Abstract
To evaluate the relative importance of ornithine (Orn) as a precursor in proline (Pro) synthesis, we isolated and sequenced a cDNA encoding the Orn-δ-aminotransferase (δ-OAT) from Arabidopsis thaliana. The deduced amino acid sequence showed high homology with bacterial, yeast, mammalian, and plant sequences, and the N-terminal residues exhibited several common features with a mitochondrial transit peptide. Our results show that under both salt stress and normal conditions, δ-OAT activity and mRNA in young plantlets are slightly higher than in older plants. This appears to be related to the necessity to dispose of an easy recycling product, glutamate. Analysis of the expression of the gene revealed a close association with salt stress and Pro production. In young plantlets, free Pro content, Δ1-pyrroline-5-carboxylate synthase mRNA, δ-OAT activity, and δ-OAT mRNA were all increased by salt-stress treatment. These results suggest that for A. thaliana, the Orn pathway, together with the glutamate pathway, plays an important role in Pro accumulation during osmotic stress. Conversely, in 4-week-old A. thaliana plants, although free Pro level also increased under salt-stress conditions, the δ-OAT activity appeared to be unchanged and δ-OAT mRNA was not detectable. Δ1-pyrroline-5-carboxylate synthase mRNA was still induced at a similar level. Therefore, for the adult plants the free Pro increase seemed to be due to the activity of the enzymes of the glutamate pathway.
One of the most common responses in all organisms for regulating internal osmolarity is the accumulation of compatible solutes such as sugars and neutral amino acids. Under stress conditions free Pro increase plays a key role for osmotic adjustment in a large number of plant species (Rhodes et al., 1986; Delauney and Verma, 1993) and in bacteria (Csonka, 1989; Whatmore et al., 1990). The potent osmoprotective role of Pro was shown first in bacteria (Csonka, 1989) and later in higher plants (Sumaryati et al., 1992; Kishor et al., 1995) by modified organisms simultaneously producing high levels of Pro and displaying an enhanced osmotolerance. In higher plants, Pro accumulation during osmotic stress appears to be essentially due to de novo synthesis (Boggess et al., 1976; Rhodes et al., 1986; Verbruggen et al., 1996). Generally, in microorganisms (Cunin et al., 1986; Davis et al., 1986), mammals (Inana et al., 1986), and plants (Kandpal and Rao, 1982; Delauney et al., 1993), both glutamate and Orn are recognized as possible precursors of Pro.
In plants formation of Pro from glutamate is similar to that established first in bacteria (Fig. 1). The glutamate is reduced into Pro by two enzymes, P5CS and P5CR. The corresponding genes were isolated in plants by functional complementation of bacterial mutants defective for the respective steps (Delauney and Verma 1990; Hu et al., 1992). Regulation of the glutamate route is relatively well documented: moth bean (Vigna aconitifolia) P5CS enzyme synthesized in Escherichia coli is allosterically inhibited by Pro (Hu et al., 1992). The stimulation of Pro biosynthesis under salt and water stress is also correlated with an increase in the levels of P5CR and P5CS mRNA (Hu et al., 1992; Verbruggen et al., 1993; Savoure et al., 1995; Yoshiba et al., 1995; Strizhof et al., 1997).
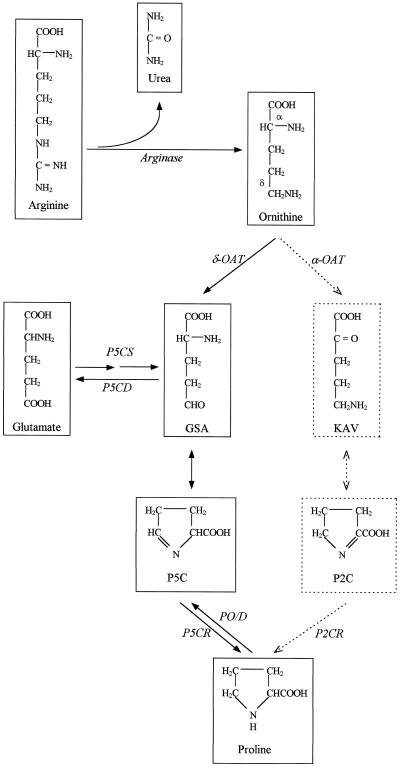
Figure 1.
Interrelation between Orn and glutamate pathways in Pro biosynthesis. GSA, Glutamic-γ-semialdehyde; KAV, α-ketoaminovalerate; P2C, Δ1-pyrroline-2-carboxylate; P2CR, Δ1-pyrroline-2-carboxylate reductase; PO/D, Pro oxidase/dehydrogenase; P5CD, P5C dehydrogenase.
It is well known from labeling experiments that Orn can also serve as a precursor to Pro in microorganisms, mammals, and higher plants (Csonka and Baich, 1983). For mammals and microorganisms, the conversion of Orn to Pro via the loss of the δ-amino group is catalyzed by δ-OAT, which produces P5C (Csonka and Baich, 1983). In plants it was established that transamination of Orn gives two possible intermediates: P5C and Δ1-pyrroline-2-carboxylate, which can both be reduced into Pro (Fig. 1; Mestichelli et al., 1979). Only the route involving the transamination of Orn into P5C was strongly indicated by the functional complementation of a defective E. coli mutant (Delauney et al., 1993).
As seen before, the role of the glutamate pathway in the Pro accumulation under stress conditions is well established. However, the importance of the relative contribution of the Orn pathway in Pro accumulation during stress is still a matter of discussion. As a matter of fact, considering the response of δ-OAT in osmotic stress conditions, there is a discrepancy between the results indicating an increase of enzymatic activity occurring in many plants (Kandpal and Rao, 1982; Hervieu et al., 1995) and the decrease of mRNA level observed in moth bean plantlets (Delauney et al., 1993). Since these studies were performed on different plant species, the discrepancy could be due to variations in the δ-OAT enzymatic activity from one plant to another or within the same plant, according to the organ or developmental stage.
Aiming to understand the role of δ-OAT in the Pro biosynthesis, our approach was to study δ-OAT in Arabidopsis thaliana, for which the enzymes of the glutamate pathway have already been cloned (Verbruggen et al., 1993; Yoshiba et al., 1995; Strizhof et al., 1997) and to determine how δ-OAT expression varies as a function of plant development after salt stress.
We report the cloning and the sequencing of full-length δ-OAT cDNA from an A. thaliana library. The expression patterns of this δ-OAT gene and the P5CS gene were compared at the RNA level in plants subjected or not to salt stress.
MATERIALS AND METHODS
Plant Growth and Salt-Stress Treatment
Arabidopsis thaliana seeds (C24 ecotype) were sown in vitro on solid Feenstra medium (Oostindiër and Feenstra, 1973) and grown in a culture room (24 ± 1°C, 16-h light/8-h dark cycle). Young (12-d-old) or older (4-week-old) plants were transferred to liquid Feenstra medium 24 h before the induction of salt stress. Salt-stress treatment consisted of replacing the Feenstra solution with the same solution containing 200 mm NaCl for 6, 24, and 72 h.
Isolation of the δ-OAT cDNA Clone Obtained from the EST Database
The A. thaliana EST clone 184H18T7 was selected by comparing the δ-OAT amino acid sequences from moth bean (Vigna aconitifolia; Delauney et al., 1993), human (Inana et al., 1986), and yeast (Saccharomyces cerevisiae; Degols, 1987) with the EMBL database using the BLAST network service.
Screening of the A. thaliana cDNA Library
DNA Probes
Template (25–50 ng) was labeled with [α32P]dCTP using a random-primed DNA-labeling kit (Boehringer Mannheim).
Homologous Radioactive Probing
Using the EST clone 184H18T7 as a radioactive probe, we screened the A. thaliana (Colombia ecotype) cDNA library from a cell-suspension culture (Höfte et al., 1993), graciously provided by Dr. T. Deprez (Institut National de la Recherche Agronomique, Versailles, France). The putative positive clones isolated from a first screening were pooled per Petri dish (13.5 cm in diameter) for elution. This elution was subjected to PCR using primers ORN2 and ORN3 with sequences chosen on the basis of the sequence of the δ-OAT EST clone. The primers ORN2 (5′-GAAGTACAAAGCGGTCTGGCTAGA-3′) and ORN3 (5′-CAGTTCCTC TCCAAGACTTGCTGA-3′) are localized in the sequence shown in Figure 2. The positive tubes from the first screening were subjected to subsequent radioactive and PCR screenings until purification.
Figure 2.
Nucleotide sequence of the δ-OAT cDNA and deduced primary structure of the δ-OAT enzyme of A. thaliana. ORN2 and ORN3 represent the primers used for the PCR screening. The putative poly(A+) signal is underlined and marked. The putative mitochondrial transit peptide is underlined and its basic (+) and hydroxylated (OH) residues are marked.
PCR Amplifications Used for Screening
PCR amplifications of the δ-OAT fragment were performed on resuspended bacterial colonies for the cDNA library. The total volume of the PCR reaction was 25 μL (5 μL of DNA template, 10 mm Tris-HCl, pH 8.3, 1.5 mm MgCl2, 50 mm KCl, 0.1 mg/mL of gelatin, 0.2 mm each deoxyribonucleotide triphosphate, 0.5 μm each primer, and 0.5 unit of Taq DNA polymerase [Boehringer Mannheim]). The optimized conditions for the reactions were 5 min of denaturation at 94°C and 35 cycles at 94°C, 1 min of denaturation, 55°C, 1 min of annealing, 72°C, 1 min of extension, and 10 min of extra extension at 72°C.
Sequencing of the DNA
Plasmids were extracted from a midi-scale culture via the alkaline lysis procedure (Maniatis et al., 1982). Deletion clones were obtained and their sequence was determined using the dideoxynucleotide chain-termination method with either radioactive (35S using Sequenase II, Amersham) or automatic laser fluorescence (Pharmacia) detection. The sequences were assembled and analyzed using the GeneCompar program (Vauterin M, Applied Maths, Kortrijk, Belgium).
Genomic DNA Extraction and Southern Hybridization
Genomic DNA was prepared from leaves of A. thaliana plantlets as described by Dellaporta et al. (1983). Twenty micrograms of genomic DNA was digested with the appropriate restriction enzymes and electrophoresed in a 0.8% agarose gel. The genomic DNA was transferred by capillary blotting onto positively charged nylon membranes (Boehringer Mannheim). The full-length δ-OAT cDNA isolated from the A. thaliana cDNA library was used as a radioactive probe. Hybridization was carried out at 65°C in the following solution: 10% SDS, pH 8.0, 0.37 g of EDTA, 67 g of Na2HPO4·2H2O, and 4 mL of H3PO4 (85% orthophosphoric acid). The membranes were washed first in 2× SSC (0.3 m NaCl, 0.03 m C6H5Na3O7·2H2O) at room temperature for 40 min and then in 0.5× SSC at 42°C for 40 min before exposing them to radiographic film (DuPont).
Total RNA Extraction and Northern Hybridization
Total RNA was isolated from 12-d-old plantlets and 4-week-old plants incubated 0, 6, 24, and 72 h under normal and salt-stress conditions (200 mm NaCl) according to the method described by Rerie et al. (1991). Twenty micrograms of RNA samples was subjected to electrophoresis in 1.5% agarose gel containing 6% formaldehyde (37%). Total RNA was transferred by capillary blotting onto positively charged nylon membranes (Boehringer Mannheim). The full-length δ-OAT cDNA isolated from the A. thaliana cDNA library was used as the first radioactive probe. The hybridization and washing were performed as described for the Southern analysis. The same experiment was performed with the P5CS cDNA (Savoure et al., 1995) used as a probe. The membrane was stained with methylene blue to verify the equal amount, loading, and transfer of RNA.
Enzymatic Assays
One gram fresh weight from 12-d-old plantlets and 4-week-old plants was extracted with 4 mL of extraction buffer as described by Hervieu et al. (1995). The activity of the δ-OAT in function of increasing substrate concentration was assayed following the measurement of the amount of P5C produced in 30 min using the ninhydrin method (Kim et al., 1994). One unit of δ-OAT velocity was defined as the micromoles of P5C produced per milligram of protein per hour. Protein determination was done following the method of Bradford (1976) using a BSA solution as the standard. Each analysis represents the mean of a minimum of three independent measures.
The direct linear plot of 1/V against S/V (where S is the substrate concentration of Orn in millimoles and V is the velocity) was used to estimate the parameters of the Michaelis-Menten equation (Cornish-Bowden and Eisenthal, 1978).
Pro Content
After 0, 6, 24, or 72 h of salt-stress treatment, Pro was extracted from 0.3 g fresh weight of material from 12-d-old plantlets and 4-week-old plants and quantitated by the method of Bates (1973). One unit of Pro concentration represents 1 μmol of Pro per gram fresh weight. Each analysis is the mean of a minimum of three independent measures.
RESULTS
Isolation, Sequencing, and Characterization of the A. thaliana δ-OAT cDNA
The determination of the full sequence of the 184H18T7 EST clone showed that it includes only part of the δ-OAT cDNA. A total length of 895 bp was obtained and included the 3′ noncoding sequence with the poly(A+) tail. About 70,000 bacterial clones from the cDNA library were screened on 10 Petri dishes. Only one positive band was detected and subjected to subsequent radioactive and PCR screenings prior to purification. The full nucleotide and the deduced amino acid sequences of the isolated A. thaliana cDNA clone are shown in Figure 2. The cDNA insert has a size of 1675 bp and contains a minimum open reading frame of 1428 nucleotides encoding a putative preprotein of 476 amino acids, assuming the translation is initiated at the first ATG codon (position 45) from the 5′ part. The polyadenylation signal AATAAA is present 15 bases upstream from the start of the poly(A+) tail.
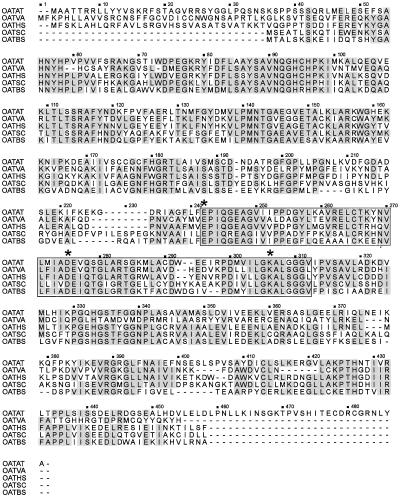
Comparison of the A. thaliana deduced δ-OAT amino acid sequence with the available plant (Delauney et al., 1993), mammalian (Inana et al., 1986), yeast (Degols, 1987), and bacterial (Bacillus subtilis; Gardan et al., 1995) sequences revealed, respectively, 65, 70, 71, and 72% similarity and 46, 52, 53, and 54% identity (Fig. 3). As reported for the moth bean δ-OAT (Delauney et al., 1993), the A. thaliana δ-OAT sequence also shows regions that have considerable homology with OAT enzymes that catalyze ω-transamination (Fig. 3), such as the region involved in the interaction with pyridoxal phosphate (Heimberg et al., 1990).
Figure 3.
Comparison of the deduced δ-OAT amino acid sequence from Arabidopsis with plant (OATVA), mammalian (OATHS), yeast (OATSC), and bacterial (OATBS) protein sequences. Shaded boxes indicate conserved amino acid residues. The domain corresponding to the putative pyridoxal phosphate-binding site is underlined. δ-Amino acids involved in the interaction with pyridoxal phosphate are marked by a star.
The similarity between the different δ-OAT enzymes is about 60% over most of the sequence. The greatest discrepancies are observed in the N-terminal region. This is because human and moth bean δ-OATs are mitochondrial enzymes (Inana et al., 1986; Delauney et al., 1993); therefore, the preprotein includes a transit peptide that is removed upon entry into the mitochondrial matrix. In contrast, bacterial and yeast δ-OAT does not have a transit peptide (Degols, 1987; Gardan et al., 1995). There is no consensus for the mitochondrial leader sequence, but the N-terminal residues deduced from our isolated cDNA exhibits several features in common with mitochondrial transit peptides, such as the absence of acidic residues, the uniform repartition, the relatively high content in positively charged residues, and the high level of hydroxylated residues (Van Heijne et al., 1989; Canel et al., 1996; Fig. 2). We propose that the δ-OAT precursor protein has a mitochondrial transit peptide.
Southern Analysis of the Genomic DNA
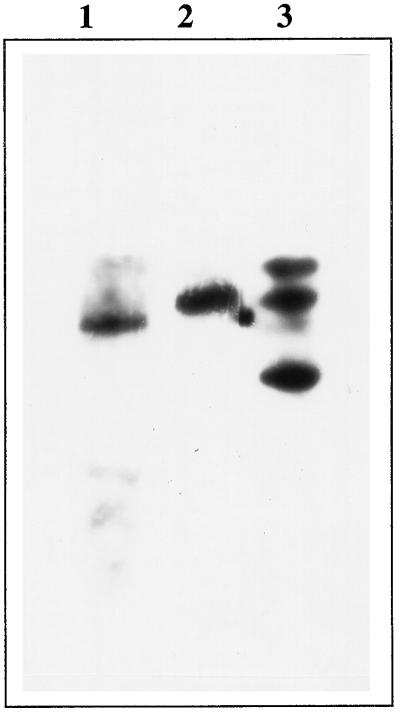
To check for the presence of gene family encoding δ-OAT in Arabidopsis, the full δ-OAT cDNA was used as a probe against the Arabidopsis genome DNA digested with various restriction enzymes (Fig. 4).
Figure 4.
Southern blot of the δ-OAT gene. A. thaliana genomic DNA was digested with different restriction enzymes: Lane 1, BamHI; lane 2, KpnI; and lane 3, SstI. Blot was hybridized with the full-length δ-OAT cDNA.
Strongly hybridizing fragments have been identified as a single band of about 5 kb in the BamHI digest, as a single band of about 6 kb in the KpnI digest, and as three bands in the SstI digest (about 2, 6, and 9 kb). These results are in accordance with the genomic δ-OAT sequence, in which no restriction sites for BamHI and KpnI are present, and two SstI restriction sites have been identified (data not shown). These results suggest that only one δ-OAT gene is present per A. thaliana haploid genome.
Effect of Salt Stress on the Expression of the δ-OAT A. thaliana Gene at the Enzyme Level
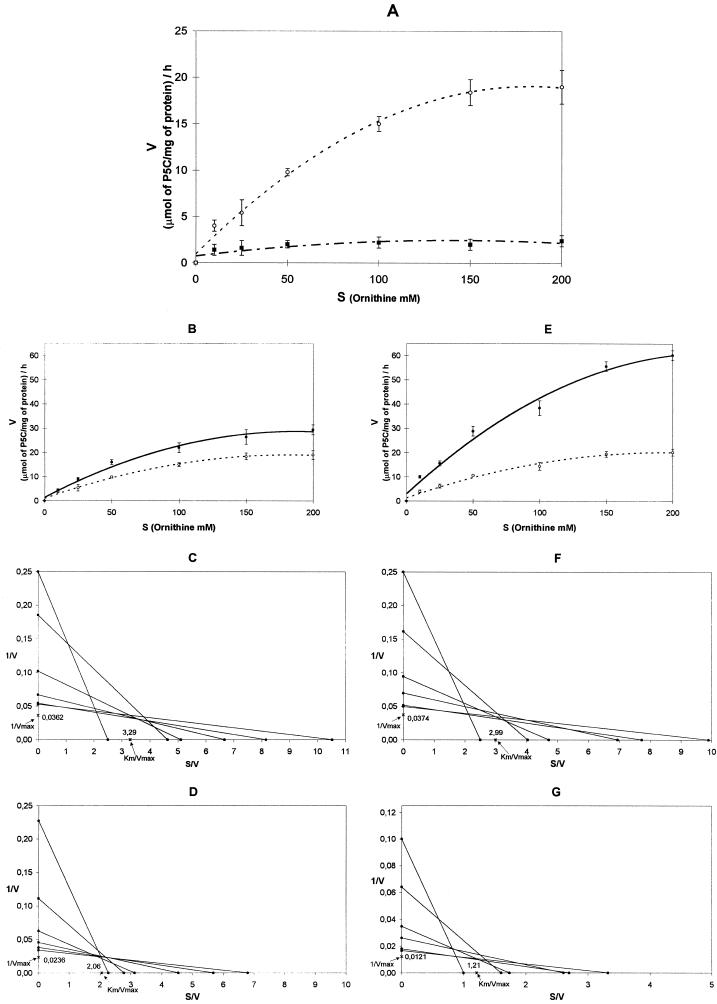
To observe how A. thaliana δ-OAT enzymatic activity responds to salt stress in plant development, enzymatic assays were performed on 12-d-old plantlets and 4-week-old plants incubated for 24 and 72 h in normal and salt-stress conditions (200 mm NaCl; Fig. 5, A, B, and E). In the control 12-d-old plantlets, the results obtained from the direct linear plot (Fig. 5, C, D, F, and G) showed the maximum velocity to be quite high (about 28 μmol P5C mg−1 protein h−1). In the salt-stress treatment, the OAT velocity had almost doubled compared with the control after 24 h (42 μmol P5C mg−1 protein h−1) and was strongly increased after 72 h (82 μmol P5C mg−1 protein h−1). In fact, the maximum velocity reached values, respectively, 1.5 and 3 times higher for 24 and 72 h of stress compared with those observed in normal conditions. These values were significantly different, as determined using the median test (Conover, 1980).
Figure 5.
A, δ-OAT Orn saturation patterns of 12-d-old plantlets (▪) and 4-week-old plants (○). B and E, Orn saturation patterns of 12-d-old plantlets in normal conditions (○; control) and salt-stress conditions (200 mm NaCl; •) after incubation for 24 h (B) and 72 h (E). The direct linear plot of 1/V against S/V (see text for explanation) was used to estimate the parameters of Km and Vmax. Km and Vmax were calculated for 12-d-old plantlets after incubation for 24 and 72 h under normal conditions (C and F, respectively) and under salt-stress conditions (D and G, respectively).
The apparent Km values were relatively similar. For enzymes extracted from plantlets incubated 24 h in normal or salt-stress conditions, these values were 91 and 87 mm Orn, respectively. For enzyme extracts prepared from plantlets incubated for 3 d, they were 80 and 100 mm Orn, respectively. These values were not significantly different as determined by the median test. In normal conditions the maximum of activity in 4-week-old plants is approximately 10 times lower (about 1–2 μmol P5C mg−1 protein h−1) than in 12-d-old plantlets (Fig. 5A). The salt stress did not seem to induce any effect in adult plants. The maximum velocity remained very low (data not shown) and the apparent Km value was not measurable with enough precision under these conditions.
Effect of the Salt Stress on the Expression of the δ-OAT A. thaliana Gene at the RNA Level
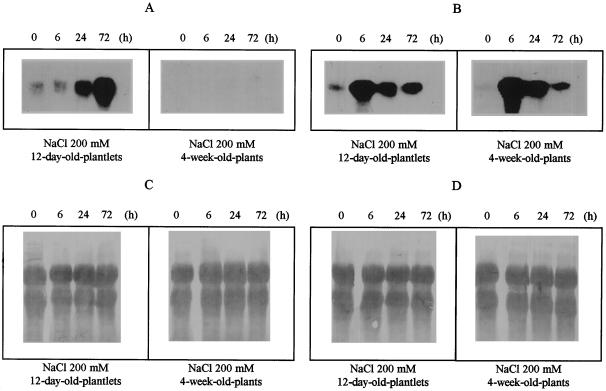
To assess the levels of δ-OAT mRNA and compare these levels with those of the P5CS mRNA, northern analysis was performed in A. thaliana on 12-d-old plantlets and 4-week-old plants using similar salt-stress conditions (Fig. 6). Probing the blot with a labeled, full-length δ-OAT cDNA showed a 1.6-kb δ-OAT transcript in all samples from the 12-d-old plantlets, whereas no clear response was detectable in the 4-week-old plants under salt-stress or normal conditions. In contrast, P5CS mRNA was observed in samples of both 12-d-old and 4-week-old plants. Moreover, after 24 h of salt stress, a significant increase in δ-OAT mRNA was observed. An extra 72 h of exposure to salt stress increased the δ-OAT mRNA level. In comparison, rapid accumulation of P5CS mRNA was observed 6 h after salt stress. After this maximum, the P5CS mRNA was still induced but at reduced transcript levels for both the 4-week-old and the 12-d-old plants.
Figure 6.
Total RNA isolated from 12-d-old plantlets and 4-week-old plants incubated for 0, 6, 24, and 72 h in 200 mm NaCl. Blots were hybridized with the full-length δ-OAT cDNA (A) or with the full-length P5CS cDNA (B). The membranes were stained with methylene blue to verify the equal amount, loading, and transfer of RNA (C and D).
Pro Accumulation in Salt-Stressed Plants
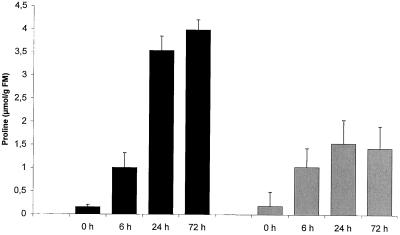
To relate the expression level of the δ-OAT under normal and salt-stress conditions with a possible consequence on Pro accumulation, analysis of the Pro content was performed on 12-d-old and 4-week-old plants (Fig. 7). This experiment shows, first, that accumulation of Pro was induced 6 h after salt-stress incubation for plant and plantlets. Second, when 12-d-old seedlings were subjected to 24 and 72 h of incubation in 200 mm NaCl, the level of free Pro reached, respectively, 3.5 and 4 μmol g−1 fresh weight, which is about 20-fold higher than in the control plantlets. In the 4-week-old plants, the free Pro level, as in young plantlets, was about 0.2 μmol Pro g−1 fresh weight in normal stress conditions, but the increase after salt-stress treatment (approximately 7-fold) was less than in young plants.
Figure 7.
Pro concentration in plant tissue after 0, 6, 24, and 72 h of 200 mm NaCl-stress treatment in 12-d-old plantlets (black bars) and 4-week-old plants (shaded bars). FM, Fresh mass.
DISCUSSION
We have isolated the cDNA encoding the full δ-OAT preprotein from A. thaliana. The similarity of the predicted δ-OAT protein with the amino acid sequences from bacteria, yeast, and mammals was high (about 70%). However, the similarity with the only plant sequence available, moth bean, was only 65%. This did not seem to be associated with the existence of different isoforms of OAT. In fact, it is well known that two forms exist, α-OAT and δ-OAT (Heimberg et al., 1990; Schultz and Coruzzi, 1995).
In plants, it has been established that transamination of Orn can happen via either α-OAT or δ-OAT (Mestichelli et al., 1979). However, until now only δ forms of OAT have been cloned in plants (Delauney et al., 1993), mammals (Inana et al., 1986), yeast (Degols, 1987), and bacteria (Gardan et al., 1995). As demonstrated for moth bean, the A. thaliana OAT is a δ-OAT. The possibility that this clone would be an α form has to be discarded because the sequence of the region involved in the interaction with the pyridoxal phosphate has no significant homology with the α-aminotransferase (Schultz and Coruzzi, 1995) but is very similar to that of all δ-aminotransferases (Heimberg et al., 1990). As for moth bean, the deduced amino acid 5′ sequence of A. thaliana δ-OAT seems to indicate the presence of a mitochondrial transit peptide. Therefore, the cloned A. thaliana δ-OAT cDNA does not seem to encode an isoenzyme localized in a distinct subcellular compartment. This is in agreement with the subcellular localization of the plant δ-OAT by measurement of its activity in isolated mitochondria (Taylor and Stewart, 1981; Polacco and Holland, 1993) and with the fact that arginase, the other enzyme of Arg catabolism, is also localized in this organelle (Taylor and Stewart, 1981).
The δ-OAT enzyme has been shown to participate in Pro biosynthesis, but its contribution relative to that of the glutamate pathway is still a matter of discussion (Kandpal and Rao, 1982; Delauney et al., 1993; Hervieu et al., 1995). This is probably because studies have been performed in different plant species and under different physiological conditions, in which the two pathways could play different roles.
Our results demonstrate that for A. thaliana in normal growth culture conditions, the δ-OAT activity and transcripts vary as a function of the age of the plant: for young seedlings (12 d after germination), the δ-OAT activity reaches values 10 times higher than those observed in 4-week-old plants. δ-OAT transcripts are detectable in only the young seedlings, whereas the Pro content does not appear to vary significantly in young tissues compared with older plants. This is in agreement with the fact that Arg is very important in seedling nitrogen nutrition.
Conversion of A. thaliana seed-storage proteins to seedling de novo synthesized proteins potentially involves Arg breakdown, which is consistent with the 10-fold increase in seedling arginase activity 6 d after germination (Zonia et al., 1995). The products of this hydrolysis are urea and Orn. Urea appears to have a function in recycling nitrogen into ammonium via urease (Zonia et al., 1995). The massively de novo synthesized Orn is transformed into glutamate semialdehyde via the δ-OAT enzyme, but this does not appear to influence the Pro levels (Polacco and Holland, 1993). In fact, glutamate semialdehyde seems to be oxidized into glutamate via P5C dehydrogenase which can then be used for transamination or to generate new carbon constituents upon entry into the Krebs cycle as a ketoglutarate (Polacco and Holland, 1993). This can explain why in our experiment, under normal conditions, the δ-OAT activity was high in the plantlets but was not related to a significant increase in Pro content.
Data about enzymatic activity in salt-stressed plants presented in the literature indicate that the contribution of the Orn pathway plays a determinant role in Pro accumulation. Production of Pro from Arg- or Orn-labeled precursors is enhanced by water stress (Boggess and Stewart, 1976; Boggess et al., 1976). The δ-OAT activity in Ragi (Eleusine coracana) leaves also increases under water stress (Kandpal and Rao, 1982). The Orn pathway contributes via an increase of δ-OAT activity to Pro biosynthesis as well as the glutamate pathway in salt-treated radish cotyledons (Hervieu et al., 1995). However, this key role is not confirmed at the transcriptional level in moth bean plantlets in which δ-OAT mRNA levels were markedly depressed by salt stress. The authors suggested that during salt stress Pro is synthesized predominantly via the glutamate pathway (Delauney et al., 1993).
Our results indicate an association between mRNA levels and enzymatic activity. We have shown that in 12-d-old plantlets δ-OAT activity is markedly increased by salt-stress treatment. Moreover, the free Pro level, the δ-OAT mRNA, and the P5CS mRNA are all increased by salt stress. However, after salt-stress treatment, δ-OAT and P5CS mRNA accumulated with different patterns. The δ-OAT mRNA increased slightly after 24 h and more markedly after 72 h, whereas the P5CS mRNA levels rapidly accumulated to a maximal value 6 h after induction and decreased until 24 h of exposure. In 4-week-old A. thaliana plants, although the free Pro level also increased under salt-stress conditions, the δ-OAT activity appeared to be unchanged and δ-OAT mRNA was not detectable. In contrast, P5CS mRNA was still induced to similar levels. Thus, for adult plants the free Pro increase seems to be due to only the activity of the enzymes of the glutamate pathway.
In conclusion, the Orn pathway could serve an important role in very young A. thaliana plantlets. In normal growth conditions the role of this pathway is related to the necessity for the plantlets to dispose of a nitrogen source but does not seem to be linked to Pro biosynthesis. On the contrary, under salt-stress conditions the main task of this pathway is devoted to Pro production and contributes, together with the glutamate pathway, to the Pro accumulation. In later stages of differentiation, the Orn pathway contributes to a lesser extent in both normal and salt-stress conditions to nitrogen availability and Pro biosynthesis, respectively. In salt-stress conditions Pro accumulation is synthesized predominantly via the glutamate pathway.
Future experiments will consist of overexpressing the δ-OAT sense and antisense cDNA in transgenic plants. This should allow evaluation of the importance of the Orn pathway in Pro formation and the validity of such approach for obtaining crops with better osmotolerance.
ACKNOWLEDGMENTS
We most gratefully thank T. Deprez for the gift of the A. thaliana cDNA library and the Laboratorium voor Genetica of Gent University for the gift of the A. thaliana P5CS cDNA. We thank V. Frankard and M. Vauterin for their critical reading of this manuscript. We particularly thank V. Frankard for her excellent guidance in molecular biology. We thank V. Bouton for her technical assistance with the automatic sequencing. We thank F. Homble for his explanations concerning the enzymatic assays. We thank C. Vermeidelen and X. Vekemans for their guidance with the statistical analysis. We thank M. De Ryck for the layout of the figures.
Abbreviations:
- δ-OAT
Orn-δ-aminotransferase
- EST
expressed sequence tag
- P5C
Δ1-pyrroline-5-carboxylate
- P5CR
Δ1-pyrroline-5-carboxylate reductase
- P5CS
Δ1-pyrroline-5-carboxylate synthase
Footnotes
N.H.C.J.R. was the recipient of a fellowship from the Fond National de la Recherche Scientifique Belge. This work was also supported by the Belgium Program of Concerted Actions (no. 92).
LITERATURE CITED
- Bates LS. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39:205–207. [Google Scholar]
- Boggess SF, Stewart CR. Contribution of arginine to proline accumulation in water-stressed barley leaves. Plant Physiol. 1976;82:890–903. doi: 10.1104/pp.58.6.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggess SF, Stewart CR, Aspinall D, Paleg L. Effect of water stress on proline synthesis from radioactive precursors. Plant Physiol. 1976;58:398–401. doi: 10.1104/pp.58.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Canel C, Bailey-Serres J, Roose M. Molecular characterization of the mitochondrial citrate synthase gene of an acidless pummelo (Citrus maxima) Plant Mol Biol. 1996;31:143–147. doi: 10.1007/BF00020613. [DOI] [PubMed] [Google Scholar]
- Conover WJ (1980) The median test. In Practical Nonparametric Statistics, Ed 2. Wiley, New York, pp 171–174
- Cornish-Bowden A, Eisenthal R. Estimation of Michaelis constant and maximum velocity from the direct linear plot. Biochim Biophys Acta. 1978;523:268–272. doi: 10.1016/0005-2744(78)90030-x. [DOI] [PubMed] [Google Scholar]
- Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN, Baich B (1983) Chapter 3. In KM Herman, RL Somerville, eds, Amino Acid Biosynthesis and Genetic Regulation. Addison Yesley, London, pp 35–48
- Cunin R, Glansdoff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev. 1986;50:280–313. doi: 10.1128/mr.50.3.280-313.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G. Eur J Biochem. 1987;169:193–200. doi: 10.1111/j.1432-1033.1987.tb13597.x. [DOI] [PubMed] [Google Scholar]
- Delauney A, Hu C, Kishor K, Verma D. Cloning of ornithine δ-aminotransferase cDNA by trans-complementation in Escherichia coli and regulation of proline biosynthesis. J Biol Chem. 1993;268:18673–18678. [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. A soybean gene encoding Δ-pyrroline-5-carboxylate reductase was isolated by functional complementation in Escherichia coli and is fund to be osmoregulated. Mol Gen Genet. 1990;221:299–305. doi: 10.1007/BF00259392. [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. Plant J. 1993;4:215–223. [Google Scholar]
- Dellaporta S, Wood J, Hicks J. A plant DNA mini preparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Gardan R, Rapoport G, Débarbouillé M. Expression of the roc DEF operon involved in the arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- Heimberg H, Boyen A, Crabeel M, Glandsdorff N. Escherichia coli and Saccharomyces cerevisiae acetylornithine aminotransferases: evolutionary relationship with ornithine aminotransferases. Gene. 1990;90:69–78. doi: 10.1016/0378-1119(90)90440-3. [DOI] [PubMed] [Google Scholar]
- Hervieu F, Le Dily L, Huault C, Billard J-P. Contribution of ornithine aminotransferase to proline accumulation in NaCl-treated radish cotyledons. Plant Cell Environ. 1995;18:205–210. [Google Scholar]
- Höfte H, Deprez T, Amselem J, Chiapello A, Caboche M. An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J. 1993;4:1051–1061. doi: 10.1046/j.1365-313x.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- Hu AC-A, Delauney AJ, Verma DPS. A bifunctional enzyme (Δ1-pyrroline-5 carboxylate synthetase) catalyses the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA. 1992;89:9354–9358. doi: 10.1073/pnas.89.19.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inana G, Totsuka S, Redmond M, Dougherty T, Nagle J, Shiono T, Ohura T, Koninami E, Katunuma N. Molecular cloning of human ornithine aminotransferase mRNA. Proc Natl Acad Sci USA. 1986;83:1203–1207. doi: 10.1073/pnas.83.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandpal RP, Rao NA. Water stress induced alterations in the properties of ornithine aminotransferase from Ragi (Eleusine coracana) leaves. Biochem Int. 1982;5:297–302. [Google Scholar]
- Kim HR, Rho HW, Park JW, Park BH, Kim JS, Lee MW. Assay of ornithine aminotransferase with ninhydrin. Anal Biochem. 1994;223:205–207. doi: 10.1006/abio.1994.1574. [DOI] [PubMed] [Google Scholar]
- Kishor KPB, Hong Z, Miao GH, Hu CAA, Verma DPS. Overexpression of Δ-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Mestichelli LJJ, Gupta RN, Spenser ID. The biosynthetic route from ornithine to proline. J Biol Chem. 1979;254:640–647. [PubMed] [Google Scholar]
- Oostindiër BF, Feenstra W. Isolation and characterization of chlorate resistant mutants of A. thaliana. Mutat Res. 1973;19:175–185. [Google Scholar]
- Polacco JC, Holland MA. Role of urease in plant cells. Int Rev Cytol. 1993;145:65–103. [Google Scholar]
- Rerie WG, Whietcross M, Higgins T. Developmental and environmental regulation of pea legumin genes in transgenic tobacco. Mol Gen Genet. 1991;225:148–157. doi: 10.1007/BF00282653. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Handa S, Bressan RA. Metabolic changes associated with adaptation of plant cells to water stress. Plant Physiol. 1986;82:890–903. doi: 10.1104/pp.82.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoure A, Jaoua S, Hua X-J, Ardiles W, Van Montagu M, Verbruggen N. Isolation, characterization, and chromosomal location of a gene encoding the Δ1-pyrroline-5 carboxylate synthetase in Arabidopsis thaliana. FEBS Lett. 1995;372:13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Coruzzi G. The aspartate aminotransferase gene family of Arabidopsis thaliana encodes isoenzymes localized to three distinct subcellular compartments. Plant J. 1995;7:61–75. doi: 10.1046/j.1365-313x.1995.07010061.x. [DOI] [PubMed] [Google Scholar]
- Strizhof N, Abraham E, Okresz L, Blickling S, Zilberstein L, Schell J, Koncz C, Szabados L. Differential expression of two P5CS genes controlling proline accumulation during salt stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- Sumaryati S, Negrutiu I, Jacobs M. Characterization and regeneration of salt- and water-stress mutants from protoplasts culture of Nicotiana plumbaginifolia (Viviana) Theor Appl Genet. 1992;83:613–619. doi: 10.1007/BF00226906. [DOI] [PubMed] [Google Scholar]
- Taylor A, Stewart G. Tissue and subcellular localization of enzymes of arginine metabolism in Pisum sativum. Biochem Biophys Res Commun. 1981;101:1281–1289. doi: 10.1016/0006-291x(81)91586-2. [DOI] [PubMed] [Google Scholar]
- Van Heijne G, Steffuhn J, Herrman RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hua XJ, May M, Van Montagu M. Environmental and developmental signals modulate proline homeostasis: evidence for negative transcriptional regulator. Proc Natl Acad Sci USA. 1996;93:8787–8789. doi: 10.1073/pnas.93.16.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Villarroel R, Van Montagu M. Osmoregulation of pyrroline-5-carboxylate reductase gene in Arabidopsis thaliana. Plant Physiol. 1993;103:771–781. doi: 10.1104/pp.103.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore AM, Chudek JA, Reed RH. J Gen Microbiol. 1990;136:2527–2535. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. Correlation between the induction of a gene for Δ-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 1995;7:751–760. doi: 10.1046/j.1365-313x.1995.07050751.x. [DOI] [PubMed] [Google Scholar]
- Zonia L, Stebbins N, Polacco J. Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol. 1995;107:1097–1103. doi: 10.1104/pp.107.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]