Abstract
How organisms adapt to the range of environments they encounter is a fundamental question in biology. Elucidating the genetic basis of adaptation is a difficult task, especially when the targets of selection are not known. Emerging sequencing technologies and assembly algorithms facilitate the genomic dissection of adaptation and population differentiation in a vast array of organisms. Here we describe the attributes of Kryptolebias marmoratus, one of two known self-fertilizing hermaphroditic vertebrates that make this fish an attractive genetic system and a model for understanding the genomics of adaptation. Long periods of selfing have resulted in populations composed of many distinct naturally homozygous strains with a variety of identifiable, and apparently heritable, phenotypes. There also is strong population genetic structure across a diverse range of mangrove habitats, making this a tractable system in which to study differentiation both within and among populations. The ability to rear K. marmoratus in the laboratory contributes further to its value as a model for understanding the genetic drivers for adaptation. To date, microsatellite markers distinguish wild isogenic strains but the naturally high homozygosity improves the quality of de novo assembly of the genome and facilitates the identification of genetic variants associated with phenotypes. Gene annotation can be accomplished with RNA-sequencing data in combination with de novo genome assembly. By combining genomic information with extensive laboratory-based phenotyping, it becomes possible to map genetic variants underlying differences in behavioral, life-history, and other potentially adaptive traits. Emerging genomic technologies provide the required resources for establishing K. marmoratus as a new model organism for behavioral genetics and evolutionary genetics research.
Introduction
The fish
The distribution of the mangrove rivulus, Kryptolebias marmoratus tracks that of the red mangrove (Rhizophora mangle) from the coastal regions of central Florida south through the Caribbean and extending to the eastern coasts of Central America and South America (Costa 2006). Kryptolebias marmoratus is an unusual vertebrate with a life history similar to that of Caenorhabditis elegans in which the majority of individuals are hermaphroditic with preferential self-fertilization (Harrington 1961). Males exist in the wild at varying frequencies (Lubinski et al. 1995; Mackiewicz et al. 2006c). To date, there is no documentation of females existing in the wild. As one of only two members of a fish clade with evidence for internal self-fertilization, K. marmoratus is uniquely suited to become a model for genomic studies.
Self-fertilization (inbreeding) has resulted in naturally homozygous individuals and the propagation of isogenic lineages in the wild (Harrington and Kallman 1968). While many wild-caught individuals are homozygous, heterozygous individuals are also found, indicating that out-crossing is a relatively common occurrence (Taylor et al. 2001), even in populations in which males have not been directly observed (Mackiewicz et al. 2006b; Tatarenkov et al. 2007). There is no current evidence of hermaphrodites fertilizing eggs from other hermaphrodites, suggesting that outcrossing occurs primarily between males and hermaphrodites. Primary and secondary males have been described (Harrington 1967). Recent microsatellite data of individuals genotyped at 35 microsatellite markers confirms a varying amount of heterozygosity in wild-caught individuals (Mackiewicz et al. 2006b).
Since the fish can be reared easily, K. marmoratus has been maintained in laboratories for ∼50 years (Harrington 1961). Due to the preferential self-fertilization, wild-caught lineages that are reared in the laboratory for several generations quickly become homozygous (Mackiewicz et al. 2006a). Over 250 distinct genetic lines are currently being maintained in laboratories. Laboratory strains are highly homozygous and, for the most part named stock lineages being maintained in laboratories internationally are very similar (Tatarenkov et al. 2010).
Lineages of K. marmoratus, identified by their sampling location, have a range of phenotypes that have been measured under field and laboratory conditions (Heuhner et al. 1985; Davis et al. 1990; Dunson and Dunson 1999; Lin and Dunson 1999; Earley et al. 2000; Taylor 2000; Hsu and Wolf 2001; Taylor et al. 2004; Grageda et al. 2005; Martin 2007; Earley and Hsu 2008; Hsu et al. 2008; Taylor et al. 2008; Molloy et al. 2011; Richards et al. 2011; Turko et al. 2011). Importantly, behavioral and life-history traits are reproducible within isogenic lineages (Edenbrow and Croft 2011; Earley et al. 2012), suggesting that many of the traits are heritable and have a genetic component. The naturally homozygous strains present distinct phenotypes that segregate between them, making this a tractable system for studying differentiation within populations and among populations. For example, Nakamura et al. (2008) found that crossing two lineages with divergent patterns of growth results in hybrid F2s with intermediate phenotypes (Nakamura et al. 2008).
Males exist at varying frequencies, from 0% to 20%, in the wild and can be induced by temperature in the laboratory (Harrington 1967). Therefore, it is possible to generate crosses between lineages that exhibit different phenotypes, which has been carried out successfully in vitro (Harrington 1971; Mackiewicz et al. 2006a; Nakamura et al. 2008). Additionally, imaging tools have been developed to utilize K. marmoratus for detailed embryological studies (Mourabit et al. 2011). Mutagenesis via N-ethyl-N-nitrosourea (ENU) has been established for identifying zygotic mutants (Moore et al. 2012)
The selfing habits and mixed-mating system of the mangrove rivulus provides genetic tractability that rivals some invertebrate (e.g., C. elegans, Daphnia) and plant (e.g., Arabidopsis) systems. This, coupled with high levels of phenotypic diversity both within and among populations, and the promise of crossing homozygous lineages possessing divergent phenotypes, make the mangrove rivulus a potentially powerful system in which to study adaptive genomics (Stapley et al. 2010).
The genomics
The dramatic reduction in sequencing costs and availability of next-generation sequencing technology makes sequencing accessible to new communities of researchers and allows the development of new genetic/genomic model systems (www.genome.gov/sequencingcosts). Sequencing technologies are continually being improved in terms of error rate and read length (Metzker 2010). The performance of assembly algorithms developed to handle various types of next-generation sequence data varies depending on the size, complexity, and similarity of the genome to already sequenced genomes (Earl et al. 2011). However, heterozygosity presents a formidable challenge to de novo assembly of the genome sequence (Vinson et al. 2005). To remove heterozygosity from de novo sequencing projects, significant effort is made to create homozygous lineages or to obtain data from haploids (Langley et al. 2011). Homozygosity improves the assembly of contigs, continuous stretches of DNA sequence created by overlapping sequence reads. It also facilitates obtaining longer scaffolds, which are sets of contigs separated by gaps, often with known approximate lengths. The natural occurrence of entirely homozygous individuals of K. marmoratus makes generating a high-quality de novo assembly of the genome a reality. Given the existing microsatellite data (Mackiewicz et al. 2006c; Tatarenkov et al. 2012), any differences detected during sequencing a single individual are likely rare, and result either from new mutations or, more probably, errors introduced during construction and sequencing of the library.
K. marmoratus is diploid with 24 chromosomes (Scheel 1972). Using initial next-generation sequencing data, we estimate the genome size of K. marmoratus to be ∼900 Megabases (Mb); which is larger than the medaka genome (700 Mb) (Takeda 2008). For a high-quality de novo assembly of a genome, multiple sequencing libraries must be used, with different insert sizes, including small-insert and large mate-pair libraries (Wetzel et al. 2011). We are taking a multi-technology approach for this de novo assembly to capitalize on different Ion Torrent and Illumina sequencing strategies, entire fragment sequencing and paired-end sequencing, respectively; the methods also have different read lengths and error models.
Microsatellite divergence between isogenic lineages and the homozygosity of such lineages in the laboratory (Tatarenkov et al. 2010), means that K. marmoratus is an attractive system for studying the genetic basis for many of the already defined phenotypes, including behavioral phenotypes (Edenbrow and Croft 2011; Earley et al. 2012). Whole-genome sequencing of many lineages will reveal the amount of shared and unique single nucleotides and the variation in insertions and deletions between lineages.
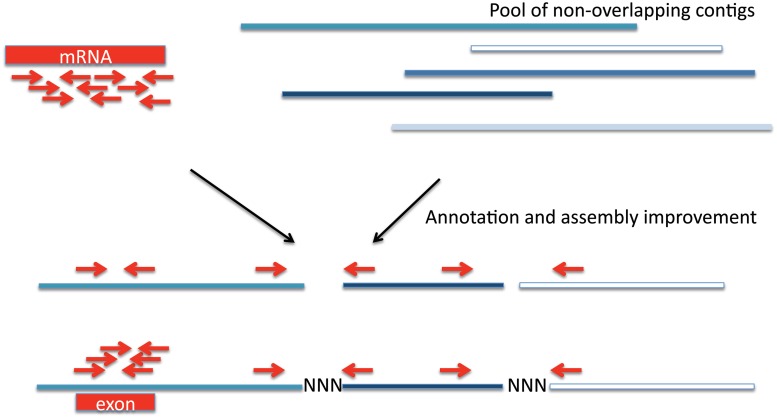
RNA-sequencing (RNA-seq) data will improve genome assembly and annotation of the K. marmoratus genome. RNA-seq is the shotgun sequencing of the transcriptome (Wang et al. 2009; Ozsolak and Milos 2011), and can be used to piece together genomic contigs or scaffolds, for increasing the length of the scaffold and for ordering and orienting contigs and scaffolds (Mortazavi et al. 2010) (Fig. 1). Annotation is an integral part of the genome assembly process (for a review see Yandell and Ence [2012]). RNA-seq data play an important role in the gene annotation of a de novo genome assembly. RNA-seq data can be used to determine exon locations and intro-exon boundaries for the transcripts expressed in the tissues from which RNA is isolated, using the program MAKER (Cantarel et al. 2008) or MAKER-2 (Holt and Yandell 2011). The programs rely on a combination of evidence-based transcripts, ab initio predictions and closely related species for training gene models and making gene annotations. For the assembly of this genome, RNA-sequencing is underway for multiple tissues. We have generated RNA-seq libraries from poly(A) tailed mRNA, to identify transcripts that will be translated into proteins, as well as from ribosomal RNA-depleted mRNA, to identify non-translated transcripts that have regulatory or other roles. We have started to use RNA-seq data to identify and annotate specific intron–exon boundaries. Additionally, we have been able to scaffold two preliminary contigs with the targeted approach of looking at the RNA-seq data (B.C. Ring et al., unpublished data).
Fig. 1.
Cartoon of how RNA-sequencing data and genomic contigs and/or scaffolds may be combined to increase scaffold length and order contigs and/or scaffolds. Additionally, the annotation of an exon using RNA-sequencing data is shown.
The improvement of sequencing technologies coupled with the potential ease of assembly due to the homozygous nature of the fish means that a high-quality draft assembly is possible in the near future and, indeed, is currently being generated. Sequencing and de novo genome assembly, coupled with gene annotation, will propel research on K. marmoratus forward and in new directions by providing a genetic foundation to ask questions about adaptation, divergence, and the genetic basis of traits of interest in K. marmoratus. Next-generation sequencing techniques, both of genomes and transcriptomes, provide tools for finding the genes underlying traits of interest, especially for new systems (Luikart et al. 2003; Stinchcombe and Hoekstra 2008; Gilad et al. 2009). These techniques have been used to identify genes underlying phenotypic variation between marine and freshwater populations of sticklebacks (Hohenlohe et al. 2010; Jones et al. 2012), adaptation to serpentine soils by Arabidopsis (Turner et al. 2010), and morphological differentiation in lake trout (Goetz et al. 2010) and whitefish (Jeukens et al. 2009). By having genomic information, researchers will be able to query specific loci of interest, as well as look at general patterns of diversity and divergence in the K. marmoratus genome.
Conclusion
Kryptolebias marmoratus is an ideal organism to develop as a genomic model for many phenotypes of interest. For example, significant differences among lineages in aggression and in responses to fighting experience have been shown (Earley and Hsu 2008). Kryptolebias marmoratus can survive for weeks out of water (Taylor et al. 2008), with major physiological changes associated with emersion, including major remodeling of the gills (Leblanc et al. 2010). There has been considerable effort put into studying the genes involved in tumorigenesis and in the metabolism of toxins to develop K. marmoratus as a model for tumor development (Lee et al. 2008; Rhee et al. 2009). The opportunity with K. marmoratus lies in the fact that lineages are highly homozygous and highly differentiated, both genetically (microsatellite markers) and phenotypically (life-history and behavioral traits), which means that crossing lineages to create F2 (or Fn) recombinant inbred lines will facilitate understanding the genetic basis of these traits and the manner in which they segregate.
Funding
National Institutes of Health Conference Grant from the Eunice Kennedy Shriver National Institute of Child Health & Human Development [R13HD070622]; Society of Integrative and Comparative Biology through the Division of Comparative Endocrinology, Division of Comparative Physiology and Biochemistry, Division of Animal Biology and the C. Ladd Prosser Fund; and the College of Agriculture and Natural Resources, University of Maryland. This work was also supported by a National Institutes of Health National Research Service Award postdoctoral fellowship [GM087069 to J.L.K.].
Acknowledgments
We would like to thank Carole Kelley for extensive comments and questions on the article.
References
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–96. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa WJEM. Redescription of Kryptolebias ocellatus (Hensel) and K. caudomarginatus (Seegers) (Teleostei: Cyprinodontiformes: Rivulidae), two killifishes from mangroves of south-eastern Brazil. J Ichthyol Aquatic Biol. 2006;11:5–12. [Google Scholar]
- Davis WP, Taylor DS, Turner BJ. Field observations of the ecology and habits of mangrove rivulus (Rivulus marmoratus) in Belize and Florida (Teleostei: Cyprinodontiformes: Rivulidae) Ichthyol Explor Freshwaters. 1990;1:123–34. [Google Scholar]
- Dunson WA, Dunson DB. Factors influencing growth and survival of the killifish, Rivulus marmorats, held inside enclosures in mangrove swamps. Copeia. 1999;1999(3):661–8. [Google Scholar]
- Earl D, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2011;21:2224–41. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley RL, Hanninen AF, Fuller A, Garcia MJ, Lee EA. Phenotypic plasticity and integration in the mangrove rivulus (Kryptolebias marmoratus) Integ Comp Biol. 2012 doi: 10.1093/icb/ics118. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley RL, Hsu Y. Reciprocity between endocrine state and contest behavior in the killifish, Kryptolebias marmoratus. Horm Behav. 2008;53:442–51. doi: 10.1016/j.yhbeh.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Earley RL, Hsu Y, Wolf LL. The use of standard aggression testing methods to predict combat behaviour and contest outcome in Rivulus marmoratus dyads (Teleostei: Cyprinodontidae) Ethology. 2000;106:743–61. [Google Scholar]
- Edenbrow M, Croft DP. Behavioural types and life history strategies during ontogeny in the mangrove killifish, Kryptolebias marmoratus. Anim Behav. 2011;82:731–41. [Google Scholar]
- Gilad Y, Pritchard JK, Thornton K. Characterizing natural variation using next-generation sequencing technologies. Trends Genet. 2009;25:463–71. doi: 10.1016/j.tig.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz F, Rosauer D, Sitar S, Goetz G, Simchick C, Roberts S, Johnson R, Murphy C, Bronte CR, Mackenzie S. A genetic basis for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush) Mol Ecol. 2010;19(Suppl. 1):176–96. doi: 10.1111/j.1365-294X.2009.04481.x. [DOI] [PubMed] [Google Scholar]
- Grageda MVC, Sakakura Y, Minamimoto M, Hagiwara A. Differences in life-history traits in two clonal strains of the self-fertilizing fish Rivulus marmoratus. Environ Biol Fish. 2005;73:427–36. [Google Scholar]
- Harrington RW., Jr Oviparous hermaphroditic fish with internal self-fertilization. Science. 1961;134:1749–50. doi: 10.1126/science.134.3492.1749. [DOI] [PubMed] [Google Scholar]
- Harrington RW., Jr Environmentally controlled induction of primary male gonochorists from eggs of the self-fertilizing hermaphroditic fish, Rivulus marmoratus. Biol Bull. 1967;132:174–99. doi: 10.2307/1539887. [DOI] [PubMed] [Google Scholar]
- Harrington RW., Jr How ecological and genetic factors interact to determine when self-fertilizing hermaphrodites of Rivulus marmoratus change into functional secondary males, with a reappraisal of the modes of intersexuality among fishes. Copeia. 1971;1971:389–432. [Google Scholar]
- Harrington RW, Jr, Kallman KD. The homozygosity of clones of the self-fertilizing hermaphroditic fish Rivulus marmoratus Poey (Cyprinodontidae, Atheriniformes) Am Nat. 1968;102:337–43. [Google Scholar]
- Heuhner MK, Schramm ME, Hens MD. Notes on the behavior and ecology of the killifish Rivulus marmoratus Poey 1880 (Cyprinodontidae) Florida Sci. 1985;48:3–7. [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y, Lee S-P, Chen M-H, Yang S-Y, Cheng K-C. Switching assessment strategy during a contest: fighting in killifish Kryptolebias marmoratus. Animal Behav. 2008;75:1641–9. [Google Scholar]
- Hsu Y, Wolf LL. The winner and loser effect: what fighting behaviors are influenced? Anim Behav. 2001;61:777–86. [Google Scholar]
- Jeukens J, Bittner D, Knudsen R, Bernatchez L. Candidate genes and adaptive radiation: insights from transcriptional adaptation to the limnetic niche among coregonine fishes (Coregonus spp., Salmonidae) Mol Biol Evol. 2009;26:155–66. doi: 10.1093/molbev/msn235. [DOI] [PubMed] [Google Scholar]
- Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Crepeau M, Cardeno C, Corbett-Detig R, Stevens K. Circumventing heterozygosity: sequencing the amplified genome of a single haploid Drosophila melanogaster embryo. Genetics. 2011;188:239–46. doi: 10.1534/genetics.111.127530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc DM, Wood CM, Fudge DS, Wright PA. A fish out of water: gill and skin remodeling promotes osmo- and ionoregulation in the mangrove killifish Kryptolebias marmoratus. Physiol Biochem Zool. 2010;83:932–49. doi: 10.1086/656307. [DOI] [PubMed] [Google Scholar]
- Lee JS, Raisuddin S, Schlenk D. Kryptolebias marmoratus (Poey, 1880): a potential model species for molecular carcinogenesis and ecotoxicogenomics. J Fish Biol. 2008;72:1871–89. [Google Scholar]
- Lin HC, Dunson WA. Phenotypic plasticity in the growth of the self-fertilizing hermaphroditic fish Rivulus marmoratus. J Fish Biol. 1999;54:250–66. [Google Scholar]
- Lubinski BA, Davis WP, Taylor DS, Turner BJ. Outcrossing in a natural population of self-fertilizing hermaphroditic fish. J Hered. 1995;86:469–73. doi: 10.1093/oxfordjournals.jhered.a111610. [DOI] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 2003;4:981–94. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Tatarenkov A, Perry A, Martin JR, Elder JF, Jr, Bechler DL, Avise JC. Microsatellite documentation of male-mediated outcrossing between inbred laboratory strains of the self-fertilizing mangrove killifish (Kryptolebias marmoratus) J Hered. 2006a;97:508–13. doi: 10.1093/jhered/esl017. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Tatarenkov A, Taylor DS, Turner BJ, Avise JC. Extensive outcrossing and androdioecy in a vertebrate species that otherwise reproduces as a self-fertilizing hermaphrodite. Proc Natl Acad Sci USA. 2006b;103:9924–8. doi: 10.1073/pnas.0603847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Tatarenkov A, Turner BJ, Avise JC. A mixed-mating strategy in a hermaphroditic vertebrate. Proc Biol Sci. 2006c;273:2449–52. doi: 10.1098/rspb.2006.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SB. Association behaviour of the self-fertilizing Kryptolebias marmoratus (Poey): the influence of microhabitat use on the potential for a complex mating system. J Fish Biol. 2007;71:1383–92. [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Molloy PP, Nyboer EA, Côté IM. Male–male competition in a mixed-mating fish. Ethology. 2011;117:586–96. [Google Scholar]
- Moore GL, Sucar S, Newsome JM, Ard ME, Bernhardt L, Bland MJ, Ring BC. Establishing developmental genetics in a self-fertilizing fish (Krytolebias marmoratus) Integ Comp Biol. 2012;52:781–91. doi: 10.1093/icb/ics052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Schwarz EM, Williams B, Schaeffer L, Antoshechkin I, Wold BJ, Sternberg PW. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res. 2010;20:1740–7. doi: 10.1101/gr.111021.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourabit S, Edenbrow M, Croft DP, Kudoh T. Embryonic development of the self-fertilizing mangrove killifish Kryptolebias marmoratus. Dev Dyn. 2011;240:1694–704. doi: 10.1002/dvdy.22668. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Suga K, Sakakura Y, Sakamoto T, Hagiwara A. Genetic and growth differences in the outcrossings between two clonal strains of the self-fertilizing mangrove killifish. Can J Zool. 2008;86:976–82. [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JS, Lee YM, Raisuddin S, Lee JS. Expression of R-ras oncogenes in the hermaphroditic fish Kryptolebias marmoratus, exposed to endocrine disrupting chemicals. Comp Biochem Physiol C Toxicol Pharmacol. 2009;149:433–9. doi: 10.1016/j.cbpc.2008.10.102. [DOI] [PubMed] [Google Scholar]
- Richards TM, Krebs JM, McIvor CC. Microhabitat associations of a semi-terrestrial fish, Kryptolebias marmoratus (Poey 1880) in a mosquito-ditched mangrove forest, west-central Florida. J Exp Mar Biol Ecol. 2011;401:48–56. [Google Scholar]
- Scheel JJ. Rivuline Karyotypes and their Evolution (Rivulinae, Cyprinodontidae, Pisces) J Zool Syst Evol Res. 1972;10:180–209. [Google Scholar]
- Stapley J, Reger J, Feulner PG, Smadja C, Galindo J, Ekblom R, Bennison C, Ball AD, Beckerman AP, Slate J. Adaptation genomics: the next generation. Trends Ecol Evol. 2010;25:705–12. doi: 10.1016/j.tree.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–70. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Takeda H. Draft genome of the medaka fish: a comprehensive resource for medaka developmental genetics and vertebrate evolutionary biology. Dev Growth Differ. 2008;50(Suppl. 1):S157–66. doi: 10.1111/j.1440-169X.2008.00992.x. [DOI] [PubMed] [Google Scholar]
- Tatarenkov A, Earley RL, Taylor DS, Avise JC. Microevolutionary distribution of isogenicity in a self-fertilizing fish (Kryptolebias marmoratus) in the Florida Keys. Integr Comp Biol. 2012;52:743–52. doi: 10.1093/icb/ics075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarenkov A, Gao H, Mackiewicz M, Taylor DS, Turner BJ, Avise JC. Strong population structure despite evidence of recent migration in a selfing hermaphroditic vertebrate, the mangrove killifish (Kryptolebias marmoratus) Mol Ecol. 2007;16:2701–11. doi: 10.1111/j.1365-294X.2007.03349.x. [DOI] [PubMed] [Google Scholar]
- Tatarenkov A, Ring BC, Elder JF, Bechler DL, Avise JC. Genetic composition of laboratory stocks of the self-fertilizing fish Kryptolebias marmoratus: a valuable resource for experimental research. PLoS One. 2010;5:e12863. doi: 10.1371/journal.pone.0012863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DS. Biology and ecology of Rivulus marmoratus: new insights and a review. Florida Sci. 2000;63:242–55. [Google Scholar]
- Taylor DS, Davis WP, Turner BJ. Groveling in the mangroves: 16 years in pursuit of the cyprinodont fish Rivlus marmoratus on the Belize Cays. Atoll Res Bull. 2004;525:1–14. [Google Scholar]
- Taylor DS, Fisher MT, Turner BJ. Homozygosity and Heterozygosity in three populations of Rivulus marmoratus. Environ Biol Fish. 2001;61:455–9. [Google Scholar]
- Taylor DS, Turner BJ, Davis WP, Chapman BB. A novel terrestrial fish habitat inside emergent logs. Am Nat. 2008;171:263–6. doi: 10.1086/524960. [DOI] [PubMed] [Google Scholar]
- Turko AJ, Earley RL, Wright PA. Behaviour drives morphology: voluntary emersion patterns shape gill structure in genetically identical mangrove rivulus. Animal Behav. 2011;82:39–47. [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet. 2010;42:260–3. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Vinson JP, et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–35. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel J, Kingsford C, Pop M. Assessing the benefits of using mate-pairs to resolve repeats in de novo short-read prokaryotic assemblies. BMC Bioinformatics. 2011;12:95. doi: 10.1186/1471-2105-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M, Ence D. A beginner's guide to eukaryotic genome annotation. Nat Rev Genet. 2012;13:329–42. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]