Background: Microarrays of plant-derived oligosaccharides are potentially powerful tools for the high throughput discovery and screening of antibodies, enzymes, and carbohydrate-binding proteins.
Results: Oligosaccharide microarrays were produced, and their utility was demonstrated in several applications.
Conclusion: A new generation of oligosaccharide microarrays will make an important contribution to plant glycomic research.
Significance: High throughput screening technology enables the more effective production of carbohydrate active enzymes and molecular probes.
Keywords: Glycobiology, Glycoconjugate, Glycomics, Microarray, Plant Cell Wall, Carbohydrate-binding Modules, Oligosaccharides
Abstract
Microarrays are powerful tools for high throughput analysis, and hundreds or thousands of molecular interactions can be assessed simultaneously using very small amounts of analytes. Nucleotide microarrays are well established in plant research, but carbohydrate microarrays are much less established, and one reason for this is a lack of suitable glycans with which to populate arrays. Polysaccharide microarrays are relatively easy to produce because of the ease of immobilizing large polymers noncovalently onto a variety of microarray surfaces, but they lack analytical resolution because polysaccharides often contain multiple distinct carbohydrate substructures. Microarrays of defined oligosaccharides potentially overcome this problem but are harder to produce because oligosaccharides usually require coupling prior to immobilization. We have assembled a library of well characterized plant oligosaccharides produced either by partial hydrolysis from polysaccharides or by de novo chemical synthesis. Once coupled to protein, these neoglycoconjugates are versatile reagents that can be printed as microarrays onto a variety of slide types and membranes. We show that these microarrays are suitable for the high throughput characterization of the recognition capabilities of monoclonal antibodies, carbohydrate-binding modules, and other oligosaccharide-binding proteins of biological significance and also that they have potential for the characterization of carbohydrate-active enzymes.
Introduction
Glycans are crucial for plant life and are used for storage, defense, and signaling and as structural cell wall components (1–6). Plant oligo- and polysaccharides are also important components of food and feed and have numerous industrial applications. Starch is the most common carbohydrate in the human diet, whereas plant cell walls provide bulk materials including timber, paper, and cloth, as well as fine chemicals, food ingredients, and biofuel feedstocks (1, 6–8). The complexity and diversity of plant polysaccharides underpin their biological roles and many of their industrially important characteristics, but also produce challenges for research and optimal utilization. A detailed knowledge of the structures, functions, interactions, and occurrence of plant glycans is essential for understanding their complex contributions to plant life and to fully exploit their commercial potential. However, unlike proteins and nucleotides, complex carbohydrates are not readily amenable to sequencing or synthesis, and existing biochemical techniques for glycan analysis, although powerful, are usually low throughput (3, 9).
The development of rapid genome sequencing methods and improvements in protein expression techniques enable the production of large numbers of carbohydrate-active enzymes and carbohydrate-binding proteins including carbohydrate-binding modules (CBMs).2 There has been an exponential increase in the number of entries of these proteins in the Carbohydrate-Active enZYmes (CAZy) database (10), but this has not been matched by structural analysis or determination of their biochemical activities (11). Similarly, monoclonal antibodies (mAb) are immensely valuable molecular probes for carbohydrate research, but their usefulness is dependent on knowledge of the epitopes they recognize (12–15). The rate-limiting step in CBM, mAb, and enzyme production is often a lack of efficient methods for screening their specificities. There is therefore a clear need in plant biology for high throughput (HTP) and high resolution techniques for the analysis of carbohydrate-active proteins including enzymes.
Microarray technology has underpinned the development of multiplexed assays that have revolutionized the HTP analysis of nucleotides, proteins, and increasingly, carbohydrates (16–18). Using microarrays, the abundance of, and interactions between, hundreds or thousands of molecules can be assessed simultaneously using very small amounts of analytes (16–18). Carbohydrate microarrays were first produced in 2002, and a variety of approaches has been developed for the printing and immobilization of oligo- and polysaccharides (19–26). However, the representation of glycomes on these arrays is generally far less comprehensive than is the coverage of transcriptomes/genomes and proteomes by nucleotide and protein arrays, respectively. The primary reason for this is the lack of facile methods for the production of sets of homogeneous, sequence-defined plant oligosaccharide structures (27, 28). In contrast, partially defined polysaccharides are relatively easy to obtain, and microarrays and enzyme-linked immunosorbent assays (ELISAs) populated with such samples have shown potential for HTP screening (29–31). However, most plant polysaccharides and especially those from plant cell walls are complex heteropolymers and, even if pure, will typically accommodate a range of smaller oligosaccharide substructures (or epitopes), and so, polysaccharide-based assays, whether they be microarrays or ELISAs, lack analytical resolution (30–32).
We have developed a new generation of glycan microarrays for plant research based on defined oligosaccharide structures produced either by isolation from polysaccharides or by de novo chemical synthesis. Once coupled to bovine serum albumin (BSA), these neoglycoprotein sets are highly versatile, and microarrays can be printed on a variety of slides and membranes. Most of the oligosaccharides we describe here are derived from, or based on, cell wall polysaccharides that are among the most complex in nature and present particular challenges for HTP analysis, but we have also included starch-related oligosaccharides and novel synthesized structures.
EXPERIMENTAL PROCEDURES
Oligosaccharide Samples
Oligosaccharides were produced either by partial enzymatic or by chemical hydrolysis of source polysaccharides followed by fractionation and purification or were prepared by chemical synthesis. For detailed information on all oligosaccharide samples, see supplemental Table 1.
MALDI-TOF Analysis of Conjugation Efficiency
Analysis was performed as described in Ref. 33.
Monoclonal Antibodies and CBMs
Previously characterized cell wall-directed rat monoclonal antibodies used in this study included LM5 (34), LM6 (35), LM13 and LM16 (36), LM10 and LM11 (37), LM15 (38), and LM21 and LM22 (39). Novel rat monoclonal antibodies were obtained as follows. LM23 was derived subsequent to immunization with a complex pectic immunogen from apple fruits, and this antibody binds to xylosyl residue in a range of antigenic contexts including xylogalacturonan and xylan. LM24 and LM25 were derived subsequent to immunization with a neoglycoprotein generated from a mixture of XXLG and XLLG xyloglucan oligosaccharides (Megazyme, Bray, Ireland). LM12 was derived subsequently to immunization with a neoglycoprotein generated with oligosaccharide structure 16 (see Fig. 1). In all these cases, immunization and hybridoma isolation protocols were carried out as described (35). CBMs were produced as described (40, 41).
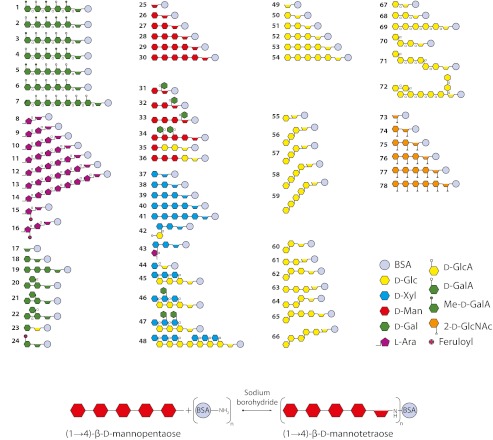
FIGURE 1.
A library of plant oligosaccharides. Upper panel, structures 1–6, 19, 20, 21, 22, and 72 were chemically synthesized, whereas all other oligosaccharides were produced by fractionation of polysaccharides followed by separation and purification. The oligosaccharides are shown after coupling to BSA by reductive amination as shown in the lower panel. α glycosidic linkages are indicated by the small α symbols on the structures, and all other linkages are β. The structures shown are the predominant structures in that sample, and the number codes of the structures are consistent throughout this study. Further details of the oligosaccharides are provided in supplemental Table S1. Note that the structures illustrated in this figure correspond to those listed in supplemental Table S1 but after conjugation and opening of the reducing end sugar.
BSA Conjugation Reaction
Oligosaccharides were conjugated to BSA essentially as described (42). Briefly, BSA was mixed with oligosaccharides and NaCNBH3 (Sigma-Aldrich) in a borate buffer at pH 8. The reaction was allowed to progress at ambient temperature for 96 h. Where necessary, conjugates were purified from reaction mixtures using a spin column with a 10-kDa cut off (Pall, Lund, Sweden). The conjugates were stored in the reaction buffer at 20 °C before use and were stable for at least 6 months whereby CN− from the reducing agent presumably acts as a preservative.
Microarray Printing
Carbohydrate microarrays were printed using two types of microarrays robot, a pin-based MicroGrid II (Digilab/Genomic Solutions, Huntingdon, UK) and a piezoelectric Sprint (Arrayjet, Roslin, UK). For printing on the MicroGrid II, microarrays were printed using four split pins or four solid pins (Digilab), and oligosaccharides were diluted to a 2 mg/ml and a 0.04 mg/ml concentration in deionized water immediately before use and transferred to a 96-well microplate for printing. Microarrays were printed at 16 °C at 35% humidity with one deposit per spot. The same procedure was used to print onto nitrocellulose membrane with a pore size of 0.45 μm (Whatman, Maidstone, UK), FAST Slides (Whatman) nitrocellulose-coated glass slides (Schott, Mainz, Germany), and a range of other surface-modified glass slides (Schott). For printing on the Arrayjet Sprint, the Sprint microarrayer was equipped with a 12-sample high capacity JetSpyder sample pick-up device. Microarrays were printed at 19 °C at 55% humidity, using six drops per spots when printing on nitrocellulose membrane and two drops per spot when printing on all glass slide types. Samples were printed in 55.2% glycerol, 44% water, 0.8% Triton X-100, and the same slides and nitrocellulose as for the MicroGrid II were used.
Microarray Probing
Arrays were blocked by incubation for 1 h in PBS (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.7 mm KH2PO4, pH 7.5) containing 5% w/v low fat milk powder (MPBS) for nitrocellulose probing, 0.05% Tween 20 for FAST Slides, and Schott nitrocellulose-coated glass slides and 0.5 m borate buffer containing 50 mm ethanolamine, pH 8.5, for all other slide types. Arrays were probed for 2 h with antibodies diluted 1/10 in PBS containing 5% w/v low fat milk powder for nitrocellulose membrane or in PBS containing 0.05% Tween 20 for all other slide types. After washing with PBS (all microarray types), nitrocellulose microarrays were incubated for 2 h in either anti-rat or anti-mouse secondary antibodies conjugated to alkaline phosphatase (Sigma, Poole, UK) diluted 1/5000 in 5% MPBS. After further washing in PBS, microarrays were developed using a substrate containing 5-bromo-4-chloro-3-indolylphosphate and nitro blue tetrazolium in alkaline phosphatase buffer (100 mm NaCl, 5 mm MgCl2, 100 mm diethanolamine, pH 9.5). All glass-based slides were incubated for 2 h in either anti-rat or anti-mouse secondary antibodies conjugated to Alexa Fluor 555 (Invitrogen, Life Technologies Europe BV Nærum, Denmark) and subsequently washed in PBS and then de-ionized water.
Scanning and Analysis
Microarrays on nitrocellulose membrane were scanned using a flatbed scanner (Cannon 8800, Søborg, Denmark) and converted to 16-bit grayscale TIFFs. Slides were scanned using a slide scanner (GenePix 4100, Molecular Devices, Sunnyvale, CA). The output from all scanning was analyzed using microarray analysis software (ImaGene 6.0, BioDiscovery, El Segundo, CA). Output from the analysis was further processed as necessary using Excel (Microsoft Denmark, Hellerup, Denmark) and presented as heat maps in which color intensity is correlated to mean spot signals.
Oligosaccharide Microarray Analysis of Phosphorylase Activity
The array was blocked by incubation for 1 h in PBS (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.7 mm KH2PO4, pH 7.5) containing 5% w/v BSA powder. After blocking, the array was washed three times for 5 min with 100 mm MOPS, pH 7.0. Following the washes, the array was probed with 1 mg/ml rabbit muscle phosphorylase a (P1261, Sigma) and 50 kBq of [14C]glucose 1-phosphate (GE Healthcare) for 2 h at 25 °C under continuous shaking (25 rpm). After incubation, the array was washed with PBS for 15 min. This was followed by a two washes with 2× PBS with 0.05% v/v Tween 20 for each 5 min. After this, a sample from the washing buffer was taken, and the radioactivity was analyzed. If radioactivity was reduced to base-line levels, the membrane was left drying and subsequently analyzed on a PhosphorImager system (445 SI, GE Healthcare).
Immunofluorescent Labeling of Tobacco Sections
The cell wall imaging of xyloglucan mAb binding to tobacco stem pith parenchyma cell walls pretreated with pectate lyase was performed as described (38).
RESULTS
A Library of Plant Oligosaccharides
A set of oligosaccharides was assembled either by the enzymatic cleavage of polysaccharides followed by purification of constituent oligosaccharides or by chemical synthesis (Fig. 1, upper panel). Each structure shown in Fig. 1, upper panel has been assigned a code number (bolded to distinguish them from reference numbers), which is consistent throughout the text. Some of the oligosaccharides have been previously described, whereas four chemically synthesized galactosyl oligosaccharides were newly produced (structures 19, 20, 21, and 22). Details of all oligosaccharides are provided in supplemental Table 1. The purities of the oligosaccharides were determined by a variety of methods that have been described previously and referenced in supplemental Table 1. The purity of oligosaccharides produced from polysaccharides was generally at least 90%, and for chemically synthesized oligosaccharides, it was ∼99%. Oligosaccharides were coupled to BSA by reductive amination with sodium cyanoborohydride, which produced a ring-opened sugar residue between the oligosaccharide and the BSA (Fig. 1, lower panel). MALDI-TOF MS analysis of 20 selected neoglycoproteins indicated that on average 8.4 oligosaccharides were attached to each BSA molecule (supplemental Table 2).
Construction and Reproducibly of Oligosaccharide Microarrays
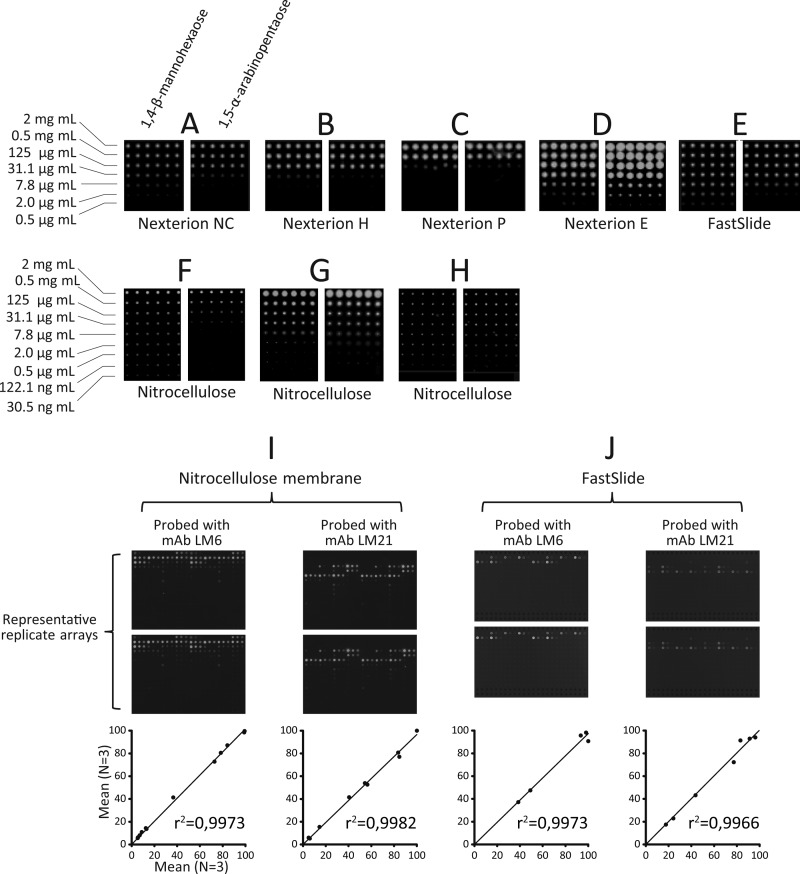
Several types of substrate surface were tested including a range of surface-treated glass slides (Fig. 2, A–C), glass slides coated with nitrocellulose (Fig. 2, D and E), and nitrocellulose membrane (Fig. 2, F–H). Two types of spotting robot were used, one piezoelectric (Arrayjet Sprint, Fig. 2, A–F) and one pin-based (MicroGrid II, Fig. 2, G and H). The test prints shown were made using BSA conjugated with (1→4)-β-d-mannohexaose and (1→5)-α-l-arabinopentaose printed in sextuplet and at 10 concentrations from 2 mg/ml to 7.6 ng/ml (Fig. 2, A–H). The arrays were probed with the anti-mannan or anti-arabinan mAbs LM21 and LM6, respectively. The oligosaccharides were successfully presented for recognition on all the surfaces used, but there were differences in detection limits and spot morphologies. Of the glass slides, the nitrocellulose-coated Nexterion E slide (Fig. 2D) and the FAST Slide (Fig. 2E) yielded the most sensitive detection (2 μg/ml for (1→4)-β-d-mannohexaose), and the least sensitive detection was obtained using the Nexterion P slide (125 μg/ml for (1→4)-β-d-mannohexaose) (Fig. 2C). Although similar in detection limits, arrays produced on the FAST Slide (Fig. 2E) were superior to those produced on the Nexterion E slide because they had a more consistent spot size across the concentration range, and this is an important consideration when quantifying spot signals. Nitrocellulose membrane was also a suitable substrate for microarray production using both the pin-based and the piezoelectric printers. With the piezoelectric Arrayjet printer, the detection limit on nitrocellulose was 122.1 ng/ml (Fig. 2F), and using the pin-based printer, the detection limit was 2 μg/ml for split pins (Fig. 2G) and 0.5 μg/ml for solid pins (Fig. 2H). These data demonstrated that the neoglycoprotein oligosaccharides are a versatile resource for the manufacture of microarrays on diverse surfaces. The reproducibility of microarrays produced on the piezoelectric Arrayjet robot was tested by producing 12 separate copies of arrays both on nitrocellulose membrane (Fig. 2I) and on slides (Fig. 2J), probing with selected antibodies, and then quantifying the spot signals from these replicate experiments (Fig. 2, I and J). For both nitrocellulose and slides, six arrays were probed with the anti-mannan mAbs LM21, six were probed with the anti-arabinan mAb LM6, and mean spot signals from three arrays were compared with the corresponding three replicate arrays. The data sets were plotted against each other, and r2 values were calculated (Fig. 2, I and J). In all cases, there was a low level of variability in the arrays, with r2 values of greater than 0.9 in all cases. Similarly high levels of reproducibility have been previously reported for arrays produced using the pin-based MicroGrid robot (32).
FIGURE 2.
Microarray printing surfaces and reproducibility. Coupling oligosaccharides to protein enabled printing on a wide variety of slides and membranes commonly used for microarray production. (1→4)-β-d-Mannohexaose and (1→5)-α-l-arabinopentaose microarrays were printed in sextuplet and at 10 concentrations from 2 mg/ml to 30.5 ng/ml. The arrays were probed with the anti-mannan or anti-arabinan monoclonal antibodies LM21 and LM6, respectively. Microarrays were printed using a noncontact piezoelectric robot (A–F) or a pin-based robot (G and H). Arrays were printed on surface-modified glass slides (A–C), nitrocellulose-coated glass slides (D and E), and nitrocellulose membrane (F–H). Reproducibility of the microarrays was tested by printing 12 copies of arrays on both nitrocellulose membrane (I) and nitrocellulose-coated glass FAST Slides (J). Arrays were probed with the anti-mannan mAbs LM21 or the anti-arabinan mAb LM6. Representative probed replicate arrays are shown, as well as graphs of mean spot signals from three arrays plotted against each other. Axes on the graphs are relative mean spot signals.
Most of the applications envisaged for oligosaccharide microarrays involve the interrogation of arrayed samples by a number of different glycan-binding proteins, ligands, or enzymes, which must be kept separate during probing. To achieve this, we designed our array production to be compatible with multipad slides that fit into a probing apparatus that forms a seal around each pad. An example of a typical array setup used is shown in supplemental Fig. 1A, where each of the 16 pads on each slide accommodates at least 324 spots in an area 6 × 6 mm, and each oligosaccharide is usually represented by four spots (two replicates at two different concentrations). As shown in supplemental Fig. 1B, the probing apparatus accommodates four such slides so that in a single probing experiment, 64 distinct mAbs, CBMs, etc. can be individually screened simultaneously against at least 80 oligosaccharides printed in quadruplet. 50 μl of probing solution is required to probe one of the 16 pads. The composite image in supplemental Fig. 1C shows the binding of five different mAbs to their respective epitopes on multiple copies of the array shown in supplemental Fig. 1A.
Specificity Screening of Monoclonal Antibodies
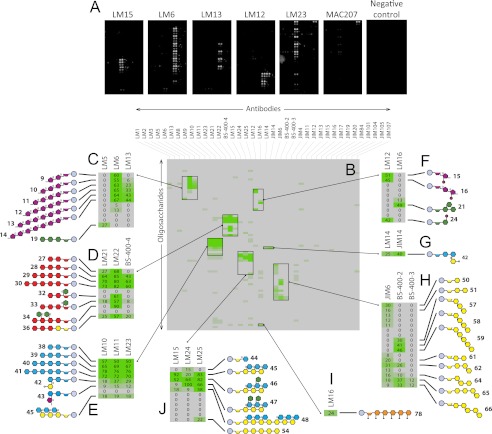
We tested the microarrays for mAb screening by probing microarrays with a selection of 38 mAbs, some with previously well characterized specificities and some with new specificities (Fig. 3). Examples of representative arrays are shown in Fig. 3A. The heat map shown in Fig. 3B is an overview of the whole data set, and the expanded heat maps (Fig. 3, C–J) provide more detailed information about the binding of selected mAbs. Oligosaccharide structures are only shown where binding produced a mean spot signal of at least 15% of the highest mean signal in the entire data set.
FIGURE 3.
Specificity screening of mAbs. Oligosaccharide microarrays were probed with a set of 38 monoclonal antibodies, and selected examples of probed arrays are shown in A. The mean spot signals obtained from three experiments are presented in heat maps in which color intensity is correlated to signal (B–I). Expanded heat maps (C–I) provide more detailed information about the binding of selected mAbs. The highest signal in the entire data set was set to 100, and all other values were normalized accordingly. Oligosaccharide structures are only shown where binding produced a mean spot signal of at least 15% of the highest mean signal in the entire data set. Further details about the oligosaccharides are provided in supplemental Table S1.
The microarray profiles obtained for mAb binding were consistent with data previously obtained by other techniques. For example, mAbs LM5 (34) and LM6 (35) bound predominantly to (1→4)-β-d-galactan (structure 19) and (1→5)-α-l-arabinans (structures 9, 10, 11, 12, 13, and 14) respectively (Fig. 3C). As previously shown (39), mAbs LM21 and LM22 bound to mannan-containing oligosaccharides, and LM22, but not LM21, bound strongly to galactomannan-derived oligosaccharides (structures 32, 33, and 34) (Fig. 3D). The inclusion of oligosaccharides with different degrees of polymerization provided information about the epitope sizes recognized by some mAbs. For example, LM6 bound strongly to an arabinan dimer (structure 9) and similarly to arabinans with degrees of polymerization up to 7 (Fig. 3C). In contrast, LM13 binding was restricted to longer oligosaccharides, and LM13 did not bind to an arabinan dimer (structure 9) and only very weakly to an arabinan trimer (structure 10), confirming and extending the previous analysis of the recognition of soluble oligosaccharides by competitive inhibition ELISAs (36) (Fig. 3C). Similarly, although LM22 (39) bound strongly to a mannan dimer (structure 27), mAb BS-400-4 (43) did not, and the minimum epitope size for this mAb appears to be at least 3 degrees of polymerization (Fig. 3D). LM12, a novel rat mAb derived subsequent to immunization with a feruloylated arabinosyl-BSA immunogen, displayed recognition of feruloyl residues attached to a range of sugars (Fig. 3F). In contrast to the previously described LM9, which is specific to feruloylated galactan (44), LM12 bound to oligosaccharides containing feruloylated arabinosyl residues (structures 15 and 16) as well as feruloylated galactosyl residues (structure 24) (Fig. 3F). The anti-RGI mAb LM16 bound with greatest avidity to a new synthetic galactotriose (structure 21) (Fig. 3F) and also showed unexpected binding to hexaamino-(1→4)-β-d-glucohexaose (structure 78, Fig. 3I), although this cross-reactivity is unlikely to present problems for plant research because this polymer is unknown in the plant kingdom. A novel aldouronic acid epitope (glucoronyl-(1→2)-α-(1→4)-β-d-xylotriose, structure 42) recognized by anti-arabinogalactan protein (AGP) mAbs, LM14 and JIM14, was also identified (Fig. 3G). As expected, the anti-(1→3),(1→4)-β-d-glucan mAb BS-400-3 bound to the (1→3),(1→4)-β-d-glucan structures 65 and 66 and did not cross-react with the (1→3)-β-d-glucan oligosaccharides 57, 58, and 59 (45), and the anti-(1→3)-β-d-glucan mAb BS-400-2 also showed its expected binding to β-glucans containing 1,3-linkages (46) (Fig. 3H). mAb JIM6 showed broad specificity for β-glucans containing 1,4-linkages but did not bind to β-glucans containing 1,3-linkages (Fig. 3H).
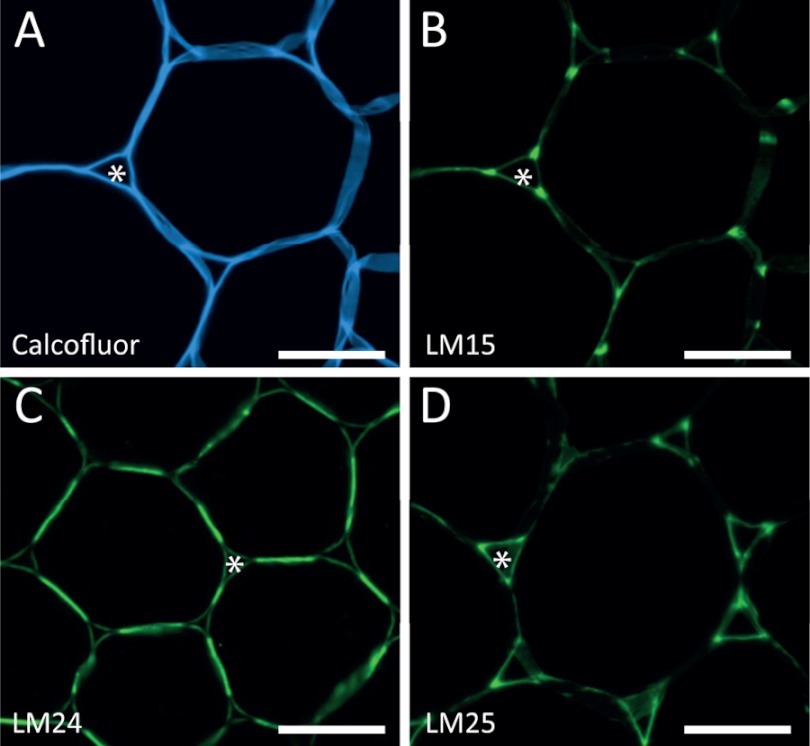
Five antibodies directed to xyloglucan displayed subtle distinctions in recognition of xyloglucan-derived oligosaccharides with novel mAbs LM24 and LM25 displaying wider recognition of galactosylated xyloglucan oligomers (and in the case of LM25 weak binding to unsubstituted β-glucan) than the previously characterized mAb LM15 (38) (Fig. 3J). The differences in the anti-xyloglucan mAb array binding profiles were reflected in differing binding profiles when applied to pectate lyase-treated transverse sections of tobacco stem pith parenchyma as shown in Fig. 4. Previous work had demonstrated that after pectic homogalacturonan removal, the LM15 xyloglucan epitope is revealed abundantly at the corners of intercellular spaces (Marcus et al. (38)) (Fig. 4B). In equivalent material, the LM24 epitope was most abundant in adhered cell walls between intercellular spaces (Fig. 4C), and the LM25 epitope was localized in cell walls lining intercellular spaces (Fig. 4D).
FIGURE 4.
Localization of xyloglucan epitopes in tobacco stems. Immunofluorescence imaging of transverse sections of tobacco stem pith parenchyma cell walls with anti-xyloglucan mAb after a pectate lyase pretreatment is shown. A, Calcofluor fluorescence showing all cell walls. B, the same section as A with immunofluorescence labeling with mAb LM15 showing abundant binding to cell walls at the corners of intercellular spaces (*). C, equivalent section showing immunofluorescence labeling with mAb LM24 indicating most abundant labeling in regions of adhered cell walls between intercellular spaces. D, equivalent section showing immunofluorescence labeling with mAb LM25, which binds abundantly to cell walls lining intercellular spaces. Scale bars = 10 μm.
Specificity Screening of CBMs
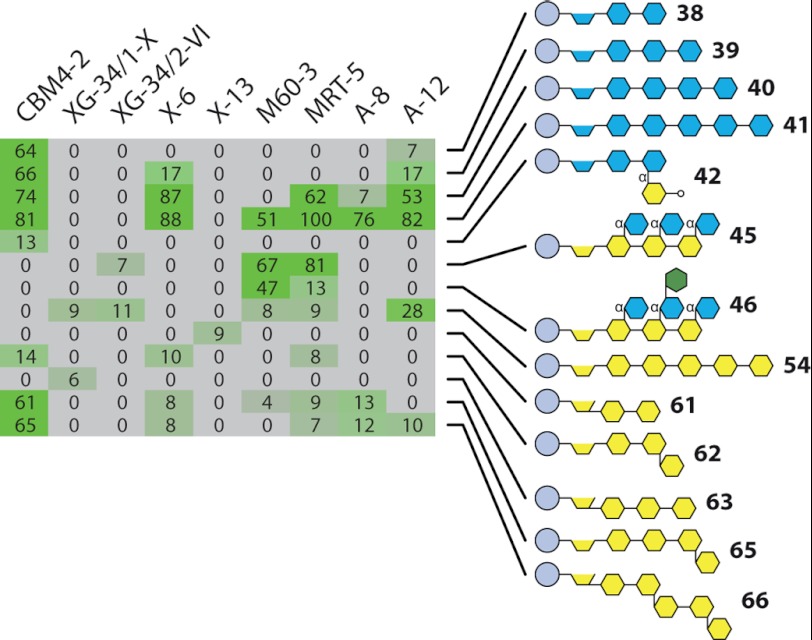
We tested the oligosaccharide microarrays for CBM screening by probing arrays with a set of CBMs that were produced by mutation of CBM4-2 from Rhodothermus marinus (47–49) and with a variety of lectins (Fig. 5). Wild type CBM4-2 is a xylan-binding CBM, and as expected, it bound to xylobiose (structure 38), xylotriose (structure 39), xylotetraose (structure 40), and xylopentaose (structure 41) (Fig. 5). CBM4-2 has also been reported to cross-react with certain (1→3)(1→4)-β-d-glucans, and semiquantitative affinity electrophoresis indicated a similar level of binding of CBM4-2 to birch xylan as to barley (1→3),(1→4)-β-d-glucan (50). The array data were in agreement with this because in addition to the xylan oligosaccharides, CBM4-2 also bound to (1→3),(1→4)-β-d-glucotetraose and (1→3),(1→4)-β-d-glucopentaose (structures 65 and 66), but its much weaker binding to (1→3),(1→4)-β-d-glucotriose (structure 62) provides insight into the minimum degrees of polymerization required for optimal binding. X-6 showed a greater specificity for xylan oligosaccharides than CBM4-2 and bound with greatest avidity to xylopentaose (structure 41) and xylotetraose (structure 40) and weakly to xylotriose (structure 39). X-6 appears to have a requirement for at least three xylose residues for binding because it did not bind to xylobiose (structure 38). A-12 showed a similar specificity to X-6 but also cross-reacted with the linear (1→4)-β-linked glucopentose (structure 54). A-8 displayed an even more restricted binding profile to xylan oligomers than X-6 and bound strongly to xylopentaose (structure 41) but only weakly to xylotetraose (structure 40). The mutant CBMs M60-3 and MRT-5 also bound to the xylopentaose (structure 41), and MRT-5, but not M60-3, bound to the xylotetraose (structure 40). However, both CBMs had new cross-reactivities when compared with wild type CBM4-2 and bound to xyloglucan oligosaccharides (structure 45) and (structure 46). The binding to MRT-5 appeared to be inhibited by the presence of galactose because this CBM bound only weakly to structure 46 but strongly to structure 45, which is identical apart from the galactose substitution of one xylose residue. Together, these data indicate that oligosaccharide microarrays are useful tools for the rapid screening of the binding profiles of CBMs. However, in contrast to most antibodies, CBMs often display moderate binding affinities, and this may limit the application of these arrays for some CBM studies.
FIGURE 5.
Specificity screening of carbohydrate-binding modules and lectins. Oligosaccharide microarrays were probed with the xylan-binding CBM CBM4-2 and also mutant variants of CBM4-2: XG-34/1-X; XG-34/2-VI; X-6; X-13; M60-3; MRT-5; A-8; and A-12. The highest signal in the entire data set was set to 100, and all other values were normalized accordingly. Further details about the oligosaccharides are provided in supplemental Table S1.
Competitive Inhibition Assays
One potential concern of assays based on immobilized targets is that the binding of antibodies, CBMs, or other probes might be either inhibited or promoted by the immobilization itself or by the coupling of the oligosaccharides to the carrier molecule, in this case BSA. To test for this, we also used the arrays in a competitive inhibition format whereby arrays were probed in the presence of soluble unconjugated oligosaccharides used at a range of concentrations (supplemental Fig. 2). A set of mAbs and CBMs was tested, and in all cases, the results obtained were consistent with those obtained using immobilized neoglycoproteins. For example, the binding of the anti-AGP LM14 to glucoronyl-(1→2)-α-(1→4)-β-d-xylotriose-BSA was inhibited by the unconjugated version of this structure but not by β-(1→4)-d-xylotetraose (structure 39). Similarly, the binding of CBM4-2 to both (1→4)-β-d-xylotriose-BSA and 3′-β-d-glucosyl-(1→4)-β-d-glucobiose-BSA was inhibited by unconjugated (1→4)-β-d-xylohexaose, and the binding of mAbs LM21 and LM22 was inhibited by appropriate haptens (supplemental Fig. 2).
Oligosaccharide Microarrays as Multiplexed Acceptors for Enzyme Assays
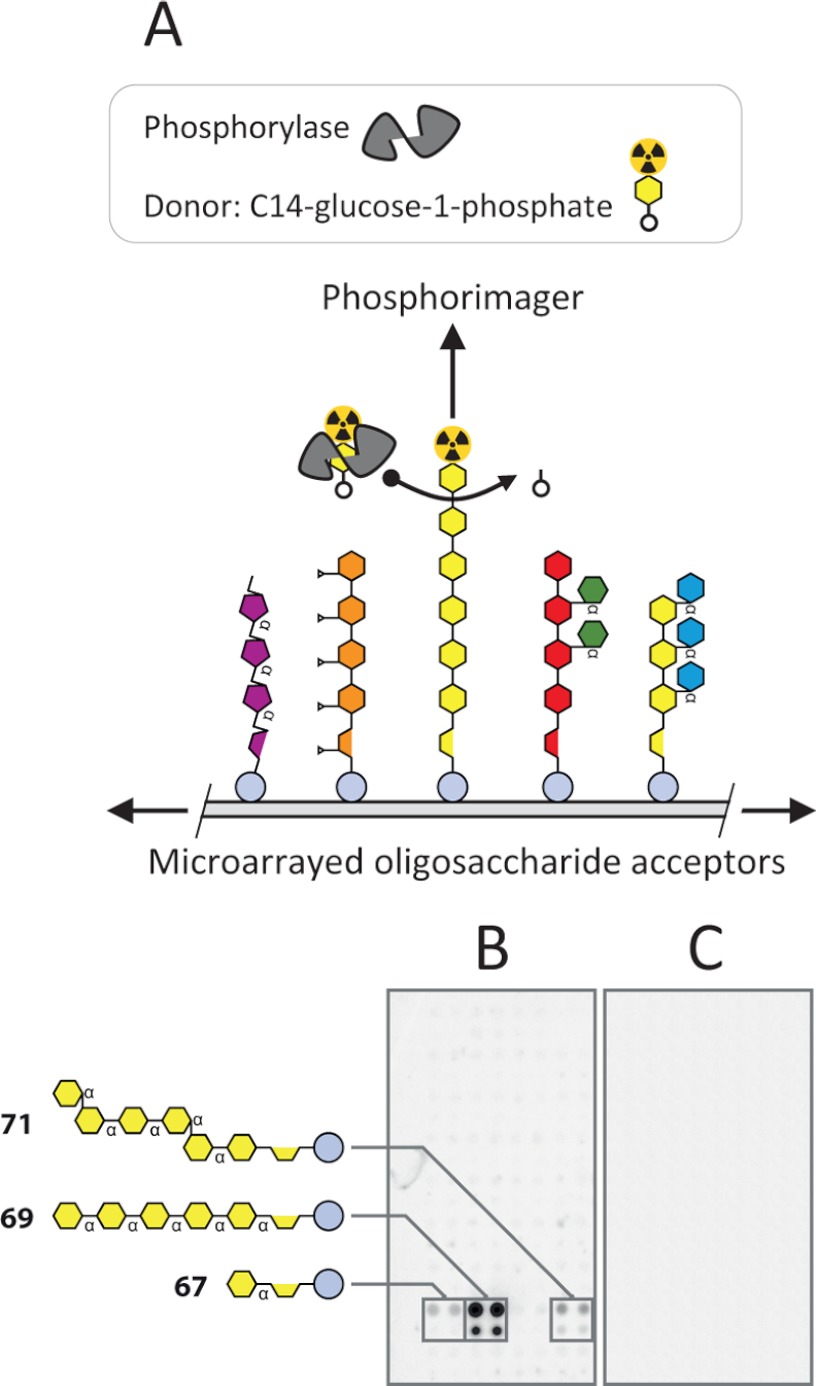
Microarrays of immobilized sets of substrates or acceptors are potentially useful tools for HTP enzyme screening and characterization that enable multiplexed analysis of activities. Arrays of polysaccharides (51) and limited numbers of oligosaccharides (52) have been used to explore glycosyltransferase activities, and we were interested to test the applicability of our arrays for similar purposes. As proof of concept, we tested rabbit muscle phosphorylase using [14C]glucose 1-phosphate as the glycosyl donor (Fig. 6). The general mechanism of the method is shown in Fig. 6A. Scanning the arrays with a PhosphorImager after incubation with enzyme and glycosyl donor revealed specific 14C-labeled spots representing the transfer of [14C]glucose onto α-linked glucan oligosaccharides that were present on this particular microarray (structures 67, 69, and 71) (Fig. 6B). A control array that had been treated with enzyme inactivated by boiling did not show incorporation of [14C]glucose (Fig. 6C).
FIGURE 6.
Oligosaccharide microarrays as multiplexed acceptors for enzyme assays. A, schematic showing the experimental setup used to assay the activity of rabbit muscle phosphorylase using a [14C]glucose 1-phosphate donor and an array of immobilized oligosaccharide acceptor. B, an oligosaccharide microarray was used to assay the activity of rabbit muscle phosphorylase a with [14C]glucose 1-phosphate as a glycosyl donor. The incorporation of 14C was detected using a PhosphorImager system. The control array in C was probed with boiled phosphorylase a under the same conditions. The structures of the oligosaccharides that were the most effective acceptors are shown. Further details about the oligosaccharides are provided in supplemental Table S1.
DISCUSSION
It is generally recognized that there is a widening gap between our ability to discover genes and proteins and to understand their roles in plant glycobiology. For example, it is estimated that we can safely predict the activities of no more than 20% of the proteins within the CAZy database (11). We show here that carbohydrate microarrays can make a valuable contribution to the HTP analysis of a variety of carbohydrate-protein interactions, and although oligosaccharide microarrays have been described for use in medical animal and microbial research, equivalent technology has not previously been developed for plant research.
The use of defined oligosaccharides rather than polysaccharides is important for obtaining detailed information about carbohydrate-interacting proteins. A library of cell wall-derived oligosaccharides is a significant resource in itself, and once coupled to protein, it can be used to produce microarrays on diverse surfaces. The versatility of being able to print microarrays on both nitrocellulose and slides is important. Although slide-based microarrays can only be analyzed using specialized scanning equipment, membrane-based arrays can be used by non-experts and scanned using an ordinary office scanner and are thus ideal for wider distribution to researchers. Several linkers have been developed for the covalent and noncovalent attachment of oligosaccharides to substrates. For example, coupling to lipids has also been shown to be a highly effective method for oligosaccharide microarray production (19, 20, 25, 26, 53). One reason for choosing BSA as a carrier molecule was because BSA-based neoglycoproteins are a multifunctional resource that can be used not just for microarray production but also as immunogens and as components of other assays for which immobilization is required. Nevertheless, any coupling procedure that involves modification of reducing ends is likely to interfere with the activity of reducing end-acting probes or enzymes, and this may be exacerbated by the large size of the BSA molecule. We found that BSA-coupled oligosaccharides arrayed as described previously (see “Experimental Procedures”) were effective substrates for several exo-acting glycosyl hydrolases but not for endo-acting enzymes (data not shown). Presumably, the exo-acting enzymes were nonreducing-end acting, and the lack of activity of the endo-acting enzymes was a result of steric hindrance from the BSA.
This study highlighted some important technical aspects of oligosaccharide microarray production including the relative merits of different microarray robot printers. The pin-based MicroGrid II printer was suited for the production of microarrays on nitrocellulose membrane with larger spot sizes. However, we found that array quality often decreased with longer print runs such that some spots were missing or not properly printed, and this is likely to result from the inevitable wear of the pins that occurs with contact printing. This drawback is avoided with noncontact printers such as the Arrayjet Sprint that dispel samples by a highly reproducible piezo-actuation process. Another major advantage of the Arrayjet Sprint is its much greater speed, which is important not just to increase throughput but because the evaporation of sample buffer with a concomitant concentration of samples can be highly problematic during long microarray print runs. Importantly, by printing arrays on multipad slides (as shown in supplemental Fig. S1), such that each pad is isolated by a gasket during probing, it is possible to simultaneously assess the binding of large numbers of mAbs, CBMs, etc. against many immobilized samples. For example, using ten 16-pad slides, it is possible to simultaneously screen 160 antibodies, each against 400 immobilized glycans.
The primary goal of this work was to develop plant oligosaccharide microarray technology per se, but we also obtained new epitope-level information about mAb specificities. For example, mAbs LM14 and JIM14 have previously been described as binding to unknown epitopes occurring on AGPs (32, 54). The oligosaccharide microarrays demonstrated that both mAbs bind with high specificity to glucoronyl-(1→2)-α-(1→4)-β-d-xylotriose (structure 42) (Fig. 3G), which is a constituent of glucoronoxylan and glucoronoarabinoxylans, and this finding is therefore interesting because it implies that these two mAbs may bind to an epitope not usually associated with AGPs in addition to binding to glucoronyl residues decorating arabinogalactan structures. The synthetic galactosyl structures 20, 21, and 22 are not known to occur on any plant cell wall polysaccharide, and it was therefore surprising that mAb LM16 bound strongly to 6′-β-d-galactosyl-(1→4)-β-d-galactotriose (structure 21). LM16 has been described previously as binding to an epitope occurring on sugar beet RGI that is generated by arabinofuranosidase treatment and is galactosidase-labile (36). Sugar beet arabinan side chains can in some cases be attached to RGI backbones via short galactosyl motifs, and it is possible that LM16 recognizes this structure once exposed by arabinofuranosidase (36). It has been shown that such short galactan stubs can be substituted with ferulic acid at the C6 position, but substitution with another sugar has not been reported (55). LM16 does not bind to galactosyl residues per se because it does not bind to linear galactan, galactomannan, or galactoxyloglucans. The strong binding of LM16 to both structure 21 and native sugar beet pectin therefore raises the intriguing possibility of a novel RGI epitope.
The importance of obtaining detailed information about epitope structures was clearly illustrated by the anti-xyloglucan mAbs LM15, LM24, and LM25. Oligosaccharide array analysis revealed subtle differences in the binding profiles of these mAbs, which most likely would not have been discriminated using previous polysaccharide-based ELISA or microarrays. Immunolabeling of tobacco sections with these mAbs showed that despite their relatively small differences in structure, the epitopes recognized had distinct cellular locations. Although the biological significance of these findings is unclear at present, they show that a detailed evaluation of epitope structures that oligosaccharide arrays can provide is important for the subsequent interpretation of data produced in antibody studies.
Supplementary Material
Acknowledgment
We thank Mohammed Saddik Motawie for oligosaccharide 72.
This work was supported by Danish Research Council grants (to W. G. T. W.) and in part by Grant 2/0011/09 from the Scientific Grant Agency (VEGA) (Slovakia) (to V. F.). This work was also supported by the Danish Research Council for Strategic Research (to M. C. F. A., M. H. C., and W. G. T. W.).

This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- CBM
- carbohydrate-binding module
- HTP
- high throughput
- AGP
- arabinogalactan protein
- RGI
- rhamnogalacturonan I.
REFERENCES
- 1. Bacic A., Harris A. J., Stone B. A. (1988) in The Biochemistry of Plants (Preiss J., ed) pp. 52–108, Academic Press, New York [Google Scholar]
- 2. De Lorenzo G., Ferrari S. (2002) Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 5, 295–299 [DOI] [PubMed] [Google Scholar]
- 3. Fry S. C. (2004) Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol. 161, 641–675 [DOI] [PubMed] [Google Scholar]
- 4. Van den Ende W., De Coninck B., Van Laere A. (2004) Plant fructan exohydrolases: a role in signaling and defense? Trends Plant Sci. 9, 523–528 [DOI] [PubMed] [Google Scholar]
- 5. Zeeman S. C., Kossmann J., Smith A. M. (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234 [DOI] [PubMed] [Google Scholar]
- 6. Lee K. J., Marcus S. E., Knox J. P. (2011) Cell wall biology: perspectives from cell wall imaging. Mol. Plant 4, 212–219 [DOI] [PubMed] [Google Scholar]
- 7. Willats W. G., Knox J. P., Dalgaard Mikkelsen J. (2006) Pectin: new insights into an old polymer are starting to gel. Trends Food Sci. Technol. 17, 97–104 [Google Scholar]
- 8. Pauly M., Keegstra K. (2010) Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 13, 305–312 [DOI] [PubMed] [Google Scholar]
- 9. Albersheim P., Darvill A., Roberts K., Sederoff R., Staehelin A. (2011) in Plant Cell Walls (Masson S., ed) pp. 52–61, Garland Science, Taylor and Francis Publishing Group, LLC, New York and Abingdon, U.K [Google Scholar]
- 10. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilbert H. J. (2010) The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knox J. P. (1997) The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int. Rev. Cytol. 171, 79–120 [DOI] [PubMed] [Google Scholar]
- 13. Willats W. G., McCartney L., Mackie W., Knox J. P. (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol. Biol. 47, 9–27 [PubMed] [Google Scholar]
- 14. Knox J. P. (2008) Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 11, 308–313 [DOI] [PubMed] [Google Scholar]
- 15. Pattathil S., Avci U., Baldwin D., Swennes A. G., McGill J. A., Popper Z., Bootten T., Albert A., Davis R. H., Chennareddy C., Dong R., O'Shea B., Rossi R., Leoff C., Freshour G., Narra R., O'Neil M., York W. S., Hahn M. G. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153, 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schena M., Shalon D., Davis R. W., Brown P. O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 [DOI] [PubMed] [Google Scholar]
- 17. Ekins R., Chu F. W. (1999) Microarrays: their origins and applications. Trends Biotechnol. 17, 217–218 [DOI] [PubMed] [Google Scholar]
- 18. McWilliam I., Chong Kwan M., Hall D. (2011) Inkjet printing for the production of protein microarrays. Methods Mol. Biol. 785, 345–361 [DOI] [PubMed] [Google Scholar]
- 19. Feizi T. (2000) Progress in deciphering the information content of the 'glycome'–a crescendo in the closing years of the millennium. Glycoconj. J. 17, 553–565 [DOI] [PubMed] [Google Scholar]
- 20. Fukui S., Feizi T., Galustian C., Lawson A. M., Chai W. (2002) Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat. Biotechnol. 20, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 21. Wang D., Liu S., Trummer B. J., Deng C., Wang A. (2002) Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat. Biotechnol. 20, 275–281 [DOI] [PubMed] [Google Scholar]
- 22. Willats W. G., Rasmussen S. E., Kristensen T., Mikkelsen J. D., Knox J. P. (2002) Sugar-coated microarrays: a novel slide surface for the high-throughput analysis of glycans. Proteomics 2, 1666–1671 [DOI] [PubMed] [Google Scholar]
- 23. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Printed covalent glycan array for ligand profiling of diverse glycan-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feizi T., Chai W. (2004) Oligosaccharide microarrays to decipher the glyco code. Nat. Rev. Mol. Cell Biol. 5, 582–588 [DOI] [PubMed] [Google Scholar]
- 25. Park S., Lee M. R., Shin I. (2008) Carbohydrate microarrays as powerful tools in studies of carbohydrate-mediated biological processes. Chem. Commun. (Camb.) 4389–4399 [DOI] [PubMed] [Google Scholar]
- 26. Smith D. F., Song X., Cummings R. D. (2010) Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 480, 417–444 [DOI] [PubMed] [Google Scholar]
- 27. Feizi T., Fazio F., Chai W., Wong C. H. (2003) Carbohydrate microarrays — a new set of technologies at the frontiers of glycomics. Curr. Opin. Struct. Biol. 13, 637–645 [DOI] [PubMed] [Google Scholar]
- 28. Liu Y., Palma A. S., Feizi T. (2009) Carbohydrate microarrays: key developments in glycobiology. Biol. Chem. 390, 647–656 [DOI] [PubMed] [Google Scholar]
- 29. Sørensen I., Willats W. G. (2008) Plant cell walls: new insights from an ancient species. Plant Signal. Behav. 3, 743–745 [Google Scholar]
- 30. Klopffleisch K., Phan N., Augustin K., Bayne R. S., Booker K. S., Botella J. R., Carpita N. C., Carr T., Chen J. G., Cooke T. R., Frick-Cheng A., Friedman E. J., Fulk B., Hahn M. G., Jiang K., Jorda L., Kruppe L., Liu C., Lorek J., McCann M. C., Molina A., Moriyama E. N., Mukhtar M. S., Mudgil Y., Pattathil S., Schwarz J., Seta S., Tan M., Temp U., Trusov Y., Urano D., Welter B., Yang J., Panstruga R., Uhrig J. F., Jones A. M. (2011) Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sørensen I., Willats W. G. (2011) Screening and characterization of plant cell walls using carbohydrate microarrays. Methods Mol. Biol. 715, 115–121 [DOI] [PubMed] [Google Scholar]
- 32. Moller I., Marcus S. E., Haeger A., Verhertbruggen Y., Verhoef R., Schols H., Ulvskov P., Mikkelsen J. D., Knox J. P., Willats W. (2008) High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 25, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clausen M. H., Madsen R. (2003) Synthesis of hexasaccharide fragments of pectin. Chemistry 9, 3821–3832 [DOI] [PubMed] [Google Scholar]
- 34. Jones L., Seymour G. B., Knox J. P. (1997) Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-d-galactan. Plant Physiol. 113, 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willats W. G., Marcus S. E., Knox J. P. (1998) Generation of monoclonal antibody specific to (1→5)-α-l-arabinan. Carbohydr. Res. 308, 149–152 [DOI] [PubMed] [Google Scholar]
- 36. Verhertbruggen Y., Marcus S. E., Haeger A., Verhoef R., Schols H. A., McCleary B. V., McKee L., Gilbert H. J., Knox J. P. (2009) Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J. 59, 413–425 [DOI] [PubMed] [Google Scholar]
- 37. McCartney L., Marcus S. E., Knox J. P. (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53, 543–546 [DOI] [PubMed] [Google Scholar]
- 38. Marcus S. E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J. J., Farkas V., Pedersen H. L., Willats W. G., Knox J. P. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marcus S. E., Blake A. W., Benians T. A., Lee K. J., Poyser C., Donaldson L., Leroux O., Rogowski A., Petersen H. L., Boraston A., Gilbert H. J., Willats W. G., Knox J. P. (2010) Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 64, 191–203 [DOI] [PubMed] [Google Scholar]
- 40. Cicortas Gunnarsson L., Nordberg Karlsson E., Albrekt A. S., Andersson M., Holst O., Ohlin M. (2004) A carbohydrate binding module as a diversity-carrying scaffold. Protein Eng. Des. Sel. 17, 213–221 [DOI] [PubMed] [Google Scholar]
- 41. von Schantz L., Gullfot F., Scheer S., Filonova L., Cicortas Gunnarsson L., Flint J. E., Daniel G., Nordberg-Karlsson E., Brumer H., Ohlin M. (2009) Affinity maturation generates greatly improved xyloglucan-specific carbohydrate binding modules. BMC Biotechnol. 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy R., Katzenellenbogen E., Jennings H. J. (1984) Improved procedures for the conjugation of oligosaccharides to protein by reductive amination. Can. J. Biochem. Cell Biol. 62, 270–275 [DOI] [PubMed] [Google Scholar]
- 43. Pettolino F. A., Hoogenraad N. J., Ferguson C., Bacic A., Johnson E., Stone B. A. (2001) A (1→4)-β-mannan-specific monoclonal antibody and its use in the immunocytochemical location of galactomannans. Planta 214, 235–242 [DOI] [PubMed] [Google Scholar]
- 44. Clausen M. H., Ralet M. C., Willats W. G., McCartney L., Marcus S. E., Thibault J. F., Knox J. P. (2004) A monoclonal antibody to feruloylated-(1→4)-β-d-galactan. Planta 219, 1036–1041 [DOI] [PubMed] [Google Scholar]
- 45. Meikle P. J., Bonig I., Hoogenraad N. J., Clarke A. E., Stone B. A. (1991) The location of (1→3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-β-glucan-specific monoclonal antibody. Planta 185, 1–8 [DOI] [PubMed] [Google Scholar]
- 46. Meikle P. J., Hoogenraad N. J., Bonig I., Clarke A. E., Stone B. A. (1994) A (1→3,1→4)-β-glucan-specific monoclonal antibody and its use in the quantitation and immunocytochemical location of (1→3,1→4)-β-glucans. Plant J. 5, 1–9 [DOI] [PubMed] [Google Scholar]
- 47. Gunnarsson L. C., Zhou Q., Montanier C., Karlsson E. N., Brumer H., 3rd, Ohlin M. (2006) Engineered xyloglucan specificity in a carbohydrate-binding module. Glycobiology 16, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 48. Johansson R., Gunnarsson L. C., Ohlin M., Ohlson S. (2006) Thermostable carbohydrate-binding modules in affinity chromatography. J. Mol. Recognit. 19, 275–281 [DOI] [PubMed] [Google Scholar]
- 49. Cicortas Gunnarsson L., Montanier C., Tunnicliffe R. B., Williamson M. P., Gilbert H. J., Nordberg Karlsson E., Ohlin M. (2007) Novel xylan-binding properties of an engineered family 4 carbohydrate-binding module. Biochem. J. 406, 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abou Hachem M., Nordberg Karlsson E., Bartonek-Roxâ E., Raghothama S., Simpson P. J., Gilbert H. J., Williamson M. P., Holst O. (2000) Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression, and binding studies. Biochem. J. 345, 53–60 [PMC free article] [PubMed] [Google Scholar]
- 51. Kosík O., Auburn R. P., Russell S., Stratilová E., Garajová S., Hrmova M., Farkas V. (2010) Polysaccharide microarrays for high-throughput screening of transglycosylase activities in plant extracts. Glycoconj. J. 27, 79–87 [DOI] [PubMed] [Google Scholar]
- 52. Shipp M., Nadella R., Gao H., Farkas V., Sigrist H., Faik A. (2008) Glyco-array technology for efficient monitoring of plant cell wall glycosyltransferase activities. Glycoconj. J. 25, 49–58 [DOI] [PubMed] [Google Scholar]
- 53. Palma A. S., Liu Y., Muhle-Goll C., Butters T. D., Zhang Y., Childs R., Chai W., Feizi T. (2010) Multifaceted approaches including neoglycolipid oligosaccharide microarrays to ligand discovery for malectin. Methods Enzymol. 478, 265–286 [DOI] [PubMed] [Google Scholar]
- 54. Yates E. A., Valdor J. F., Haslam S. M., Morris H. R., Dell A., Mackie W., Knox J. P. (1996) Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6, 131–139 [DOI] [PubMed] [Google Scholar]
- 55. Ralet M. C., André-Leroux G., Quéméner B., Thibault J. F. (2005) Sugar beet (Beta vulgaris) pectins are covalently cross-linked through diferulic bridges in the cell wall. Phytochemistry 66, 2800–2814 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.