TβRI. HepG2 cell lysate was used in IP with control rabbit IgG or an anti-TβRI antibody followed by IB with an anti-Tollip antibody. C, TGF-β1 treatment enhances Tollip association with TβRI. HEK293T cells were transfected with the plasmids as indicated and then treated with 2 ng/ml TGF-β1 for 24 h. The cell lysate was subjected to IP and IB using the antibodies as indicated. D, Smad7 knockdown abolished the interaction between Tollip and caTβRI. After infection with lentivirus containing nonspecific shRNA or Smad7-specific shRNA, HEK293T cells were transfected with the plasmids as indicated and then used in IP and IB using the antibodies as indicated. E, Smad7 enhances Tollip-caTβRI interaction. HEK293T cells were transfected with the plasmids as indicated followed by IP and IB using the antibodies as indicated. F, Smad7 increases colocalization of Tollip and caTβRI. HeLa cells were transiently transfected with the plasmids as indicated followed by immunofluorescence staining and confocal analysis. The arrows indicate apparent colocalization of Tollip with Smad7 and caTβRI. The quantitative analysis is shown in supplemental Fig. S5A. The nuclei were stained with Hoechst 33342.
To establish the role of Smad7 in the Tollip/TβRI interaction, we used shRNA to knock down Smad7 and observed that the Tollip/TβRI interaction was significantly abolished with Smad7 down-regulation (Fig. 4D). Consistently, Smad7 depletion in Smad7-depleted mouse embryonic fibroblasts could significantly reduce colocalization of Tollip with caTβRI (supplemental Fig. S4).
We next investigated the effect of overexpressed Smad7 on the interaction of Tollip with TβRI. In a co-immunoprecipitation experiment, we found that overexpression of Smad7 could profoundly enhance the association between Tollip and TβRI (Fig. 4E). The importance of Smad7 on Tollip/TβRI interaction was further analyzed by a confocal experiment. When Tollip and caTβRI were coexpressed, there was little colocalization of Tollip with caTβRI in the cell (Fig. 4F, upper panel). However, overexpression of Smad7 could markedly enhance the co-localization of Tollip with caTβRI (Fig. 4F, lower panel and supplemental Fig. S5A), further supporting our findings that Smad7 is required for the interaction of Tollip with caTβRI and that the inhibitory effect of Tollip on TGF-β signaling is dependent on Smad7 (Fig. 2E).
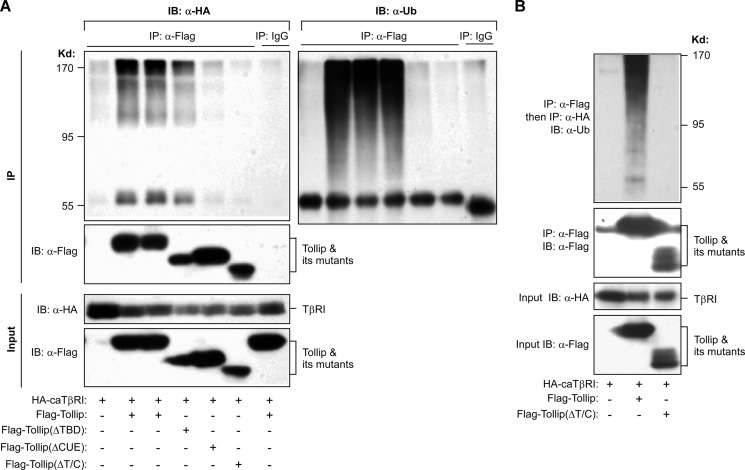
Tollip Interacts with Ubiquitinated TGF-β Type I Receptor through Its Ubiquitin Binding Domains
Previous studies have revealed that both of the TBD and CUE domains of Tollip are involved in interaction with ubiquitinated proteins (12–14). As both the TBD and CUE domains are required for the interaction of Tollip with TβRI (Fig. 4A), we therefore postulated that Tollip is able to bind ubiquitinated TβRI. To test this hypothesis, we co-expressed caTβRI with Tollip and investigated the ubiquitination status of TβRI. As shown in Fig. 5A, caTβRI could be efficiently pulled down by wild type Tollip and its mutant TollipΔTBD but not by other mutants that lacked CUE (TollipΔCUE) or both TBD and CUE (TollipΔC/T). Interestingly, Tollip interacted more efficiently with the high molecular weight forms of TβRI (Fig. 5A, top left panel). Importantly, the high molecular weight forms of TβRI contained ubiquitin (Fig. 5A, top right panel). The ubiquitination status of caTβRI associated with Tollip was further analyzed by a two-step coimmunoprecipitation experiment. Both caTβRI and Tollip were coexpressed in HEK293T cells, and the FLAG-tagged Tollip was first immunoprecipitated by an anti-FLAG antibody. The immunoprecipitated proteins were dissociated from the beads by incubation with FLAG peptide and subjected to second round immunoprecipitation to isolate caTβRI that was associated with Tollip. Consistently, we found that the Tollip-associated caTβRI did contain ubiquitin (Fig. 5B). On the other hand, deletion of both the CUE and TBD domain lost the capacity to associate with ubiquitinated caTβRI. Together, these data indicate that Tollip preferentially associates with ubiquitinated caTβRI.
FIGURE 5.
Tollip interacts with ubiquitinated TβRI. HEK293T cells were cotransfected with the plasmids as indicated, and the cell lysate was used in immunoprecipitation (IP) and immunoblotting (IB) using the antibodies as indicated. Note that a second IP was used for B after the Tollip/TβRI complex was dissociated from the beads by a free FLAG peptide.
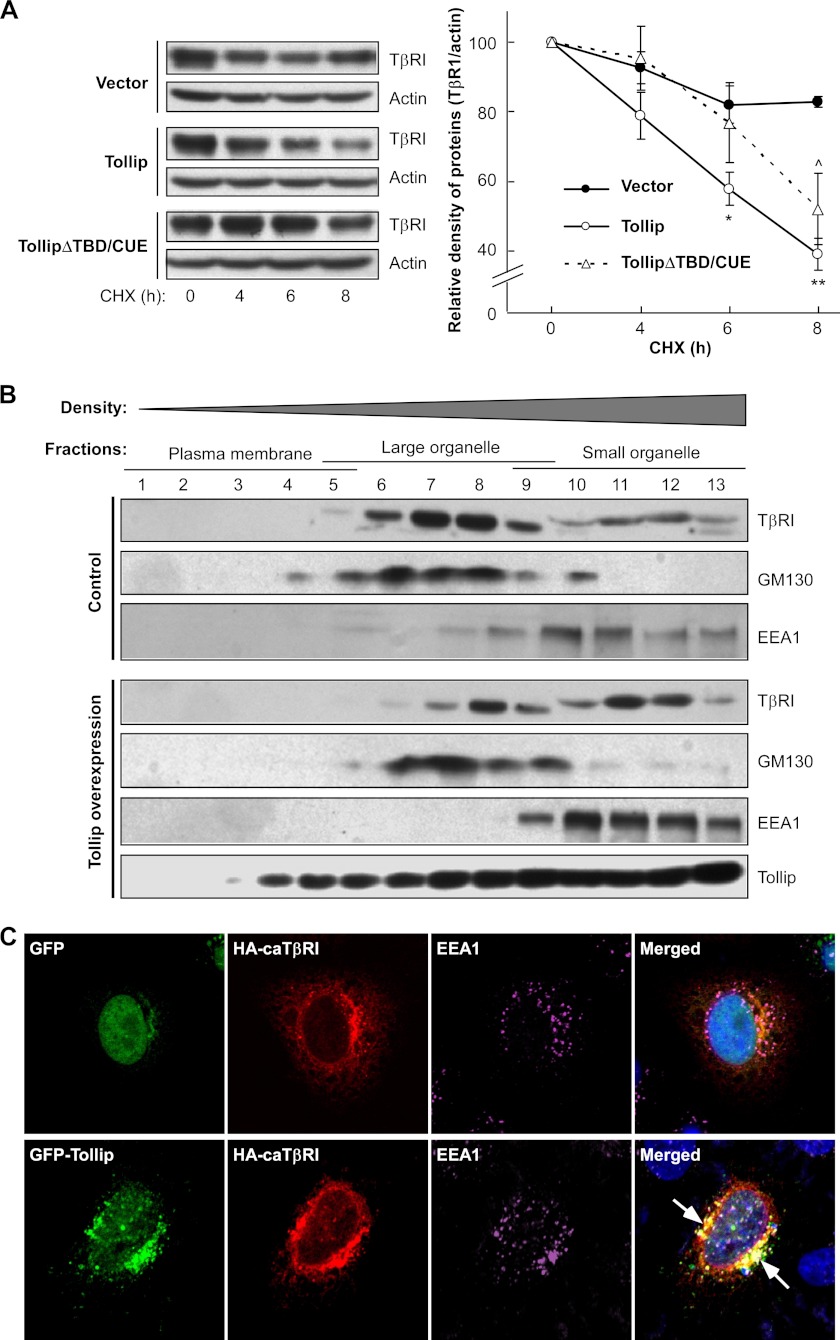
Tollip Accelerates the Degradation of TβRI and Alters Its Subcellular Compartmentalization
Ubiquitination of TβRI is commonly associated with protein degradation (5, 6). As Tollip is able to associate with ubiquitinated caTβRI, we hypothesized that such interaction would facilitate degradation of caTβRI. To address this hypothesis, we examined the half-life of caTβRI in the presence or absence of Tollip overexpression. The cells were treated with cycloheximide to block new protein biosynthesis. Consistent with a previous report (27), the half-life of caTβRI was very long when expressed alone (Fig. 6A). However, when it was co-expressed with Tollip, degradation of caTβRI was significantly accelerated (Fig. 6A). Intriguingly, deletion of both TBD and CUE domains abrogated the pro-degradation effect of Tollip on caTβRI, consistent with our findings that these two domains are required for the association of Tollip with ubiquitinated caTβRI (Fig. 4A). Because Smad7 recruits Smurfs to ubiquitinate activated TβRI for its degradation, we examined the half-life of caTβRI with overexpression of Smad7, Smurf1, and Tollip (supplemental Fig. S6). As expected, overexpression of Smad7 or Tollip alone could elevate degradation of caTβRI. Coexpression of Smad7 and Smurf1 could further increase caTβRI degradation. Interestingly, Tollip and Smad7 had a synergistic effect on caTβRI degradation, indicating that Tollip can cooperate with Smad7 to accelerate TβRI degradation. However, Tollip had no effect to further enhance Smad7/Smurf1-mediated caTβRI degradation. In addition, we observed that Tollip did not alter the interaction of Smad7 with Smurf1 (supplemental Fig. S7). These data, therefore, indicate that Tollip does not affect Smad7-mediated recruitment of E3 ubiquitin ligases and Smurf-mediated ubiquitination of TβR1.
FIGURE 6.
Tollip accelerates degradation of TβRI and alters compartmentalization of TβRI. A, Tollip accelerates turnover of TβRI. HEK293T cells were transfected with constitutively active TβR1 (HA-tagged), Tollip, and mutant Tollip as indicated. At 24 h after transfection, the cells were treated with 50 μg/ml cycloheximide (CHX) for various times and then harvested for immunoblotting with antibodies against HA (to detect TβRI) or actin. The result was quantified by Bandscan software and is shown on the right (mean ± S.D.). *, p < 0.05; **, p < 0.01 between Tollip and control groups; ^, p < 0.05 between TollipΔTBD/CUE and control groups. B, Tollip alters compartmentalization of TβRI. HEK293T cells were transfected with HA-tagged constitutively active TβRI (HA-caTβRI) and FLAG-tagged Tollip as indicated followed by fractionation with iodixanol gradients as described under “Experimental Procedures.” Equal volumes of each fraction were analyzed by immunoblotting with antibodies against HA (to detect HA-caTβRI), EEA1, and GM130. C, Tollip tethers caTβRI to early endosomes. HeLa cells were transiently transfected with the plasmids as indicated followed by immunofluorescence staining and confocal analysis. The arrows indicate colocalization of Tollip with TβRI at early endosomal structures. The quantitative analysis is shown in supplemental Fig. S5B. The early endosomes were stained with EEA1 antibody. The nuclei were stained with Hoechst 33342.
As protein degradation is commonly associated with alteration in subcellular compartmentalization (28, 29), we used gradient fractionation to examine the effect of Tollip on subcellular distribution of caTβRI. According to previous reports (24, 25), the lower density fractions mainly contains plasma membrane, whereas the middle density fractions contain large organelles including Golgi apparatus and the higher density fractions mainly consist of small organelles including endosomal structures. When caTβRI was expressed alone, it was mainly distributed in the fractions containing large subcellular organelles (Fig. 6B). However, when co-expressed with Tollip, caTβRI was partially redistributed from the middle density fractions to the higher density fractions that mainly contain endosomes (Fig. 6B).
It is noteworthy that Tollip contains an endosome anchoring domain, i.e. the C2 domain (12). We, therefore, hypothesized that Tollip might facilitate TβRI degradation by targeting the receptor to the endosomes. To address this issue, we analyzed the co-localization of caTβRI with an early endosome marker EEA1. When caTβRI was expressed alone, it barely had any colocalization with EEA1 (Fig. 6C, top panel). However, when caTβRI was coexpressed with Tollip, both of the proteins were apparently colocalized with EEA1 (Fig. 6C, lower panel, and supplemental Fig. S5B), furthering indicating that Tollip is able to enhance compartmentalization of TβRI to the endosomes.
DISCUSSION
The duration and intensity of TGF-β signaling is precisely controlled at different levels from ligand availability and receptor stability to Smads activity (30). The activity of TGF-β receptor is tightly regulated by a set of their interacting proteins. One of the key negative regulators, Smad7, antagonizes TGF-β signaling mainly by inhibiting receptor-restrictive Smads activation (4) and inducing degradation of TβRI via recruitment of E3 ubiquitin ligases (5, 6). In this study we found that Tollip is able to interact with Smad7 and inhibit TGF-β signaling and action. Tollip is also able to interact with ubiquitinated TβRI and Smad7 facilitates such interaction. Consistently, two ubiquitin-associated domains of Tollip including CUE and TBD domains are required for the interaction between Tollip and TβRI. Furthermore, we demonstrated that Tollip is able accelerate degradation, alter subcellular distribution, and elevate endosome localization of TβRI. Collectively, our studies indicate that Tollip acts an adaptor protein that regulates the compartmentalization and degradation of TβRI.
Our findings are consistent with the known functions of Tollip in regulating the signaling of IL-1 receptor. Tollip was initially identified as an important constituent of the IL-1 receptor signaling pathway (11). Recent studies have revealed that Tollip is implicated in the ubiquitination and endocytosis of IL-1 receptor (15). Tollip is a modular protein that contains three functional domains: an N-terminal TBD, a central C2 domain, and a C-terminal CUE domain. The CUE domain of Tollip directly binds ubiquitinated proteins, whereas the C2 domain is responsible for the association with endosomal membranes. On the other hand, Tom1 is also able to associate with ubiquitinated proteins. Therefore, Tollip is able to bind ubiquitinated proteins directly via its CUE domain or indirectly via its TBD domain. Because of the properties of its functional domains, Tollip has been found to be an important player in regulating endosomal trafficking of ubiquitinated proteins (12, 15).
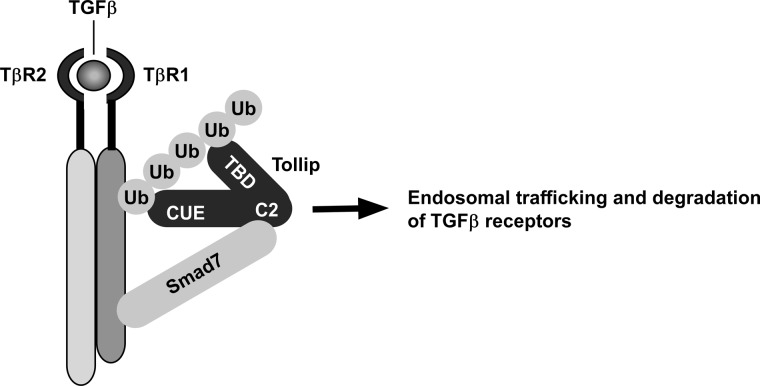
Based on our findings, we propose a model to depict the functional roles of Tollip in modulating TGF-β signaling (Fig. 7). The C2 domain of Tollip interacts with the MH2 domain of Smad7. Through such interaction, Tollip is recruited to the activated TβRI. Via the two ubiquitin-associated domains (CUE and TBD), Tollip is able to form a stable complex with ubiquitinated TβRI. Subsequently, the Tollip-TβRI complex is routed to endosomal compartment and subjected to protein degradation. Based on this model, Smad7 plays two important roles in facilitating degradation of TβRI. On the one hand, Smad7 can recruit E3 ligases (Smurfs) to TβRI and initiate the ubiquitination of the receptor. On the other hand, Smad7 functions as an adaptor protein to assist in recruitment of Tollip to the ubiquitinated TβRI and facilitate the subsequent endosomal trafficking and degradation of the receptor. However, a few issues with this model need to be clarified in the future. First, it is not clear how endosomal trafficking of TβRI is coupled to protein degradation. It is likely that the Tollip-mediated compartmentalization of TβRI is later sorted to late endosomes or lysosomes for degradation. Second, it is unclear whether Tollip plays a role in the crosstalk between IL-1 and TGF-β signaling pathways, as Tollip is a known modulator of IL-1 signaling (16). A few studies have reported IL-1 can influence TGF-β signaling (31, 32). Interleukin-1 receptor antagonist-deficient mice exhibit suppressed TGF-β signaling, whereas IL-1 signaling is enhanced (33). If the intracellular amount of Tollip were a rate-limiting factor, recruitment of Tollip to the IL-1 receptor would prevent it from interacting with TβRI, leading to reduced degradation of TβRI and enhanced TGF-β signaling. In contrast, binding of Tollip to TβRI would also limit its availability to IL-1 receptor. Third, what is the physiological importance of Tollip in regulating TGF-β functions? The mice depleted of Tollip have dysregulated inflammatory responses (19). As TGF-β plays a pivotal role in modulating immune response, it will be important to elucidate in the future whether the dysregulated inflammatory responses in Tollip-ablated mice is partially mediated by alterations in TGF-β signaling pathway.
FIGURE 7.
A model to depict Tollip cooperating with Smad7 to mediate degradation of the TGF-β receptor complex. Tollip is recruited to TGF-β receptor complex by its interaction with Smad7 via its C2 domain. Tollip then forms a complex with ubiquitinated TβRI via its two ubiquitin-associated domains: CUE and TBD. The complex is trafficked to the endosomes through the endosomes-targeting domain of Tollip and then undergoes protein degradation.
Supplementary Material
Acknowledgment
We thank Dr. Yeguang Chen (Tsinghua University, Beijing, China) for TβRI cDNA and its constitutively active clone.
This work was supported by research grants from National Natural Science Foundation of China (30830037, 81021002, and 81130077 (to Y. C.) and 30971660 (to Y. P.)), Ministry of Science and Technology of China (2012CB524900 (to Y. C.) and 2010CB529506 (to Y. P. and Z. W.)), and Chinese Academy of Sciences (KSCX2-EW-R-08 (to Y. C.)).

This article contains supplemental Figs. S1–S7.
Y. Chen, unpublished data.
- TβRI
- TGF-β type I receptor
- caTβRI
- constitutively active TβRI
- CUE
- coupling of ubiquitin to endoplasmic reticulum degradation
- EEA1
- early endosomal autoantigen-1
- IL-1
- interleukin-1
- MH
- Mad-homology domain
- TBD
- target of Myb1 (Tom1) binding domain
- C2
- protein kinase C conserved region 2.
REFERENCES
- 1. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 2. Wrana J. L., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X. F., Massagué J. (1992) TGF β signals through a heteromeric protein kinase receptor complex. Cell 71, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 3. Wrana J. L., Attisano L. (2000) The Smad pathway. Cytokine Growth Factor Rev. 11, 5–13 [DOI] [PubMed] [Google Scholar]
- 4. Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P. (1997) Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 5. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 6. Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. (2001) Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276, 12477–12480 [DOI] [PubMed] [Google Scholar]
- 7. Shi W., Sun C., He B., Xiong W., Shi X., Yao D., Cao X. (2004) GADD34-PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. J. Cell Biol. 164, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imamura T., Takase M., Nishihara A., Oeda E., Hanai J., Kawabata M., Miyazono K. (1997) Smad6 inhibits signalling by the TGF-β superfamily. Nature 389, 622–626 [DOI] [PubMed] [Google Scholar]
- 9. Di Guglielmo G. M., Le Roy C., Goodfellow A. F., Wrana J. L. (2003) Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nat. Cell Biol. 5, 410–421 [DOI] [PubMed] [Google Scholar]
- 10. Chen Y. G. (2009) Endocytic regulation of TGF-β signaling. Cell Res. 19, 58–70 [DOI] [PubMed] [Google Scholar]
- 11. Burns K., Clatworthy J., Martin L., Martinon F., Plumpton C., Maschera B., Lewis A., Ray K., Tschopp J., Volpe F. (2000) Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat. Cell Biol. 2, 346–351 [DOI] [PubMed] [Google Scholar]
- 12. Katoh Y., Shiba Y., Mitsuhashi H., Yanagida Y., Takatsu H., Nakayama K. (2004) Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J. Biol. Chem. 279, 24435–24443 [DOI] [PubMed] [Google Scholar]
- 13. Shih S. C., Prag G., Francis S. A., Sutanto M. A., Hurley J. H., Hicke L. (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 22, 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamakami M., Yoshimori T., Yokosawa H. (2003) Tom1, a VHS domain-containing protein, interacts with tollip, ubiquitin, and clathrin. J. Biol. Chem. 278, 52865–52872 [DOI] [PubMed] [Google Scholar]
- 15. Brissoni B., Agostini L., Kropf M., Martinon F., Swoboda V., Lippens S., Everett H., Aebi N., Janssens S., Meylan E., Felberbaum-Corti M., Hirling H., Gruenberg J., Tschopp J., Burns K. (2006) Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr. Biol. 16, 2265–2270 [DOI] [PubMed] [Google Scholar]
- 16. Zhang G., Ghosh S. (2002) Negative regulation of toll-like receptor-mediated signaling by Tollip. J. Biol. Chem. 277, 7059–7065 [DOI] [PubMed] [Google Scholar]
- 17. Bulut Y., Faure E., Thomas L., Equils O., Arditi M. (2001) Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein. Role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J. Immunol. 167, 987–994 [DOI] [PubMed] [Google Scholar]
- 18. Li T., Hu J., Li L. (2004) Characterization of Tollip protein upon Lipopolysaccharide challenge. Mol. Immunol. 41, 85–92 [DOI] [PubMed] [Google Scholar]
- 19. Didierlaurent A., Brissoni B., Velin D., Aebi N., Tardivel A., Käslin E., Sirard J. C., Angelov G., Tschopp J., Burns K. (2006) Tollip regulates proinflammatory responses to interleukin-1 and lipopolysaccharide. Mol. Cell Biol. 26, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qin X. F., An D. S., Chen I. S., Baltimore D. (2003) Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. U.S.A. 100, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Jiang X., Qin X., Ye D., Yi Z., Liu M., Bai O., Liu W., Xie X., Wang Z., Fang J., Chen Y. (2010) RKTG inhibits angiogenesis by suppressing MAPK-mediated autocrine VEGF signaling and is down-regulated in clear-cell renal cell carcinoma. Oncogene 29, 5404–5415 [DOI] [PubMed] [Google Scholar]
- 22. Feng L., Xie X., Ding Q., Luo X., He J., Fan F., Liu W., Wang Z., Chen Y. (2007) Spatial regulation of Raf kinase signaling by RKTG. Proc. Natl. Acad. Sci. U.S.A. 104, 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harada J., Kokura K., Kanei-Ishii C., Nomura T., Khan M. M., Kim Y., Ishii S. (2003) Requirement of the co-repressor homeodomain-interacting protein kinase 2 for ski-mediated inhibition of bone morphogenetic protein-induced transcriptional activation. J. Biol. Chem. 278, 38998–39005 [DOI] [PubMed] [Google Scholar]
- 24. Chen X. W., Inoue M., Hsu S. C., Saltiel A. R. (2006) RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J. Biol. Chem. 281, 38609–38616 [DOI] [PubMed] [Google Scholar]
- 25. Xie X., Gong Z., Mansuy-Aubert V., Zhou Q. L., Tatulian S. A., Sehrt D., Gnad F., Brill L. M., Motamedchaboki K., Chen Y., Czech M. P., Mann M., Krüger M., Jiang Z. Y. (2011) C2 domain-containing phosphoprotein CDP138 regulates GLUT4 insertion into the plasma membrane. Cell Metab. 14, 378–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagarajan R. P., Chen F., Li W., Vig E., Harrington M. A., Nakshatri H., Chen Y. (2000) Repression of transforming-growth-factor-beta-mediated transcription by nuclear factor κB. Biochem. J. 348, 591–596 [PMC free article] [PubMed] [Google Scholar]
- 27. Yan X., Lin Z., Chen F., Zhao X., Chen H., Ning Y., Chen Y. G. (2009) Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-β signaling. J. Biol. Chem. 284, 30097–30104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katzmann D. J., Babst M., Emr S. D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 29. Bonifacino J. S., Traub L. M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 30. Lönn P., Morén A., Raja E., Dahl M., Moustakas A. (2009) Regulating the stability of TGFβ receptors and Smads. Cell Res. 19, 21–35 [DOI] [PubMed] [Google Scholar]
- 31. Fan J. M., Huang X. R., Ng Y. Y., Nikolic-Paterson D. J., Mu W., Atkins R. C., Lan H. Y. (2001) Interleukin-1 induces tubular epithelial-myofibroblast transdifferentiation through a transforming growth factor-β1-dependent mechanism in vitro. Am. J. Kidney Dis. 37, 820–831 [DOI] [PubMed] [Google Scholar]
- 32. Shephard P., Martin G., Smola-Hess S., Brunner G., Krieg T., Smola H. (2004) Myofibroblast differentiation is induced in keratinocyte-fibroblast co-cultures and is antagonistically regulated by endogenous transforming growth factor-β and interleukin-1. Am. J. Pathol. 164, 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ishida Y., Kondo T., Kimura A., Matsushima K., Mukaida N. (2006) Absence of IL-1 receptor antagonist impaired wound healing along with aberrant NF-κB activation and a reciprocal suppression of TGF-β signal pathway. J. Immunol. 176, 5598–5606 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.