Background: The inositol 1,4,5-trisphosphate receptor (InsP3R) is a ubiquitous intracellular calcium release channel.
Results: CaMKII phosphorylates the InsP3R at Ser-150 and modulates the intrinsic calcium channel activity.
Conclusion: InsP3R-mediated calcium release is negatively feedback-modulated by CaMKII-mediated phosphorylation.
Significance: Elucidating the regulation of the InsP3R is crucial for understanding calcium-mediated signaling events.
Keywords: Calcium Signaling; CaMKII; Inositol 1,4,5-Trisphosphate; Phosphorylation; Receptor Regulation
Abstract
InsP3-mediated calcium release through the type 2 inositol 1,4,5-trisphosphate receptor (InsP3R2) in cardiac myocytes results in the activation of associated CaMKII, thus enabling the kinase to act on downstream targets, such as histone deacetylases 4 and 5 (HDAC4 and HDAC5). The CaMKII activity also feedback modulates InsP3R2 function by direct phosphorylation and results in a dramatic decrease in the receptor-channel open probability (Po). We have identified S150 in the InsP3R2 core suppressor domain (amino acids 1–225) as the specific residue that is phosphorylated by CaMKII. Site-directed mutagenesis reveals that S150 is the CaMKII phosphorylation site responsible for modulation of channel activity. Nonphosphorylatable (S150A) and phosphomimetic (S150E) mutations were studied in planar lipid bilayers. The InsP3R2 S150A channel showed no decrease in activity when treated with CaMKII. Conversely, the phosphomimetic (S150E) channel displayed a very low Po under normal recording conditions in the absence of CaMKII (2 μm InsP3 and 250 nm [Ca2+]FREE) and mimicked a WT channel that has been phosphorylated by CaMKII. Phopho-specific antibodies demonstrate that InsP3R2 Ser-150 is phosphorylated in vivo by CaMKIIδ. The results of this study show that serine 150 of the InsP3R2 is phosphorylated by CaMKII and results in a decrease in the channel open probability.
Introduction
Intracellular calcium signaling plays an important role in the physiology of the cell, impinging upon a multitude of cellular events ranging from fertilization and cellular proliferation to cellular death (4). One mechanism in which calcium is mobilized from intracellular stores is via the activation of inositol 1,4,5-trisphosphate receptors (InsP3Rs)3 by the second messenger, inositol 1,4,5-trisphosphate (InsP3). InsP3 is produced when one of many extracellular factors bind G protein and tyrosine kinase-coupled receptors on the plasma membrane, resulting in the activation of phospholipase C and hydrolysis of phosphatidylinositol 4,5-bisphosphate (5). InsP3-mediated activation of InsP3Rs leads to release of calcium into the cyto- and nucleoplasm of the cell, where it can transduce a large number of signaling cascades mediating local and global cellular events. InsP3Rs have a ubiquitous tissue and cellular distribution. However, within a given cell type, multiple homologues of the receptor may be expressed. In heart the predominant isoform expressed in cardiac myocytes is the type 2 InsP3R (6). Atrial myocytes express considerably higher levels of receptor than that of ventricular myocytes (7). Recent studies have shown that in both cell types the activation of InsP3R can contribute to the modulation of excitation contraction coupling and increase the susceptibility for arrhythmogenic Ca2+ release events and nuclear Ca2+ signaling events (for review, see Ref. 8).
It is imperative for a cell to detect and decode a Ca2+ signal for downstream signaling processes. One such enzyme activated by a rise in intracellular [Ca2+] is Ca2+/calmodulin-dependent protein kinase II (CaMKII). CaMKII is a multifunctional serine/threonine protein kinase involved in many signaling pathways in various cell types (9). The enzyme is composed of homomultimers or heteromultimers of 6–12 subunits, with the subunit isoforms being derived from a family of four closely related genes (α, β, δ, and γ) (10). All isoforms contain an amino-terminal catalytic domain where ATP binds, a central regulatory domain containing an autoinhibitory and calmodulin binding sites, and a carboxyl-terminal association domain. The δ and γ isoforms of CaMKII are the predominant isoforms found in the heart where they have been implicated in regulating cardiac gene expression (2, 11). Recent results from our laboratory have shown that the predominant InsP3R isoform in the heart, the InsP3R2, is targeted primarily to the nuclear envelope in ventricular myocytes (1, 6, 11). Here it forms a macromolecular complex with the nuclear-localized cardiac isoform of CaMKII, CaMKIIδb. Upon stimulation of InsP3 production, Ca2+ released through the InsP3R2 activates CaMKIIδb, allowing it to act on downstream targets, such as histone deacetylase 4 and 5 (HDAC4 and HDAC5) (2, 12). Additionally, CaMKII appears to feedback-modulate InsP3R2 function by direct phosphorylation and results in a significant decrease in the channel open probability (Po) (1). The results of this study and others (2, 13, 14) suggest that the activity of InsP3Rs can be inhibited by CaMKII-mediated phosphorylation. Furthermore, the amino-terminal 1078 amino acids of the InsP3R2 have been shown to interact with, as well as be phosphorylated by CaMKII (1). However, it has not yet been established what amino acid(s) or if additional downstream residues of the InsP3Rs are implicated in channel regulation by CaMKII phosphorylation events.
In this study, we use expressed fragments of the InsP3R2 to show that CaMKII can phosphorylate the InsP3R2 at Ser-150. Upon phosphorylation, the channel open probability is significantly decreased in planar lipid bilayers. This could be reversed by addition of a specific CaMKII inhibitor (KN-93) and two protein phosphatases (PP1 and PP2A). Single channel studies of the receptor in which the phosphorylation site was mutated to an alanine (S150A) resulted in channels with properties similar to those of WT InsP3R2; however, they failed to respond to CaMKII addition and the Po remained unchanged. A phosphomimetic mutation (S150E) displayed a constitutively low open probability and mimicked an InsP3R2 phosphorylated by CaMKII. These results strongly suggest that Ser-150 is the site impinged upon by CaMKII and results in negative modulation of the InsP3R.
EXPERIMENTAL PROCEDURES
Expression Plasmid Construction
The construction of the full-length type 2 InsP3R protein expression vector (pInsP3R-T2) was described previously (15). Briefly, the expression plasmid was assembled using overlapping cDNA clones originally isolated from a rat brain library (16). The full-length expressed protein includes amino acid residues 1–2701 from the rat type 2 cDNA (accession number X61677).
The pIP3R2-Stopl078 construct is a mammalian expression vector of the first 1078 amino acids of the type-2 InsP3 receptor. This sequence is followed by the 12 carboxyl-terminal amino acids of the 116,000 subunit of the proton pump and was described previously (16). Briefly, pIP3R2-Stopl078 was constructed by cloning a 2.45-kb EcoRI-KpnI fragment followed by the 1.04-kb KpnI-PstI fragment of InsP3R2 into pCMV2 followed by an oligonucleotide encoding the carboxyl-terminal proton pump epitope (16). The fully expressed protein includes amino acid residues 1–1078 from the rat type 2 cDNA.
Regions cloned into the pCMV-3Tag-1a vector were PCR-amplified using pInsP3R2 as the template to create three mammalian expression vectors of the InsP3R2 carboxyl-terminal amino acids 1074–1640, 1635–2118, and 2114–2701. PCR products were digested with SalI, XhoI, and/or EcoRI and inserted into a similarly digested pCMV-3Tag-1a vector (Stratagene) creating new protein fusions that contained three copies of the FLAG epitope on the amino terminus of the InsP3R2 protein fragment.
Regions cloned into the bacterial expression plasmid pET-3a were PCR-amplified using pInsP3R2 as template and specific primers engineered to contain an amino-terminal NdeI restriction site and a carboxyl-terminal BglII site in addition to a methionine inserted after the NdeI site and a termination codon directly before the BglII site. PCR products were then digested with NdeI/BglII and ligated into NdeI/BamHI-digested pET-3a plasmid.
Construction of the full-length InsP3R2 S150 mutants (pInsP3R2-S150A and pInsP3R2-S150E) was done as follows. A plasmid coding for amino acids 1–1078 of the type 2 InsP3R was used as the template along with primers for mutagenesis of Ser-150 to alanine (5′-GAATGCCATGCGTGTGGCCCTGGATGCTGCAGGG-3′) or glutamate (5′-GAATGCCATGCGTGTGGAACTGGATGCTGCAGGG-3′). Mutagenesis of Ser-150 to either alanine or glutamate was confirmed by DNA sequencing. The region containing S150A or S150E of 1–1078 vector was cut from the plasmid with NdeI and AfeI and ligated into similarly digested pInsP3R2, thus creating the full-length InsP3R2 expression construct pInsP3R2-S150A or pInsP3R2-S150E. Mutagenesis was performed using Change-IT Multiple Mutation Site-Directed Mutagenesis Kit (USB Corp.). Constructs were verified by DNA sequence analysis using a commercial facility at University of California Davis (Davis Sequencing) using the Applied Biosystems Big Dye Terminator V3.0 sequencing chemistry.
CaMKIIδB/C plasmids were a kind gift from Dr. Joan Heller-Brown (University of California San Diego, La Jolla, CA). Adenoviral constructs containing the CaMKIIδB/C inserts were derived from these plasmids and prepared by the Cell and Molecular Physiology departmental adenovirus core (Loyola University Medical Center, Maywood, IL) in Adeasy vector backbones.
COS-1 Cell Transfection
COS-1 cells were transiently transfected with expression plasmids for pInsP3R2 using a DEAE-dextran method as described previously (17). Following the expression period, the COS cells were washed with phosphate-buffered saline (PBS), harvested in IP buffer (50 mm HEPES, pH 7.6, 1.0% bovine serum albumin, 10 mm magnesium acetate, 50 mm NaCl, 0.5 mm CaCl2, 1 mm dithiothreitol (DTT), 0.1 mm phenylmethylsulfonyl fluoride (PMSF), 100 μg/ml soybean trypsin inhibitor, 10 μm leupeptin, and 10 μm pepstatin) and lysed by 20–40 passages through a 27-gauge needle. The membranes were pelleted by centrifugation at 135,000 × gmax 4 °C for 10 min, and the supernatant containing any soluble protein fraction was removed from the microsomes. To solubilize any membrane-bound proteins, the membrane fraction was resuspended in IP buffer containing 1.0% Triton X-100 and incubated on ice with stirring for 1–2 h followed by removal of insoluble material by centrifugation at 135,000 × gmax at 4 °C.
Bacterial Cell Expression
Rosetta 2 (DE3) competent cells (Novagen) were transformed with the bacterial expression plasmids and grown at 37 °C with shaking in liquid culture until the A600 reached 0.6. Protein expression was induced with 1 mm isopropyl-β-d-thiogalactopyranoside, and the culture was incubated for 3 h at 37 °C. Cells were then pelleted at 3220 × gmax for 20 min and resuspended in lysis buffer (8% sucrose, 0.5% Triton X-100, 10 mm Tris-Cl, pH 8.0, 50 mm EDTA, 1 mm PMSF) with 10 mg/ml lysozyme (Sigma) and incubated at room temperature with agitation for 20 min. The lysate was then homogenized, and 1 mg/ml DNase and 100 mg/ml RNase were added, and the solution was incubated at room temperature for 25 min with agitation. The sample was then spun at 12,000 × gmax for 10 min, and the pellet was washed with 0.5 m NaCl, 0.5% Triton X-100, 1 mm PMSF. This was repeated twice, with the final wash containing 1 m urea. The resulting inclusion body pellet was then solubilized in 8 m urea, 150 mm NaCl, 50 mm Tris-Cl, pH 7.4, 3 mm 2-mercaptoethanol, 3 mm DTT, and insoluble material was pelleted by centrifugation at 106,000 × gmax for 10 min. Soluble material was dialyzed against a 4000-fold excess of 150 mm NaCl, 50 mm Tris-Cl, pH 7.4, 2 mm 2-mercaptoethanol. Recovered material was cleared by centrifugation at 106,000 × gmax for 10 min and was stored at 4 °C.
Sf9 Cell Culture and Baculovirus Infection
Sf9 cells were grown in 75-cm2 tissue culture flasks in culture medium containing TNM-FH medium (Cellgro) and 10% fetal bovine serum. A baculovirus expressing the full-length rat type 2 InsP3R (16, 18) was kindly provided by Dr. Ilya Bezprozvanny (University of Texas Southwestern Medical Center, Dallas, TX). Virus was added to the medium at a multiplicity of infection of 1, and cells were incubated at 27 °C for 72 h to allow for expression of recombinant protein.
Immunoprecipitation of Recombinant Proteins
Recombinant InsP3R2 proteins from either mammalian or bacterial expression were immunoprecipitated as follows. Proteins were divided into equal volume aliquots, and 10 μg of the appropriate primary antibodies was added and the samples incubated overnight at 4 °C with agitation. Then 30 μl of a 10% slurry of protein A-Sepharose CL-4B beads (Amersham Biosciences) was added and incubated for 2 h at 4 °C with agitation. Immune complexes were then washed three times with IP buffer before addition of kinase. Following kination the samples were treated with SDS-PAGE sample buffer, boiled 5 min, and resolved on SDS-PAGE.
CaMKII Phosphorylation of Recombinant Proteins
IP complexes were used as substrates for in vitro CaMKII phosphorylation 32P incorporation assays. Immune complexes were incubated at 30 °C for 20 min with 500 units of exogenous preactivated CaMKII (New England Biolabs) or with 10 μm concentrations of the specific CaMKII inhibitor, KN-93 (Seikagaku Corp.) CaMKII enzyme was activated by incubation in reaction buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 2 mm DTT, 0.1 mm Na2EDTA, 2 mm CaCl2, 1.2 μm calmodulin, 200 μm ATP) for 10 min at 37 °C. Following CaMKII activation, the reaction buffer was supplemented with [γ-32P]ATP to a final specific activity of 200 μCi/μmol for visualization of phosphorylated proteins via autoradiography.
The in vivo phosphorylation of InsP3R2 experiments used COS-1 cells that were singly or co-transfected with InsP3R2 and the CaMKII plasmids. Phosphorylation of endogenous InsP3R2 was achieved by adenoviral expression of CaMKIIδC and its dominant negative derivative (DN) in neonatal rat ventricular myocytes. The cells were harvested 24 h after infection, and protein extracts were subjected to Western immunoblotting with the phospho- and non-phospho-specific antibodies. Neonatal rat ventricular myocytes were prepared in the Cell and Molecular Physiology myocyte core (Loyola University Medical Center, Maywood, IL).
Antibodies
The affinity-pure InsP3R2-specific amino-terminal (T2NH) and carboxyl-terminal (T2C) antibodies were directed against the sequences CPDYRDAQNEGKTVRDGELP (residues 320–338) and CNKQRLGFLGSNTPH ENHHMPPH (residues 2679–2701) of the rat InsP3R2, respectively (19). The Ser-150-T2 and phospho-specific Ser(P)-150-T2 antibodies were directed against CKNAMRVSLDAAG (residues 144–155). For Ser(P)-150-T2 rabbits were immunized with phosphopeptide where Ser-150 was phosphorylated. Both antibodies were affinity-purified using immunogenic peptide. Anti-FLAG antibody was purchased from Affinity BioReagents and is a rabbit polyclonal against the peptide DYKDDDDKC.
SDS-PAGE and Immunoblotting
SDS-PAGE and Western blotting were performed as described previously (17) using 5, 7.5, or 10% SDS-polyacrylamide gels. Visualization was accomplished using ECL reagents (GE Healthcare).
Preparation of Microsomes for Lipid Bilayers
Microsomal fractions from Sf9 and COS-1 cells were prepared for use in planar lipid bilayers as described previously (20). Briefly, COS-1 or Sf9 cells were harvested in homogenization buffer (50 mm Tris-HCl, pH 8.3, 1 mm EDTA, 5 mm sodium azide, 0.25 mm PMSF, 10 μm leupeptin, 10 μm pepstatin, 100 μg/ml trypsin inhibitor) and lysed with a glass/Teflon Potter-Elvehjem tissue grinder. An equal volume of buffer containing 0.5 m sucrose was added, and the material was rehomogenized and centrifuged for 5 min at 1,200 × gmax. The supernatant containing the microsomes was recovered, and KCl and disodium pyrophosphate were added to a final concentration of 0.6 m and 20 mm, respectively. The material was homogenized and mixed for 30 min at 4 °C. Following a final homogenization, the microsomes were centrifuged for 5 min at 200 × gmax, and the supernatant was recovered. Finally, the microsomes were pelleted at 100,000 × gmax for 10 min, resuspended in storage buffer containing 10% sucrose, and snap-frozen in liquid nitrogen.
Planar Lipid Bilayers
Single channel recordings of recombinant InsP3R2 activities were performed by fusing microsomes into planar lipid bilayers. Bilayers were formed across a 150-μm-diameter hole in the wall of a Delrin cup using a 7:3 lipid mixture of phosphatidylethanolamine and phosphatidylcholine (50 mg/ml in decane; Avanti Polar Lipids, Alabaster, AL). The bilayer separated two pools (cis and trans). The microsomes were added to the cis-side of the bilayer. Standard solution contained 20 mm HEPES-Tris, pH 7.4, 1 mm EGTA, [Ca2+]FREE = 250 nm, and 220 mm CsCH3SO3 in the cis-chamber (20 mm, trans). Free calcium concentration was calculated using MaxChelator software. 2 μm InsP3 and 10 μm ryanodine were used. The trans pool was held at virtual ground. The channels were positively identified by their sensitivity to InsP3 and heparin. Open probability was determined by using the half-threshold crossing from 3-min records at 0 mV. Unitary currents were recorded using a conventional patch clamp amplifier (Axopatch 200B, Axon Instruments, Union City, CA). The current signal was digitized at 10 kHz with a 32-bit AD/DA converter (Digidata 1322A; Axon Instruments) and filtered at 1 kHz with a low pass eight-pole Bessel filter. Data acquisition, unitary current measurement, statistical analysis, and data processing were performed using commercially available software packages (pClamp V10.2, Axon Instruments and Origin, Microcal).
Data Presentation
Statistical comparisons were made using Student's t test for paired and unpaired data, with statistical significance from wild-type control InsP3R2 channels set at p < 0.05. Bar graphs are presented as average open probabilities ± S.E. of n measurements, where n refers to the number of channels. All Western blot data and channel traces shown are representative of three or more identical experiments.
RESULTS
CaMKII Modulation of InsP3R2 Channel Activity Is Reversible with Phosphatase Treatment
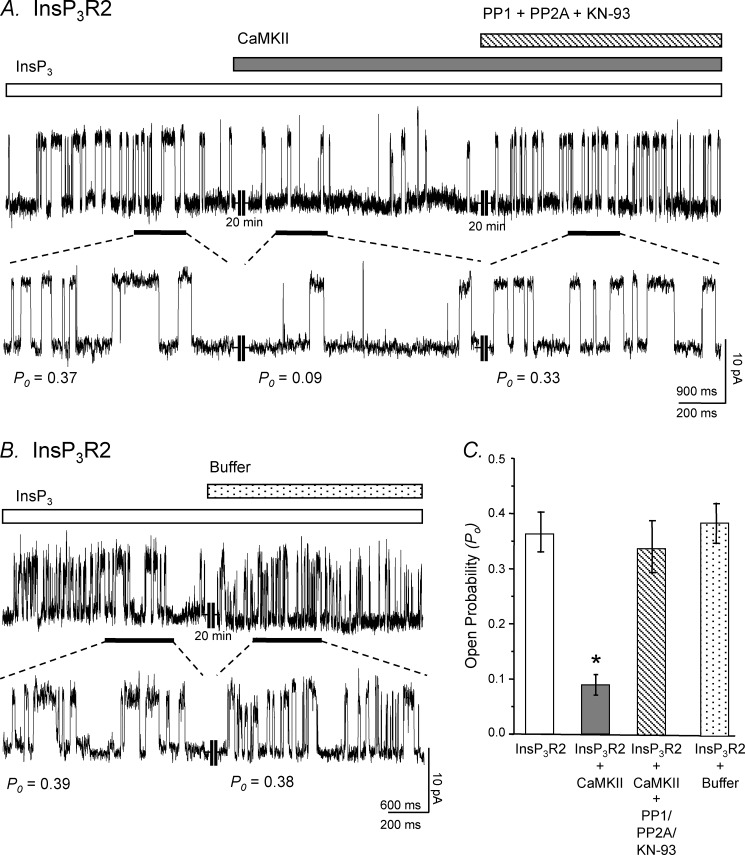
Previous data from our laboratory have shown that the InsP3R2 and CaMKII form a macromolecular complex in cardiac ventricular myocytes and that CaMKII can modulate InsP3R2 channel activity (1). To investigate further the role of CaMKII in mediating InsP3R2 activity, we fused microsomes from Sf9 insect cells infected with a baculovirus coding for the rat InsP3R2 into planar lipid bilayers for channel analysis. In Fig. 1A, a single InsP3R2 channel is recorded under normal conditions (2 μm InsP3, [Ca2+]FREE = 250 nm) and displayed a Po of ∼0.37. Active exogenous CaMKII (1500 units) was then added to the cis side of the bilayer and channel recording continued. After ∼20 min in the presence of the CaMKII, the channel Po decreased ∼4-fold to 0.09. The addition of PP1 and PP2A and the CaMKII inhibitor KN-93 were able to mitigate the CaMKII-mediated inhibition of the channel activity, and the Po returned to approximately starting level (0.33). The control experiment was performed by adding all components present in the CaMKII activation mixture except the kinase to the bath solution during recording of an InsP3R2 single channel (Fig. 1B). These had no effect on InsP3R2 activity and suggest that the diminution of InsP3R2 channel activity was a consequence of phosphorylation events mediated by CaMKII.
FIGURE 1.
Modulation of recombinant InsP3R2 activity by CaMKII is reversible with phosphatase treatment. Recordings are representative of InsP3R2 single channels from InsP3R2 baculovirus-infected Sf9 cell microsomes. A, a channel Po of 0.36 was recorded under normal conditions. To the bath solution, active CaMKII enzyme was then added, and recording continued after 20 min. This treatment resulted in a decrease in the Po to 0.09. The addition of protein phosphatases (10 units of PP1 and 50 ng of PP2A) and KN-93 (30 μm) was able to relieve CaMKII-mediated regulation of the channel activity, and the Po returned to approximately starting level (0.33) after 20 min with treatment. B, a channel Po of 0.38 was recorded under normal conditions. To the bath solution, CaMKII reaction buffer alone was added, and recording continued after 20 min. This treatment resulted in no change in the recorded open probability of the channel. Both A and B were taken as representative windows having Po reflecting the 10-min recordings used for the actual determination of Po. Channel openings are shown as upward deflections from the zero current level. C, bar graphs summarize the planar lipid bilayer studies (n = 5 for all conditions; *, p < 0.001; error bars, S.E.).
A summary of the planar lipid bilayer results is shown in Fig. 1C. These data, together with the previous data from Bare et al. (1), support the hypothesis that CaMKII-mediated phosphorylation of the channel is responsible for the decrease in Po seen upon CaMKII addition. Additionally, these results indicate that the inhibitory site on the receptor is likely localized to the large cytoplasmic region of the protein oriented on the cis side of the bilayer membrane, as that was the side the kinase was added. Of note, addition of active CaMKII to the trans side of the bilayer had no effect on the channel properties (data not shown). This is consistent with our previous in vitro phosphorylation results indicating that a fragment encompassing the amino-terminal 1078 residues of the receptor is phosphorylated by CaMKII.
CaMKII Phosphorylates the Full-length InsP3R2 on Ser-150
To determine the specific residue(s) of the InsP3R2 phosphorylated by CaMKII, we used expressed fragments of the InsP3R2 in combination with in vitro CaMKII-dependent phosphorylation assays. Receptor subfragments spanning residues 1–1078, 1074–1640, 1635–2118, and 2114–2701 (Fig. 2) were expressed in COS-1 cells and immunoprecipitated with an amino-terminal antibody (T2NH, for 1–1078) or anti-FLAG antibody (for 1074–1640, 1635–2118, and 2114–2701). The immune complexes were then incubated with [γ-32P]ATP and either preactivated CaMKII (500 units) or the CaMKII inhibitor KN-93 (10 μm), resolved on 7.5% SDS-PAGE, transferred to nitrocellulose membrane, and autoradiographed. Following autoradiography the membranes were Western blotted with either a InsP3R2 amino-terminal (T2NH) or anti-FLAG antibodies to visualize the protein bands. As shown in Fig. 2, the fragment spanning amino acids 1–1078 was the only fragment that showed consistent 32P incorporation upon treatment with CaMKII. The incorporation of the 32P label in this fragment was blocked with KN-93 and is in agreement with our previous results demonstrating CaMKII-mediated phosphorylation of this span of amino acids (1). These results suggest that the only detectable CaMKII phosphorylation site on the InsP3R2 is localized to the amino-terminal 1078 amino acids.
FIGURE 2.
CaMKII-dependent phosphorylation of InsP3R2(1–1078) fragment. CaMKII phosphorylation assays performed using immunoprecipitations of InsP3R2 subfragments expressed in COS-1 cells as substrates. Upper panel, with the addition of CaMKII, the 1–1078 construct showed significant incorporation of 32P, whereas no signal could be detected with the 1074–1640, 1635–2118, or 2114–2701 constructs. Lower panel, Western blotting of the membrane with T2NH (1–1078) or anti-FLAG (1074–1640, 1635–2118, and 2114–2701) antibody confirmed the success and uniformity of immunoprecipitations used in the phosphorylation reactions. Diagrams of the fragments used in this experiment are shown at the bottom with amino-terminal FLAG tags on fragments 1074–1640, 1635–2118, and 2114–2701.
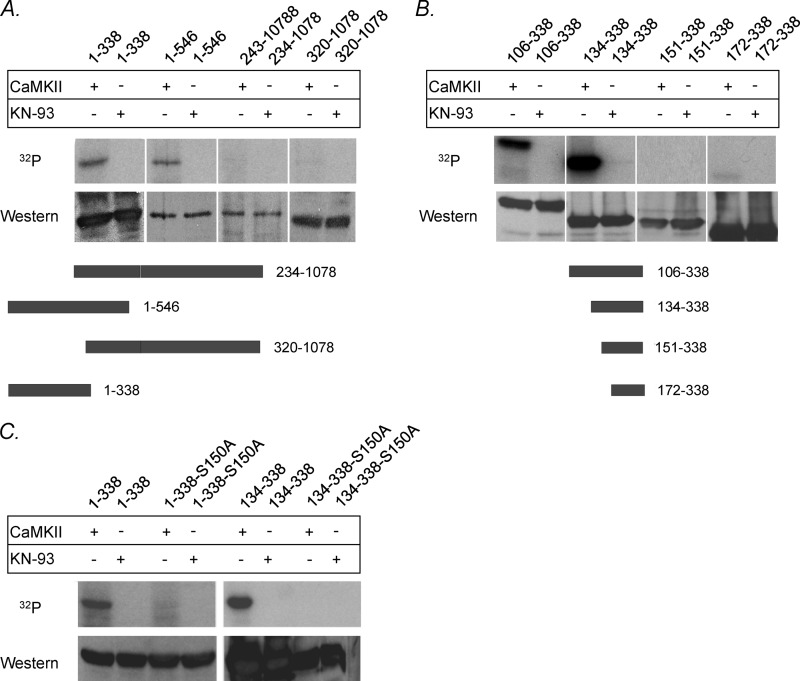
To elucidate further the site of CaMKII-dependent phosphorylation on the InsP3R2, we used eight bacterially expressed subfragments spanning the amino-terminal 1078 residues. Fragments spanning residues 1–338, 1–546, 234–1078, and 320–1078 were subjected to in vitro kinase assays. Expression products from constructs spanning residues 1–338 and 1–546 showed significant incorporation of 32P after CaMKII treatment, whereas no signal could be detected with the 234–1078 and 320–1078 constructs (Fig. 3A). The incorporation of label for the 1–338 and 1–546 constructs was inhibited with 10 μm KN-93, demonstrating CaMKII-specific 32P incorporation. These results suggest that the site of CaMKII phosphorylation on the InsP3R2 lies within the amino-terminal 234 residues. Some very weak label incorporation could be observed for the two fragments encompassed by residues 234–1078, and this was interpreted as nonspecific trapping of label although it cannot be ruled out that the fragments were misfolded and possible sites were inaccessible to CaMKII.
FIGURE 3.
CaMKII-dependent phosphorylation of bacterially expressed InsP3R2 fragments. CaMKII phosphorylation assays were performed using immunoprecipitations of bacterially expressed InsP3R2 subfragments as substrates. A, with the addition of CaMKII, the 1–546 and 1–338 constructs showed significant incorporation of 32P, whereas no signal could be detected with the 234–1078 and 320–1078 constructs. B, the 106–338 and 134–338 constructs showed significant incorporation of 32P with the addition of CaMKII. No significant signal could be detected with the 151–338 or the 172–338 constructs (A and B, upper panels). Western blotting of the membrane with T2NH antibody confirmed the success and uniformity of immunoprecipitations used in the phosphorylation reactions (A and B, lower panels). Diagrams of the fragments used in this experiment are shown at the bottom. C, mutation of Ser-150 → Ala could abolish CaMKII-mediated phosphorylation of fragments 1–338-S150A and 134–338-S150A. Upper panel, fragments 1–338 and 134–338 not containing the mutation showed significant incorporation of 32P when treated with CaMKII. Lower panel, Western blotting of the membrane with T2NH antibody confirmed the presence of similar protein amounts within samples used in the phosphorylation reactions.
The localization of the phosphorylation site was further delineated by preparing amino-terminal deletions that preserve the NH2-terminal antibody epitope (T2NH, amino acids 320–338) used for protein immunoprecipitation. These include residues 106–338, 134–338,151–338, and 172–338. As shown in Fig. 3B, the 106–338 and 134–338 constructs showed robust incorporation of 32P after treatment with CaMKII, which was sensitive to inhibition with 10 μm KN-93. No significant signal could be detected with the CaMKII-treated 151–338 or 172–338 expression products. These results imply that the CaMKII phosphorylation site lies within amino acids 134–150 of the InsP3R2.
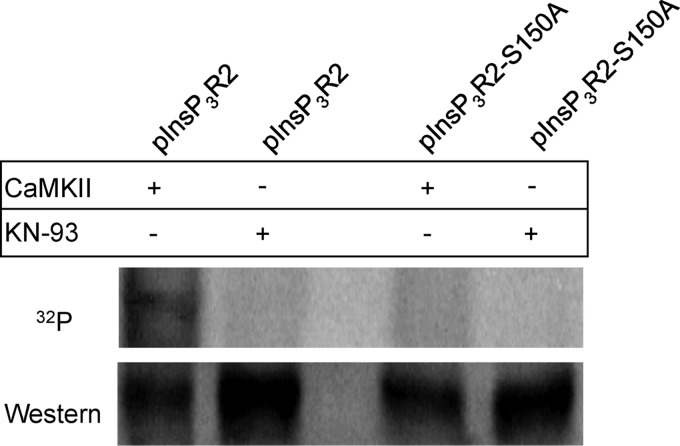
Within this span of amino acids, there is one potential CaMKII site (either serine or threonine residue) located at Ser-150. This serine is part of an S-X-D amino acid motif (RVSLDAAGN) in the InsP3R2. This motif was shown to be the CaMKII-dependent phosphorylation site on the RyR2 (21). Mutagenesis of the serine 150 to alanine in the two fragments (1–338 and 134–338) resulted in no appreciable CaMKII-dependent 32P incorporation for either fragment (Fig. 3C). Site-directed mutagenesis was also performed to mutate the serine at residue 150 to alanine (S150A) in the full-length InsP3R2 protein. Upon treatment with CaMKII, the InsP3R2-S150A showed no 32P incorporation compared with the InsP3R2-WT protein that could be phosphorylated with exogenously supplied CaMKII (Fig. 4). These results clearly identify Ser-150 as the CaMKII phosphorylation site on the InsP3R2. Furthermore, it can be concluded that at the levels of sensitivity of this assay, this is likely the only CaMKII phosphorylation site present on the InsP3R2 because mutagenesis of this site abolished CaMKII-mediated phosphorylation of the full-length protein.
FIGURE 4.
CaMKII can phosphorylate the full-length InsP3R2 on Ser-150. Upper panel, CaMKII phosphorylation assays performed using full-length InsP3R2 expressed in COS-1 cells as substrates show that mutation of Ser-150 → Ala could abolish CaMKII-mediated phosphorylation of full-length InsP3R2-S150A. Full-length InsP3R2 not containing the mutation showed significant incorporation of 32P when treated with CaMKII. Lower panel, Western blotting of the membrane with T2NH antibody confirmed the presence of similar protein amounts within samples used in the phosphorylation reactions.
Homology of CaMKII Phosphorylation Sites
Serine 150 is part of an S-X-D motif that was also found to be a target for CaMKII phosphorylation in the RyR2 (21). The conservation of this region in the other isoforms of the InsP3R and other CaMKII substrates (3, 21, 22) is shown in Table 1. Homology of the CaMKII phosphorylation site in the InsP3R2 with the RyR2 and can be seen with the actual phosphorylation site (S-X-D) along with two alanine residues directly following the S-X-D motif. Furthermore, the valine directly before Ser-150 is also conserved in the RyR2 and the InsP3R1 and InsP3R3. Interestingly, Ser-150 is conserved as a threonine in the type 1 and type 3 InsP3R homologues. We have observed that CaMKII can phosphorylate the InsP3R1 on the first 1081 amino acids (data not shown). This supports the hypothesis that Thr-150 of the InsP3R1 is a potential CaMKII phosphorylation site. This provides a potential mechanism of differential regulation between isoforms given that one is a serine and the other is a threonine. CaMKII has not been observed to show any preferential phosphorylation of either residue.
TABLE 1.
Homology of CaMKII phosphorylation sites
CaMKII phosphorylation site alignment with the three isoforms of the InsP3R and from various species (mouse, rat, human, bovine) shows that Ser-150 of the type 2 receptor is conserved as a threonine in the type 1 and type 3 receptors. The region surrounding Ser-150/Thr-150 is also highly homologous in the three isoforms and among species. The CaMKII phosphorylation sites for human RyR2 and HDAC4 are also shown to illustrate the homology the InsP3R2 Ser-150 site shares with these known CaMKII phosphorylation sites on other cardiac proteins.

Ser-150 Is the Site of CaMKII-dependent Modulation of the InsP3R2 Channel
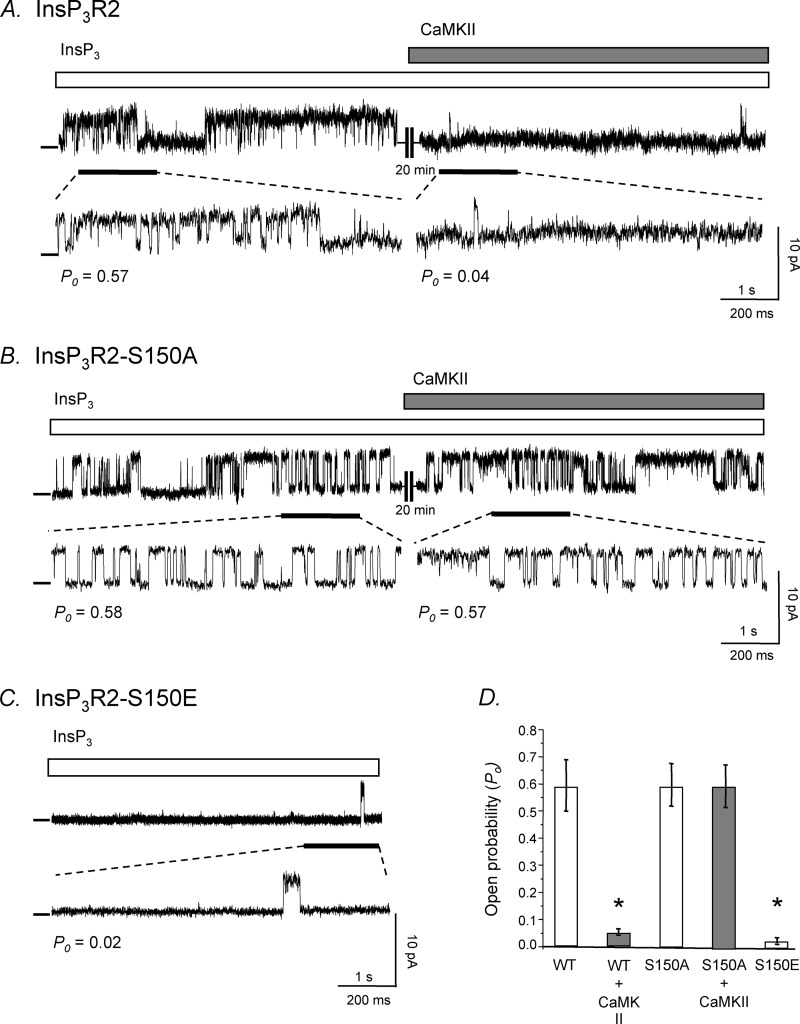
In addition to the InsP3R2-S150A nonphosphorylatable mutation, a InsP3R2-S150E phosphomimetic mutation changing Ser-150 to a glutamate (S150E) was made for use in planar lipid bilayer studies. Wild type and the two mutant (S150A and S150E) constructs were transiently transfected into COS-1 cells for expression of the proteins. Microsomal fractions from the three samples were prepared for use in channel recordings from planar lipid bilayers. In Fig. 5, a sample recording from WT microsomes shows that upon CaMKII treatment, the open probability (Po) of the channel is significantly decreased from 0.57 to 0.04. Conversely, the channel trace shown in Fig. 5B from InsP3R2-S150A microsomes did not show any decrease in open probability when active CaMKII enzyme was added to the bath solution (Po = 0.58 and 0.57, respectively), indicating that the channel was insensitive to the effect of CaMKII-dependent phosphorylation. Finally, Fig. 5C is a representative channel trace from InsP3R2-S150E microsomes showing that this channel protein exhibits a constitutively low open probability (Po = 0.02), thus mimicking an InsP3R2 that has been phosphorylated by CaMKII. Furthermore, it confirms that replacing Ser-150 with a glutamate did not alter the ability of the receptor to form a functional tetrameric channel. Fig. 5D is a summary bar graph showing the open probability of the three InsP3R2 channels with or without CaMKII treatment. There may be other CaMKII sites on the receptor that we were unable to detect; however, here we show that Ser-150 is a site that is phosphorylated by CaMKII and the site that is responsible for the channel modulation by CaMKII.
FIGURE 5.
Representative single channel recordings of WT, S150A, and S150E InsP3R2. A, for WT-InsP3R2 single channels, a channel Po of 0. 57 was recorded. Active CaMKII enzyme was added to the bath solution and recording continued. This treatment resulted in a decrease in the Po of 0.04 after 20 min. B, for S150A channels, a channel Po of 0.58 was recorded. Active CaMKII enzyme was added to the bath solution and recording continued. This treatment resulted in no change in the recorded Po of the channel after 20 min. C, for S150E channels, a channel Po of 0.02 was recorded. A, B, and C were taken as representative windows having Po values reflecting the 10-min recordings used for the actual determination of Po. Channel openings are shown as upward deflections from the zero current level. D, bar graphs summarize the planar lipid bilayer studies (n = 5 for all conditions; *, p < 0.001; error bars, S.E.)
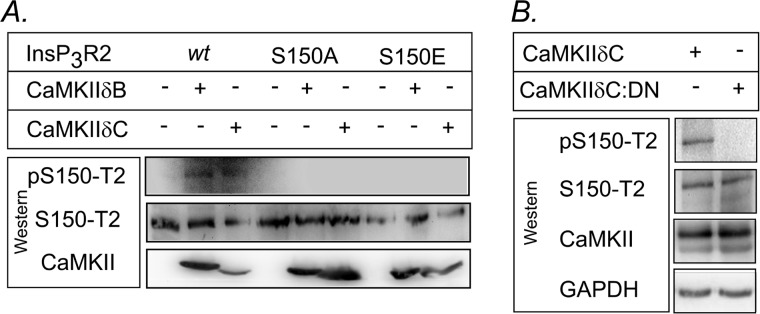
InsP3R2 Is Phosphorylated in Vivo by CaMKII at Ser-150 in COS Cells and Neonatal Rat Ventricular Myocytes
In support of the planar lipid bilayer studies demonstrating the functional consequences of Ser-150 phosphorylation to channel activity, we examined whether the InsP3R2 was phosphorylated in vivo as a consequence of CaMKII activity. COS 1 cells were transfected with either the WT or S150A or S150E mutations, with and without CaMKIIδB and δC then analyzed by Western blotting using antibodies directed against the Ser-150 region. As shown in Fig. 6A, an antibody directed against phosphorylated Ser-150 (Ser(P)-150-T2) detected the InsP3R2 only in samples which were co-expressing the WT InsP3R2 and either of the CaMKIIδ plasmids. No receptor was detected in the S150A or S150E mutants with the phospho-specific (Ser(P)-150-T2) antibody; however, robust signals were observed in all samples using an antibody directed against the nonphosphorylated Ser-150 region (Ser-150-T2).
FIGURE 6.
In vivo phosphorylation of InsP3R2 by CaMKIIδ. A, Western immunoblotting of COS-1 cells transfected with WT, S150A, or S150E InsP3R2 and CaMKIIδB or δC using the phospho-specific (pS150-T2) and nonphosphorylated (S150-T2) antibodies. Note the detection of phospho-specific signals for both CaMKIIδB and δC in the WT InsP3R only and no detectable signal in either of the Ser-150 mutants. B, Western blots of endogenous InsP3R2 expressed in acutely isolated neonatal rat ventricular myocytes that were infected with CaMKIIδC or a dominant negative (CaMKIIδC:DN) adenovirus. Note the significant phospho-InsP3R2 signal seen in the CaMKIIδC-infected cells compared with cells expressing the dominant negative mutant. GAPH is used as control.
Additionally, we examined the endogenous InsP3R2 in acutely isolated neonatal rat ventricular myocytes infected with adenovirus expressing CaMKIIδC and a dominant negative mutation (CaMKIIδC:DN). Western blots (Fig. 6B) using Ser(P)-150-T2 reveal a strong phospho-InsP3R2 signal in CaMKIIδC infected cells and little if any signal in the cells expressing CaMKIIδC:DN. Together these results demonstrate that phosphorylation of InsP3R2 at S150 by CaMKIIδ occurs in vivo.
DISCUSSION
The InsP3R2 is localized predominantly to the nuclear envelope and to a lesser extent in the SR of cardiac myocytes (1, 23). InsP3R2s are intracellular Ca2+ release channels that play a role in the regulation of Ca2+ signaling in the cardiac myocyte. Their role in the regulation of ET-1-induced positive inotropy in both atrial and ventricular myocytes has been recently well characterized, along with their part in a nuclear signal transduction cascade involving CaMKII-mediated activation of gene expression (2, 7, 23). Thus, CaMKII-dependent phosphorylation of the InsP3R2 is an important method of physiological regulation of the channel in a cellular environment. This is the first report of the specific site of CaMKII-mediated phosphorylation of the InsP3R2 and the functional affects of this post-translational modification on the channel activity. The results demonstrate that serine-150 of the rat InsP3R2 is the site of modulation of the channel activity by CaMKII-dependent phosphorylation. Phosphorylation leads to inhibition of the InsP3R2, and this functional effect can be abolished by mutation of Ser-150 to alanine. Furthermore, a phosphomimetic InsP3R2 channel (S150E) exhibits a constitutively low open probability very similar to that of a WT channel phosphorylated by CaMKII.
Functional Consequences of CaMKII Phosphorylation of the InsP3R2
Type 2 InsP3R-mediated signals have a sigmoidal Ca2+ dependence, meaning termination of this signal is not intrinsically controlled by a Ca2+-dependent inactivation mechanism (15). Of significance in cardiac tissues where type 2 receptors are present, this sigmoidal Ca2+ dependence makes the InsP3R2 resistant to the RyR-mediated Ca2+ signals driving the cardiac contractile cycle and allows InsP3-dependent intracellular signaling cascades to operate independent of the global Ca2+ fluxes associated with contraction. We show a potential mechanism of termination of InsP3R2 calcium release by which CaMKII directly phosphorylates the InsP3R2 at Ser-150 and inhibits channel activity, thus terminating InsP3-mediated SR and nuclear Ca2+ release.
CaMKII-mediated phosphorylation of InsP3R2 most likely has different physiological effects based on which population of receptors is being phosphorylated. Data show that knock-out of InsP3R2 in mouse myocytes abolished the positive inotropic and arrhythmogenic effects of ET-1 in cardiac myocytes (24) and the ET-1 induced HDAC5 nuclear translocation in ventricular myocytes (2). This HDAC5 pathway seems to be isolated from the beat-to-beat global fluctuations in [Ca2+]i, creating an elegant system that is dependent on very local control of [Ca2+]. InsP3R2s localized to the nuclear envelope release Ca2+ into the nucleoplasm, activating CaMKII which can phosphorylate HDAC5 and cause its translocation out of the nucleus (2). Furthermore, it has been proposed that the activated CaMKII can feedback-inhibit the InsP3R to close the channel and terminate the InsP3R-mediated release of Ca2+ from the nuclear envelope into the nucleoplasm (1, 2). The results presented here strengthen this hypothesis and show that CaMKII feedback phosphorylates the InsP3R2 at Ser-150, and this phosphorylation results in a decrease of the channel open probability which is an important part of signaling pathway leading to hypertrophy and heart failure (2).
In heart failure, the InsP3R2 protein is up-regulated (25). Functionally, the majority of experimental evidence indicates that in cardiac myocytes the modest inotropic effect of the InsP3R2s also comes with the considerable ability to disrupt ECC at normal SR Ca2+ load (23). Despite this, the up-regulation of the InsP3R2 seen in heart failure may be an attempt to preserve and enhance the inotropic response of ET-1 in this condition. In addition to the increased InsP3R2 protein levels, the levels of the RyR are decreased in heart failure (25). The increased InsP3R2 protein level may contribute to arrhythmogenesis and the mishandling of Ca2+ regulation also seen in heart failure, or the InsP3R2s may create an environment where this increased expression in the SR can help potentiate Ca2+-induced calcium release from the decreased population of RyRs.
InsP3R2s co-localize with RyRs in both atrial and ventricular cells and can modulate the activity of the RyR (7, 23, 26). InsP3-induced calcium release can potentiate Ca2+-induced calcium release from RyRs leading to enhanced systolic SR Ca2+ release and positive inotropy. Due to the relatively low expression levels of InsP3R2s compared with RyRs (27), SR Ca2+ release from InsP3R2s alone does not affect the beat-to-beat global Ca2+ transient during action potential stimulation in the cell. However, it can influence RyR-mediated SR Ca2+ release which does affect the global SR Ca2+ release (7, 23, 26). Thus, it seems the effect of CaMKII-mediated phosphorylation of the SR-localized InsP3R2s would have an indirect effect on global SR Ca2+ release through the RyR.
Possible Mechanism of Inhibition
InsP3 binds to a distinct region of the InsP3R, located at the amino terminus of each subunit, known as the ligand binding domain. This region is composed of an amino-terminal suppressor domain and an adjacent InsP3-binding core (28–30). The suppressor domain reduces the affinity of InsP3 for the InsP3-binding core and is required for channel gating, as it was shown that removal of suppressor domain increased affinity for InsP3 but did not form functional channels (31). It is possible that phosphorylation of Ser-150 localized within the suppressor domain could perturb InsP3 binding to the InsP3-binding core or that phosphorylation impedes the interactions necessary for channel gating, thus preventing ligand-induced activation.
In summary, we have identified Ser-150 of the InsP3R2 as the target of CaMKII-mediated phosphorylation. This event results in the negative modulation of the InsP3R2 receptors intrinsic calcium channel activity.
Acknowledgments
We thank Drs. Ilya Bezprozvanny for the generous gift of the InsP3R2 baculovirus expression construct and Ademuyiwa Aromolaran for expert technical assistance with the initial single-channel recordings; Drs. Donald M. Bers, Lothar Blatter, and Pieter deTombe for helpful discussions during the study and Jennifer L. Ashfield for assistance with the preparation of the manuscript.
This work supported, in whole or in part, by National Institutes of Health Grant P01HL080101 (to G. A. M.).
- InsP3R
- inositol 1,4,5-trisphosphate receptor
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- DN
- dominant negative
- ET-1
- endothelin 1
- HDAC
- histone deacetylase
- Po
- open probability
- PP1 and PP2A
- protein phosphatases 1 and 2A
- RyR2
- ryanodine receptor 2
- SR
- sarcoplasmic reticulum.
REFERENCES
- 1. Bare D. J., Kettlun C. S., Liang M., Bers D. M., Mignery G. A. (2005) Cardiac type 2 inositol 1,4,5-trisphosphate receptor: interaction and modulation by calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 280, 15912–15920 [DOI] [PubMed] [Google Scholar]
- 2. Wu X., Zhang T., Bossuyt J., Li X., McKinsey T. A., Dedman J. R., Olson E. N., Chen J., Brown J. H., Bers D. M. (2006) Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J. Clin. Invest. 116, 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang T., Kohlhaas M., Backs J., Mishra S., Phillips W., Dybkova N., Chang S., Ling H., Bers D. M., Maier L. S., Olson E. N., Brown J. H. (2007) CaMKIIδ isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J. Biol. Chem. 282, 35078–35087 [DOI] [PubMed] [Google Scholar]
- 4. Berridge M. J., Lipp P., Bootman M. D. (2000) The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell. Biol. 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 5. Mignery G. A., Johnston P. A., Südhof T. C. (1992) Mechanism of Ca2+ inhibition of inositol 1,4,5-trisphosphate (InsP3) binding to the cerebellar InsP3 receptor. J. Biol. Chem. 267, 7450–7455 [PubMed] [Google Scholar]
- 6. Perez P. J., Ramos-Franco J., Fill M., Mignery G. A. (1997) Identification and functional reconstitution of the type 2 inositol 1,4,5-trisphosphate receptor from ventricular cardiac myocytes. J. Biol. Chem. 272, 23961–23969 [DOI] [PubMed] [Google Scholar]
- 7. Domeier T. L., Zima A. V., Maxwell J. T., Huke S., Mignery G. A., Blatter L. A. (2008) IP3 receptor-dependent Ca2+ release modulates excitation-contraction coupling in rabbit ventricular myocytes. Am. J. Physiol. Heart Circ. Physiol. 294, H596–604 [DOI] [PubMed] [Google Scholar]
- 8. Kockskämper J., Zima A. V., Roderick H. L., Pieske B., Blatter L. A., Bootman M. D. (2008) Emerging roles of inositol 1,4,5-trisphosphate signaling in cardiac myocytes. J. Mol. Cell. Cardiol. 45, 128–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maier L. S., Bers D. M. (2002) Calcium, calmodulin, and calcium-calmodulin kinase II: heartbeat to heartbeat and beyond. J. Mol. Cell. Cardiol. 34, 919–939 [DOI] [PubMed] [Google Scholar]
- 10. Hudmon A., Schulman H., Kim J., Maltez J. M., Tsien R. W., Pitt G. S. (2005) CaMKII tethers to L-type Ca2+ channels, establishing a local and dedicated integrator of Ca2+ signals for facilitation. J. Cell Biol. 171, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zima A. V., Bare D. J., Mignery G. A., Blatter L. A. (2007) IP3-dependent nuclear Ca2+ signalling in the mammalian heart. J. Physiol. 584, 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Little G. H., Bai Y., Williams T., Poizat C. (2007) Nuclear calcium/calmodulin-dependent protein kinase IIδ preferentially transmits signals to histone deacetylase 4 in cardiac cells. J. Biol. Chem. 282, 7219–7231 [DOI] [PubMed] [Google Scholar]
- 13. Matifat F., Hague F., Brûlé G., Collin T. (2001) Regulation of InsP3-mediated Ca2+ release by CaMKII in Xenopus oocytes. Pflugers Arch. 441, 796–801 [DOI] [PubMed] [Google Scholar]
- 14. Zhu D. M., Tekle E., Chock P. B., Huang C. Y. (1996) Reversible phosphorylation as a controlling factor for sustaining calcium oscillations in HeLa cells: involvement of calmodulin-dependent kinase II and a caliculin A-inhibitable phosphatase. Biochemistry 35, 7214–7223 [DOI] [PubMed] [Google Scholar]
- 15. Ramos-Franco J., Bare D., Caenepeel S., Nani A., Fill M., Mignery G. (2000) Single-channel function of recombinant type 2 inositol 1,4,5-trisphosphate receptor. Biophys. J. 79, 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Südhof T. C., Newton C. L., Archer B. T., 3rd, Ushkaryov Y. A., Mignery G. A. (1991) Structure of a novel InsP3 receptor. EMBO J. 10, 3199–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mignery G. A., Newton C. L., Archer B. T., 3rd, Südhof T. C. (1990) Structure and expression of the rat inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 265, 12679–12685 [PubMed] [Google Scholar]
- 18. Tu H., Wang Z., Nosyreva E., De Smedt H., Bezprozvanny I. (2005) Functional characterization of mammalian inositol 1,4,5-trisphosphate receptor isoforms. Biophys. J. 88, 1046–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramos-Franco J., Caenepeel S., Fill M., Mignery G. (1998) Single channel function of recombinant type-1 inositol 1,4,5-trisphosphate receptor ligand binding domain splice variants. Biophys. J. 75, 2783–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaznacheyeva E., Lupu V. D., Bezprozvanny I. (1998) Single-channel properties of inositol (1,4,5)-trisphosphate receptor heterologously expressed in HEK-293 cells. J. Gen. Physiol. 111, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wehrens X. H., Lehnart S. E., Reiken S. R., Marks A. R. (2004) Ca2+/calmodulin-dependent protein kinase II phosphorylation regulates the cardiac ryanodine receptor. Circ. Res. 94, e61–70 [DOI] [PubMed] [Google Scholar]
- 22. Backs J., Song K., Bezprozvannaya S., Chang S., Olson E. N. (2006) CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116, 1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mackenzie L., Bootman M. D., Laine M., Berridge M. J., Thuring J., Holmes A., Li W. H., Lipp P. (2002) The role of inositol 1,4,5-trisphosphate receptors in Ca2+ signalling and the generation of arrhythmias in rat atrial myocytes. J. Physiol. 541, 395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X., Zima A. V., Sheikh F., Blatter L. A., Chen J. (2005) Endothelin-1-induced arrhythmogenic Ca2+ signaling is abolished in atrial myocytes of inositol 1,4,5-trisphosphate (IP3) receptor type 2-deficient mice. Circ. Res. 96, 1274–1281 [DOI] [PubMed] [Google Scholar]
- 25. Go L. O., Moschella M. C., Watras J., Handa K. K., Fyfe B. S., Marks A. R. (1995) Differential regulation of two types of intracellular calcium release channels during end-stage heart failure. J. Clin. Invest. 95, 888–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zima A. V., Blatter L. A. (2004) Inositol 1,4,5-trisphosphate-dependent Ca2+ signalling in cat atrial excitation-contraction coupling and arrhythmias. J Physiol. 555, 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moschella M. C., Marks A. R. (1993) Inositol 1,4,5-trisphosphate receptor expression in cardiac myocytes. J. Cell Biol. 120, 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bosanac I., Alattia J. R., Mal T. K., Chan J., Talarico S., Tong F. K., Tong K. I., Yoshikawa F., Furuichi T., Iwai M., Michikawa T., Mikoshiba K., Ikura M. (2002) Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature 420, 696–700 [DOI] [PubMed] [Google Scholar]
- 29. Bosanac I., Yamazaki H., Matsu-Ura T., Michikawa T., Mikoshiba K., Ikura M. (2005) Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor. Mol. Cell 17, 193–203 [DOI] [PubMed] [Google Scholar]
- 30. Seo M. D., Velamakanni S., Ishiyama N., Stathopulos P. B., Rossi A. M., Khan S. A., Dale P., Li C., Ames J. B., Ikura M., Taylor C. W. (2012) Structural and functional conservation of key domains in InsP3 and ryanodine receptors. Nature 483, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uchida K., Miyauchi H., Furuichi T., Michikawa T., Mikoshiba K. (2003) Critical regions for activation gating of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 278, 16551–16560 [DOI] [PubMed] [Google Scholar]