Abstract
The mangrove rivulus Kryptolebias marmoratus and a closely related species are the world’s only vertebrates that routinely self-fertilize. Such uniqueness presents a model for understanding why this reproductive mode, common in plants and invertebrates, is so rare in vertebrates. A survey of 32 highly polymorphic loci in >200 specimens of mangrove rivulus from multiple locales in the Florida Keys, USA, revealed extensive population-genetic structure on microspatial and micro-temporal scales. Observed heterozygosities were severely constrained, as expected for a hermaphroditic species with a mixed-mating system and low rates of outcrossing. Despite the pronounced population structure and the implied restrictions on effective gene flow, isogenicity (genetic identity across individuals) within and among local inbred populations was surprisingly low even after factoring out probable de novo mutations. Results indicate that neither frequent bottlenecks nor directional genetic adaptation to local environmental conditions were the primary driving forces impacting multilocus population-genetic architecture in this self-fertilizing vertebrate species. On the other hand, a high diversity of isogenic lineages within relatively small and isolated local populations is consistent with the action of diversifying selection driven by the extreme spatio-temporal environmental variability that is characteristic of mangrove habitats.
Introduction
Reproduction by self-fertilization is common in many hermaphroditic animals and plants (Avise 2011; Jarne and Auld 2006). When repeated across successive generations, selfing leads to a rapid decay of heterozygosity and can eventuate in isogenic lineages composed of multiple individuals that are genetically identical (barring de novo mutation) and that in effect offer targets for natural selection operating at the level of the whole genome. Isogenic lines that are well adapted to local environmental conditions should increase in frequency, eventually replacing less adapted lineages until only one or a few favored genotypes predominate at a given locale. On the other hand, sharply reduced genetic variation in homozygous lineages may prevent their adaptation to changing environments and lead to quick termination. Perhaps as a result of such contradictory evolutionary pressures, many plants and invertebrate animals show “mixed-mating” strategies in which both selfing and outcrossing coexist in a population in various proportions (Goodwillie et al. 2005; Jarne and Auld 2006; Escobar et al. 2011; Winn et al. 2011). Indeed, although selfing is common in many hermaphroditic species, few known organisms seem to have employed selfing as an exclusive reproductive tactic for long periods of evolutionary time.
The impact of selfing on the structure of populations and dynamics of isogenic lineages is of interest because it provides a model for understanding how environmental selection operates on microevolutionary scales (Charlesworth and Wright 2001; Charlesworth 2003; Epinat and Lenormand 2009). An extensive literature on the population genetics of self-fertilizing plants and invertebrates has demonstrated that strong differentiation often exists on a local scale, sometimes between sites only meters apart (Bomblies et al. 2010); in a few notable cases such differentiation was associated with specific habitat characteristics (Clegg and Allard 1972; Hamrick and Allard 1972; Selander and Hudson 1976). In the latter examples, specific multilocus genotypes proved to be associated with mesic versus xeric environments on local and regional geographic scales, presumably attributable to natural selection acting either through coadapted, multilocus gene complexes, or perhaps via genetic hitchhiking (Hedrick and Holden 1979). Of course, strong population subdivision on a local scale also can arise by genetic drift and low gene flow in the absence of natural selection per se (Jarne 1995; Charlesworth 2003; Bomblies et al. 2010).
Outcrossing also takes place in many plant and animal species with predominant selfing at rates known to be extremely heterogeneous in such mixed-mating species (Igic and Kohn 2006). For example, in the predominantly selfing mustard Arabidopsis thaliana, outcrossing rates sometimes reach nearly 20%, and reported rates of selfing/outcrossing in populations of the closely related Arabidopsis lyrata cover the full spectrum of 0–100 percent (Bomblies et al. 2010; Willi and Määttänen 2010).
A small cyprinodontiform fish—the mangrove rivulus, Kryptolebias marmoratus—is one of only two vertebrate species known to self-fertilize (the other being its close relative Kryptolebias hermaphroditus, formerly known as Kryptolebias ocellatus [Costa 2011]). Although selfing predominates in the mangrove rivulus, outcrossing takes place as well, meaning that the species actually has a mixed-mating system (Mackiewicz et al. 2006c). Outcrossing rates are known to vary from near-zero in the Bahamas and some Floridian locales to almost 60% in Belize (Mackiewicz et al. 2006b; Tatarenkov et al. 2009; Ellison et al. 2011). Outcrossing presumably occurs via spawning between hermaphrodites and males, as suggested by the fact that pure males constitute nearly 25% of the population in some localities in Belize but are much less frequent (<1%) in Florida (Turner et al. 1992). The co-presence of males and hermaphrodites means that K. marmoratus also provides an example of androdioecy, an extremely rare reproductive system otherwise known in only a few dozen extant plant and animal species (Pannell 2002; Weeks et al. 2006; Avise 2011).
The uniqueness of its reproductive mode makes mangrove rivulus a model system for understanding why self-fertilization, common in plants and invertebrates, is so rare in vertebrates. Previous studies of the mangrove rivulus have documented heterogeneity in outcrossing rates and population-genetic composition at scales of tens to hundreds of kilometers (Tatarenkov et al. 2007, 2011). Little is known about local population-genomic structure in this secretive species with a cryptic lifestyle, yet such knowledge should be germane to understanding any ecological forces that might drive selfing/outcrossing rates in this, and other, mixed-mating species. One particular aspect that has been missing in previous works (due largely to limited sampling) is the spatial distribution of isogenic lineages (multilocus genotypes) and their persistence over time. Meanwhile, distribution pattern of isogenic lineages can potentially be highly informative about adaptive significance of self-fertilization in hermaphroditic taxa. Here, we examine microevolutionary population-genetic patterns at 32 highly polymorphic microsatellite loci in >200 specimens of K. marmoratus from the Florida Keys, USA. Sampling across multiple years and locales allowed us to assess variation in outcrossing rates as well as examine the temporal and spatial distributions of isogenic lineages in these highly selfing populations.
Materials and methods
Study area
K. marmoratus together with its possible sibling species K. hermaphroditus has a broad distribution stretching from Florida and the Bahamas through the Antilles and along the Central and South American coastlines to south-eastern Brazil (Taylor et al. 1995; Costa 2011; Tatarenkov et al. 2011). We chose here to study the Florida Keys for three reasons: (1) this region was poorly represented in our previous analyses of population structure on various geographic scales (Tatarenkov et al. 2007, 2009), (2) oceanic currents suggest that the Keys might be an important source population for locations lying further north at the periphery of the species' range (primarily the eastern coast of mainland Florida), and (3) earlier studies indicated high rates of selfing in this area, so many individuals in these populations are expected to be homozygous, thus giving us a favorable opportunity to address questions about the temporal stability and spatial distribution of particular multilocus, isogenic genotypes.
Samples
A total of 201 specimens of K. marmoratus was collected from 12 locales in the Florida Keys on several occasions in 2007, 2010, and 2011; a few locales were sampled repeatedly (Fig. 1; Table 1). Some samples were rather small to be included in population-genetic analyses, but they were informative on the distribution of genetically identical individuals. Fish were captured from temporary pools or the burrows of great land crabs (Cardisoma guanhumi) using cup traps, wire minnow traps, hook and line, or dip nets (Florida Fish and Wildlife Conservation Commission Permit #SAL-09-1132B-SR to RLE and United States Fish and Wildlife National Key Deer Refuge Permit No. 2010-008 to DST). K. marmoratus is secretive, making it extremely difficult to catch. Over 1000 traps were set to collect fish for this study. The traps were checked on average every 2.5 h during daytime. Overnight, 170 traps were set, but resulted in 8 fish only. On average, it required about 76.9 trap hours to capture one mangrove rivulus in its natural habitat (mangrove forest). In comparison, in the same habitat only 6.1 trap hours were needed on average to capture other fish such as Fundulus confluentus, Gambusia sp., Adinia xenica, Poecilia latipinna, or Cyprinodon variegatus.
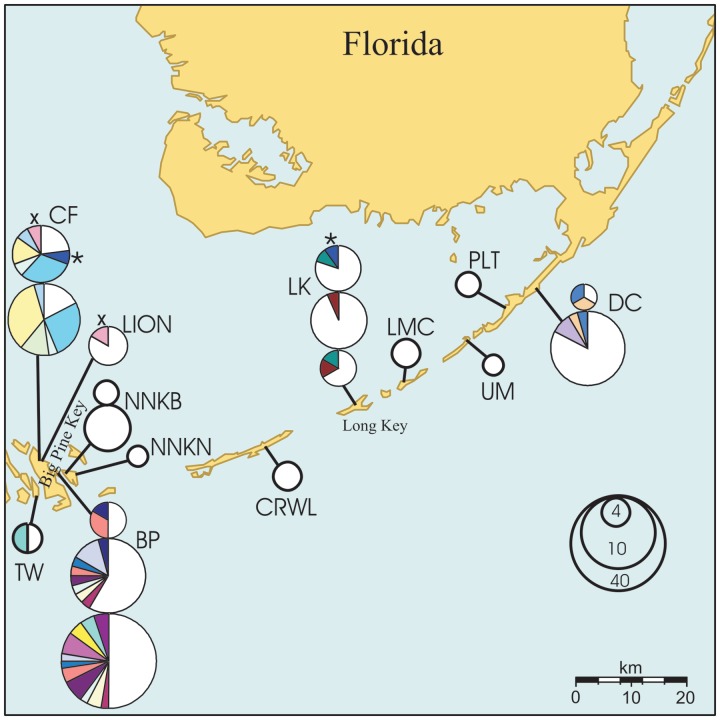
Fig. 1.
Map showing studied sites of K. marmoratus in the Florida Keys. Each circle denotes a collection from 1 year; circle sizes are proportional to sample sizes. Samples from different years at the same location are stacked with earlier years shown at bottom. Sections of different colors correspond to groups of genetically identical (isogenic) individuals (except for white, which represents pooled singletons or genetically unique individuals). The asterisks and crosses indicate pairs of genetically identical individuals from different sites.
Table 1.
Collections of K. marmoratus from the Florida Keys, 2007–2011
| Site designation | Location | Year collected | Coordinates | Sample size |
|---|---|---|---|---|
| TW | Little Torch Key | 2011 | N24°39′11.6″, W081°23′12.4″ | 4 |
| BP | Big Pine / Bogie Rd. | 2007 | N24°41′48.1″, W081°20′51.3″ | 40 |
| 2010 | N24°41′48.1″, W081°20′51.3″ | 24 | ||
| 2011 | N24°41′48.1″, W081°20′51.3″ | 6 | ||
| CF | Big Pine / “Chickenfoot” | 2010 | N24°43′32.9″, W081°23′14.5″ | 23 |
| 2011 | N24°43′32.9″, W081°23′14.5″ | 13 | ||
| LION | Big Pine / Lion’s Club | 2010 | N24°43′32.9″, W081°23′15.0″ | 6 |
| NNKB | No Name Key / Bridge | 2007 | N24°41′51.75″, W81°20′21.13″ | 10 |
| 2010 | N24°41′51.75″, W81°20′21.13″ | 3 | ||
| NNKN | No Name Key / North | 2010 | N24°41′52.07″, W81°19′14.77″ | 2 |
| CRWL | Crawl Key | 2011 | N24°44′55.0″, W080°58′41.2″ | 4 |
| LK | Long Key / Goshen Marine Lab | 2007 | N24°49′23.3″, W080°48′40.3″ | 6 |
| 2010 | N24°49′23.3″, W080°48′40.3″ | 15 | ||
| 2011 | N24°49′23.3″, W080°48′40.3″ | 10 | ||
| LMC | Lower Matecumbe Key | 2011 | N24°51′21.1″, W080°43′56.6″ | 4 |
| UM | Upper Matecumbe Key | 2011 | N24°56′16.5″, W080°36′49.1″ | 2 |
| PLT | Plantation Key | 2011 | N24°59′24.8″, W080°33′04.7″ | 3 |
| DC | Tavernier / Dove Creek | 2010 | N25°01′45.64″, W080°29′49.24″ | 23 |
| 2011 | N25°01′45.64″, W080°29′49.24″ | 3 | ||
| Total | 201 |
The sex of each specimen (hermaphrodite versus pure male) was assessed using phenotypic characters, especially body coloration and presence/absence of a black ocellus on the caudal fin (Turner et al. 2006). Preliminary data from 56 specimens collected in 2007 previously were reported by Tatarenkov et al. (2009).
Isolation of DNA and genotyping of microsatellites
Genomic DNA was extracted from fin clips of fish preserved in DMSO solution (Seutin et al. 1991) using proteinase-K tissue digestion followed by phenol-chloroform-isoamyl extraction and ethanol precipitation (Milligan 1998).
Genetic markers employed in this study were from a set of 32 microsatellite loci developed for K. marmoratus originating from a location in Florida (Mackiewicz et al. 2006a). For each locus, one PCR primer in each pair was labeled with a fluorescent dye (HEX, 6-FAM, or NED) and DNA was amplified in several multiplex reactions. The following multiplex combinations were formed: (1) R93Hex, R86Fam, R103Fam, and R90Ned; (2) B10Hex, B86Hex, and R112Fam; (3) R1Fam, R10Ned, R26Hex, R5Fam, R9Hex, R25Hex, and R27Fam; (4) R19Hex, R3Fam, R28Ned, R37Hex, R17Fam, and R23Ned; (5) R4Hex, R38Fam, and R22Ned; (6) R11Hex, R18Fam, and R34Ned; (7) R7Fam, R30Ned, R35Hex, and R6Hex; and (8) R33Fam and R92Hex. The PCR cocktail (final volume 10 µL) consisted of 1X GoTaq reaction buffer (which included 1.5 mM MgCl2), 0.25 µg BSA, 0.2 mM each dNTP, 0.25 µM each primer, 0.4 units GoTaq DNA polymerase (Promega), and 1 µL genomic DNA. Amplifications were conducted under an initial denaturation step at 95°C for 5 min, followed by 32 cycles of denaturation at 95°C for 40 s, annealing at 55°C for 40 s, and extension at 72°C for 1 min, with a final extension at 72°C for 7 min. Multiplexed PCR products were diluted 10–20-fold, after which 1 µL of diluted product was mixed with 9.6 µL of deionized formamide and 0.4 µL size standard GS500 (ROX labeled; Applied Biosystems), denatured for 4 min at 95°C, and electrophoresed on an ABI 3100. Alleles were scored using Genemapper 4.0 (Applied Biosystems). Binning of alleles follows the system of Tatarenkov et al. (2010).
Statistical analyses
Basic descriptive statistics (HE, HO, FIS) were calculated in GDA (Lewis and Zaykin 2001); this software was also used to find 95% confidence intervals for FIS by bootstrapping of loci (Weir 1996). Rates of selfing (S) and outcrossing (T = 1 − S) were estimated from the empirical fixation index using the equation S = 2FIS/(1 + FIS) (Wright 1969, 195). Pairwise and overall estimates of population differentiation FST were computed in FSTAT (version 2.9.3.2) (Goudet 1995). Significance of FST was evaluated by performing 1000 permutations of genotypes among samples using the same software. Simultaneous estimations of differentiation between populations and temporal variation within populations were conducted in HIERFSTAT (Goudet 2005). In this analysis, all variations are decomposed into three levels: (1) between individuals of the same year and site (FIS); (2) between years within sites (FYears/Site); and (3) between sites (FSites/Total). Furthermore, overall differentiation (FST) between all samples is calculated. Statistical significance of each level was assessed with 1000 randomizations in HIERFSTAT (for FYears/Site and FSites/Total) and FSTAT (for FIS and FST). The number of heterozygous loci for each individual was counted with the help of Microsatellite Analyzer (MSA) (Dieringer and Schlötterer 2003). The theoretical distributions of individual heterozygosities under mixed mating and under random mating were calculated as described by Mackiewicz et al. (2006b).
Results
Among the 201 individuals examined, we identified 150 different multilocus genotypes. These genotypes differed from one another at least at one locus (Table 2), and 125 of them (62.2%) were singletons, meaning that each was represented by a single assayed specimen. The remaining 25 multilocus genotypes were present in multiple (2–10) individuals each. All genotypes present in multiple specimens were fully homozygous, because progeny only of homozygous individuals may be genetically identical in a sexually reproducing species. The overall number of fully homozygous individuals was 150, as also indicated in the leftmost column in the histogram showing the distribution of K. marmoratus individuals according to their heterozygosities (Fig. 2). All studied populations, even those assessed on the basis of a few individuals, contained some genetically distinct individuals. In other words, no population was monotypic (i.e., fixed for a single multilocus genotype). In our collection, only two fish (from the LK and LION sites) were judged to be males based on phenotypic coloration. As determined by the multilocus, genetic evidence, neither of those fish sired any other specimen in our collection.
Table 2.
Distribution of 150 distinct multilocus genotypes observed in 201 K. marmoratus specimens in the Florida Keys
| Site | Year | Shared multilocus genotypes |
Singletons | Total | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | ||||
| TW | 2011 | 2 | 2 | 4 | ||||||||||||||||||||||||
| BP | 2007 | 1 | 2 | 1 | 3 | 2 | 1 | 1 | 3 | 2 | 2 | 2 | 20 | 40 | ||||||||||||||
| 2010 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 14 | 24 | ||||||||||||||||||
| 2011 | 2 | 1 | 3 | 6 | ||||||||||||||||||||||||
| CF | 2010 | 6 | 1 | 3 | 8 | 1 | 4 | 23 | ||||||||||||||||||||
| 2011 | 1 | 4 | 1 | 2 | 1 | 1 | 3 | 13 | ||||||||||||||||||||
| LION | 2010 | 1 | 5 | 6 | ||||||||||||||||||||||||
| LK | 2007 | 1 | 1 | 4 | 6 | |||||||||||||||||||||||
| 2010 | 1 | 14 | 15 | |||||||||||||||||||||||||
| 2011 | 1 | 1 | 8 | 10 | ||||||||||||||||||||||||
| DC | 2010 | 2 | 1 | 1 | 19 | 23 | ||||||||||||||||||||||
| 2011 | 1 | 1 | 1 | 3 | ||||||||||||||||||||||||
| NNKB | 2007 | 10 | 10 | |||||||||||||||||||||||||
| 2010 | 3 | 3 | ||||||||||||||||||||||||||
| NNKN | 2010 | 2 | 2 | |||||||||||||||||||||||||
| PLT | 2011 | 3 | 3 | |||||||||||||||||||||||||
| UM | 2011 | 2 | 2 | |||||||||||||||||||||||||
| CRWL | 2011 | 4 | 4 | |||||||||||||||||||||||||
| LMC | 2011 | 4 | 4 | |||||||||||||||||||||||||
The body of the table shows the observed numbers of fish from each locale displaying a given multilocus genotype. Twenty-five shared genotypes (i.e., genotypes that were observed in at least two individuals collected in the same or different years) arranged vertically are labeled with upper-case alphabet letters. Singletons are genotypes each detected in only one individual (but pooled here into a single column).
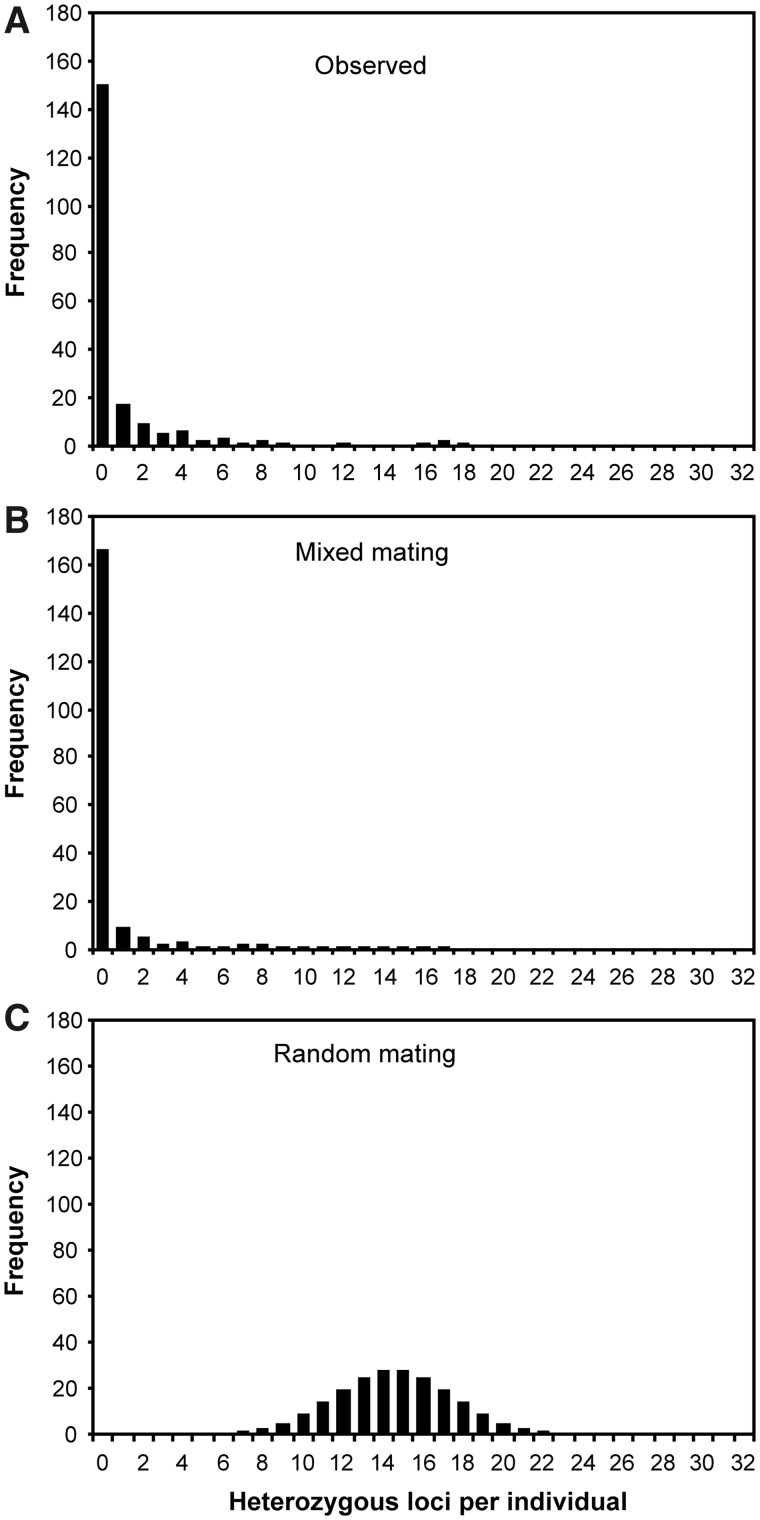
Fig. 2.
Frequency distribution of 201 K. marmoratus from the Florida Keys as a function of the observed number of heterozygous loci each possesses (A). Also shown are the distributions expected under mixed-mating (B) and random-mating (C) scenarios, assuming the same mean level of gene diversity. An empirically estimated average outcrossing rate of 0.037 was used in calculating the expected distribution under mixed mating (B).
Spatial and temporal distribution of isogenic individuals (clones)
At five sites (BP, CF, NNKB, LK, and DC), our collections spanned 2–3 years, and in four such cases (the exception being NNKB) genetically identical individuals were observed multiple times both within and across years (Fig. 1; Table 2). For example, at site BP on Big Pine Key, seven distinct genotypes (among 20 individuals) were common between the years 2007 and 2010. One of these genotypes (“E”) was also detected in 2011, despite the fact that only six individuals were sampled then. Similarly, site LK on Long Key harbored one genotype (“M”) common to years 2007 and 2010, and another genotype (“N”) in both years 2007 and 2011. Several instances of identical genotypes were detected between 2010 and 2011 at sites CF, BP (both on Big Pine Key), and DC (Tavernier) (Table 2). Some multilocus genotypes (four in BP2007, one in CF2010, and one in DC2010) were present in several copies in only 1 year. Although the total genotypes shared by multiple specimens sometimes accounted for a considerable portion (ranging from 16% at site LK on Long Key to 81% at site CF on Big Pine Key) of a population sample, any particular genotype typically represented only a small fraction of the population in any particular year (and overall). The one notable exception to this pattern involved site CF, where genotypes “P” and “S” reached substantial frequencies (26% and 35% of the population, respectively) (Table 2). Individuals displaying these genotypes were also found in the subsequent year at frequencies of 31% and 15%.
Despite the geographic proximity of sites on Big Pine Key, Little Torch Key, and No Name Key, only two pairs of identical individuals were found at different sites. One such pair (genotype “U”) involved single specimens at locales CF2011 and LION2010 separated by 220–350 m, and another two-specimen genotype (O) was found in 2011 at Big Pine Key (CF) and Long Key (LK), about 65 km apart.
Temporal variation within populations
Our collections included several samples from different years within sites. To avoid potential bias associated with estimating temporal heterogeneity from small sample sizes, only samples with N ≥ 10 from three sites (BP in 2007 and 2010, CF in 2010 and 2011, and LK in 2010 and 2011) were included in our statistical analyses. The extent of differentiation and its significance were obtained using an approach implemented in HIERFSTAT (Table 3). Overall FST among these samples was high (0.197), but most of the differentiation was among sites (0.196), whereas differentiation among years within sites was low (0.002) and statistically nonsignificant (P = 0.21). Selfing rates for these samples were compared using 95% confidence intervals obtained by bootstrapping of loci in GDA. The 95% confidence intervals for pairs of yearly samples from the three sites were broadly overlapping, indicating no significant variation in selfing rates within focal sites over the considered time periods of 1–3 years. Taking into account that local populations were stable both in frequencies of alleles and in rates of selfing, for the following analyses we pooled samples within the same site from different years.
Table 3.
Three-level hierarchical F-statistics
| Hierarchical level | F-statistic | P-value |
|---|---|---|
| FYears/Site | 0.002 | 0.214 (n.s.) |
| FSites/Total | 0.196 | 0.007 |
| FST | 0.197 | <0.001 |
| FIS | 0.939 | <0.001 |
Only samples with N ≥10 are used.
“/” means “within”; FST, overall differentiation; FIS, coefficient of inbreeding.
Diversity and rates of selfing
Table 4 summarizes basic statistics characterizing genetic variation in the studied populations. Gene diversity (i.e., heterozygosity expected under random mating) ranged from 0.206 to 0.522, with values at the lower end likely to be biased due to small sample size (for sample sizes of ≥10 individuals, HE varied from 0.390 to 0.511). Observed heterozygosity (HO) was much lower (0.000–0.076) than was gene diversity (suggesting strong heterozygote deficiency, as expected under selfing), and yielded positive and highly significant coefficients of inbreeding (FIS) at all sites (0.84–1.00, P < 0.001), which in turn translated into estimated rates of selfing ranging from 91% to 100%. Under such conditions, many fish have become completely homozygous after several reproductive cycles of selfing. Figure 2 shows the distribution of HO in Florida Keys, where 150 of the 201 collected fish (74.6%) were fully homozygous and another 17 were heterozygous at one locus only.
Table 4.
Summary of genetic variation in 12 local populations of K. marmoratus from the Florida Keys
| Population | N | P | NA | AR | HE | HO | FIS | S |
|---|---|---|---|---|---|---|---|---|
| LK | 31 | 0.84 | 4.2 | 3.8 | 0.486 | 0.061 | 0.875 | 0.93 |
| NNKB | 13 | 0.88 | 3.9 | 3.9 | 0.522 | 0.038 | 0.929 | 0.96 |
| NNKN | 2 | 0.63 | 1.6 | nc | 0.417 | 0 | 1 | 1.00 |
| BP | 70 | 0.81 | 4.9 | 4.0 | 0.491 | 0.024 | 0.951 | 0.97 |
| LION | 6 | 0.75 | 2.3 | nc | 0.424 | 0 | 1 | 1.00 |
| CF | 36 | 0.78 | 3.7 | 3.1 | 0.390 | 0.004 | 0.989 | 0.99 |
| DC | 26 | 0.81 | 4.4 | 3.9 | 0.451 | 0.076 | 0.835 | 0.91 |
| LMC | 4 | 0.72 | 2.0 | nc | 0.381 | 0.055 | 0.874 | 0.93 |
| PLT | 3 | 0.69 | 1.9 | nc | 0.404 | 0.042 | 0.916 | 0.96 |
| TW | 4 | 0.47 | 1.5 | nc | 0.206 | 0.008 | 0.967 | 0.98 |
| UM | 2 | 0.47 | 1.5 | nc | 0.323 | 0.031 | 0.933 | 0.97 |
| CRWL | 4 | 0.56 | 1.8 | nc | 0.323 | 0.008 | 0.979 | 0.99 |
| Mean | 0.70 | 2.8 | nc | 0.402 | 0.029 | 0.933 | 0.97 |
N, number of individuals; P95, proportion of polymorphic loci (95% criterion); NA, average number of alleles; AR, allelic richness; HE, gene diversity; HO, observed heterozygosity; FIS, coefficient of inbreeding; S, selfing rate. All FIS values are significant (P < 0.001) as determined by randomization tests in FSTAT (Goudet 1995). AR is based on sample size of 13 individuals; it was not calculated (nc) for smaller samples.
Geographic differentiation
Genetic differentiation between sites can be quantified by the fixation index, FST. We were concerned that small sample sizes at some sites might bias FST (likely by inflating it) and, consequently, conducted analyses using all samples as well as using only samples with N ≥ 13. The results were similar and indicated strong differentiation between sites. In the area of Florida Keys stretching from Little Torch Key in the south to Tavernier in the north, overall FST values were 0.19–0.20 (depending on whether small samples were included or not) (Table 5), and were highly significant. When all samples were considered, pairwise FST values varied from 0.008 to 0.470. For the subset of large samples, the range of pairwise differentiation was narrower but still large, ranging from FST = 0.061 between BP and NNKB to FST = 0.258 between CF and LK.
Table 5.
Tests of genotypic differentiation among sites
| Populations included | FST | P-value | ||
|---|---|---|---|---|
| Florida Keys | ||||
| All 12 populations | 0.195 | <0.001 | ||
| Five populations (N ≥ 13) | 0.189 | <0.001 | ||
| Big Pine Key Area | ||||
| Six populations | 0.178 | <0.001 | ||
| Three populations (N ≥ 13) | 0.176 | <0.001 | ||
Yearly samples from the same site are pooled.
The area around Big Pine Key (including Little Torch Key and No Name Key) was particularly well sampled, allowing us further to evaluate heterogeneity at a microgeographic scale. Several sites in this area showed some of the smallest pairwise FST values (<0.10) of the entire study. However, not all sites in close proximity exhibited low genetic heterogeneity. For example, LION (which was separated from CF by a distance of only 220–350 m) differed substantially from the latter (FST = 0.151) and, furthermore, simultaneous consideration of all six sites in the Big Pine Key area (or three sites with large sample sizes) resulted in FST values of about 0.18 (i.e., nearly as high as when all Florida Keys were considered) (Table 5). Finally, considering all 12 populations, the Mantel test suggested no effect of isolation by distance (P = 0.15).
Discussion
K. marmoratus and its sibling species K. hermaphroditus are unique among vertebrates in that they reproduce by self-fertilization. Such uniqueness presents a model for understanding why this mode of reproduction is so rare in vertebrates, compared to plants and invertebrates. The cryptic lifestyle and relative rarity of K. marmoratus has hindered studies of its population structure and dynamics. Previous works established that the mangrove rivulus combines selfing with occasional outcrossing and that the frequency of these reproductive modes varies considerably among distant geographic areas (such as Belize and Florida). However, much less is known about the variation of selfing rates and its impact on population structure on a smaller spatial scale. However, such knowledge may be crucial for understanding proximate factors (such as parasites or various environmental conditions) that might impact selfing rates. Another aspect that has been absent in previous works is the spatial and temporal distribution of isogenic lineages (multilocus genotypes). Here we address these issues by employing a set of highly polymorphic microsatellite loci to study mangrove rivulus populations in the Florida Keys.
Genetic differentiation between populations
Population structure in the Florida Keys was pronounced, with the average fixation index FST reaching nearly 20% (range 1–47%) between rivulus populations up to 112 km apart. Although the smallest FST values were found between some neighboring locations (BP, NNKB, NNKN, and LION), some similarly close or even closer pairs of locations (CF versus other sites on Big Pine and No Name Key) showed considerable differentiation (15–21%). Conversely, some distant pairs of populations displayed relatively low genetic divergence (FST < 10%). As a result of such mosaic structure, we detected no isolation by distance in the region (P = 0.15), suggesting that rates of gene exchange in K. marmoratus are not a function of geographic separation. Instead, it seems that exchange by migrants happens only rarely and when it does, the immigrant is perhaps as likely to come from a distant location as from a nearby one. This is further supported by the observation that neighboring sites generally did not share identical or closely similar genotypes. Indeed, we found only one case in which a pair of genetically identical individuals belonged to neighboring populations (CF and LION); there was one other case in which identical individuals were from relatively distant populations (CF and LK, about 65 km apart).
Considering that particular multilocus genotypes persisted across 3–4 years at some locales, we were surprised to find a near-complete absence of multilocus genotypes shared between sites. Perhaps rare cases of dispersal in this species are associated with exceptional circumstances (such as might occur during hurricanes when some fish or their eggs are dislodged and transferred to new locations). Alternatively, perhaps dispersal happens routinely but the migrants survive poorly in new locations (such that high dispersal does not translate into high gene flow). The latter explanation, however, seems unlikely in view of the high genotypic diversity in most populations. If the environment exerted strong power on the survival of particular genotypes, then each population probably would become dominated by a single lineage, or few lineages, most fit to the given set of environmental conditions.
Genetic diversity within populations and outcrossing rates
Gene diversity in populations of K. marmoratus in the Florida Keys was relatively high (HE = 0.40) on average. Furthermore, the variation in gene diversity between populations was quite modest (ranging between 0.39 and 0.52 for samples with ≥10 individuals). This overall pattern of high, yet uniform, gene diversity stands in contrast to the situation in many predominantly selfing plants with similar rates of outcrossing, in which local stands often are monotypic or dominated by a single multilocus genotype (Bomblies et al. 2010).
Average effective outcrossing for the Florida Keys was estimated at 3.5%, but there was some variation between populations, with DC in Tavernier showing as high as 9% and CF in Big Pine Keys as low as 1% (not counting localities with small sample sizes). These estimates are similar to those obtained from the western coast of Florida, which was reasonably well sampled, but they are noticeably lower than previous estimates for the Florida Keys (∼20%) based on very small numbers of fish (Mackiewicz et al. 2006b). Overall, outcrossing rates in Florida are in the range 1–9%, considerably lower than in Belize where they were as high as 60% at Twin Cayes and about 24% on a cay in Turneffe atoll (Mackiewicz et al. 2006b; Ellison et al. 2011).
Our data suggest that selfing rates remained stable over a period of up to 4 years. Such constancy is somewhat puzzling, taking into consideration the low frequency and seemingly erratic occurrence in males in Florida (which are thought to be necessary for outcrossing). Turner et al. (1992) reported a frequency of males at about 1% in southern Florida. We obtained a similar estimate in this study, with only two males being identified among the 201 fish collected (∼1%). The relative constancy of deduced selfing rates in the Florida Keys suggests that males are generated at a low but consistent rate. Alternatively, hermaphrodites might mate with one another on occasion, but mating between hermaphrodites has never been reported in aquaria or in nature (and behaviorally, hermaphrodites prefer to associate with males rather than with other hermaphrodites) (Martin 2007).
In our current study area, 75% of the fish were fully homozygous and another 15% were heterozygous at three or fewer of the surveyed loci. For the latter fish, some of the heterozygosity could in principle be due to de novo mutation and some to residual or ancestral heterozygosity tracing back to the last outcross event. Regardless, the overall genetic signature clearly signifies that selfing predominates in these local populations. By way of comparison, if mating instead had been at random, individual fish should be heterozygous on average at more than a dozen surveyed loci (from calculations based on observed levels of gene diversity in these populations) (Fig. 2). At least four generations of selfing would be necessary to reach the complete homozygosity that we observed in the majority of fish from the Florida Keys, and the reduced levels of heterozygosity in most of the remaining fish are consistent with them being a product of one or more recent generations of selfing. Indeed, the distribution of observed heterozygosities (Fig. 2) indicates that only five individuals (<2.5%) in our total sample were the products of outcrossing in the prior generation.
Overall, our study documents extensive spatial genotypic diversity in local populations of K. marmoratus, thus implying considerable constraints on effective gene flow. Strong genetic subdivision in principle should favor the development of local genetic adaptations, which in the case of highly-selfing organisms could be manifest as the predominance of certain selectively favored homozygous lineages at particular locations. Although isogenic individuals sometimes were rather common within locales, and some isogenic lineages persisted across all 4 years of our survey, no isogenic assemblages greatly predominated at any of our surveyed locales in the Florida Keys. Thus, the high genotypic diversity that we observed in the mangrove rivulus is likely an indication that local genetic adaptation resulting from directional selection is not the primary driving force impacting multilocus population-genetic architecture in this self-fertilizing species. Observation of high genetic diversity, combined with strong population structure, also suggests that local populations of K. marmoratus were relatively stable and that they did not experience major recent reductions of effective population size. On the other hand, the high diversity of multilocus genotypes itself may be promoted to some extent by extreme spatial and temporal variability in environmental conditions characteristic for mangroves. Detailed studies comparing physiological performances of distinct genetic lineages of the mangrove rivulus will be helpful in elucidating the role of microhabitats in promoting genetic variation.
Funding
This work was supported by funds from University of California at Irvine; by an NIH Conference Grant R13HD070622 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development; SICB through the DCE, DCPB, DAB and the C. Ladd Prosser Fund; and the College of Agriculture and Natural Resources, University of Maryland.
Acknowledgments
We would like to thank Phillip Hughes (U.S. Fish & Wildlife Service; National Key Deer Refuge) and Kyle Miller (Florida Fish & Wildlife Conservation Commission) for their assistance with permitting and Yvonne Wielhouwer (Monroe County, Lower Keys Mosquito Control) for her invaluable assistance scouting mosquito-ditch field sites. Thanks also to Amanda Hanninen, Andrew Turko, Yuying Hsu, Mark Garcia, Benjamin Perlman, Yu-Yun Huang, and Ching Chang for assistance with the field collections. We are grateful to personnel of the Keys Marine Laboratory (Long Key, FL) for the logistical support.
References
- Avise JC. Hermaphroditism: a primer on the biology, ecology, and evolution of dual sexuality. New York: Columbia University Press; 2011. p. 232. [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim ST, Hollister JD, Warthmann N, Fitz J, Weige D. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. Plos Genet. 2010;6:e1000890. doi: 10.1371/journal.pgen.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Effects of inbreeding on the genetic diversity of populations. Philos Trans R Soc B Biol Sci. 2003;358:1051–70. doi: 10.1098/rstb.2003.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr Opin Genet Dev. 2001;11:685–90. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- Clegg MT, Allard RW. Patterns of genetic differentiation in slender wild oat species Avena barbata. Proc Natl Acad Sci U S A. 1972;69:1820–24. doi: 10.1073/pnas.69.7.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa WJEM. Identity of Rivulus ocellatus and a new name for a hermaphroditic species of Kryptolebias from south-eastern Brazil (Cyprinodontiformes: Rivulidae) Ichthyol Explor Freshw. 2011;22:185–92. [Google Scholar]
- Dieringer D, Schlötterer C. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes. 2003;3:167–69. [Google Scholar]
- Ellison A, Cable J, Consuegra S. Best of both Worlds? Association between outcrossing and parasite loads in a selfing fish. Evolution. 2011;65:3021–26. doi: 10.1111/j.1558-5646.2011.01354.x. [DOI] [PubMed] [Google Scholar]
- Epinat G, Lenormand T. The evolution of assortative mating and selfing with in- and outbreeding depression. Evolution. 2009;63:2047–60. doi: 10.1111/j.1558-5646.2009.00700.x. [DOI] [PubMed] [Google Scholar]
- Escobar JS, Auld JR, Correa AC, Alonso JM, Bony YK, Coutellec MA, Koene JM, Pointier JP, Jarne P, David P. Patterns of mating-system evolution in hermaphroditic animals: correlations among selfing rate, inbreeding depression, and the timing of reproduction. Evolution. 2011;65:1233–53. doi: 10.1111/j.1558-5646.2011.01218.x. [DOI] [PubMed] [Google Scholar]
- Goodwillie C, Kalisz S, Eckert CG. The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst. 2005;36:47–79. [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–86. [Google Scholar]
- Goudet J. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes. 2005;5:184–86. [Google Scholar]
- Hamrick JL, Allard RW. Microgeographical variation in allozyme frequencies in Avena barbata. Proc Natl Acad Sci USA. 1972;69:2100–4. doi: 10.1073/pnas.69.8.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW, Holden L. Hitch-hiking: an alternative to coadaptation for the barley and slender wild oat examples. Heredity. 1979;43:79–86. [Google Scholar]
- Igic B, Kohn JR. The distribution of plant mating systems: study bias against obligately outcrossing species. Evolution. 2006;60:1098–103. [PubMed] [Google Scholar]
- Jarne P. Mating system, bottlenecks and genetic polymorphism in hermaphroditic animals. Genet Res. 1995;65:193–207. [Google Scholar]
- Jarne P, Auld JR. Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution. 2006;60:1816–24. doi: 10.1554/06-246.1. [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D. Genetic data analysis: computer program for the analysis of allelic data. 2001 Version 1.0 (d16c) ( http://www.eeb.uconn.edu/people/plewis/software.php) [Google Scholar]
- Mackiewicz M, Tatarenkov A, Perry A, Martin JR, Elder JF, Bechler DL, Avise JC. Microsatellite documentation of male-mediated outcrossing between inbred laboratory strains of the self-fertilizing mangrove killifish (Kryptolebias marmoratus) J Hered. 2006a;97:508–13. doi: 10.1093/jhered/esl017. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Tatarenkov A, Taylor DS, Turner BJ, Avise JC. Extensive outcrossing and androdioecy in a vertebrate species that otherwise reproduces as a self-fertilizing hermaphrodite. Proc Natl Acad Sci USA. 2006b;103:9924–28. doi: 10.1073/pnas.0603847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Tatarenkov A, Turner BJ, Avise JC. A mixed-mating strategy in a hermaphroditic vertebrate. Proc R Soc B. 2006c;273:2449–52. doi: 10.1098/rspb.2006.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SB. Association behaviour of the self-fertilizing Kryptolebias marmoratus (Poey): the influence of microhabitat use on the potential for a complex mating system. J Fish Biol. 2007;71:1383–92. [Google Scholar]
- Milligan BG. Total DNA isolation. In: Hoelzel AR, editor. Molecular genetic analysis of populations: a practical approach. Oxford: Oxford University Press; 1998. pp. 28–64. [Google Scholar]
- Pannell JR. The evolution and maintenance of androdioecy. Annu Rev Ecol Syst. 2002;33:397–425. [Google Scholar]
- Selander RK, Hudson RO. Animal population structure under close inbreeding: the land snail Rumina in Southern France. Am Nat. 1976;110:695–718. [Google Scholar]
- Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analyses. Can J Zool. 1991;69:2–90. [Google Scholar]
- Tatarenkov A, Gao H, Mackiewicz M, Taylor DS, Turner BJ, Avise JC. Strong population structure despite evidence of recent migration in a selfing hermaphroditic vertebrate, the mangrove killifish (Kryptolebias marmoratus) Mol Ecol. 2007;16:2701–11. doi: 10.1111/j.1365-294X.2007.03349.x. [DOI] [PubMed] [Google Scholar]
- Tatarenkov A, Lima SMQ, Taylor DS, Avise JC. Long-term retention of self-fertilization in a fish clade. Proc Natl Acad Sci U S A. 2009;106:14456–9. doi: 10.1073/pnas.0907852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarenkov A, Ring BC, Elder JF, Bechler DL, Avise JC. Genetic composition of laboratory stocks of the self-fertilizing fish Kryptolebias marmoratus: a valuable resource for experimental research. Plos One. 2010;5:e12863. doi: 10.1371/journal.pone.0012863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarenkov A, Lima SMQ, Avise JC. Extreme homogeneity and low genetic diversity in Kryptolebias ocellatus from southeastern Brazil suggest a recent foundation for this androdioecious fish population. J Fish Biol. 2011;79:2095–105. doi: 10.1111/j.1095-8649.2011.03155.x. [DOI] [PubMed] [Google Scholar]
- Taylor DS, Davis WP, Turner BJ. Rivulus marmoratus: ecology of distributional patterns in Florida and the central Indian River Lagoon. Bull Mar Sci. 1995;57:202–7. [Google Scholar]
- Turner BJ, Davis WP, Taylor DS. Abundant males in populations of a selfing hermaphrodite fish, Rivulus marmoratus, from some Belize Cays. J Fish Biol. 1992;40:307–10. [Google Scholar]
- Turner BJ, Fisher MT, Taylor DS, Davis WP, Jarrett BL. Evolution of ‘maleness’ and outcrossing in a population of the self-fertilizing killifish, Kryptolebias marmoratus. Evol Ecol Res. 2006;8:1475–86. [Google Scholar]
- Weeks SC, Benvenuto C, Reed SK. When males and hermaphrodites coexist: a review of androdioecy in animals. Integr Comp Biol. 2006;46:449–64. doi: 10.1093/icb/icj048. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II: methods for discrete population genetics data. Sunderland, MA: Sinauer Associates; 1996. p. 445. [Google Scholar]
- Willi Y, Määttänen K. Evolutionary dynamics of mating system shifts in Arabidopsis lyrata. J Evol Biol. 2010;23:2123–31. doi: 10.1111/j.1420-9101.2010.02073.x. [DOI] [PubMed] [Google Scholar]
- Winn AA, Elle E, Kalisz S, Cheptou PO, Eckert CG, Goodwillie C, Johnston MO, Moeller DA, Ree RH, Sargent RD, et al. Analysis of inbreeding depression in mixed-mating plants provides evidence for selective interference and stable mixed mating. Evolution. 2011;65:3339–59. doi: 10.1111/j.1558-5646.2011.01462.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations. II. The theory of gene frequencies. Chicago: University of Chicago Press; 1969. p. 511. [Google Scholar]